Abstract
Germ line mutations in the breast and ovarian cancer susceptibility gene BRCA1 are responsible for the majority of cases involving hereditary breast and ovarian cancer. While all truncating mutations are considered as functionally deleterious, most of the missense variants identified to date cannot be readily distinguished either as disease-associated mutations or as benign polymorphisms. The C-terminal domain of BRCA1 displays an intrinsic transactivation activity and mutations linked to disease predisposition have been shown to confer loss of such activity in yeast and mammalian cells. In an attempt to clarify the functional importance of the BRCA1 C-terminus as a transcription activator in cancer predisposition, we have characterized the effect of C-terminal germ-line variants identified in Scandinavian breast and ovarian cancer families. Missense variants A1669S, C1697R, R1699W, R1699Q, A1708E, S1715R, G1738E and a truncating mutation, W1837X, were characterized using yeast- and mammalian-based transcription assays. In addition, four additional missense variants (V1665M, D1692N, S1715N, and D1733G) and one in-frame deletion (V1688del) were included in the study. Our findings demonstrate that transactivation activity may reflect a tumor suppressing function of BRCA1 and further support the role of BRCA1 missense mutations in disease predisposition. We also report a discrepancy between results from yeast- and mammalian-based assays indicating that it might not be possible to unambiguously characterize variants with the yeast assay alone. We show that transcription-based assays can aid in the characterization of deleterious mutations in the C-terminal part of BRCA1 and may form the basis of a functional assay.
INTRODUCTION
Germ-line mutations in BRCA1 (OMIM #113705) predispose carriers to early onset breast and breast-ovarian cancer (1,2) and it is estimated that ~5% of all breast cancer cases are caused by inherited mutations in dominant disease genes. The majority of familial cases with both breast and ovarian cancer and a substantial part of families with breast cancer alone, involve mutations in BRCA1 (3). The BRCA1 gene product is an 1863 amino acid phosphoprotein with a RING-finger motif at its N-terminus and two BRCT domains at its C-terminus (1,4). With the exception of these domains, BRCA1 displays no similarity to other known proteins. The BRCT domains are mainly found in proteins involved in DNA repair, recombination and cell cycle control (5,6). Early findings suggest that BRCA1 is a tumor suppressor because loss of the wild-type allele was observed in familial breast and ovarian cancer cases (7). Although the function of BRCA1 remains unclear, there is increasing support for a role in DNA repair and transcription activation (reviewed in 8, 9).
BRCA1 interacts with large protein complexes involved in DNA repair such as Rad51/BRCA2 (10,11) and Rad50/Mre11/p95 (12,13). Importantly, BRCA1 becomes hyperphosphorylated and disperses from Rad51-containing nuclear foci in response to DNA damage (14,15). In mice, Brca1 is required for transcription-coupled repair of oxidative DNA damage (16) and Brca1−/− embryonic cells accumulate genetic aberrations (17). However, no direct mechanism of action has been described which explains how BRCA1 exerts its functions.
A transactivation activity was first ascribed to BRCA1 by demonstrating that when fused to a heterologous DNA binding domain, the C-terminus of BRCA1 acts as a transcription activator (18,19). BRCA1 associates in vivo with RNA polymerase II (Pol II) holoenzyme as well as with the core pol II (20–22) and modulates transcription mediated by several transcription factors (9 and references therein).
The discovery of a transactivation activity revealed a testable function of BRCA1 and yeast-based assays have been proposed as a means of characterizing missense variants because disease-associated mutations abolish this activity (23,24). Numerous mutations in BRCA1 have been described and established as disease-associated (Breast Cancer Information Core database, BIC). Such mutations are located throughout the gene and typically result in premature translation termination. Apart from a handful of clearly linked or strongly suspected disease-associated mutations, most amino acid substitutions reported hitherto cannot readily be distinguished as either disease-associated or benign polymorphisms and are classified as variants of uncertain significance (BIC), posing a very relevant problem in genetic counseling. Nevertheless, although the precise biochemical function of the protein remains unknown, increasing knowledge of the structural properties and biological roles of BRCA1 provides support in discriminating these alterations eventually allowing functional assays to be developed (24,25). Yeast-based assays have been able to discriminate between disease-associated mutations and benign polymorphisms in the C-terminus of BRCA1 (18,24,26,27). Therefore, it is tempting to suggest that the transactivation activity reflects a tumor suppressing function of BRCA1 in vivo. Here we use a transcription activation assay to characterize the effect of unique germ-line variants identified in Scandinavian breast and ovarian cancer families. Seven of the included variants are of missense type and one is of nonsense type. In addition, we analyzed five C-terminal BRCA1 variants reported by others (BIC).
RESULTS
Analysis of hereditary breast and ovarian cancer has revealed several novel as well as previously described variants of BRCA1. Patients have been screened for mutations in BRCA1 and BRCA2 as previously described (28). Here we analyze missense variants and one truncating mutation that localize to the C-terminal region of BRCA1 (Figure 1). These variants were not found in 50 healthy Swedish control individuals (No screen has been done for G1738E). Moreover, over 450 index cases with familial history of breast-ovarian cancer have been screened for mutations in BRCA1 and the variants reported here have only been found in their respective family, indicating that they represent rare variants.
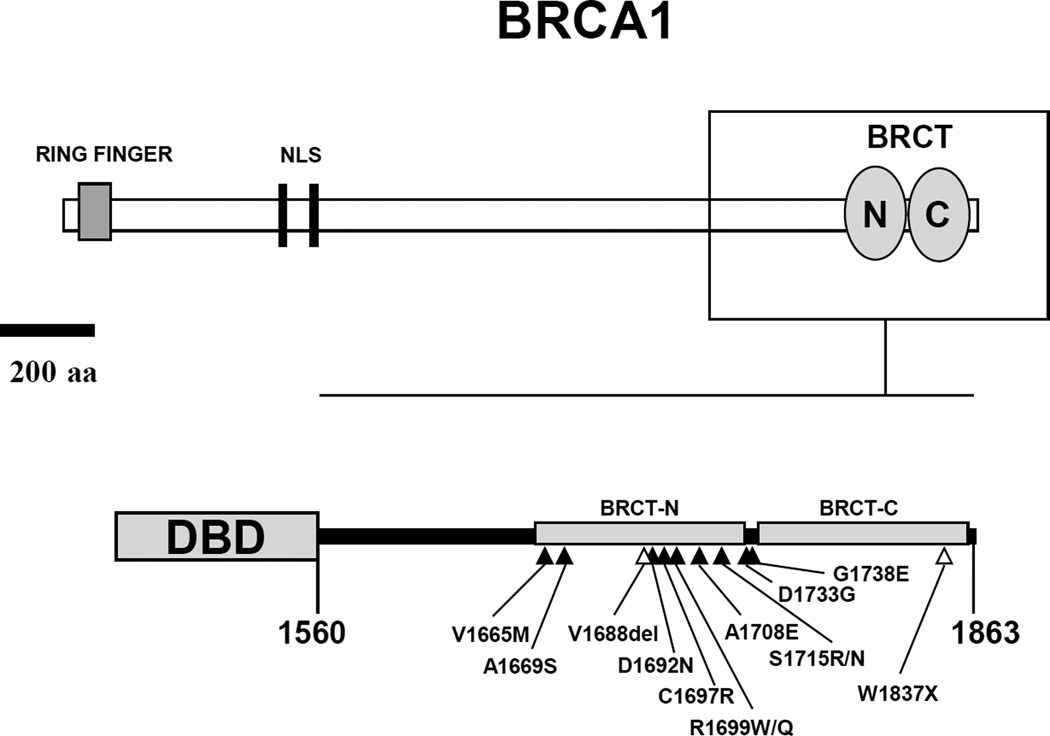
Figure 1. Domain structure of BRCA1.
Top panel. Schematic representation of full length BRCA1 protein featuring the RING domain in the N-terminal region and the BRCT domains in the C-terminal region. The region analyzed in this study (aa 1560–1863) is contained in the box, which is enlarged and represented in the bottom panel. Gray circles represent the two BRCT domains, BRCT-N (aa 1649–1736) and BRCT-C (aa 1756–1855). NLS, nuclear localization signals. Bottom panel. GAL4- and LexA- DNA binding domain (DBD) fusions to BRCA1 C-terminal region (aa 1560–1863). Mutations analyzed in this study are depicted as black (missense) or open (nonsense and in-frame deletion) triangles.
We introduced the variants in constructs containing the fusion GAL4 DNA-binding domain (DBD):BRCA1 (aa 1560–1863) (Figure 1) (18,26). In order to assess their transactivation activity these constructs were transformed into two Saccharomyces cerevisiae strains, HF7c and SFY526, containing reporter genes under the control of GAL1 UAS, recognized by GAL4 DBD. Wild type BRCA1 (aa 1560–1863) was used as a positive control and vector without insert was used as a negative control. Results were comparable in both yeast strains in a semi-quantitative assay (Table 1).
Table 1.
Transcriptional activity of BRCA1 variants identified in Scandinavian breast-ovarian cancer families and variants obtained from the Breast Cancer Information Core database.
| Transcriptional activity | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family/ source |
Exon | Mutation | Doga | Mouseb | Ratc | Nucleotided | Base change |
Probable secondary structure elementse | HF7c (His)f |
SFY526 (β-gal)g |
EGY48 (β-gal)h |
| Lund 321 | 17 | A1669S | A | A | A | 5124 | G to T | α-helix 1 of BRCT-N. | + | + | + |
| Lund 275 | 18 | C1697R | C | C | C | 5208 | T to C | α-helix 2 of BRCT-N. | − | − | − |
| Lund 279 | 18 | R1699W | R | R | R | 5214 | C to T | α-helix 2 of BRCT-N. | + | + | + |
| Lund 488 | 18 | R1699Q | R | R | R | 5215 | G to A | α-helix 2 of BRCT-N. | + | +i | + |
| Lund 20 | 18 | A1708E | A | A | A | 5242 | C to A | α-helix 2 /β-strand 4 loop of BRCT-N. | −j | −j | n.d.k |
| Lund 184 | 18 | S1715R | S | S | S | 5262 | A to C | β-strand 4/ α-helix 3 loop of BRCT-N. | − | − | − |
| Lund 32 | 20 | G1738E | G | G | G | 5332 | G to A | BRCT-N/BRCT-C interval. | − | − | −l |
| Lund 190 | 24 | W1837X | W | W | W | 5630 | G to A | α-helix 3 of BRCT-C. Conserved W in BRCT domains. | − | − | − |
| BIC | 17 | V1665M | V | V | V | 5112 | G to A | α-helix 1 of BRCT-N. | + | + | + |
| BIC | 17 | V1688del | I | I | I | 5181 | delGTT | β-strand 3 of BRCT-N. | − | − | − |
| BIC | 17 | D1692N | D | D | D | 5193 | G to A | β-strand 3/α-helix 2 of BRCT-N. | + | + | + |
| BIC | 18 | S1715N | S | S | S | 5263 | G to A | β-strand 4/ α-helix 3 loop of BRCT-N. | − | − | − |
| BIC | 20 | D1733G | D | E | E | 5317 | A to G | BRCT-N/BRCT-C interval. | + | + | +l |
Amino acid correspond to predicted translation from canine Brca1 cDNA deposited in GenBank accession # U50709.
Amino acid correspond to predicted translation from murine Brca1 cDNA deposited in GenBank accession # U68174.
Amino acid correspond to predicted translation from rat Brca1 cDNA deposited in GenBank accession # AF036760.
Nucleotide numbering corresponds to human BRCA1 cDNA deposited in GenBank accession #U14680. Alignement was performed using Vector NTI Multiple Sequence Alignement v.1.0.1.1.
According to a BRCA1 BRCT model from Zhang et al. 1998.
36 individual colonies were streaked on solid SD medium lacking tryptophan and histidine and scored for growth after two days at 30°C. A positive score (+) was noted if growth was visually identical to the positive control (wild type BRCA1 aa 1560–1863) and a negative score (−) was noted if growth was visually identical to the negative control (vector with no insert).
36 individual colonies were streaked on filter overlaid on solid SD medium and assayed for β-gal activity after two days at 30°C. A positive score (+) was noted if the activity was visually identical to the positive control (wild type BRCA1 aa1560–1863) and a negative score (−) was noted if the activity was visually identical to the negative control (vector with no insert). Clones were scored 6 hr after addition of X-gal.
At least 6 individual colonies were streaked on filter overlaid on solid SD medium lacking tryptophan and uracyl and assayed for β-gal activity the next day at 30°C. A positive score (+) was noted if the activity was visually identical to the positive control (wild type BRCA1 aa1560–1863) and a negative score (−) was noted if the activity was visually identical to the negative controls (M1775R and Y1853X). Clones were scored 2 hr after addition of X-gal.
partly reduced β-gal activity.
published results (Monteiro et al. 1996).
not done.
published results (Hayes et al. 2000).
Analysis of variants identified in Lund families
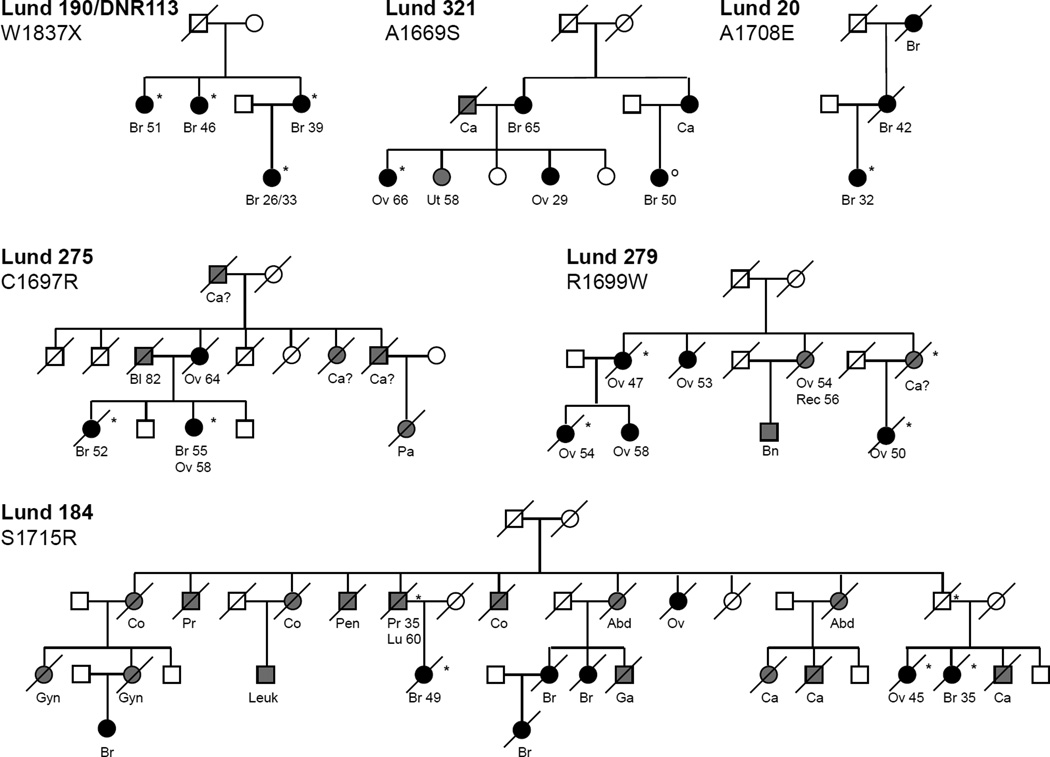
Variants A1669S, R1699W and R1699Q, displayed wild-type activity suggesting that they represent benign polymorphisms (Table 1). For variant A1669S, the data from functional assays are in agreement with the pedigree analysis (Figure 2). One of the affected family members did not carry the mutation and cases of uterine and very early onset ovarian cancer indicate involvement of predisposing genes other than BRCA1 or BRCA2. Additional clinical data should provide insight regarding A1669S and will serve as measurement of the prediction provided by the assay. Interestingly, pedigree analysis seemed to indicate that the R1699W is a cancer-predisposing allele (Figure 2, Lund 279). Disease association is further emphasized by other findings where R1699W was found in a large pedigree in which several women, through three generations and across four degrees of relatedness, diagnosed with ovarian cancer carried the variant (Frank TS and Scalia J, manuscript in preparation). Disease association is less clear for R1699Q, found in a patient diagnosed with breast cancer at the age of 39 but without familial history of disease. Others found this variant in an unaffected individual, whose mother was diagnosed with premenopausal breast cancer and considered to be an obligate carrier of the R1699Q variant, and whose grandmother was diagnosed with ovarian cancer at the approximate age of 60 years but without known mutation status (Frank TS, personal communication). The apparent discrepancy between the family and functional data prompted further examination of the R1699 variants (see below).
Figure 2. Scandinavian breast and breast-ovarian cancer families with germline BRCA1 C-terminal missense or truncating mutations.
Shown are cancer types, age at diagnosis, and mutation status (*mutation; °confirmed from blood sample not to carry mutation; ^determined from paraffin embedded tumor tissue not to carry mutation). Cancer type abbreviations: Br (breast), Ov (ovary), Ut (uterus), Gyn (gynecological), Pr (prostate), Co (colon), Rec (rectal), Leuk (leukemia), Lu (lung), Bn (brain), Bl (bladder), Pa (pancreas), Ga (gastric), Pen (Penile), Abd (Abdominal) Ca (cancer of unknown type), Ca? (possibly affected).
Variant W1837X results in a truncated protein lacking the last 27 residues and displayed loss of activity. Smaller truncations (11 residues) have been linked to disease (2) and shown to confer loss of function in transcription and in small colony phenotype assays (18,27). Our result is in agreement with pedigree analysis in which the mutation segregates with the disease (Figure 2, Lund 190).
Variants C1697R, A1708E, S1715R and G1738E displayed loss of activity suggesting that they represent disease-associated mutations (Table 1), an observation in agreement with pedigree analysis for mutations C1697R, A1708E and S1715R (Figure 2). The amino acid substitution C1697R is a rather dramatic one, from a nonpolar residue capable of forming disulfide linkages to a positively charged residue, located in a critical α-helix based on the structure of XRCC1 BRCT (29). Furthermore, the residue in question is strictly conserved in other BRCA1 homologues (Table 1)(30,31). In addition to family Lund 275, in which it segregates with the disease, the C1697R variant has been found in three other breast cancer patients. One case had multicentric disease at age 35 and a family history of breast cancer (sister and mother), whereas the other two cases had bilateral disease at age 41 and 44 and mothers with breast/skin cancer and cancer of unknown origin respectively (Bergthorsson et al, unpublished data). Thus, combined clinical data indicates association between the variant and breast cancer. Variant A1708E is reported to the BIC database 14 times including our finding in Lund 20. It has been previously shown to cause loss of function in different assays (18,19,27) and the presence of A1708E in Lund 20 further demonstrates the variant as a disease-associated mutation. S1715 is an evolutionarily conserved residue. However, the disease pattern in Lund 184 (harboring a S1715R substitution; variant S1715N was also analyzed, see below) is not satisfactorily explained by a mutation in BRCA1 alone because it presents an uncharacteristic phenotype. Multiple cases of colon cancer might suggest the involvement of a mismatch repair gene defect. However, co-segregation between the mutation and breast and ovarian cancer is observed and these cancer forms are predominant among women in the pedigree (Figure 2). We recently found the G1738E variant, which displayed loss of transactivation activity in our assays, in a young patient affected with bilateral breast cancer and a family history of disease. In addition, others found the variant in a family with a strong pattern of hereditary disease in which the patient carrying the alteration suffered from breast cancer at an early age (T. Frank, personal communication). These findings strengthen the correlation between disease-predisposition and predictions made by the transcription assay (24).
Analysis of variants in the BIC database
Variants V1665M, D1692N and D1733G displayed wild-type activity suggesting that they represent benign polymorphisms (Table 1). The V1665M variant affects a residue close to A1669, in the predicted BRCT conformation (29), which also displayed wild-type activity (Table 1 and Figure 3) suggesting that this small stretch is tolerant to mutations. Variant D1692N affects the residue predicted to form a salt bridge with S1715 therefore stabilizing the interactions between BRCT α2 and α4 regions (29). Still, D1692N displayed wild type transactivation activity suggesting that the predicted salt bridge is not important for the transactivation ability of BRCA1 (Table 1 and Figure 3). The D1733G variant is a conserved acidic residue located in the BRCT-N/BRCT-C interval. However, more information is still needed for a reliable characterization of this variant.
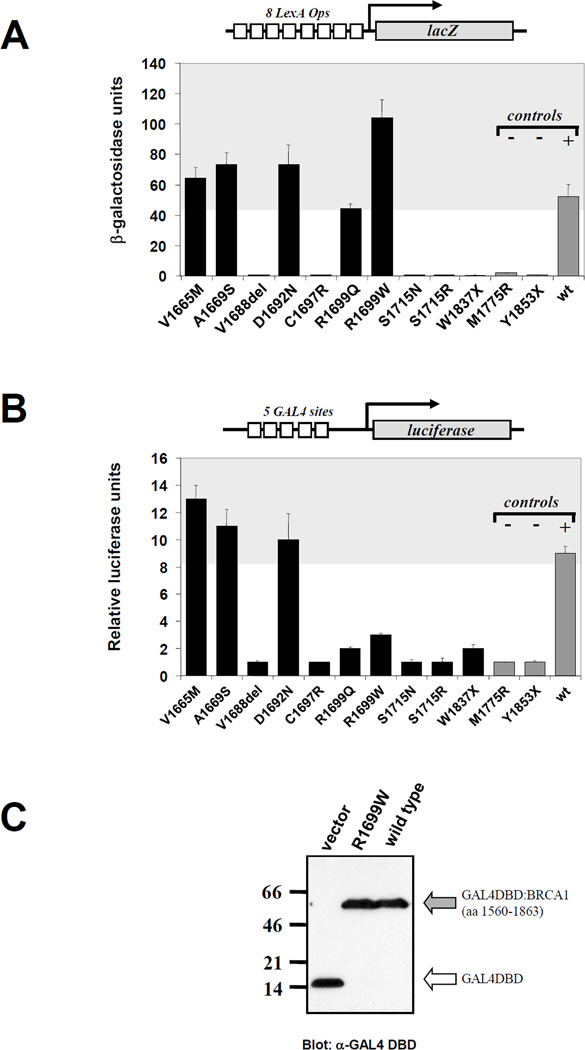
Figure 3. Transcriptional activity of BRCA1 variants.
A. Activity in yeast cells. Structure of the reporter plasmid is depicted on top of the graph. Variants (black bars) are in order of location in the structure of BRCA1 with the exception of last three constructs (gray bars), which correspond to negative (M1775R and Y1853X) and positive (wild type) controls. Shaded area represents range of activity equal or higher than wild type. B. Activity in human cells. Structure of the reporter plasmid is depicted on top of the graph. Variants (black bars) are in order of location in the structure of BRCA1 with the exception of last three constructs (gray bars), which correspond to negative (M1775R and Y1853X) and positive (wild type) controls. Shaded area represents range of activity equal or higher than wild type. C. Mutant R1699W is expressed at the same level as wild type (gray arrow). White arrow indicates expression of the GAL4 DBD moiety in the absence of any fusion fragment.
Variants V1688del and S1715N displayed loss of activity suggesting that they represent cancer-associated mutations (Table 1). Alteration V1688del is an in-frame deletion of a conserved hydrophobic residue predicted to be part of β3 in the BRCT-N domain (29). Previous mutation analysis has underscored the importance of hydrophobic residues for the function of BRCA1 (24). Similar to S1715R (Lund 184) the substitution S1715N (BIC) resulted in loss of activity in the assay.
Fusion protein and promoter stringency do not influence assay outcome
To rule out the possibility that the results obtained with the GAL4 DBD fusions were dependent on the DNA binding domain, we also performed the experiments using fusions to LexA DBD in the Saccharomyces cerevisiae strain EGY48 (24). A fusion of wild type BRCA1 (aa 1560–1863) was used as a positive control and two mutants defined by genetic linkage as disease-associated, M1775R and Y1853X were used as negative controls (1,2). Results from the LexA-based and GAL4-based assays were comparable (Table 1).
The reporter genes used in the yeast experiments contain multiple binding sites in their promoters (8 for LexA; 4 for GAL4) raising the possibility that variants with partial loss of function could score as wild type in the semi-quantitative filter β-galactosidase assay. This could be particularly important in the case of R1699W variant for which we found a contradiction between the family data and transcription activity. Therefore, EGY48 experiments with the R1699W variant were performed with the LacZ reporter under control of one, two or eight LexA operators (32). In all cases, R1699W was indistinguishable from the wild-type allele (not shown).
Quantitative assessment of transcription activation
Despite the fact that we saw no difference that could be attributed to promoter stringency, it was still possible that variants with partial loss of activity could only be differentiated using quantitative liquid β-galactosidase assay. However, results were comparable to the semi-quantitative assays (Figure 3A and Table 1). Interestingly, R1699W was approximately two-fold more active than the wild-type control. In conclusion, the contradiction found for variant R1699W was not due to a partial loss of function undistinguishable from the wild type in semi-quantitative assays.
Analysis in mammalian cells
To further examine the transcription activity of the variants we performed assays in mammalian cells. With the exception of variants R1699W and R1699Q, transcription activation was comparable between yeast and mammalian cells (Figure 3A,B). In 293T cells, variants R1699W and R1699Q displayed loss of function phenotype in accordance with pedigree analysis, suggesting that these variants are indeed cancer-associated mutations. Protein levels of R1699W and wild type were similar ruling out increased instability of the protein as the cause for the loss of function phenotype (Figure 3C).
DISCUSSION
The notion that cancer-predisposing mutations in tumor suppressor genes cause a loss-of-function phenotype is a key concept in cancer genetics. Here we utilized a functional assay to characterize clinically relevant BRCA1 variants. Our rationale was that transactivation activity of BRCA1 might mirror a functionally important feature of the protein in vivo and form the basis for a functional assay. Several lines of evidence have called attention to BRCA1 as a transcription regulator and it has been demonstrated that disease-associated mutations abolish the transactivation by BRCA1 in different experimental systems (reviewed in 9). Importantly, BRCA1 alleles carrying benign polymorphisms retain wild-type activity (24,26). Thus, relevant functional information might be gained from characterizing the effect of BRCA1 mutations on transcription activation. In addition, development of a functional assay for BRCA1 will fill a gap within the field directed at providing risk assessment information for counseling. The main difference between the present and past studies (24,25) is that this study is combined with pedigree and segregation analysis, providing a background to validate the results.
As demonstrated by our results in Table 1, the effect of an introduced BRCA1 mutation on transcription activation in the yeast-based assay is not affected by the DBD of the fusion protein or the promoter context of the reporter gene. Problems in interpreting results might nevertheless arise when characterizing variants that do not affect protein function in yeast. This is exemplified by the R1699 variants (Table 1). While the clinical data indicates that R1699W is likely to predispose carriers to ovarian cancer (Figure 2) our yeast-based tests revealed a wild-type activity, an apparent divergence between disease predisposition in vivo and the transcription activation assay (Table 1). This disagreement could not be explained by vector background or by differences in promoter stringency. However, we found that in the mammalian cell-based assay, transactivation activity of the R1699 variants was reduced in a fashion comparable to the negative controls. In fact, all variants presented here with the exception of R1699W and R1699Q behave similarly in the yeast and mammalian-based assay. Thus, it is possible that specific protein alterations that have an effect on in vivo phenotype remain undetected in the simplified yeast model. We are currently investigating the reasons for this difference. Consequently, at this time we cannot unambiguously characterize variants that do not disrupt transcription activation in yeast as benign polymorphisms. Using a mammalian-based assay to supplement results from the yeast assay might provide needed scrutiny to exclude or confirm disease predisposition of a certain variant. Similarly, mutations that affect mRNA processing in vivo might also be erroneously scored as a benign polymorphism because our assay is based on expression from an artificial cDNA. This could be the case for variant D1692N because the alteration affects a conserved guanine at a splice donor site and its potential effects on mRNA have not been examined. Conceivably, false negative results (i.e. benign polymorphisms that behave as loss-of-function mutants) can also occur when a particular variant causes message or protein instability in yeast. By extending our analysis using mammalian cells we should be able to distinguish those variants.
Considering the excellent correspondence between genetic alterations associated with breast and ovarian cancer in families and those that abolish transactivation, we tentatively characterized several additional BRCA1 unclassified variants. We propose that variants V1665M, D1692N, and D1733G represent benign polymorphisms and variants V1668del and S1715N represent disease-associated mutations. Final characterization of these variants must await independent confirmation.
Our findings, taken together with previously published data (18,24,26) demonstrate a correlation between loss of transactivation activity and disease predisposition and it will be interesting to see whether future data will corroborate the predictions made here. Our results indicate that yeast-based assays can aid in the characterization of deleterious mutations in the C-terminal part of BRCA1 but it may be unable to unambiguously characterize benign polymorphisms. This is exemplified by mutations at residue R1699, for which we report a discrepancy in effect on transcription between yeast and mammalian cells. Thus, our study underlines the importance of analyzing the effect of putative disease-causing mutations in mammalian-based assays and taking into account data from population-based studies. In summary, we show that transcription activation may reflect the tumor suppressing function of BRCA1 and provide further support for the role of missense mutations in disease predisposition.
MATERIALS AND METHODS
Yeast strains
Three Saccharomyces cerevisiae strains were used: HF7c (MATa, ura3-52, his3-200, lys2-801, ade2-101, trp1-901, leu2-3,112, gal4-542, gal80-538, LYS2::GAL1-HIS3, URA3::(GAL4 17-mers)3-CYC1-lacZ)(33); SFY526 (MATa, ura3-52, his 3-200, ade 2-101, lys 2-801, trp 1-901, leu 2-3, 112, canr, gal4-542, gal80-538, URA3::GAL1-lacZ (34) and EGY48 (MATa, ura3, trp1, his3, 6 lexA operator-LEU2)(35). HF7c and SFY526 contain reporter genes under the control of GAL1 UAS, which is recognized by GAL4 DBD. When activated, the reporter gene in SFY526 will produce β-galactosidase and HF7c will grow in minimal medium lacking histidine. EGY48 cells were transformed with plasmid reporters under control of LexA operators (pSH18-34, pJK103 or pRB1840) that when activated produce β-galactosidase (35).
Yeast expression constructs
A fusion construct containing GAL4 DBD:BRCA1 (aa 1560–1863) in pGBT9 (Clontech, Palo Alto, CA) used as a wild type control and as a backbone to introduce mutations was described previously (18). Specific alterations in BRCA1 were introduced by Quick-change site-directed mutagenesis (Stratagene, La Jolla, CA) according to manufacturer’s instructions. In short, primers containing the alteration were used in a PCR reaction to copy wild type constructs produced in a methylation competent bacterial strain and amplification was performed using Pfu polymerase. DpnI was subsequently added to digest the parental plasmid, leaving only cDNAs with introduced mutations to be transformed into bacteria. Confirmation of the introduced mutations was obtained by direct sequencing of the BRCA1 (aa 1560–1863) insert using two primers: GAL4 DNA-BD, 5´-TCATCGGAAGAGAGTAG- 3´(17-mer) (Clontech) and pGBT9 M13 REV, 5´-TGTAAAACGACGGCCCGTTTTAAAACCTAAGAGTCAC- 3´. For experiments in EGY48, BRCA1 inserts with mutations were subcloned into pLex9 (35) in frame with the DBD of LexA. Both pGBT9 and pLex9 have TRP1 as a selectable marker allowing growth in medium lacking tryptophan.
Yeast Transformation
Transformations were performed using the yeast transformation system based on lithium acetate (Clontech). Briefly, a single colony was inoculated in YPD medium for 16–18 hours to produce a saturated culture. Cells were transferred to fresh medium and grown for 3 hours, centrifuged, washed, resuspended in TE/LiAc (10 mM Tris-HCl, 1 mM EDTA, 100 mM lithium acetate, pH 7.5) solution and used immediately for transformation. Competent cells were incubated in polyethylene glycol/LiAc (40% PEG 4000, 10 mM Tris-HCl, 1 mM EDTA, 100 mM lithium acetate, pH 7.5) solution at 30°C for 30 minutes with appropriate vector and carrier DNA. DMSO was added to 10% final concentration and the mix was heat shocked at 42°C for 15 minutes. Cells were subsequently chilled, centrifuged and resuspended in TE (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). Cells were plated on synthetic dropout medium (SDM) and incubated at 30°C to select for transformants.
Yeast growth assay
Thirty-six individual HF7c clones for each variant were streaked on solid SDM lacking tryptophan and on SDM lacking both tryptophan and histidine and growth was scored after two days. A positive score (+) or a negative score (−) was noted if growth was identical to the positive control (wild type BRCA1 aa1560–1863) or to the negative control (vector with no insert), respectively.
β-galactosidase assays
Thirty-six individual SFY526 clones and at least six individual EGY48 clones for every variant were streaked on filter paper overlaid on solid SDM lacking tryptophan (or tryptophan and uracil for EGY48). Plates were incubated at for 2 days (SFY526) or 24 hr (EGY48) and cells growing on the filter paper were lysed by freeze-thawing in liquid nitrogen and assayed for β-galactosidase activity in 2.5ml Z buffer (16 g/l Na2HPO4•7H2O, 5.5 g/l NaH2PO4•H2O, 0.75 g/l KCl, 0.246 g/l MgSO4•7H2O, pH 7.0) containing 40 µl of X-gal solution (20mg/ml in N,N-dimethylformamide) and 6.6 µl β-mercaptoethanol. For SFY526, a positive score (+) or a negative score (−) was noted if the activity was identical to the positive control (wild type BRCA1 aa1560–1863) or to the negative control (vector with no insert), respectively. For EGY48 a positive score (+) was noted if the activity was identical to the positive control (wild type BRCA1 aa1560–1863) and a negative score (−) was noted if the activity was identical to the negative controls (M1775R and Y1853X). Clones were scored 6 hr (SFY526) or 2 hr (EGY48) after addition of X-gal. Liquid assays were performed as described previously (36). At least three separate transformants were assayed and each was performed in triplicate.
Transcription assay in mammalian cells
GAL4 DBD:BRCA1 fusions were subcloned into pCDNA3 (Invitrogen, Carlsbad, CA). We used pG5Luc, which contains a firefly luciferase gene under the control of five GAL4 binding sites (37). Transfections were normalized with an internal control, pRL-TK, which contains a Renilla luciferase gene under a constitutive TK basal promoter using a dual luciferase system (Promega). Human 293T cells were cultured in DMEM supplemented with 10% calf serum and plated in 24-well plates at ~60% confluence the day before transfection. Transfections were performed in triplicates using Fugene 6 (Roche, Indianapolis, IN) and harvested 24 hr post-transfection.
Western Blot
Cells were lysed in RIPA (150 mM NaCl, 10 mM Tris-Cl [pH 7.4], 5mM EDTA, 0.1% sodium dodecyl sulfate, 1% Triton X-100, 0.1% sodium deoxycholate), boiled in sample buffer and separated on a 10% SDS-PAGE. Gels were electroblotted on a wet apparatus to a PVDF membrane. The blots were blocked overnight with 5% skim milk using TBS-tween, and incubated with α-GAL4 DBD monoclonal antibody (Clontech) using 0.5% BSA in TBS-tween. The blots were subsequently incubated with the α-mouse IgG-horseradish peroxidase conjugate in 1% skim milk in TBS-tween and developed using an enhanced chemiluminescent reagent (NEN, Boston, MA).
Acknowledgments
We thank Niklas Dahl for providing information from family L321, Robert Coyne for sharing unpublished results and Pär-Ola Bendahl for help with statistics and presentation of data. This study was supported by the Swedish Cancer Society, the Lund Family American Cancer Society Grant, the Julia Murtha Fund, U.S. Army award DAMD17-99-1-9389 and the following foundations: Mrs. Berta Kamprads; Gunnar Arvid & Elisabeth Nilsson; Franke & Margareta Bergqvist; Hospital of Lund; King Gustav V:s Jubilee; Irving Weinstein; Lee Kaplan; and the AMDeC Foundation of New York City.
Abbreviations
- X-gal
5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside
- BRCA1
breast and ovarian cancer susceptibility gene 1
- BRCT
BRCA1 C-terminal domain
- UAS
upstream activating sequence
- PVDF
polyvinylidene difluoride
Footnotes
Electronic Database Information
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/omim
Breast Cancer Information Core (BIC): The Breast Cancer Information Core is an online database of mutations in breast cancer susceptibility genes hosted by the National Human Genome Research Institute and can be accessed at http://www.nhgri.nih.gov/Intramural_research/Lab_transfer/Bic/
REFERENCES
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Friedman LS, Ostermeyer EA, Szabo CI, Dowd P, Lynch ED, Rowell SE, King MC. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat. Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 3.Ford D, Easton DF, Peto J. Estimates of the gene frequency of BRCA1 and its contribution to breast and ovarian cancer incidence. Am. J. Hum. Genet. 1995;57:1457–1462. [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin EV, Altschul SF, Bork P. BRCA1 protein products … Functional motifs…. Nat. Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 6.Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 7.Smith SA, Easton DF, Evans DG, Ponder BA. Allele losses in the region 17q12–21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat. Genet. 1992;2:128–131. doi: 10.1038/ng1092-128. [DOI] [PubMed] [Google Scholar]
- 8.Welcsh PL, Owens KN, King MC. Insights into the functions of BRCA1 and BRCA2. Trends Genet. 2000;16:69–74. doi: 10.1016/s0168-9525(99)01930-7. [DOI] [PubMed] [Google Scholar]
- 9.Monteiro AN. BRCA1: exploring the links to transcription. Trends Biochem. Sci. 2000;25:469–474. doi: 10.1016/s0968-0004(00)01632-7. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol.Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 11.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 12.Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 14.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 15.Thomas JE, Smith M, Tonkinson JL, Rubinfeld B, Polakis P. Induction of phosphorylation on BRCA1 during the cell cycle and after DNA damage. Cell Growth Differ. 1997;8:801–809. [PubMed] [Google Scholar]
- 16.Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 17.Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan XY, Ried T, Deng CX. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 18.Monteiro AN, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc. Natl. Acad. Sci. U S A. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman MS, Verma IM. Transcriptional activation by BRCA1. Nature. 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 20.Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, Livingston DM, Parvin JD. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. U S A. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 22.Schlegel BP, Green VJ, Ladias JA, Parvin JD. BRCA1 interaction with RNA polymerase II reveals a role for hRPB2 and hRPB10alpha in activated transcription. Proc. Natl. Acad. Sci. U S A. 2000;97:3148–3153. doi: 10.1073/pnas.070452397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monteiro AN, Humphrey JS. Yeast-based assays for detection and characterization of mutations in BRCA1. Breast Disease. 1998;10:61–70. doi: 10.3233/bd-1998-101-208. [DOI] [PubMed] [Google Scholar]
- 24.Hayes F, Cayanan C, Barilla D, Monteiro AN. Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res. 2000;60:2411–2418. [PMC free article] [PubMed] [Google Scholar]
- 25.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 26.Monteiro AN, August A, Hanafusa H. Common BRCA1 variants and transcriptional activation. Am. J. Hum. Genet. 1997;61:761–762. doi: 10.1086/515515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humphrey JS, Salim A, Erdos MR, Collins FS, Brody LC, Klausner RD. Human BRCA1 inhibits growth in yeast: potential use in diagnostic testing. Proc. Natl. Acad. Sci. U S A. 1997;94:5820–5825. doi: 10.1073/pnas.94.11.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakansson S, Johannsson O, Johansson U, Sellberg G, Loman N, Gerdes AM, Holmberg E, Dahl N, Pandis N, Kristoffersson U, et al. Moderate frequency of BRCA1 and BRCA2 germ-line mutations in Scandinavian familial breast cancer. Am. J. Hum. Genet. 1997;60:1068–1078. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Morera S, Bates PA, Whitehead PC, Coffer AI, Hainbucher K, Nash RA, Sternberg MJ, Lindahl T, Freemont PS. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J. 1998;17:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett LM, Brownlee HA, Hagavik S, Wiseman RW. Sequence analysis of the rat Brca1 homolog and its promoter region. Mamm. Genome. 1999;10:19–25. doi: 10.1007/s003359900935. [DOI] [PubMed] [Google Scholar]
- 31.Szabo CI, Wagner LA, Francisco LV, Roach JC, Argonza R, King MC, Ostrander EA. Human, canine and murine BRCA1 genes: sequence comparison among species. Hum. Mol. Genet. 1996;5:1289–1298. doi: 10.1093/hmg/5.9.1289. [DOI] [PubMed] [Google Scholar]
- 32.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feilotter HE, Hannon GJ, Ruddell CJ, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel P, Chien CT, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 35.Golemis EA, Gyuris J, Brent R. Two-hybrid system/interaction traps. In: Ausubel FM, Brent R, Kingston R, Moore D, Seidman J, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York: John Wiley & Sons; 1994. pp. 13.14.1–13.14.17. [Google Scholar]
- 36.Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 37.Seth A, Gonzalez FA, Gupta S, Raden DL, Davis RJ. Signal transduction within the nucleus by mitogen-activated protein kinase. J. Biol. Chem. 1992;267:24796–24804. [PubMed] [Google Scholar]