Abstract
Sirt1 is the most prominent and extensively studied member of sirtuins, the family of mammalian class III histone deacetylases heavily implicated in health span and longevity. Although primarily a nuclear protein, Sirt1’s deacetylation of Peroxisome proliferator-activated receptor Gamma Coactivator-1α (PGC-1α) has been extensively implicated in metabolic control and mitochondrial biogenesis, which was proposed to partially underlie Sirt1’s role in caloric restriction and impacts on longevity. The notion of Sirt1’s regulation of PGC-1α activity and its role in mitochondrial biogenesis has, however, been controversial. Interestingly, Sirt1 also appears to be important for the turnover of defective mitochondria by mitophagy. I discuss here evidences for Sirt1’s regulation of mitochondrial biogenesis and turnover, in relation to PGC-1α deacetylation and various aspects of cellular physiology and disease.
Keywords: mitochondria, mitochondrial biogenesis, mitophagy, PGC-1α, Sirt1
INTRODUCTION
Protein deacetylases counter the action of protein acetyl transferases (Verdin and Ott, 2015) by removing acetyl groups added to lysine residues by the latter. This large family of enzymes is often referred to as histone deacetylases (HDACs), but histones are certainly not their only substrates. In mammals, HDACs are broadly divided into four classes (I–IV) (Yang and Seto, 2008). Class I HDACs (HDACs 1, 2, 3 and 8) are homologous with the yeast Reduced Potassium Deficiency 3 (RPD3), are ubiquitously expressed, and are primarily localized to the nucleus. Class II HDACs (namely HDACs 4, 5, 6, 7, 9 and 10), which are homologues of yeast Histone DeAcetylase 1 (HDA1), shuttle between the nucleus and the cytoplasm, and are more tissue specific in terms of expression. HDAC11 has only fairly low homology to both RPD3 and HDA1, and has been proposed to constitute a separate class IV (Gregoretti et al., 2004) so as to be distinguishable from the class III HDACs, or sirtuins.
Sirtuins are a family of proteins with homologies to the Silence Information Regulator 2 (SIR2) gene of the yeast S. cerevisiae (Brachmann et al., 1995), and are not structurally homologous to the other HDACs. Lysine deacetylation by sirtuins is coupled to the cleavage of a key intermediate of cellular energy metabolism, nicotinamide adenine dinucleotide (NAD+), into nicotinamide and 1′-O-acetyl-ADP-ribose (Tanner et al., 2000) or 2′- and 3′-O-acetyl-ADP-ribose (Jackson and Denu, 2002). The activities of sirtuins are thus obligatorily dependent on cellular NAD+, inhibited by the reaction product nicotinamide, and effectively influenced by cellular metabolic and redox states. Homologues of yeast SIR2 exist in lower model organisms, such as the nematode C. elegans sir-2.1 and the fruit fly D. melanogaster Sir2. The mammalian genome has seven sirtuin paralogues (Sirt1-7), with Sirt1 having the highest homology to yeast Sir2 (Haigis and Guarente, 2006; Haigis and Sinclair, 2010). In yeast, Sir2p regulates chromatin silencing, and reduces the accumulation of rDNA circles during yeast replicative aging (Sinclair and Guarente, 1997). SIR2 and its metazoan orthologues have been prominently associated with lifespan extension (Howitz et al., 2003; Tissenbaum and Guarente, 2001), in particular that induced by caloric or dietary restriction (CR/DR) (Rogina and Helfand, 2004; Wood et al., 2004). A large number of studies have connected Sirt1 expression and/or activation by compounds such as resveratrol, with increase health span and lifespan (Guarente, 2011). However, the notion that Sirt1 is a mediator of CR effects and a longevity factor in mammals and humans has been controversial in two major ways. There were doubts about the specificity and effectiveness of natural and synthetic Sirt1 activators such as resveratrol and SRT1720 (Kaeberlein et al., 2005; Pacholec et al., 2010). Sirt1, at least in certain contexts (Longo, 2009; Tang, 2006), may also promote aging. Sirt1’s role in longevity has been more extensively investigated in yeast and lower invertebrate models, and although the notion of Sirt1 as a longevity gene has a great deal of support, there were also controversies and evidence to the contrary (Burnett et al., 2011). The demonstration of Sirt1’s role in lifespan extension in mammalian models proved difficult. Sirt1’s knockout phenotype was difficult to interpret, as the mice often died early due largely to developmental defects (Cheng et al., 2003; McBurney et al., 2003). Earlier work on transgenic over-expression of Sirt1 in mice did not result in a significant increase in lifespan (Herranz et al., 2010). More recent findings have, however, provided some reconciliation of the contrasting results and opinions. The mode of action of sirtuin-activating compounds (STACs), possibly via an allosteric mechanism, has been better clarified (Hubbard et al., 2013). Additionally, both the transgenic expression of Sirt1 (Satoh et al., 2013) and the specific activator SRT1720 (Mitchell et al., 2014) have eventually been shown to increase lifespan in mice.
Indeed, dissection of Sirt1’s role in CR and aging is difficult because Sirt1 has multiple physiological functions as well as wide-ranging roles in pathological settings. This is due to its astonishingly large repertoire of targets, many of which are key transcription factors associated with important aspects of cell survival and metabolism. Prominent amongst these are the tumor suppressor TP53 (Vaziri et al., 2001), the Forkhead bOX class O (FoxO) family members (Brunet et al., 2004), and nuclear factor κB (NF-κB) (Yeung et al., 2004), all key regulators of cell death and survival. Sirt1 represses a key metabolic regulator, Peroxisome Proliferator-Activated Receptor gamma (PPARγ) (Picard et al., 2004; Sugden et al., 2010), via deacetylation of PPARγ coactivator-1α (PGC-1α) (Gerhart-Hines et al., 2007; Lagouge et al., 2006; Nemoto et al., 2005; Rodgers et al., 2005), which regulates energy metabolism in skeletal muscle, adipose tissues and the liver. Sirt1 has also been implicated in cellular metabolism through its activation of AMP-dependent kinase (AMPK) via liver kinase B1 (LKB1) (Hou et al., 2008; Lan et al., 2008; Zu et al., 2010), as well as suppression of the mammalian target of rapamycin (mTOR) pathway (Ghosh et al., 2010; Hong et al., 2014; Liu et al., 2010).
Of the seven mammalian sirtuins, three (Sirt3, Sirt4 and Sirt5) have a primary mitochondrial localization (Michishita et al., 2005). Sirt1, on the other hand, is primarily nuclear, but its activities have large bearings on mitochondrial biogenesis and turnover. In the ensuing paragraphs, we shall discuss how Sirt1 is functionally associated with the mitochondria. We first look at some evidence for the predominantly nuclear Sirt1 being physically localized at the mitochondria.
SIRT1’S MITOCHONDRIAL LOCALIZATION
Sirt1 is primarily found in the nucleus of most cell types (Michishita et al., 2005), and it is not just a bona fide histone deacetylase (Vaquero et al., 2006) but also deacetylates histone modifying enzymes such as the histone methyl-transferase SUV39H1 (Vaquero et al., 2007). Both Sirt1 and SUV39H1 are required for the epigenetic control of the rDNA locus for rRNA synthesis (Murayama et al., 2008), and Sirt1 may also be nucleolar in localization. Befitting its nuclear localization, a majority of Sirt1’s known function is associated with deacetylation of transcription factors, as mentioned in the section above. As per all nuclear proteins, it is synthesized in the cytoplasm and imported into the nucleus. Sirt1, however, appears to possess both nuclear localization signals and nuclear export signals, and could shuttle between the cytoplasm and the nucleus (Tanno et al., 2007). Sirt1’s cytoplasmic appearance has been associated with enhanced apoptotic cell death (Jin et al., 2007), and also shown to be due to an increased protein stability via oncogenic signaling through the insulin-like growth factor-1-phosphoinositide 3-kinase (IGF-1/PI3 kinase) axis in cancer cells (Byles et al., 2010). On the other hand, Sirt1 does have cytosolic substrates whose deacetylation by Sirt1 results in distinct physiological or functional consequences. For instance, the cytoplasmic acetyl-CoA synthetase (Hallows et al., 2006) and the actin binding protein cortactin (Zhang et al., 2009) are both deacetylated by Sirt1, and these influence cytosolic fatty acid synthesis and cytoskeletal changes during cell migration, respectively.
Sirt1 and its substrate, PGC-1α, regulate aspects of energy metabolism through the mitochondria, but this was initially thought to occur through their influence on nuclear localized transcription. A rather interesting aspect of Sirt1’s activity and function that is less well defined is its extranuclear localization, particularly at the mitochondria (Aquilano et al., 2012). This was first shown by Aquilano and colleagues using both confocal imaging and subcellular fractionation analysis (Aquilano et al., 2010). Both Sirt1 and the nuclear transcription factor PGC-1α could be found in the mitochondria of human cell lines and platelets, as well as in various mouse organs. Within the mitochondria, both deacetylase and its substrate are associated with the mitochondrial DNA (mtDNA) nucleoids (Bogenhagen, 2012), as well as with the mitochondrial transcription factor A (TFAM), a key mitochondrial gene transcription factor and regulator of mtDNA copy number (Campbell et al., 2012). These findings are fascinating, as they suggest that Sirt1 and PGC-1α may also directly affect mitochondrial transcription. However, the degree of mitochondrial Sirt1 and PGC-1α’s functionality in this regard is still uncertain. In the sections that follow, we shall outline findings that point to how Sirt1 mediates aspects of mitochondrial biogenesis and turnover, as well as the controversies associated with these findings.
SIRT1’S DEACETYLATION OF PGC - 1α - METABOLIC REGULATION AND MITOCHONDRIAL BIOGENESIS
Mitochondria are dynamic organelles, and inheritance occurs by partitioning existing mitochondria between daughter cells in dividing cells (Mishra and Chan, 2014). However, within all cells, either dividing or terminally differentiated, the mitochondria population is constantly renewed, and the steady state number of this organelle represents an equilibrium between mitochondrial biogenesis and eventual degradation through the process of mitophagy (Stotland and Gottlieb, 2015; Vega et al., 2015). Mitochondrial biogenesis involves the transcription of both nuclear and mtDNA-encoded genes. Mitochondrial biogenesis is orchestrated by the Peroxisome proliferator-activated receptor Gamma Coactivator-1 (PGC-1) family of transcriptional coactivators (Austin and St-Pierre, 2012). The PGC-1 family consists of three members, namely, PGC-1α, PGC-1β and the PGC related coactivator (PRC). PGC-1α, in particular, is often cited as a master regulator of this process. PGC-1α co-activates the transcription of Nuclear Respiratory Factor (NRF) 1 and 2, which, in turn, regulate the transcription of TFAM. TFAM translocates to mitochondrial matrix where it stimulates mitochondrial DNA replication and mitochondrial gene expression.
PGC-1α undergoes several modes of post-translational modification, which includes acetylation and phosphorylation. Acetylation of PGC-1α occurs at several of its lysine residues and is catalyzed by the ubiquitous histone acetyl transferase General control of amino acid synthesis 5 (GCN5) (Dominy et al., 2010). Acetylation alters PGC-1α’s localization within the nucleus, and inhibits its transcriptional activity. Conversely, deacetylation of PGC-1α was shown by several studies to be dependent on Sirt1 activity (Amat et al., 2009; Dominy et al., 2010; Gerhart-Hines et al., 2007; Gurd, 2011; Lagouge et al., 2006; Nemoto et al., 2005; Olmos et al., 2013; Rodgers et al., 2005), which increases PGC-1α’s transcriptional activity. PGC-1α is phosphorylated by both Mitogen-activated protein kinase p38 (Barger et al., 2001), and the AMP-dependent kinase (AMPK) in skeletal muscle (Leick et al., 2010), which also stabilizes the protein and increases its activity. On the other hand, phosphorylation of PGC-1α by AKT/protein kinase B (PKB), downstream of insulin signaling in the liver, is known to decrease its stability and transcriptional activity (Li et al., 2007).
Given the importance of its acetylation status to PGC-1α activity, the connection between Sirt1 and PGC-1α has been a focus of investigations into metabolic regulation and mitochondrial biogenesis. Finkel’s group has provided clear biochemical evidence that Sirt1 physically and functionally interacts with PGC-1α (Nemoto et al., 2005), and this interaction could be distinguished by a single amino acid mutation in the putative ADP-ribosyltransferase domain of Sirt1. This mutation abolishes Sirt1’s interaction with PGC-1α, but not with two other major substrates, p53 and Foxo3a. In what contexts would this interaction occur? A rather simplified view in this regard is that Sirt1 acts a metabolic and redox sensor of changes in nutrient and energy status, such as those occurring during CR (Cantó and Auwerx, 2009). CR has been shown to significantly increase mitochondrial biogenesis in multiple tissues in mice (Nisoli et al., 2005) and in human muscles (Civitarese et al., 2007). Early work from Puigserver’s group showed that Sirt1 is induced during fasting, and induced Sirt1 interacts with PGC-1α to increase the expression of hepatic gluconeogenic genes (Rodgers et al., 2005). The group also showed that fasting induced PGC-1α deacetylation by Sirt1 in skeletal muscle, which is required for the activation of mitochondrial fatty acid oxidation genes (Gerhart-Hines et al., 2007). Auwerx’s group, on the other hand, showed that resveratrol treatment of mice increased running time and oxygen consumption by the skeletal muscles, which is accompanied by the induction of genes for oxidative phosphorylation and mitochondrial biogenesis (Lagouge et al., 2006). The authors attributed these changes to activation of Sirt1, and a consequential decrease in PGC-1α acetylation and activity. The authors also showed that resveratrol treatment protected mice against diet-induced-obesity and insulin resistance. A more specific Sirt1 activator, SRT1720, likewise enhanced muscle endurance and protected mice from diet-induced obesity and insulin resistance by enhancing oxidative metabolism in skeletal muscle, liver, and brown adipose tissue (Feige et al., 2008). Auwerx and colleagues also found that deletion of the poly(ADP-ribose) polymerase-1(PARP-1), a major NAD+-consuming enzyme, resulted in increased Sirt1 activity. PARP-1 knockout mice have a higher mitochondrial content, an increase in energy expenditure, and were protected against metabolic disease (Bai et al., 2011). The work of Anderson and colleagues (Anderson et al., 2008) provided dynamical insights as to how Sirt1 may regulate PGC-1α action. The latter’s nuclear transcription activity is enhanced by Sirt1-dependent nuclear accumulation and is counter regulated by glycogen synthase kinase beta (GSK3β), which targets PGC-1α for intranuclear proteasomal degradation.
Despite the apparently convincing functional connection between Sirt1 and PGC-1α, whether Sirt1 is critically or directly involved in mitochondrial biogenesis has been controversial. Gurd and colleagues (Gurd et al., 2009) examined the relationship between Sirt1 levels and activity, PGC-1α and markers of mitochondrial density (such as Cytochrome c Oxidase subunit IV, COXIV) in skeletal and heart muscles, and surprisingly found a largely negative correlation between Sirt1 levels and the rest of the parameters. In fact, overexpression of Sirt1 in muscle downregulated PGC-1α and mtTFA levels, and while muscle stimulation and the AMPK activator AICAR upregulated PGC-1α and COX IV, Sirt1 level was downregulated. The group further showed that while nuclear Sirt1 activity correlated with indices of mitochondrial density, nuclear Sirt1 protein levels were negatively correlated in skeletal and heart muscles (Gurd et al., 2011). While training did not alter muscle or nuclear Sirt1 protein content in human muscle, it did increase muscle and nuclear PGC-1α and Sirt1 activity. Thus, it is an increased nuclear Sirt1 activity that may actually contribute to exercise-stimulated mitochondrial biogenesis in muscle.
From a more fundamental perspective, Hancock and colleagues (Hancock et al., 2011) have reexamined the claim that CR resulted in dramatic increases in mitochondrial biogenesis in multiple mouse tissues (Nisoli et al., 2005). In contrast to previous work, the authors found no elevation in PGC-1α levels after CR, and no evidence of any significant elevation in mitochondrial biogenesis markers in heart, brain, skeletal muscle, liver and adipose tissues. Although the authors did not report any measurement of Sirt1 level/activity status or PGC-1α’s acetylation status, the study does challenge the reproducibility of the CR-induced mitochondrial phenomenon. Another study directly questioned the role of Sirt1 in exercise-induced mitochondrial biogenesis through PGC-1α decetylation (Philp et al., 2011). The authors generated muscle-specific Sirt1 knockout mice (Sirt1 exon 4 was ‘floxed’ and crossed with Cre driven by muscle creatine kinase promoter), and found that skeletal muscle endurance, mitochondrial oxidative metabolism, and mitochondrial biogenesis were all not impaired. Moreover, mice deficient in muscle Sirt1 are similar to wild-type mice with regard to PGC-1α expression, nuclear translocation, activity, and deacetylation status. The authors further suggest that it is really the changes in GCN5 acetyltransferase activity that are important in regulating PGC-1α activity after exercise, and Sirt1 may not be relevant. Another recent study added to the controversy by showing that resveratrol feeding had no effect on mitochondrial proteins in muscle (Higashida et al., 2013), while high levels of resveratrol activated AMPK in C2C12 myotubes in culture, which did increase mitochondrial protein levels. The group further showed that Sirt1 silencing or a dominant negative mutant did increase PGC-1α acetylation, and over-expression of Sirt1 reduced it. However, the latter actually suppressing, instead of enhanced PGC-1α’s co-activator activity, with a consequential reduction in mitochondrial proteins.
THE CONNECTION BETWEEN SIRT1, AMPK AND PGC-1α IN REGULATING MITOCHONDRIAL FUNCTION
The discussion above highlights a particularly controversial point pertaining to the effect of resveratrol on PGC-1α and mitochondrial biogenesis, as well as the mode of involvement of the key cellular nutrient and energy status sensor AMPK (Hardie, 2011). Using an inducible knockout model, Sinclair’s group has shown that intact Sirt1 in mice is required for resveratrol’s activity on mitochondrial biogenesis and function (Price et al., 2012). The authors also showed that with moderate levels of resveratrol, AMPK activation in the system was Sirt1-dependent. Although higher levels of resveratrol could activate AMPK in a Sirt1-independent manner, improvement in mitochondrial function in both cases of moderate and high dosages of resveratrol was not observed in the absence of Sirt1. That Sirt1 may act upstream of AMPK has support in a previous report, which showed that Sirt1 could potentially deacetylate and activate the major AMPK activating kinase, Serine/threonine kinase 11 (STK11) (or liver kinase B1, LKB1) (Lan et al., 2008).
Resveratrol’s pharmacological effect is multifaceted, and amongst its known effects is the direct activation of AMPK (Dasgupta and Milbrandt, 2007) and inhibition of cAMP-degrading phosphodiesterases (Park et al., 2012). The work of Dasgupta and Milbrandt indicates that resveratrol could directly activate AMPK, at least in certain cell types, in a manner that is completely independent of Sirt1 activity (Dasgupta and Milbrandt, 2007). Another report has also shown that AMPK deficient mice are fairly resistant to the metabolic improvement effect of resveratrol (Um et al., 2010). A role for AMPK in mitochondrial biogenesis in skeletal muscle and other tissues has been extensively documented (Garcia-Roves et al., 2008; Kukidome et al., 2006; Reznick and Shulman, 2006; Reznick et al., 2007; Zong et al., 2002). In skeletal muscle, AMPK has been shown to directly phosphorylate PGC-1α (Jäger et al., 2007), and the phosphorylation of Thr-177 and Ser-538 is required for the PGC-1α-dependent induction of its own promoter. It was also demonstrated that a single administration of the AMPK activator AICAR increased the mRNA expression of all PGC-1α isoforms (Tadaishi et al., 2011). It is therefore clear that any compound or physiological regimes that elevate AMPK levels may also influence PGC-1α levels/activity and consequent mitochondrial biogenesis, in a manner that may be largely independent of Sirt1.
While some results, such as those of Price et al. (2012) are in support of Sirt1 acting upstream of AMPK, others have demonstrated just the opposite. Auwerx’s group has shown that AMPK could at least exert an influence on Sirt1 activity by increasing cellular NAD+ levels (Cantó et al., 2009; 2010). The work of Park et al. (2012) indicated that resveratrol inhibits cAMP phosphodiesterases, thus leading to elevated cAMP levels and the activation of AMPK and cAMP-inducible factors such as the Rap Exchange factor directly activated by cAMP 1 (Epac1). The authors showed that Epac1 is required for resveratrol-mediated increases in NAD+ levels, which were abolished by silencing of Epac1. What could be discerned from these remarkably different reports is that the connection between Sirt1, AMPK and PGC-1α is nonlinear, is definitely cell/tissue-type dependent and is influenced by energy and nutrient status.
SIRT1 AND MITOPHAGY
Given Sirt1’s role in mitochondrial biogenesis discussed above, it might appear paradoxical that Sirt1 has also been recently implicated in the opposite process, namely, the destruction of damaged or aged mitochondria via mitophagy (Yoshii and Mizushima, 2015). A detailed examination of mitophagy and its mechanism is beyond the scope of this review, and the reader is referred to several excellent recent review articles on this topic (Durcan and Fon, 2015; Eiyama and Okamoto, 2015; Hamacher-Brady and Brady, 2015; Yoshii and Mizushima, 2015). Mitophagy is critically dependent on two factors - the PTEN-induced putative kinase 1 (PINK1) and the E3 ubiquitin ligase Parkin. These proteins act in sensing the functional and health statuses of mitochondria, and mark damaged mitochondria for autophagic disposal (Eiyama and Okamoto, 2015). Sirt1’s regulation of general macroautophagy is well known (Ng and Tang, 2013) and is broadly viewed as a cellular protective mechanism against stress and death insults (Ou et al., 2014; Suzuki and Bartlett, 2014). A connection between Sirt1 and mitophagy was first shown by Hwang’s group, when the authors found that nicotinamide treatment of primary human fibroblasts extended their replicative lifespan apparently by accelerating autophagic degradation of mitochondria (Kang and Hwang, 2009). Nicotinamide decreased mitochondrial mass and increased mitochondrial membrane potential, with elevations of the autophagosome marker light chain 3 (LC3)-II and levels of proteins that regulate mitochondrial fusion and fission.
Nicotinamide can be converted to NAD+ by the salvage pathway of NAD+ synthesis. A subsequent paper from the Hwang lab indeed suggested that the mitophagic effect of nicotinamide is mediated through an increase of the NAD+/NADH ratio and Sirt1 activation (Jang et al., 2012), and the mitochondrial phenotype induced by nicotinamide could be mimicked with the Sirt1 activator SRT1720. Mitochondrial dysfunction in Xeroderma Pigmentosum group A (XPA), a nucleotide excision DNA repair disorder, is characterized by defective mitophagy (Fang et al., 2014). This is apparently due to hyperactivation of the DNA damage sensor PARP-1, a major NAD+ consuming enzyme, and the consequent lowering of Sirt1 activity. Another recent report indicated that the Sirt1 activator resveratrol acts through a signaling axis that involves Sirt1-Sirt3 and PINK1/Parkin-mediated mitophagy, resulting in cardio-protection (Das et al., 2014). Furthermore, loss of Sirt1 in the luminal epithelium in human prostate cancer delayed Parkin translocation to the mitochondria and reduced mitophagy (Di Sante et al., 2015).
SIRT1, PGC-1α AND NEURODEGENERATIVE DISEASES
A particularly interesting aspect of the findings discussed above is Sirt1’s known role in protection against neurodegenerative disorders (Ng et al., 2015; Tang, 2009; Zhang et al., 2011). Over the years, Sirt1 activity has been extensively associated with neuroprotection, and autophagy (or more specifically, mitophagy) induction could be part of its protective repertoire. Sirt1 activity is known to attenuate α-synuclein aggregation and toxicity, a hallmark of Parkinson’s disease (PD) (Albani et al., 2009; Donmez et al., 2012; Sampaio-Marques et al., 2012). The mitophagy regulators PINK1 and Parkin are both encoded by PD susceptibility genes that are mutated in patients with juvenile or early-onset Parkinsonism (Mullin and Schapira, 2015). Interestingly, a genetic screen in Drosophila found that ectopic expression of the fly Sir2 rescued mitochondrial defects in PINK1-null mutants (but not parkin mutants) (Koh et al., 2012).
A better known connection between Sirt1 and neurodegenerative diseases is not a mitophagy factor, but rather PGC-1α (Róna-Vörös and Weydt, 2010; Tsunemi and La Spada, 2012). PGC-1α suppression promotes α-synuclein accumulation in cellular models of PD (Ebrahim et al., 2010). The Zn-finger protein Parkin-interacting substrate (PARIS, or ZNF746) accumulates in cellular models of Parkin inactivation and in the human PD brain (Shin et al., 2011). PARIS represses the expression of PGC-1α and NRF-1, and transgenic overexpression of PARIS leads to the selective loss of dopaminergic neurons in the substantia nigra, the disease-susceptible neuron group in PD. Another report indicated that loss of Parkin in cells from patients with early-onset PD increased PGC-1α, but the elevated PGC-1α appeared transcriptionally non-functional (Pacelli et al., 2011). Meta-analysis of gene expression profiles and molecular pathways clearly implicated PGC-1α-responsive genes amongst those downregulated in PD (Zheng et al., 2010). Co-transduction of adenovirus carrying human PGC-1α in rat E17 mid-brain neuron cultures attenuated dopaminergic neuron loss induced by either mutant α-synuclein, or the mitochondrial respiratory chain Complex I-inhibitor rotenone. Transgenic mice that over-express PGC-1α in substantia nigra’s dopaminergic neurons are resistant to degeneration induced by the neurotoxin MPTP (Mudò et al., 2012).
Loss of PGC-1α is also a well-known feature in Huntington’s disease (HD), a hereditary disorder where aggregation of mutant Huntingtin protein with an extended polyglutamine tract causes striatal neuron loss (Ross and Tabrizi, 2011). Nuclear accumulation of mutant huntingtin repressed PGC-1α gene transcription by associating with its promoter (Cui et al., 2006), and over-expression of PGC-1α could partially reverse the toxicity of mutant huntingtin in cultured striatal neurons. Interestingly, nicotinamide could upregulate PGC-1α and the neuroprotective brain-derived neurotrophic factor (BDNF) in a transgenic mouse model of HD, and improve motor deficits (Hathorn et al., 2011). Another study with inducible PGC-1α expression in HD mice not only rescued the HD phenotype and spared neurons, but could result in significant elimination of mutant Huntingtin aggregates (La Spada, 2012). Given PGC-1α’s beneficial effect on a multitude of neurodegenerative pathologies, it is likely that the neuroprotective effect of Sirt1 may, at least in part, be exerted through activation of PGC-1α.
While all of the above results appear to attest to the promise of PGC-1α as a drug target in PD and HD, one must avoid generalizing PGC-1α’s potential beneficial effects on other neurodegenerative diseases. In particular, a recent report has shown that crossing of an Alzheimer’s disease (AD) mouse model (Tg19959 mice) with transgenic strains over-expressing human PGC-1α exacerbated amyloid and tau accumulation (Dumont et al., 2014).
OTHER SIRT1-REGULATED FACTORS/SUBSTRATES IN MITOCHONDRIAL FUNCTION
Sirt1’s protective activity against the detriments of mitochondrial dysfunction involves other Sirt1 substrates, particularly transcription factors that influence mitochondrial activity. One such group of factors are members of the Forkhead box O (FoxO) transcription factor family (Webb and Brunet, 2014). Sirt1 is known to deacetylate members of the FoxO family, particularly in response to stress (Brunet et al., 2004). Drosophila Sir2 expression prevented the demise of fly dopaminergic neurons in PINK1-null mutants in a dFOXO-dependent manner (Koh et al., 2012). Activation of FoxO3a, in particular, has been shown to inhibit mitochondrial gene expression, causing a reduction in mitochondrial DNA copy number and respiratory activity (Ferber et al., 2012). In fact, a declined in nuclear NAD+, and a consequentially diminished Sirt1 activity, was also found to underlie a specific loss of mitochondrial-encoded subunits of the oxidative phosphorylation system (Gomes et al., 2013).
Interestingly, FoxO3a could translocate into the mitochondria where it could potentially be a substrate of mitochondrial Sirt3 (Jacobs et al., 2008). FoxO3a mitochondrial translocation was also shown to occur during GTP-induced erythroid differentiation (Meshkini and Yazdanparast, 2012). Recent work in C. elegans has indicated that worm sir2.1 mediates longevity via the mitochondrial unfolded protein response (UPRmt) and the activation of the worm FoxO orthologue DAF-16 (Mouchiroud et al., 2013). It was further shown that inhibition of the mitochondrial chaperone TRAP1 generates a FoxO-dependent retrograde cell protective signal from the mitochondria to the nucleus (Kim et al., 2015). Sirt1’s modulation of FoxO activity could therefore influence mitochondria function via gene expression in the nucleus, as well as affecting retrograde signaling from the mitochondria to the nucleus.
A few other Sirt1-modified transcription factors could also have a role, either directly or indirectly, in mitochondrial activity and function. Sirt1 deacetylates and represses the activity of the hypoxia inducible factor 1-alpha (HIF-1α) (Lim et al., 2010). HIF-1α is known to repress mitochondrial function and oxygen consumption by inducing pyruvate dehydrogenase kinase 1 (PDK1) (Papandreou et al., 2006), and Sirt1 activity may thus influence mitochondrial respiratory function via the former. One prominent transcription factor substrate of Sirt1 is the tumor suppressor TP53 (Vaziri et al., 2001), and Sirt1 deacetylation of p53 suppresses its transactivation capacity and promotes cell survival (Luo et al., 2001). Interestingly, it has been shown that mouse embryonic stem (mES) cells produces endogenous reactive oxygen species (ROS), which causes p53 translocation into the mitochondria in the presence of Sirt1, but induces p53 nuclear translocation in Sirt1-null mES cells (Han et al., 2008). Sirt1 activity may therefore influence mitochondria-mediated apoptosis by influencing mitochondrial translocation of factors such as p53. Another transcription factor that is deacetylated by Sirt1 which has known roles in mitochondrial physiology is the Nuclear factor erythroid 2-related factor 2 (Nrf2) (Kawai et al., 2011). Nrf2 and its inhibitor, the Kelch-like ECH associated protein 1 (Keap1), are master regulators of the cellular anti-oxidative response, and have key roles in cellular bioenergetics (Dinkova-Kostova et al., 2015). It is clear from these examples that Sirt1 activity exerts a complex and multi-effector mediated influence on mitochondrial functions.
CONCLUSION
In this review, I discussed evidence for Sirt1’s regulation of mitochondrial biogenesis and turnover, particularly in relation to PGC-1α (see Fig. 1). The emerging view in the field is that Sirt1 activity could exert important influence on mitochondrial function, but the degree of influence is likely cell type- and physiological context-dependent. Sirt1 activation could promote mitochondrial biogenesis in conditions of energy deficiency associated with disease and injury. On the other hand, it could also have an important role in triggering the demise or turnover of damaged mitochondria. It is conceivable that both pathways would occur in parallel. Should one pathway occur preferentially over the other, which pathway will predominate would likely depend on the nature of disease/injury, the relevant Sirt1 substrates available, and the cell’s energetic and metabolic status, such as the availability of NAD+. For example, factors that result in direct mitochondrial damage may induce a higher rate of mitophagy. In terminally differentiated cells with acute energy requirements such as neurons, sustaining a steady supply of mitochondria and salvaging whatever that remains functional could be important to ensure survival. In dividing cells, a quick mitophagic clearance of damaged mitochondria may be more urgent so that these are not passed down to daughter cells during division. On the whole, Sirt1’s regulation of mitochondrial biogenesis and mitophagy could thus act in concert for mitochondria quality maintenance. The large regulatory repertoire of Sirt1 could potentially be exploited for therapeutics, but a better understanding of the molecular factors it influences and their dynamic interactions would be necessary.
Fig. 1.
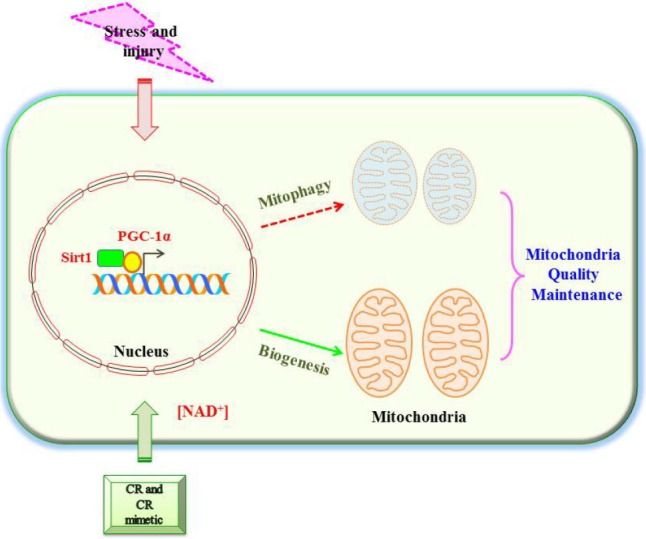
A schematic diagram illustrating a role for Sirt1 in both mitochondrial biogenesis and mitophagy. Caloric restriction (CR) and energetic status that elevates Sirt1 levels or NAD+ levels could promote accumulation of PGC-1α in the nucleus, which results in the transcription of genes that are necessary for mitochondrial function and biogenesis. How Sirt1 could promote mitophagy is less clear. However, nicotinamide, which increases NAD+ levels and Sirt1 activity, could trigger mitophagy. Sirt1’s regulation of mitochondrial biogenesis and mitophagy could function in concert for mitochondria quality maintenance.
REFERENCES
- Albani D., Polito L., Batelli S., De Mauro S., Fracasso C., Martelli G., Colombo L., Manzoni C., Salmona M., Caccia S., et al. The SIRT1 activator resveratrol protects SK-N-BE cells from oxidative stress and against toxicity caused by alpha-synuclein or amyloid-beta (1–42) peptide. J. Neurochem. 2009;110:1445–1456. doi: 10.1111/j.1471-4159.2009.06228.x. [DOI] [PubMed] [Google Scholar]
- Amat R., Planavila A., Chen S.L., Iglesias R., Giralt M., Villarroya F. SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma Co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J. Biol. Chem. 2009;284:21872–21880. doi: 10.1074/jbc.M109.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M., Barger J.L., Edwards M.G., Braun K.H., O’Connor C.E., Prolla T.A., Weindruch R. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquilano K., Vigilanza P., Baldelli S., Pagliei B., Rotilio G., Ciriolo M.R. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J. Biol. Chem. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquilano K., Baldelli S., Pagliei B., Ciriolo M.R. Extranuclear localization of SIRT1 and PGC-1α: an insight into possible roles in diseases associated with mitochondrial dys-function. Curr. Mol. Med. 2012;13:140–154. [PubMed] [Google Scholar]
- Austin S., St-Pierre J. PGC1α and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J. Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- Bai P., Cantó C., Oudart H., Brunyánszki A., Cen Y., Thomas C., Yamamoto H., Huber A., Kiss B., Houtkooper R.H., et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger P.M., Browning A.C., Garner A.N., Kelly D.P. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: a potential role in the cardiac metabolic stress response. J Biol. Chem. 2001;276:44495–44501. doi: 10.1074/jbc.M105945200. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D.F. Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta. 2012;1819:914–920. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Sherman J.M., Devine S.E., Cameron E.E., Pillus L., Boeke J.D. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Hu L.S., Cheng H.L., Jedrychowski M.P., Gygi S.P., Sinclair D.A., Alt F.W., Greenberg M.E. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Burnett C., Valentini S., Cabreiro F., Goss M., Somogyvári M., Piper M.D., Hoddinott M., Sutphin G.L., Leko V., McElwee J.J., et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byles V., Chmilewski L.K., Wang J., Zhu L., Forman L.W., Faller D.V., Dai Y. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int. J. Biol. Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.T., Kolesar J.E., Kaufman B.A. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim. Biophys. Acta. 2012;1819:921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Cantó C., Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol. Metab. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Jiang L.Q., Deshmukh A.S., Mataki C., Coste A., Lagouge M., Zierath J.R., Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.L., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civitarese A.E., Carling S., Heilbronn L.K., Hulver M.H., Ukropcova B., Deutsch W.A., Smith S.R., Ravussin E., CALERIE Pennington Team Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Dasgupta B., Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc. Natl. Acad. Sci. USA. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova A.T., Baird L., Holmström K.M., Meyer C.J., Abramov A.Y. The spatiotemporal regulation of the Keap1-Nrf2 pathway and its importance in cellular bioenergetics. Biochem. Soc. Trans. 2015;43:602–610. doi: 10.1042/BST20150003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy J.E., Lee Y., Gerhart-Hines Z., Puigserver P. Nutrient-dependent regulation of PGC-1alpha’s acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim. Biophys. Acta. 2010;1804:1676–1683. doi: 10.1016/j.bbapap.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G., Arun A., Chung C.Y., McLean P.J., Lindquist S., Guarente L. SIRT1 protects against α-synuclein aggregation by activating molecular chaperones. J. Neurosci. 2012;32:124–132. doi: 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dumont M., Stack C., Elipenahli C., Jainuddin S., Launay N., Gerges M., Starkova N., Starkov A.A., Calingasan N.Y., Tampellini D., Pujol A., Beal M.F. PGC-1α overexpression exacerbates β-amyloid and tau deposition in a transgenic mouse model of Alzheimer’s disease. FASEB J. 2014;28:1745–1755. doi: 10.1096/fj.13-236331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim A.S., Ko L.W., Yen S.H. Reduced expression of peroxisome-proliferator activated receptor gamma coactivator-1alpha enhances alpha-synuclein oligomerization and down regulates AKT/GSK3beta signaling pathway in human neuronal cells that inducibly express alpha-synuclein. Neurosci. Lett. 2010;473:120–125. doi: 10.1016/j.neulet.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiyama A., Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr. Opin. Cell Biol. 2015;33:95–101. doi: 10.1016/j.ceb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Fang E.F., Scheibye-Knudsen M., Brace L.E., Kassahun H., SenGupta T., Nilsen H., Mitchell J.R., Croteau D.L., Bohr V.A. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J.N., Lagouge M., Canto C., Strehle A., Houten S.M., Milne J.C., Lambert P.D., Mataki C., Elliott P.J., Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Ferber E.C., Peck B., Delpuech O., Bell G.P., East P., Schulze A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 2012;19:968–979. doi: 10.1038/cdd.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Roves P.M., Osler M.E., Holmström M.H., Zierath J.R. Gain-of-function R225Q mutation in AMP-activated protein kinase gamma3 subunit increases mitochondrial biogenesis in glycolytic skeletal muscle. J. Biol. Chem. 2008;283:35724–35734. doi: 10.1074/jbc.M805078200. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z., Rodgers J.T., Bare O., Lerin C., Kim S.H., Mostoslavsky R., Alt F.W., Wu Z., Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh H.S., McBurney M., Robbins P.D. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One. 2010;5:e9199. doi: 10.1371/journal.pone.0009199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A.P., Price N.L., Ling A.J.Y., Moslehi J.J., Montgomery M.K., Rajman L., White J.P., Teodoro J.S., Wrann C.D., Hubbard B.P., et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti I.V., Lee Y.M., Goodson H.V. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011;76:81–90. doi: 10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- Gurd B.J. Deacetylation of PGC-1α by SIRT1: importance for skeletal muscle function and exercise-induced mitochondrial biogenesis. Appl. Physiol. Nutr. Metab. 2011;36:589–597. doi: 10.1139/h11-070. [DOI] [PubMed] [Google Scholar]
- Gurd B.J., Yoshida Y., Lally J., Holloway G.P., Bonen A. The deacetylase enzyme SIRT1 is not associated with oxidative capacity in rat heart and skeletal muscle and its over-expression reduces mitochondrial biogenesis. J. Physiol. 2009;587:1817–1828. doi: 10.1113/jphysiol.2008.168096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurd B.J., Yoshida Y., McFarlan J.T., Holloway G.P., Moyes C.D., Heigenhauser G.J.F., Spriet L., Bonen A. Nuclear SIRT1 activity, but not protein content, regulates mitochondrial biogenesis in rat and human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;301:R67–R75. doi: 10.1152/ajpregu.00417.2010. [DOI] [PubMed] [Google Scholar]
- Haigis M.C., Guarente L.P. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Haigis M.C., Sinclair D.A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows W.C., Lee S., Denu J.M. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.K., Song E.K., Guo Y., Ou X., Mantel C., Broxmeyer H.E. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock C.R., Han D.H., Higashida K., Kim S.H., Holloszy J.O. Does calorie restriction induce mitochondrial biogenesis? A reevaluation. FASEB J. 2011;25:785–791. doi: 10.1096/fj.10-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G. Sensing of energy and nutrients by AMP-activated protein kinase. Am J. Clin. Nutr. 2011;93:891S–8916. doi: 10.3945/ajcn.110.001925. [DOI] [PubMed] [Google Scholar]
- Hathorn T., Snyder-Keller A., Messer A. Nicotinamide improves motor deficits and upregulates PGC-1α and BDNF gene expression in a mouse model of Huntington’s disease. Neurobiol. Dis. 2011;41:43–50. doi: 10.1016/j.nbd.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D., Muñoz-Martin M., Cañamero M., Mulero F., Martinez-Pastor B., Fernandez-Capetillo O., Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat. Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashida K., Kim S.H., Jung S.R., Asaka M., Holloszy J.O., Han D.H. Effects of resveratrol and SIRT1 on PGC-1α activity and mitochondrial biogenesis: A reevaluation. PLoS Biol. 2013;11:e1001603. doi: 10.1371/journal.pbio.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Zhao B., Lombard D.B., Fingar D.C., Inoki K. Cross-talk between sirtuin and mammalian target of rapamycin complex 1 (mTORC1) signaling in the regulation of S6 kinase 1 (S6K1) phosphorylation. J. Biol. Chem. 2014;289:13132–13141. doi: 10.1074/jbc.M113.520734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Xu S., Maitland-Toolan K.A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T.J., et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J. Biol. Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz K.T., Bitterman K.J., Cohen H.Y., Lamming D.W., Lavu S., Wood J.G., Zipkin R.E., Chung P., Kisielewski A., Zhang L.L., et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Hubbard B.P., Gomes A.P., Dai H., Li J., Case A.W., Considine T., Riera T.V., Lee J.E., E S.Y., Lamming D.W., et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.D., Denu J.M. Structural identification of 2′-and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- Jacobs K.M., Pennington J.D., Bisht K.S., Aykin-Burns N., Kim H.S., Mishra M., Sun L., Nguyen P., Ahn B.H., Leclerc J., et al. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int. J. Biol. Sci. 2008;4:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S.Y., Kang H.T., Hwang E.S. Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. J. Biol. Chem. 2012;287:19304–19314. doi: 10.1074/jbc.M112.363747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Yan T., Ge X., Sun C., Shi X., Zhai Q. Cytoplasm-localized SIRT1 enhances apoptosis. J. Cell. Physiol. 2007;213:88–97. doi: 10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McDonagh T., Heltweg B., Hixon J., Westman E.A., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., et al. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kang H.T., Hwang E.S. Nicotinamide enhances mitochondria quality through autophagy activation in human cells. Aging Cell. 2009;8:426–438. doi: 10.1111/j.1474-9726.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- Kawai Y., Garduño L., Theodore M., Yang J., Arinze I.J. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J. Biol. Chem. 2011;286:7629–7640. doi: 10.1074/jbc.M110.208173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Yang J., Kim M.J., Choi S., Chung J.R., Kim J.M., Yoo Y.H., Chung J., Koh H. Tumor necrosis factor receptor-associated protein 1 (TRAP1) mutation and TRAP1 inhibitor gamitrinib-triphenylphosphonium (G-TPP) induce a fork-head box O (FOXO)-dependent cell protective signal from mitochondria. J. Biol. Chem. 2015. (in press) [DOI] [PMC free article] [PubMed]
- Koh H., Kim H., Kim M.J., Park J., Lee H.J., Chung J. Silent information regulator 2 (Sir2) and Forkhead box O (FOXO) complement mitochondrial dysfunction and dopaminergic neuron loss in Drosophila PTEN-induced kinase 1 (PINK1) null mutant. J. Biol. Chem. 2012;287:12750–12758. doi: 10.1074/jbc.M111.337907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukidome D., Nishikawa T., Sonoda K., Imoto K., Fujisawa K., Yano M., Motoshima H., Taguchi T., Matsumura T., Araki E. Activation of AMP-activated protein kinase reduces hyperglycemia-induced mitochondrial reactive oxygen species production and promotes mitochondrial biogenesis in human umbilical vein endothelial cells. Diabetes. 2006;55:120–127. [PubMed] [Google Scholar]
- La Spada A.R. PPARGC1A/PGC-1α, TFEB and enhanced proteostasis in Huntington disease: defining regulatory linkages between energy production and protein-organelle quality control. Autophagy. 2012;8:1845–1847. doi: 10.4161/auto.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lan F., Cacicedo J.M., Ruderman N., Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J. Biol. Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leick L., Fentz J., Biensø R.S., Knudsen J.G., Jeppesen J., Kiens B., Wojtaszewski J.F.P., Pilegaard H. PGC-1{alpha} is required for AICAR-induced expression of GLUT4 and mitochondrial proteins in mouse skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2010;299:E456–E465. doi: 10.1152/ajpendo.00648.2009. [DOI] [PubMed] [Google Scholar]
- Li X., Monks B., Ge Q., Birnbaum M.J. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- Lim J.H., Lee Y.M., Chun Y.S., Chen J., Kim J.E., Park J.W. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol. Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Liu M., Wilk S.A., Wang A., Zhou L., Wang R.H., Ogawa W., Deng C., Dong L.Q., Liu F. Resveratrol inhibits mTOR signaling by promoting the interaction between mTOR and DEPTOR. J. Biol. Chem. 2010;285:36387–36394. doi: 10.1074/jbc.M110.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V.D. Linking sirtuins, IGF-I signaling, and starvation. Exp. Gerontol. 2009;44:70–74. doi: 10.1016/j.exger.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- McBurney M.W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J.R., Lansdorp P.M., Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshkini A., Yazdanparast R. Foxo3a targets mitochondria during guanosine 5′-triphosphate guided erythroid differentiation. Int. J. Biochem. Cell Biol. 2012;44:1718–1728. doi: 10.1016/j.biocel.2012.06.023. [DOI] [PubMed] [Google Scholar]
- Michishita E., Park J.Y., Burneskis J.M., Barrett J.C., Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Chan D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.J., Martin-Montalvo A., Mercken E.M., Palacios H.H., Ward T.M., Abulwerdi G., Minor R.K., Vlasuk G.P., Ellis J.L., Sinclair D.A., et al. The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell Rep. 2014;6:836–843. doi: 10.1016/j.celrep.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y.S., Viswanathan M., Schoonjans K., et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudò G., Mäkelä J., Di Liberto V., Tselykh T.V., Olivieri M., Piepponen P., Eriksson O., Mälkiä A., Bonomo A., Kairisalo M., et al. Transgenic expression and activation of PGC-1α protect dopaminergic neurons in the MPTP mouse model of Parkinson’s disease. Cell. Mol. Life Sci. 2012;69:1153–1165. doi: 10.1007/s00018-011-0850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin S., Schapira A. The genetics of Parkinson’s disease. Br Med. Bull. 2015;114:39–52. doi: 10.1093/bmb/ldv022. [DOI] [PubMed] [Google Scholar]
- Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K., et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Ng F., Tang B.L. Sirtuins’ modulation of autophagy. J. Cell. Physiol. 2013;228:2262–2270. doi: 10.1002/jcp.24399. [DOI] [PubMed] [Google Scholar]
- Ng F., Wijaya L., Tang B.L. SIRT1 in the brain-connections with aging-associated disorders and lifespan. Front. Cell Neurosci. 2015;9:64. doi: 10.3389/fncel.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E., Tonello C., Cardile A., Cozzi V., Bracale R., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Olmos Y., Sánchez-Gómez F.J., Wild B., García-Quintans N., Cabezudo S., Lamas S., Monsalve M. SirT1 regulation of antioxidant genes is dependent on the formation of a FoxO3a/PGC-1α complex. Antioxid. Redox Signal. 2013;19:1507–1521. doi: 10.1089/ars.2012.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Lee M.R., Huang X., Messina-Graham S., Broxmeyer H.E. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells. 2014;32:1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacelli C., De Rasmo D., Signorile A., Grattagliano I., di Tullio G., D’Orazio A., Nico B., Comi G.P., Ronchi D., Ferranini E., et al. Mitochondrial defect and PGC-1α dysfunction in parkin-associated familial Parkinson’s disease. Biochim. Biophys. Acta. 2011;1812:1041–1053. doi: 10.1016/j.bbadis.2010.12.022. [DOI] [PubMed] [Google Scholar]
- Pacholec M., Bleasdale J.E., Chrunyk B., Cunningham D., Flynn D., Garofalo R.S., Griffith D., Griffor M., Loulakis P., Pabst B., et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I., Cairns R.A., Fontana L., Lim A.L., Denko N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Park S.J., Ahmad F., Philp A., Baar K., Williams T., Luo H., Ke H., Rehmann H., Taussig R., Brown A.L., et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp A., Chen A., Lan D., Meyer G.A., Murphy A.N., Knapp A.E., Olfert I.M., McCurdy C.E., Marcotte G.R., Hogan M.C., et al. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) deacetylation following endurance exercise. J. Biol. Chem. 2011;286:30561–30570. doi: 10.1074/jbc.M111.261685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M.W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N.L., Gomes A.P., Ling A.J., Duarte F.V., Martin-Montalvo A., North B.J., Agarwal B., Ye L., Ramadori G., Teodoro J.S., et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick R.M., Shulman G.I. The role of AMP-activated protein kinase in mitochondrial biogenesis. J. Physiol. 2006;574:33–39. doi: 10.1113/jphysiol.2006.109512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick R.M., Zong H., Li J., Morino K., Moore I.K., Yu H.J., Liu Z.X., Dong J., Mustard K.J., Hawley S.A., et al. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab. 2007;5:151–156. doi: 10.1016/j.cmet.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rogina B., Helfand S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Róna-Vörös K., Weydt P. The role of PGC-1α in the pathogenesis of neurodegenerative disorders. Curr. Drug Targets. 2010;11:1262–1269. doi: 10.2174/1389450111007011262. [DOI] [PubMed] [Google Scholar]
- Ross C.A., Tabrizi S.J. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- Sampaio-Marques B., Felgueiras C., Silva A., Rodrigues M., Tenreiro S., Franssens V., Reichert A.S., Outeiro T.F., Winderickx J., Ludovico P. SNCA (α-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy. 2012;8:1494–1509. doi: 10.4161/auto.21275. [DOI] [PubMed] [Google Scholar]
- Satoh A., Brace C.S., Rensing N., Cliften P., Wozniak D.F., Herzog E.D., Yamada K.A., Imai S.I. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.H., Ko H.S., Kang H., Lee Y., Lee Y.I., Pletinkova O., Troconso J.C., Dawson V.L., Dawson T.M. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D.A., Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Stotland A., Gottlieb R.A. Mitochondrial quality control: Easy come, easy go. Biochim. Biophys. Acta. 2015;1853:2802–2811. doi: 10.1016/j.bbamcr.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden M.C., Caton P.W., Holness M.J. PPAR control: it’s SIRTainly as easy as PGC. J. Endocrinol. 2010;204:93–104. doi: 10.1677/JOE-09-0359. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Bartlett J.D. Sirtuin1 and autophagy protect cells from fluoride-induced cell stress. Biochim. Biophys. Acta. 2014;1842:245–255. doi: 10.1016/j.bbadis.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadaishi M., Miura S., Kai Y., Kawasaki E., Koshinaka K., Kawanaka K., Nagata J., Oishi Y., Ezaki O. Effect of exercise intensity and AICAR on isoform-specific expressions of murine skeletal muscle PGC-1α mRNA: a role of β2-adrenergic receptor activation. Am. J. Physiol. Endocrinol. Metab. 2011;300:E341–E349. doi: 10.1152/ajpendo.00400.2010. [DOI] [PubMed] [Google Scholar]
- Tang B.L. SIRT1, neuronal cell survival and the insulin/IGF-1 aging paradox. Neurobiol. Aging. 2006;27:501–505. doi: 10.1016/j.neurobiolaging.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Tang B.L. Sirt1’s complex roles in neuroprotection. Cell Mol. Neurobiol. 2009;29:1093–1103. doi: 10.1007/s10571-009-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner K.G., Landry J., Sternglanz R., Denu J.M. Silent information regulator 2 family of NAD- dependent his-tone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- Tissenbaum H.A., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tsunemi T., La Spada A.R. PGC-1α at the intersection of bioenergetics regulation and neuron function: from Huntington’s disease to Parkinson’s disease and beyond. Prog Neurobiol. 2012;97:142–151. doi: 10.1016/j.pneurobio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um J.H., Park S.J., Kang H., Yang S., Foretz M., McBurney M.W., Kim M.K., Viollet B., Chung J.H. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59:554–563. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A., Scher M.B., Lee D.H., Sutton A., Cheng H.L., Alt F.W., Serrano L., Sternglanz R., Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Erdjument-Bromage H., Tempst P., Serrano L., Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vega R.B., Horton J.L., Kelly D.P. Maintaining Ancient Organelles: Mitochondrial Biogenesis and Maturation. Circ. Res. 2015;116:1820–1834. doi: 10.1161/CIRCRESAHA.116.305420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Webb A.E., Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends. Biochem. Sci. 2014;39:159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.G., Rogina B., Lavu S., Howitz K., Helfand S.L., Tatar M., Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yang X.J., Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F., Hoberg J.E., Ramsey C.S., Keller M.D., Jones D.R., Frye R.A., Mayo M.W. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii S.R., Mizushima N. Autophagy machinery in the context of mammalian mitophagy. Biochim. Biophys. Acta. 2015;1853:2797–2801. doi: 10.1016/j.bbamcr.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang M., Dong H., Yong S., Li X., Olashaw N., Kruk P.A., Cheng J.Q., Bai W., Chen J., et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28:445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang S., Gan L., Vosler P.S., Gao Y., Zigmond M.J., Chen J. Protective effects and mechanisms of sirtuins in the nervous system. Prog. Neurobiol. 2011;95:373–395. doi: 10.1016/j.pneurobio.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Liao Z., Locascio J.J., Lesniak K.A., Roderick S.S., Watt M.L., Eklund A.C., Zhang-James Y., Kim P.D., Hauser M.A., et al. PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci. Transl. Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H., Ren J.M., Young L.H., Pypaert M., Mu J., Birnbaum M.J., Shulman G.I. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc. Natl. Acad. Sci. USA. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y., Liu L., Lee M.Y.K., Xu C., Liang Y., Man R.Y., Vanhoutte P.M., Wang Y. SIRT1 promotes proliferation and prevents senescence through targeting LKB1 in primary porcine aortic endothelial cells. Circ. Res. 2010;106:1384–1393. doi: 10.1161/CIRCRESAHA.109.215483. [DOI] [PubMed] [Google Scholar]


