Abstract
Single-cell RNA-sequencing methods are now robust and economically practical and are becoming a powerful tool for high-throughput, high-resolution transcriptomic analysis of cell states and dynamics. Single-cell approaches circumvent the averaging artifacts associated with traditional bulk population data, yielding new insights into the cellular diversity underlying superficially homogeneous populations. Thus far, single-cell RNA-sequencing has already shown great effectiveness in unraveling complex cell populations, reconstructing developmental trajectories, and modeling transcriptional dynamics. Ongoing technical improvements to single-cell RNA-sequencing throughput and sensitivity, the development of more sophisticated analytical frameworks for single-cell data, and an increasing array of complementary single-cell assays all promise to expand the usefulness and potential applications of single-cell transcriptomic profiling.
Keywords: Single-cell RNA-sequencing, single-cell transcriptomic profiling
Introduction
The advent of next-generation sequencing over a decade ago spurred the development of a host of sequencing-based technologies 1 for probing genomic variation and dynamics. Of these methods, RNA-sequencing (RNA-seq) enabled transcriptomic profiling at unprecedented sensitivity and breadth, leading to the discovery of new RNA species and deepening our understanding of transcriptome dynamics 2, 3. In recent years, low-input RNA-seq methods have been adapted to work in single cells 4. These single-cell RNA-seq (scRNA-seq) technologies can quantify intra-population heterogeneity and enable study of cell states and transitions at very high resolution, potentially revealing cell subtypes or gene expression dynamics that are masked in bulk, population-averaged measurements 5, 6. In this review, we will discuss recent advancements and current limitations of scRNA-seq methodologies and highlight major applications of scRNA-seq in biological research.
scRNA-seq technologies: overview and recent advancements
Over the past six years, numerous scRNA-seq protocols have been developed 4, 7– 21. Currently published scRNA-seq protocols all follow the same general workflow: single cells are isolated; cells are lysed, and the RNA is captured for reverse transcription into cDNA; and the cDNA is pre-amplified and then used to prepare libraries for sequencing and downstream analysis. Kolodziejczyk et al. 22 provide a comprehensive review of individual scRNA-seq protocols and their relative strengths and weaknesses.
Although cDNA pre-amplification is necessary because only minute amounts of RNA are captured from each cell 23, amplification bias arising during pre-amplification limits the quantitative accuracy of scRNA-seq. Unique molecular identifiers (UMIs) can be used to barcode individual RNA molecules during the reverse transcription step, allowing direct transcript counting 24– 29, and many of the newer scRNA-seq protocols use UMIs to improve transcript quantitation 9, 16– 19. Alternatively, exogenous RNA standards such as those from the External RNA Control Consortium (ERCC) can be “spiked in” with cellular RNA to map between relative and absolute transcript counts 20, 30. Stegle et al. 31 provide a more detailed discussion of methods for scRNA-seq transcript quantitation and highlight some of the analytical challenges unique to single-cell data.
scRNA-seq methods have also been improving in terms of throughput and scalability. Whereas most earlier methods have been limited to measuring hundreds or thousands of cells at a time, recent advancements in microwell 17 and droplet-based 18, 19 cell-barcoding strategies have enabled the analysis of tens of thousands of cells in a single experiment. The high-throughput capacity of these new technologies will increase the resolution of single-cell experiments, improving their ability to detect rare cell subtypes or transitional states.
Challenges and limitations of scRNA-seq
Current scRNA-seq technologies still face a number of challenges. Collectively, existing scRNA-seq methods have low capture efficiency. Because only a small fraction of each cell’s transcript complement (approximately 10% for many protocols 9) is represented in the final sequencing libraries, scRNA-seq has limited sensitivity and is unable to reliably detect low-abundance transcripts 9, 32, 33. The low amount of input material for scRNA-seq libraries also leads to high levels of technical noise, which complicates data analysis and can mask underlying biological variation 22, 34– 37. Methods for modeling technical variation in scRNA-seq data have been proposed 35– 37; however, most approaches use the sample-to-sample variation in ERCC read counts to model and control for technical noise in the single-cell data and thus can be used only with experiments incorporating spike-in controls. Moreover, these approaches assume that the spike-in transcripts are treated the same as cellular RNA during library prep. However, naked spike-in RNA does not pass through cellular lysis and is not in complex with ribosomes or RNA-binding proteins. Thus, although spike-in procedures serve as useful indicators of transcript frequency and sensitivity in an experiment, there are many sources of variability that remain difficult to control in scRNA-seq.
Another potential source of bias stems from procedures to isolate and capture individual cells. Although micromanipulation or laser dissection techniques can isolate single cells from known locations within a cell population or tissue, these methods are labor-intensive or require specialized equipment 22, 33, 38. Most scRNA-seq protocols—and all of the existing high-throughput methods—first dissociate tissues to form a single-cell suspension before capturing individual cells. This cell dissociation step is often non-trivial, and enzymatic treatments used to break down tissues may impact cell viability, potentially affecting cells’ transcriptional profiles 22. To avoid biases stemming from such enzymatic treatments, Grindberg et al. have developed techniques for performing RNA-seq directly on single nuclei 39, 40, which can be isolated without using harsh protease treatments.
For most single-cell isolation procedures, information about cells’ original spatial context and cellular environment is lost. Recently, computational methods have been developed to infer a cell’s original position in three-dimensional space from its transcriptional profile by using a reference gene expression map built from existing in situ data 41, 42. However, these methods rely on the existence of spatial expression data for a panel of reference genes in the tissue of interest. Alternatively, emerging in situ sequencing strategies are able to capture and amplify RNA within the original tissue context, although current methods can measure up to only a few dozen genes per cell 43– 45. These methods sequence RNA directly inside unlysed cells: cDNA amplicons are generated and circularized, amplified via rolling circle amplification, and then sequenced by ligation in situ by using the SOLiD platform 44, 45. Such in situ sequencing approaches are distinct from fluorescence in situ hybridization (FISH) strategies (discussed further below), which detect transcripts through the binding of fluorescently labeled probes. However, although in situ sequencing methods preserve spatial information and can measure RNA expression patterns at subcellular resolution, these approaches are currently limited in throughput and require specialized tools which may not be widely accessible.
Finally, the bulk of scRNA-seq literature has focused solely on polyadenylated mRNAs; almost all published scRNA-seq protocols isolate cellular RNA by using poly-T priming, which captures only polyadenylated transcripts. Consequently, current methods are ill suited to investigate non-polyadenylated transcript classes, such as regulatory non-coding RNA (e.g. microRNAs 46, 47, lncRNAs 48, or circular RNAs 49, 50) or bacterial RNA 21. Random hexamer priming has been suggested as a strategy to simultaneously capture both polyadenylated and non-polyadenylated transcripts in single cells 20, 21, and computationally selected “not-so-random” primers could potentially be used to capture poly(A)+ and poly(A)– species while depleting for ribosomal RNA 51. Incorporating these alternative priming strategies into existing scRNA-seq technologies would enable the exploration of a wider spectrum of transcript types, broadening the scope and applicability of scRNA-seq.
Complementary single-cell technologies
Although scRNA-seq alone is a powerful tool for dissecting cell populations and processes, combining scRNA-seq with other single-cell technologies supplements transcriptomic data with complementary information that helps to paint a more complete picture of each cell. RNA FISH, in which individual transcripts are labeled with fluorescent probes and then detected via high-resolution microscopy, provides an orthogonal method of quantifying transcript levels and is often used to independently validate results from scRNA-seq data 52. Unlike scRNA-seq, single-cell FISH preserves the spatial context of assayed transcripts and can localize molecules down to subcellular resolution 53, 54. RNA localization and trafficking dynamics often play a crucial role in regulating protein translation and cellular function 55; used in conjunction with scRNA-seq, single-cell FISH could supplement the global transcriptomic snapshots of scRNA-seq with information on the spatial dynamics of selected transcripts. Whereas spectral overlap between fluorophores still limits the number of transcripts that can be simultaneously assayed, new approaches using super-resolution microscopy and combinatorial labeling schemes can measure up to thousands of transcripts in each cell 53, 54, 56.
Single-cell genome sequencing has been developing alongside scRNA-seq and has been used successfully to map genetic variation at single-cell resolution and to infer cell lineages 57– 61. Moreover, in the past year, methods have been developed to sequence both the genome and the transcriptome of the same cell 62, 63, enabling direct comparison of genetic and gene expression variation within a single cell. This integrated, parallel-sequencing approach shows great promise for uncovering genotype-phenotype relationships and has already been used to demonstrate strong correlations between gene copy number and gene expression levels 62, 63.
Over the past few years, methods have also been developed to assay the epigenetic landscape of single cells: both bisulfite sequencing 64– 67 (measuring DNA methylation) and assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) 68, 69 (measuring chromatin accessibility) have been adapted to work with single cells. These methods offer insight into the epigenetic heterogeneity within cell populations, and paired epigenomic and transcriptomic data could deepen our understanding of the mechanisms underlying gene expression regulation. Although direct comparison of a cell’s epigenomic and transcriptomic profiles is not currently possible, combining single-cell bisulfite sequencing or single-cell ATAC-seq with scRNA-seq from the same cell could enable such analyses in the future. Similarly, integrating scRNA-seq with single-cell proteomic methods 70, 71 would provide insight into post-transcriptional gene regulation and the degree to which mRNA expression is reflected at the protein level.
Applications of scRNA-seq
Recent studies have demonstrated high cell-to-cell transcriptomic variation 10, 72– 74, even within genetically homogenous cell populations 75. Consequently, bulk measurements can mask important cellular heterogeneity 5, 76 and lead to averaging artifacts 6. One major advantage of scRNA-seq is its ability to detect such cell-to-cell heterogeneity and capitalize upon it to uncover population structure and cell dynamics hidden at the group level.
scRNA-seq has been used to dissect heterogeneous cell populations and complex tissues, such as intestine 77, spleen 16, lung 78, or brain 42, 79– 83. Clustering methods 16, 75, 77 or dimensionality reduction techniques 78 can be used directly on single-cell expression data to group cells by transcriptomic similarity and to detect the underlying population structure in an unsupervised manner ( Figure 1A). Cell subgroups identified from such analyses can often be matched to known cell types via previously established marker genes 16, 52, 78, 81, 82; however, structural analysis of single-cell data has also led to the discovery of novel cell subtypes 79, 83, 84 as well as the identification of new marker genes for known cell types 78, 84, 85. In the context of cancer, scRNA-seq analyses have been used to characterize intra-tumoral heterogeneity and to classify tumor subpopulations 86– 88. scRNA-seq profiling can also detect variation among cell states within a seemingly homogenous population, such as differences in cell cycle stage 89 or differential signaling responses to an outside stimulus 52, 75, 90.
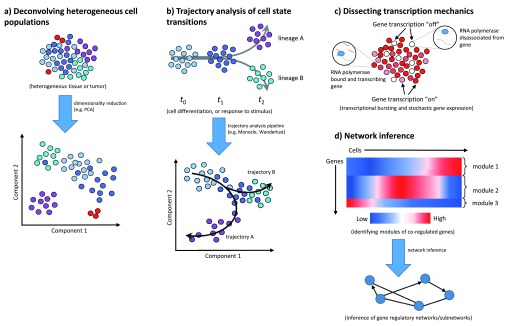
Figure 1. Common applications of single-cell RNA sequencing.
( a) Deconvolving heterogeneous cell populations. Clustering by single-cell transcriptomic profiles can reveal population substructure and enable the identification of cell subtypes and rare cell species (e.g. red cells above). Clusters may be tight and well defined (purple, red) or diffuse (blue). ( b) Trajectory analysis of cell state transitions. Single-cell RNA sequencing time-series data can be used to map cell developmental trajectories over the course of dynamic processes such as differentiation or signaling responses to an external stimulus. Some computational suites (e.g. Monocle 6) can also accommodate branching trajectories, enabling identification of lineage-specific gene expression and key genes that drive branching events. ( c) Dissecting transcription mechanics. Genes’ expression profiles across many cells can be compared to study transcriptional bursting and to model the kinetics of stochastic gene expression. ( d) Network inference. Genes can be clustered by expression profile to identify modules of putatively co-regulated genes, and gene-gene covariation relationships can be used to infer gene regulatory networks or subnetworks.
scRNA-seq is also commonly used to study cellular transitions between different states and to map cell trajectories through processes like differentiation ( Figure 1B). Several analytical frameworks have been proposed for inferring such trajectories: Monocle introduced the concept of “pseudotime” as a quantitative measure of “progress through a biological process” and uses techniques from computational geometry to order cells in pseudotime on the basis of their transcriptomic profiles 6. Wanderlust uses an entirely different algorithm based on local topological clustering to place cells along a developmental trajectory 91 by using single-cell proteomic measurements. More recently, Shin et al. 92 and Moignard et al. 93 have outlined additional strategies for reconstructing cell trajectories. Once cells have been ordered along a trajectory, gene expression patterns over the course of the established developmental trajectory can be analyzed to identify key regulators and genes with “switch-like” behavior 6, 72, 91. Sensitivity for identifying intermediate differentiation states can also be improved by using latent variable models to account for potential confounding factors (such as cell cycle) in the expression data prior to applying trajectory analysis techniques 94.
Growing evidence suggests that genes are not transcribed continuously but rather undergo short bursts of transcription interspersed with silent intervals 95. Transitions between “on” and “off” states are governed by several stochastic processes 96, 97, and this phenomenon of “transcriptional bursting” is a major source of gene expression heterogeneity between cells. scRNA-seq can be used to explore transcriptional mechanics and to model the kinetics of stochastic gene transcription 96, 98, 99 ( Figure 1C). Recent studies have also reported instances of cells preferentially expressing a single allele 32 or a single splice isoform 75; however, the low mRNA capture efficiency of scRNA-seq makes it difficult to draw definitive conclusions about allele-specific or isoform-specific expression at the single-cell level.
The inherent gene expression variability between cells in scRNA-seq data can be used to infer gene regulatory networks (GRNs) 100– 102. Most commonly, genes are grouped into co-regulated “modules” on the basis of expression profile similarity 16, 52, 75, 86, 87, 103 ( Figure 1D). Network inference from scRNA-seq data poses several challenges. Owing to low capture efficiency and stochastic gene expression, gene dropout (where gene expression is zero in a given cell) is quite common, leading to zero-inflated expression data 104. Although zero-inflated distributions can be used to accommodate expected dropout 104– 106, such models also have a greater number of parameters and can be more difficult to fit than a simpler model, particularly when sample size is limited. As previously mentioned, scRNA-seq data are very noisy, and separating biological variation from technical noise remains a non-trivial problem 35, 36. Additionally, the number of model parameters to be estimated (genes and gene interactions) usually greatly exceeds the number of sample observations (cells measured), and this disparity poses challenges for parameter estimation 107, 108. Simplifying the model on the basis of prior knowledge or focusing on only a small subnetwork of key players may be necessary to make parameter estimation feasible 107– 110. Finally, experimentally validating inferred GRNs can be very difficult; whereas knocking out a single gene is relatively straightforward, disrupting interactions between two proteins or between a protein and its target sequence can be much harder, and very few hypothesized models have been rigorously tested thus far.
Conclusions
scRNA-seq technologies have advanced significantly since their inception, improving in terms of both transcript quantitation and experimental throughput. Whereas low capture efficiency and high levels of technical noise limit the sensitivity and accuracy of scRNA-seq, more sophisticated analytical frameworks are emerging to facilitate the interpretation of scRNA-seq data 35– 37. Pairing single-cell transcriptomic data with spatial information 41, 42, 54 or orthogonal single-cell genomic assays 62, 63, 65, 68 also promises to provide new insights into transcriptional dynamics and the mechanisms underlying gene regulation.
scRNA-seq has been very effective at dissecting complex, heterogeneous cell populations, enabling unsupervised learning of population structure and the discovery of novel subtypes and rare cell species 79, 84. In the context of dynamic processes, cell trajectories reconstructed from single-cell transcriptomic data have provided insight into transient intermediate cell states and have helped to identify key regulator genes 6, 91. Finally, scRNA-seq also shows great potential for elucidating stochastic transcriptional kinetics and inferring gene regulatory networks. However, network inference from scRNA-seq data is computationally challenging and difficult to validate; inferred network models should thus be critically evaluated and experimentally tested where possible.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Roger S Lasken, J. Craig Venter Institute, La Jolla, CA, 92121, USA
Sara B Linker, Laboratory of Genetics, The Salk Institute for Biological Studies, La Jolla, CA, USA
Sten Linnarsson, Unit of Molecular Neurobiology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden
Funding Statement
CT is supported by an NIH DP2 HD088158 grant, an Alfred P. Sloan Fellowship, and a Dale F. Frey Award for Breakthrough Scientists from the Damon Runyon Cancer Research Foundation. SL is supported by an NSF IGERT grant DGE-1258485.
[version 1; referees: 2 approved]
References
- 1. Soon WW, Hariharan M, Snyder MP: High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9:640. 10.1038/msb.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Z, Gerstein M, Snyder M: RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 3. Ozsolak F, Milos PM: RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. 10.1038/nrg2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tang F, Barbacioru C, Wang Y, et al. : mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Wills QF, Livak KJ, Tipping AJ, et al. : Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat Biotechnol. 2013;31(8):748–52. 10.1038/nbt.2642 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Trapnell C, Cacchiarelli D, Grimsby J, et al. : The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–6. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Islam S, Kjällquist U, Moliner A, et al. : Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21(7):1160–7. 10.1101/gr.110882.110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Islam S, Kjällquist U, Moliner A, et al. : Highly multiplexed and strand-specific single-cell RNA 5' end sequencing. Nat Protoc. 2012;7(5):813–28. 10.1038/nprot.2012.022 [DOI] [PubMed] [Google Scholar]
- 9. Islam S, Zeisel A, Joost S, et al. : Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11(2):163–6. 10.1038/nmeth.2772 [DOI] [PubMed] [Google Scholar]
- 10. Hashimshony T, Wagner F, Sher N, et al. : CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2(3):666–73. 10.1016/j.celrep.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 11. Ramsköld D, Luo S, Wang YC, et al. : Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–82. 10.1038/nbt.2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Picelli S, Björklund ÅK, Faridani OR, et al. : Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–8. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- 13. Bhargava V, Ko P, Willems E, et al. : Quantitative transcriptomics using designed primer-based amplification. Sci Rep. 2013;3:1740. 10.1038/srep01740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sasagawa Y, Nikaido I, Hayashi T, et al. : Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14(4):R31. 10.1186/gb-2013-14-4-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura T, Yabuta Y, Okamoto I, et al. : SC3-seq: a method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res. 2015;43(9):e60. 10.1093/nar/gkv134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. : Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776–9. 10.1126/science.1247651 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Fan HC, Fu GK, Fodor SP: Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347(6222):1258367. 10.1126/science.1258367 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Macosko EZ, Basu A, Satija R, et al. : Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–14. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Klein AM, Mazutis L, Akartuna I, et al. : Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–201. 10.1016/j.cell.2015.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Fan X, Zhang X, Wu X, et al. : Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015;16(1):148. 10.1186/s13059-015-0706-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang Y, Norris MH, Zarzycki-Siek J, et al. : Transcript amplification from single bacterium for transcriptome analysis. Genome Res. 2011;21(6):925–35. 10.1101/gr.116103.110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Kolodziejczyk AA, Kim JK, Svensson V, et al. : The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58(4):610–20. 10.1016/j.molcel.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 23. Wang Y, Navin NE: Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58(4):598–609. 10.1016/j.molcel.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hug H, Schuler R: Measurement of the number of molecules of a single mRNA species in a complex mRNA preparation. J Theor Biol. 2003;221(4):615–24. 10.1006/jtbi.2003.3211 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Casbon JA, Osborne RJ, Brenner S, et al. : A method for counting PCR template molecules with application to next-generation sequencing. Nucleic Acids Res. 2011;39(12):e81. 10.1093/nar/gkr217 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Kivioja T, Vähärautio A, Karlsson K, et al. : Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2012;9(1):72–4. 10.1038/nmeth.1778 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Shiroguchi K, Jia TZ, Sims PA, et al. : Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci U S A. 2012;109(4):1347–52. 10.1073/pnas.1118018109 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Fu GK, Hu J, Wang PH, et al. : Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc Natl Acad Sci U S A. 2011;108(22):9026–31. 10.1073/pnas.1017621108 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Fu GK, Wilhelmy J, Stern D, et al. : Digital encoding of cellular mRNAs enabling precise and absolute gene expression measurement by single-molecule counting. Anal Chem. 2014;86(6):2867–70. 10.1021/ac500459p [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Jiang L, Schlesinger F, Davis CA, et al. : Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21(9):1543–51. 10.1101/gr.121095.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stegle O, Teichmann SA, Marioni JC: Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16(3):133–45. 10.1038/nrg3833 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Deng Q, Ramsköld D, Reinius B, et al. : Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343(6167):193–6. 10.1126/science.1245316 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Saliba AE, Westermann AJ, Gorski SA, et al. : Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42(14):8845–60. 10.1093/nar/gku555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marinov GK, Williams BA, McCue K, et al. : From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24(3):496–510. 10.1101/gr.161034.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brennecke P, Anders S, Kim JK, et al. : Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10(11):1093–5. 10.1038/nmeth.2645 [DOI] [PubMed] [Google Scholar]
- 36. Grün D, Kester L, van Oudenaarden A: Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11(6):637–40. 10.1038/nmeth.2930 [DOI] [PubMed] [Google Scholar]
- 37. Ding B, Zheng L, Zhu Y, et al. : Normalization and noise reduction for single cell RNA-seq experiments. Bioinformatics. 2015;31(13):2225–7. 10.1093/bioinformatics/btv122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lovatt D, Ruble BK, Lee J, et al. : Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11(2):190–6. 10.1038/nmeth.2804 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Grindberg RV, Yee-Greenbaum JL, McConnell MJ, et al. : RNA-sequencing from single nuclei. Proc Natl Acad Sci U S A. 2013;110(49):19802–7. 10.1073/pnas.1319700110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Grindberg RV, et al. : Using single nuclei for RNA-Seq to capture the transcriptome of postmortem neurons. Nat Protoc.[in press]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Satija R, Farrell JA, Gennert D, et al. : Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Achim K, Pettit JB, Saraiva LR, et al. : High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;33(5):503–9. 10.1038/nbt.3209 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Ke R, Mignardi M, Pacureanu A, et al. : In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10(9):857–60. 10.1038/nmeth.2563 [DOI] [PubMed] [Google Scholar]
- 44. Lee JH, Daugharthy ER, Scheiman J, et al. : Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343(6177):1360–3. 10.1126/science.1250212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee JH, Daugharthy ER, Scheiman J, et al. : Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10(3):442–58. 10.1038/nprot.2014.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He L, Hannon GJ: MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 47. Cai Y, Yu X, Hu S, et al. : A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–54. 10.1016/S1672-0229(08)60044-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang L, Duff MO, Graveley BR, et al. : Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12(2):R16. 10.1186/gb-2011-12-2-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Memczak S, Jens M, Elefsinioti A, et al. : Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Jeck WR, Sharpless NE: Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Armour CD, Castle JC, Chen R, et al. : Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods. 2009;6(9):647–9. 10.1038/nmeth.1360 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Shalek AK, Satija R, Shuga J, et al. : Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510(7505):363–9. 10.1038/nature13437 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Battich N, Stoeger T, Pelkmans L: Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Methods. 2013;10(11):1127–33. 10.1038/nmeth.2657 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Chen KH, Boettiger AN, Moffitt JR, et al. : RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233):aaa6090. 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Buxbaum AR, Haimovich G, Singer RH: In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16(2):95–109. 10.1038/nrm3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lubeck E, Cai L: Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Methods. 2012;9(7):743–8. 10.1038/nmeth.2069 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Evrony GD, Cai X, Lee E, et al. : Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151(3):483–96. 10.1016/j.cell.2012.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Falconer E, Hills M, Naumann U, et al. : DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat Methods. 2012;9(11):1107–12. 10.1038/nmeth.2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zong C, Lu S, Chapman AR, et al. : Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–6. 10.1126/science.1229164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McConnell MJ, Lindberg MR, Brennand KJ, et al. : Mosaic copy number variation in human neurons. Science. 2013;342(6158):632–7. 10.1126/science.1243472 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Navin N, Kendall J, Troge J, et al. : Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Dey SS, Kester L, Spanjaard B, et al. : Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol. 2015;33(3):285–9. 10.1038/nbt.3129 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Macaulay IC, Haerty W, Kumar P, et al. : G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12(6):519–22. 10.1038/nmeth.3370 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Guo H, Zhu P, Wu X, et al. : Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23(12):2126–35. 10.1101/gr.161679.113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Smallwood SA, Lee HJ, Angermueller C, et al. : Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11(8):817–20. 10.1038/nmeth.3035 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Farlik M, Sheffield NC, Nuzzo A, et al. : Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10(8):1386–97. 10.1016/j.celrep.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Gravina S, Ganapathi S, Vijg J: Single-cell, locus-specific bisulfite sequencing (SLBS) for direct detection of epimutations in DNA methylation patterns. Nucleic Acids Res. 2015;43(14):e93. 10.1093/nar/gkv366 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Cusanovich DA, Daza R, Adey A, et al. : Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348(6237):910–4. 10.1126/science.aab1601 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Buenrostro JD, Wu B, Litzenburger UM, et al. : Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523(7561):486–90. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 70. Han L, Qiu P, Zeng Z, et al. : Single-cell mass cytometry reveals intracellular survival/proliferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry A. 2015;87(4):346–56. 10.1002/cyto.a.22628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Amir el-AD, Davis KL, Tadmor MD, et al. : viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–52. 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tang F, Barbacioru C, Bao S, et al. : Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6(5):468–78. 10.1016/j.stem.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Buganim Y, Faddah DA, Cheng AW, et al. : Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150(6):1209–22. 10.1016/j.cell.2012.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Kumar RM, Cahan P, Shalek AK, et al. : Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature. 2014;516(7529):56–61. 10.1038/nature13920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shalek AK, Satija R, Adiconis X, et al. : Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–40. 10.1038/nature12172 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. de Vargas Roditi L, Claassen M: Computational and experimental single cell biology techniques for the definition of cell type heterogeneity, interplay and intracellular dynamics. Curr Opin Biotechnol. 2015;34:9–15. 10.1016/j.copbio.2014.10.010 [DOI] [PubMed] [Google Scholar]
- 77. Grün D, Lyubimova A, Kester L, et al. : Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–5. 10.1038/nature14966 [DOI] [PubMed] [Google Scholar]
- 78. Treutlein B, Brownfield DG, Wu AR, et al. : Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–5. 10.1038/nature13173 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Luo Y, Coskun V, Liang A, et al. : Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 2015;161(5):1175–86. 10.1016/j.cell.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Llorens-Bobadilla E, Zhao S, Baser A, et al. : Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17(3):329–40. 10.1016/j.stem.2015.07.002 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Zeisel A, Muñoz-Manchado AB, Codeluppi S, et al. : Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–42. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Pollen AA, Nowakowski TJ, Shuga J, et al. : Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32(10):1053–8. 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Usoskin D, Furlan A, Islam S, et al. : Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–53. 10.1038/nn.3881 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Mahata B, Zhang X, Kolodziejczyk AA, et al. : Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep. 2014;7(4):1130–42. 10.1016/j.celrep.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Spaethling JM, Sanchez-Alavez M, Lee J, et al. : Single-cell transcriptomics and functional target validation of brown adipocytes show their complex roles in metabolic homeostasis. FASEB J. 2016;30(1):81–92. 10.1096/fj.15-273797 [DOI] [PubMed] [Google Scholar]
- 86. Patel AP, Tirosh I, Trombetta JJ, et al. : Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. Min JW, Kim WJ, Han JA, et al. : Identification of Distinct Tumor Subpopulations in Lung Adenocarcinoma via Single-Cell RNA-seq. PLoS One. 2015;10(8):e0135817. 10.1371/journal.pone.0135817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wills QF, Mead AJ: Application of single-cell genomics in cancer: promise and challenges. Hum Mol Genet. 2015;24(R1):R74–84. 10.1093/hmg/ddv235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Leng N, Chu LF, Barry C, et al. : Oscope identifies oscillatory genes in unsynchronized single-cell RNA-seq experiments. Nat Methods. 2015;12(10):947–50. 10.1038/nmeth.3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Avraham R, Haseley N, Brown D, et al. : Pathogen Cell-to-Cell Variability Drives Heterogeneity in Host Immune Responses. Cell. 2015;162(6):1309–21. 10.1016/j.cell.2015.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bendall SC, Davis KL, Amir el-AD, et al. : Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157(3):714–25. 10.1016/j.cell.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shin J, Berg DA, Zhu Y, et al. : Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17(3):360–72. 10.1016/j.stem.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 93. Moignard V, Woodhouse S, Haghverdi L, et al. : Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol. 2015;33(3):269–76. 10.1038/nbt.3154 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 94. Buettner F, Natarajan KN, Casale FP, et al. : Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155–60. 10.1038/nbt.3102 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 95. Suter DM, Molina N, Gatfield D, et al. : Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332(6028):472–4. 10.1126/science.1198817 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Kim JK, Marioni JC: Inferring the kinetics of stochastic gene expression from single-cell RNA-sequencing data. Genome Biol. 2013;14(1):R7. 10.1186/gb-2013-14-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Raj A, van Oudenaarden A: Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135(2):216–26. 10.1016/j.cell.2008.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 98. Hey KL, Momiji H, Featherstone K, et al. : A stochastic transcriptional switch model for single cell imaging data. Biostatistics. 2015;16(4):655–69. 10.1093/biostatistics/kxv010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Daigle BJ, Jr, Soltani M, Petzold LR, et al. : Inferring single-cell gene expression mechanisms using stochastic simulation. Bioinformatics. 2015;31(9):1428–35. 10.1093/bioinformatics/btv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Padovan-Merhar O, Raj A: Using variability in gene expression as a tool for studying gene regulation. Wiley Interdiscip Rev Syst Biol Med. 2013;5(6):751–9. 10.1002/wsbm.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hurley D, Araki H, Tamada Y, et al. : Gene network inference and visualization tools for biologists: application to new human transcriptome datasets. Nucleic Acids Res. 2012;40(6):2377–98. 10.1093/nar/gkr902 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 102. Bansal M, Belcastro V, Ambesi-Impiombato A, et al. : How to infer gene networks from expression profiles. Mol Syst Biol. 2007;3:78. 10.1038/msb4100120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xue Z, Huang K, Cai C, et al. : Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500(7464):593–7. 10.1038/nature12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kharchenko PV, Silberstein L, Scadden DT: Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11(7):740–2. 10.1038/nmeth.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. McDavid A, Finak G, Chattopadyay PK, et al. : Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29(4):461–7. 10.1093/bioinformatics/bts714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pierson E, Yau C: ZIFA: Dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol. 2015;16:241. 10.1186/s13059-015-0805-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang YX, Huang H: Review on statistical methods for gene network reconstruction using expression data. J Theor Biol. 2014;362:53–61. 10.1016/j.jtbi.2014.03.040 [DOI] [PubMed] [Google Scholar]
- 108. Stark J, Brewer D, Barenco M, et al. : Reconstructing gene networks: what are the limits? Biochem Soc Trans. 2003;31(Pt 6):1519–25. 10.1042/bst0311519 [DOI] [PubMed] [Google Scholar]
- 109. Geier F, Timmer J, Fleck C: Reconstructing gene-regulatory networks from time series, knock-out data, and prior knowledge. BMC Syst Biol. 2007;1:11. 10.1186/1752-0509-1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Angelini C, Costa V: Understanding gene regulatory mechanisms by integrating ChIP-seq and RNA-seq data: statistical solutions to biological problems. Front Cell Dev Biol. 2014;2:51. 10.3389/fcell.2014.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]