Abstract
Prokaryotes form ubiquitin (Ub)-like isopeptide bonds on the lysine residues of proteins by at least two distinct pathways that are reversible and regulated. In mycobacteria, the C-terminal Gln of Pup (prokaryotic ubiquitin-like protein) is deamidated and isopeptide linked to proteins by a mechanism distinct from ubiquitylation in enzymology yet analogous to ubiquitylation in targeting proteins for destruction by proteasomes. Ub-fold proteins of archaea (SAMPs, small archaeal modifier proteins) and Thermus (TtuB, tRNA-two-thiouridine B) that differ from Ub in amino acid sequence, yet share a common β-grasp fold, also form isopeptide bonds by a mechanism that appears streamlined compared with ubiquitylation. SAMPs and TtuB are found to be members of a small group of Ub-fold proteins that function not only in protein modification but also in sulfur-transfer pathways associated with tRNA thiolation and molybdopterin biosynthesis. These multifunctional Ub-fold proteins are thought to be some of the most ancient of Ub-like protein modifiers.
Keywords: pupylation, sampylation, TtuB, sulfur, tRNA thiolation, molybdopterin
INTRODUCTION
Ubiquitin (Ub) is a 76–amino acid protein modifier that represents one of the most highly conserved polypeptides in eukaryotes and was likely present in the last eukaryotic common ancestor (57). Ub has a compact globular β-grasp fold structure common to a Ub-fold protein superfamily that is of ancient origin and phylogenetically widespread (13–16, 49). Ub is covalently attached through isopeptide bonds to lysine residues of proteins by a tightly regulated cascade of E1-E2-E3 enzymes in a process named ubiquitylation (54, 71). Proteins modified by Ub are recognized by specific Ub-binding proteins and altered in function, turnover, and interactions including subcellular localization (54). Ubiquitylation is essential to cell growth and viability (80, 110, 111) and is important in cell-cycle regulation, DNA repair, morphogenesis, signaling, and immune function (51, 75, 86, 121, 125).
Prokaryotes do not encode Ub, but they do synthesize two types of small protein modifiers that are linked onto protein targets by Ub-like isopeptide bonds. The first type is represented by the intrinsically disordered Pup (prokaryotic ubiquitin-like protein) of actinobacteria, which is covalently linked to proteins by a mechanism named pupylation (5, 8, 105). Pupylation tags proteins for degradation by proteasomes but is distinct from ubiquitylation in enzymology (9, 116). Proteins representing the second group include the archaeal SAMPs (small archaeal modifier proteins) and Thermus TtuB (tRNA-two-thiouridine B), which differ from Ub in sequence but share a common compact globular β-grasp fold (27, 77–79). These Ub-fold proteins are linked by isopeptide bonds to lysine residues of protein targets by mechanisms that appear to be simple versions of ubiquitylation in their requirement for E1 (but not E2 or E3) enzyme homologs. SAMPs and TtuB also function as sulfur carriers to form biomolecules such as thiolated wobble uridine tRNA and molybdopterin (79), with sulfur mobilization a common function of most prokaryotic Ub-fold proteins (42, 55). This review highlights both types of systems that form Ub-like isopeptide bonds in prokaryotes.
PUPYLATION
Pupylation is a posttranslational tagging system conserved in Actinobacteria and Nitrospirae (50) that mediates the covalent attachment of Pup to the lysine residues of target proteins (99). Biological roles of this tagging system include targeting proteins for destruction by proteasomes (7, 12, 99, 115) and the disassembly of complexes into monomers (28). Pupylation shares analogous features with ubiquitylation. In both systems, the protein modifiers (Pup and Ub) are small, cleaved at their C-terminus by posttranslational processing, activated by ATP-dependent mechanisms, covalently linked by isopeptide bonds to lysine residues of substrate proteins, and used to target proteins to proteasomes for destruction (105, 116). However, pupylation differs from ubiquitylation in its narrow phylogenetic distribution, the type of isopeptide bond formed, the structure of the protein modifier, and the enzymes and reaction mechanism used to mediate the modification (105, 116). Unlike Ub and related Ub-fold proteins, which are conjugated to proteins by means of successive E1-E2-E3 enzyme–mediated trans-thiolation reactions involving their C-terminal glycine α-carboxylate, Pup is covalently linked to target lysines via the γ-carboxylate of its C-terminal glutamate that is exposed after Gln deamidation by enzymes of the carboxylate-amine/ammonia ligase superfamily (105, 116). Thus, the reaction series and type of covalent bonds formed differ between the pathways of pupylation and ubiquitylation.
Discovery of Pupylation
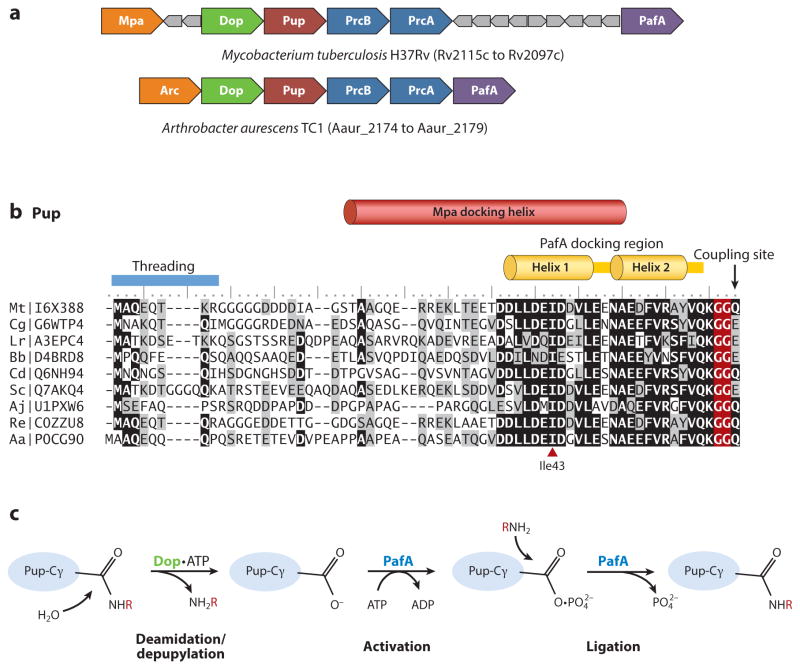
Pup was identified as a covalent ubiquitin-like protein modifier by study of mycobacteria (99). At the time, Mpa (Mycobacterium proteasomal ATPase) and PafA (proteasome accessory factor A) were known to be essential for virulence and resistance to nitric oxide stress (26), with Mpa identical to Rhodococcus ARC [AAA ATPase forming ring-shaped complexes; a distant homolog of AAA ATPases (134) important for the function of eukaryotic (35) and archaeal (132, 137) proteasomes]. However, the biological mechanism for targeting proteins for destruction by actinobacterial proteasomes was not known. Based on genomic sequence comparison, 20S proteasome genes were found connected in gene neighborhoods with Mpa, PafA, and a small open reading frame (encoding Pup) of unknown function (23, 56, 72, 120) (Figure 1a).
Figure 1.
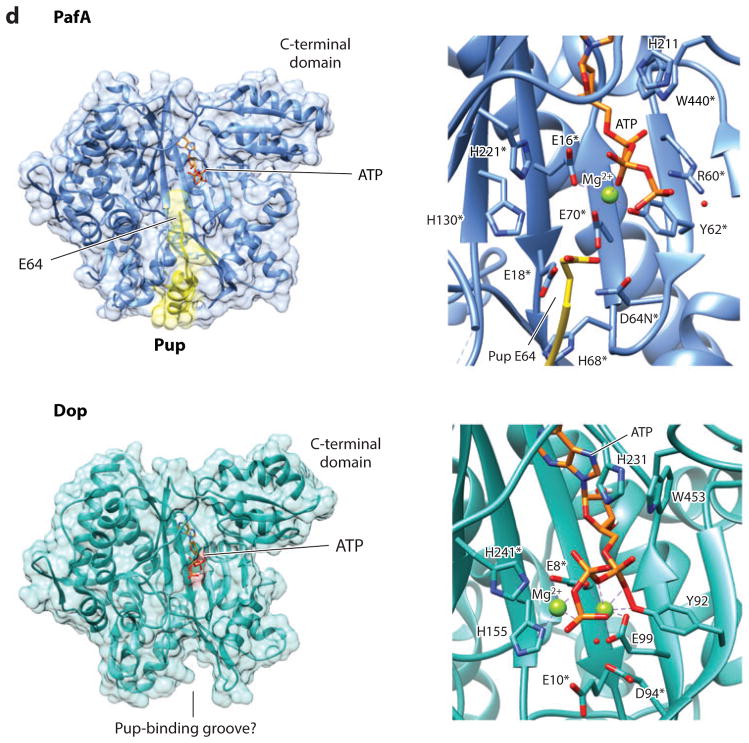
Pupylation and formation of ubiquitin-like isopeptide bonds. (a) Pup gene neighborhoods. Pup-, Dop-, Mpa-, and 20S proteasome (PrcA/B)-encoding genes are organized in gene neighborhoods in genomes of the phyla Nitrospirae and Actinobacteria. Genes are depicted as arrows, with the arrowheads pointing in the 5′ to 3′ direction. Representative gene neighborhoods are labeled below, including the name of the organism and gene locus tag numbers. (b) Multiple amino acid sequence alignment of representative Pup proteins. Identical and similar residues are highlighted in black and gray, respectively. UniProtKB/TrEMBL accession numbers are included on the left. Critical regions required for Pup activity as described in the text are indicated. (c) Schematic representation of reactions catalyzed by Dop and PafA. Dop mediates the deamidation of Pup-GGQ as well as the depupylation of pupylated proteins, whereas PafA activates and ligates Pup-GGE to target lysines. (d) PafA ligase and Dop depupylase/deamidase are structurally related to carboxylate-amine ligases. PafA and Dop structures are based on Protein Data Bank ID 4BJR and 4B0S, respectively. The Pup C-terminal tail that lines a groove on the surface of PafA that leads to the putative active site is highlighted in yellow. Mg-ATP and residues (*) required for catalytic activity based on site-directed mutagenesis and substrate analog trap are also indicated.
The breakthrough in pupylation came when Darwin and colleagues (99) used a bacterial two-hybrid system with Mpa as the bait and a series of coexpression studies to further define the proteasome system in Mycobacterium tuberculosis. Mpa was found to interact noncovalently with Pup and a Pup-derived peptide spanning the last 26 amino acids of its C-terminus, suggesting Pup was associated with proteasome function (99). Expression of Pup with FabD (malonyl Co-A acyl carrier protein transacylase), a presumed proteasomal substrate based on its enhanced steady state levels in ΔpafA, Δmpa, and proteasome-inhibited strains (98), revealed the two proteins could form a covalent complex (99). Whereas Pup was not predicted to have a Ub-fold structure, the diglycine motif preceding the C-terminal Q64 of Pup was noted to be common with Ub (99). Thus, the Pup-FabD complex was analyzed for Ub-like bonds by tandem mass spectrometry (MS/MS), and Pup was found to be isopeptide linked to K173 of FabD by a mechanism that likely involved deamidation of the Pup C-terminal glutamine to glutamate prior to its attachment to FabD (99). Later nuclear magnetic resonance (NMR) and biochemical experiments provided evidence that target lysine residues are coupled through the side chain γ-carboxylate of Pup E64 of the GGE motif (not the α-carboxylate) (118).
PafA Is the Pup Ligase, and Dop Is the Deamidase of Pup
Early work in mycobacteria implicated PafA in the degradation of proteasomal substrates: The steady state levels of certain proteins [i.e., Mpa, FabD, and PanB (ketopantoate hydroxymethyl-transferase)] were found to be increased by deletion of pafA and proteasome inhibition (30, 98). Thus, PafA function was examined and found essential for detection of Pup conjugates in mycobacteria, including for the attachment of Pup to target lysines of the proteasomal substrates FabD and PanB (99). Further bioinformatic study, using sensitive sequence profile searches with the PSI-BLAST program and HMMer package, revealed that PafA and a PafA homolog (now named Dop) are related to carboxylate-amine ligases (e.g., γ-glutamyl-cysteine synthetase and glutamine synthetase) that are commonly encoded near Pup, Mpa/ARC, and 20S proteasomal genes of Actinobacteria (50) (Figure 1a). Thus, a PafA family member was predicted to catalyze the ATP-dependent peptide ligase reaction needed to attach Pup to lysine residues on target proteins (50). In recognition that the Pup deamidation and ligation reactions were likely related, PafA or a nonspecific amidase was also speculated to deamidate the C-terminal glutamine before proceeding with the ligation reaction (50). To further define the pupylation pathway, Pup-decorated beads were used as bait in pull-down experiments with mycobacterial cell lysate as prey (117), analogous to studies that identified E1-E2-E3 enzymes of ubiquitylation (21, 22, 41). The results were rewarding: PafA and Dop were found to be Pup interaction partners (117). By use of purified components, Dop was demonstrated to deamidate the C-terminal Gln of Pup in the presence of nonhydrolyzable ATP, whereas ATP-hydrolysis was required for PafA-mediated conjugation of deamidated Pup (Pup-GGE) to protein substrate (i.e., FabD) (117). A follow-up in vivo study supported the role of Dop in deamidation based on the finding that mycobacterial dop mutant strains had reduced levels of Pup-modified proteins and were complemented by trans-expression of Pup-GGE but not Pup-GGQ (47). Thus, Dop was proposed to deamidate Pup using ATP as a cofactor, whereas PafA was thought to mediate the energy-dependent ligation of Pup-GGE to lysine targets via a phosphorylated Pup-GGE intermediate.
Dop Has Depupylase and Deamidase Activities
Comparative genomics reveals some lineages within the Nitrospirae and Actinobacteria phyla harbor homologs of Dop and Pup with a C-terminal Glu, suggesting Dop has another function in addition to deamidation of Pup (50, 117). Deamidation of Pup has mechanistic similarities to reactions used to cleave the isopeptide bond between Ub/Ub-fold proteins and target lysine residues in eukaryotic cells (31, 103). Thus, Dop was examined by multiple groups (6, 48) for a possible depupylating activity that may reverse the modification of proteins by Pup and, thus, regulate pupylation. Using purified components, researchers found that Dop specifically cleaves the isopeptide bond linking Pup to the lysine residues of protein targets (6, 48) and not a linear peptide bond linking Pup to the N-terminus of proteins by expression from a genetic fusion (48). Similar to the deamidase activity, Dop-mediated depupylation requires ATP as a cofactor (6, 48). This ATP requirement is supported by the finding that Dop E10A, with a point mutation in the predicted ATP-binding motif (47), is inactive in depupylation (6, 48). Dop-mediated depupylation is stimulated by Mpa/ARC (6, 48), and based on in vitro assay, this stimulation is no longer observed if pupylated substrates can no longer be unfolded by Mpa or if Mpa is a translocation-deficient variant (F341A) (48). Together these results reveal that Dop functions as a depupylase using ATP as a cofactor and that Mpa can stimulate this activity, most likely by serving to unfold pupylated proteins and render isopeptide bonds accessible for hydrolysis by Dop. Deamidation of Pup C-terminal Gln by Dop is thought to counter this activity by promoting PafA-mediated ligation of Pup to target lysines.
Distinct Features of Pup
Pup differs markedly in amino acid sequence and structure from Ub. Like Ub, Pup is a small protein modifier only 60–70 amino acids in length with a conserved C-terminal diglycine motif. However, only the C-terminal half of Pup is well conserved among Actinobacteria and Nitrospirae species (Figure 1b), and its diglycine motif is followed by an additional Glu or Gln residue [–GG(E/Q)] that is required at the C-terminus for pupylation (118). By contrast, Ub is highly conserved in amino acid sequence among eukaryotes (e.g., only 3 amino acids distinguish yeast Ub from human Ub), and mature forms of Ub require the diglycine motif at the extreme C-terminus for recognition in conjugation and deconjugation reactions (52, 130). Interestingly, Pup does not have the extremely compact and tightly hydrogen-bonded structure of Ub and is instead intrinsically disordered in its unbound state (19, 66, 74, 119). Whereas the conserved C-terminal region of Pup has hydrophobic-hydrophilic sequence patterns characteristic of formation of coiled coils, the monomer displays only weak helix formation (119).
PafA and Dop Are Related to Carboxylate-Amine Ligases
PafA and Dop are not related to E1-E2-E3 enzymes of ubiquitylation and are instead members of the carboxylate-amine/ammonia ligase (CAL) superfamily (50). Members of this superfamily include glutamine synthetase, γ-glutamylcysteine synthetase, and tRNA-dependent GatCAB amidotransferases that proceed by a two-step reaction mechanism in which a γ-glutamyl carboxylate is phosphorylated using ATP as a phosphoryl group donor, forming an intermediate that undergoes nucleophilic attack by ammonia or an α-amino group (44, 59, 67, 82, 96, 129). Similarly to other CAL enzymes, PafA catalyzes the turnover of ATP to ADP associated with formation of a phosphorylated γ-glutamyl carboxylate followed by condensation of this intermediate with an amino group (38, 117) (Figure 1c). However, in contrast to other CAL enzymes that form a C-N bond between the γ-glutamyl carboxylate of free glutamate or the glutamate moiety on glutamyl-tRNAGln and ammonia or the α-amino group of free cysteine, PafA forms a C-N bond via ligation of the γ-glutamyl carboxylate of a glutamate residue of a protein (Pup E64) to the ε-amino group of target lysines (38, 99, 117). Although the C-terminal E64 residue of the ligation-competent PupE features two carboxylates (i.e., the C-terminal α-carboxylate and the γ-carboxylate of the glutamyl side chain), only the γ-carboxylate is used to form the isopeptide bond between Pup and the target lysine, based on an NMR study with PanB substrate (118). In contrast to PafA, Dop catalyzes hydrolysis (depupylation and deamidation of Pup) and not condensation of C-N bonds, with ATP binding alone sufficient for Dop activity (6, 48, 117) (Figure 1c).
Atomic-level structures of PafA and Dop have been found to be similar to those of enzymes of the CAL superfamily. PafA and Dop have an N-terminal domain (~400 amino acids) with a fold characteristic of CAL enzymes, with an additional C-terminal domain (70 amino acids) that is unique to the PafA/Dop homologs (97). The CAL domain features a twisted antiparallel β-sheet, denoted as a β-sheet cradle, that is centrally positioned and surrounded by a cluster of α-helices (97) (Figure 1d, left panels). Buried deep within this β-sheet cradle, ATP is bound in a pocket by its adenine moiety and positioned with its triphosphate chain toward the apparent active site located on the concave surface of the cradle and opening to a groove on the enzyme surface (4, 97). Based on the crystal structure of a PafA D64N dimer solved with the C-terminally fused PupE38-E64 fragment reciprocally provided in trans (4), the C-terminal tail of Pup lines this groove and leads to the putative active site (Figure 1d, upper panels). Pup E64 is positioned near the triphosphate chain of ATP and surrounded by PafA residues required for pupylation (4, 97), with analogous residues required for Dop activity (11, 97) (Figure 1d, right panels).
PafA and Dop Catalyze a Related Step of Nucleophilic Attack
Even though PafA and Dop catalyze opposing reactions (condensation versus hydrolysis of C-N bonds), both enzymes use analogous residues to mediate the nucleophilic attack of the C-terminal carbonyl carbon of Pup (Figure 1c). The differences are in the type of nucleophile and the derivative of Pup used as a substrate. In Dop-mediated reactions, water is thought to act as the nucleophile in attack of the carbonyl carbon of Pup substituting (a) the amino group of glutamine with release of ammonia in deamidation (117) and (b) the Pup-modified lysine residues with liberation of the protein target in depupylation (6, 48). By contrast, PafA catalyzes attack of the carbonyl carbon of the phosphorylated Pup intermediate by use of the ε-amino group of lysine residues as a nucleophile with subsequent release of inorganic phosphate (38). Recently, an aspartate residue near the active site of Dop (analogous to D94, Figure 1d, lower panel) was found chemically modified by a substrate analog trap of Pup and, thus, was suggested to function as a direct nucleophile forming a unique anhydride intermediate or as part of a catalytic center with polarized water as the nucleophile (11). This aspartate is strictly conserved in Dop/PafA homologs and required for enzyme activity of both enzymes (4, 97). Thus, PafA may also use this conserved aspartate (analogous to D64, Figure 1d, upper panel) as the catalytic base in activating the nucleophilic ε-amino group of target lysine residues for attack of the phospho-Pup intermediate (116).
PafA Is Regulated
The activity of PafA is suggested to be regulated by Mpa and the availability of target proteins requiring degradation by the proteasome. PafA has a low KM for Pup-GGE and ATP (low micromolar range) (38) and, thus, appears relatively insensitive to the levels of deamidated Pup and the energy charge of the cell. When all substrates are present at saturating concentrations, the rate-limiting step of PafA catalysis is the activation of Pup-GGE (38). Once activated, phosphorylated Pup-GGE and ADP are stably retained on PafA and await nucleophilic attack by a lysine residue of the target protein with several orders of magnitude difference in the specificity constant between bona fide and model substrates of conjugation (38). Thus, PafA is suggested to be regulated by the availability of nucleophilic substrates that possess the correct specificity determinants (i.e., target lysine residues) (38). Recent evidence suggests PafA functions as a dimer or higher-order oligomer that cooperatively binds target proteins and is allosterically controlled by these target proteins (95). PafA also associates with Mpa in a complex proposed to serve as a modular protein-tagging and degradation machine that would couple pupylation to proteasome function (32). Mpa stimulates PafA-mediated pupylation based on in vitro assay (32). Thus, PafA appears to be regulated by Mpa and the availability of target proteins.
Pup Adopts Different Folds Depending on Its Interaction Partner
Pup is intrinsically disordered in its unbound state (19, 66, 74, 119). Upon binding to the N-terminal coiled-coil domains of Mpa, Pup residues 21–51 form an α-helix (119, 128) (Figure 1b). By contrast, Pup residues 38–58 adopt a helix H1-linker-helix H2 conformation when bound to PafA (4) (Figure 1b). Dop surface residue conservation suggests at least the region of Pup that forms helix H1 in the PafA-bound form may also associate with Dop (4). Pup residue Ile43 (Figure 1b) within helix H1 of the PafA-bound form and the α-helix of the Mpa-bound form is necessary for efficient binding to Mpa and to PafA (4, 32). Fluorescently labeled Pup probing suggests the C-terminal 26–amino acid sequence of Pup-GGE (residues 39–64) is the minimal motif needed for recognition by PafA, with specific hydrophobic residues within this sequence important for Pup interactions with Mpa and PafA (114). As the regions of Pup that form the interface with its different interaction partners overlap, a partner exclusion model is proposed in which Mpa and Dop compete for Pup-modified proteins to control the destruction versus depupylation of Pup-modified proteins, respectively (4).
Pupylation and Proteasomes
Pup targets proteins to proteasomes for degradation (7, 116). Cell lysates of wild type (but not a proteasome mutant) efficiently degrade Pup-modified proteins (10), and inactivation of pupylation sites (e.g., FabD K173R) stabilizes target proteins (99). The disordered N-terminal region of Pup promotes substrate entry into the pore of the Mpa ATPase and translocation of Pup, together with the conjugated substrate, into the 20S proteolytic chamber (12, 115, 128). Pup residues 21–51, which transition to an ordered α-helix upon docking with Mpa, are proposed to serve in a binding-induced folding mechanism that stabilizes interaction of the pupylated substrate with Mpa or the translocation of the substrate through Mpa into the proteasome for destruction (12, 128). Thus, Pup provides a two-part degron to proteasome substrates: the disordered N-terminus of Pup is required for degradation and the portion that forms an α-helix mediates attachment to Mpa (12). Analysis by in-cell NMR (74) suggests that transient binding of Mpa to proteasomal core particles (CPs) controls the fate of pupylated substrates, whereas Pup (Pup-GGQ) is found to interact only weakly with Mpa in the absence of CPs and to bind strongly to Mpa when CPs are present (74).
Pupylation not only targets proteins for degradation by proteasomes but can also regulate proteasome activity. In particular, covalent attachment of Pup to the C-terminal tail of Mpa (Lys591) prevents association of the Mpa ATPase with proteasomal CPs (28). Pupylation triggers the disassembly of Mpa from the hexameric ring into monomers (28). Mpa activity and degradation of pupylated proteins by the proteasome are restored after depupylation of Mpa by Dop (28). Thus, pupylation plays a direct role in regulating proteolysis through posttranslational modification of the proteasomes themselves.
Substrates of Pupylation
To date, several hundred proteins are found to be modified by the Pup system (17, 29, 100). Pupylated proteins and modification sites defined by these MS-based proteomic studies are integrated with corresponding atomic structures and functional annotations into a publically accessible database, PupDB (123). Computational programs designed to predict pupylation sites have also been developed, including GPS-PUP (70) and PupPred (18). The factors that provide specificity to substrate recognition are not fully defined but are likely minimal based on the finding that pupylation can be reconstituted in Escherichia coli by heterologous expression of PafA and Pup-GGE alone (17).
Pupylated proteins are from a variety of pathways, including respiration, intermediate metabolism, virulence, and detoxification (17, 29, 100) and are clearly linked to the physiology and pathogenesis of M. tuberculosis (5, 25). Although not essential outside of the host (26), pupylation and proteasome-associated factors are important for survival of M. tuberculosis in the host (33, 34, 61). Inhibition of the proteasome is bactericidal in nonreplicating M. tuberculosis (68, 69). Thus, the identity of proteins targeted to the Pup-proteasome system is of general interest.
PROKARYOTIC FORMS OF UBIQUITYLATION
Prokaryotic forms of ubiquitylation have recently been identified in which small Ub-fold proteins are isopeptide linked to lysine residues of protein targets by a mechanism that appears to be a simple version of ubiquitylation based on use of E1 (but not E2 or E3) homologs. Prokaryotic Ub-fold protein modifiers include archaeal SAMP1–3 (46, 85) and Thermus TtuB (tRNA-two-thiouridine B) (112), which function not only in protein modification but also in steps of sulfur transfer associated with molybdopterin (MPT) biosynthesis and/or wobble uridine tRNA thiolation (84, 113). These Ub-fold proteins are commonly attached to enzymes of tRNA thiolation (46, 112), revealing an ancient form of posttranslational modification conserved between archaea and thermophilic bacteria. The archaeal Ub-fold proteins also modify proteins that appear targeted for destruction by proteasomes (46, 85). The following sections highlight the history and details of these systems.
Archaeal SAMPs and Thermus TtuB of the Ub-Fold Protein Superfamily
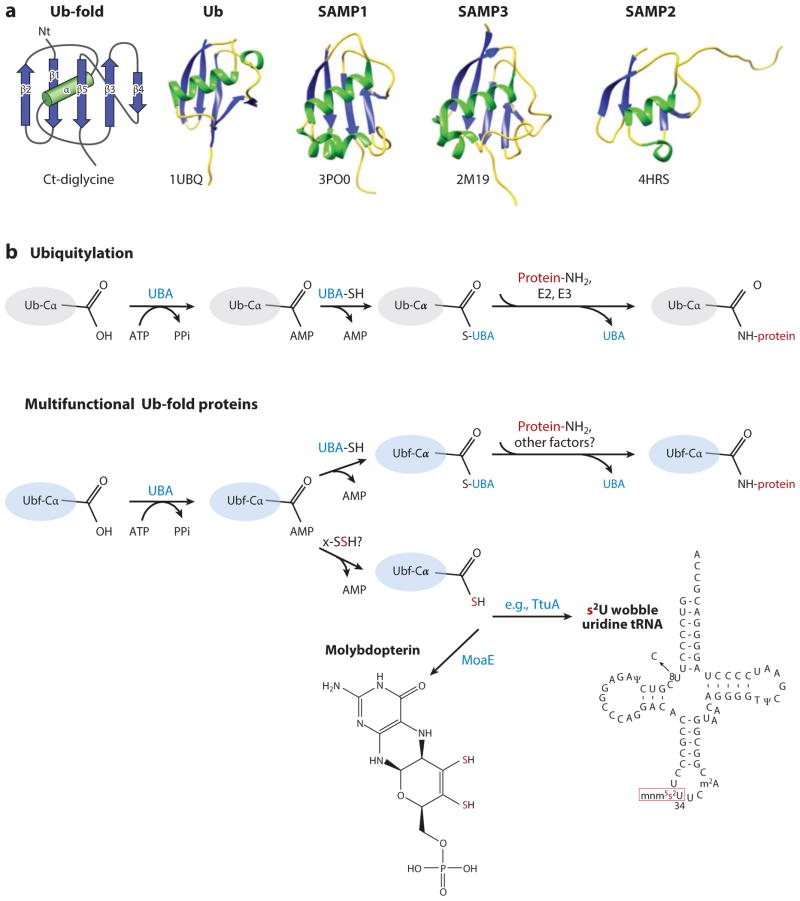
Archaeal SAMPs and Thermus TtuB are of the Ub-fold superfamily of proteins that adopt a compact globular β-grasp fold, similarly to Ub; this superfamily of proteins has members in all domains of life (13–16, 49, 90, 91). The β-grasp or Ub-fold (Figure 2a) is formed by an approximately 70–amino acid core that comprises five β-strands (S) wrapped around an α-helix (H) in an SSHSSS configuration and is typically associated with a C-terminal diglycine motif (127). Eukaryotic Ub-fold proteins are isopeptide linked to protein targets, are covalently bound to lipids (124, 126), or serve in sulfur transfer to form biomolecules including MPT (64, 81) and thiolated tRNA (45, 62, 88, 89, 92, 107, 108). Most bacterial Ub-fold proteins function in sulfur transfer, including in biosynthetic steps associated with formation of MPT (60), thiamine (135), thiolated wobble-position uridine tRNAs (113), secondary siderophores (36), and, in rare examples, the amino acids methionine and cysteine (1, 58, 95). Archaeal SAMP1/2 and Thermus TtuB are multifunctional. SAMP2 and TtuB are covalently attached to protein lysine residues (46, 112) and used in sulfur transfer reactions to form thiolated wobble uridine tRNA (84, 113). SAMP1 is also a protein modifier (40) and is associated with MPT biosynthesis (84). By contrast, only a role in protein modification has been identified for SAMP3 (85).
Figure 2.
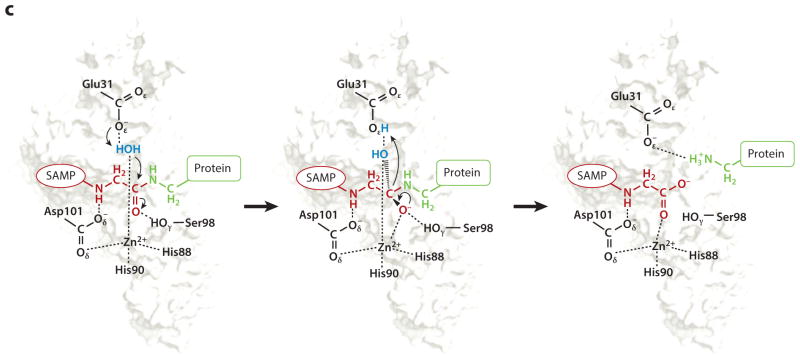
Sampylation ubiquitin (Ub)-fold protein modification system. (a) Ub-fold protein superfamily. The β-grasp or Ub-fold comprises five β-strands (S, blue) wrapped around an α-helix (H, green) in an SSHSSS configuration followed by a C-terminal (Ct) diglycine motif as indicated. Ub [Protein Data Base (PBD) ID 1UBQ] and SAMP1–3 (PDB IDs 3PO0, 4HRS, and 2M19) are members of this superfamily. (b) Schematic representation of ubiquitylation (Ub, upper panel) and prokaryotic Ub-fold proteins (Ubf, lower panel) that function in sulfur transfer and protein modification pathways. (c) HvJAMM1 desampylase. Schematic representation of the catalytic mechanism proposed for HvJAMM1 in cleaving small archaeal modifier protein (SAMP)-modified proteins. SAMP (red) conjugated to the target protein (green), the nucleophilic water (blue) coordinated to the catalytic Zn2+, and conserved active-site residues (black) are indicated. Abbreviations: UBA, ubiquitin-activating E1-type enzyme; x-SSH, cysteinyl persulfide of rhodanese or cysteine desulfurase. Figure adapted from Reference 40.
Whereas E1-E2-E3 enzymes typically mediate the covalent attachment of Ub and Ub-fold proteins to their targets in eukaryotes, sulfur-transfer pathways require only an E1-like enzyme from this cascade for Ub-fold protein activation. In both systems, the Ub or Ub-fold protein is activated through adenylation of its C-terminal α-carboxylate by a ubiquitin-activating E1-like enzyme (60, 65, 109) (Figure 2b). The ATP-dependent adenylation reaction readies the Ub-fold protein for E2-E3 transfer to protein targets (71) or acceptance of sulfur from cysteinyl persulfides formed on rhodanese domain proteins and cysteine desulfurases (24, 76). The Ub-fold protein accepts sulfur via thiocarboxylation at its C-terminal Gly and is then ready for use in biosynthetic reactions (14, 79). E2 and E3 are not used in thiocarboxylation. Instead, downstream components associated with each pathway provide the specificity needed for incorporation of sulfur into the appropriate biomolecule (e.g., Ub-fold MoaD associates with MoaE to form MPT) (131). Archaeal SAMPs and Thermus TtuB require an E1-like enzyme for covalent attachment to target lysine residues and sulfur transfer activities (84, 85, 112, 113). However, the genomes of these organisms do not encode canonical E2 or E3 enzymes. Thus, these prokaryotic systems appear to be streamlined forms of ubiquitylation that use only an E1-like enzyme for protein modification, with the E1 also needed for sulfur transfer.
Discovery of Archaeal Sampylation
Facilitating the discovery of sampylation were the findings that Ub-fold and E1-like protein homologs are widespread in archaeal genome sequences (13–16, 49, 90, 91); that eukaryotes mediate E3-independent monoubiquitylation (43); and that a single type of Ub-fold protein can serve in protein modification and in sulfur transfer, based on study of eukaryotic ubiquitin-related modifier 1 (Urm1) (45, 62, 88, 89, 92, 107, 108). The breakthrough came upon completion of the genome sequences of archaea with established genetic systems (63). In particular, the genome of the genetically amenable halophilic archaeon Haloferax volcanii was sequenced (39). Several small Ub-fold proteins with C-terminal diglycine motifs and an E1-like enzyme homolog were identified that could be studied for biological function (46). The Ub-fold proteins (now named SAMPs) were expressed with N-terminal FLAG-tag fusions and monitored for formation of protein conjugates in Hfx. volcanii (46). Two of the three Ub-fold proteins (SAMP1/2) were found to be covalently attached to proteins through nonthiol linkages by immunoblotting, and the C-terminal α-carboxylate of SAMP2 was demonstrated to be covalently linked to the ε-amino group of lysine residues of target proteins by MS/MS analysis (46). Later evidence revealed the other two Ub-fold proteins SAMP1/3 formed isopeptide bonds on target lysine residues (40, 85). Thus, all three Ub-fold proteins of Hfx. volcanii that harbor diglycine motifs are linked to target lysines by Ub-like isopeptide bonds.
E1-Like UbaA Is Required for Sampylation
Most archaea encode only a single Ub-activating E1-type enzyme homolog and have no apparent E2 or E3 homologs. In Hfx. volcanii, the ubaA gene (encoding the archaeal ubiquitin-activating E1 enzyme homolog) can be deleted, and the cells are viable (84). Comparison of ΔubaA and wild-type strains shows that UbaA is required for formation of the SAMP conjugates observed by immunoblotting and MS/MS analysis (84, 85). Consistent with this finding, a close homolog of UbaA (ELSA, E1-like SAMP activator of Methanosarcina acetivorans) activates archaeal Ub-fold proteins in an ATP-dependent manner (102). Activation occurs by formation of an adenylate at the C-terminus of the Ub-fold protein that is detectable by electrospray ionization mass spectrometry (102). Thus, sampylation is thought to function by an ATP-dependent E1-type mechanism.
Substrates of Sampylation
To date, numerous proteins are found covalently modified by the SAMP system. Substrates of sampylation in Hfx. volcanii include homologs of transcription, stress response, and metabolism proteins; they total 29 proteins (36 sites) [9 protein (11 lysine residue) targets of samp2ylation (46), 23 protein (11 lysine residue) targets of samp3ylation (85), and 1 protein (2 lysine residue) target of samp1ylation (40)], with some target protein overlap. Unlike the ψ KxE/D motif (where ψ represents a large hydrophobic residue) indicative of SUMO modification (104) and the KEN motif that serves as a Ub target for substrate destruction during mitotic progression (83), a consensus sequence for sampylation is not evident in these target proteins. Similarly to Ub, homo-polymeric SAMP chains have been detected, including K58-linked SAMP2 (46) and K18-, K55-, and K62-linked SAMP3 (85).
SAMPs are found isopeptide linked to a number of proteins associated with sulfur chemistry, including homologs of MoCo biosynthesis, tRNA thiolation, methionine-S-sulfoxide reductase, and rhodanese domain proteins (40, 84, 85). One protein target commonly modified by SAMP1/3 is MoaE, a homolog of the large subunit of MPT synthase (40, 85). In particular, the conserved active site residues of MoaE (K240 and K247) are covalently bonded to SAMP1/3, suggesting sampylation negatively regulates MPT synthesis (40, 85). Consistent with this biological role, MoaE is the major substrate of samp1ylation under aerobic growth conditions when the demand for MPT is low (85). Thus, sampylation may provide an autoregulatory negative feedback loop for Ub-fold-dependent sulfur transfer pathways.
Sampylation Is Dependent on Growth Conditions
Hfx. volcanii is a facultative aerobe that does not fix nitrogen. When cells are grown under aerobic or anaerobic conditions, addition of the terminal electron acceptor dimethylsulfoxide (DMSO) is found to alter the levels and types of proteins modified by SAMP1–3 (84, 85). Likewise, growth in the presence of alanine versus ammonium chloride as a nitrogen source causes an increase in SAMP1/2 conjugate levels (46). The influence of DMSO on sampylation is speculated to be associated with protein oxidation or regulation of MPT levels for DMSO reductase activity, whereas the association of sampylation with altered nitrogen availability may be linked to protein turnover and amino acid pools.
Sampylation Is Associated with Proteasome Function
Whereas SAMP3 conjugates appear unperturbed by proteasome function (85), evidence suggests that SAMP1 and SAMP2 are associated with targeting of proteins for destruction by proteasomes. In particular, the steady state levels of SAMP1 conjugates are significantly higher in proteasomal mutants compared with wild type (46), and chemical inhibition of 20S proteasome activity causes an increase in the levels of proteins modified by SAMP1/2 (most notably SAMP2) (85). Based on NMR spectroscopy, SAMP1 is found to bind an N-terminal peptide of the proteasome-associated nucleotidase (PAN)-B/2 (residues 1–74) with weak affinity over a wide range of salt concentrations, even those that disrupt SAMP1 structure (136). Thus, the PANs may form weak associations with the SAMPs alone. Far Western experiments demonstrate that the full-length PAN-A/1 binds the sampylation target MoaE only when covalently linked to SAMP1 (101). Thus, at least PAN-A/1 appears to have a higher affinity for SAMP1 when covalently bound to a target protein compared with SAMP1 alone, suggesting sampylation triggers association of protein targets with the proteasomal ATPase.
Sampylation Is Reversible
Archaea harbor deubiquitylating enzyme homologs (DUBs) of the JAB1/MPN/MOV34 metal-loenzyme (JAMM/MPN+) subfamily (3, 53). Of these homologs, Archaeoglobus fulgidus AfJAMM has been characterized at the atomic level, and although it is inactive in hydrolyzing Ub derivatives, resofurin-labeled casein, and D-alanine compounds, it is found to coordinate Zn2+ in a putative active-site configuration (2, 122). Similarly, Hfx. volcanii HvJAMM1 and HvJAMM2 do not hydrolyze unmodified proteins, diglycine, or the amide bond that links the fluorescent reporter 7-amino-4-methylcoumarin (AMC) to Ub (40). However, HvJAMM1 catalyzes the removal of SAMP1–3 from isopeptide- and linear-linked protein targets and, thus, functions as a desampylase (40, 85).
Based on biochemical characterization and 3-D modeling (40), HvJAMM1 is predicted to have a catalytic mechanism analogous to AMSH (Figure 2c). AMSH is a eukaryotic JAMM/MPN+ isopeptidase that specifically cleaves K63-linked polyubiquitin chains (106). HvJAMM1 is shown to coordinate a catalytic Zn2+, and site-directed mutagenesis suggests the Zn2+ is tetrahedrally coordinated by residues H88, H90, and D101 and a water molecule. HvJAMM1 E31 is important for function and likely to serve in a general base mechanism to polarize the water for nucleophilic attack of the carbonyl carbon of the scissile bond of SAMP conjugates (40). HvJAMM1 S98 is also important for activity and may serve to stabilize the negatively charged tetrahedral intermediate (40). Ultimately, HvJAMM1 releases the SAMPs from the sampylated protein to form desampylated protein product, an activity that is likely regulated to avoid a futile cycle with the E1-like UbaA. In the cell, HvJAMM1 is speculated to provide a proofreading mechanism to ensure the proper proteins are modified by sampylation and maintain SAMP homeostasis. In addition, HvJAMM1 is thought to reactivate enzymes that are transiently modified by SAMPs, remodel polySAMP chains, and remove/recycle SAMPs from sampylated proteins as these modified proteins undergo destruction by proteasomes.
Ubiquitin-Fold Protein Modification System in Thermus
Recently a Ub-fold protein conjugation system was identified in the bacterium Thermus thermophilus (112). This system includes the Ub-fold protein TtuB, known to function as a sulfur carrier in tRNA thiouridine synthesis (113), and TtuC, an E1 homolog responsible for the synthesis of thiolated wobble uridine tRNA, MPT, and thiamin (113). In vitro, TtuC can activate TtuB as an acyl-adenylate prior to its thiocarboxylation by cysteine desulfurases and can also form a thiol-dependent linkage with TtuB (113). In T. thermophilus, TtuB is found covalently attached to target proteins by a mechanism that requires TtuC (112). In particular, lysine residues (K137, K226, and K229) of TtuA are found isopeptide linked to the C-terminal α-carboxylate of TtuB (112). TtuA is a tRNA thiouridine synthetase that transfers the sulfur atom of thiocarboxylated TtuB to tRNA (113). Based on location of the target lysine residues in a crystal structure of TtuA, the catalytic activity of TtuA is proposed to be regulated by TtuB conjugation (87). Deletion of the single JAMM/MPN+ homolog of T. thermophilus (Ttc1133) results in an ~50% decrease in tRNA thiouridine levels, which may be due to a reduction in the level of TtuB C-terminal tails available for thiocarboxylation (112). Thus, thiouridine synthesis is proposed to be regulated by TtuB conjugation.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
Our understanding of Ub-like protein modification in prokaryotes has come full circle. Early reports of Ub in bacteria (37) and archaea (133) were found to be contamination from the Ub derived from the yeast extract of the growth medium. Thus, the name ubiquitin was considered a misnomer because the protein was not as ubiquitous as was initially thought (20). We now understand that prokaryotes have the capacity to form Ub-like isopeptide bonds on the lysine residues of proteins by at least two distinct pathways: mycobacterial pupylation and archaeal/thermophilic bacterial Ub-fold protein modification systems. At least in mycobacteria and archaea, the systems appear linked to targeting of proteins for destruction by proteasomes, similar to the ubiquitylation pathway of eukaryotic cells. The systems also appear to have nonproteolytic roles in covalent attachment of the protein modifier to target lysine residues to disrupt complex assembly (e.g., pupylation of Mpa ATPase) and enzyme activity (e.g., sampylation of MPT synthase subunit MoaE). Much is yet to be learned regarding prokaryotic Ub-like protein modification systems, and, based on recent comparative genomic studies (15, 16, 73, 93), the future holds promise that certain lineages will provide new insight into novel mechanisms of Ub-fold protein modification.
Acknowledgments
Thank you to the scientists in the field who have worked to provide insight into prokaryotic Ub-like protein modification systems. I apologize for not citing all references that have contributed to this review owing to space limitations. Work in the author’s laboratory is funded in part by grants from the National Institutes of Health (GM57498) and the Department of Energy Office of Basic Energy Sciences (DE-FG02-05ER15650).
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Agren D, Schnell R, Oehlmann W, Singh M, Schneider G. Cysteine synthase (CysM) of Mycobacterium tuberculosis is an O-phosphoserine sulfhydrylase: evidence for an alternative cysteine biosynthesis pathway in mycobacteria. J Biol Chem. 2008;283:31567–74. doi: 10.1074/jbc.M804877200. [DOI] [PubMed] [Google Scholar]
- 2.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Barandun J, Delley CL, Ban N, Weber-Ban E. Crystal structure of the complex between prokaryotic ubiquitin-like protein and its ligase PafA. J Am Chem Soc. 2013;135:6794–97. doi: 10.1021/ja4024012. [DOI] [PubMed] [Google Scholar]
- 5.Barandun J, Delley CL, Weber-Ban E. The pupylation pathway and its role in mycobacteria. BMC Biol. 2012;10:95. doi: 10.1186/1741-7007-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns KE, Cerda-Maira FA, Wang T, Li H, Bishai WR, Darwin KH. “Depupylation” of prokaryotic ubiquitin-like protein from mycobacterial proteasome substrates. Mol Cell. 2010;39:821–27. doi: 10.1016/j.molcel.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns KE, Darwin KH. Pupylation: a signal for proteasomal degradation in Mycobacterium tuberculosis. Subcell Biochem. 2010;54:149–57. doi: 10.1007/978-1-4419-6676-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns KE, Darwin KH. Pupylation versus ubiquitylation: tagging for proteasome-dependent degradation. Cell Microbiol. 2010;12:424–31. doi: 10.1111/j.1462-5822.2010.01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns KE, Darwin KH. Pupylation: proteasomal targeting by a protein modifier in bacteria. Methods Mol Biol. 2012;832:151–60. doi: 10.1007/978-1-61779-474-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE. Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–75. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns KE, McAllister FE, Schwerdtfeger C, Mintseris J, Cerda-Maira F, et al. Mycobacterium tuberculosis prokaryotic ubiquitin-like protein-deconjugating enzyme is an unusual aspartate amidase. J Biol Chem. 2012;287:37522–29. doi: 10.1074/jbc.M112.384784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns KE, Pearce MJ, Darwin KH. Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J Bacteriol. 2010;192:2933–35. doi: 10.1128/JB.01639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burroughs AM, Balaji S, Iyer LM, Aravind L. A novel superfamily containing the β-grasp fold involved in binding diverse soluble ligands. Biol Direct. 2007;2:4. doi: 10.1186/1745-6150-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the β-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burroughs AM, Iyer LM, Aravind L. Structure and evolution of ubiquitin and ubiquitin-related domains. Methods Mol Biol. 2012;832:15–63. doi: 10.1007/978-1-61779-474-2_2. [DOI] [PubMed] [Google Scholar]
- 16.Burroughs AM, Iyer LM, Aravind L. The natural history of ubiquitin and ubiquitin-related domains. Front Biosci. 2012;17:1433–60. doi: 10.2741/3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerda-Maira FA, McAllister F, Bode NJ, Burns KE, Gygi SP, Darwin KH. Reconstitution of the Mycobacterium tuberculosis pupylation pathway in Escherichia coli. EMBO Rep. 2011;12:863–70. doi: 10.1038/embor.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Qiu JD, Shi SP, Suo SB, Liang RP. Systematic analysis and prediction of pupylation sites in prokaryotic proteins. PLoS ONE. 2013;8:e74002. doi: 10.1371/journal.pone.0074002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ. Prokaryotic ubiquitin-like protein Pup is intrinsically disordered. J Mol Biol. 2009;392:208–17. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 21.Ciechanover A, Elias S, Heller H, Hershko A. “Covalent affinity” purification of ubiquitin-activating enzyme. J Biol Chem. 1982;257:2537–42. [PubMed] [Google Scholar]
- 22.Ciechanover A, Heller H, Katz-Etzion R, Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc Natl Acad Sci USA. 1981;78:761–65. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 24.Dahl JU, Urban A, Bolte A, Sriyabhaya P, Donahue JL, et al. The identification of a novel protein involved in molybdenum cofactor biosynthesis in Escherichia coli. J Biol Chem. 2011;286:35801–12. doi: 10.1074/jbc.M111.282368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darwin KH. Prokaryotic ubiquitin-like protein (Pup), proteasomes and pathogenesis. Nat Rev Microbiol. 2009;7:485–91. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–66. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 27.Darwin KH, Hofmann K. SAMPyling proteins in archaea. Trends Biochem Sci. 2010;35:348–51. doi: 10.1016/j.tibs.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delley CL, Striebel F, Heydenreich FM, Özcelik D, Weber-Ban E. Activity of the mycobacterial proteasomal ATPase Mpa is reversibly regulated by pupylation. J Biol Chem. 2012;287:7907–14. doi: 10.1074/jbc.M111.331124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, et al. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis [corrected] PLoS ONE. 2010;5:e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Festa RA, Pearce MJ, Darwin KH. Characterization of the proteasome accessory factor (paf) operon in Mycobacterium tuberculosis. J Bacteriol. 2007;189:3044–50. doi: 10.1128/JB.01597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forer N, Korman M, Elharar Y, Vishkautzan M, Gur E. Bacterial proteasome and PafA, the Pup ligase, interact to form a modular protein tagging and degradation machine. Biochemistry. 2013;52:9029–35. doi: 10.1021/bi401017b. [DOI] [PubMed] [Google Scholar]
- 33.Gandotra S, Lebron MB, Ehrt S. The Mycobacterium tuberculosis proteasome active site threonine is essential for persistence yet dispensable for replication and resistance to nitric oxide. PLoS Pathog. 2010;6:e1001040. doi: 10.1371/journal.ppat.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–20. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–23. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 36.Godert AM, Jin M, McLafferty FW, Begley TP. Biosynthesis of the thioquinolobactin siderophore: an interesting variation on sulfur transfer. J Bacteriol. 2007;189:2941–44. doi: 10.1128/JB.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guth E, Thommen M, Weber-Ban E. Mycobacterial ubiquitin-like protein ligase PafA follows a two-step reaction pathway with a phosphorylated Pup intermediate. J Biol Chem. 2011;286:4412–19. doi: 10.1074/jbc.M110.189282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartman A, Norais C, Badger J, Delmas S, Haldenby S, et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS ONE. 2010;5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hepowit NL, Uthandi S, Miranda HV, Toniutti M, Prunetti L, et al. Archaeal JAB1/MPN/MOV34 metalloenzyme (HvJAMM1) cleaves ubiquitin-like small archaeal modifier proteins (SAMPs) from protein-conjugates. Mol Microbiol. 2012;86:971–87. doi: 10.1111/mmi.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–14. [PubMed] [Google Scholar]
- 42.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–29. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoeller D, Hecker CM, Wagner S, Rogov V, Dötsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–98. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Horiuchi KY, Harpel MR, Shen L, Luo Y, Rogers KC, Copeland RA. Mechanistic studies of reaction coupling in Glu-tRNAGln amidotransferase. Biochemistry. 2001;40:6450–57. doi: 10.1021/bi002599l. [DOI] [PubMed] [Google Scholar]
- 45.Huang B, Lu J, Byström AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–94. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Humbard M, Miranda H, Lim J, Krause D, Pritz J, et al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, et al. Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol. 2010;75:744–54. doi: 10.1111/j.1365-2958.2009.07013.x. [DOI] [PubMed] [Google Scholar]
- 48.Imkamp F, Striebel F, Sutter M, Ozcelik D, Zimmermann N, et al. Dop functions as a depupylase in the prokaryotic ubiquitin-like modification pathway. EMBO Rep. 2010;11:791–97. doi: 10.1038/embor.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like β-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 52.Kanda F, Sykes DE, Yasuda H, Sandberg AA, Matsui S. Substrate recognition of isopeptidase: specific cleavage of the epsilon-(alpha-glycyl)lysine linkage in ubiquitin-protein conjugates. Biochim Biophys Acta. 1986;870:64–75. doi: 10.1016/0167-4838(86)90009-9. [DOI] [PubMed] [Google Scholar]
- 53.Katz EJ, Isasa M, Crosas B. A new map to understand deubiquitination. Biochem Soc Trans. 2010;38:21–28. doi: 10.1042/BST0380021. [DOI] [PubMed] [Google Scholar]
- 54.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 55.Kessler D. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol Rev. 2006;30:825–40. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 56.Knipfer N, Shrader TE. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol. 1997;25:375–83. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- 57.Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krishnamoorthy K, Begley TP. Protein thiocarboxylate-dependent methionine biosynthesis in Wolinella succinogenes. J Am Chem Soc. 2011;133:379–86. doi: 10.1021/ja107424t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krishnaswamy PR, Pamiljans V, Meister A. Activated glutamate intermediate in the enzymatic synthesis of glutamine. J Biol Chem. 1960;235:PC39–40. [PubMed] [Google Scholar]
- 60.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–29. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 61.Lamichhane G, Raghunand TR, Morrison NE, Woolwine SC, Tyagi S, et al. Deletion of a Mycobacterium tuberculosis proteasomal ATPase homologue gene produces a slow-growing strain that persists in host tissues. J Infect Dis. 2006;194:1233–40. doi: 10.1086/508288. [DOI] [PubMed] [Google Scholar]
- 62.Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–32. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 63.Leigh JA, Albers SV, Atomi H, Allers T. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol Rev. 2011;35:577–608. doi: 10.1111/j.1574-6976.2011.00265.x. [DOI] [PubMed] [Google Scholar]
- 64.Leimkühler S, Freuer A, Araujo JA, Rajagopalan KV, Mendel RR. Mechanistic studies of human molybdopterin synthase reaction and characterization of mutants identified in group B patients of molybdenum cofactor deficiency. J Biol Chem. 2003;278:26127–34. doi: 10.1074/jbc.M303092200. [DOI] [PubMed] [Google Scholar]
- 65.Leimkühler S, Wuebbens MM, Rajagopalan KV. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J Biol Chem. 2001;276:34695–701. doi: 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- 66.Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009;422:207–15. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- 67.Liaw SH, Eisenberg D. Structural model for the reaction mechanism of glutamine synthetase, based on five crystal structures of enzyme-substrate complexes. Biochemistry. 1994;33:675–81. doi: 10.1021/bi00169a007. [DOI] [PubMed] [Google Scholar]
- 68.Lin G, Chidawanyika T, Tsu C, Warrier T, Vaubourgeix J, et al. N,C-Capped dipeptides with selectivity for mycobacterial proteasome over human proteasomes: role of S3 and S1 binding pockets. J Am Chem Soc. 2013;135:9968–71. doi: 10.1021/ja400021x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin G, Li D, de Carvalho LP, Deng H, Tao H, et al. Inhibitors selective for mycobacterial versus human proteasomes. Nature. 2009;461:621–26. doi: 10.1038/nature08357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Z, Ma Q, Cao J, Gao X, Ren J, Xue Y. GPS-PUP: computational prediction of pupylation sites in prokaryotic proteins. Mol Biosyst. 2011;7:2737–40. doi: 10.1039/c1mb05217a. [DOI] [PubMed] [Google Scholar]
- 71.Lorenz S, Cantor AJ, Rape M, Kuriyan J. Macromolecular juggling by ubiquitylation enzymes. BMC Biol. 2013;11:65. doi: 10.1186/1741-7007-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lupas A, Zwickl P, Baumeister W. Proteasome sequences in eubacteria. Trends Biochem Sci. 1994;19:533–34. doi: 10.1016/0968-0004(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 73.Makarova KS, Koonin EV. Archaeal ubiquitin-like proteins: functional versatility and putative ancestral involvement in tRNA modification revealed by comparative genomic analysis. Archaea. 2010;2010:710303. doi: 10.1155/2010/710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maldonado AY, Burz DS, Reverdatto S, Shekhtman A. Fate of Pup inside the Mycobacterium proteasome studied by in-cell NMR. PLoS ONE. 2013;8:e74576. doi: 10.1371/journal.pone.0074576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marchese A, Trejo J. Ubiquitin-dependent regulation of G protein-coupled receptor trafficking and signaling. Cell Signal. 2013;25:707–16. doi: 10.1016/j.cellsig.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marelja Z, Mullick Chowdhury M, Dosche C, Hille C, Baumann O, et al. The L-cysteine desulfurase NFS1 is localized in the cytosol where it provides the sulfur for molybdenum cofactor biosynthesis in humans. PLoS ONE. 2013;8:e60869. doi: 10.1371/journal.pone.0060869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maupin-Furlow J. Proteasomes and protein conjugation across domains of life. Nat Rev Microbiol. 2012;10:100–11. doi: 10.1038/nrmicro2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maupin-Furlow JA. Archaeal proteasomes and sampylation. Subcell Biochem. 2013;66:297–327. doi: 10.1007/978-94-007-5940-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maupin-Furlow JA. Ubiquitin-like proteins and their roles in archaea. Trends Microbiol. 2013;21:31–38. doi: 10.1016/j.tim.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McGrath JP, Jentsch S, Varshavsky A. UBA1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–36. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mendel RR, Kruse T. Cell biology of molybdenum in plants and humans. Biochim Biophys Acta. 2012;1823:1568–79. doi: 10.1016/j.bbamcr.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 82.Midelfort CF, Rose IA. A stereochemical method for detection of ATP terminal phosphate transfer in enzymatic reactions. Glutamine synthetase. J Biol Chem. 1976;251:5881–87. [PubMed] [Google Scholar]
- 83.Min M, Mayor U, Lindon C. Ubiquitination site preferences in anaphase promoting complex/cyclosome (APC/C) substrates. Open Biol. 2013;3:130097. doi: 10.1098/rsob.130097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miranda H, Nembhard N, Su D, Hepowit N, Krause D, et al. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc Natl Acad Sci USA. 2011;108:4417–22. doi: 10.1073/pnas.1018151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miranda HV, Antelmann H, Hepowit N, Chavarria NE, Krause DJ, et al. Archaeal ubiquitin-like SAMP3 is isopeptide-linked to proteins via a UbaA-dependent mechanism. Mol Cell Proteomics. 2014;13:220–39. doi: 10.1074/mcp.M113.029652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moffat JM, Mintern JD, Villadangos JA. Control of MHC II antigen presentation by ubiquitination. Curr Opin Immunol. 2013;25:109–14. doi: 10.1016/j.coi.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 87.Nakagawa H, Kuratani M, Goto-Ito S, Ito T, Katsura K, et al. Crystallographic and mutational studies on the tRNA thiouridine synthetase TtuA. Proteins. 2013;81:1232–44. doi: 10.1002/prot.24273. [DOI] [PubMed] [Google Scholar]
- 88.Nakai Y, Harada A, Hashiguchi Y, Nakai M, Hayashi H. Arabidopsis molybdopterin biosynthesis protein Cnx5 collaborates with the ubiquitin-like protein Urm11 in the thio-modification of tRNA. J Biol Chem. 2012;287:30874–84. doi: 10.1074/jbc.M112.350090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem. 2008;283:27469–76. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 90.Nercessian D, de Castro RE, Conde RD. Ubiquitin-like proteins in halobacteria. J Basic Microbiol. 2002;42:277–83. doi: 10.1002/1521-4028(200208)42:4<277::AID-JOBM277>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 91.Nercessian D, Marino Buslje C, Ordóñez MV, de Castro RE, Conde RD. Presence of structural homologs of ubiquitin in haloalkaliphilic Archaea. Int Microbiol. 2009;12:167–73. [PubMed] [Google Scholar]
- 92.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–52. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–23. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ofer N, Forer N, Korman M, Vishkautzan M, Khalaila I, Gur E. Allosteric transitions direct protein tagging by PafA, the prokaryotic ubiquitin-like protein (Pup) ligase. J Biol Chem. 2013;288:11287–93. doi: 10.1074/jbc.M112.435842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O’Leary SE, Jurgenson CT, Ealick SE, Begley TP. O-phospho-L-serine and the thiocarboxylated sulfur carrier protein CysO-COSH are substrates for CysM, a cysteine synthase from Mycobacterium tuberculosis. Biochemistry. 2008;47:11606–15. doi: 10.1021/bi8013664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Orlowski M, Meister A. Partial reactions catalyzed by γ-glutamylcysteine synthetase and evidence for an activated glutamate intermediate. J Biol Chem. 1971;246:7095–105. [PubMed] [Google Scholar]
- 97.Özcelik D, Barandun J, Schmitz N, Sutter M, Guth E, et al. Structures of Pup ligase PafA and depupylase Dop from the prokaryotic ubiquitin-like modification pathway. Nat Commun. 2012;3:1014. doi: 10.1038/ncomms2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH. Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J. 2006;25:5423–32. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–7. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Poulsen C, Akhter Y, Jeon AH, Schmitt-Ulms G, Meyer HE, et al. Proteome-wide identification of mycobacterial pupylation targets. Mol Syst Biol. 2010;6:386. doi: 10.1038/msb.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Prunetti L, Reuter CJ, Hepowit NL, Wu Y, Barrueto L, et al. Structural and biochemical properties of an extreme ‘salt-loving’ proteasome activating nucleotidase from the archaeon Haloferax volcanii. Extremophiles. 2014;18:283–93. doi: 10.1007/s00792-013-0615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ranjan N, Damberger FF, Sutter M, Allain FH, Weber-Ban E. Solution structure and activation mechanism of ubiquitin-like small archaeal modifier proteins. J Mol Biol. 2011;405:1040–55. doi: 10.1016/j.jmb.2010.11.040. [DOI] [PubMed] [Google Scholar]
- 103.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–97. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–59. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 105.Samanovic MI, Li H, Darwin KH. The Pup-proteasome system of Mycobacterium tuberculosis. Subcell Biochem. 2013;66:267–95. doi: 10.1007/978-94-007-5940-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–62. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 107.Schlieker CD, van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci USA. 2008;105:18255–60. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schmitz J, Chowdhury MM, Hänzelmann P, Nimtz M, Lee EY, et al. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–89. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- 109.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–31. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Seufert W, Jentsch S. Yeast ubiquitin-conjugating enzymes involved in selective protein degradation are essential for cell viability. Acta Biol Hung. 1991;42:27–37. [PubMed] [Google Scholar]
- 111.Seufert W, McGrath JP, Jentsch S. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 1990;9:4535–41. doi: 10.1002/j.1460-2075.1990.tb07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shigi N. Posttranslational modification of cellular proteins by a ubiquitin-like protein in bacteria. J Biol Chem. 2012;287:17568–77. doi: 10.1074/jbc.M112.359844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shigi N, Sakaguchi Y, Asai S, Suzuki T, Watanabe K. Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. EMBO J. 2008;27:3267–78. doi: 10.1038/emboj.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smirnov D, Dhall A, Sivanesam K, Sharar RJ, Chatterjee C. Fluorescent probes reveal a minimal ligase recognition motif in the prokaryotic ubiquitin-like protein from Mycobacterium tuberculosis. J Am Chem Soc. 2013;135:2887–90. doi: 10.1021/ja311376h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Striebel F, Hunkeler M, Summer H, Weber-Ban E. The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup’s N-terminus. EMBO J. 2010;29:1262–71. doi: 10.1038/emboj.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Striebel F, Imkamp F, Özcelik D, Weber-Ban E. Pupylation as a signal for proteasomal degradation in bacteria. Biochim Biophys Acta. 2014;1843:103–13. doi: 10.1016/j.bbamcr.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 117.Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16:647–51. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- 118.Sutter M, Damberger FF, Imkamp F, Allain FH, Weber-Ban E. Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J Am Chem Soc. 2010;132:5610–12. doi: 10.1021/ja910546x. [DOI] [PubMed] [Google Scholar]
- 119.Sutter M, Striebel F, Damberger FF, Allain FH, Weber-Ban E. A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009;583:3151–57. doi: 10.1016/j.febslet.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 120.Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, et al. The first characterization of a eubacterial proteasome: the 20S complex of. Rhodococcus Curr Biol. 1995;5:766–74. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- 121.Teixeira LK, Reed SI. Ubiquitin ligases and cell cycle control. Annu Rev Biochem. 2013;82:387–414. doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- 122.Tran HJ, Allen MD, Löwe J, Bycroft M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry. 2003;42:11460–65. doi: 10.1021/bi035033g. [DOI] [PubMed] [Google Scholar]
- 123.Tung CW. PupDB: a database of pupylated proteins. BMC Bioinformatics. 2012;13:40. doi: 10.1186/1471-2105-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–57. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 125.Verlhac MH, Terret ME, Pintard L. Control of the oocyte-to-embryo transition by the ubiquitin-proteolytic system in mouse and C. elegans. Curr Opin Cell Biol. 2010;22:758–63. doi: 10.1016/j.ceb.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 126.Vierstra RD. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 2012;160:2–14. doi: 10.1104/pp.112.200667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 Å resolution. J Mol Biol. 1987;194:531–44. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 128.Wang T, Darwin KH, Li H. Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat Struct Mol Biol. 2010;17:1352–57. doi: 10.1038/nsmb.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wilcox M. γ-Glutamyl phosphate attached to glutamine-specific tRNA. A precursor of glutaminyl-tRNA in Bacillus subtilis. Eur J Biochem. 1969;11:405–12. doi: 10.1111/j.1432-1033.1969.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 130.Wilkinson KD, Audhya TK. Stimulation of ATP-dependent proteolysis requires ubiquitin with the COOH-terminal sequence Arg-Gly-Gly. J Biol Chem. 1981;256:9235–41. [PubMed] [Google Scholar]
- 131.Williams MJ, Kana BD, Mizrahi V. Functional analysis of molybdopterin biosynthesis in mycobacteria identifies a fused molybdopterin synthase in Mycobacterium tuberculosis. J Bacteriol. 2011;193:98–106. doi: 10.1128/JB.00774-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wilson HL, Ou MS, Aldrich HC, Maupin-Furlow J. Biochemical and physical properties of the Methanococcus jannaschii 20S proteasome and PAN, a homolog of the ATPase (Rpt) subunits of the eucaryal 26S proteasome. J Bacteriol. 2000;182:1680–92. doi: 10.1128/jb.182.6.1680-1692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wolf S, Lottspeich F, Baumeister W. Ubiquitin found in the archaebacterium Thermoplasma acidophilum. FEBS Lett. 1993;326:42–44. doi: 10.1016/0014-5793(93)81757-q. [DOI] [PubMed] [Google Scholar]
- 134.Wolf S, Nagy I, Lupas A, Pfeifer G, Cejka Z, et al. Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J Mol Biol. 1998;277:13–25. doi: 10.1006/jmbi.1997.1589. [DOI] [PubMed] [Google Scholar]
- 135.Xi J, Ge Y, Kinsland C, McLafferty FW, Begley TP. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein–protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc Natl Acad Sci USA. 2001;98:8513–18. doi: 10.1073/pnas.141226698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ye K, Liao S, Zhang W, Fan K, Zhang X, et al. Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii. Protein Sci. 2013;22:1174–82. doi: 10.1002/pro.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem. 1999;274:26008–14. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]