Abstract
While ADP-ribosyltransferase diphtheria toxin-like 1 (ARTD1, formerly PARP1) and its enzymatic activity have been shown to be important for reprogramming and differentiation of cells, such as during adipogenesis, their role and mechanism in regulating osteoclastogenesis and bone homeostasis are largely unknown. Here, in cell culture-based RANKL-induced osteoclastogenesis models, we show that silencing of ARTD1 or inhibition of its enzymatic activity enhances osteoclast differentiation and function. As a consequence of ARTD1 silencing or inhibition, the recruitment of p65/RelA to the IL-1β promoter, which is associated with transcriptionally active histone marks, IL-1β expression and inflammasome-dependent secretion of IL-1β are enhanced. This subsequently promotes sustained induction of the transcription factor Nfatc1/A and osteoclastogenesis in an autocrine manner via the IL-1 receptor. In vivo, Artd1-deficient mice display significantly decreased bone mass as a consequence of increased osteoclast differentiation. Accordingly, the expression of osteoclast markers is enhanced in mutant compared to wild-type mice. Together, these results indicate that ARTD1 controls osteoclast development and bone remodelling via its enzymatic activity by modulating the epigenetic marks surrounding the IL-1β promoter and expression of IL-1β and subsequently also Nfatc1/A.
ADP-ribosyltransferase diphtheria toxin-like 1 (ARTD1, formerly called PARP1) belongs to the family of ADP-ribosyltransferases (ARTs) and utilizes NAD+ for the synthesis of ADP-ribose polymers on acceptor proteins. The human ARTD family consists of 18 proteins with either mono- or poly-ADP-ribosyltransferase activity that all share the same ARTD signature motif in their homologous catalytic domain. Several inhibitors have been developed that inhibit ARTD1 activity, but also other ARTD family members, because of their similarity and because they mostly mimic β-NAD+ 1. Since the discovery of ARTD1, most studies have focused on its role in DNA damage detection and repair responses2. However, over the past decade, ARTD1 has gained increasing attention for its role in gene regulation3,4,5,6,7. Previous studies have shown that ARTD1 is the now long forgotten RNA polymerase II basal transcription factor TFIIC, which represses nick-dependent transcription in biochemical assays8. The positive or negative impact of ARTD1 on gene expression results primarily from its cooperation with transcription factors such as NF-κB, and chromatin remodelling and modification9.
Accumulating data are now linking ARTD1 activity with cell differentiation and cell fate commitment. The change from preadipocytes to adipocytes triggered by the nuclear receptor PPARγ is a prime example of the role of nuclear ADP-ribosylation reactions in cellular differentiation. ARTD1-dependent PAR formation increases during differentiation of 3T3L1 preadipocytes and is required for sustained expression of PPARγ target genes10. Similarly, adipogenic differentiation is reduced in adipose-derived stromal cells isolated from Artd1 knockout mice11. This function of ARTD1 is specific for adipogenesis, as osteogenic differentiation of adipose-derived stromal cells is not affected by Artd1 deletion10,11. A recent study has shown that the methylcytosine dioxygenase ten-eleven translocation 1 (TET1) interacts with PPARγ in an ADP-ribosylation-dependent manner during adipogenesis12, suggesting a model of active, PAR-dependent DNA demethylation of key adipocyte-specific genes by TET112. Moreover, ADP-ribosylation positively regulates TET1 expression by maintaining a permissive chromatin state at the gene locus.
Bone is a highly dynamic tissue that undergoes continuous remodelling, being dependent on an intricate array of factors, including cytokines/chemokines, hormones, and mechanical stimuli13,14. Normal bone turnover is maintained through the coordinated actions of osteoblasts and osteoclasts on bone formation and resorption respectively15. While osteoblasts develop from mesenchymal stem cells, osteoclasts are of myeloid origin16. The differentiation of osteoclast precursors (monocytes/macrophages) into fully functional osteoclasts depends on the stimulation of the receptor activator of nuclear factor kappa-B (NF-κB) (RANK) by its ligand (RANKL) and comprises of several steps: fate commitment, early phase multinucleation and late phase fusion with other committed cells, which finally gives rise to the large and functionally active bone-resorbing cells17.
At the molecular level, RANKL stimulation triggers the induction of the NF-κB heterodimer p65 (RelA)/p50 (NF-κB1), which induces the expression of nuclear factor of activated T-cells (NFATc1), a transcription factor that regulates the terminal RANKL-induced differentiation of osteoclasts18. NFATc1 drives the formation of bone-resorbing cells and regulates expression of osteoclast-specific genes such as that of cathepsin K (Ctsk) and tartrate-resistant acid phosphatase (Tracp)18. In addition, the RANKL-induced NF-κB pathway controls the transcription of a wide range of pro-inflammatory cytokines, chemokines and growth factors19. Some of these cytokines such as TNFα, IL-1, IL-17, IL-4, and IFNs have been well documented to modulate the maturation of functionally active osteoclasts in an autocrine feedback loop20,21.
Two recent papers have shown that ARTD1 promotes osteoblast differentiation of mesenchymal stem cells in vitro, suggesting a possible involvement of ARTD1 in bone remodelling22,23. Furthermore, ARTD1 has also been reported to repress the gene expression of bone-resorbing factors such as Tracp and the a3 isoform of V-ATPase subunit (Tcirg1) during RANKL-induced osteoclastogenesis24,25, suggesting that ARTD1 might also regulate osteoclastogenesis and thereby bone remodelling, although the detailed molecular mechanism by which ARTD1 is thought to regulate osteoclastogenesis and bone formation is unknown.
These findings and our observation that ARTD1 is a co-factor of NF-κB-dependent gene expression prompted us to elucidate the molecular mechanisms responsible for the involvement of ARTD1 in osteoclastogenesis and bone homeostasis. Here, we report that silencing or inhibition of Artd1 epigenetically enhances RANKL-induced and NF-κB-dependent expression of IL-1β that subsequently drives NFATc1-dependent osteoclastogenesis. The epigenetic regulation of IL-1β expression strongly depends on the enzymatic activity of ARTD1. Furthermore, Artd1-deficiency impairs bone mass, which is likely due to alterations in osteoclastogenesis.
Results
ARTD1 activity represses RANKL-induced osteoclast differentiation
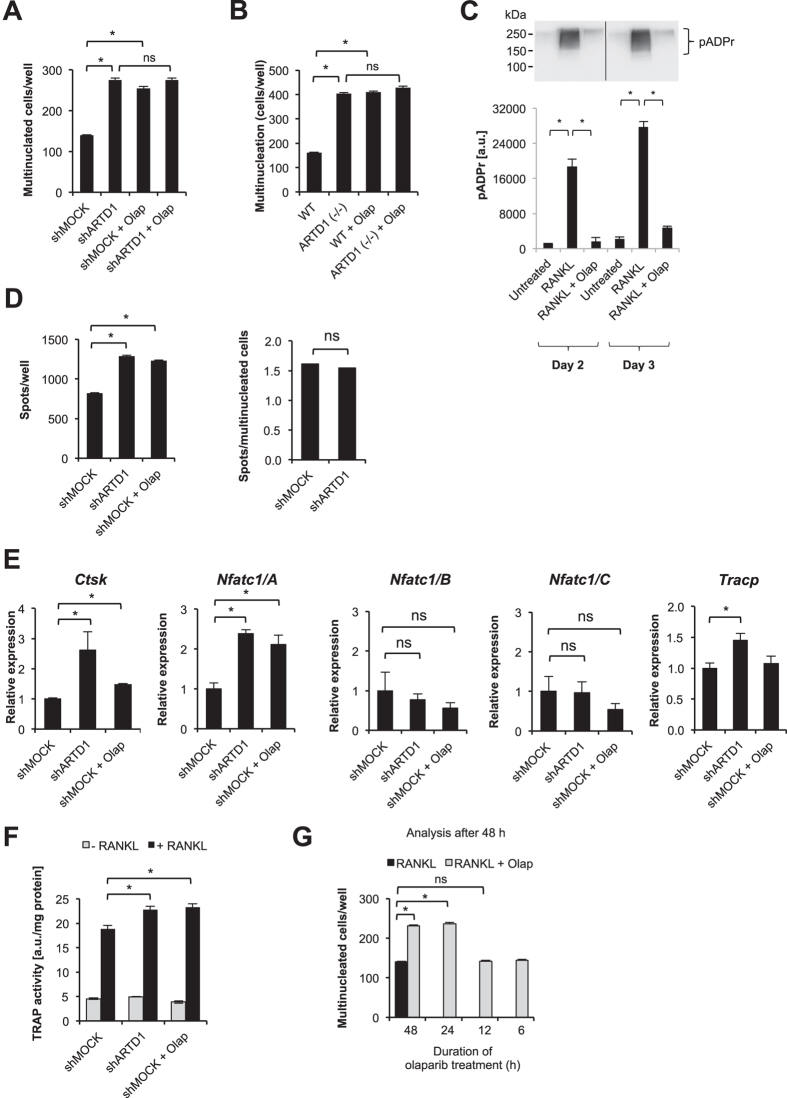
To study the role of ARTD1 in osteoclastogenesis, RAW 264.7 macrophage-derived cells that overexpress an shRNA by retroviral transduction against a control sequence (shMock) or against Artd1 (shARTD1) (Suppl. Fig. 1A,B) were stimulated with RANKL and allowed to differentiate into osteoclasts (see Suppl. Fig. 1C for overall experimental design). As expected, early multinucleation was observed in RAW 264.7 cells expressing shMock on day 2 (D2) after RANKL administration, and osteoclast expansion on day 3 (D3) (Suppl. Fig. 1C). Remarkably, silencing of Artd1 in RAW 264.7 cells strongly enhanced the RANKL-induced number of multinucleated cells when compared to RANKL-stimulated shMOCK-expressing RAW 264.7 cells (Fig. 1A), indicating that ARTD1 represses osteoclastogenesis. The same experiment with stable knockdown of p65 expression in RAW 264.7 cells caused reduced matrix dissolution and multinucleation (Suppl. Fig. 1 A,B,D), confirming the NF-κB-dependency of the system. An even more prominent effect of ARTD1 was found between RANKL-stimulated bone marrow-derived macrophages (BMDM) isolated from WT mice compared to Artd1-deficient (−/−) mice (Fig. 1B), confirming the results obtained with the engineered RAW 264.7 cells. RANKL treatment of shMock RAW 264.7 macrophages was associated with an induction of PAR formation on day 2 and 3 of differentiation (early development), which was inhibited by co-treatment of the cells with the ADP-ribosylation inhibitor olaparib (Fig. 1C). The presence of olaparib for the entire duration of osteoclast differentiation using shMOCK RAW 264.7 cells or WT BMDM phenocopied the enhanced multinucleation observed in RANKL-stimulated shARTD1-treated or Artd1 (−/−) macrophages (cf. Fig. 1A,B), indicating that ADP-ribosylation is important for repressing osteoclastogenesis. Since inhibition with olaparib did not further increase multinucleation in ARTD1-depleted or -deficient cells (Fig. 1A,B), the effect of olaparib on ARTD1-proficient cells was likely primarily due to inhibition of ARTD1. Multinucleated osteoclasts derived from Artd1-silenced or olaparib-treated RAW 264.7 macrophages were fully functional, as demonstrated by the dissolution of a mineral-coated surface (Fig. 1D, Suppl. Fig. 1E). At the molecular level, Artd1 silencing or inhibition led to increased expression of the osteoclast marker gene Nfatc1/A (but not Nfatc1/B or C) (Fig. 1E), as well as increased TRAP enzymatic activity (Fig. 1F) 72 h after RANKL stimulation of RAW 264.7 macrophages, while transcription of Ctsk and Tracp was only affected by ARTD1 silencing and not inhibition.
Figure 1. ARTD1 silencing or inhibition enhances osteoclast differentiation.
The multinucleation capacity of (A) RAW264.7 macrophages expressing basal levels of Artd1 (shMOCK) and Artd1 knock-down (shARTD1), as well as of (B) bone marrow-derived macrophages (BMDM) isolated from wild-type (WT) and Artd1-deficient mice was quantified 48 h after RANKL (50 ng/ml) administration. The ADP-ribosylation inhibitor olaparib (1 μM) was added to shMOCK and WT BMDM cell cultures simultaneously with RANKL and was present throughout the course of differentiation. (C) The accumulation of ADP-ribosylated proteins in untreated BMDM (osteoclast precursors) and in 2 and 3 day old osteoclasts differentiated in the presence and absence of olaparib was studied with western blot. Densitometry was employed for quantification of ADP-ribosylation. (D) The resorbing activity of osteoclasts differentiated for 72 h in the presence of RANKL from RAW 264.7 cells with silenced and inhibited ARTD1 were determined based on matrix dissolution (left panel). The number of matrix dissolution spots was normalized to the number of multinucleated cells (right panel). (E) The gene expression of osteoclast markers was analysed in purified RAW 264.7-derived 3-day-old osteoclasts. (F) TRAP activity was quantified in RAW264.7 cells differentiated in the presence of RANKL for 48 h. (G) Olaparib was administered at selected time points after induction of osteoclastogenesis, and multinucleated cells were counted after 48 h.
To test at which time point during RANKL-induced multinucleation ARTD1 and its enzymatic activity are important, multinucleation was induced with RANKL for 48 (Fig. 1G) or 72 h (Suppl. Fig. 2A) and olaparib added at different time points following RANKL-treatment. A significant increase in multinucleation could be observed when olaparib was added in both settings from the beginning (48 h and 72 h, respectively) or for at least 24 h after RANKL treatment (24 h and 48 h respectively), while supplementation for a shorter time did not enhance multinucleation, suggesting that ARTD1 and its enzymatic activity enhance osteoclastogenesis during the time period between 24 and 48 h of differentiation, and not during the initial RANKL signalling (i.e. the first 24 h after RANKL treatment).
Together, these results demonstrate that ARTD1 and its enzymatic activity regulate RANKL-induced multinucleation and expression of the osteoclast differentiation driver NFATc1/A after initiation of the differentiation process and that the enhanced differentiated osteoclasts are fully functional.
ARTD1 activity is induced during osteoclast differentiation in a topoisomerase II-dependent manner
Since our data indicated that the enzymatic activity of ARTD1 regulates osteoclastogenesis, we next investigated the mechanism by which the enzymatic activity of ARTD1 is induced. Since ROS have been described to strongly activate ARTD126, we investigated whether the presence of the potent ROS scavenger N-acetylcysteine affect the differentiation potential of differentiating osteoclasts. However, no significant effects of N-acetylcysteine on cell multinucleation were observed at the concentration tested (Suppl. Fig. 3A). We have recently provided evidence that activation of ARTD1 and PAR formation during adipogenesis is dependent on topoisomerase II (topo II) activity27. Interestingly, treatment of RAW 264.7 cells during osteoclastogenesis with the topo II inhibitor merbarone enhanced multinucleation to the same level as observed with Artd1 silencing (Suppl. Fig. 3B), suggesting that the observed ARTD1-mediated repression of cell multinucleation is regulated by topo II. The same observation was made with osteoclast progenitors from WT and Artd1 (−/−) mice. Treatment of progenitors with merbarone enhanced osteoclastogenesis in WT but not Artd1 (−/−) cells, suggesting that indeed topoisomerase activity induces ARTD1 activity, which subsequently represses osteoclastogenesis (Suppl. Fig. 3C).
ARTD1 silencing or inhibition enhances osteoclastogenesis by stimulating IL-1β secretion and autocrine cell stimulation
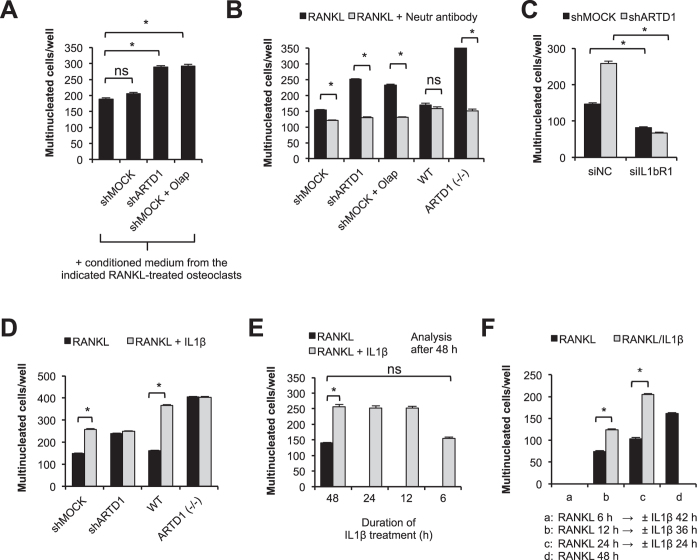
As autocrine or paracrine acting cytokines are known to regulate the differentiation of osteoclasts, we investigated whether ARTD1 controls osteoclastogensis via a secreted cytokine. Cultivation of RANKL-treated RAW264.7 macrophages with conditioned medium of either shARTD1 or olaparib-treated shMOCK, but not shMOCK osteoclasts, increased the multinucleation of RAW 264.7 macrophages through a secreted factor (Fig. 2A). Since IL-1β plays a major role in bone metabolism under physiological conditions28,29, we tested whether IL-1β could be involved in the enhanced multinucleation induced by ARTD1 depletion, inhibition or deficiency. Repeated addition of IL-1β neutralizing antibody to medium (every 8 h) from RANKL-induced Artd1-silenced or inhibited RAW 264.7 macrophages, as well as Artd1 (−/−) BMDM substantially reduced the ability of the medium to enhance multinucleation of RANKL-treated RAW264.7 macrophages (Fig. 2B), indicating that IL-1β is indeed the secreted factor responsible for the observed enhanced multinucleation of Artd1 (−/−), knock-downs or olaparib-treated cells. The transient silencing of IL-1 receptor 1 (IL-1r1) in shMOCK or shARTD1 macrophages phenocopied the effect of IL-1β neutralizing antibody on the differentiation potential of macrophages (Fig. 2C, Suppl. Fig. 3D), further supporting the idea that the observed effect is mediated by IL-1β. Differentiation of RAW 264.7 and BMDM with a mixture of RANKL and recombinant IL-1β (10 ng/ml every 8 h30) led to an increase in the number of osteoclasts formed comparable to the level of RANKL-stimulated shARTD1-treated and Artd1-deficient macrophages, respectively (Fig. 2D). To further strengthen the importance of IL-1β in RANKL-induced osteoclastogenesis during the initial/sustained phase of osteoclastogenesis, shMOCK RAW264.7 macrophages were treated with either RANKL alone or RANKL together with IL-1β for different periods of times. RANKL alone was able to induce multinucleation only when added for at least 48 h, but not for a shorter time period (Fig. 2E). Addition of IL-1β at different time points to 48 h RANKL-treated macrophages significantly enhanced multinucleation for all tested time points, but 6 h of IL-1β (Fig. 2E), although analysis of the multinucleation for this sample after an additional 24 h (72 h after RANKL) also revealed significantly enhanced multinucleation (Suppl. Fig. 2B), suggesting that IL-1β is able to enhance RANKL-induced multinucleation throughout the analysed osteoclastogenesis, but required at least 12 h of incubation. Interestingly, the contribution of IL-1β to osteoclast formation was not limited to enhancement of multinucleation, since after a certain stage of osteoclast commitment (between 6 and 12 h after RANKL stimulation) and subsequent removal of RANKL, IL-1β drove osteoclast differentiation even in the absence of RANKL (Fig. 2F), indicating that IL-1β is important for sustaining osteoclastogenesis, once RANKL has initiated the differentiation process.
Figure 2. Inhibition of ADP-ribosylation as well as Artd1 knock-out and knock-down are responsible for IL-1β overexpression-induced increase in osteoclast differentiation.
(A) The pro-osteoclastogenic potential of media conditioned for 24 h by 3-day-old osteoclasts derived from RAW264.7 macrophages with normal ARTD1 levels as well as ARTD1 silenced and inhibited cells was compared. RANKL was added to the cell culture in combination with 30% conditioned medium and multinucleation was quantified after 48 h. (B) IL-1β neutralizing antibody (100 ng) was added every 8 h to RAW 264.7 and BMDM induced with RANKL for differentiation. (C) 24 h after transfection of shMOCK and shARTD1 RAW 264.7 macrophages with siRNA targeting IL-1β receptor 1 (IL-1r1) expression, cells were supplemented with RANKL and the role of IL-1R1 in the differentiation process was determined based on multinucleation. (D) To test the impact of IL-1β on osteoclastogenesis, RANKL was added to RAW 264.7 and BMDM cultured in the presence or absence of IL-1β (10 ng/ml) and the number of osteoclasts was counted on day 2. (E) IL-1β (2 ng/ml) was added at the indicated time points to shMOCK RAW 264.7 macrophages cultured in the presence of RANKL, and multinucleated cells were quantified 48 h after RANKL administration. (F) shMOCK RAW 264.7 macrophages were stimulated with RANKL for the indicated periods of time. RANKL was then washed away and the differentiation was allowed to continue for a total time of 48 h in the presence (grey bars) or absence (black bars) of IL-1β (10 ng/ml).
Inflammasome activity is required for IL-1β secretion, and subsequent expression of the osteoclastogenic transcription factor NFATc1/A
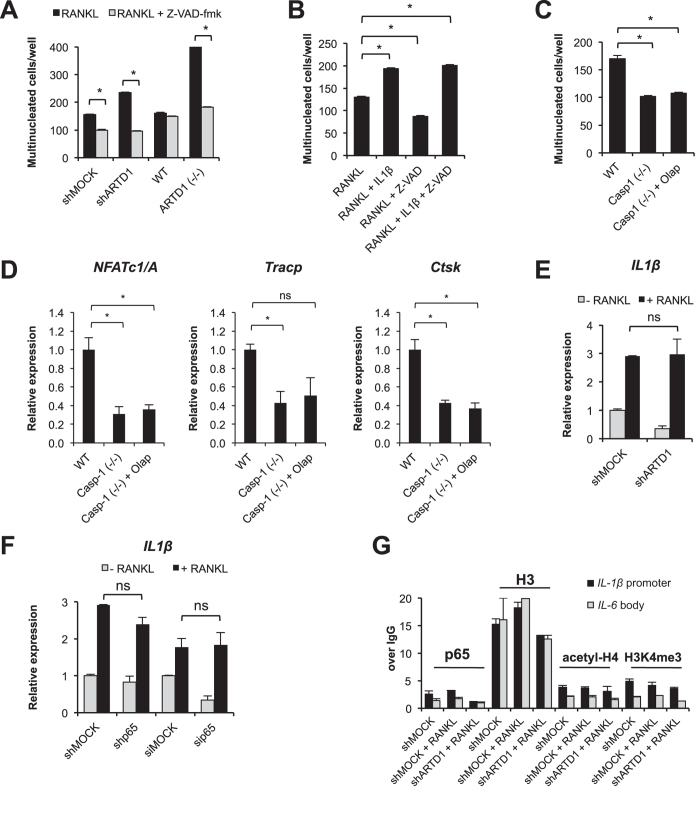
Caspase-1 is a subunit of the inflammasome complex, which cleaves pro-IL-1β, thereby causing the release of the active cytokine. We thus tested whether the pan-caspase inhibitor Z-VAD-fmk prevents the enhanced osteoclast formation observed in Artd1-silenced RAW 264.7 and Artd1-deficient BMDM (Fig. 3A). Indeed, Z-VAD-fmk added to the cell culture together with RANKL reduced the number of shARTD1-treated or Artd1-deficient osteoclasts to the level of shMOCK-treated and WT BMDM treated with caspase inhibitor, similar to the effect of IL-1β neutralizing antibody, indicating that caspase activity was required for osteoclast differentiation. IL-1β repeatedly added to the culture of differentiating cells was able to overcome the Z-VAD-fmk-induced repression of osteoclast formation (Fig. 3B), suggesting that caspase inhibition prevented processing of IL-1β. BMDM derived from Casp-1-deficient mice showed a reduced differentiation potential upon stimulation with RANKL when compared to WT BMDM, and very interestingly, olaparib failed to enhance osteoclast formation from Casp-1-deficient BMDM, further confirming that caspase-1 is indispensible for mediating the effect of ARTD1 silencing or inhibition on osteoclastogenesis (Fig. 3C). RANKL-stimulated Casp-1-deficient BMDM also expressed Nfatc1/A, Tracp and Ctsk mRNA to a significantly reduced level, which were not affected by treatment with olaparib (Fig. 3D), further supporting that the inflammasome-induced activation of caspase-1 and subsequent processing of IL-1β is required to enhance osteoclast differentiation due to ARTD1 deficiency or inhibition and stimulate the expression of Nfatc1/A.
Figure 3. Inhibition of ADP-ribosylation as well as Artd1 knock-out and knock-down are responsible for enhanced IL-1β expression during osteoclast differentiation.
(A) The contribution of the inflammasome to osteoclast differentiation was studied by comparing cell multinucleation in RANKL-induced RAW 264.7 macrophage cultures supplemented or not with pan-caspase inhibitor Z-VAD-fmk (10 μM). (B) IL-1β (2 ng/ml) was added every 8 h to RANKL-induced RAW 264.7 macrophages (shMOCK) cultured in the presence or absence of Z-VAD-fmk. (C) The multinucleation of BMDM isolated from WT or Casp-1-deficient mice differentiated in the presence of RANKL and co-treated with olaparib or not for 2 d. (D) The expression of osteoclast markers in 3-day-old osteoclasts isolated from the RANKL-induced culture of WT, Casp-1-deficient BMDM and Casp-1-deficient BMDM incubated with olaparib was compared using real-time PCR. (E) IL-1β expression was compared between shMOCK and shARTD1 RAW264.7 macrophages stimulated with RANKL for 3 h. (F) The expression of IL-1β was compared between shMOCK or siMOCK RAW 264.7 macrophages and cells treated with shRNA or siRNA against p65 after 3 h cell stimulation with RANKL. (G) The association of p65 with the promoter of IL-1β as well as H3 occupancy, H3K4 trimethylation and H4 acetylation was determined by ChIP in RAW 264.7 macrophages (shMOCK, shARTD1) incubated with RANKL for 1 h.
ARTD1 does not control expression of IL-1β upon initial RANKL signalling
To further elucidate the mechanism underlying the ARTD1 involvement in the regulation of IL-1β transcription during the differentiation of osteoclasts, we analysed the expression of IL-1β in RAW 264.7 macrophages after initial (3 h) RANKL stimulation (Fig. 3E). Although the level of IL-1β mRNA was increased 3 h after RANKL treatment, Artd1 silencing had no effect on the expression of IL-1β in these cells. As the canonical NF-κB pathway is known to promote IL-1β production, we tested whether IL-1β expression is regulated by p65 during the initial RANKL-stimulation of RAW 264.7 macrophages. While RANKL stimulation for 3 h triggered IL-1β transcription, the level of IL-1β mRNA remained the same in shp65/sip65-silenced cells compared to the corresponding shMOCK/siMOCK-treated cells, suggesting that the initial RANKL-induced expression of IL-1β in macrophages is independent of the canonical NF-κB pathway (Fig. 3F). ChIP experiments confirmed a lack of p65 recruitment to the IL-1β promoter and a lack of chromatin remodelling within this sequence after short-term RANKL treatment (Fig. 3G). Furthermore, silencing of Artd1 did not influence p65 binding, which agrees with our previous finding that ARTD1 does not regulate IL-1β transcription induced by short-term RANKL treatment (Fig. 3E).
ARTD1 limits expression of IL-1β during sustained osteoclastogenesis by reducing NF-κB recruitment to the IL-1β promoter and maintaining a transcriptionally silent chromatin state around the p65-binding site
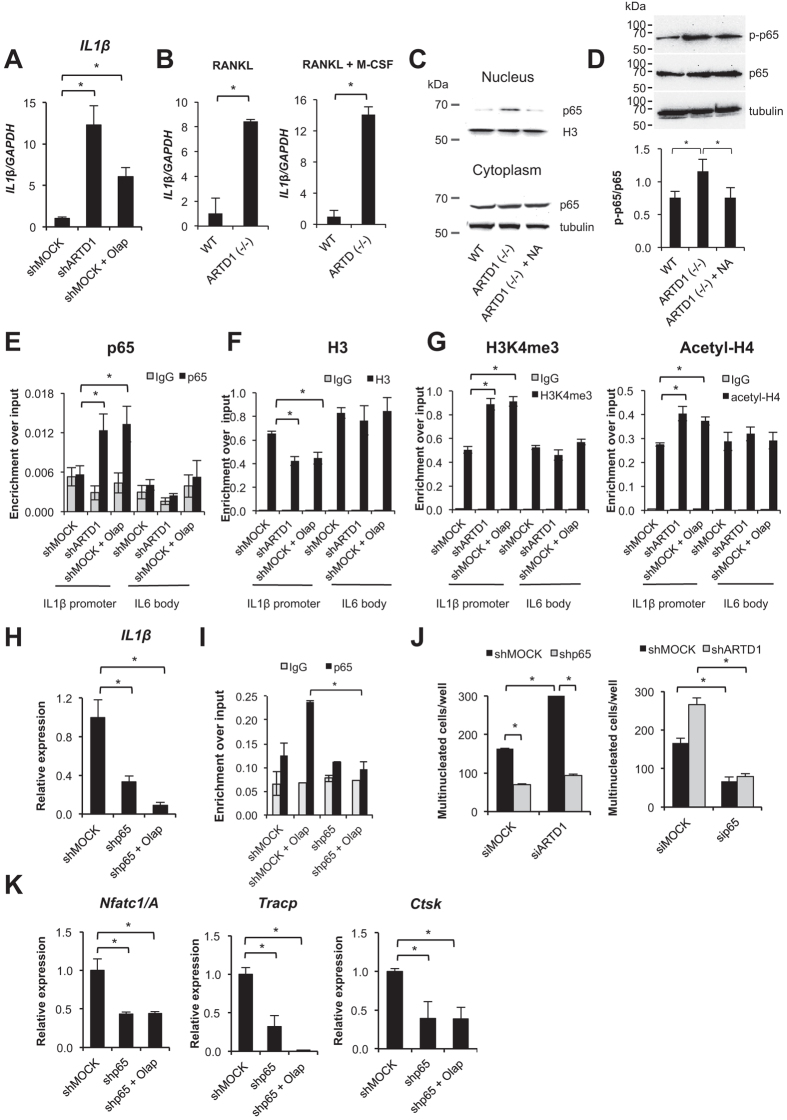
In contrast to the above described short-term experiments (i.e., the initial RANKL-induced phase), ARTD1 silencing or inhibition significantly augmented IL-1β expression after 3 d of RANKL-induced osteoclastogenesis (Fig. 4A), suggesting that RANKL-induced osteoclast differentiation paves the way for ARTD1 to control the regulation of IL-1β expression. We also analysed the expression of IL-1β in differentiating osteoclast precursors isolated from Artd1 (−/−) mice, confirming that IL-1β expression is indeed significantly upregulated in the absence of ARTD1 (Fig. 4B, left panel). This effect was independent of whether M-CSF was present during the RANKL-induced differentiation or not (Fig. 4B, right panel). To see whether the canonical NF-κB pathway is activated in 3 d differentiated osteoclasts that lack ARTD1, we analysed nuclear translocation of p65 by Western blot analysis. Indeed, p65 was increased in the nuclear extract of day 3 differentiated pre-osteoclasts derived from Artd1 (−/−) mice compared to their WT counterparts (Fig. 4C). Furthermore, p65 phosphorylation at Ser536 was also enhanced in day 3 differentiated pre-osteoclasts derived from Artd1 (−/−) mice compared to their WT counterparts (Fig. 4D). Addition of neutralizing IL-1β antibody reversed both the enhanced nuclear translocation of RelA and its phosphorylation at Ser536 in the cells from Artd1 (−/−) mice (Fig. 4C,D). These results provide strong evidence that the observed enhanced RelA activation is driven by an autocrine (i.e. local) IL-1β loop.
Figure 4. ARTD1 silencing or inhibition of ADP-ribosylation facilitates p65 recruitment to the promoter of IL-1β and chromatin remodelling at this site.
(A) IL-1β expression was compared between 3-day-old osteoclasts derived from shMOCK and shARTD1 RAW264.7 macrophages. (B) The expression of IL-1β was quantified in 3-day-old osteoclasts derived from WT and Artd1 (−/−) BMDM by real-time PCR. (C,D) The activation of NF-κB was analysed by measuring nuclear translocation (C) and Ser536 phosphorylation (D) of p65/RelA in 3-day-old osteoclasts derived from BMDM isolated from WT and Artd1 (−/−) by western blotting. IL-1β neutralizing antibody (NA) was added to the medium of differentiating Artd1 (−/−) every 8 h. (E–G) The association of p65 with the promoter of IL-1β as well as H3 occupancy, H3K4 trimethylation and H4 acetylation was determined by ChIP in RAW 264.7 macrophages-derived osteoclasts differentiated in the presence of RANKL for 72 h. As a negative control, the body of IL6 was used. (H) The expression of IL-1β was compared between 3-day-old osteoclasts derived from shMOCK and shp65 RAW 264.7 macrophages treated or not with olaparib. (I) Recruitment of p65 to the IL-1β promoter was assessed in 3-day-old osteoclasts derived from shMOCK and shp65 RAW 264.7 macrophages treated or not with olaparib. (J) The multinucleation of double-silenced RAW 264.7 macrophages (left panel: siARTD1/shp65, right panel: sip65/shARTD1) was quantified 48 h after induction of the differentiation with RANKL (72 h after cell transfection with siRNA). (K) The expression of osteoclast markers in 3-day-old shMOCK and shp65 ( ± olaparib) osteoclasts was quantified using real-time PCR.
Furthermore, ChIP experiments with ARTD1-silenced or inhibited cells revealed facilitated p65 recruitment to the IL-1β promoter in multinucleated cells (Fig. 4E), the eviction of histone H3 (Fig. 4F), as well as trimethylation of histone H3 and acetylation of H4 (Fig. 4G), histone marks of transcriptionally active chromatin. Stable silencing of p65 resulted in decreased IL-1β expression in 3 d differentiated osteoclasts even in the presence of olaparib, indicating that during the sustained phase of differentiation, ARTD1-mediated ADP-ribosylation represses IL-1β transcription in osteoclasts in a p65-dependent manner (Fig. 4H). Furthermore, enhanced recruitment of p65 to the IL-1β promoter by olaparib treatment was inhibited in cells with stably knocked-down p65 (Fig. 4I). Interestingly, ARTD1 activity, but not the protein itself was required to control expression of IL-1β, because olaparib phenocopied the effect of ARTD1 without leading to dissociation of the enzyme from the chromatin (Suppl. Fig. 3F).
To confirm the contribution of canonical NF-κB signalling in the enhanced IL-1β-mediated osteoclast formation after 3 d by Artd1-silencing or inhibition, we compared the differentiation capacity between Artd1 and p65 single and double knock-downs (Fig. 4J, Suppl. Fig. 3G). The targeting of Artd1 in shp65 RAW 264.7 macrophages with siRNA did not intensify the differentiation process, whereas silencing of p65 dramatically reduced the osteoclastogenic potential of shARTD1 RAW 264 macrophages, suggesting that when p65 is knocked-down, ARTD1 does not repress osteoclastogenesis. Moreover, the presence of olaparib in differentiating shp65 osteoclasts had no influence on the differentiation process (Suppl. Fig. 2C). The change in osteoclast differentiation was also not observed in differentiating cells supplemented with conditioned medium from 3-d-old shp65 osteoclasts cultured in the presence or absence of olaparib (Suppl. Fig. 2D), providing further evidence that IL-1β is not released from p65-silenced osteoclasts even upon inhibition of ARTD1. Importantly, expression levels of the osteoclast markers Nfatc1/A (but not of Nfatc1/B and Nfatc1/C), Ctsk and Tracp were all dependent on p65, but remained unchanged after additional olaparib treatment of differentiating p65-silenced cells, further supporting that the presence of p65 is also required downstream of ARTD1-regulated IL-1β expression (Fig. 4K, Suppl. Fig. 3E).
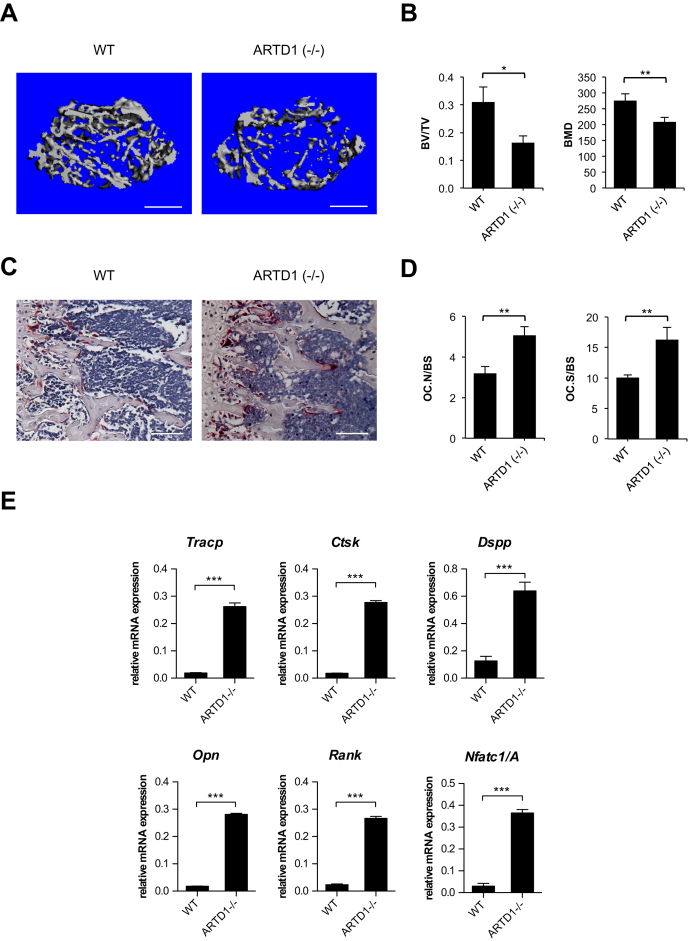
Artd1-deficient mice display a low bone mass phenotype owing to increased osteoclast differentiation
Our cell culture experiments provided strong evidence that ARTD1 represses the sustained phase of osteoclastogenesis. Mice lacking ARTD1 are thus to be expected to have altered bone structure due to enhanced osteoclast activity. To investigate whether ARTD1 indeed regulates bone homeostasis in vivo, long bones of age- and sex-matched wild-type (WT) and Artd1-deficient (−/−) mice were collected and analysed by micro-CT and histology. At 8 weeks of age, male Artd1 (−/−) mice exhibited a significant decrease in bone mass (BV/TV, Fig. 5A,B), along with decreases in bone mineral density (BMD, Fig. 5B), trabecular number (Tb.N) and a significant increase in trabecular spacing (Tb.Sp) (Suppl. Fig. 4A). Also the cortical envelope is osteopenic in Artd1 (−/−) mice compared to WT littermates (Suppl. Fig. 4B). The abnormal bone phenotype in Artd1 (−/−) mice is likely due to alterations in osteoclastogenesis, since the number of osteoclasts is significantly increased in mutant mice (Fig. 5C,D) while the number of osteoblasts was comparable between both mouse strains (Suppl. Fig. 4C). To strengthen the notion that the altered trabecular structure is the result of increased osteoclast differentiation, we analysed the expression levels of osteoclast specific gene markers in femurs (Fig. 5E). Indeed, the expression levels of Tracp, Ctsk, and Rank were all significantly upregulated in the bones of Artd1 (−/−) mice. The expression of the dentin sialophosphoprotein gene Dspp, the osteopontin gene Opn, and Nfatc1/A was also upregulated in Artd1 (−/−) mice (Fig. 1E), strongly supporting that the observed bone phenotype in Artd1 (−/−) mice is due to enhanced osteoclast formation.
Figure 5. ARTD1 deficiency results in osteopenia and increased osteoclastogenesis.
Femurs of 8-week-old Artd1-deficient (−/−) mice (n = 4) and their wild-type littermates (n = 4) were analysed by μCT. (A) Representative 3D reconstruction. Size bars = 500 μm. (B) Bone volume density (BV/TV) and bone mineral density (BMD) were quantified. (C) TRAP staining of tibial sections. Size bars = 100 μm. (D) Number of osteoclast (stained in red)/bone surface (Oc.N/BS) and osteoclast surface/bone surface (Oc.S/BS) were quantified. Values are mean ± SD. **p < 0.01. (E) Expression profiles of osteoclastogenic markers were analysed by qRT-PCR of RNA isolated from the proximal tibial epiphysis and part of the diaphysis of wt (n = 4) and Artd1 (−/−) (n = 5).
Discussion
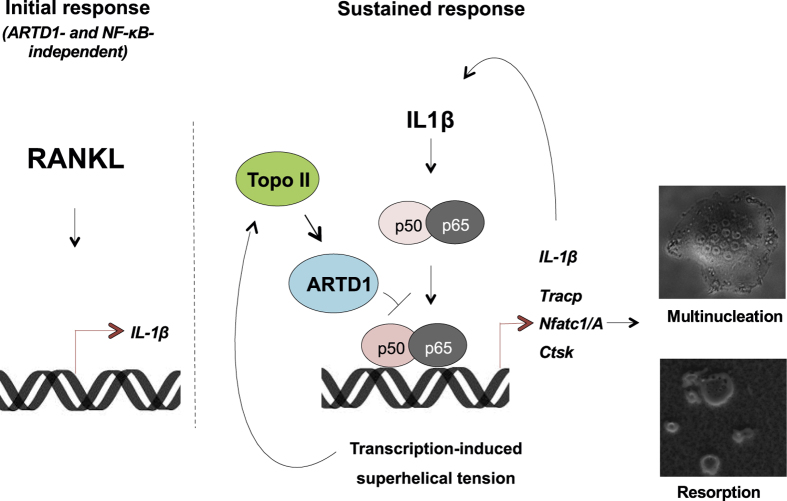
Osteoclast deregulation is associated with several diseases, either through deficient osteoclast function, resulting in osteopetrosis31, or aberrant hyperactivation, giving rise to generalized bone loss as observed in osteoporosis32 and rheumatoid arthritis33. Moreover, osteoclasts cause bone complications in cancers, including multiple myeloma34, prostate and breast cancers35. Thus, understanding the molecular mechanisms that govern osteoclast differentiation and function is of major importance in the development of more effective therapeutic strategies to combat these diseases. The differentiation of osteoclasts from peripheral blood macrophages in cell culture allows studying osteoclastogenesis in great detail36. Our results show that ARTD1 regulates NF-κB-induced IL-1β expression in osteoclasts, leading to autocrine regulation of the expression of osteoclast markers (e.g. NFATc1/A). This is achieved through ARTD1’s ability to enzymatically alter the epigenetic status of the IL-1β promoter during sustained osteoclastogenesis. Thus, ARTD1 through the regulation of IL-1β expression has the ability to affect bone homeostasis (Fig. 6).
Figure 6. Graphical representation of the functional role of ARTD1 in RANKL-induced osteoclastogenesis.
IL-1β is a multifunctional cytokine that regulates various cellular tissue functions including osteoclastogenesis37,38. In our analysis, IL-1β was initially induced in a RANKL-dependent but NF-κB-independent manner when tested after 3 h after RANKL treatment. It remains to be investigated, through which signalling cascade and transcription factor the (short term) expression of IL-1β is initiated. Besides canonical NF-κB, RANKL binding to its surface receptor also activates non-canonical NF-κB, AP1 and NFATc139. All these factors possess the consensus sequence for the interaction with the IL-1β promoter, and may therefore be responsible for the observed expression of this cytokine. Moreover, the lack of promoter responsiveness to RANKL stimulation may be a result of the type of osteoclast precursors employed in this study. Bone marrow-derived macrophages require alternative cell polarization with M-CSF, which directs them toward anti-inflammatory responses, while RAW 234.7 macrophages express surface markers of type 2 macrophages (M2)39. Therefore, the differentiation process associated with extensive chromatin dynamics may prime IL-1β regulatory elements to effective IL-1β transcription, as well as place the ADP-ribosylation process in the centre of IL-1β expression regulatory events. In contrast, RANKL treatment for 3 days revealed that ARTD1 fully regulates IL-1β expression at this later time point in a NF-κB-independent manner, suggesting that RANKL induces IL-1β, which subsequently, in an autocrine loop (local manner), sustains its expression in an NF-κB- and ARTD1-dependent manner. IL-1β was able to enhance RANKL-induced multinucleation, but its action required at least 12 h of incubation. Interestingly, the contribution of IL-1β to osteoclast formation was not limited to enhancement of multinucleation, since after a certain stage of osteoclast commitment (between 6 and 12 h after RANKL stimulation) and subsequent removal of RANKL, IL-1β drove osteoclast differentiation even in the absence of RANKL.
Under physiological conditions, osteoclastogenesis is thought to be dependent on M-CSF and RANKL. However, IL-1β has been reported to induce osteoclast formation directly from M-CSF-induced osteoclast precursors overexpressing c-Fos or Nfatc140, thus supporting the important function of IL-1β during osteoclastogenesis. Similarly, overexpression of type 1 IL-1R in BMDM is sufficient to drive osteoclastogenesis when RANKL is neutralized with a RANK–Fc fusion protein, suggesting that IL-1β can also act independently of RANKL30,41. Indeed, our studies indicate that IL-1β is important for sustaining osteoclastogenesis in an autocrine manner by inducing the sustained expression levels of important osteoclast markers such as NFATc1/A. The ARTD inhibitor olaparib did not completely phenocopy the effect of ARTD1 silencing on Ctsk and Tracp, as it did in the case of Nfatc1/A, but rather corresponded to IL1β expression, which is more strongly increased in shARTD1 than in shMOCK + Olap. Moreover, expression of these genes in differentiating osteoclasts is controlled by the RANKL–IL1β–p65 axis, as shown in Figs 3D and 4K. Therefore, their transcription is affected indirectly by ARTD1 and the p65–IL1β–p65 autocrine feedback loop. The olaparib-induced increase in IL1β expression is sufficient to achieve maximum osteoclast differentiation, although concomitant Nfatc1/A transcription might not be sufficient to augment Tracp and Ctsk expression.
The importance of IL-1β is further underlined by the observation that IL-1β-deficient mononuclear cells have a decreased RANKL responsiveness, although the response to TNF-α is maintained42,43. Molecular characterization of osteoclast precursors revealed a decreased expression of RANK as the underlying cause for a blunted response of IL-1β-deficient cells to RANKL, whereas other surface and functional markers such as c-fms, cathepsin K, TRAP, and MMP9 were similarly expressed.
Since olaparib also inhibits other ARTD family members, we cannot per se exclude that other ARTDs also contribute to osteoclastogensis. However, co-treatment of shARTD1-treated or Artd1-deficient cells with RANKL and olaparib did not cause a further increase in the number of osteoclasts developed, strongly suggesting that the inhibitory effect is primarily due to inhibition of the enzymatic activity of ARTD1. Our experiments using the topo II inhibitor merbarone suggest that topo II is responsible for the activation of ARTD1 during IL-1β-induced/sustained osteoclastogenesis, leading to the repression of fully functional osteoclast formation. The differentiation process is accompanied by extensive transcription of genes contributing to conditioning of osteoclast function and phenotype. The transcription machinery requires topo II activity, which may activate ARTD1, placing topo II upstream of ARTD1. A comparable mode of action has already been described for adipogenesis27, although the actual molecular mechanism remains to be elucidated. However, it is most likely that the enzymatic activity of topo II and the subsequent structural changes of the chromatin contribute to this process. In our in vitro model of osteoclast differentiation, topo II serves as a major component of the transcription–differentiation–transcription feedback loop, as transcription-induced DNA gyrase activity represses transcription of osteoclastogenic genes by activating ARTD1. Although investigating topo II recruitment to the IL-1β promoters in osteoclasts would be interesting, the lack of suitable antibodies currently prevents this type of analysis.
The impact of ARTD1 on gene expression remains mostly undefined, as its function is cell type-specific and depends e.g. on the local histone variants, interacting partner and ADP-ribosylation acceptor protein, and may therefore differ after differentiation. Moreover, ARTD1 has recently been shown to function in various aspects of the transcription process through a variety of mechanisms, including roles as a modulator of chromatin and a regulator of DNA methylation4,5. PARylation of KDM5B, a histone lysine demethylase that acts on H3 lysine 4 trimethyl (H3K4me3), has been shown to block the binding of KDM5B to chromatin and inhibit its demethylase activity44. Our results are thus in line with recent data showing that inhibition of the enzymatic activity of ARTD1 regulates the activity of KDM544. It remains to be elucidated whether ARTD1 auto-modification is sufficient to promote these changes, or whether other proteins (histone modifiers or histone themselves) are also modified.
Of note, we cannot preclude that ARTD1 itself acts as a cofactor in the transcription machinery assembled with other transcription factors activated upon RANK stimulation, such as NFATc1 or AP1 independent of its enzymatic activity; however, this requires further analysis. Moreover, our results provide evidence that the inflammasome NOD-like receptor (NLR) family, pyrin domain-containing 3 (NLRP3) is activated upon RANKL stimulation to process pre-IL-1β, which is followed by subsequent secretion of IL-1β, although an inflammasome-independent source of IL-1β has already been described to also provoke inflammation and osteolytic bone disease45. Based on our results, inflammasome-dependent IL-1β would signal to the osteoclast in an autocrine manner, which is consistent with previous findings that osteoclasts express IL-1β receptor46. Restriction of an activating D301N mutation in Nlrp3 in myeloid cells of mice, induced systemic inflammation and severe osteopenia similar to mice globally expressing the knock-in mutation47,48. Mice expressing D301N only in osteoclasts show the same phenotype, due to increased osteolysis, without changing the number of osteoclasts or inducing systemic inflammation. Aside from its role in IL-1β maturation, Nlrp3D301N expression enhances osteoclast bone resorbing ability through reorganization of the actin cytoskeleton, while promoting the degradation of ARTD1 at the same time. Thus, NLRP3 inflammasome activation is not restricted to the production of pro-inflammatory mediators only, but also leads to intracellular autonomous responses.
Interestingly, analysis of the bones from Artd1 (−/−) mice showed a severely decreased BV/TV as a result of a decreased bone mineral density (BMD), number of trabeculae (Tb.N) and increased trabecular space (Tb.Sp). The cortical envelope in Artd1 (−/−) mice was also osteopenic compared to wild-type littermates. Histological analysis of the bone samples by histology revealed the low bone mass phenotype is likely caused by deregulated osteoclastogenesis, since the number of osteoclasts is increased in mutant mice, while that of osteoblasts is unaffected, strongly supporting that the observed bone phenotype in Artd1 (−/−) mice is due to enhanced osteoclast formation and activity.
Taken together, our results provide important novel insights into the mechanisms underlying the fine-tuning of the transcriptional processes involved in macrophage-to-osteoclast differentiation. Most importantly, ARTD1 and its enzymatic activity repress the expression of osteoclastogenesis-driving genes, including several osteoclast-specific genes through controlling the expression of IL-1β. Secondly, our results reinforce the essential function of ARTD1’s enzymatic activity for the role of NF-κB as a direct regulator of osteoclast formation. ARTD1 and its enzymatic activity thus keep the detrimental expression of IL-1β in osteoclasts in check by altering the chromatin structure and subsequently the accessibility of transcription factors such as NF-κB.
Materials and Methods
Antibodies and reagents
p65/RelA antibody (sc-372), α-tubulin antibody (H-300, sc-5546), and IgG (sc-2027) was from Santa Cruz Biotechnology, PARP1 antibody, histone H3 antibody (D1H2, #4499), and Cell Fractionation Kit (#9038) from Cell Signaling Technology, growth media, FBS, antibiotics, and Lipofectamine RNAiMAX from Life Technologies, IL-1β, IL6, TNFα and Z-VAD-fmk from Sigma Aldrich, H3K4me3 and acetyl-H4 antibody from Millipore, H3 antibody from Abcam, RANKL from Peprotech, siRNA from Qiagen. Corning Osteo Assay plates were from Corning. IL-1β neutralizing antibody was from R&D.
Animals
Male Artd1-deficient C57BL/6J mice and aged-matched wild-type littermates were sacrificed at 6 and 8 weeks of age for retrieving bones for BMDM isolation and bone structure analysis, respectively. The Artd1-deficient mice were originally kindly provided by Zhao-Qi Wang49, and all mice were bred at the animal facility of the University of Zurich. We have focused on male C57BL/6J mice, because they exhibit higher bone mass (BV/TV) at baseline than female mice. All animal experiments were carried out in accordance with the Swiss and EU ethical guidelines and have been approved by the local animal experimentation committee of the Canton of Zurich under license #2012207 and following the 3R guidelines.
Isolation of bone marrow-derived macrophages (BMDM)
Bone marrow was isolated from femurs and tibiae of 6-week-old male mice and after lysis of erythrocytes, cells were cultivated for one week in RPMI supplemented with 10% heat inactivated FBS, antibiotics (50 U/ml penicillin, 50 μg/ml streptomycin) and 20% M-CSF-containing medium conditioned by L929 fibroblasts. Over the following two weeks conditioned medium free RPMI was gradually replaced by DMEM.
Cell culture condition
Both RAW 264.7 and BMDM were cultured in DMEM supplemented with 10% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1% L-glutamine (growth medium) at 37°C in a humidified atmosphere of 5% CO2.
Osteoclast differentiation
The differentiation of BMDM and RAW 264.7 macrophages was initiated by the addition of 50 ng/ml RANKL one day after cell seeding. For osteoclast quantification and determination of TRAP activity at the earliest possible time point, cells were plated at the density of 8,100 per cm2 and analysed 48 h after supplementation of the cell culture with RANKL (D2). For all other experiments, which required more osteoclasts, cells were seeded at the density of 32,400 per cm2 and analysed 72 h after RANKL administration (D3). The undifferentiated cells were removed from strongly attached osteoclasts by successive washing with DMEM (1x) and PBS (3x).
Osteoclast formation and determination of their activity
Two-day-old cultures of differentiating cells were stained for TRAP activity using Acid Phosphatase, Leukocyte (TRAP) Kit (Sigma Aldrich) according to the manufacturer’s instructions. TRAP-positive, multinucleated cells (carrying 3 ≥ nuclei) were counted as osteoclasts and data shown represent the number of multinucleated cells derived from initially 2,500 bone marrow or RAW 264.7-derived macrophages stimulated with RANKL.
For quantification of TRAP activity, stained cells were dissolved in DMSO, solution was acidified with 3% HCl, absorbance was measured at 540 nm and normalized to the protein content, which was quantified with Lowry assay. Osteoclast resorbing activity was evaluated in the culture of cells differentiated for 3 days by counting matrix dissolution spots on the Corning® Osteo Assay plates (Corning Life Sciences). Undifferentiated cells were removed by washing and remaining osteoclasts were trypsinised for 15 min and washed off. After drying, images of plate surface were acquired on an Olympus inverted microscope.
Generation of stable ARTD1 and p65 knock-downs and siRNA transfection
shRNA constructs specific for ARTD1 and for p65/RelA (see Suppl. Table 1) in pSUPER retroviral vector were packed in HEK-293cell line into invasive viruses. RAW 264.7 macrophages were infected with generated retroviruses for 8 h. After another 48 h, puromycin (5 μg/ml) was added to select the transduced cells. For transient silencing Artd1 and p65 were targeted with siRNA using Lipofectamine RNAiMAX as a carrier. Efficiency of the silencing was monitored by Western blot analysis and real-time PCR. In order to analyse the impact of ARTD1 and p65 in osteoclast development, RANKL was added to cells 24 h after their transfection with siRNA and multinucleation was quantified on day 2 (D2).
Gene expression
RNA was extracted with TRI Reagent, reversed transcribed (High Capacity cDNA Reverse Transcription Kit, Life Technologies) and cDNA was quantified by real-time PCR (Rotor-Gene 3000, Qiagen) using primers listed in Suppl. Table 1 and SYBR Green (Kapa Biosystems). Gapdh was used as a housekeeping control gene.
Chromatin immunoprecipitation
Recruitment of p65 to promoter sequences, H3 and ARTD1 occupation as well as H3K4 trimethylation and H4 acetylation was studied with ChIP according to50. In brief, cells were cross-linked with 1% formaldehyde solution, isolated chromatin was sheared with the Bioruptor (Diagenode) and after incubation with antibody-conjugated protein G magnetic Dynabeads (Life Technologies), chromatin was isolated with phenol:chlorophorm:isoamyl alcohol. Extracted DNA was further analysed with real-time PCR. The list of primers used for IL-1β promoter and IL6 body are listed in Suppl. Table 1.
Isolation of RNA from mouse bone
Total RNA was isolated from a region encompassing the proximal tibial epiphysis and part of the diaphysis of WT (n = 4) and Artd1−/− (n = 5) mice using TRIzol reagent, and treated with TURBO DNase (Life Technologies) as previously described51.
Bone mass and microstructure
Femurs from 8-week-old male mice were stabilized in 2% agarose gel, and analysed by micro-computed tomography (μCT) system (μCT 40; Scanco Medical AG, Zurich, Switzerland) as described previously48. The volume of interest analysed was located just proximal to the growth plate, spanning a height of 350 μm each for the metaphyseal region.
Histology and histomorphometry
Tibiae were fixed in 10% formalin, decalcified in 14% (w/v) EDTA pH 7.2 for 10–14 days at room temperature, embedded in paraffin, sectioned at 5 μm thickness and mounted on glass slides. Sections stained with haematoxylin and eosin or for TRAP were used for the analysis of osteoblasts and osteoclasts, respectively, as described previously48. Photographs were taken and cell number counted using nanozoomer (Hamamatsu).
Statistical analysis
Bars in the figures represent mean ± standard error of the mean (SEM). Student’s t-test was used to compare the difference between two means. *P < 0.05, **P < 0.01, ***P < 0.001. Results of the in vitro model of differentiation represent data from three independent biological replicates (with three technical replicates each).
Additional Information
How to cite this article: Robaszkiewicz, A. et al. ARTD1 regulates osteoclastogenesis and bone homeostasis by dampening NF-κB-dependent transcription of IL-1β. Sci. Rep. 6, 21131; doi: 10.1038/srep21131 (2016).
Supplementary Material
Acknowledgments
We thank Alan Valaperti for help with bone isolation. We thank Stephan Christen and the other members of the Department of Molecular Mechanisms of Disease (University of Zurich, Switzerland) for help and discussions during the preparation of this manuscript. This work was financed by the Science Exchange Program [12.193 to A.R.], the Kanton of Zurich, Swiss National Science Foundation [310030B_138667 and 310030_157019 to M.O.H.] and by the Mäxi Foundation (for consumables). The research in the laboratory of A.R. is funded by Polish National Science Centre grant UMO-2013/11/D/NZ2/00033, and that in the laboratory of G.M. by NIH/NIAMS R01-AR064755.
Footnotes
Author Contributions A.R. planned and performed the experiments, evaluated the data and wrote the manuscript. P.J.R. and A.M. isolated RNA from bones. F.K. analysed gene expression in bone. E.W. and T.P. analysed gene expression in BMDM. A.R., P.C., C.Q. and G.M performed the μCT experiments, and C.Q. and G.M additionally performed histological analyses of the bones. MOH supervised experiments and wrote the manuscript. All authors have read and approved the manuscript.
References
- Hottiger M. O. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem. 84, 227–263 (2015). [DOI] [PubMed] [Google Scholar]
- D’Amours D., Desnoyers S., D’Silva I. & Poirier G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 (1999). [PMC free article] [PubMed] [Google Scholar]
- Ji Y. & Tulin A. V. The roles of PARP1 in gene control and cell differentiation. Curr. Opin. Genet. Dev . 20, 512–518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W. L. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R. & Kraus W. L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzell K. M. et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J. Biol. Chem. 284, 33926–33938 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X. & Kraus W. L. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 26, 417–432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery E., Dignam J. D., Matsui T. & Roeder R. G. Purification and analysis of a factor which suppresses nick-induced transcription by RNA polymerase II and its identity with poly(ADP-ribose) polymerase. J. Biol. Chem. 258, 5955–5959 (1983). [PubMed] [Google Scholar]
- Kraus W. L. & Hottiger M. O. PARP-1 and gene regulation: progress and puzzles. Mol. Aspects Med. 34, 1109–1123 (2013). [DOI] [PubMed] [Google Scholar]
- Erener S., Hesse M., Kostadinova R. & Hottiger M. O. Poly(ADP-ribose)polymerase-1 (PARP1) controls adipogenic gene expression and adipocyte function. Mol. Endocrinol. 26, 79–86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S. et al. ARTD1 deletion causes increased hepatic lipid accumulation in mice fed a high-fat diet and impairs adipocyte function and differentiation. FASEB J. 26, 2631–2638 (2012). [DOI] [PubMed] [Google Scholar]
- Fujiki K. et al. PPARgamma-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat. Commun. 4, 2262 (2013). [DOI] [PubMed] [Google Scholar]
- Suda T. et al. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 20, 345–357 (1999). [DOI] [PubMed] [Google Scholar]
- Teitelbaum S. L. Bone resorption by osteoclasts. Science 289, 1504–1508 (2000). [DOI] [PubMed] [Google Scholar]
- Walsh M. C. et al. Osteoimmunology: interplay between the immune system and bone metabolism. Annu. Rev. Immunol. 24, 33–63 (2006). [DOI] [PubMed] [Google Scholar]
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 13, 27–38 (2012). [DOI] [PubMed] [Google Scholar]
- Boyle W. J., Simonet W. S. & Lacey D. L. Osteoclast differentiation and activation. Nature 423, 337–342 (2003). [DOI] [PubMed] [Google Scholar]
- Takayanagi H. et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3, 889–901 (2002). [DOI] [PubMed] [Google Scholar]
- Hayden M. S., West A. P. & Ghosh S. NF-kappaB and the immune response. Oncogene 25, 6758–6780 (2006). [DOI] [PubMed] [Google Scholar]
- Boyce B. F., Schwarz E. M. & Xing L. Osteoclast precursors: cytokine-stimulated immunomodulators of inflammatory bone disease. Curr. Opin. Rheumatol. 18, 427–432 (2006). [DOI] [PubMed] [Google Scholar]
- Lee S. K. & Lorenzo J. Cytokines regulating osteoclast formation and function. Curr. Opin. Rheumatol. 18, 411–418 (2006). [DOI] [PubMed] [Google Scholar]
- Robaszkiewicz A. et al. Hydrogen peroxide-induced poly(ADP-ribosyl)ation regulates osteogenic differentiation-associated cell death. Free Radic. Biol. Med. 53, 1552–1564 (2012). [DOI] [PubMed] [Google Scholar]
- Robaszkiewicz A. et al. The role of p38 signaling and poly(ADP-ribosyl)ation-induced metabolic collapse in the osteogenic differentiation-coupled cell death pathway. Free Radic. Biol. Med. 76, 69–79 (2014). [DOI] [PubMed] [Google Scholar]
- Beranger G. E., Momier D., Rochet N., Carle G. F. & Scimeca J. C. Poly(adp-ribose) polymerase-1 regulates Tracp gene promoter activity during RANKL-induced osteoclastogenesis. J. Bone Miner. Res. 23, 564–571 (2008). [DOI] [PubMed] [Google Scholar]
- Beranger G. E. et al. RANKL treatment releases the negative regulation of the poly(ADP-ribose) polymerase-1 on Tcirg1 gene expression during osteoclastogenesis. J. Bone Miner. Res. 21, 1757–1769 (2006). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Vargas J. M. et al. ROS-induced DNA damage and PARP-1 are required for optimal induction of starvation-induced autophagy. Cell Res. 22, 1181–1198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M. et al. ARTD1-induced poly-ADP-ribose formation enhances PPARgamma ligand binding and co-factor exchange. Nucleic Acids Res. 43, 129–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. M., Fujikado N., Manaka H., Yasuda H. & Iwakura Y. IL-1 plays an important role in the bone metabolism under physiological conditions. Int. Immunol. 22, 805–816 (2010). [DOI] [PubMed] [Google Scholar]
- Scianaro R. et al. Deregulation of the IL-1beta axis in chronic recurrent multifocal osteomyelitis. Pediatr Rheumatol Online J 12, 30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H. et al. The mechanism of osteoclast differentiation induced by IL-1. J. Immunol. 183, 1862–1870 (2009). [DOI] [PubMed] [Google Scholar]
- Tolar J., Teitelbaum S. L. & Orchard P. J. Osteopetrosis. N. Engl. J. Med. 351, 2839–2849 (2004). [DOI] [PubMed] [Google Scholar]
- Rachner T. D., Khosla S. & Hofbauer L. C. Osteoporosis: now and the future. Lancet 377, 1276–1287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. L., Wolfe F. & Huizinga T. W. Rheumatoid arthritis. Lancet 376, 1094–1108 (2010). [DOI] [PubMed] [Google Scholar]
- Mundy G. R., Raisz L. G., Cooper R. A., Schechter G. P. & Salmon S. E. Evidence for the secretion of an osteoclast stimulating factor in myeloma. N. Engl. J. Med. 291, 1041–1046 (1974). [DOI] [PubMed] [Google Scholar]
- Yoneda T. Cellular and molecular mechanisms of breast and prostate cancer metastasis to bone. Eur. J. Cancer 34, 240–245 (1998). [DOI] [PubMed] [Google Scholar]
- Nicholson G. C. et al. Induction of osteoclasts from CD14-positive human peripheral blood mononuclear cells by receptor activator of nuclear factor kappaB ligand (RANKL). Clin. Sci. 99, 133–140 (2000). [PubMed] [Google Scholar]
- Garlanda C., Dinarello C. A. & Mantovani A. The interleukin-1 family: back to the future. Immunity 39, 1003–1018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E., Stashenko P. P., Mole J. E. & Tsurumachi T. Purification and partial sequence of human osteoclast-activating factor: identity with interleukin 1 beta. J. Immunol. 135, 2562–2568 (1985). [PubMed] [Google Scholar]
- Souza P. P. & Lerner U. H. The role of cytokines in inflammatory bone loss. Immunol. Invest. 42, 555–622 (2013). [DOI] [PubMed] [Google Scholar]
- Yao Z., Xing L., Qin C., Schwarz E. M. & Boyce B. F. Osteoclast precursor interaction with bone matrix induces osteoclast formation directly by an interleukin-1-mediated autocrine mechanism. J. Biol. Chem. 283, 9917–9924 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Kitaura H., Zhou P., Ross F. P. & Teitelbaum S. L. IL-1 mediates TNF-induced osteoclastogenesis. J. Clin. Invest. 115, 282–290 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzer K. et al. Interleukin-1 is essential for systemic inflammatory bone loss. Ann. Rheum. Dis. 69, 284–290 (2010). [DOI] [PubMed] [Google Scholar]
- Zwerina J. et al. TNF-induced structural joint damage is mediated by IL-1. Proc. Natl. Acad. Sci. USA 104, 11742–11747 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R. & Kraus W. L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens J. R. et al. Critical role for inflammasome-independent IL-1beta production in osteomyelitis. Proc. Natl. Acad. Sci. USA 111, 1066–1071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimi E. et al. Activation of NF-kappaB is involved in the survival of osteoclasts promoted by interleukin-1. J. Biol. Chem. 273, 8799–8805 (1998). [DOI] [PubMed] [Google Scholar]
- Bonar S. L. et al. Constitutively activated NLRP3 inflammasome causes inflammation and abnormal skeletal development in mice. PLoS ONE 7, e35979 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu C. et al. NLRP3 mediates osteolysis through inflammation-dependent and -independent mechanisms. FASEB J. 29, 1269–1279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Q. et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9, 509–520 (1995). [DOI] [PubMed] [Google Scholar]
- Saccani S., Pantano S. & Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat. Immunol. 3, 69–75 (2002). [DOI] [PubMed] [Google Scholar]
- Mirsaidi A. et al. Therapeutic potential of adipose-derived stromal cells in age-related osteoporosis. Biomaterials 35, 7326–7335 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.