Summary
Plant architecture, a complex of the important agronomic traits that determine grain yield, is a primary target of artificial selection of rice domestication and improvement. Some important genes affecting plant architecture and grain yield have been isolated and characterized in recent decades; however, their underlying mechanism remains to be elucidated. Here, we report genetic identification and functional analysis of the PLANT ARCHITECTURE AND YIELD 1 (PAY1) gene in rice, which affects plant architecture and grain yield in rice. Transgenic plants over‐expressing PAY1 had twice the number of grains per panicle and consequently produced nearly 38% more grain yield per plant than control plants. Mechanistically, PAY1 could improve plant architecture via affecting polar auxin transport activity and altering endogenous indole‐3‐acetic acid distribution. Furthermore, introgression of PAY1 into elite rice cultivars, using marker‐assisted background selection, dramatically increased grain yield compared with the recipient parents. Overall, these results demonstrated that PAY1 could be a new beneficial genetic resource for shaping ideal plant architecture and breeding high‐yielding rice varieties.
Keywords: PAY1, plant architecture, grain yield, polar auxin transport, rice, Oryza sativa
Significance Statement
This study describes the identification and functional analysis of the PLANT ARCHITECTURE AND YIELD 1 (PAY 1) gene in rice, which affects plant architecture and grain yield in rice. PAY 1 can optimize plant architecture through altering auxin polar transport and distribution, leading to more desirable plant architecture and increased grain yield in rice.
Introduction
Plant architecture, usually referred to as the three‐dimensional organization of the aerial part of a plant, is mainly determined by factors including branching (tillering) pattern, plant height, leaf shape and arrangement, and inflorescence morphology (Reinhardt and Kuhlemeier, 2002; Wang and Li, 2006). Plant architecture is the best means of identifying a plant species, and is also of major agronomic importance as it determines plant survival ability under environmental stress, the suitability of a plant for cultivation, its harvest index and potential grain yield (Peng et al., 1999; Reinhardt and Kuhlemeier, 2002). During the process of crop domestication and improvement, desirable plant architecture was the main selection direction for obtaining high‐yielding varieties. For example, the Green Revolution led to dramatic increases in worldwide agricultural productivity since the 1960s from cultivation of lodging‐resistant semi‐dwarf varieties of wheat and rice (Peng et al., 1999). Therefore, understanding the mechanism underlying plant architecture will facilitate the breeding of crop varieties with high‐yield potential.
Rice (Oryza sativa L.) is the world's most important cereal crop and feeds half of the world's population. As arable land decreases and global population increases, looking for ways to raise grain yield has become a priority. To meet this challenge, new elite rice varieties with ideal plant architecture that can produce much higher grain yield need to be developed. Rice plant architecture, a comprehensive reflection of important agronomic traits, is mainly determined by tilling pattern, plant height and panicle morphology, and has a decisive effect on grain yield (Wang and Li, 2005, 2008; Xing and Zhang, 2010). In recent years, many important genes/quantitative trait loci controlling plant architecture and grain yield have been isolated and functionally characterized. For example, artificial selection for the PROSTRATE GROWTH 1 (PROG1) mutant during rice domestication led to the transition from the plant architecture of wild rice to that of domesticated rice, resulting in erect growth, greater grain number and higher grain yield (Jin et al., 2008; Tan et al., 2008). The DWARF genes, including D3, D10, D14, D17, D27 and D53, have been shown to be involved in the synthesis or signaling pathway of strigolactones, and influence rice tiller number and plant height (Ishikawa et al., 2005; Zou et al., 2005; Arite et al., 2007, 2009; Lin et al., 2009; Jiang et al., 2013; Zhou et al., 2013). Ghd7 was reported to function in rice growth, development and environmental response, thus regulating rice grain yield, plant height and heading date (Xue et al., 2008; Weng et al., 2014). The IDEAL PLANT ARCHITECTURE1 (IPA1) gene, which encodes OsSPL14 (SOUAMOSA PROMOTER BINDING PROTEIN‐LIKE 14) and is regulated by microRNA (miRNA) OsmiR156, controls rice plant architecture and substantially enhances grain yield (Jiao et al., 2010; Miura et al., 2010). Molecular characterization of genes controlling rice plant architecture and grain yield will not only strengthen our understanding of regulatory mechanisms of these traits, but also aid genetic improvement and breeding of high‐yielding rice.
Here, we report the identification of the PLANT ARCHITECTURE AND YIELD 1 (PAY1) gene in rice, which affects plant architecture and grain yield in rice. Our analyses suggest that PAY1 could affect auxin polar transport and distribution and, as a consequence, optimize plant architecture and increase grain yield in rice. Most interestingly, introduction of PAY1 can further improve plant architecture and significantly increase grain yield in the background of high‐yielding varieties. This study could enhance our understanding of rice plant architecture, and findings concerning PAY1 will be of value for breeding high‐yielding rice.
Results and Discussion
The PAY1 mutant displays pleiotropic phenotypes
To identify new regulators of rice plant architecture, a wild rice introgression line YIL55, which displays short plant height, high tillering, thin stems, fewer grains and low yield, was mutagenized with ethyl methane sulfonate to generate a library for genetic screening of mutants with altered plant architecture. We identified a mutant with greatly changed plant architecture, referred to as PLANT ARCHITECTURE AND YIELD 1 (PAY1). Compared with YIL55, the PAY1 mutant exhibited greater plant height, lower tiller number, smaller tiller angle, thicker stems and larger panicles (Figures 1 and S1). In‐depth analysis revealed that the PAY1 mutant had much longer internodes I–V (Figures 1c and S1a). Microscopy revealed that the elongation of the PAY1 mutant stem was likely mainly due to an increase in cell size (Figure S1b). Furthermore, PAY1 mutant showed compact plant architecture with a narrower tiller angle from jointing stage to filling stage, whereas YIL55 had a tiller‐spreading phenotype with a wider tiller angle (Figure S1c,d). Further observation showed more vascular bundles in stems of the PAY1 mutant than of YIL55 (Figure 1e). Interestingly, statistical analysis indicated that the panicles of PAY1 mutant produced more panicle branches, especially secondary branches (Figure S1e). Most importantly, the PAY1 mutant had significantly more grains per panicle (73.7%, P < 0.01) and grain yield per plant (27.8%, P < 0.01) (Figure 1f).
Figure 1.
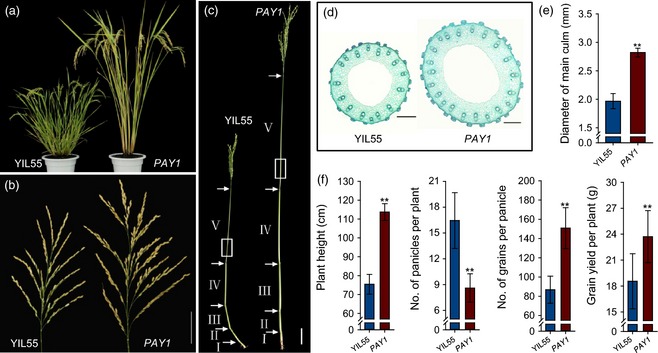
Phenotype of wild‐type (YIL55) and PAY1 mutant. (a) Introgression line YIL55 and the PAY1 mutant at maturity stage. (b) Main panicle of YIL55 and PAY1 mutant. Scale bar, 5 cm. (c) Stem structure of YIL55 and PAY1 mutant. The interval between two arrows showed the length of internode. (d) Longitudinal sections of the fifth internode (marked by white squares in (c)) between YIL55 and PAY1 mutant. Scale bars, 200 μm. (e) The diameter of the fifth internode between YIL55 and PAY1 mutant. Data are means ± standard deviation (SD) (n = 20). (f) Comparison of plant height, number of panicles per plant, grain number per panicle and grain yield per plant between YIL55 and PAY1 mutant plants. Data are means ± SD (n = 30). In (e) and (f), the double asterisks represent a significant difference determined by Student's t‐test at P < 0.01.
Cloning and characterization of PAY1
The F1 plants from the cross between PAY1 and YIL55 showed a similar phenotype to the PAY1 mutant (Figure S2). Of 400 F2 plants, 304 had a PAY1‐like phenotype, suggesting a segregation rate of the PAY1 mutant and YIL55 plants fitting a 3:1 ratio (χ2 = 0.015; P = 0.726). These results indicated that the PAY1 mutant phenotype was controlled by a single dominant gene.
To clone PAY1, we generated an F2 population of 1100 plants derived from the cross between the PAY1 mutant and a japonica variety Nipponbare, and mapped PAY1 between the single sequence repeat markers RM339 and RM223 on the long arm of chromosome 8 (Figure 2a). Upon analyses of an additional 4340 F2 individuals, we further delimited PAY1 within a 51‐kb region between the sp5 and sp7 markers (Figure 2b). Within this region, there were seven predicted genes in the Nipponbare genome (TIGR Rice Genome Annotation Database) (Figure 2c and Table S4). Sequencing the 51‐kb mapping region of wild‐type YIL55 and mutant PAY1 revealed a single nucleotide change, G to A, at position +1244 in exon 4 of LOC_Os08g31470. This resulted in a single amino acid substitution from glutamine (Q) in YIL55 to arginine (R) in the PAY1 mutant (Figures 2d and S3).
Figure 2.
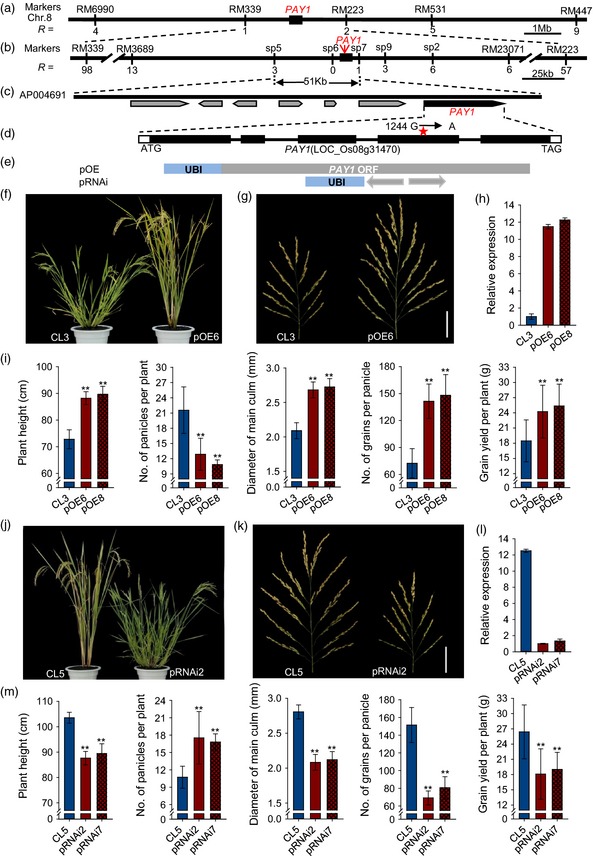
Molecular identification of PAY1. (a) PAY1 was mapped in the interval of RM339 and RM223 on the long arm of chromosome 8. R is the number of recombinants. (b) PAY1 was delimited to a 51‐kb region between the sp5 and sp7 markers. (c) Annotation of the 51‐kb region harboring PAY1 on Nipponbare BAC AP004691. (d) PAY1 structure and the mutation site in PAY1 mutant. The white boxes represent the 5′‐ and 3′‐UTR, the black boxes represent the coding sequences and lines between boxes represent introns. The red asterisk indicates the PAY1 mutation site. (e) Gene structure of PAY1 and constructs used in PAY1 function investigation. pOE contains PAY1 ORF (mutation allele) used for overexpression; pRNAi denoted the RNA interference constructs. UBI is a maize Ubiquitin promoter. (f) The phenotype of control plant (CL3) harboring an empty plasmid and PAY1‐overexpression transgenic plant (pOE6). (g) Comparison of the main panicle between control plant (CL3) and PAY1‐overexpression transgenic plants (pOE6). Scale bar, 5 cm. (h) Relative expression levels of PAY1 in PAY1‐overexpression transgenic plants leaves (pOE6 and pOE8) using RT‐qPCR analysis. (i) Comparison of plant height, number of panicles per plant, diameter of main culm, number of grains per panicle and grain yield per plant between control plant (CL3) and PAY1‐overexpression transgenic plants (pOE6 and pOE8). Data are means ± standard deviation (SD) (n = 30). (j) The phenotype of control plant (CL5) harboring an empty plasmid and RNAi transgenic plant (pRNAi2). (k) Comparison of the main panicle between control (CL5) and RNAi transgenic plants (pRNAi2). Scale bar, 5 cm. (l) Relative expression levels of PAY1 in RNAi transgenic plant leaves (pRNAi2 and pRNAi7). (m) Comparison of plant height, number of panicles per plant, diameter of main culm, number of grains per panicle and grain yield per plant between control (CL5) and RNAi transgenic plants (pRNAi2 and pRNAi7). Data are means ± SD (n = 30). In (i) and (m), the double asterisks represent a significant difference determined by Student's t‐test at P < 0.01.
To verify whether the altered plant architecture was caused by the single nucleotide change in the LOC_Os08g31470 gene, we generated transgenic YIL55 plants with overexpression of LOC_Os08g31470 cDNA of the PAY1 mutant (Figure 2e). Real‐time quantitative PCR (RT‐qPCR) analysis showed that the expression levels of PAY1 were much higher in transgenic than in control plants (Figure 2h). Consistent with increased PAY1 expression, all the tested independent over‐expressing transgenic lines had similar phenotypes to the PAY1 mutant–greater plant height, reduced tillers, smaller tiller angle, thicker culms, more panicle branches, more grains per panicle and enhanced grain yield–compared with the control transgenic plants harboring an empty vector (Figures 2f,g,i and S3a).
We further transformed the PAY1 mutant with the pRNAi construct (Figure 2e). RT‐qPCR analysis showed that the expression levels of PAY1 were down‐regulated in RNAi transgenic compared with control plants (Figure 2l). All tested independent RNAi transgenic lines had more tillers, greater tiller angle and marked reductions in plant height, diameter of culms, panicle branches, grains per panicle and grain yield compared with control transgenic plants (Figures 2j,k,m and S3b).
Both the genetic evidence and results of transformation demonstrated that the LOC_Os08g31470 gene corresponded to PAY1, and it controlled plant architecture and grain yield in rice.
Sequence analysis of 5′‐ and 3′‐RACE (random amplification of cDNA ends) cDNA products indicated that the PAY1 cDNA was 2172‐bp long, with an open reading frame (ORF) of 1773 bp, a 172‐bp 5′‐untranslated region (UTR) and a 227‐bp 3′‐UTR (Figure S4). Further sequence analysis indicated that PAY1 encoded a protein of 590 amino acids, which contained a peptidase S64 domain, and shared high identities with deduced proteins in other plant species including monocots and dicots, such as maize, wheat, barley, tomato and Arabidopsis (Figures S4 and S5). The transient expression experiment in tobacco epidermal cells showed that the PAY1–GFP (green fluorescent protein) fusion protein was specifically localized in the nucleus (Figure 3a).
Figure 3.
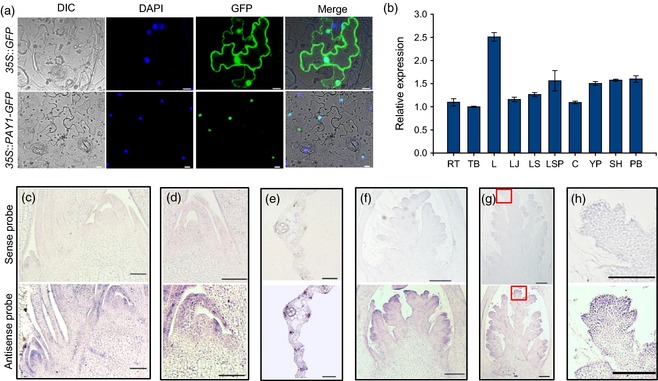
Subcellular localization and expression pattern analysis of PAY1. (a) PAY1 subcellular localization. 35S::GFP (top) and 35S::PAY1–GFP fusion gene (bottom) were transiently expressed in tobacco epidermal cells. The PAY1–GFP fusion protein was exclusively expressed in the nucleus. Scale bars, 100 μm. (b) The relative expression levels of PAY1 in various organs. RT, root; TB, tiller base; L, leaf; LJ, lamina joint; LS, leaf sheath; LSP, leaf‐sheath pulvinus; C, culm; YP, young panicle; SH, spikelet hull; PB, panicle branch. (c–h) PAY1 expression patterns revealed by mRNA in situ hybridization. The top panels are sense probes as negative controls, and the bottom panels are antisense probes. (h) was enlarged from (g) marked by red square. Scale bars, 200 μm.
Expression patterns of PAY1
To study the tissue specificity of PAY1 expression, we introduced a construct consisting of a 1911‐bp fragment of the PAY1 promoter region fused to the GUS reporter gene into the japonica cultivar Zhonghua17. GUS staining of transgenic plants indicated that PAY1 was expressed in almost all organs, including root, leaf, leaf sheath, node, culm, leaf‐sheath pulvinus, spikelet hull and panicle branch (Figure S6a–e). Consistent with GUS staining data, RT‐qPCR analysis also showed that PAY1 was ubiquitously expressed in various rice organs, especially leaves (Figure 3b). RNA in situ hybridization showed that PAY1 was predominantly expressed in the leaf primordia, shoot apical meristem, tiller buds, the primordia of primary and secondary branches, and developing spikelets (Figure 3c–h), suggesting that PAY1 may play critical roles in outgrowth of tiller buds, panicle development and grain production in rice.
The PAY1 mutant shows reduced polar auxin transport activity
Several decades of studies in different plant species have fully demonstrated that auxin is a determinant of plant architecture, and polar auxin transport (PAT) plays a key role in the regulation of many aspects of plant growth and development (Leyser, 2003; Gallavotti, 2013). To investigate whether PAY1 was involved in PAT, we compared the basipetal and acropetal indole‐3‐acetic acid (IAA) transport in etiolated coleoptiles of wild‐type and PAY1 plants. The basipetal IAA transport in PAY1 was reduced to approximately 41% of that in the wild‐type, whereas basipetal transport of 3H‐IAA treated with the PAT inhibitor N‐1‐naphthylphtalamic acid (NPA) showed no differences between wild‐type and mutant plants (Figure 4a).
Figure 4.
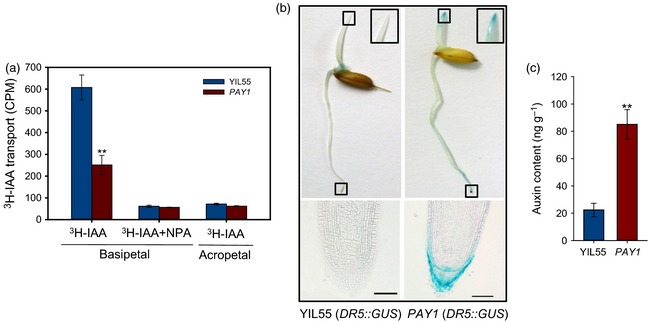
Comparison of auxin biosynthesis and transport between wild‐type (YIL55) and PAY1 plants. (a) Comparison of PAT between YIL55 and PAY1 mutant in dark‐grown coleoptiles. The acropetal auxin transport measurement was used as a negative control. Values are means ± standard deviation (SD) (n = 5). (b) DR5::GUS expression patterns in dark‐grown coleoptiles and roots. Left, DR5::GUS within YIL55 background; right, DR5::GUS within PAY1 mutant background. The smaller square in the upper half of panels showed stem apexes of YIL55 and PAY1 plants, while the larger squares immediately next to it show an enlarged drawing, respectively. The panels below are cross‐sections of the root tips shown in panels above, respectively. Scale bars, 100 μm. (c) Comparison of auxin content in the tip of dark‐grown coleoptiles between YIL55 and PAY1 mutant. Coleoptile fragments 2 mm in length from the tip were used for detection. In (a) and (c), the double asterisks represent a significant difference determined by Student's t‐test at P < 0.01.
To determine whether the reduced basipetal IAA transport activity caused by the mutation in PAY1 affected the distribution of endogenous IAA, we investigated the endogenous auxin distribution by analyzing GUS expression levels in the transgenic plants for the auxin reporter DR5::GUS. Higher GUS expression was detected in the apexes of coleoptiles and root tips in the PAY1 mutant, but not in YIL55. GUS expression in YIL55 was mainly enriched in the basal part of coleoptiles (Figure 4b). Consistent with the DR5::GUS expression pattern, the endogenous auxin (IAA) in the apexes of coleoptiles in PAY1 mutant was nearly four‐fold that in YIL55 (Figure 4c). These results indicated that reduced basipetal IAA transport activity led to altered endogenous IAA distribution in PAY1 mutant plants, and so affected plant architecture.
PAY1 could potentially be used for high‐yield breeding
To evaluate the PAY1 potential application for optimizing rice plant architecture and increasing grain yields, we introduced the PAY1 allele into Teqing (TQ) and 9311, two high‐yielding indica cultivars who harbor the same pay1 allele to YIL55, to generate the near isogenic lines (NILs) TQ‐PAY1‐NIL and 9311‐PAY1‐NIL, respectively. Compared with recipient plants (TQ and 9311), both TQ‐PAY1‐NIL and 9311‐PAY1‐NIL showed enhanced apical dominance (greater plant height, less tiller number, smaller tiller angle, thicker culms, more secondary branches and larger panicles) and significantly increased grain number per panicle (57.9 and 40.5%, respectively, P < 0.01) and grain yield per plant (16.8 and 23.2%, respectively, P < 0.05) (Figures 5 and S7). These results indicated that PAY1 could further enhance the grain yield of currently cultivated rice varieties, and that PAY1 is a useful allele for shaping ideal plant architecture and increasing grain yield in rice.
Figure 5.
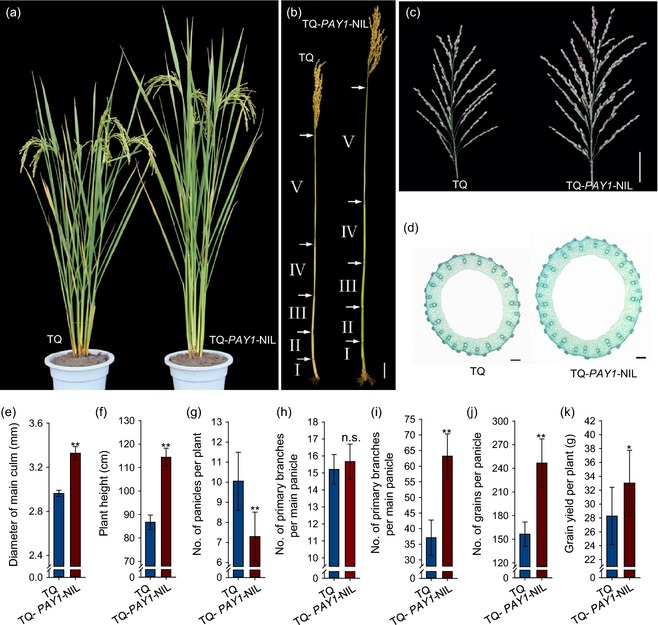
Phenotype of Teqing (TQ) and TQ‐PAY1‐ NIL plants. (a) Gross morphologies of TQ and TQ‐PAY1‐ NIL plants at the maturity stage. (b) Stem structure of TQ (left) and TQ‐PAY1‐ NIL (right) plants. The interval between two arrows showed the length of internode. (c) Comparison of the main panicle between TQ and TQ‐PAY1‐ NIL plants. Scale bar, 5 cm. (d) Cross‐sections of the fifth internode between TQ and TQ‐PAY1‐ NIL plants. Scale bars, 200 μm. (e) The diameter of the fifth internode between TQ and TQ‐PAY1‐ NIL plants. Data are means ± standard deviation (SD) (n = 20). (f–k) Comparison of plant height (f), number of panicles per plant (g), number of primary branches per main panicle (h), number of primary branches per main panicle (i), grain number per panicle (j) and grain yield per plant (k) between TQ and TQ‐PAY1‐NIL plants. Data are means ± SD (n = 30). **, Significant at 1% level; *, significant at 5% level; n.s., not significant.
In this study, we characterized the PAY1 gene, which improves plant architecture and enhances grain yield in rice. The dominant PAY1 gene could significantly optimize plant architecture and increase grain yield in rice, due to altered auxin polar transport and distribution. It is well known that auxin is a central regulator of plant growth and plays a critical role in a wide variety of developmental processes, including embryogenesis, maintenance of apical dominance and formation of lateral organs (Leyser, 2003; McSteen and Leyser, 2005; Vanneste and Friml, 2009). Auxin, produced mainly in the shoot apex, young leaves and root apex, is transported basipetally in the PAT stream, and then inhibits the growth of axillary buds (McSteen and Leyser, 2005; Zhao, 2010; Mashiguchi et al., 2011). Disruption of the auxin gradient, by changing auxin biosynthesis, transport or signaling, will alter organ growth patterns and change plant architecture. Bennett et al. (2006) reported that the branching phenotype of max mutants in Arabidopsis was caused by increased auxin transport capacity. Arabidopsis MAP KINAS EKINASE7 (MKK7) negatively regulates PAT, thus affecting the formation of plant architecture (Dai et al., 2006). Loss of function of LAZY1 enhanced PAT and affected lateral auxin transport, thus altering endogenous auxin distribution and resulting in reduced shoot gravitropism and a tiller‐spreading phenotype (Li et al., 2007). The enhanced PAT in d27 mutant led to increased shoot branching and reduced plant height in rice (Lin et al., 2009). Consistently, the results of the present study revealed that PAY1 reduced PAT and altered endogenous auxin distribution, leading to a phenotype of enhanced apical dominance with reduced tiller number and angle, and increased plant height in the PAY1 mutant. The identification of PAY1 strengthens our understanding of the mechanism by which auxin inhibits the growth of lateral branches and regulates plant architecture in rice.
Improving grain yield has been the primary goal of rice breeding. Rice plant architecture – mainly determined by plant height, tiller number and angle, and panicle morphology – plays a vital role in grain yield formation (Wang and Li, 2008; Xing and Zhang, 2010). Although tremendous progress has been made in characterizing the genes controlling plant architecture and grain yield in rice, the molecular mechanisms for generating ideal plant architecture and increasing grain yield remain to be elucidated. In the present study, the PAY1 mutant showed characteristics of ideal plant architecture including reduced tiller number and angle, increased plant height, stem thickness and grain number per panicle compared with the wild‐type YIL55. The NILs with Teqing or 9311 genetic background both demonstrated that PAY1 could shape better plant architecture and enhance grain yield of rice. Overall, our findings show that PAY1 is an important dominant regulator of rice plant architecture and would be useful for rice genetic improvement and breeding of new varieties with increased grain yield, thus contributing to global food security.
Experimental Procedures
Plant materials
YIL55, the PAY1 mutant and the segregation population for mapping were grown in paddy fields in Beijing in summer or in Hainan Province in winter. The NILs were generated using continuous backcrossing between PAY1, as the donor, and elite indica varieties Teqing and 9311, as the recurrent parent. Field‐grown selfed progeny of third backcross (BC3F3)‐generation plants were used for phenotype analysis.
Primers
The primers used in this study are listed in Tables S1–S3.
Genetic confirmation
The entire coding sequence of PAY1 cDNA (mutation allele), a 1773‐bp fragment, was inserted into the vector pCAMBIA1301 driven by the maize Ubiquitin promoter to form the overexpression construct pOE. The construct was introduced to Agrobacterium tumefaciens strain EHA105 and subsequently transferred into YIL55. There were 18 independent transgenic lines (T3 generation) harvested, and two lines (pOE6 and pOE8) were used for phenotypic evaluation. The construct pRNAi was generated by the insertion of a hairpin sequence with two 356‐bp cDNA inverted repeat fragments targeting the sequence of PAY1 into pJL1460, driven by the maize Ubiquitin promoter. We harvested 13 independent RNAi transgenic lines (T3 generation) and phenotypically evaluated two of these: pRNAi2 and pRNAi7.
Subcellular localization of PAY1
The construct p35S::PAY1–GFP contained PAY1 fused with GFP, driven by a 35S promoter, and introduced into leaf cells of Nicotiana benthamiana (a wild Australian tobacco) using Agrobacterium tumefaciens‐mediated transient transformation by infiltration (Sparkes et al., 2006). Then GFP was visualized in leaf epidermal cells using a Carl Zeiss LSM510 Meta confocal laser scanning microscope (Carl Zeiss AG, http://www.zeiss.com/).
Tissue localization and RNA in situ hybridization
The pPAY1::GUS construct was transformed into the japonica variety Zhonghua17, and the resulting transgenic plants were analyzed by a standard GUS staining assay (Scarpella et al., 2003). RNA in situ hybridization assays were performed according to Javelle et al. (2011) with some modifications. Shoot apexes and leaves of rice seedlings at the four‐leaf stage were fixed with 4% (w/v) paraformaldehyde at 4°C overnight, followed by a series of dehydration and infiltration, and embedded in paraffin (Tissue Path Paraplast Plus, Fisher Scientific). The tissues were sliced into 8‐mm sections with a microtome (Leica RM2155; Leica, http://www.leica.com). The 505‐bp 3′‐region of PAY1 was subcloned into the pMD18‐T vector and used as templates to generate sense and antisense RNA probes. Digoxigenin‐labeled RNA probes were prepared using a DIG Northern Starter Kit (Cat. No. 2039672; Roche, http://www.roche.com), according to the manufacturer's instructions.
RT‐qPCR and RACE analysis
For RT‐qPCR analysis, total RNA was extracted from various samples using TRIzol reagent (Invitrogen, http://www.lifetechnologies.com) and was purified using the RNeasy Micro Kit (Qiagen, http://www.qiagen.com). First‐strand cDNA was synthesized to using oligo(dT)18 primer (TaKaRa) and SuperScript® III Reverse Transcriptase (Invitrogen) from 3 μg of total RNA. The expression levels of PAY1 and other genes were analyzed using a CFX96 Real‐Time System (Bio‐Rad, http://www.bio-rad.com) and rice Ubiquitin gene as an internal control. Each set of experiments was repeated three times, and the relative quantification method () used to evaluate quantitative variation. 5′‐ and 3′‐RACE was carried out using a 5′‐Full RACE Kit with tobacco acid pyrophosphatase (TAP) and 3′‐Full RACE Core Set with PrimeScript™ reverse transcriptase (RT) (TaKaRa, http://www.takara.com) following the manufacturer's instructions.
PAT assay
PAT assays were performed according to Li et al. (2007) with some minor modifications. Five groups of six 5‐day‐old dark‐grown coleoptile segments (0.2 cm away from the tip) of 2 cm length were used for the assay. The segments were incubated in 1/2 Murashige and Skoog (MS; pH 5.8) liquid medium with shaking at 100 rpm for 2 h to remove endogenous IAA. The apical or basal ends of the coleoptile segments (for basipetal or acropetal transport assays, respectively) were then placed in a 200‐μl microcentrifuge tube with one end submerged in 10 μl of ½MS liquid medium containing 0.35% phytogel, 500 nm [3H] IAA and 500 nm free IAA in darkness at room temperature for 2 h. NPA was applied to the media as indicated. The unsubmerged ends of the segments (0.5 cm in length) were excised and washed three times with ½MS liquid medium. After 20 h incubation in 2 ml of scintillation liquid, the radioactivity of each section was counted by a liquid scintillation counter (1450 MicroBeta TriLux; Perkin‐Elmer, http://www.perkinelmer.com).
Measurement of free IAA content
IAA extraction and measurement were performed using liquid chromatography‐mass spectrometry (LC‐MS) according to Kowalczyk and Sandberg (2001) with some modifications. The tips (0.2 cm in length) of 3‐day‐old dark‐grown coleoptiles were harvested and used for the assay. After extraction and purification, the samples were subjected to LC‐MS analysis using a liquid scintillation counter (1450 MicroBeta TriLux, http://www.perkinelmer.com).
Accession numbers
Data deposition: The PAY1 sequence reported in this paper was deposited in the GenBank database accession no. KP233774 (full‐length cDNA).
Supporting information
Figure S1. Phenotype of wild‐type (YIL55) and PAY1 mutant.
Figure S2. Phenotypic characterization of F1 plants derived from the cross between YIL55 and PAY1 mutant.
Figure S3. Full‐length cDNA of pay1 from wild‐type (YIL55) and the deduced amino acid sequence.
Figure S4. Phenotype of control plant and transgenic plant.
Figure S5. Phylogeny of the PAY1 protein family.
Figure S6. Expression pattern of PAY1.
Figure S7. Phenotype of 9311 and 9311‐PAY1‐NIL plants.
Table S1. Primers used for mapping of PAY1.
Table S2. Primer sequences used to generate constructs.
Table S3. Primers used for RACE, RT‐qPCR and RNA in situ hybridization.
Table S4. Information on predicted genes in the fine mapping region of PAY1.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 91335202, No. 30930057), the China National High‐tech Research and Development (‘863’) Program (No. 2012AA10A301).
References
- Arite, T. , Iwata, H. , Ohshima, K. , Maekawa, M. , Nakajima, M. , Kojima, M. , Sakakibara, H. and Kyozuka, J. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite, T. , Umehara, M. , Ishikawa, S. , Hanada, A. , Maekawa, M. , Yamaguchi, S. and Kyozuka, J. (2009) d14, a strigolactone‐insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424. [DOI] [PubMed] [Google Scholar]
- Bennett, T. , Sieberer, T. , Willett, B. , Booker, J. , Luschnig, C. and Leyser, O. (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16, 553–563. [DOI] [PubMed] [Google Scholar]
- Dai, Y. , Wang, H. , Li, B. , Huang, J. , Liu, X. , Zhou, Y. , Mou, Z. and Li, J. (2006) Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis . Plant Cell, 18, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti, A. (2013) The role of auxin in shaping shoot architecture. J. Exp. Bot. 64, 2593–2608. [DOI] [PubMed] [Google Scholar]
- Ishikawa, S. , Maekawa, M. , Arite, T. , Onishi, K. , Takamure, I. and Kyozuka, J. (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46, 79–86. [DOI] [PubMed] [Google Scholar]
- Javelle, M. , Marco, C.F. and Timmermans, M. (2011) In situ hybridization for the precise localization of transcripts in plants. J. Vis. Exp. 57, e3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Liu, X. , Xiong, G. et al. (2013) DWARF53 acts as a repressor of strigolactone signalling in rice. Nature, 504, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y. , Wang, Y. , Xue, D. et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Jin, J. , Huang, W. , Gao, J.P. , Yang, J. , Shi, M. , Zhu, M.Z. , Luo, D. and Lin, H.X. (2008) Genetic control of rice plant architecture under domestication. Nat. Genet. 40, 1365–1369. [DOI] [PubMed] [Google Scholar]
- Kowalczyk, M. and Sandberg, G. (2001) Quantitative analysis of indole‐3‐acetic acid metabolites in Arabidopsis . Plant Physiol. 127, 1845–1853. [PMC free article] [PubMed] [Google Scholar]
- Leyser, O. (2003) Regulation of shoot branching by auxin. Trends Plant Sci. 8, 541–545. [DOI] [PubMed] [Google Scholar]
- Li, P. , Wang, Y. , Qian, Q. , Fu, Z. , Wang, M. , Zeng, D. , Li, B. , Wang, X. and Li, J. (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17, 402–410. [DOI] [PubMed] [Google Scholar]
- Lin, H. , Wang, R. , Qian, Q. et al. (2009) DWARF27, an iron‐containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell, 21, 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi, K. , Tanaka, K. , Sakai, T. et al. (2011) The main auxin biosynthesis pathway in Arabidopsis . Proc. Natl Acad. Sci. USA, 108, 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen, P. and Leyser, O. (2005) Shoot branching. Annu. Rev. Plant Biol. 56, 353–374. [DOI] [PubMed] [Google Scholar]
- Miura, K. , Ikeda, M. , Matsubara, A. , Song, X.J. , Ito, M. , Asano, K. , Matsuoka, M. , Kitano, H. and Ashikari, M. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Peng, J. , Richards, D.E. , Hartley, N.M. et al. (1999) ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature, 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D. and Kuhlemeier, C. (2002) Plant architecture. EMBO Rep. 3, 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella, E. , Rueb, S. and Meijer, A. (2003) The RADICLELESS1 gene is a required for vascular pattern formation in rice. Development, 130, 645–658. [DOI] [PubMed] [Google Scholar]
- Sparkes, I. , Runions, J. , Kearns, A. and Hawes, C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Tan, L. , Li, X. , Liu, F. et al. (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. [DOI] [PubMed] [Google Scholar]
- Vanneste, S. and Friml, J. (2009) Auxin: a trigger for change in plant development. Cell, 136, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Wang, Y. and Li, J. (2005) The plant architecture of rice (Oryza sativa). Plant Mol. Biol. 59, 75–84. [DOI] [PubMed] [Google Scholar]
- Wang, Y. and Li, J. (2006) Genes controlling plant architecture. Curr. Opin. Biotechnol. 17, 123–129. [DOI] [PubMed] [Google Scholar]
- Wang, Y. and Li, J. (2008) Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. [DOI] [PubMed] [Google Scholar]
- Weng, X. , Wang, L. , Wang, J. , Hu, Y. , Du, H. , Xu, C. , Xing, Y. , Li, X. , Xiao, J. and Zhang, Q. (2014) Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 164, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y. and Zhang, Q. (2010) Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. [DOI] [PubMed] [Google Scholar]
- Xue, W. , Xing, Y. , Weng, X. et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. (2010) Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F. , Lin, Q. , Zhu, L. et al. (2013) D14‐SCFD3‐dependent degradation of D53 regulates strigolactone signaling. Nature, 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Chen, Z. , Zhang, S. , Zhang, W. , Jiang, G. , Zhao, X. , Zhai, W. , Pan, X. and Zhu, L. (2005) Characterizations and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.). Planta, 222, 604–612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phenotype of wild‐type (YIL55) and PAY1 mutant.
Figure S2. Phenotypic characterization of F1 plants derived from the cross between YIL55 and PAY1 mutant.
Figure S3. Full‐length cDNA of pay1 from wild‐type (YIL55) and the deduced amino acid sequence.
Figure S4. Phenotype of control plant and transgenic plant.
Figure S5. Phylogeny of the PAY1 protein family.
Figure S6. Expression pattern of PAY1.
Figure S7. Phenotype of 9311 and 9311‐PAY1‐NIL plants.
Table S1. Primers used for mapping of PAY1.
Table S2. Primer sequences used to generate constructs.
Table S3. Primers used for RACE, RT‐qPCR and RNA in situ hybridization.
Table S4. Information on predicted genes in the fine mapping region of PAY1.


