Abstract
X-ray crystallography remains the most robust method to determine protein structure at the atomic level. However, the bottlenecks of protein expression and purification often discourage further study. In this chapter, we address the most common problems encountered at these stages. Based on our experiences in expressing and purifying antimicrobial efflux proteins, we explain how a pure and homogenous protein sample can be successfully crystallized by the vapor diffusion method. We present our current protocols and methodologies for this technique. Case studies show step-by-step how we have overcome problems related to expression and diffraction, eventually producing high quality membrane protein crystals for structural determinations. It is our hope that a rational approach can be made of the often anecdotal process of membrane protein crystallization.
Keywords: Vapor diffusion, X-ray crystallography, membrane protein crystallization, antimicrobial efflux, multidrug resistance
Introduction
As of this writing, nearly 100,000 protein structures have been deposited to the Protein Data Bank (PDB). Approximately 90% were solved by X-ray crystallography—by far the most successful technique used to study protein structure. However, of more than 30,000 unique X-ray structures, only approximately 1% represent membrane proteins (http://blanco.biomol.uci.edu/mpstruc/). In contrast to their share of the known structures, membrane proteins are estimated to comprise approximately 30% of genes.1,2 They play the critical roles of gatekeepers, mediating messages and materials moving into and out of the cell. For example, G-protein coupled receptors, which are responsible for vision, taste, smell and immune response in eukaryotes;3,4 multidrug efflux transporters, which confer antibiotic resistance to pathogenic bacteria;5 voltage-gated ion channels, such as those responsible for neuron firing and muscle contraction;6,7 and porins, through which bacteria, mitochondria, and chloroplasts exchange material, such as nutrients, with the extracellular environment.8,9 These proteins comprise at least 60% of all drug targets.10,11
Unfortunately, understanding the structures and action mechanisms of these important integral membrane proteins is often hampered by the difficulties associated with expression, purification, and crystallization. There are at least two non-trivial tasks in the way of straightforward application of X-ray crystallography to membrane proteins: i) obtaining a sufficient quantity of purified protein and ii) producing high quality crystals.12 The latter will be the focus of this chapter. However, as any practicing membrane protein crystallographer can attest to, obtaining diffraction-quality crystals can be a formidable task, which requires a pure and homogeneous protein sample. In this respect, sample preparation and crystallization are inseparable. Thus, a significant portion of this review will be devoted to the topics of membrane protein expression and purification. In addition, extensive examples and detailed discussions, based on our experiences in expressing, purifying, and crystallizing antimicrobial efflux proteins, will be provided. Hopefully, by introducing the available methodologies and protocols, a rational approach can be made to the often anecdotal process of membrane protein crystallization.
Membrane Protein Expression
With few exceptions, membrane proteins are not abundant in their native environment. For example, Escherichia coli cells were found to contain only eight transcripts per cell corresponding to the inner membrane copper transporter CusA, even after induction.13 Those few exceptions include the rhodopsins, porins, ATPases, photosynthetic reaction centers, and other light-harvesting complexes.14 Coincidentally, the first membrane protein structure determined by X-ray crystallography was the photosynthetic reaction center of Rhodopseudomonas viridis.15 It wasn’t until 1998, more than a decade later, that a membrane protein structure was obtained from a non-native, i.e. recombinant, source.16,17 To date, recombinant overexpression is the most common approach to obtain proteins of interest. Often, this step is the first bottleneck that must be overcome in order to produce the milligram quantities of pure protein necessary for a single crystallization experiment.12,14
Host
Thus far, E. coli remains the favorite host for heterologous protein expression. It grows quickly, tolerates a high cell density, and does so on a relatively lean diet.18 The protein yield from E. coli cells is also particularly high, compared to other organisms. In one experiment, the expression of 20 membrane proteins in six host organisms (three prokaryotic, including E. coli, and three eukaryotic) was measured.12 Overall, the highest yields were achieved with E. coli cells. Further, only two membrane proteins tested were not successfully overexpressed in E. coli.
Bacterial proteins do tend to express better in E. coli, in general, but it is not a cure-all for heterologous membrane protein expression. Especially for eukaryotic membrane proteins, other host organisms must be considered.12,19,20 Often, eukaryotic proteins contain post-translational modifications, such as phosphorylation, glycosylation, methylation, or acetylation, which a non-native host may be incapable of reproducing.21 E. coli and other bacterial hosts, in particular, tend to lack these post-translational capabilities.18
Translational machinery will also vary between the host and native organism. E. coli cells exhibit a clear codon bias, with the population of tRNA approximately proportional to the chromosomal frequency.22 Occurrence of rare codons or many minor codons in the target protein sequence has a marked effect on expression levels. The reason for this is apparent when considering that an overexpressed membrane protein may represent a significant fraction of the total cellular protein. In some cases, even a single rare codon can be responsible for reduced expression levels.22 Accordingly, optimal expression of a protein might be achieved by optimizing the codons for the particular host. This can be a time consuming process. For example, the Neisseria gonorrhoeae gene norM, encoding the multidrug efflux pump NorM, contains nine rare codons for E. coli. To overexpress the N. gonorrhoeae NorM membrane protein in E. coli TOP10 cells, our lab had to spend a few months fixing each of these codons.23 Even the E. coli gene acrD, which produces the aminoglycoside efflux pump AcrD, has 15 rare codons. Thus, it is incompatible with E. coli host cells. We had to correct each of these 15 codons in order to overexpress this membrane protein in E. coli BL21(DE3) cells. Fortunately, several companies, such as Integrated DNA Technologies (IDT), offer protein codon optimization tools to correct for the codon bias encountered in many host cells.
Upon translation, membrane proteins must then be chaperoned from the cytoplasm and properly inserted into the corresponding membrane. This is of particular importance to outer membrane proteins of Gram-negative bacteria. A proper signaling peptide is essential to guide the expressed protein across the inner membrane and anchor to the outer membrane. Differences in the lipid bilayer structure and composition, as well as differences in insertion machinery, of the native organism compared to the host determine whether this process results in an active membrane protein or an intractable aggregate.24,25
Finally, properly overexpressed, folded, and active membrane proteins tend to be toxic to their host.12 The reason for this is obvious in the case of membrane channels or transporters, which directly control the permeability and delicate intracellular chemistry of their host, and can have direct roles in metabolism. In an experiment conducted with Rhodopseudomonas blastica, the overexpression of porins resulted directly in lysis of the host cell.26,27 Thus, the amount of inducer, such as isopropyl β-D-1-thiogalactopyranoside or arabinose, as well as the expression time and temperature, are often found to be critical variables for optimal protein expression.
Various strains of E. coli have been commercially developed to address the challenges encountered in membrane protein overexpression. Among those used in our lab, the BL21(DE3) strain (Novagen) is deficient in OmpT and lon proteases, which allows for increased protein stability;18 mutations in the lac promoter render C41(DE3) and C43(DE3) strains far less sensitive to the toxicity of membrane protein overexpression;28,29 and the BL21-CodonPlus strain (Stratagen) contains additional copies of rare E. coli tRNAs.
In addition to using these specialized strains of E. coli, our lab also generates ΔacrB knockout strains by removing the acrB gene, which encodes the multidrug efflux pump AcrB. Deletions of abundant native membrane proteins have been shown to facilitate the purification process and produce a more homogenous protein.27 In many cases, without this specific deletion, our attempts at crystallizing membrane proteins would result in crystals of AcrB, instead.
Vector
In general, it is not easy to predict whether an expression vector is suitable for the protein of interest. Choosing a vector that is compatible with both the membrane protein target and host organism, and one that is tailored to the desired expression and purification systems, is typically a trial-and-error process. Luckily, many powerful expression vectors are commercially available. In particular, our lab likes the pET-type vectors (Novagen) for the expression of α-helical transmembrane proteins. For β-barrel transmembrane proteins, we have inordinate success with the pBAD-type vectors (Life Technologies).
If a particular protein cannot be expressed with any combination of vectors and host cells, one may want to think about expressing the protein fused with green fluorescent protein (GFP) or maltose-binding protein (MBP). In many cases, the expression levels of these fusion proteins are much higher than the parental ones.20 It has also been reported that protein expression can be improved simply by single-point mutagenesis. To this end, Molina et al. subjected eight bacterial and one human membrane proteins to cycles of random mutagenesis.29 In five of these proteins, expression levels could be increased, as much as 40-fold, by one round of mutagenesis corresponding to approximately 6 mutations per 1000 residues. Further, the structure of the first β1-adrenergic receptor was solved with one mutation, cysteine-to-lysine, which reportedly improved expression.30
Membrane Protein Purification
Successfully overexpressed membrane proteins must be extracted and purified from the host’s lipid membrane before crystallization, with the intention of providing as similar an environment as possible to the native bilayer (Fig. 1). At the same time, we wish to leave out all the properties that make the bilayer unsuitable for crystallization, such as heterogeneity, polarity, charge, etc.24 Ideally, these should be replaced by the most important indicators of crystallizability: monodispersity, and stability.31 This is a difficult task, indeed. As stated by Bowie, membrane proteins are “delicate weaklings, unable to withstand the rigors of life outside the safety of the bilayer.”32 While detergent-based solubilization is the most popular and effective technique to replace the bilayer, most membrane proteins are only marginally stable even in the best available detergent.33
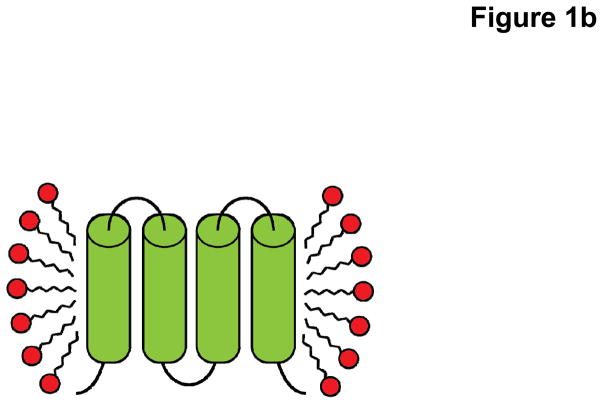
Fig. 1.
Cartoon representation cross-section of a membrane protein solubilized in (a) lipid bilayer of the host cell and (b) detergent introduced during the purification process. Upon removal of the protein from the native lipid (blue) environment, the protein-detergent complex consists of a uniform disordered ring of detergent monomers (red) making hydrophobic contacts with the surface of the protein (green).
The reason for their weakness is twofold: i) detergent, often smaller than the lipid environment it is replacing, may not completely cover the largely hydrophobic surface of the membrane protein and ii) membrane proteins, of diverse function, tend to exhibit much conformational flexibility, with some conformations more stable than others. Without the ordering imposed by the bilayer, they are free to adopt those conformations that may favor aggregation.33
Detergent
Beginning with the first membrane protein crystal structure in 1980, detergents have been rationally designed especially for the purpose of facilitating protein purification and crystallization. The number of these detergents is now estimated in the 100s.17 Nonetheless, only a small subset have been successful. The search for a suitable detergent benefits greatly from internet-based databases, which compile the crystallization conditions of all membrane proteins to date. For example, “Membrane Proteins of Known Structure” (http://blanco.biomol.uci.edu/mpstruc/) is particularly helpful.
At least structurally, all detergents are similar—a hydrophilic headgroup joined to a hydrophobic tail. This amphipathic character allows the detergent monomers to interact with both the hydrophobic surfaces of membrane proteins and solvent, as well as each other.34 All detergents used for membrane protein crystallization are alkyl-chain, between 7 and 14 carbons in length, with varying headgroups.17 A detergent falls into one of three categories based on the properties of the headgroup: ionic, nonionic, or zwitterionic (Fig. 2). Importantly, the most effective families of mild detergents for purification and crystallization have non-ionic or, less commonly, zwitterionic headgroups. Besides solubilization, other detergent effects on membrane proteins can include inactivation, denaturation, or aggregation.35 In these cases, harsh ionic detergents are often responsible. For example, the ionic detergent sodium dodecyl sulfate (SDS) is used in gel electrophoresis for exactly the purpose of protein denaturation. As of the publication of their paper in 2008, Newstead et al. reported that no outer membrane proteins to date had been crystallized with ionic detergent.36
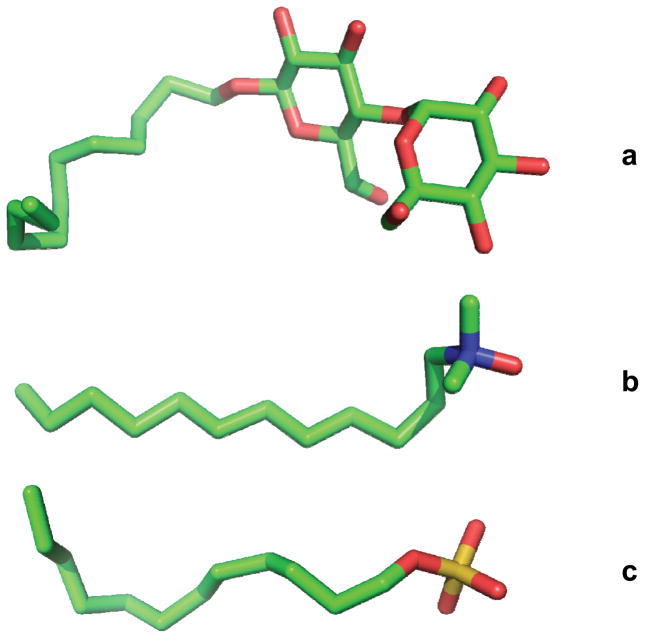
Fig. 2.
Representative structures of commonly used detergents of three classes. (a) n-Dodecyl-β-D-maltoside (DDM); non-ionic (b) Lauryldimethylamine-oxide (LDAO); zwitterionic. (c) Sodium dodecyl sulfate (SDS); anionic. Atoms are colored C, green; O, red; N, blue; and S, yellow.
To determine the effect of non-ionic and zwitterionic detergents on stability, Sonoda et al. used six common detergents to solubilize 24 prokaryotic and eukaryotic membrane transporters.37 The average stability measured was greatest for the two longest chain lengths of detergent, n-dodecyl-β-D-maltoside (DDM) and dodecyl nonaethylene glycol ether (C12E9), both non-ionic. In a similar study, Mancusso et al. examined 20 homologous lipid phosphatases.31 All 20 purified membrane proteins tended to be unstable in DDM solution, and all precipitated when concentrated over 1 mg/mL. When DDM was exchanged for other detergents, both the stability and monodispersity of the sample varied. In one of the tested samples, LP-1, both these traits improved when solubilized with 6-cyclohexyl-1-hexyl-β-D-maltoside (CYMAL-6). The size-exclusion chromatography peaks were sharper and the protein could be concentrated to 4 mg/mL. Taken together, these results indicate the preference of membrane proteins for long-chain, non-ionic detergents. For instance, DDM, with its 12 alkyl-carbons, is well known to stabilize membrane proteins and is often used for both solubilization and crystallization.38
Protein-Detergent Complex
After purification in the best available detergent, the membrane protein carries between 40 and 200% of its own weight in detergent.34,39 Save one or two molecules, all of these detergent coats are believed to be unstructured.40 Their shapes are also known, based on the low-resolution neutron structures of two reaction centers, from Rhodopseudomonas viridis and Rhodobacter sphaeroides.41,42 Remarkably, the shape of detergent surrounding each protein molecule is somewhat similar and is independent of the detergent molecules (n-dodecyl-N,N-dimethylamine-N-oxide (LDAO) and n-octyl-β-D-glucopyranoside (β-OG), respectively). In each case, the hydrophobic surface of the protein was covered in detergent molecules, wrapping the protein in a toroidal ring. Approximately 200 molecules cover 40% of the surface area of each reaction center. The shear amount of detergent in each protein-detergent complex, the building blocks of the crystal lattice, demonstrates that the detergent is at least as important as the protein itself in crystallization.
Within the solvent surrounding the protein-detergent complex, detergent monomers exist which self-associate and bind to the membrane protein surface in a concentration-dependent manner, with a minimum concentration equal to the critical micelle concentration (CMC). For example, at 0.3% (w/v) β-OG, each monomer of human prostaglandin synthase was found to bind at least 40 molecules of detergent. At 0.7% (w/v) β-OG, the number of bound detergent molecules roughly doubled.34 Crystallization is mostly done at detergent concentrations greater than the CMC, where equilibrium exists between detergent monomers, micelles, and detergent involved with the protein-detergent complex, with the concentration of monomers approximately equal to the CMC.17 As a higher CMC detergent will have more monomers in solution, it is expected that those monomers will exchange more readily between detergent micelles and the protein molecule surface.34
Membrane Protein Crystallization via Vapor Diffusion
Many techniques exist to crystallize membrane proteins, including the recently developed lipidic cubic phase,43,44 bicelle,45 and vesicle fusion46 methods. This chapter will focus on the vapor diffusion approach. The most common and successful method of membrane protein crystallization, to date, this approach has allowed researchers in our lab to determine the crystal structures of a number of membrane proteins. These include the inner membrane efflux pumps CusA,47 AcrB,48 and MtrD,49 the outer membrane channels CusC,50 MtrE,51 and CmeC,52 as well as the CusBA adaptor-transporter efflux complex.53
If not for the necessary presence of detergent, the crystallization of membrane proteins via vapor diffusion would be exactly similar to that of soluble proteins. While protein-protein interactions dominate the crystallization of soluble proteins, both detergent-detergent and protein-detergent interactions are additional considerations for the membrane protein crystallographer. Unlike crystallization of water-soluble proteins, choosing the right detergent has been the key success for our membrane protein crystallization efforts. In our opinion, this is the most important factor for obtaining high quality membrane protein crystals suitable for X-ray diffraction. Thus, the rule of thumb is that more efforts should be made to screen a variety of mild detergents than to optimize other parameters when crystallizing a new membrane protein.
In a simple crystallization experiment, a precipitant solution, containing at least buffer, precipitant, and salt components, is mixed with a protein solution, containing the purified protein-detergent complexes. This forces the protein solution into a state of supersaturation. To escape, the protein-detergent complex has two routes: i) aggregation and ii) crystallization. If the crystallization solution is not too harsh and the protein-detergent complex has enough time to make specific interactions with its neighbors, the latter results. In general, a slow introduction of the crystallization solution to the protein-detergent complex facilitates this, resulting in larger crystals with fewer imperfections.54
Compared to other techniques, the conditions for crystallizing membrane protein via vapor diffusion tend to be very mild, involving a mixture of relatively low concentrations of precipitant, salt, and buffer. In short, a small quantity of the purified protein solution is mixed, usually in equal ratio, with the crystallization solution contained in the well (precipitant solution). This drop, now half precipitant solution and half protein solution, is either mounted in the well itself (sitting-drop) (Fig. 3a) or inverted above the well (hanging-drop) (Fig. 3b), and the chamber is made airtight. Vapor diffusion describes the evaporation of volatile species in the drop into the chamber, followed by diffusion across the chamber into the well. The end result of this process is chemical equilibrium between the drop and well. Because the well is much bigger, the equilibrium concentrations of drop components are approximated by the concentrations in the well.
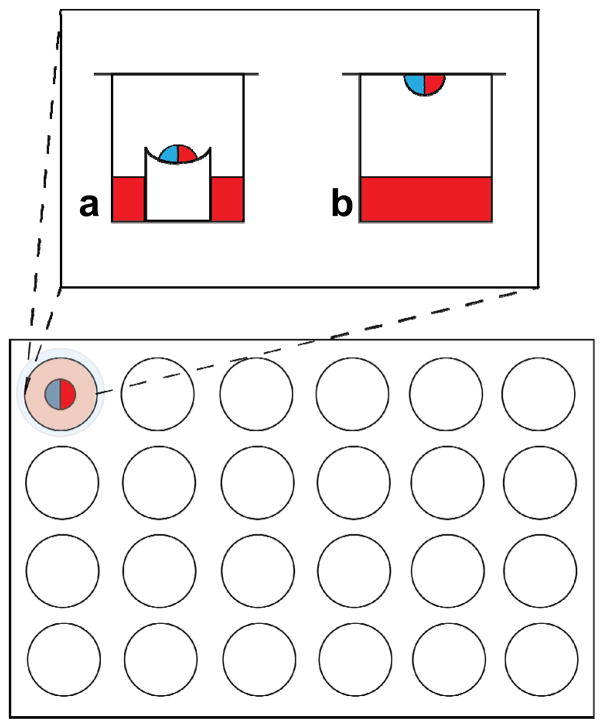
Fig. 3.
A 24-well plate in which either (a) sitting-drop or (b) hanging-drop vapor diffusion is done. The well solution (colored red) is mixed in equal proportion with protein solution (colored blue). In the sitting-drop technique, the resulting mixture sits on a pedestal above the well solution and the top of the chamber is sealed with grease and glass. In the hanging-drop technique, the mixed drop is included on the glass slide itself, inverted and sealed with grease atop the chamber.
For simplicity, it is generally assumed that only water is exchanged between the drop and well.55 However, any volatile species can evaporate into the chamber and exchange between the drop and well. When it reaches equilibrium, or close to it, the hanging or sitting drop will manifest different types and degrees of phase behavior. In addition to the protein, the detergent and the interactions between the two must be considered. The phase transition of detergent is relatively complex. It can crystallize, aggregate into micelles, or separate into detergent-rich, and detergent-poor phases.25,56 Thus, searching for the right crystallization condition involves exploring point-by-point a complicated many-dimensional phase diagram for both protein and detergent.
The dimensions of this phase space include, at least, both protein and detergent concentrations, detergent type, precipitant concentration and type, buffer pH and type, salt concentration and type, and additive concentration and type. Further, the optimal crystallization conditions occupy only a very small volume of this phase space. Finally, the contribution of these factors to crystallization is non-linear. In general, the effect of one component on the solubility of the protein is coupled to the effect of another.57 Even with the advent of robotic screening technology, capable of testing up to 40,000 conditions per day,58 crystallization cannot be approached naively.
There are two general approaches to navigating the phase space of a protein: i) a ‘grid search,’ by systematic screening of what are believed to be the important variables,59 or ii) using a ‘coarse matrix screen,’ heavily biased on known crystallization conditions of other proteins.60 To date, many coarse matrix screens designed specifically for membrane proteins, including MemGold (Molecular Dimensions) and MembFac (Hampton Research), have been made commercially available. In many cases, these screening kits are instrumental in the search for initial crystallization conditions. The individual components of these conditions and their roles in the crystallization process will be discussed in the following sections, with an emphasis on the most common and successful ones.
Detergent
Some say that the biggest hurdle in membrane protein crystallization is not obtaining the crystals, but optimizing them for high-resolution X-ray diffraction.37 In this respect, no other variable is more influential in the success of membrane protein crystallization than detergent.
Although no single detergent category can be generally applied to membrane protein crystallization, statistical evidence suggests that some are more useful than others. Almost all successfully used detergents are nonionic alkyl sugars with C7-C14 alkyl chains. By far, the most crystal structures have been solved using the alkylmaltoside family of detergents—nearly one half of all α-helical proteins.61 This family includes the single most popular detergent, DDM, which contains 12 carbons in the main alkyl chain. By contrast, the alkylglucoside family of detergents are most commonly used to crystallize outer membrane proteins.37 They include the second most popular detergent, β-OG, which is an 8 carbon alkyl-chained detergent.
Recently, the development of new detergents has emerged as a tool for handling difficult membrane proteins.62 Non-conventional approaches, such as amphipols,63 nanodiscs,63 hemifluorinated surfactants,63,64 cholate acid-based amphiphiles,65 and the new family of maltose-neopentyl glycol amphiphiles66 have been proven to aid in the solubilization of membrane proteins, while retaining their structure and function. This new generation of amphiphiles is particularly promising, and may eventually have a large role in the solubilization, stabilization, and crystallization of membrane proteins for high resolution structural analysis.
Precipitant
Based on their mechanism of action, precipitants can be divided into three categories: i) salts, ii) organic solvents, and iii) long-chain polymers. As will be discussed in the following section, salt acts as a precipitant by either dehydrating the protein molecules or altering the ionic strength.
Organic solvents, such as ethanol or isopropanol, generally act to reduce the dielectric constant of the crystallization solution. This makes electrostatic interactions weaker and reduces the solubility of ionic compounds slightly. Interestingly, one of the first membrane proteins to be crystallized, the hydrophobic seed protein crambin, was initially solubilized in ethanol and crystallized by slowly adding water.34,67 At high concentrations, most of these compounds are denaturing. However, 2-Methyl-2,4-pentanediol (MPD) has been remarkably successful for outer membrane protein crystallization.36
The third and most successful category of precipitants comprises the long-chain polymers, including polyethylene glycols (PEGs). More than 80% of all membrane proteins have been successfully crystallized with various molecular weights of PEGs36,61 and 100% of all membrane proteins in our lab. The mechanism of action of these polymers is attributed to volume exclusion effects.57 Unlike other components of the crystallization solution, PEGs have no consistent conformation, resembling a flail. PEGs with molecular weights between 400 and 20000 Da have been effective for membrane protein crystallization, with the larger ones obviously being more forceful.
Salt
Salt ions, both cationic and anionic, can alter the solubility of proteins. This effect was first discovered by Franz Hofmeister.68 The action mechanism is not entirely clear. However, it is thought to be due to specific interactions between ions, protein macromolecules, and water molecules in the solvent layer surrounding the protein. The effect of anions are usually ordered:
The order of cations is usually given as:
Early members of the series tend to increase solvent surface tension and decrease the solubility of nonpolar molecules. They strengthen the hydrophobic interaction and this effect is often referred to “salting out”. In this way, ammonium sulfate precipitation is commonly used in protein purification. Additionally, more than 10% of membrane protein crystals reported in the PDB are crystallized with the help of this salt.61 By contrast, later ions in the series tend to increase the solubility of nonpolar molecules and decrease the order of water molecules. They weaken the hydrophobic interaction and this effect is called “salting in”. Typically, ions that have a strong “salting in” effect are strong denaturants, because they interact much more strongly with the unfolded protein than with its native folded form.
Buffer
The actual net charge of the protein is determined not only by its composite amino acids, but by the pH of the solvent.69 For each amino acid, its charge is determined by its pKa; at a pH below the pKa, an amino acid is positively charged and vice versa. Therefore, altering the pH of the crystallization solution will alter the degree of electrostatic interaction, mainly, between the protein-detergent complexes.
At a pH equal to the pI, which is an average of the pKa of the surface residues of the protein, the net charge is neutralized and solubility drops dramatically. While this is usually too strong a condition for crystallization, it is common to find the optimal crystallization pH nearby.69 The pH determines the charge of the zwitterionic detergents in a similar way.
Although no direct correlation appears to exist between predicted pI and the pH at which crystallization was reported, there is a good correlation between their difference, pH-pI, and pI. Specifically, acidic proteins tend to crystallize between 0 and 2.5 pH units above their pI, while basic proteins crystallize between 0.5 and 3 pH units below their pI.70
Case Studies
Crystallization of the E. coli CusA Heavy-Metal Efflux Pump
Initially, we cloned the E. coli cusA gene into pET15b to form the expression vector pET15bΩcusA. This expression vector was then used to transform E. coli BL21(DE3) cells and overexpress the full-length CusA membrane protein, containing a 6xHis tag at the N-terminus. The expressed protein was purified using a Ni2+-affinity column to >95% purity. The final purified protein was then dialyzed and concentrated to 20 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5) and 0.05% DDM for crystallization trials. The original protein crystals crystallized fairly quickly, and these crystals diffracted X-rays to 4 Å resolution. However, mass spectrometry identified these as crystals of the E. coli AcrB protein, which was probably co-purified with the CusA protein as a contaminant.
To avoid further contamination with the AcrB protein, we made an E. coli knockout strain BL21(DE3)ΔacrB, which harbors a deletion in the chromosomal acrB gene. The CusA transporter was then overexpressed using the transformed cells BL21(DE3)ΔacrB/pET15bΩcusA and purified using a Ni2+-affinity column. The purified protein was then dialyzed, concentrated to 20 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5) and 0.05% DDM, and subjected to crystallization trials using sitting-drop vapor diffusion. This time, mass spectrometry confirmed that the crystals were composed of CusA molecules. However, these crystals diffracted X-rays poorly, to ~9 Å resolution. After extensive screening, with as many salts, PEGs, pHs, temperatures, additives, and detergents available to us, we finally obtained plate-shaped crystals by adding 0.05% CYMAL-6 to the initial crystallization condition. Although these plate-shaped crystals were much better in quality, they diffracted X-rays anisotropically to a resolution of ~5 Å in one dimension and ~8 Å in the other.
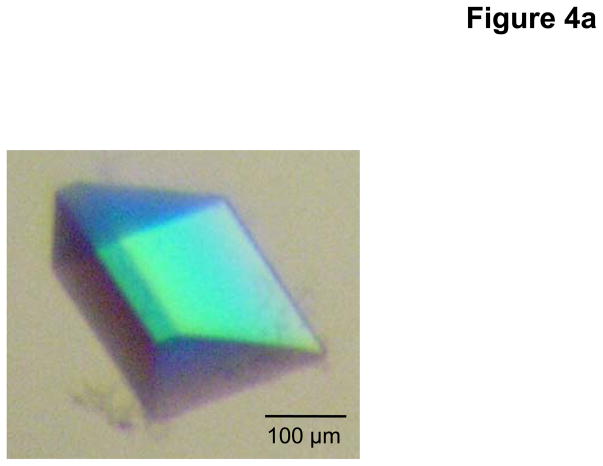
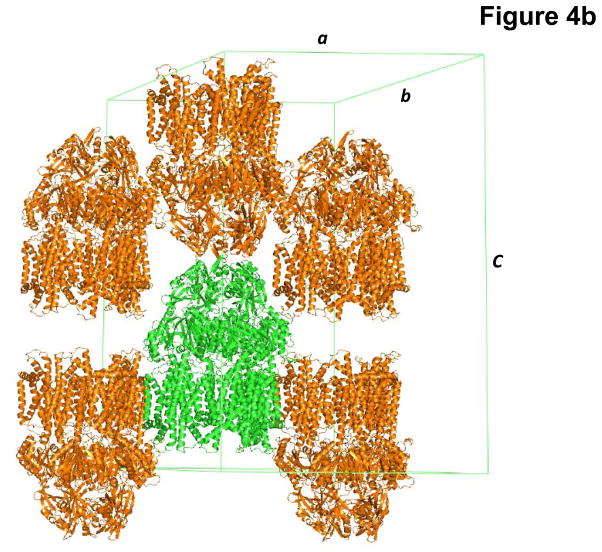
As the addition of CYMAL-6 significantly improved the crystal quality, we decided to use CYMAL-6 to purify and crystallize the CusA protein. During this process, we also found that the additives Jeffamine M-600 (JM-600) and glycerol further improved their quality. Thus, the final crystallization condition was 2 μl protein solution (containing 20 mg/ml CusA protein in 20 mM Na-HEPES (pH 7.5) and 0.05% (w/v) CYMAL-6) mixed with 2 μl of reservoir solution containing 10% PEG 3350, 0.1 M NaMES (pH 6.5), 0.4 M (NH4)2SO4, 1% JM-600, and 10% glycerol. The resulting mixture was equilibrated against 500 μl of the reservoir solution. Typically, the dimensions of the crystals were 0.2 mm × 0.2 mm × 0.2 mm (Fig. 4a). Crystals of CusA were found to be trigonal with space group R32 (a = b = 178.4 Å, c = 285.8 Å) (Fig. 4b).47 Based on the molecular weight of CusA (115.72 kDa), it was found that a single molecule occupied the asymmetric unit with a solvent content of 67.5%. The final crystal structure of the CusA transporter was determined to a resolution of 3.5 Å.
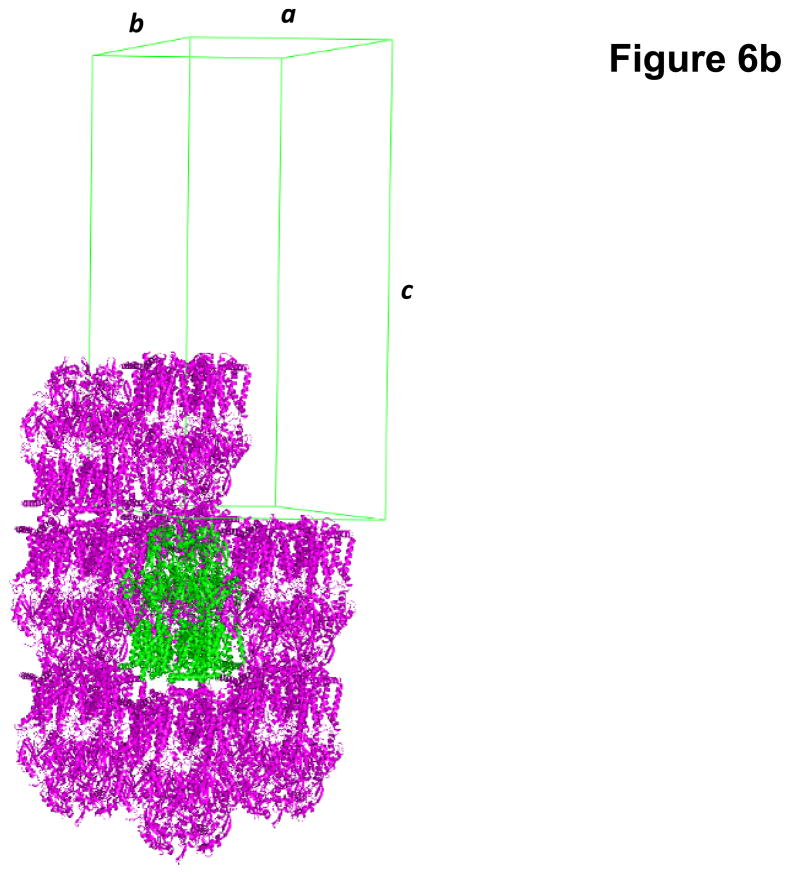
Fig. 4.
Crystal structure of the E. coli inner membrane heavy metal efflux pump CusA. (a) A single crystal of CusA. (b) Packing diagram of the CusA crystal viewed orthogonal to the long axis of the unit cell.
Crystallization of the E. coli CusBA Adaptor-Transporter Complex
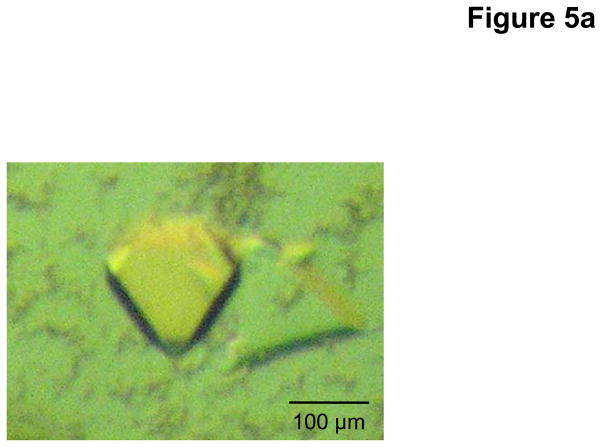
As of this article’s publication, CusBA remains the only adaptor-transporter complex for which a high resolution X-ray structure is available. For purification of this important complex, the individual proteins of the CusA transporter and CusB71 adaptor were individually overexpressed in E. coli cells BL21(DE3)ΔacrB/pET15bΩcusA and BL21(DE3)ΔacrB/pET15bΩcusB, respectively. The CusA protein was solubilized with 2% CYMAL-6 and purified using Ni2+-affinity chromatography, as mentioned previously. The CusB protein was also purified using a Ni2+-affinity column, without the addition of any detergent. Co-crystals of the CusBA complex were obtained using sitting-drop vapor diffusion. After extensive optimization, the CusBA crystals were grown at room temperature in 24-well plates with a final crystallization condition of 2 μl protein solution (containing 0.1 mM CusA and 0.1 mM CusB in 20 mM Na-HEPES (pH 7.5) and 0.05% (w/v) CYMAL-6) mixed with 2 μl of reservoir solution containing 10% PEG 6000, 0.1 M Na-HEPES (pH 7.5), 0.1 M ammonium acetate, and 20% glycerol. The resulting mixture was equilibrated against 500 μl of the reservoir solution. Co-crystals of CusBA grew to a full size in the drops within two months. Typically, the dimensions of the crystals were 0.1 mm × 0.1 mm × 0.1 mm (Fig. 5a).
Fig. 5.
Crystal structure of the E. coli inner membrane heavy metal efflux pump CusA in complex with the periplasmic membrane fusion protein CusB. (a) A single crystal of the CusBA complex. (b) Packing diagram of the CusBA cocrystal viewed orthogonal to the long axis of the unit cell.
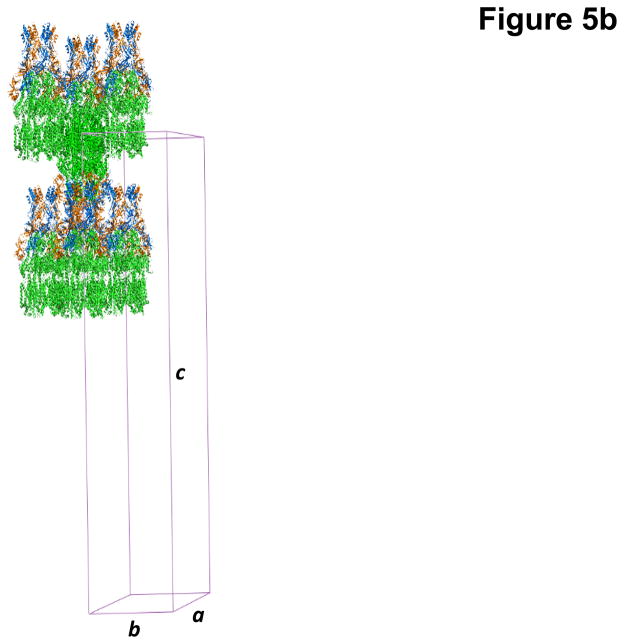
The co-crystals of the CusBA adaptor-transporter complex took a trigonal space group R32 with unit cell parameters a = b = 160.2 Å, c = 682.7 Å (Fig. 5b).53 Based on the molecular weights of CusA (115.7 kDa) and CusB (42.3 kDa), the asymmetric unit contained one CusA and two CusB molecules with a solvent content of 70.8%. As it turns out, the presence of CusB drastically improved crystal quality in comparison with the crystals of CusA alone. The reason for this may be that CusB enlarges that hydrophilic surface of the membrane protein, thereby providing additional surface for crystal contacts. The final structure of the CusBA adaptor-transporter complex was determined to a resolution of 2.9 Å.
Crystallization of the N. gonorrhoeae MtrD Multidrug Efflux Pump
It took us a few years of effort to crystallize the MtrD membrane protein. First, two different constructs, based on the protein sequence of MtrD in N. gonorrhoeae strain FA19, were used to express the MtrD protein, containing a 6xHis tag at the N- or C-terminus, respectively. The open reading frame (ORF) of mtrD from N. gonorrhoeae FA19 was cloned into pET15b to form each expression vectors. After screening of expression conditions, only the C-terminal 6xHis tagged MtrD protein could be expressed in E. coli C43(DE3)ΔacrB cells, which harbor a deletion in the chromosomal acrB gene. Using these conditions, we then purified the MtrD protein using a Ni2+-column. Optimization of the initial crystals with respect to primary and secondary detergent, salt, precipitant, pH and temperature, yielded only showers of microcrystals. Unfortunately, these microcrystals did not even diffract X-rays to low resolution.
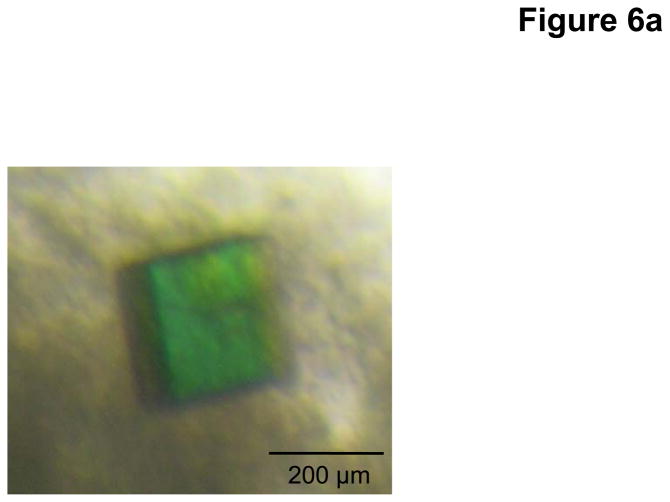
Based on protein sequence alignments of different N. gonorrhoeae strains, we decided to focus on the homologous MtrD efflux pump from N. gonorrhoeae strain PID332. The alignment indicated that MtrD of strain PID332 is 11 amino acids shorter at the C-terminus in comparison with FA19 MtrD. We then cloned the N. gonorrhoeae PID332 mtrD gene into pET15b to generate the pET15bΩmtrD expression vector. This recombinant plasmid encoding the N. gonorrhoeae PID332 MtrD protein, with a 6xHis at the C-terminus, was then transformed into C43(DE3)ΔacrB cells to overexpress this protein. Upon expression, MtrD was solubilized in 2% CYMAL-6 and subjected to purification using Ni2+-affinity chromatography. Initial crystallization trials, using 0.05% CYMAL-6 as a primary detergent, did not yield any crystals. However, small cubic shaped crystals appeared in the drops after adding sucrose monododecanoate (SM) as a secondary detergent. Subsequent crystallization trials were carried out using both CYMAL-6 and SM detergents. After extensive optimization, the final MtrD crystals were grown at room temperature using sitting-drop vapor diffusion with the following procedures. A 2 μl protein solution containing 0.2 mM MtrD in buffer solution (20 mM Na-HEPES (pH 7.5), 0.05% (w/v) CYMAL-6, and 0.5% (w/v) SM) was mixed with 2 μl of reservoir solution, containing 30% PEG 400, 0.1 M Na-Bicene (pH 8.5), 0.1 M NH4SO4, 0.05 M BaCl2, and 9% glycerol. The resulting mixture was equilibrated against 500 μl of the reservoir solution. Typically, the dimensions of the crystals were 0.2 mm × 0.2 mm × 0.2 mm (Fig. 6a). Crystals of MtrD belonged to space group R32 (a = b = 153.0 Å, c = 360.7 Å) (Fig. 6b).49 Analysis of the Matthews coefficient indicated the presence of one MtrD molecule (113.69 kDa) per asymmetric unit, with a solvent content of 66.7%. The final crystal structure of the MtrD transporter was determined to a resolution of 3.5 Å.
Fig. 6.
Crystal structure of the N. gonorrhoeae inner membrane multidrug efflux pump MtrD. (a) A single crystal of MtrD. (b) Packing diagram of the MtrD crystal viewed orthogonal to the long axis of the unit cell.
Crystallization of the E. coli CusC Heavy-Metal Efflux Channel
The CusC outer membrane channel is one of the most well-studied proteins in our laboratory. Its structures have allowed us to unmask the sequential transition of conformations leading to the folding and membrane insertion of this channel. To express CusC, the ORF of cusC from E. coli K12 genomic DNA was cloned into pBAD22 to produce the expression vector pBAD22ΩcusC. This expression system included a CusC signaling peptide at the N-terminus as well as a 6xHis tag at the C-terminal end. The N-terminal signaling peptide was needed to guide the channel protein to express at the outer membrane. It should be noted that this signaling peptide would be removed automatically when the CusC protein was matured in the cell. This recombinant plasmid was then used to transform the C43(DE3)ΔacrB cells to overexpress the channel protein. To ensure that the harvested CusC protein was attached to or anchored the E. coli outer membrane, and not the inner membrane, we performed a pre-extraction procedure using 0.5% sodium lauroyl sarcosinate to selectively dissolve and remove proteins of the inner membrane. The CusC outer membrane protein was then solubilized in 2 % (w/v) DDM. The extracted protein was purified with a Ni2+-affinity column to >95% purity. The purified protein was then dialyzed and concentrated to 15 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5), 200 mM NaCl, and 0.05% (w/v) DDM for crystallization trials.
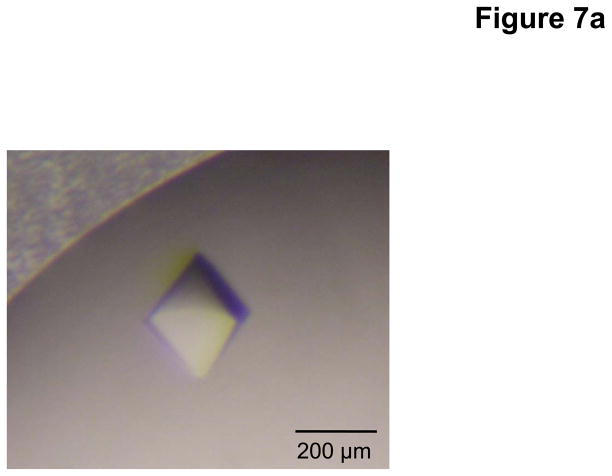
Sitting-drop vapor diffusion was employed for initial crystal screening. During this screening, we found that the full-length CusC channel could be crystallized only in the presence of 2% (w/v) β-OG, which served as a secondary detergent to DDM. It was also found that the addition of a small amount of JM 600 could improve the quality of the crystals. The best crystals of CusC were grown at room temperature with a reservoir solution containing 8% PEG 3350, 0.05 M sodium acetate (pH 4.0), 0.2 M (NH4)2SO4, 1% JM 600, and 2% β-OG. Typically, the dimensions of the crystals were 0.2 mm × 0.2 mm × 0.2 mm (Fig. 7a).
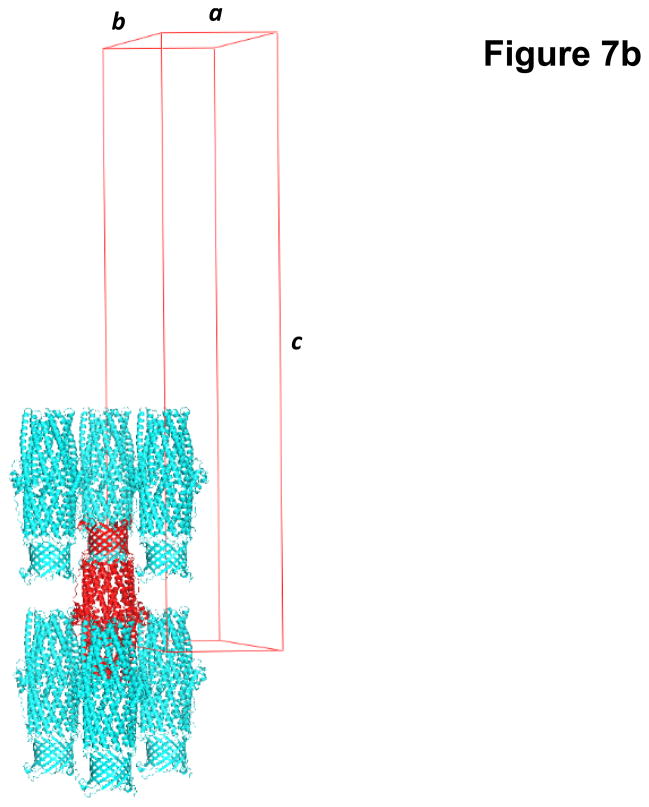
Fig. 7.
Crystal structure of the E. coli outer membrane channel CusC. (a) A single crystal of CusC. (b) Packing diagram of the CusC crystal viewed orthogonal to the long axis of the unit cell.
Crystals of the full-length CusC channel took the space group R32 with the unit-cell parameters a = b = 88.5 Å, c = 474.42 Å (Fig. 7b).50 Based on the molecular weight of CusC (49.30 kDa), only one CusC molecule occupied the asymmetric unit with a solvent content of 67.8%. The final model was refined to a resolution of 2.1 Å.
It is interesting to note that a single point mutation in the first cysteine residue of CusC resulted in a dramatically different conformation. Instead of the four-stranded beta-sheet present in the wild-type CusC monomer, the corresponding residues in this mutant form two independent random loops. By mutating or deleting this key residue, we obtained a different structure entirely, which corresponds to the state immediately before membrane insertion.50 In this membrane protein, even the modification of one residue can prevent anchoring and insertion into the bilayer. Therefore, one should be cautious when crystallizing membrane proteins with any mutations or truncated sequences.
Crystallization of the N. gonorrhoeae MtrE Multidrug Efflux Channel
The construct used to express the N. gonorrhoeae MtrE outer membrane channel was similar to that of E. coli CusC. However, after several trials, we found that the signaling peptide sequence of Pseudomonas aeruginosa OprM was the best peptide for expressing the N. gonorrhoeae MtrE heterogeneously in E. coli. In short, the ORF of mtrE from N. gonorrhoeae FA19 was cloned into pBAD22 to form the expression vector pBAD22ΩmtrE, which included the P. aeruginosa OprM signaling peptide at the N-terminus and a 6xHis tag at the C-terminus. The full-length MtrE protein was then expressed in E. coli C43(DE3) cells possessing this vector. The procedures for protein extraction and purification were the same as those for the CusC membrane protein. The purified protein was dialyzed and concentrated to 15 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5), 200 mM NaCl, and 0.05% (w/v) DDM for crystallization trials.
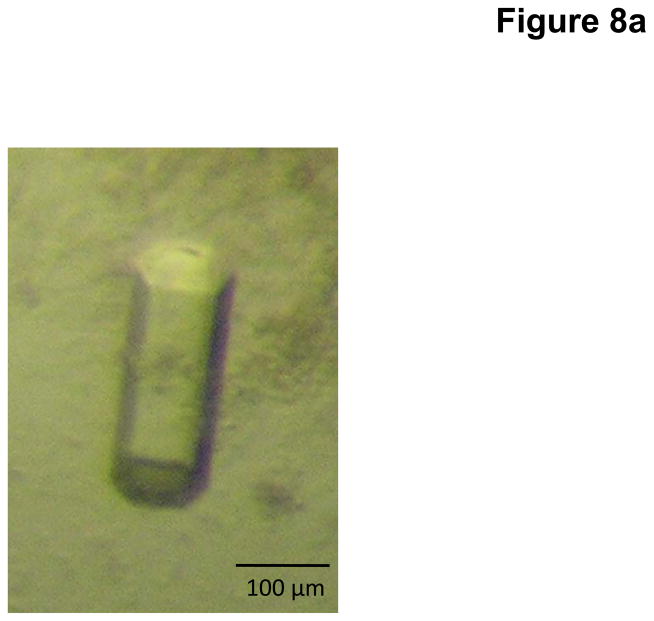
Crystals of MtrE were obtained using sitting-drop vapor diffusion. Based on crystal screening, the following procedure was adopted. A 2 μl protein solution containing 15 mg/ml MtrE protein in 20 mM Na-HEPES (pH 7.5), 200 mM NaCl, and 0.05% (w/v) DDM was mixed with a 2 μl of reservoir solution containing 20% PEG 400, 0.2 M sodium acetate (pH 4.6), 0.25 M MgSO4, and 2% (w/v) β-OG. The resultant mixture was equilibrated against 500 μl of the reservoir solution at room temperature. Crystals of MtrE grew to a full size in the drops within two weeks. Typically, the dimensions of the crystals were 0.1 mm × 0.1 mm × 0.2 mm (Fig. 8a).
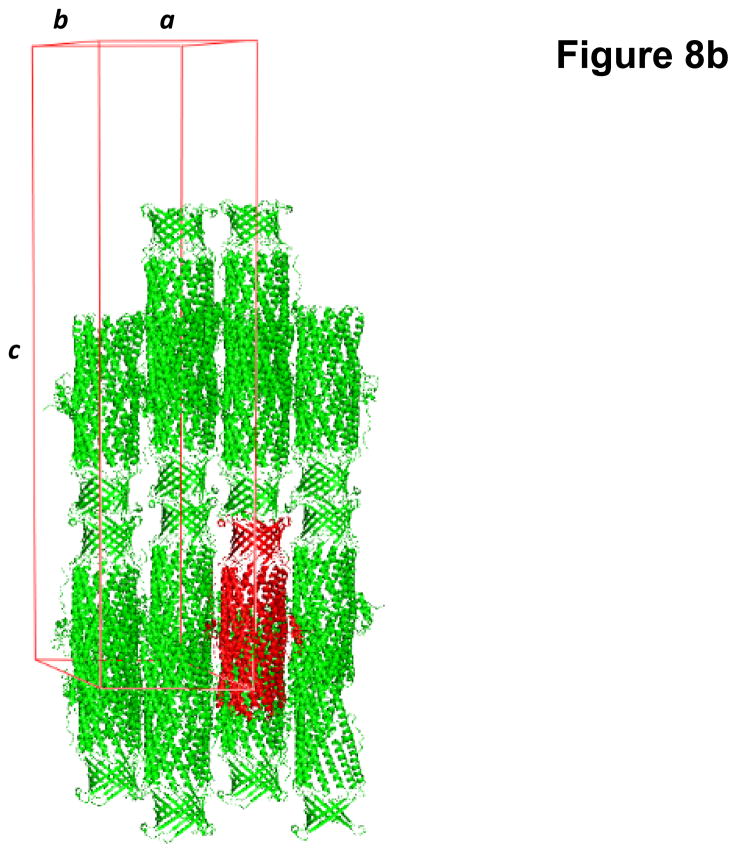
Fig. 8.
Crystal structure of the N. gonorrhoeae outer membrane channel MtrE. (a) A single crystal of MtrE. (b) Packing diagram of the MtrE crystal viewed orthogonal to the long axis of the unit cell.
Crystals of the MtrE channel protein belonged to the space group P6322 (a = b = 93.9 Å, c = 391.5 Å) (Fig. 8b).51 Analysis of the Matthews coefficient indicated the presence of one MtrE protomer (49.29 kDa) per asymmetric unit, with a solvent content of 75.8%. The final structural model was resolved to a resolution of 3.3 Å.
Crystallization of the C. jejuni CmeC Multidrug Efflux Channel
The construct used to express the C. jejuni CmeC outer membrane channel was similar to that of N. gonorrhoeae MtrE. Briefly, the full-length CmeC membrane protein containing a 6xHis tag at the C-terminus was overproduced in E. coli C43(DE3)/pBAD22bΩcmeC cells. This expression system included an OprM signaling peptide at the N-terminus and a 6xHis tag at the C-terminus. The purified protein was dialyzed and concentrated to 15 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5), 200 mM NaCl and 0.05% DDM.
Initial crystallization trials were not successful and did not yield even poor quality crystals. These trials were followed by extensive screening of secondary detergents. Fortunately, hexagonal shaped CmeC crystals were obtained, but only in drops containing the detergent C8E4. Further optimization of the crystallization conditions eventually produced well diffracting crystals.
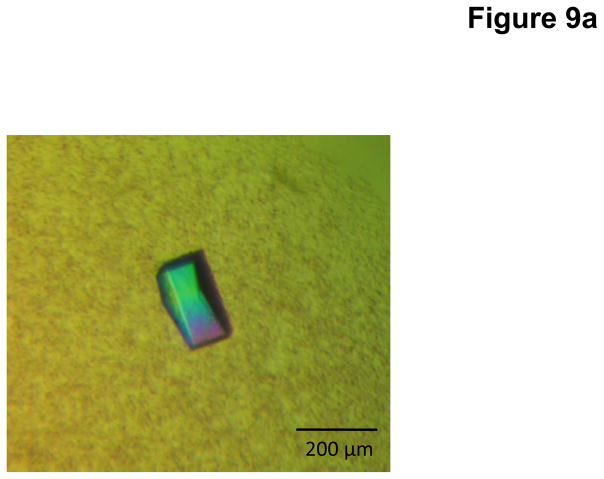
Crystals of CmeC were obtained using sitting-drop vapor diffusion at room temperature in 24-well plates with the following procedures. A 2 μl protein solution containing 15 mg/ml CmeC protein in 20 mM Na-HEPES (pH 7.5), 200 mM NaCl, and 0.05% (w/v) DDM was mixed with 2 μl of reservoir solution containing 18% PEG 400, 0.1 M sodium acetate (pH 4.0), 0.3 M (NH4)2SO4, and 2% C8E4. The resulting mixture was equilibrated against 500 μl of the reservoir solution. Crystals of CmeC grew to a full size in the drops within two weeks. Typically, the dimensions of the crystals were 0.1 mm × 0.2 mm × 0.2 mm (Fig. 9a).
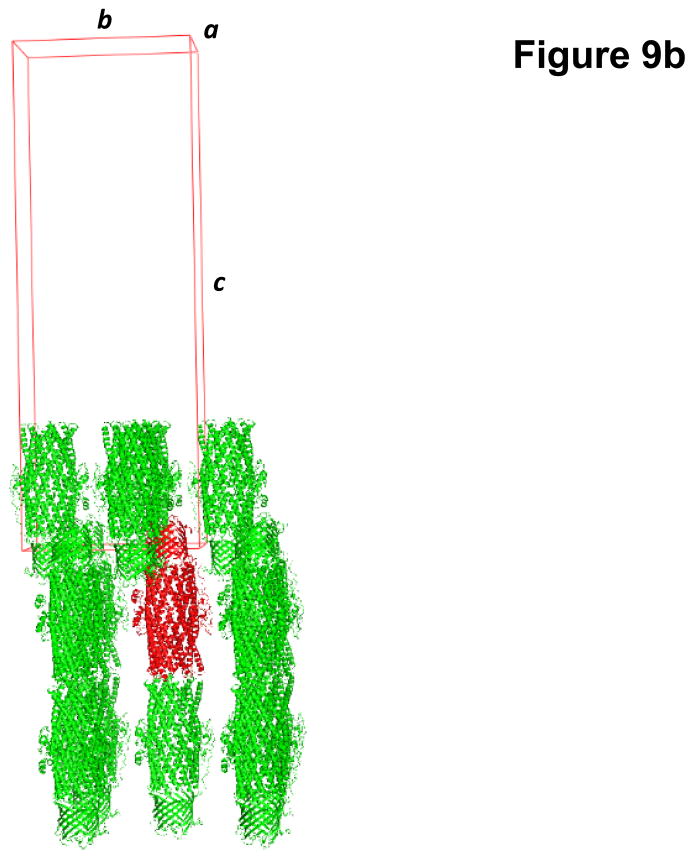
Fig. 9.
Crystal structure of the C. jejuni outer membrane channel CmeC. (a) A single crystal of CmeC. (b) Packing diagram of the CmeC crystal viewed orthogonal to the long axis of the unit cell.
Crystals of the CmeC outer membrane channel belonged to the space group C2221 with unit-cell parameters: a = 92.38 Å, b = 147.35 Å, c = 420.43 Å (Fig. 9b).52 Based on the molecular weight of CmeC (54.26 kDa), three molecules were found in the asymmetric unit with a solvent content of 80.6%. These three molecules assembled to form a trimeric channel within the unit cell. The final crystal structure was determined to a resolution of 2.4 Å.
Concluding Remarks
For the routine availability of three-dimensional membrane protein structures to become possible, there is still a mountain to climb. Like the state of soluble protein crystallization 20 years ago, initial data is still being acquired to facilitate the large-scale investigations demanded today. Only then can more lofty goals of crystallization be achieved. For example, no atomic-resolution model of a drug efflux complex, spanning both the inner and outer bilayer, has been reported to date. In Gram-negative bacteria, such complexes are responsible for conferring resistance to commonly used antibiotics, a problem that is exacerbated by the increased use of these drugs. While bacterial infections remain a leading cause of death worldwide, invaluable structural information leading to antibiotic resistance mechanisms would provide a platform to produce new drugs and inhibitors that increase the efficacy of our weakened therapies.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Science. 1998;7(4):1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane topology with a hidden markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 3.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. Beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248(4962):1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-couple receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 5.Saier MH, Jr, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Molecular Microbiology. 1994;11(5):841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 6.Lee S, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450(7168):376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 8.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 9.Weiss MS, Abele U, Weckesser J, Welte W, Schiltz E, Schulz GE. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991;254(5038):1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- 10.Drew D, Fröderberg L, Baars L, de Gier JL. Assembly and overexpression of membrane proteins in Escherichia coli. Biochimica et Biophysica Acta. 2003;1610(1):3–10. doi: 10.1016/s0005-2736(02)00707-1. [DOI] [PubMed] [Google Scholar]
- 11.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nature Reviews Drug Discovery. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 12.Bernaudat F, Frelet-Barrand A, Pochon N, Dementin S, Hivin P, Boutigny S, Rioiux J, Salvi D, Seigneurin-Berny D, Richaud P, Joyard J, Pignol D, Sabaty M, Desnos T, Pebay-Peyroula E, Darrouzet E, Vernet T, Rolland N. Heterologous expression of membrane proteins: choosing the appropriate host. PLoS ONE. 2011;6:e29191. doi: 10.1371/journal.pone.0029191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thieme D, Neubauer P, Nies DH, Grass G. Sandwich hybridization assay for sensitive detection of dynamic changes in mRNA transcript levels in crude Escherichia coli cell extracts in response to copper ions. Applied and Environmental Microbiology. 2008;74(24):7463–7470. doi: 10.1128/AEM.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bill RM, Henderson PJF, Iwata S, Kunji ERS, Michel H, Neutze R, Newstead S, Poolman B, Tate CG, Vogel H. Overcoming barriers to membrane protein structure determination. Nature Biotechnology. 2011;29(4):335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- 15.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature. 1985;318(6047):618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 16.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conductivity and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 17.Wiener MC. A pedestrian guide to membrane protein crystallization. Methods. 2004;34(3):364–372. doi: 10.1016/j.ymeth.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 18.Sahdev S, Khattar SK, Saini KS. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Molecular and Cellular Biochemistry. 2008;307(1):249–264. doi: 10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- 19.Schertler GFX. Overproduction of membrane proteins. Current Opinion in Structural Biology. 1992;2(4):534–544. [Google Scholar]
- 20.Pryor KD, Leiting B. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expression and Purification. 1997;10(3):309–319. doi: 10.1006/prep.1997.0759. [DOI] [PubMed] [Google Scholar]
- 21.Gabrielsen M, Gardiner AT, Fromme P, Cogdell RJ. Membrane protein crystallization: approaching the problem and understanding the solutions. Current Topics in Membranes. 2009;63:127–149. [Google Scholar]
- 22.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Current Opinion in Biotechnology. 1995;6(8):494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 23.Long F, Rouquette-Loughlin C, Shafer WM, Yu EW. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli. Antimicrobial Agents and Chemotherapy. 2008;52(9):3052–3060. doi: 10.1128/AAC.00475-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caffrey M. Membrane protein crystallization. Journal of Structural Biology. 2003;142(1):108–132. doi: 10.1016/s1047-8477(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 25.Loll PJ. Membrane protein structural biology: the high throughput challenge. Journal of Structural Biology. 2003;142(1):144–153. doi: 10.1016/s1047-8477(03)00045-5. [DOI] [PubMed] [Google Scholar]
- 26.Schmid B, Krömer M, Schulz GE. Expression of porin from Rhodopseudomonas blastica in Escherichia coli inclusion bodies and folding into exact native structure. Federation of European Biochemical Societies Letters. 1996;381(1):111–114. doi: 10.1016/0014-5793(96)00080-4. [DOI] [PubMed] [Google Scholar]
- 27.Bannwarth M, Schulz GE. The expression of outer membrane proteins for crystallization. Biochimica et Biophysica Acta. 2003;1610(1):37–45. doi: 10.1016/s0005-2736(02)00711-3. [DOI] [PubMed] [Google Scholar]
- 28.Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Högbom M, van Wijk KJ, Slotboom DJ, Persson JO, de Gier J. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molina DM, Cornvik T, Eshaghi S, Haeggström JZ, Nordlund P, Sabet MI. Engineering membrane protein overproduction in Escherichia coli. Protein Science. 2008;17:673–680. doi: 10.1110/ps.073242508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454(7203):486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mancusso R, Karpowich N, Czyzewski B, Wang D. Simple screening method for improving membrane protein thermostability. Methods. 2011;55(4):324–329. doi: 10.1016/j.ymeth.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowie JU. Stabilizing membrane proteins. Current Opinion in Structural Biology. 2001;11(4):397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y, Bowie JU. Building a thermostable membrane protein. Journal of Biological Chemistry. 2000;275(10):6975–6979. doi: 10.1074/jbc.275.10.6975. [DOI] [PubMed] [Google Scholar]
- 34.Garavito RM, Picot D, Loll PJ. Strategies for crystallizing membrane proteins. Journal of Bioenergetics and Biomembranes. 1996;28(1):13–27. [PubMed] [Google Scholar]
- 35.De Grip WJ. Thermal stability of rhodopsin and opsin in some novel detergents. Methods in Enzymology. 1982;81:256–265. doi: 10.1016/s0076-6879(82)81040-9. [DOI] [PubMed] [Google Scholar]
- 36.Newstead S, Hobbs J, Jordan D, Carpenter EP, Iwata S. Insights into outer membrane protein crystallisation. (2010) Molecular Membrane Biology. 2008;25(8):631–638. doi: 10.1080/09687680802526574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonoda Y, Newstead S, Hu N, Alguel Y, Niji E, Beis K, Yashiro S, Lee C, Leung J, Cameron AD, Byrne B, Iwata S, Drew D. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure. 2011;19(1):17–25. doi: 10.1016/j.str.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang HJ, Lee C, Drew D. Breaking the barriers in membrane protein crystallography. The International Journal of Biochemistry and Cell Biology. 2013;45(3):636–644. doi: 10.1016/j.biocel.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 39.Møller J, Le Maire M. Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. Journal of Biological Chemistry. 1993;268(25):18659–18672. [PubMed] [Google Scholar]
- 40.Deisenhofer J, Michel H. The photosynthetic reaction center from the purple bacterium Rhodopseudomonas viridis. Science. 1989;245(4925):1463–1473. doi: 10.1126/science.245.4925.1463. [DOI] [PubMed] [Google Scholar]
- 41.Roth M, Lewit-Bentley A, Michel H, Deisenhofer J, Huber R, Oesterhelt D. Detergent structure in crystals of a bacterial photosynethic reaction centre. Nature. 1989;340:659–662. [Google Scholar]
- 42.Roth M, Arnoux B, Ducruix A, Reiss-Husson F. Structure of the detergent phase and protein-detergent interactions in crystals of the wild-type (strain Y) Rhodobacter sphaeroides photochemical reaction center. Biochemistry. 1991;30(39):9403–9413. doi: 10.1021/bi00103a003. [DOI] [PubMed] [Google Scholar]
- 43.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(25):14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nollert P, Navarro J, Landau EM. Crystallization of membrane proteins in cubo. Methods in Enzymology. 2002;343:183–199. doi: 10.1016/s0076-6879(02)43135-7. [DOI] [PubMed] [Google Scholar]
- 45.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. Journal of Molecular Biology. 2002;316(1):1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 46.Takeda K, Sato H, Hino T, Kono M, Fukuda K, Sakurai I, Okada T, Kouyama T. A novel three-dimensional crystal of bacteriorhodopsin obtained by successive fusion of the vesicular assemblies. Journal of Molecular Biology. 1998;283(2):463–474. doi: 10.1006/jmbi.1998.2103. [DOI] [PubMed] [Google Scholar]
- 47.Long F, Su C, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467(7314):484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su C, Li M, Gu R, Takatsuka Y, McDermott G, Nikaido H, Yu EW. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. Journal of Bacteriology. 2006;188(20):7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolla JR, Su C, Do SV, Radhakrishnan A, Kumar N, Long F, Chou T, Delmar JA, Lei H, Rajashankar KR, Shafer WM, Yu EW. Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS ONE. 2014;9(6):e97903. doi: 10.1371/journal.pone.0097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei H, Bolla JR, Bishop NR, Su C, Yu EW. Crystal structures of CusC review conformational changes accompanying folding and transmembrane channel formation. Journal of Molecular Biology. 2014;426(2):403–411. doi: 10.1016/j.jmb.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lei H, Chou T, Su C, Bolla JR, Kumar N, Radhakrishnan A, Long F, Delmar JA, Do SV, Rajashankar KR, Shafer WM, Yu EW. Crystal structure of the open state of the Neisseria gonorrhoeae MtrE outer membrane channel. PLoS ONE. 2014;9:e97475. doi: 10.1371/journal.pone.0097475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su C, Radhakrishnan A, Kumar N, Long F, Bolla JR, Lei H, Delmar JA, Do SV, Chou T, Rajashankar KR, Zhang Q, Yu EW. Crystal structure of the Campylobacter jejuni CmeC outer membrane channel. Protein Science. 2014;23(7):954–961. doi: 10.1002/pro.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su C, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011;470(7335):558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benvenuti M, Mangani S. Crystallization of soluble proteins in vapor diffusion for x-ray crystallography. Nature Protocols. 2007;2(7):1633–1651. doi: 10.1038/nprot.2007.198. [DOI] [PubMed] [Google Scholar]
- 55.Fowlis WW, DeLucas LJ, Twigg PJ, Howard SB, Meehan EJ, Jr, Baird JK. Experimental and theoretical analysis of the rate of solvent equilibration in the hanging drop method of protein crystal growth. Journal of Crystal Growth. 1988;90:117–129. [Google Scholar]
- 56.Newby ZER, O’Connell JD, III, Gruswitz F, Hays FA, Harries WEC, Harwood IM, Ho JD, Lee JK, Savage DF, Miercke LJW, Stroud RM. A general protocol for the crystallization of membrane proteins for X-ray structural determination. Nature Protocols. 2009;4(5):619–637. doi: 10.1038/nprot.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McPherson A. Protein crystallization in the structural genomics era. Journal of Structural and Functional Genomics. 2004;5(1):3–12. doi: 10.1023/B:JSFG.0000029199.43875.92. [DOI] [PubMed] [Google Scholar]
- 58.Stevens RC. High-throughput protein crystallization. Current Opinion in Structural Biology. 2000;10:558–563. doi: 10.1016/s0959-440x(00)00131-7. [DOI] [PubMed] [Google Scholar]
- 59.McPherson A. Introduction to the crystallization of biological macromolecules. Current Topics in Membranes. 2009;63:5–23. [Google Scholar]
- 60.Jancarik J, Kim SH. Sparse matrix sampling: a screening method for the crystallization of proteins. Journal of Applied Crystallography. 1991;24:409–411. [Google Scholar]
- 61.Newstead S, Ferrandon S, Iwata S. Rationalizing α-helical membrane protein crystallization. Protein Science. 2008;17(1):466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolla JR, Su C, Yu EW. Biomolecular membrane protein crystallization. Philosophical Magazine (Abingdon) 2012;92(19):2648–2661. doi: 10.1080/14786435.2012.670734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Popot JL. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annual Review of Biochemistry. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 64.Breyton C, Gabel F, Abla M, Pierre Y, Lebaupain F, Durand G, Popot JL, Ebel C, Pucci B. Micellar and biochemical properties of (hemi)fluorinated surfactants are controlled by the size of the polar head. Biophysical Journal. 2009;97(4):1077–1086. doi: 10.1016/j.bpj.2009.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. Designing facial amphiphiles for the stabilization of integral membrane proteins. Angewandte Chemie International Edition. 2007;46(37):7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 66.Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland SJ, Pierre Y, Drew D, Popot JL, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization, and crystallization of membrane proteins. Nature Methods. 2010;7(12):1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teeter M. Water structure of a hydrophobic protein at atomic resolution: pentagon rings of water molecules in crystals of crambin. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(19):6014–6018. doi: 10.1073/pnas.81.19.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hofmeister F. Zur lehre von der wirkung der saize. Archiv für Experimentelle Pathologie und Pharmakologie. 1888;24(4):247–260. [Google Scholar]
- 69.Rosenberger F. Protein crystallization. Journal of Crystal Growth. 1996;166(1):40–54. [Google Scholar]
- 70.Kantardjieff KA, Rupp B. Protein isoelectric point as a predictor for increased crystallization screening efficiency. Bioinformatics. 2004;20(14):2162–2168. doi: 10.1093/bioinformatics/bth066. [DOI] [PubMed] [Google Scholar]
- 71.Su C, Yang F, Long F, Reyon D, Routh MD, Kuo DW, Mokhtari AK, Van Ornam JD, Rabe KL, Hoy JA, Lee YJ, Rajashankar KR, Yu EW. Crystal structure of the membrane fusion protein CusB from Escherichia coli. Journal of Molecular Biology. 2009;393(2):342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]