Abstract
Near-full-length 18S and 28S rRNA gene sequences were obtained for 33 nematode species. Datasets were constructed based on secondary structure and progressive multiple alignments, and clades were compared for phylogenies inferred by Bayesian and maximum likelihood methods. Clade comparisons were also made following removal of ambiguously aligned sites as determined using the program ProAlign. Different alignments of these data produced tree topologies that differed, sometimes markedly, when analyzed by the same inference method. With one exception, the same alignment produced an identical tree topology when analyzed by different methods. Removal of ambiguously aligned sites altered the tree topology and also reduced resolution. Nematode clades were sensitive to differences in multiple alignments, and more than doubling the amount of sequence data by addition of 28S rRNA did not fully mitigate this result. Although some individual clades showed substantially higher support when 28S data were combined with 18S data, the combined analysis yielded no statistically significant increases in the number of clades receiving higher support when compared to the 18S data alone. Secondary structure alignment increased accuracy in positional homology assignment and, when used in combination with paired-site substitution models, these structural hypotheses of characters and improved models of character state change yielded high levels of phylogenetic resolution. Phylogenetic results included strong support for inclusion of Daubaylia potomaca within Cephalobidae, whereas the position of Fescia grossa within Tylenchina varied depending on the alignment, and the relationships among Rhabditidae, Diplogastridae, and Bunonematidae were not resolved.
Keywords: Bayesian inference, Daubaylia, Fescia, maximum likelihood, molecular phylogenetics, multiple sequence alignment, progressive alignment, Rhabditida, ribosomal RNA, secondary structure
The first molecular phylogeny that included a representative sample of the phylum Nematoda was based on a single gene (18S rRNA) and had 53 terminal taxa (Blaxter et al., 1998). Since then, numerous molecular phylogenetic hypotheses for nematodes have been published. These include many based on one genetic locus, such as the 18S rRNA gene from more than 1,200 species (van Megen et al., 2009) or analysis of 12 protein-coding genes from the mitochondrial genomes of up to 100 species (Kang et al., 2009; Park et al., 2011; Sun et al., 2014; Kim et al., 2015). The application of multilocus data to estimate nematode phylogeny has so far been focused on taxonomically restricted questions (Nadler and Hudspeth, 2000; Nadler et al., 2006a; Kiontke et al., 2007, 2011; Subbotin et al., 2008; Mayer et al., 2009). Whether focused on single genes or multiple loci, optimal estimates of evolutionary history require consideration of many factors that influence phylogenetic analysis of sequence data. For example, methods for producing multiple alignments of ribosomal sequences and decisions to include or exclude aligned sites because of ambiguity in positional homology can have profound effects on inferred nematode relationships (Smythe et al., 2006).
Ribosomal RNA genes remain the most commonly sequenced and analyzed molecules for phylogenetics of nematodes, with studies usually focused on either the near-complete 18S rRNA gene or partial sequences of the 28S rRNA gene (e.g., D2-D3 or D1-D2-D3 domains). Because these two genes are from the same locus, they are non-independent estimators of the same underlying gene tree. Differences in topology between phylogenies inferred using 18S and 28S sequences for the same taxa occur, but they are best explained as stochastic inference errors for the individual datasets. Analysis of near-complete 18S or partial 28S sequences do not provide sufficient resolution for certain phylogenetic questions involving nematodes (Smythe and Nadler, 2007; Holovachov et al., 2011; Shokoohi et al., 2013), but their continued use is justified because of the large database of comparative information available in GenBank and the relative ease of rRNA PCR amplification and sequencing, even from individual nematodes.
Structural information from rRNA has been frequently used to improve alignments of 18S and 28S genes in evolutionary studies of nematodes that address questions at broad taxonomic levels (Aleshin et al., 1998; Blaxter et al., 1998; Holterman et al., 2006, 2008a, 2008b; Meldal et al., 2007; Holovachov et al., 2009, 2012, 2013b; van Megen et al., 2009; Bik et al., 2010) or within individual orders (Chilton et al., 2006; Kiontke et al., 2007; Bert et al., 2008; Holterman et al., 2009; Holovachov et al., 2013a). Some of these studies divided the alignment into “stem” (paired sites) and “loop” (non-paired sites) partitions with separate substitution models for each (Holterman et al., 2006, 2008a, 2008b, 2009; Bert et al., 2008; van Megen et al., 2009; Bik et al., 2010; Holovachov et al., 2012, 2013a, 2013b), but few used paired-site substitution models in consideration of compensatory substitutions (Bert et al., 2008; Holovachov et al., 2012, 2013a, 2013b). There are also few studies of nematode relationships above the family level that use nearly full-length 18S and 28S rRNA sequences for phylogeny inference (e.g., Chilton et al., 2006; Kiontke et al., 2007), and none that combine such sequences with paired-site substitution models for both genes.
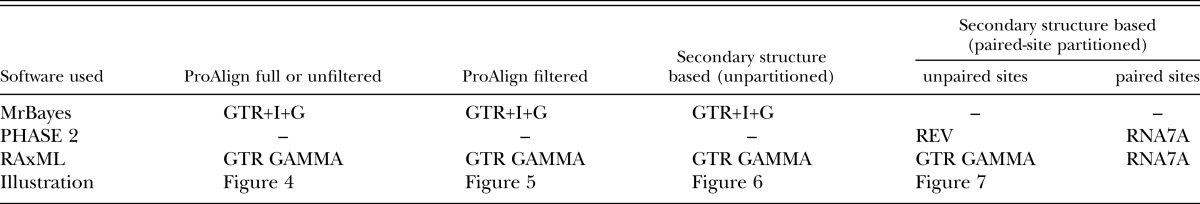
In phylogenetic analyses based on sequence data, there are several factors that can influence the outcome, that is, the tree topology and its branch lengths. These include (i) multiple sequence alignment (Smythe et al., 2006); (ii) the number of genes included in the analysis (Rokas and Carroll, 2005); (iii) the length of the sequences used for analysis (Rosenberg and Kumar, 2001); (iv) the choice of inference method and associated models (Kelchner and Thomas, 2006); and (v) selection of model parameter values. Herein, we explored factors i, iii, and iv in some detail for a group of Rhabditida (Table 1), evaluating if combined analysis of near-full-length 18S and 28S rRNA sequences yields increased clade resolution and support over analysis of 18S data alone. In addition, we evaluated if phylogenetic analyses based on secondary structure alignments, either alone or combined with paired-site substitution models, are advantageous when compared to those based on computer-based progressive multiple alignment. Finally, we evaluated the phylogenetic effect of removing characters judged as alignment ambiguous from the progressive alignment dataset.
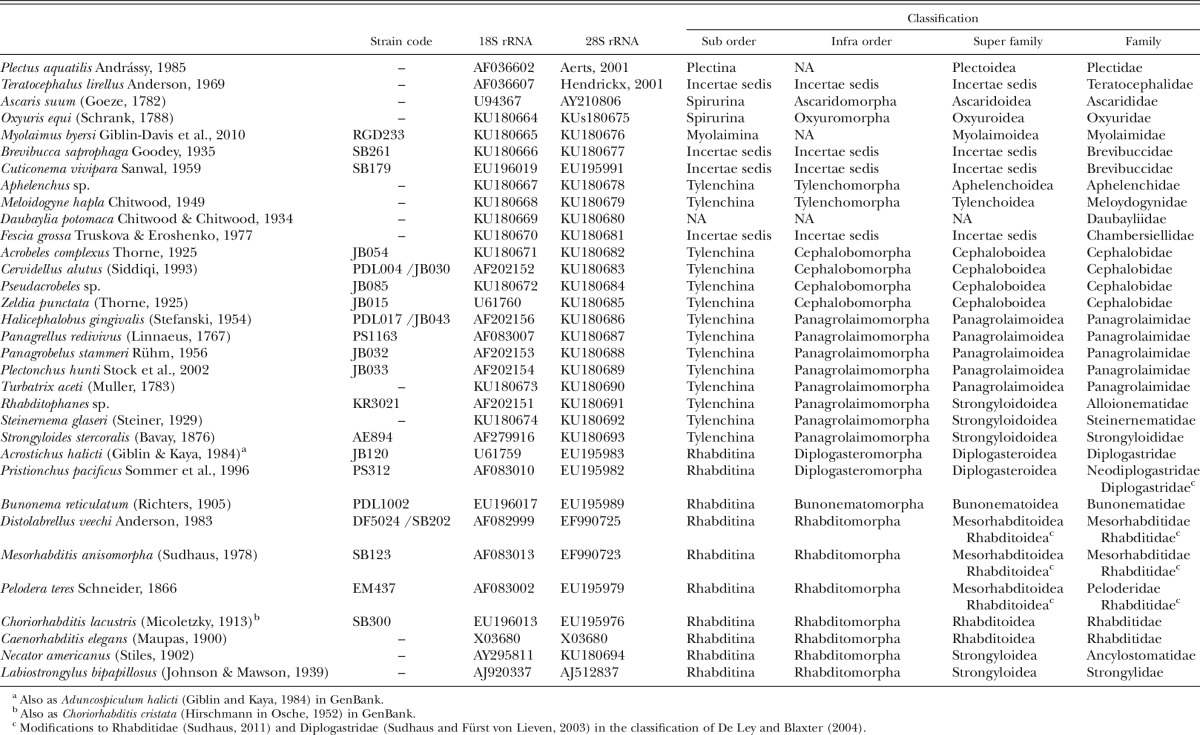
Table 1.
GenBank accession numbers or references for sequences of nematode species used in the phylogenetic analysis and their position in the classification of De Ley and Blaxter (2004).
Materials and Methods
Species selection:
When selecting species to be included in the analysis, we considered the availability and taxonomic representation of nearly complete 18S and 28S rRNA sequences in GenBank or published literature, as well as the possibility to sequence these genes de novo. GenBank lists many species from the superfamilies Diplogasteroidea, Mesorhabditoidea, Rhabditoidea, and Strongyloidea for which both near-full-length 18S and 28S rRNA genes are available, but only selected representatives of each superfamily were included in our analysis. A minimum of two species per superfamily were chosen, with priority given to species for which published secondary structure annotation models were available and species with fuller-length sequences (Table 1).
DNA amplification and sequencing: For nematodes that could be cultured, DNA was extracted from pools of 10 to 30 individuals using commercial kits (DNAzol; Molecular Research Center Inc. or MasterPure; Epicentre Technologies). For species obtained and identified directly from environmental or host samples, DNA was extracted from 1 to 10 individuals using the same kits. Nuclear 18S (small-subunit) ribosomal DNA (rDNA) was amplified by PCR and sequenced directly using previously described methods and primers (Nadler et al., 2007). Near-complete nuclear 28S (26S/28S or large subunit) rDNA was amplified and sequenced in three overlapping pieces. Two 28S rRNA regions representing ∼80% of the gene were amplified using three PCR reactions. Approximately 1,000 bp of the 28S 5′-end was amplified using forward PCR primer 391 (5′-AGCGGAGGAAAAGAAACTAA) and reverse primer 501 (5′-TCGGAAGGAACCAGCTACTA). Approximately 1,600 bp of the remaining 28S rRNA was amplified in two overlapping pieces. These two amplicons were normally obtained using forward primer 527 (5′-CTAAGGAGTGTGTAACAACTCACC) in combination with reverse primer 532 (5′-AATGACGAGGCATTTGGCTACCTT), and forward primer 537 (5′-GATCCGTAACTTCGGGAAAAGGAT) with reverse primer 531 (5′-CTTCGCAATGATAGGAAGAGCC). If amplification using primers 527/531 failed, the alternative forward primer 563 (5′-ACCCGAAAGATGGTGATCTAT) was used for PCR in combination with either primer 532 or primer 531. PCR reactions (25 µl) consisted of 0.5 µM of each primer, 200 µM deoxynucleoside triphosphates, and MgCl2 ranging from 1.5 to 3.0 mM as needed for specific and robust amplification. Proofreading polymerase (0.5 units, Finnzymes DNAzyme EXT; MJ Research) was used for PCR; the range of PCR cycling parameters included denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 30 sec, 50°C to 60°C for 30 sec, and 72°C for 60 to 80 sec, followed by a post-amplification extension at 72°C for 7 min.
PCR products were prepared for direct sequencing as described previously (Nadler et al., 2006b). For instances when PCR products could not be directly sequenced, the amplicons were cloned and sequenced using methods detailed in the work of Nadler et al. (2006b). CodonCode Aligner (5.0.2) and Phred base calling were used in assembly of contigs, and polymorphisms were evaluated as described previously (Nadler et al., 2006b).
ProAlign alignment:
Sequences of nematode species listed in the Table 1 were aligned using ProAlign Version 0.5 (Loytynoja and Milinkovitch, 2003). For each alignment (18S and 28S), a ProAlign guide tree was constructed using corrected pairwise distances, and this tree was used to estimate the hidden Markov model parameters (δ and ɛ) for progressive multiple alignment. Program memory and bandwidth were increased empirically to permit alignment program completion. The average minimum posterior probability (PP) of sites was used as the criterion for detecting and removing unreliably aligned sequences, since this value is strongly correlated with correctness as determined by simulation studies (Loytynoja and Milinkovitch, 2003). To reduce the likelihood of excluding correctly aligned sites, the filter threshold was set to 60% minimum PP, a value intermediate between the threshold of PP for correctly versus incorrectly aligned sites in most simulation results (Loytynoja and Milinkovitch, 2003). The 18S dataset included 1,807 aligned characters, and the filtered (60% PP) 18S dataset had 1,231 characters (Fig. 1). The 28S dataset included 4,000 aligned characters, and the filtered (60% PP) 28S dataset had 1,848 characters (Figs. 2,3). Therefore, the combined 18S + 28S datasets included 5,807 characters (full datasets) or 3,079 characters (filtered datasets).
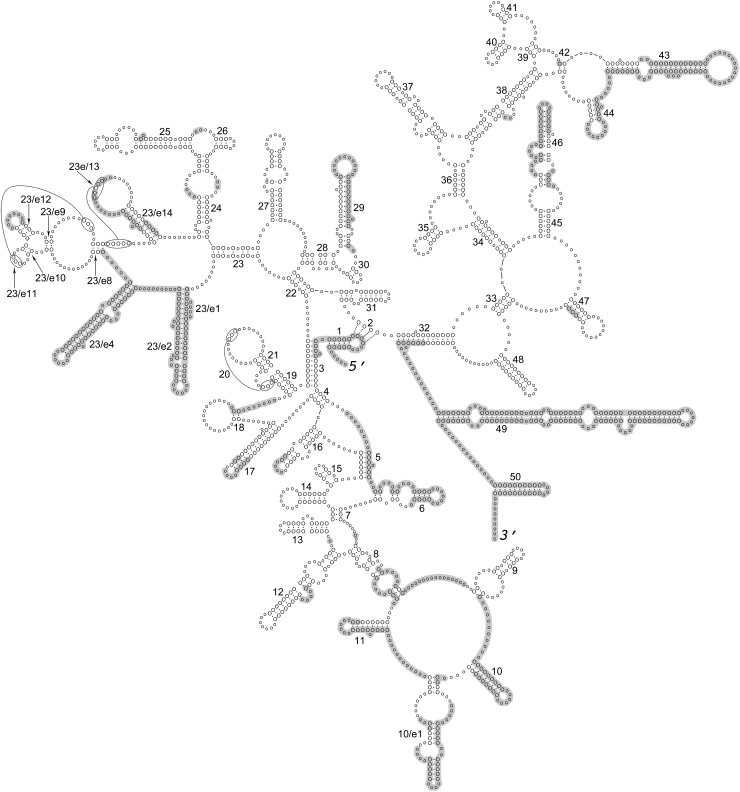
Fig. 1.
Generalized secondary structure model for the 18S rRNA based on individual secondary structure models of nematode species listed in Table 1. Helices are numbered according to Wuyts et al. (2002). Shaded areas mark nucleotide positions excluded in the ProAlign progressive posterior-probability-filtered alignment.
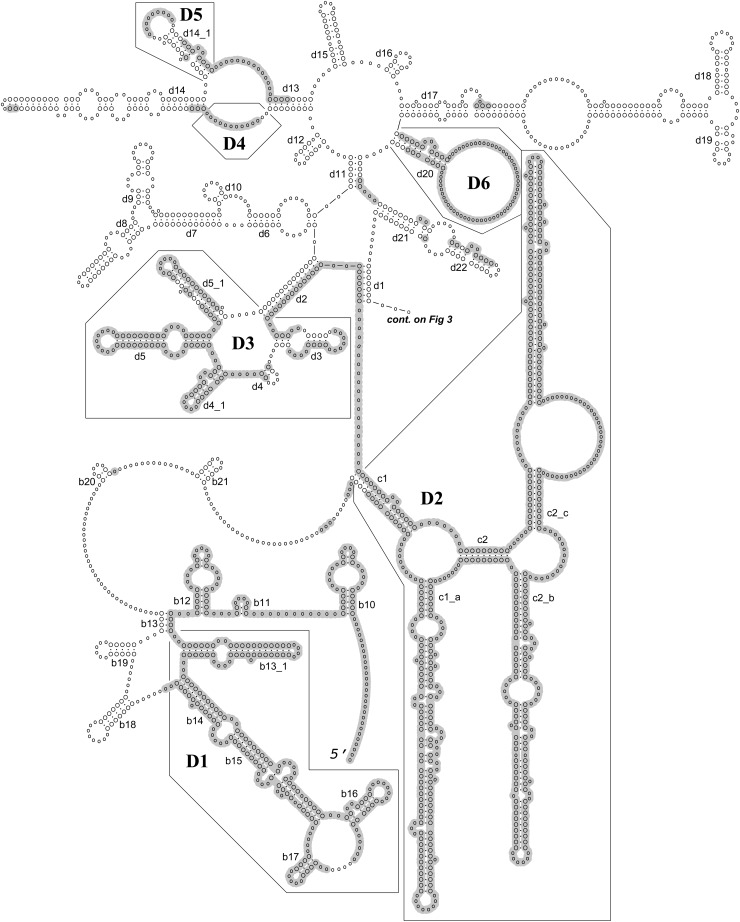
Fig. 2.
Generalized secondary structure model for the 28S rRNA based on individual secondary structure models of nematode species listed in Table 1. Helices are numbered according to Wuyts et al. (2001) and Chilton et al. (2003). Twelve D domains (expansion segments) are defined according to Ellis et al. (1986) and Chilton et al. (2003). Shaded areas mark nucleotide positions excluded in the ProAlign progressive posterior-probability-filtered alignment. Continued on Figure 3.
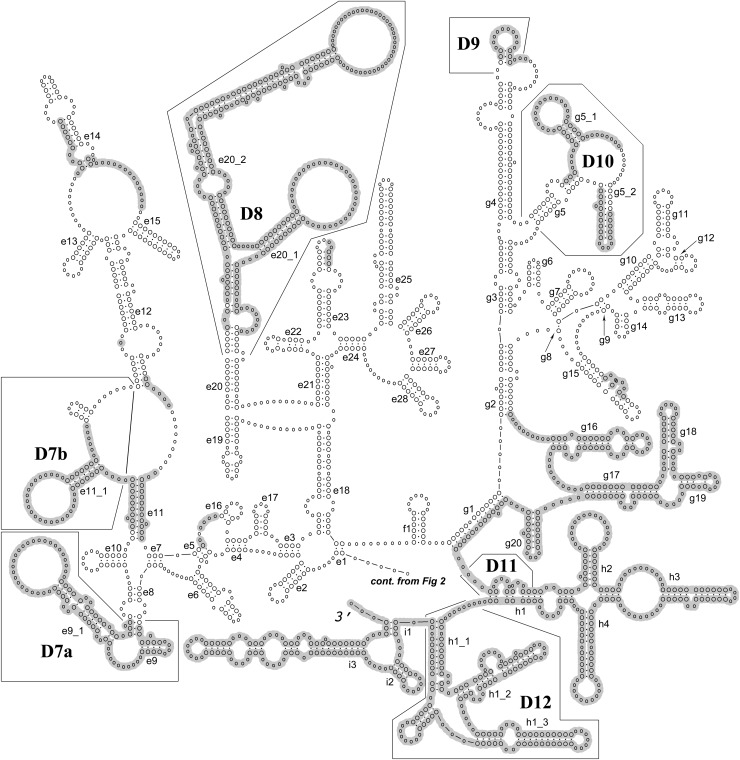
Fig. 3.
Continued from Figure 2. Generalized secondary structure model for the 28S rRNA based on individual secondary structure models of nematode species listed in Table 1. Helices are numbered according to Wuyts et al. (2001) and Chilton et al. (2003). Twelve D domains (expansion segments) are defined according to Ellis et al. (1986) and Chilton et al. (2003). Shaded areas mark nucleotide positions excluded in the ProAlign progressive posterior-probability filtered alignment.
Secondary structure-based alignment:
The secondary structure alignment was created based on existing secondary structure models of 18S (Fig. 1) and 28S (Figs. 2,3) rRNA genes. Secondary structure annotation of 18S rRNA was based on the European ribosomal RNA Database (Wuyts et al., 2004). The entire available nematode dataset that includes over 100 aligned sequences from various nematode taxa was downloaded from the European ribosomal RNA Database before the database was archived and its annotation was converted from the unsupported DCSE format (De Rijk and De Wachter, 1993) to a contemporary format compatible with the JAVA-based editor 4SALE (Seibel et al., 2006). The European Ribosomal RNA Database already included 18S secondary structure annotation for Acrostichus halicti (Giblin and Kaya, 1984), Ascaris suum (Goeze, 1782), Caenorhabditis elegans (Maupas, 1900), Cervidellus alutus (Siddiqi, 1993), Distolabrellus veechi Anderson, 1983, Halicephalobus gingivalis (Stefanski, 1954), Mesorhabditis anisomorpha (Sudhaus, 1978), Panagrellus redivivus (Linnaeus, 1767), Panagrobelus stammeri Rühm, 1956, Pelodera teres Schneider, 1866, Plectus aquatilis Andrássy, 1985, Plectonchus hunti Stock et al., 2002, Pristionchus pacificus Sommer et al., 1996, Teratocephalus lirellus Anderson, 1969, and Zeldia punctata (Thorne, 1925). The remaining 18S sequences (Table 1) were added to this dataset and aligned as described below.
Published models of the secondary structure of the 28S rRNA gene for C. elegans, Labiostrongylus bipapillosus (Johnson and Mawson, 1939), P. aquatilis, and T. lirellus (Ellis et al., 1986; Gutell and Fox, 1988; Aerts, 2001; Hendrickx, 2001; Chilton et al., 2003) were used as a basis for alignment and annotation of the 28S rRNA sequences of the remaining 29 species (Table 1).
Secondary structure annotation was manually added to all non-annotated sequences using the JAVA-based editor 4SALE (Seibel et al., 2006), and all sequences were manually aligned to maximize apparent positional homology of nucleotides. Complementary base pairings in stem regions was manually verified for all sites.
Phylogenetic analyses using Bayesian (BI) and maximum likelihood (ML) methods:
Phylogenetic trees were rooted using P. aquatilis as the out-group in all analyses. MrModeltest v2.3 (Nylander, 2004) was used to determine the best-fit model of nucleotide substitution for individual genes from each alignment; GTR+I+G was optimal for all datasets based on the Akaike information criterion. MrBayes v3.1.2 (Ronquist and Huelsenbeck, 2003), as implemented by the CIPRES web portal (Miller et al., 2010), was used to infer trees for all datasets. These nine datasets included three for both the 18S and 28S rRNA genes (ProAlign alignment, ProAlign filtered alignment, and secondary structure alignment), plus three for the combined datasets (ProAlign combined, ProAlign filtered combined, and the secondary structure combined). Separate character partitions were invoked for the two genes in the combined 18S + 28S datasets. The GTR+I+G model was implemented for each dataset (Table 2), but the model-based estimates of gamma shape and proportion of invariable sites were not fixed.
Table 2.
Software and substitution models used for analyses of both 18S and 18S + 28S rRNA datasets.
Two independent runs with four Markov Chain Monte Carlo chains were executed in MrBayes for 4 million generations, sampling trees every 4,000 generations. The average standard deviation of split frequencies (<0.01) was used to confirm that a sufficient number of generations had been run for each dataset. Two approaches were used to determine burn-in values. Following all analyses, the log likelihood values were plotted against generation times, and trees sampled before stationarity of Ln values were discarded. Stationarity of the remainder of the parameters was examined using Tracer v1.6 (http://tree.bio.ed.ac.uk/software/tracer/; Rambaut et al., 2014). Based on these methods, the first 10% of trees were then discarded as burn-in for each dataset. The remaining trees were then used to construct 50% majority-rule consensus trees with clades representing the PP distribution for each dataset.
Maximum likelihood analysis of the nine datasets was performed using RAxML version 7.0.3 (Stamatakis et al., 2008) and implemented using the CIPRES web portal (Miller et al., 2010). In RAxML, the GTR GAMMA model was used for finding the best ML tree (Table 2), and the GTR CAT approximation was used for ML bootstrapping to increase efficiency for the number of replicates we performed. In combined analyses, each gene was treated as an independent partition, with model parameters estimated separately for each partition. Bootstrap ML analysis was performed using the rapid bootstrapping option with 1,000 iterations.
Phylogenetic analysis using structural models (paired-site substitution models):
The alignment based on secondary structure annotation was analyzed with Bayesian phylogenetic inference using the “mcmcphase” program in the PHASE package (Version 2.0; Gowri-Shankar and Jow, 2006). For this analysis, the dataset was partitioned into “stems” (paired sites) and “loops” (non-paired sites) to account for the potential phylogenetic importance of compensatory substitutions. The REV nucleotide substitution model (Tavare, 1986) was used for non-paired sites, whereas RNA7A (Higgs, 2000), RNA7D (Tillier and Collins, 1998), or RNA16A (Gowri-Shankar and Jow, 2006) nucleotide substitution models were used for paired sites in three independent analyses. All three analyses produced identical topologies, among which the analysis using RNA7A model (Table 2) showed the highest PP and was chosen for subsequent discussion. The 18S, 28S, and combined 18S + 28S datasets were analyzed; for the combined datasets, separate partitions were assigned to 18S and 28S rRNA genes. Model parameters were estimated independently for all four sub-partitions (non-paired and paired sites of 18S and 28S). For each model combination, two independent runs were performed, and for each run, the chains were allowed to burn in for 500,000 generations, followed by 5 million generations (5.5 million generations in total) during which tree topologies, branch length, and model parameters were sampled every 200 generations. An additional run with burn in for 1,000,000 generations and 10,000,000 sampling generations (total 11 million generations) was performed for the combined 18S + 28S dataset to determine if the longer run provided better resolution or clade support, but the resulting tree showed only minimal (0.01 Bayesian posterior probability, BPP) increase in support for two clades.
Maximum likelihood analysis of the partitioned dataset was performed using RAxML version 8.0.24 (Stamatakis, 2014) and implemented using CIPRES (Miller et al., 2010). The GTR GAMMA nucleotide substitution model was used for non-paired sites, whereas the RNA7A (Higgs, 2000) substitution model was used for paired sites (Table 2). Bootstrap ML analysis was performed using the rapid bootstrapping option with 1,000 iterations.
One-tailed sign test:
A one-tailed binomial sign test with a cumulative probability significance level of p = 0.05 was used to test if the number of clades (Tables 3,4) receiving increased support was greater for the combined 18S + 28S data than for the 18S data alone. To be included in the sign test calculation, a clade in one of the two compared datasets had to achieve a minimum ML bootstrap of ≥70% or a BPP value of ≥0.70. Clades meeting these criteria were scored as tied (equal support in both analyses), win 18S (higher support in 18S analysis), or win 18S + 28S (higher support in 18S + 28S analysis). All eight dataset/inference method combinations were tested (each of the four dataset/model combinations with trees inferred by ML and Bayesian inference).
Table 3.
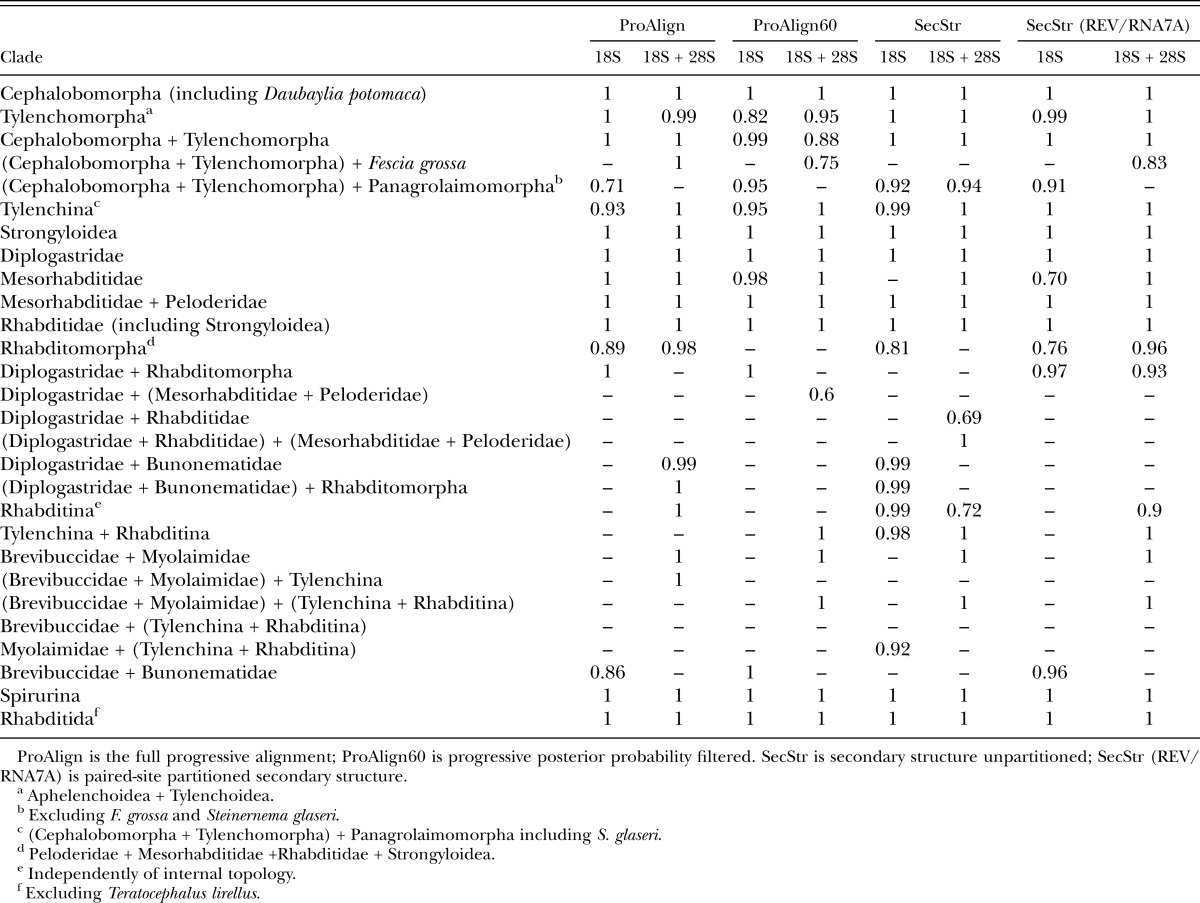
Major clade (family level and higher) presence and posterior probabilities for different alignments and datasets based on Bayesian inference.
Table 4.
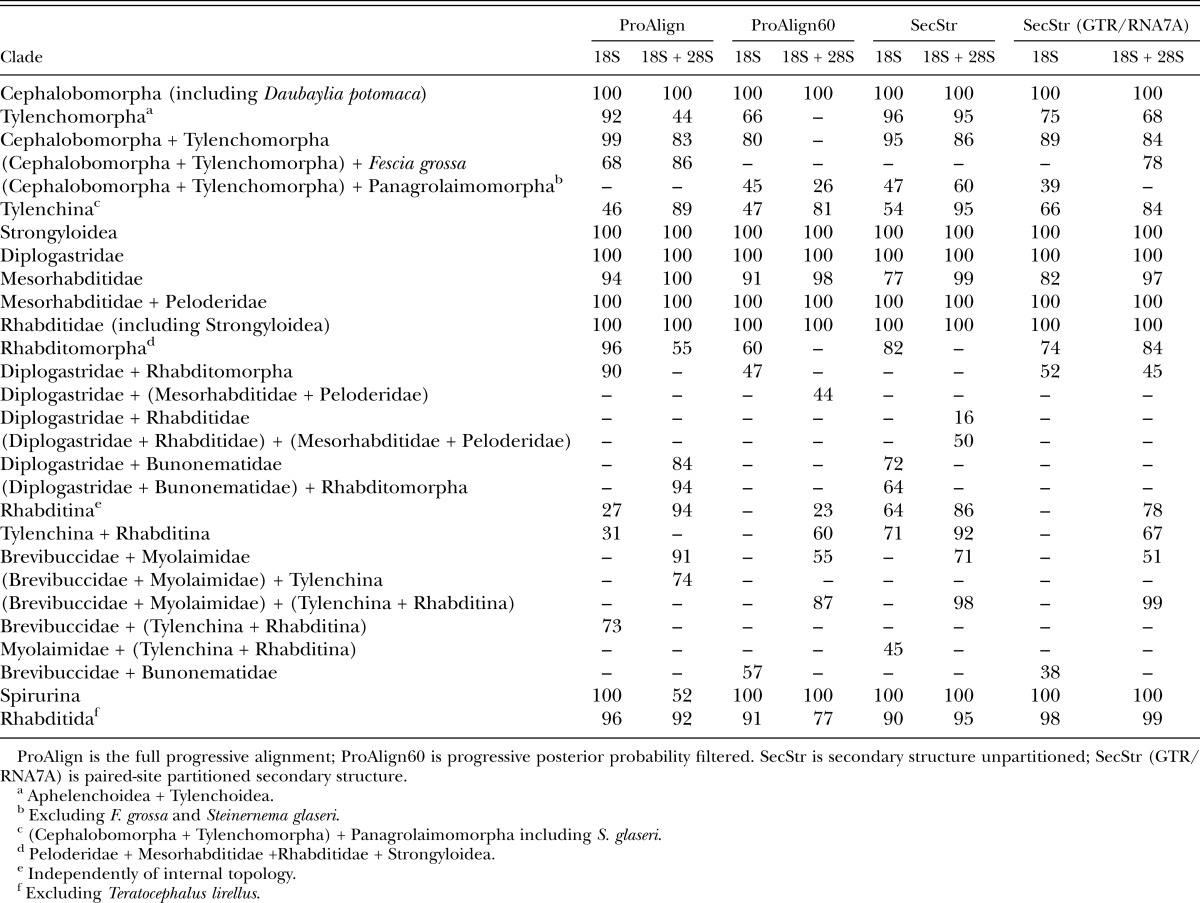
Major clade (family level and higher) presence and bootstrap values for different alignments and datasets based on maximum likelihood inference.
Results
Combined 18S + 28S datasets, general features:
Three different alignments of combined 18S + 28S sequence were used for analysis and comparison. The full-length ProAlign alignment, filtered ProAlign alignment, and a secondary structure alignment were analyzed (Table 2) using ML (RAxML) and Bayesian inference methods (MrBayes, PHASE 2). The secondary structure alignment was analyzed in two different ways; first as an unpartitioned dataset using single-nucleotide substitution models (GTR GAMMA for RAxML and GTR+I+G for MrBayes), and second as a partitioned dataset using paired-site substitution models for stem parts of the rRNA (RNA7A model for both RAxML and PHASE2) and single-nucleotide substitution models for unpaired sites (GTR GAMMA for RAxML and REV for PHASE2).
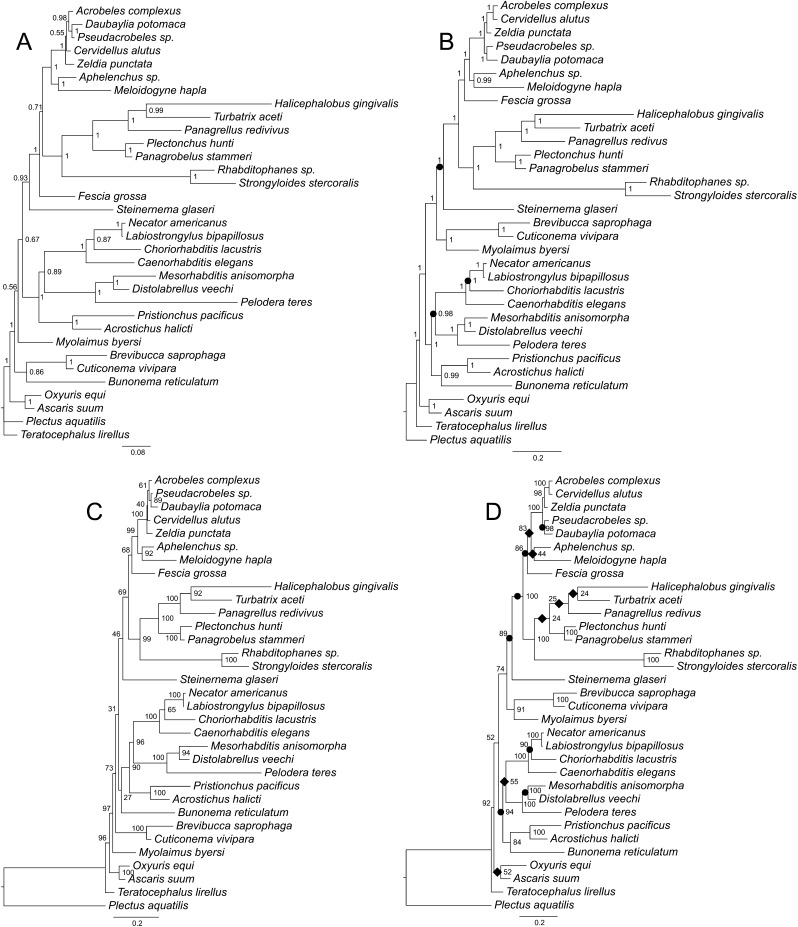
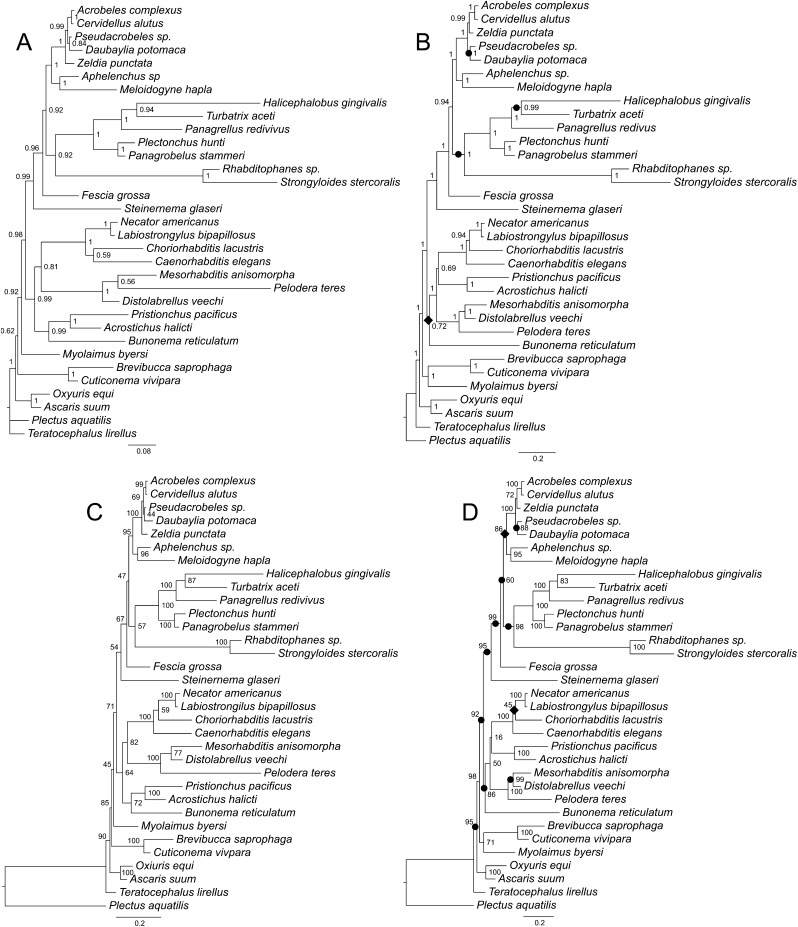
Analysis of the ProAlign (5,807 characters) alignment yielded an identical tree topology by BI and ML methods (Fig. 4B,D). Similarly, BI and ML trees for the unpartitioned secondary structure analyses (SecStr; GTR models) had the same topology (Fig. 6B,D). Finally, the partitioned paired-site analyses (Fig. 7B,D) also did not differ in topology between BI and ML methods. However, the ProAlign alignment and the two main secondary structure datasets (unpartitioned and partitioned paired-site models) yielded different tree topologies when analyzed by BI or ML methods (Figs. 4B; 6B; 7B and 4D; 6D; 7D respectively). Within the ((Cephalobomorpha + Tylenchomorpha) + Panagrolaimomorpha) clade, topological variation among these analyses only involved the position of Fescia grossa. Within the remainder of the in-group taxa, topological differences among analyses of these three datasets (excluding filtered ProAlign) involved the position of Bunonema reticulatum, and two clades, Diplogastridae and (Myolaimidae + Brevibuccidae). Although these are notable variations in the evolutionary hypotheses, differences in topology among these analyses could arguably be characterized as moderate. Thus, for these very large 18S + 28S datasets (5,760–5,807 characters, depending on alignment) and a moderate number of taxa, tree topology within each alignment was insensitive to the two inference methods, but moderately sensitive to different alignments and use of a secondary structure model.
Fig. 4.
Phylogenetic trees of the relationships of Rhabditida inferred using two different ProAlign progressive full alignments. A. Bayesian consensus tree based on the 18S data. B. Bayesian consensus tree based on the combined 18S + 28S data. C. Best maximum likelihood tree based on the 18S data. D. Best maximum likelihood tree based on the combined 18S + 28S data. Bayesian posterior probability (BPP) values and maximum likelihood bootstrap values are shown at nodes where appropriate. Individual increases and decreases in support values for clades (by 0.05 or more in BPP values or by 5% or more in bootstrap values) based on comparison of the 18S + 28S results to the 18S results are marked by black circles (increase) or rhombs (decrease) on the 18S + 28S trees.
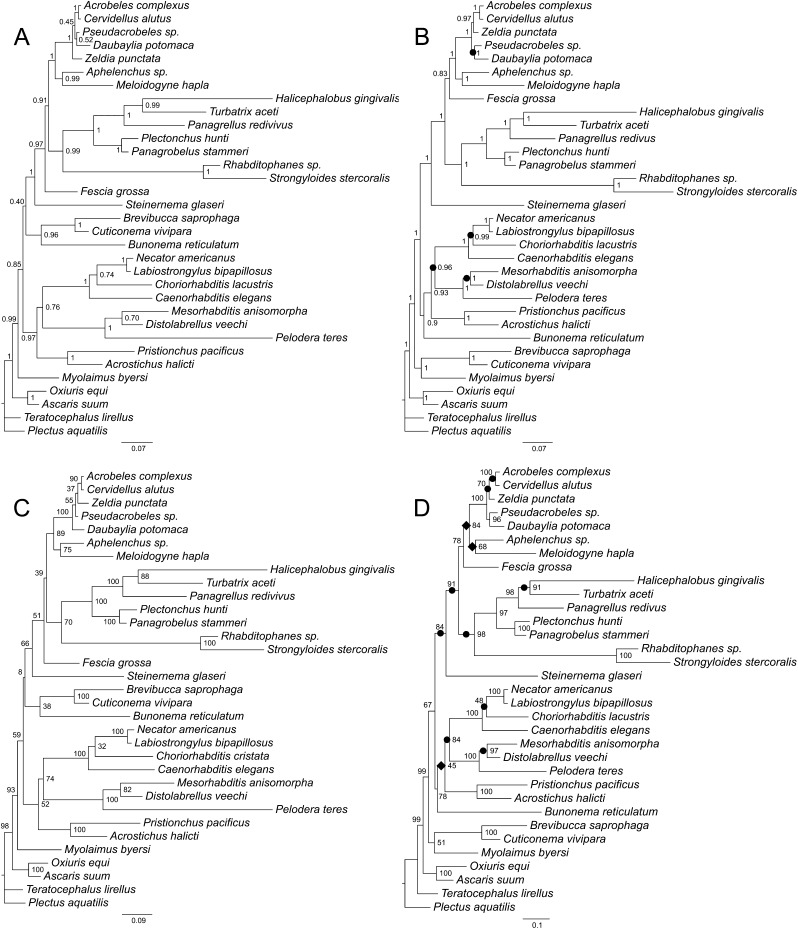
Fig. 6.
Phylogenetic trees of the relationships of Rhabditida inferred using two different unpartitioned alignments based on secondary structure models. A. Bayesian consensus tree based on the 18S data. B. Bayesian consensus tree based on the combined 18S + 28S data. C. Best maximum likelihood tree based on the 18S data. D. Best maximum likelihood tree based on the combined 18S + 28S data. Bayesian posterior probability (BPP) values and maximum likelihood bootstrap values are shown at nodes where appropriate. Individual increases and decreases in support values for clades (by 0.05 or more in BPP values or by 5% or more in bootstrap values) based on comparison of the 18S + 28S results to the 18S results are marked by black circles (increase) or rhombs (decrease) on the 18S + 28S trees.
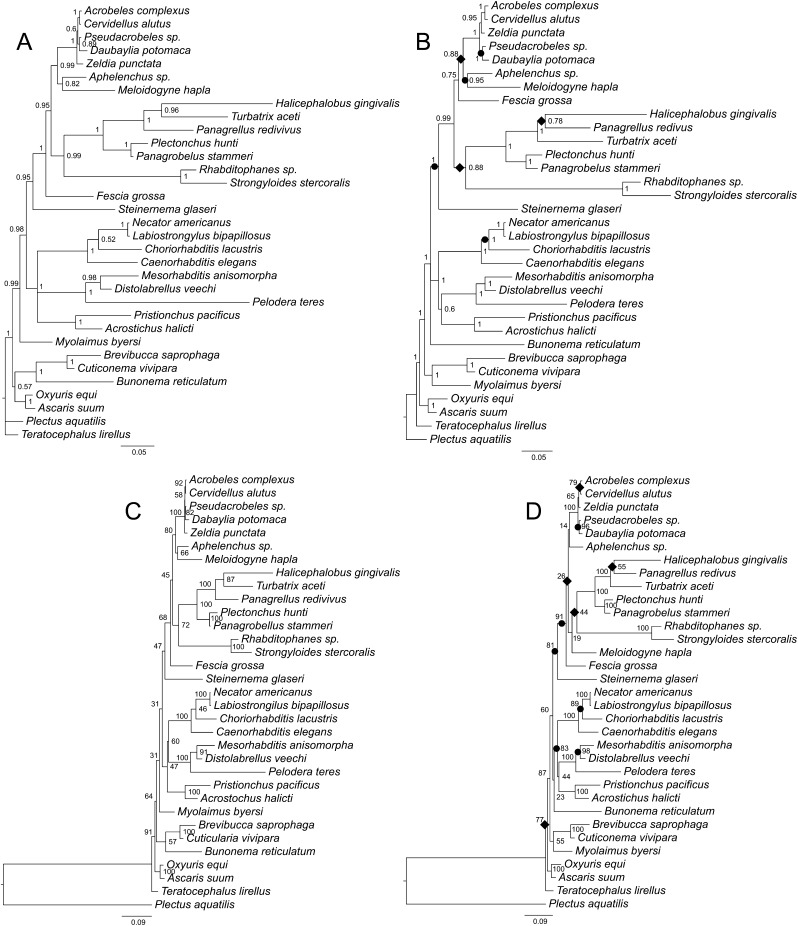
Fig. 7.
Phylogenetic trees of Rhabditida inferred using two different partitioned alignments based on secondary structure models using REV (BI) or GTR GAMMA (ML) for unpaired sites and RNA7A for paired sites. A. Bayesian consensus tree based on the 18S data. B. Bayesian consensus tree based on the combined 18S + 28S data. C. Best maximum likelihood tree based on the 18S data. D. Best maximum likelihood tree based on the combined 18S + 28S data. Bayesian posterior probability (BPP) values and maximum likelihood bootstrap values are shown at nodes where appropriate. Individual increases and decreases in support values for clades (by 0.05 or more in BPP values or by 5% or more in bootstrap values) based on comparison of the 18S + 28S results to the 18S results are marked by black circles (increase) or rhombs (decrease) on the 18S + 28S trees.
In contrast, differences between the two ProAlign datasets (5,807 characters [full] versus 3,079 characters [filtered]) involved more substantial topological changes, even for a single inference method (e.g., Fig. 4B versus 5B or Fig. 4D versus 5D). For example, within the ((Cephalobomorpha + Tylenchomorpha) + Panagrolaimomorpha) clade, topological differences were substantial for ML analyses (Fig. 4D versus 5D). Considering the remainder of the in-group taxa, both BI and ML analyses showed noticeable differences between the full and filtered ProAlign alignments. These results must be considered relative to the potential influence (and removal) of highly variable and potentially ambiguously aligned characters on the phylogenetic analyses. The full ProAlign alignment of 5,807 characters included 3,625 variable sites (62% of total sites), whereas the ProAlign filtered alignment of 3,079 characters included 1,469 variable sites, or 48% of total sites (Figs. 1–3 show filtered or removed sites).
Fig. 5.
Phylogenetic trees of the relationships of Rhabditida inferred using two different ProAlign progressive posterior probability-filtered alignments. A. Bayesian consensus tree based on the 18S data. B. Bayesian consensus tree based on the combined 18S + 28S data. C. Best maximum likelihood tree based on the 18S data. D. Best maximum likelihood tree based on the combined 18S + 28S data. Bayesian posterior-probability values and maximum likelihood bootstrap values are shown at nodes where appropriate. Individual increases and decreases in support values for clades (by 0.05 or more in BPP values or by 5% or more in bootstrap values) based on comparison of the 18S + 28S results to the 18S results are marked by black circles (increase) or rhombs (decrease) on the 18S + 28S trees.
Specific differences resulting from different datasets and alignments:
Among the four trees produced using Bayesian inference for the combined 18S + 28S data, the tree based on the filtered ProAlign alignment and the tree based on partitioned secondary structure analysis using a paired-site substitution model have similar overall topology (Figs. 5B; 7B), except for the branching of Bunonema reticulatum (an unresolved trichotomy in the filtered ProAlign alignment), the position of Diplogastridae (A. halicti and P. pacificus) relative to Rhabditidae and Strongyloidea, and the sister-group relationships within the clade consisting of H. gingivalis, P. redivivus, and T. aceti. However, for these differing sister-group relationships, one or more of the trees has low BPP for the clades involved. For example, in the filtered ProAlign analysis, the sister-group relationship of H. gingivalis and P. redivivus has a BPP of 0.78. Similarly, the relationships of Diplogastridae to Rhabditidae and Strongyloidea involve some clades with BPP < 0.95 in both trees.
The trees based on the unpartitioned secondary structure alignment (18S + 28S data) are different from others with respect to the clade that includes Rhabditidae, Diplogastridae, and Strongyloidea; their topology (Fig. 6B,D) is similar to the tree obtained by Kiontke et al. (2007) using 18S, 28S, and partial RNA polymerase II genes, albeit with different support for different dichotomies, as expected. On the other hand, the position of F. grossa in these trees and the filtered ProAlign dataset (ML) is markedly different from the other five trees, in that F. grossa is robustly placed as a part of the Panagrolaimomorpha “grade,” that is, as sister to the (Panagrolaimomorpha partim + (Cephalobomorpha + Tylenchomorpha)) clade (see Discussion).
The tree that is based on the unfiltered ProAlign alignment for combined data (Fig. 4B,D) shows the most distinct topology from other trees generated in this study, namely in the position of the (Myolaimidae + Brevibuccidae) clade and in the relationships between Bunonematidae, Diplogastridae, and Rhabditidae. Both of these different relationships are strongly supported in the tree generated using BI.
There were only two clades that were moderately or strongly supported in the combined analysis of 18S + 28S data, but were absent in analysis of 18S data. These clades are (Brevibuccidae + Myolaimidae) (Figs. 4B,D; 5B,D; 6B,D; 7B,D), and ((Tylenchina + Rhabditina) + (Brevibuccidae + Myolaimidae)) (Figs. 5B,D; 6B,D; 7B,D). Some clades that were present in analyses of the 18S dataset showed marked increases in clade support with the addition of 28S data, at least for certain inference methods (Tables 3,4). For example, ML bootstrap support for Tylenchina increased as much as 43%. Similarly, Bayesian posteriors for Tylenchomorpha increased from a nonsignificant value of 0.82 to 0.95 for the combined ProAlign filtered data. Conversely, there are some instances where analysis of the combined 18S + 28S data yielded lower clade support than for analysis of 18S alone, e.g., ProAlign filtered BPP for (Cephalobomorpha + Tylenchomorpha).
Some clades were both rare and strongly supported (Tables 3,4); this result was not exclusive to certain datasets or analysis methods although it appeared to occur more frequently with the combined datasets and Bayesian inference. An example is the clade ((Diplogastridae + Rhabditidae) + (Mesorhabditidae + Peloderidae)), which was only recovered in 2/16 analyses, and in one (combined, unpartitioned secondary structure) the BPP was 1.0. In some instances, strong support for otherwise rarely recovered clades was found in multiple dataset/analysis combinations, for example, (Diplogastridae + Bunonematideae) (Tables 3,4). Conversely, there are also some instances of the absence of a clade that is otherwise generally strongly supported. For example, Tylenchomorpha is recovered as a distinct clade in all analyses except for the filtered ProAlign dataset of 18S + 28S analyzed using ML.
Comparison of PP and bootstrap support:
It is important to note that Bayesian posteriors and bootstrap support frequencies are not equivalent measures of clade confidence (Alfaro et al., 2003). A clade with high bootstrap support is expected to be present in other analyses of datasets generated by the same fundamental process (Felsenstein, 1985), that is, bootstrap resampling measures repeatability (Berry and Gascuel, 1996). In contrast, BPP sampling is used to assess how well data support results of a fully probabilistic model of character evolution; Bayesian posteriors are results conditioned on the observed data and models employed. Empirically, bootstrap support for clades in an ML tree is often lower than BI PP for the same clades.
Another approach to evaluating results is comparing differences in BPP or ML bootstrap support values for clades. Do some datasets yield uniformly higher PP or bootstrap frequencies or do differences in such patterns vary over different parts of the phylogeny? Individual increases and decreases for all clades (≥5% difference) based on comparison of the 18S + 28S results with the 18S results are marked by symbols on the 18S + 28S trees (Figs. 4–7); values of support (BPP and ML bootstrap) for the major clades including 18S and combined 18S + 28S trees are provided in Tables 3 and 4. Does addition of the 28S data to the 18S data yield an increase in overall clade support or was there no significant effect? This question was evaluated by comparisons within each of the four separate alignments (ProAlign, ProAlign60, SecStr unpartitioned, and SecStr partitioned) and for both inference methods (BPP and ML bootstrap) using a one-tailed sign test. There were no significant differences in any of the comparisons.
For the combined (18S + 28S) datasets, BPP for clades were high and similar among the ProAlign alignment (Fig. 4B) and the two secondary structure datasets: unpartitioned (Fig. 6B) and partitioned paired-site models (Fig. 7B). For the combined secondary structure datasets, most clades had >0.95 PP, whereas few (2–4 clades per analysis) were supported at <0.95 PP. In the combined ProAlign filtered alignment (Fig. 5B), five clades were supported at <0.95 PP. A similar pattern was observed with these datasets and ML bootstrap values (summarized in Table 4). For example, the combined ProAlign alignment (Fig. 4D) had seven clades with ML bootstrap frequencies <70%, whereas the two secondary structure datasets had slightly fewer (4–5 clades <70%). In contrast, the combined ProAlign-filtered alignment showed an increase in poorly supported clades, with 10 at <70% frequency (Fig. 5D). Removal of characters judged alignment ambiguous from the combined 18S + 28S dataset showed more instances of decreased clade support than the converse (Table 4). Considering all changes in clade frequency in analyses of the combined ProAlign versus the ProAlign filtered datasets, there were 20 instances of decreases in support (summing over Bayesian posteriors and ML bootstrap results) versus 9 instances of increased clade support. A similar result was observed for analysis of the 18S dataset. There were no notable differences across datasets (except ProAlign filtered) in the number of clades with strong bootstrap or high PP values (bootstrap ≥80%, Bayesian posteriors ≥0.95).
Discussion
Unless otherwise noted, discussion of phylogenetic results is based on analysis of the largest number of sequence characters, that is, the combined analysis of 18S + 28S data.
Phylogenetic affinities of Daubaylia potomaca (family Daubayliidae):
Daubaylia potomaca Chitwood and Chitwood, 1934, like all other known members of the genus Daubaylia Chitwood and Chitwood, 1934, are obligate parasites of fresh-water invertebrates, mostly Mollusca and Hirudinea (Anderson and Bartlett, 1993). In all phylogenetic analyses, independent of alignment and inference method, D. potomaca was robustly placed within Cephalobidae as the sister taxon to Pseudacrobeles sp. (Figs. 4–7). This is the first phylogenetic analysis that includes a species of Daubaylia; the placement of this genus within Cephalobidae supports the independent evolution of parasitism within this clade of microbivores. Detailed morphological studies are required to evaluate structural similarities between Daubaylia and Cephalobidae, since published descriptions of species do not provide sufficient details. Notably, D. olsoni Poinar, 1984 is described as having an offset sac-like spermatheca in females (Poinar, 1984). Drawings of D. pearsoni Anderson and Bartlell, 1993 also show what appears to be an offset spermatheca at the oviduct-uterus junction of the gonad flexure point (Fig. 1 in Anderson and Bartlett, 1993), but this structure is not mentioned in the description. The presence of an offset spermatheca is a synapomorphic character uniting Daubaylia and Cephalobidae and is the only morphological character known to support the molecular phylogenetic result. Daubaylia and Dicelis (Drilonematoidea) are the only animal-parasitic taxa found nested within Cephalobidae based on molecular phylogenies. Ribosomal RNA sequences place the genus Dicelis Dujardin, 1845 as a sister taxon to either Z. punctata (Spiridonov et al., 2005, 2007) or Acrobeloides sp. (Holovachov et al., 2011). The drilonematid genera Siconema Timm, 1966 (family Ungellidae) and Perodira Baylis, 1943 (family Homungellidae) represent a single clade in molecular phylogenies, and either have sister-taxon relationships to Cephalobidae or belong to a polytomy with representatives of Cephalobidae (Spiridonov et al., 2007). Improved resolution of phylogenetic relationships among drilonematid genera and Daubaylia is required to determine if animal parasitism evolved once or multiple times within Cephalobidae.
Phylogenetic affinities of F. grossa (family Chambersiellidae):
Phylogenetic affinities of the family Chambersiellidae have never been clearly resolved or analyzed in detail. The structure of their sensory organs (outer labial and cephalic sensilla, amphids, and male precloacal sensilla) is considered plesiomorphic compared to Cephalobomorpha or Panagrolaimomorpha (De Ley and Blaxter, 2002; Holovachov et al., 2003), but chambersiellids have not been explicitly compared to other representatives of Rhabditida. Phylogenetic analysis of the chambersiellid F. grossa Truskova and Eroshenko, 1977 was first published by Nadler et al. (2006b) based on 28S rRNA sequences. These authors found F. grossa to be sister to the (Cephalobomorpha + Tylenchomorpha) clade. Subsequently, F. grossa and other species of the family Chambersiellidae were used as out-groups in phylogenetic studies of Cephalobomorpha (Boström et al., 2011; Holovachov et al., 2011) or Tylenchomorpha (Koshel et al., 2014). The inferred phylogenetic position of Fescia is not consistent in our study. In five of eight analyses, it is the sister taxon to (Cephalobomorpha + Tylenchomorpha) but with varying support values (Figs. 4B,D; 5B; 7B,D). However, for analyses of the unpartitioned secondary structure alignment, Fescia is part of the Panagrolaimomorpha grade, that is, a sister taxon to ((Cephalobomorpha + Tylenchomorpha) + Panagrolaimomorpha partim) with high support in both ML and BI analyses (Fig. 6B,D). The ML tree based on the filtered ProAlign dataset resolves Fescia in the same manner as the unpartitioned secondary structure trees (ML and Bayesian) and with strong bootstrap support (Fig. 5D). However, some relationships within the sister group to Fescia ((Cephalobomorpha + Tylenchomorpha) + Panagrolaimomorpha partim) do not have reliable bootstrap support from this dataset. Phylogenetic results for relationships of Chambersiellidae are thus inconsistent, varying depending upon analysis type (unpartitioned versus partitioned secondary structure models) and presence versus absence of characters deemed ambiguous in alignment (ProAlign versus ProAlign filtered datasets). The analyses we consider optimal, partitioned secondary structure models (see Discussion below), also show moderate to poor support (ML bootstrap, BPP) for a sister-group relationship of Fescia to (Cephalobomorpha + Tylenchomorpha). These results indicate that the large 18S + 28S dataset appears to contain insufficient phylogenetic signal for unambiguously resolving the relationship of F. grossa.
Phylogenetic affinities of Diplogastridae and Bunonematidae:
The family Diplogastridae occupies a different position in all four combined (18S + 28S) datasets: (i) as a sister clade to Bunonematidae in analysis of the full ProAlign dataset (Fig. 4B,D); (ii) as the sister group to (Mesorhabditidae + Peloderidae) in the analysis of the filtered ProAlign dataset (Fig. 5B,D); (iii) as the sister group to ((Choriorhabditis + Strongyloidea) + Caenorhabditis) in the analysis based on the unpartitioned secondary structure dataset (Fig. 6B,D); and (iv) as the sister taxon to entire Rhabditidae (including Strongyloidea) clade in the analyses based on the partitioned secondary structure models (Fig. 7B,D). Only some of these relationships received strong support as assessed by Bayesian posteriors or ML bootstrap resampling, and only the tree based on the unpartitioned secondary structure analysis is similar in topology to the phylogenies obtained by Kiontke et al. (2007) using multigene data and by Sudhaus (2011) based on combined molecular and morphological evidence.
The position of B. reticulatum in our analyses varied substantially, from being a sister taxon to Diplogastridae in both ML and BI analyses of the unfiltered ProAlign dataset (Fig. 4B,D), to a sister taxon of the clade containing Rhabditidae, Strongyloidea, Mesorhabditidae, Peloderidae, and Diplogastridae (with different internal topology) in both ML and BI analyses of the remaining datasets (Figs. 5D; 6B,D; 7B,D). In the Bayesian analysis of the filtered ProAlign dataset, Bunonema is part of an unresolved trichotomy (Fig. 5B). The first result (sister-group relationships between Bunonematidae and Diplogastridae) is supported by morphological studies (Fürst von Lieven, 2002), whereas the second result (including our preferred partitioned secondary structure analyses, see following section) is more in agreement with an independent molecular analysis (Kiontke et al., 2007).
Effects based on choice of alignment, inference method, and models:
The majority of inferred molecular phylogenies for Nematoda are based on rDNA sequences that encode rRNA genes, including 18S and 28S, as well as internal transcribed spacer regions. These rRNA phylogenies have been essential for testing ideas of relationships implicit in nematode classifications and developing detailed new hypotheses for nematode evolution. However, some of these phylogenies have been insufficiently qualified by consideration of the caveats involving analysis of rRNA sequences, including uncertainties involving multiple alignment of data, use of sequence evolution models that account for compensatory substitutions in base-paired stems, and choice of substitution model. These issues are in addition to the limitations of interpreting any single-locus gene tree as a reconstruction of evolutionary history. The inference methods in our analyses have been used for phylogenetics of rRNA sequences in nematodes and other organisms. These methods include secondary-structure-based multiple alignment, both with and without partitioning of base-paired rRNA stem structures in analysis; probabilistic progressive multiple alignment, both with and without removal of alignment-ambiguous characters; Bayesian and ML inference; and substitution model selection based on model fitting programs. Although there are some strongly supported clades in common to the phylogenetic trees produced by all these analyses, there are also instances of notable topological differences between analyses that are reliably supported. This outcome is not unusual for analyses of rRNA: investigators employing different alignments and analytical approaches will obtain different phylogenetic results (Smythe et al., 2006). Alternatively, some approaches to analysis might be better justified than others based on theoretical considerations. For example, the conservation of rRNA secondary structure exceeds nucleotide conservation (Gutell et al., 1994; Kjer, 1995) facilitating identification of homologous characters (alignment) as sequences diverge. In contrast, progressive multiple alignment methods use a gap penalty to align length-variable sequences, but this procedure is problematic when applied to rRNA sequences (Kjer, 1995; Kjer et al., 2006).
Many phylogenetic analyses of nematode rRNA sequences are based on datasets obtained using progressive multiple alignment rather than secondary structure; the latter are more difficult and time consuming to produce (Gardner et al., 2005). There are several theoretical advantages of alignments based on secondary structure, including greater potential accuracy of positional homology inference within features of rRNA such as stems that are conserved across distantly related species in the absence of significant sequence similarity and the ability to identify base-pairing and non-pairing positions and apply different paired-site models to these regions such as implemented in PHASE (Hudelot et al., 2003), MrBayes (Ronquist and Huelsenbeck, 2003), or RAxML (Stamatakis, 2014). Theoretically, incorporating structural information in analysis of rRNA sequences, including modeling compensatory (and non-independent) evolution of paired-site regions should increase phylogenetic accuracy (Gillespie, 2004). Although progressive multiple alignments do not preclude incorporation of structural information, determinations of paired versus non-paired sites would necessarily require additional information. Progressive alignment approaches require use of arbitrary gap-opening and gap-extension costs, and Smythe et al. (2006) showed that differences in these parameters for nematode 18S data were correlated with differences in tree topologies, that is, choice of arbitrary costs made a predictable difference in relationships. Removing alignment-ambiguous regions from 18S datasets greatly reduced this effect but at the cost of decreased tree resolution due to removal of informative character data (Smythe et al., 2006), and this effect was also observed in our combined 18S + 28S analyses. In some cases, the tree resolution after removal of alignment-ambiguous characters may be sufficient to resolve the relationships of interest. For example, all eight analyses of the combined 18S + 28S data, including the ProAlign filtered alignment (excluding ambiguously aligned sites), yielded monophyly for Cephalobomorpha, Strongyloidea, Diplogastridae, (Mesorhabditidae + Peloderidae), and Rhabditidae (including Strongyloidea). However, in most cases, the highest levels of phylogenetic resolution are desired to develop detailed hypotheses of relationships among taxa. For progressive alignments, pooling of trees from several different alignments (and gap parameters) can sample alignment space and produce well-resolved trees, but the range of alignment parameters to explore is problematic to define (Smythe et al., 2006). Thus, theoretical and analytical considerations favor use of secondary structure alignments that employ structural (paired-site) models for analysis when the need for higher levels of phylogenetic resolution outweighs concerns over potential ambiguity in positional homology. It is for these reasons that we consider secondary structure alignment to be overall more accurate than progressive multiple alignment for constructing datasets for rRNA sequences, while also recognizing that producing structural alignments is very labor intensive and not without ambiguity. Notably, in the combined analysis of 18S + 28S data, the secondary structure analyses with paired-site models (ML, GTR GAMMA/RNA7A; Bayesian, REV/RNA7A) yielded identical phylogenetic tree topologies, with many strongly supported clades. We believe these hypotheses represent the best estimates of the rRNA gene tree for the sampled taxa.
Literature Cited
- Aerts J. 2001. Sequentiebepaling van het gen voor het grote ribosomale subeenheid RNA van de nematoden Plectus aquatilis en Meloidogyne javanica. Ph.D. dissertation, Universiteit Antwerpen, Belgium.
- Aleshin VV, Kedrova OS, Milyutina IA, Vladychenskaya NS, Petrov NB. Relationships among nematodes based on the analysis of 18S rRNA gene sequences: Molecular evidence for monophyly of chromadorian and secernentian nematodes. Russian Journal of Nematology. 1998;6:175–184. [Google Scholar]
- Alfaro ME, Zoller S, Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Molecular Biology and Evolution. 2003;20:255–266. doi: 10.1093/molbev/msg028. [DOI] [PubMed] [Google Scholar]
- Anderson RC, Bartlett CM. Daubaylia pearsoni n. sp. (Nematoda: Daubayliidae) in Glyptophysa gibbosa (Planorbidae) in Australia. Journal of Parasitology. 1993;79:671–673. [Google Scholar]
- Berry V, Gascuel O. On the interpretation of bootstrap trees: Appropriate threshold of clade selection and induced gain. Molecular Biology and Evolution. 1996;13:999–1011. [Google Scholar]
- Bert W, Leliaert F, Vierstraete A, Vanfleteren JR, Borgonie G. Molecular phylogeny of the Tylenchida and evolution of the female gonoduct (Nematoda: Rhabditida) Molecular Phylogenetics and Evolution. 2008;48:728–744. doi: 10.1016/j.ympev.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Bik HM, Lambshead PJD, Thomas WK, Lunt DH. Moving towards a complete molecular framework of the Nematoda: A focus on the Enoplida and early branching clades. BMC Evolutionary Biology. 2010;10:353. doi: 10.1186/1471-2148-10-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, Vida JT, Thomas WK. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Boström S, Holovachov O, Nadler SA. Description of Scottnema lindsayae Timm, 1971 (Rhabditida: Cephalobidae) from Taylor Valley, Antarctica and its phylogenetic relationship. Polar Biology. 2011;34:1–12. [Google Scholar]
- Chilton NB, Huby-Chilton F, Gasser RB. First complete large subunit ribosomal RNA sequence for a parasitic nematode: Phylogenetic and diagnostic implications. Molecular and Cellular Probes. 2003;17:33–39. doi: 10.1016/s0890-8508(02)00107-x. [DOI] [PubMed] [Google Scholar]
- Chilton NB, Huby-Chilton F, Gasser RB, Beveridge I. The evolutionary origins of nematodes within the order Strongylida are related to predilection sites within hosts. Molecular Phylogenetics and Evolution. 2006;40:118–128. doi: 10.1016/j.ympev.2006.01.003. [DOI] [PubMed] [Google Scholar]
- De Ley P, Blaxter M. 2002. Systematic position and phylogeny. Pp. 1–30 in D. L. Lee, ed. The biology of nematodes. London and New York: Taylor & Francis.
- De Ley P, Blaxter M. A new system for Nematoda: Combining morphological characters with molecular trees, and translating clades into ranks and taxa. Nematology Monographs and Perspectives. 2004;2:633–653. [Google Scholar]
- De Rijk P, De Wachter R. DCSE, an interactive tool for sequence alignment and secondary structure research. Computer Applications in the Biosciences. 1993;9:735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Sulston JE, Coulson AR. The rDNA of C. elegans: Sequence and structure. Nucleic Acids Research. 1986;14:2345–2364. doi: 10.1093/nar/14.5.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fürst von Lieven A. The sister group of the Diplogastrina (Nematoda) Russian Journal of Nematology. 2002;10:127–137. [Google Scholar]
- Gardner PP, Wilm A, Washietl S. A benchmark of multiple sequence alignment programs upon structural RNAs. Nucleic Acids Research. 2005;33:2433–2439. doi: 10.1093/nar/gki541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ. Characterizing regions of ambiguous alignment caused by the expansion and contraction of hairpin-stem loops in ribosomal RNA molecules. Molecular Phylogenetics and Evolution. 2004;33:936–943. doi: 10.1016/j.ympev.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Gowri-Shankar V, Jow H. 2006. PHASE: A software package for phylogenetics and sequence evolution. Manchester, UK: University of Manchester.
- Gutell RR, Fox GE. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Research. 1988;16:r175–r269. doi: 10.1093/nar/16.suppl.r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiological Reviews. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A. 2001. Sequentiebepaling van het grote ribosomale subeenheid RNA van de nematoden Oscheius dolichura en Teratocephalus lirellus. Ph.D. dissertation, Universiteit Antwerpen, Antwerp, Belgium.
- Higgs PG. RNA secondary structure: Physical and computational aspects. Quarterly Review in Biophysics. 2000;33:199–253. doi: 10.1017/s0033583500003620. [DOI] [PubMed] [Google Scholar]
- Holovachov O, Boström S, Mundo-Ocampo M, Tandingan De Ley I, Yoder M, Burr AHJ, De Ley P. Morphology, molecular characterisation and systematic position of Hemiplectus muscorum Zell, 1991 (Nematoda: Plectida) Nematology. 2009;11:719–737. [Google Scholar]
- Holovachov O, Boström S, Robinson C, Tandingan De Ley I, Nadler SA. Redescription of Placodira lobata Thorne, 1937 (Rhabditida: Cephalobidae) with a discussion of the systematic position of the genus. Nematology. 2011;13:103–114. [Google Scholar]
- Holovachov O, Boström S, Tandingan De Ley I, Robinson C, Mundo-Ocampo M, Nadler SA. Morphology, molecular characterisation and systematic position of the genus Cynura Cobb, 1920 (Nematoda: Plectida) Nematology. 2013a;15:611–627. [Google Scholar]
- Holovachov O, Esquivel A, Bongers T. Free-living nematodes from nature reserves in Costa Rica. 4. Cephalobina. Nematology. 2003;5:1–15. [Google Scholar]
- Holovachov O, Fadeeva N, Tandingan De Ley I, Mundo-Ocampo M, Gingold R, De Ley P. Revision and phylogeny of Tarvaia Allgén, 1934 (Nematoda: Tarvaiidae) Nematology. 2012;14:677–708. [Google Scholar]
- Holovachov O, Rodrigues CF, Zbinden M, Duperron S. Trophomera conchicola sp. n. (Nematoda: Benthimermithidae) from chemosymbiotic bivalves Idas modiolaeiformis and Lucionoma kazani (Mollusca: Mytilidae and Lucinidae) in Eastern Mediterranean. Russian Journal of Nematology. 2013b;21:1–12. [Google Scholar]
- Holterman M, Holovachov O, van den Elsen S, van Megen H, Bongers T, Bakker J, Helder J. Small subunit ribosomal DNA-based phylogeny of basal Chromadoria (Nematoda) suggests that transitions from marine to terrestrial habitats (and vice versa) require relatively simple adaptation. Molecular Phylogenetics and Evolution. 2008a;48:758–763. doi: 10.1016/j.ympev.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Holterman M, Karssen G, van den Elsen S, van Megen H, Bakker J, Helder J. Small subunit rDNA-based phylogeny of the Tylenchida sheds light on relationships among some high-impact plant-parasitic nematodes and the evolution of plant feeding. Phytopathology. 2009;99:227–235. doi: 10.1094/PHYTO-99-3-0227. [DOI] [PubMed] [Google Scholar]
- Holterman M, Rybaczyk K, van den Elsen S, van Megen H, Mooyman P, Peña Santiago R, Bongers T, Bakker J, Helder J. A ribosomal DNA-based framework for the detection and quantification of stress-sensitive nematode families in terrestrial habitats. Molecular Ecology Resources. 2008b;8:23–34. doi: 10.1111/j.1471-8286.2007.01963.x. [DOI] [PubMed] [Google Scholar]
- Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Molecular Biology and Evolution. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- Hudelot C, Gowri-Shankar V, Jow H, Rattray M, Higgs PG. RNA-based phylogenetic methods: Application to mammalian mitochondrial RNA sequences. Molecular Phylogenetics and Evolution. 2003;28:241–252. doi: 10.1016/s1055-7903(03)00061-7. [DOI] [PubMed] [Google Scholar]
- Kang S, Sultana T, Eom KS, Park YC, Soonthornpong N, Nadler SA, Park J-K. The mitochondrial genome sequence of Enterobius vermicularis (Nematoda: Oxyurida)—an idiosyncratic gene order and phylogenetic information for chromadorean nematodes. Gene. 2009;429:87–97. doi: 10.1016/j.gene.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Kelchner SA, Thomas MA. Model use in phylogenetics: Nine key questions. Trends in Ecology and Evolution. 2006;22:87–94. doi: 10.1016/j.tree.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee S-H, Gazi M, Kim T, Jung D, Chun J-Y, Kim S, Seo T-K, Park C, Baldwin JG, Nadler SA, Park J-K. Mitochondrial genomes advance phylogenetic hypotheses for Tylenchina (Nematoda: Chromadorea) Zoologica Scripta. 2015;44:446–462. [Google Scholar]
- Kiontke K, Barriere A, Kolotuev I, Podbilewicz B, Sommer R, Fitch DH, Félix M-A. Trends, stasis, and drift in the evolution of nematode vulva development. Current Biology. 2007;17:1925–1937. doi: 10.1016/j.cub.2007.10.061. [DOI] [PubMed] [Google Scholar]
- Kiontke KC, Félix M-A, Ailion M, Rockman MV, Braendle C, Pénigault J-B, Fitch DHA. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evolutionary Biology. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer K. Use of rRNA secondary structure in phylogenetic studies to identify homologous positions: An example of alignment and data presentation from the frogs. Molecular Phylogenetics and Evolution. 1995;4:314–330. doi: 10.1006/mpev.1995.1028. [DOI] [PubMed] [Google Scholar]
- Kjer KM, Gillespie JJ, Ober KA. Structural homology in ribosomal RNA, and a deliberation on POY. Arthropod Systematics and Phylogeny. 2006;64:71–76. [Google Scholar]
- Koshel EI, Aleshin VV, Eroshenko GA, Kutyrev VV. Phylogenetic analysis of entomoparasitic nematodes, potential control agents of flea populations in natural foci of plague. BioMed Research International. 2014;2014:135218. doi: 10.1155/2014/135218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Milinkovitch MC. A hidden Markov model for progressive multiple alignment. Bioinformatics. 2003;19:1505–1513. doi: 10.1093/bioinformatics/btg193. [DOI] [PubMed] [Google Scholar]
- Mayer WE, Herrmann M, Sommer RJ. Molecular phylogeny of beetle associated diplogastrid nematodes suggests host switching rather than nematode-beetle coevolution. BMC Evolutionary Biology. 2009;9:212. doi: 10.1186/1471-2148-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldal BHM, Debenham NJ, De Ley P, Tandingan De Ley I, Vanfleteren JR, Vierstraete AR, Bert W, Borgonie G, Moens T, Tyler PA, Austen MC, Blaxter ML, Rogers AD, Lambshead PJD. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Molecular Phylogenetics and Evolution. 2007;42:622–636. doi: 10.1016/j.ympev.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA, Pp. 1–8.
- Nadler SA, Bolotin E, Stock SP. Phylogenetic relationships of Steinernema Travassos, 1927 (Nematoda: Cephalobina: Steinernematidae) based on nuclear, mitochondrial, and morphological data. Systematic Parasitology. 2006a;63:161–181. doi: 10.1007/s11230-005-9009-3. [DOI] [PubMed] [Google Scholar]
- Nadler SA, Carreno RA, Mejía-Madrid H, Ullberg J, Pagan C, Houston R, Hugot J-P. Molecular phylogeny of clade III nematodes reveals multiple origins of tissue parasitism. Parasitology. 2007;134:1421–1442. doi: 10.1017/S0031182007002880. [DOI] [PubMed] [Google Scholar]
- Nadler SA, De Ley P, Mundo-Ocampo M, Smythe AB, Stock SP, Bumbarger D, Adams BJ, Tandingan De Ley I, Holovachov O, Baldwin JG. Phylogeny of Cephalobina (Nematoda): Molecular evidence for recurrent evolution of probolae and incongruence with traditional classifications. Molecular Phylogenetics and Evolution. 2006b;40:696–711. doi: 10.1016/j.ympev.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Nadler SA, Hudspeth DSS. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: Hypotheses of structural and sequence evolution. Journal of Parasitology. 2000;86:380–393. doi: 10.1645/0022-3395(2000)086[0380:POTANA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Sweden.
- Park J-K, Sultana T, Lee S-H, Kang S, Kim HK, Min G-S, Eom KS, Nadler SA. Monophyly of clade III nematodes is not supported by phylogenetic analysis of complete mitochondrial genome sequences. BMC Genomics. 2011;12:392. doi: 10.1186/1471-2164-12-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinar GO. Daubaylia olsoni sp. n. (Daubayliidae: Rhabditida) from the leech, Dina anoculata, in California. Proceedings of the Helminthological Society of Washington. 1984;51:217–220. [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer.
- Rokas A, Carroll SB. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Molecular Biology and Evolution. 2005;22:1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosenberg MS, Kumar S. Incomplete taxon sampling is not a problem for phylogenetic inference. Proceedings of the National Academy of Sciences. USA. 2001;98:10751–10756. doi: 10.1073/pnas.191248498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M. 4SALE—a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics. 2006;7:498. doi: 10.1186/1471-2105-7-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokoohi E, Mehrabi-Nasab A, Abolafia J, Holovachov O. Study of the genus Plectus Bastina, 1865 (Nematoda: Plectidae) from Iran. Biologia. 2013;68:1142–1154. [Google Scholar]
- Smythe AB, Nadler SA. Molecular phylogeny of Acrobeloides and Cephalobus (Nematoda: Cephalobidae) reveals paraphyletic taxa and recurrent evolution of simple labial morphology. Nematology. 2007;8:819–836. [Google Scholar]
- Smythe AB, Sanderson MJ, Nadler SA. Nematode small subunit phylogeny correlates with alignment parameters. Systematic Biology. 2006;55:972–992. doi: 10.1080/10635150601089001. [DOI] [PubMed] [Google Scholar]
- Spiridonov SE, Ivanova ES, Pham VL. Two new species of Ungellidae and Homungellidae (Drilonematoidea; Rhabditida) from Vietnamese earthworms and the phylogenetic links of these families. Russian Journal of Nematology. 2007;15:101–108. [Google Scholar]
- Spiridonov SE, Ivanova ES, Wilson MJ. The nematodes of the genus Dicelis Dujardin, 1845 parasitic in earthworms: The interrelationships of four Eurasian populations. Russian Journal of Nematology. 2005;13:61–81. [Google Scholar]
- Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web-Servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Subbotin SA, Ragsdale EJ, Mullens T, Roberts PA, Mundo-Ocampo M, Baldwin JG. A phylogenetic framework for root lesion nematodes of the genus Pratylenchus (Nematoda): Evidence from 18S and D2-D3 expansion segments of 28S ribosomal RNA genes and morphological characters. Molecular Phylogenetics and Evolution. 2008;48:491–505. doi: 10.1016/j.ympev.2008.04.028. [DOI] [PubMed] [Google Scholar]
- Sudhaus W. Phylogenetic systematisation and catalogue of paraphyletic “Rhabditidae” (Secernentea, Nematoda) Journal of Nematode Morphology and Systematics. 2011;14:113–178. [Google Scholar]
- Sudhaus W, Fürst von Lieven A. A phylogenetic classification and catalogue of the Diplogastridae (Secernentea, Nematoda) Journal of Nematode Morphology and Systematics. 2003;6:43–90. [Google Scholar]
- Sun L, Zhuo K, Wang H, Song H, Chi W, Zhang L-H, Liao J. The complete mitochondrial genome of Aphelenchoides besseyi (Nematoda, Aphelenchoididae), the first sequenced representative of the subfamily Aphelenchoidinae. Nematology. 2014;16:1167–1180. [Google Scholar]
- Tavare S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lecture Notes on Mathematical Modelling in the Life Sciences. 1986;17:262–272. [Google Scholar]
- Tillier ERM, Collins RA. High apparent rate of simultaneous compensatory base-pair substitutions in ribosomal RNA. Genetics. 1998;148:1993–2002. doi: 10.1093/genetics/148.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Megen H, van den Elsen S, Holterman M, Karssen G, Mooyman P, Bongers T, Holovachov O, Bakker J, Helder J. A phylogenetic tree of nematodes based on about 1200 full length small subunit ribosomal DNA sequences. Nematology. 2009;11:927–950. [Google Scholar]
- Wuyts J, De Rijk P, Van de Peer Y, Winkelmans T, De Wachter R. The European large subunit ribosomal RNA database. Nucleic Acids Research. 2001;29:175–177. doi: 10.1093/nar/29.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Perriere G, Van de Peer Y. The European ribosomal RNA database. Nucleic Acids Research. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Van de Peer Y, Winkelmans T, De Wachter R. The European database on small subunit ribosomal RNA. Nucleic Acids Research. 2002;30:183–185. doi: 10.1093/nar/30.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]