Abstract
Inteins, also called protein introns, are self-splicing mobile elements found in all domains of life. A bioinformatic survey of genomic data highlights a biased distribution of inteins among functional categories of proteins in both bacteria and archaea, with a strong preference for a single network of functions containing replisome proteins. Many nonorthologous, functionally equivalent replicative proteins in bacteria and archaea carry inteins, suggesting a selective retention of inteins in proteins of particular functions across domains of life. Inteins cluster not only in proteins with related roles but also in specific functional units of those proteins, like ATPase domains. This peculiar bias does not fully fit the models describing inteins exclusively as parasitic elements. In such models, evolutionary dynamics of inteins is viewed primarily through their mobility with the intein homing endonuclease (HEN) as the major factor of intein acquisition and loss. Although the HEN is essential for intein invasion and spread in populations, HEN dynamics does not explain the observed biased distribution of inteins among proteins in specific functional categories. We propose that the protein splicing domain of the intein can act as an environmental sensor that adapts to a particular niche and could increase the chance of the intein becoming fixed in a population. We argue that selective retention of some inteins might be beneficial under certain environmental stresses, to act as panic buttons that reversibly inhibit specific networks, consistent with the observed intein distribution.
Keywords: evolution, replisome, Clusters of Orthologous Groups, ATPases, replicative helicase
Introduction
Inteins are intervening sequences that are transcribed and translated with flanking host protein sequences (exteins) and then autocatalytically excised in a process called protein splicing (Saleh and Perler 2006). There is much intrigue associated with intein distribution, dissemination, and potential biological function. Inteins of several types occur in all three domains of life, in unicellular organisms. Mini-inteins are relatively short and carry only protein domains essential for self-splicing, whereas split inteins are mini-inteins separated into two parts, which associate and ligate their exteins in a protein trans-splicing reaction (Saleh and Perler 2006). In contrast, bifunctional inteins have a homing endonuclease (HEN) domain interrupting the protein splicing domain. The HEN renders an intein mobile by introducing a double-strand break into an intein-less allele (Liu 2000), in a homing process that is similar to that of mobile group I introns (Jacquier and Dujon 1985; Belfort and Roberts 1997). Variations on this homing reaction are thought to be responsible for the horizontal transfer of introns and inteins (Parker et al. 1999; Koufopanou et al. 2002).
Inteins, originally discovered in the VMA1 gene encoding a vacuolar membrane H+-ATPase of the yeast Saccharomyces cerevisiae (Hirata et al. 1990; Kane et al. 1990), are found in a wide range of bacterial and archaeal species, as well as in eukaryotes, viruses, and bacteriophage (Perler et al. 1997; Perler 2002; Pedulla et al. 2003; Poulter et al. 2007; Swithers et al. 2013; Novikova et al. 2014). Many intein sequences from bacteria and archaea reside within proteins involved in DNA replication, recombination, repair, or transcription, such as DNA and RNA polymerases, helicases and topoisomerases (for review, Novikova et al. 2014). In nuclear genomes of eukaryotes, inteins occur primarily in vacuolar membrane H+-ATPases and the PRP8 protein, important for intron splicing (Butler et al. 2006; Poulter et al. 2007; Swithers et al. 2013). Inteins have also been detected in chloroplast genomes of green and cryptophyte algae where they occupy various genes, including those encoding a replicative helicase, the ClpP protease, and RNA polymerases (Wang and Liu 1997; Douglas and Penny 1999; Luo and Hall 2007; Turmel et al. 2009). In viruses and bacteriophages, terminases involved in DNA packaging are a common target for intein insertions (Pedulla et al. 2003; Dassa et al. 2009).
Little is known about intein origin, diversity, evolution or their potential roles in native host cells. Early on, it was noted that many inteins are found primarily in highly conserved domains of essential house-keeping proteins. It was hypothesized that removal of the intein sequence from a conserved domain is difficult as an imprecise deletion would lead to a protein inactivation. As a result, inteins are maintained in these conserved domains (Pietrokovski 2001; Gogarten et al. 2002; Swithers et al. 2009). The observed distribution of inteins is also directly linked to the presence of the HEN. One of the evolutionary models proposes that HEN-containing inteins undergo a “homing cycle” where the inteins spread in the population without intrinsic benefit to the host organism (Gimble and Thorner 1992; Burt and Koufopanou 2004). However, precise intein loss to provide vacant homing targets could be difficult to achieve in the face of HEN-based reinvasion (Barzel, Naor, et al. 2011). In an attempt to resolve this conundrum, it was suggested that inteins can be lost if they impose a fitness cost on their hosts (Butler et al. 2006). Modeling has revealed that population structure can ensure long-term persistence of the inteins without requiring a homing cycle. The long-term coexistence of intein-containing and intein-free alleles was shown to be achievable in completely mixed homogeneous populations (Gogarten and Hilario 2006; Yahara et al. 2009; Barzel, Obolski, et al. 2011). Finally, recent data indicate that inteins can be sensitive to the environment and play regulatory roles at the posttranslational level and thereby could confer selective advantage (Callahan et al. 2011; Topilina, Green, et al. 2015; Topilina, Novikova, et al. 2015). Nevertheless, to fully understand the evolutionary trends that shape intein distribution and diversity, comprehensive sampling and analysis of intein sequences from many species is necessary.
In the present study, we perform an in-depth analysis of thousands of sequenced bacterial, archaeal, and eukaryal species for the presence of inteins. The identified intein-containing proteins are classified and annotated. The distribution of inteins appears to be strongly biased toward proteins involved in DNA replication and repair, as well as nucleotide metabolism and transport. Moreover, the enriched gene ontology (GO) terms among intein-containing genes indicate that diverse ATPases are preferable sites of intein occupancy, even in nonorthologous proteins of equivalent function across the separate archaeal and bacterial domains. The evolutionary and functional implications of this strongly biased intein distribution are examined.
Results
Sporadic Distribution of Inteins in All Three Domains of Life
The growing number of fully sequenced and annotated genomes allows comprehensive computational mining of inteins in diverse species across kingdoms. Such screening provides insight into the overall picture of intein distribution and the evolutionary dynamics of inteins (Novikova et al. 2014). We performed primary intein mining from protein databases available at the National Center for Biotechnology Information (NCBI) (Tatusova et al. 2015), followed by verification of the intein sequence by the presence of the conserved splicing domains. A comparative analysis was then conducted (supplementary fig. S1, Supplementary Material online).
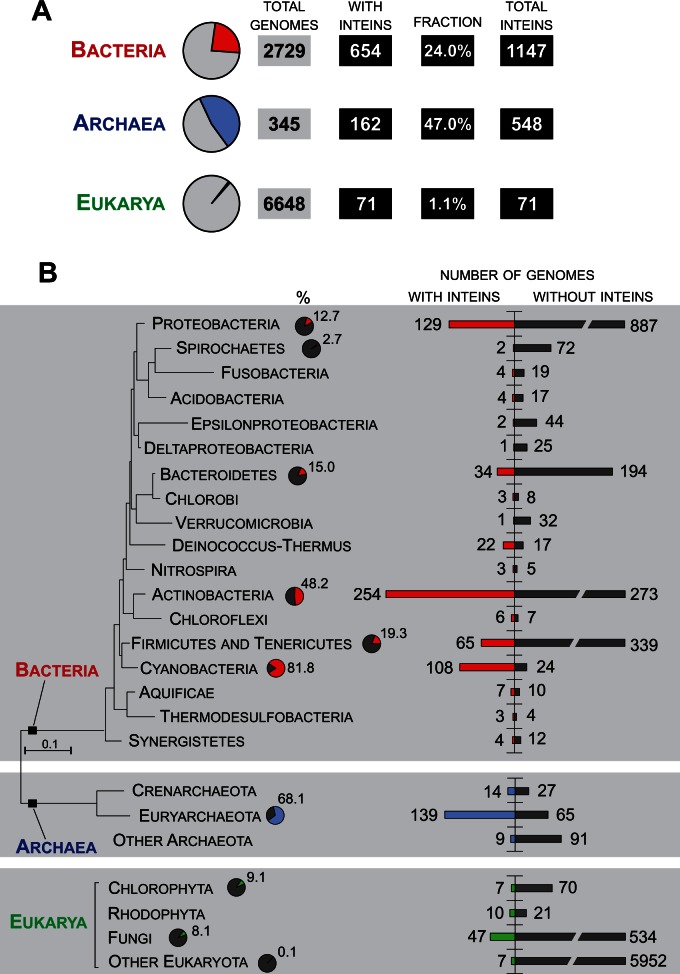
In total, 2,729 bacterial, 345 archaeal, and 6,648 eukaryotic genomes were screened for the presence of the inteins. Of these, 654 bacterial and 162 archaeal species contain inteins; many not previously reported (fig. 1A and supplementary tables S1 and S2, Supplementary Material online). Among the 71 inteins found in eukarya, all had been previously described (supplementary table S3, Supplementary Material online) (Wang and Liu 1997; Butler et al. 2006; Poulter et al. 2007; Turmel, et al. 2009; Swithers, et al. 2013; Novikova et al. 2014). Of bacterial and archaeal species, 24% and 47%, respectively, contain inteins, with many of these harboring more than one, whereas only 1.1% of eukarya contain inteins with only one per genome (fig. 1A). Here, we focus primarily on inteins from bacteria and archaea.
Fig. 1.
Distribution of intein-containing proteins is sporadic. (A) Summary of intein mining. Total number of genomes analyzed, number and fraction of genomes with inteins, and total number of inteins found are indicated. In the present study, the species and a reference genome for all the strains of a species were defined following NCBI RefSeq microbial genome collection procedure (Tatusova et al. 2015). (B) Schematic evolutionary tree for some bacterial and archaeal clades, and list of eukaryal clades. The three domains of life are indicated on the left. Results are from intein mining of genomic sequences. Horizontal bars represent the number of genomes either with (red, blue, or green) or without (black) inteins. The pie charts next to the taxon names indicate the fraction of genomes with inteins for groups with large numbers of species. The bacterial and archaeal evolutionary tree reproduced after “The All-Species Living Tree” Project with modifications (Yarza et al. 2008). Not all intein-containing bacterial clades are shown. The full list of species and their taxonomy is available in supplementary table S1 (bacteria) and table S2 (archaea), Supplementary Material online.
Our screens revealed that the distribution of inteins is sporadic among closely related species and even strains of the same species (Topilina, Novikova, et al. 2015), suggesting a high flux of inteins in the analyzed genomes (Novikova et al. 2014). The sporadic distribution of the inteins was previously reported for the Halobacteria, a class of halophilic Euryarchaeota (Soucy et al. 2014). The bacterial phylogenetic tree shows clusters of bacterial groups with many inteins (fig. 1B). Specifically, Cyanobacteria (81.8% genomes with inteins), Firmicutes and Tenericutes (19.3%), Actinobacteria (48.2%), Proteobacteria (12.7%), and Bacteriodetes (15.0%) all have inteins. Archaeal genomes are replete with inteins, with 68.1% of Euryarchaeota containing at least one intein in the genome.
The number of intein-containing proteins and the number of inteins vary greatly among analyzed genomes and also among investigated phyla (figs 1 and 2; supplementary tables S1 and S2, Supplementary Material online). However, no correlation between genome size and number of protein-coding sequences with inteins or frequency of inteins was observed (fig. 2 and supplementary fig. S2 and table S4, Supplementary Material online). For Proteobacteria, Actinobacteria and Cyanobacteria, the correlation coefficients (r values) are 0.05, 0.31 and 0.15, respectively (fig. 2A), whereas for Euryarchaeota the r value is −0.26 (fig. 2B). Also, no correlation was found between gene locations in genomic sequences and the presence of inteins. The intein-containing genes seem not to be associated with prominent chromosomal landmarks, such as the origin of replication and replication termini (data not shown).
Fig. 2.
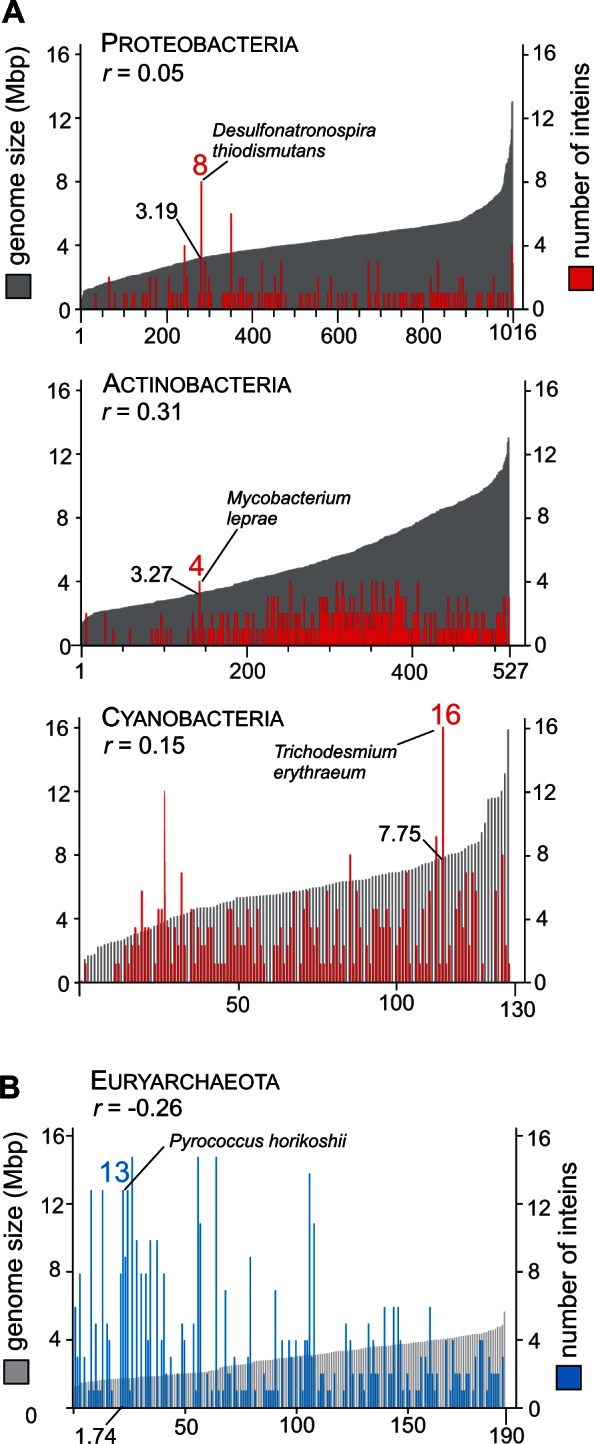
Distribution of inteins does not correlate with genome size. (A) Distribution of the genome sizes and number of inteins in three bacterial clades. The clades are Proteobacteria (1016 species), Actinobacteria (527 species), and Cyanobacteria (130 species). (B) Distribution of the genome sizes and number of inteins in Euryarchaeota (190 species). For (A) and (B), vertical axis of plots represents distribution of the genome sizes (gray) and number of inteins (red and blue) in corresponding species on the horizontal axis. A representative species, genome size, and number of inteins are indicated for each group. No strong correlation between genome size and number of inteins was found, as indicated by correlation coefficient (r) for each group. Distribution of the coding sequences and frequencies of inteins (number of inteins per 1,000 coding sequences) is available in supplementary figure S2, Supplementary Material online. Correlation coefficients are provided in supplementary table S4, Supplementary Material online.
Functional Genomics of Intein-Containing Proteins Reveals a Dramatic Bias
Inteins were found in many different proteins performing diverse functions in the cell (tables 1–3 and supplementary tables S5–S7, Supplementary Material online). There is no unified classification scheme to describe the distribution and diversity of inteins, except for the common practice of distinguishing inteins based on their extein identity and insertion point (Perler et al. 1997). Here, we use the Clusters of Orthologous Groups of proteins (COGs) to classify the proteins with intein insertions based on their function (Tatusov et al. 2003; Galperin et al. 2015). The COGs are divided into functional categories based on the biological processes in which COGs are involved (Tatusov et al. 2003; Galperin et al. 2015).
Table 1.
Top 15 Intein-Containing Proteins in Bacteria.
| Protein | Number of Inteins | COG (category) | Description: Full Name | Distribution |
|---|---|---|---|---|
| DnaB | 317 | COG0305 (L) |
Replicative DNA helicase (P-loop NTPase) |
Actinobacteria, Aquificae, Bacteroidetes, Chloroflexi, Cyanobacteria, Deinococcus–Thermus, Firmicutes, Gemmatimonadetes, Ignavibacteriae, Proteobacteria |
| RNR | 211 | COG0209 (F) | Ribonucleotide reductase | Acidobacteria, Actinobacteria, Aquificae, Armatimonadetes, Bacteroidetes, Chlorobi, Chloroflexi, Cyanobacteria, Deinococcus–Thermus, Firmicutes, Fusobacteria, Nitrospirae, Proteobacteria, Spirochetes, Synergistetes, Thermodesulfobacteria |
| PolIIIα | 116 | COG0587 (L) | DNA polymerase III alpha subunit | Acidobacteria, Actinobacteria, Aquificae, Bacteroidetes, Cyanobacteria, Deinococcus–Thermus, Firmicutes, Proteobacteria, Planctomycetes |
| RecA | 56 | COG0468 (L) |
Recombination protein (P-loop NTPase) |
Actinobacteria, Cyanobacteria, Proteobacteria |
| GyrA | 50 | COG0118 (L) | DNA gyrase, subunit A | Actinobacteria, Aquificae, Chloroflexi, Proteobacteria |
| SplB | 48 | COG1533 (L) | DNA repair photolyase or spore photoproduct lyase | Actinobacteria |
| GyrB | 33 | COG0187 (L) | DNA gyrase, subunit B | Actinobacteria, Chloroflexi, Cyanobacteria, Proteobacteria |
| RtcB | 27 | COG1690 (J) | tRNA-splicing ligase | Actinobacteria, Aquificae, Bacteroidetes, Cyanobacteria, Deinococcus–Thermus |
| TSase | 21 | COG1351 (F) | Thymidylate synthase | Actinobacteria, Cyanobacteria |
| PhoH | 21 | COG1702 (T) |
Phosphate starvation-inducible protein (P-loop NTPase) |
Actinobacteria, Deinococcus–Thermus |
| UvrD | 16 | COG0210 (L) | DNA-dependent ATPase I and helicase II (P-loop NTPase) | Acidobacteria, Actinobacteria, Deinococcus–Thermus, Firmicutes, Proteobacteria |
| PolIIIγ | 15 | COG2812 (L) | DNA polymerase III tau and gamma subunits (P-loop NTPase) | Cyanobacteria |
| HepA | 15 | COG0553 (KL) | SNF2 superfamily II DNA/RNA helicase | Actinobacteria, Bacteroidetes, Cyanobacteria, Deinococcus–Thermus, Verrucomicrobia |
| DnaG | 14 | COG0358 (L) | DNA primase | Actinobacteria, Proteobacteria |
| RecG | 12 | COG1200 (L) |
ATP-dependent DNA helicase (P-loop NTPase) |
Actinobacteria |
Table 2.
Top 15 Intein-Containing Proteins in Archaea.
| Protein | Number of Inteins | COG (category) | Description: Full Name | Distribution |
|---|---|---|---|---|
| MCM | 116 | COG0417 (L) |
Replicative DNA helicase (P-loop NTPase) |
Crenarcheota, Euryarchaeota, Other Archaea |
| PolC/DP2 | 80 | COG1933 (L) | DNA polymerase II large subunit | Euryarchaeota, Other Archaea |
| PolB | 67 | COG0417 (L) | DNA polymerase II small subunit | Euryarchaeota, Other Archaea |
| RNR | 43 | COG0209 (F) | Ribonucleotide reductase | Crenarcheota, Euryarchaeota, Other Archaea |
| RFC-S | 36 | COG0470 (L) |
Replication factor C small subunit (P-loop NTPase) |
Euryarchaeota, Other Archaea |
| Top6B | 15 | COG1389 (L) | Type II DNA topoisomerase VI subunit B | Euryarchaeota, Other Archaea |
| aIF2 | 15 | COG0532 (J) | Translation initiation factor IF-2 subunit gamma (P-loop NTPase) | Euryarchaeota, Other Archaea |
| RNA Pol | 14 | COG0086 (K) | DNA-directed RNA polymerase | Euryarchaeota |
| V-ATPase subunit A | 12 | COG1155 (C) |
V-type ATP synthase subunit A (P-loop NTPase) |
Euryarchaeota, Other Archaea |
| TopA | 11 | COG0550 (L) | archaeal DNA type IA topoisomerase | Euryarchaeota |
| Ski2 | 11 | COG1204 (L) |
Ski2-like DEAD/DEAH box RNA helicases, superfamily II helicase (P-loop NTPase) |
Euryarchaeota |
| RtcB | 10 | COG1690 (J) | tRNA-splicing ligase | Crenarcheota, Euryarchaeota, Other Archaea |
| UDP-GlcDH | 10 | COG1004 (M) | UDP-glucose/GDP-mannose_dehydrogenase | Euryarchaeota |
| LonB | 10 | COG1067 (O) |
ATP-dependent protease LA (P-loop NTPase) |
Euryarchaeota |
| RadA | 9 | COG0468 (L) | Recombination protein (P-loop NTPase) | Crenarcheota, Euryarchaeota |
Table 3.
Intein-Containing Proteins from Bacterial and Archaeal Mobilome and Secretion Systems.
| Protein | Number of Inteins | COG (category) | Description: Full Name | Distribution |
|---|---|---|---|---|
| Bacteria | ||||
| Terminase_6 | 22 | COG5362 (X) | Prophage: phage terminase, large subunit, family Terminase_6 (PF03237) | Firmicutes, Fusobacteria, Proteobacteria |
| Gp6 | 14 | COG2369 (X) | Prophage: phage portal protein, SPP1 Gp6-like protein, phage Mu F gp6 | Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria |
| XtmB | 5 | COG1783 (X) | Prophage: PBSX defective prophage terminase, large subunit | Bacteroidetes, Firmicutes, Synergistetes |
| VirB4 | 5 | COG3451 (U) | T4SS: component of the type IV secretion system virB/virD4 | Firmicutes, Proteobacteria |
| Archaea | ||||
| VirB11 | 21 | COG0630 (NU) | T2SS: P-type DNA transfer ATPase VirB11, component of the type IV secretion system virB/virD4 (P-loop NTPase) | Crenarcheota, Euryarchaeota, Other Archaea |
| CpaF | 5 | COG4962 (U) | T2SS: Flp pilus assembly protein ATPase (P-loop NTPase) | Euryarchaeota |
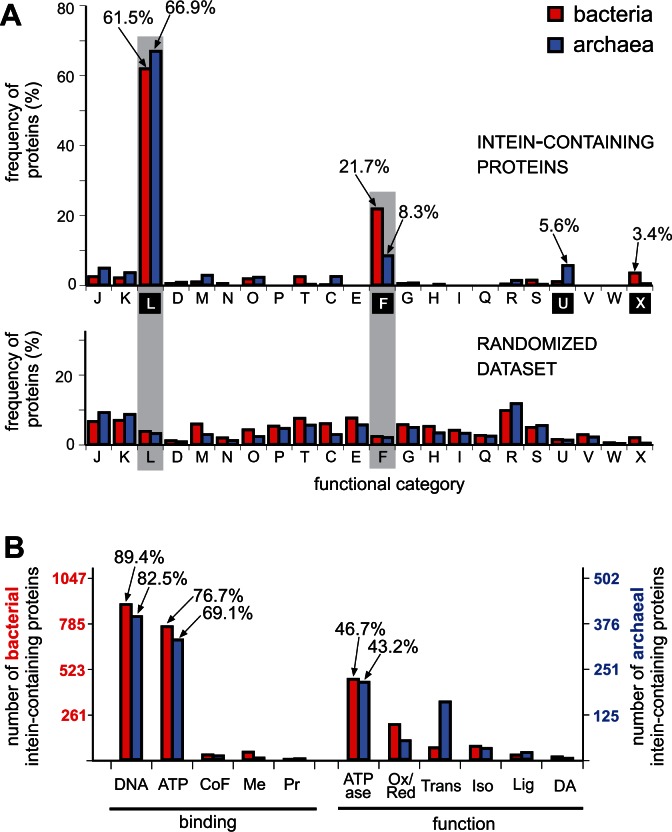
The classification of the intein-containing proteins using COGs indicates a strong bias toward proteins from functional category L (61.5% in bacteria and 66.9% in archaea), which comprise DNA replication, recombination and repair proteins, and category F (21.7% in bacteria and 8.3% in archaea), which includes proteins involved in nucleotide metabolism and transport (fig. 3A, top). Other COG functional categories are considerably less represented among the intein-containing proteins. Remarkably, this pattern holds in both bacteria and archaea despite their divergence (fig. 3A, tables 1 and 2). At the same time, categories L and F are represented by less than 3% of sequences in randomized data sets, which were prepared by randomly sampling sequences from the pools of all predicted protein sequences in all intein-containing genomes (fig. 3a, bottom).
Fig. 3.
Functional genomics of intein-containing proteins. (A) Dominant functional categories of proteins with inteins based on Clusters of Orthologous Groups (COGs). COG annotation is for bacteria (red bars) and archaea (blue bars). The frequency of proteins with inteins/COGs is shown above and the frequency of proteins/COGs for each functional category within randomized data sets of proteins from bacteria and archaea is shown below. Frequency of intein-containing proteins in the top functional categories is indicated next to arrows. Functional category L (replication, recombination and repair) and F (nucleotide transport and metabolism) are dominant among intein-containing proteins in both bacteria and archaea. Functional categories are designated based on conventional classification (Tatusov et al. 1997; Tatusov et al. 2003; Galperin et al. 2015) and are as follows: J, translation, ribosomal structure and biogenesis; K, transcription; D, cell cycle control, cell division, chromosome partitioning; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, post-translational modification, protein turnover, chaperones; P, Inorganic ion transport and metabolism; T, signal transduction mechanisms; C, energy production and conversion; E, amino acid transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; W, extracellular structures; X, mobilome: prophage, transposons. (B) GO enrichment analysis for bacterial and archaeal intein-containing proteins. GO enrichment of 1,047 bacterial (red) and 502 archaeal (blue) intein-containing proteins was performed using WEGO (Ye et al. 2006). Enriched GO terms in binding and molecular function are shown. DNA and ATP binding as well as ATPase activities are the dominant GO terms among the intein-containing proteins from both bacteria and archaea. The percentage of the associated proteins is indicated on the top for dominant categories. CoF, cofactor; Me, metal clusters; Pr, protein; Ox/Red, oxidoreductase; Trans, transferase; Iso, isomerase; Lig, ligase; DA, deaminase.
We utilized the GO annotations, complementary to COGs, to emphasize the larger biological role or process with which inteins are associated (Harris et al. 2004). The GO enrichment analysis is widely used to identify biological processes and has played an important role in the functional analysis of the large data sets derived from high-throughput studies. Using GO enrichment analysis, we were able to obtain valuable insight into the collective biological function underlying the set of intein-containing proteins identified in bacteria and archaea. Interestingly, but not surprisingly, this analysis showed that the majority of intein-containing proteins are characterized as DNA binding proteins with ATP-hydrolyzing activity (fig. 3B). More than 80% of intein-containing proteins from both bacteria (89.4%) and archaea (82.5%) bind DNA (GO:0003677). ATP binding (GO:0005524) is a feature of 76.7% of bacterial and 69.1% of archaeal proteins with inteins.
Functionally Equivalent but Unrelated Proteins of the Replisome Carry Inteins in Bacteria and Achaea
Strikingly, there is not only consistency in the distribution of intein-containing protein COGs among functional categories and GO enrichment between bacteria and archaea but also considerable overlap in actual protein functions, despite the divergent nature of bacteria and archaea, which contain many nonorthologous proteins (fig. 4). Both COG and GO identify many intein-containing proteins as members of cellular processes including DNA replication, recombination, and repair as well as nucleotide metabolism. The replisome itself is an example of the tight network of intein-containing proteins in both bacteria and archaea (fig. 4, tables 1 and 2).
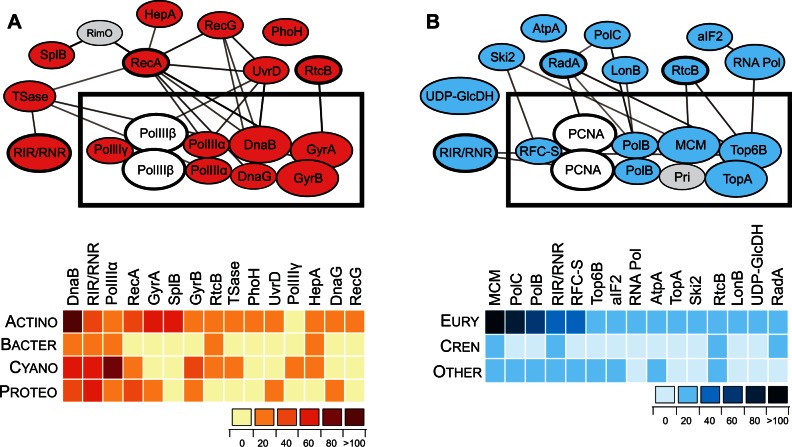
Fig. 4.
Intein-containing proteins are often members of the same complexes and networks. (A) Top 15 bacterial intein-containing proteins, their interactions, and intein distribution. Proteins of the DNA replication fork are boxed at the center of the network. Network was reconstructed using STRING database of known and predicted protein interactions (http://string-db.org/; Szklarczyk et al. 2015). The list of the proteins, their full names, and description is available in table 1. Critical proteins with no inteins are shown in gray as follows: PolIIIβ, DNA polymerase III beta subunit; RimO, 2-methylthioadenine synthetase. The heatmap reflects distribution of the inteins among listed proteins (top) of four bacterial clades (side): Actinobacteria (Actino), Bacteroidetes (Bacter), Cyano (Cyanobacteria), and Proteobacteria (Proteo). For full network and list of the proteins, see supplementary figure S3 and table S5, Supplementary Material online. (B) Top 15 archaeal intein-containing proteins, their interactions, and intein distribution. Proteins of the DNA replication fork are boxed at the center of the network. Network was reconstructed as in panel (A) and the proteins listed in table 2. Critical proteins with no inteins are shown in gray as follows: PCNA, proliferating cell nuclear antigen or DNA clamp; Pri, primase. The heatmap is displayed as in (A) with three groups indicated on the side: Euryarchaeota (Eury), Crenarchaeota (Cren), and other archaea (Other). For full network and list of the proteins, see supplementary figure S4 and table S6, Supplementary Material online.
Although bacteria and archaea typically share such characteristic as chromosome organization, transcription regulation and cotranscriptional translation, many traits of chromosome replication, genome segregation and cell division processes in archaea resemble those in eukaryotes. DNA polymerase sliding clamp subunits, PolIIIβ (DNA polymerase III subunit beta) in bacteria and PCNA in archaea, DNA ligases, flap nucleases, clamp loader ATPases, and topoisomerases I of bacterial and archaeal replisomes display considerable conservation (Leipe et al. 1999). Other protein components are either only distantly related or completely nonorthologous. Nevertheless, the replisome in both bacteria and archaea is highly enriched for inteins (fig. 4). Intein-containing replicative helicases, replicative DNA-dependent DNA polymerases, clamp loaders, recombinases and topoisomerases involved in replication, recombination and repair were found to contain inteins in both bacteria and archaea, indicating that particular functional categories are enriched for intein-containing proteins (fig. 4, tables 1 and 2, supplementary tables S1, S2, S5, and S6, Supplementary Material online). Remarkably the top 15 intein-containing proteins in archaea and bacteria are in a single network, containing most replisome proteins.
Replicative helicases DnaB and minichromosome maintenance protein (MCM) are at the top of our lists for intein-containing proteins from bacteria and archaea, with 317 inteins identified in DnaB and 116 inteins found in MCM. Performing equivalent functions, these proteins are only distantly related and structurally distinct (fig. 5, tables 1 and 2, supplementary tables S1, S2, S5, and S6, Supplementary Material online). Additionally, diverse topoisomerases, which are involved in cellular DNA topology management and aid helicases in DNA replication and repair, were found to contain inteins. These include bacterial topoisomerase-primase DnaG, bacterial DNA gyrase subunit A (GyrA) and subunit B (GyrB), archaeal topoisomerases such as GyrB (Soucy et al. 2014), TopA and Top6B (fig. 4), and a hyperthermophile-specific reverse gyrase (supplementary table S5, Supplementary Material online) (Forterre and Gadelle 2009).
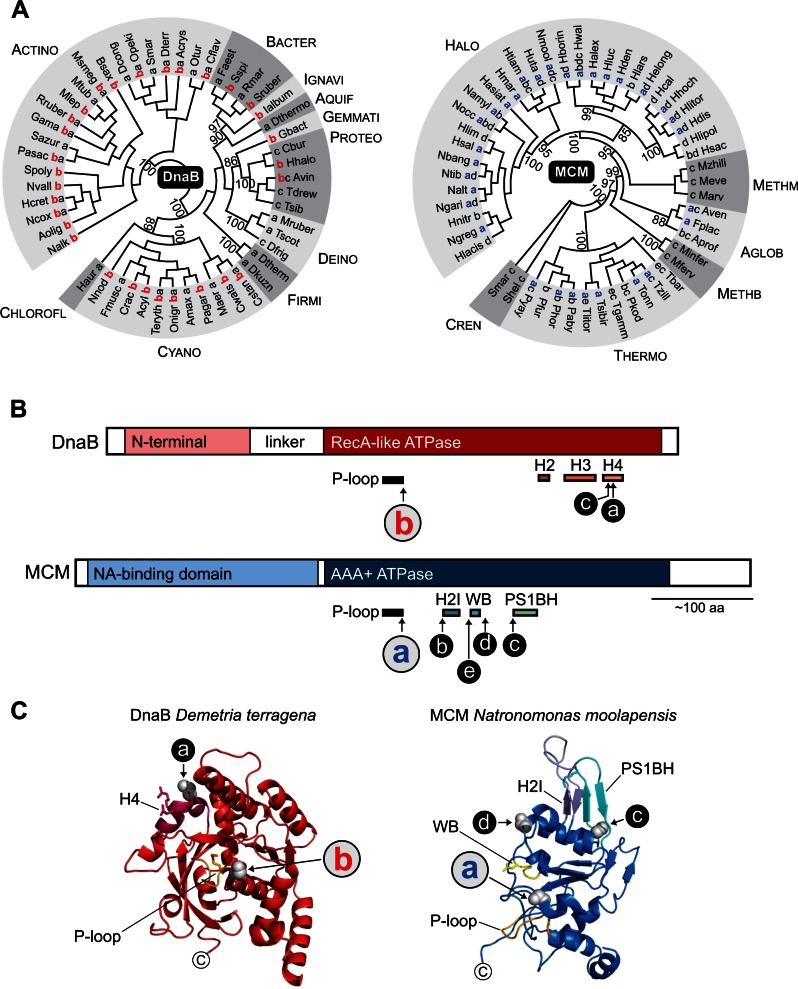
Fig. 5.
Replicative helicases DnaB and MCM are the most common intein-containing proteins. (A) Distribution of inteins in DnaB and MCM. Phylogenetic trees for bacterial replicative helicase DnaB and archaeal helicase MCM were reconstructed based on the extein amino acid sequences (ATPase domain) using the ML algorithm with the WAG (Whelan and Goldman)+G+I models; 50 representatives covering major bacterial and archaeal diversity were chosen among both DnaB and MCM proteins for tree reconstructions. Statistical support for the tree was evaluated by the nonparametric version of approximate likelihood-ratio test (SH-aLRT); however, only values for critical nodes, which were higher than 85%, are shown. The intein insertion point(s) a–e and abbreviated species names are shown next to branches. Letters for insertion points in DnaB and MCM do not correspond to each other (see B). The trees with full-length species names are available in supplementary figures S5 and S7, Supplementary Material online; the trees reconstructed based on the extended data sets including intein-containing and intein-less proteins are available in supplementary figures S6 and S8, Supplementary Material online. Although DnaB and MCM are functionally equivalent counterparts in bacteria and archaea, these proteins are only distantly related. Bacterial clades as follows: Cyano, Cyanobacteria; Chlorofl, Chloroflexi; Firmi, Firmicutes; Deino, Deionococcus–Thermus; Aquif, Aquificae; Bacter, Bacteroidetes; Ignavi, Ignavibacteriae; Gemmati, Gemmatimonadetes; Proteo, Proteobacteria; Actino, Actinobacteria. Archaeal clades as follows: Halo, Halobacteria; Methm, Methanomicrobia; Aglob, Archaeolglobi; Methb, Methanobacteria; Thermo, Thermococci; Cren, Crenarchaeota. (B) Intein insertion points. Intein locations are shown along DnaB and MCM relative to structural and functional domains. The ATPase domain has multiple intein insertion points in both DnaB and MCM. The important conserved structural motifs within the ATPase domain are shown on the bottom for each protein. The insertion point b in DnaB (red) and insertion point a in MCM (blue) are in functionally equivalent motifs which correspond to P-loops in ATPase. Three other structurally and functionally important motifs are shown for DnaB (H2–H4). Other motifs shown for MCM are: WB, Walker B motif; H2I, β–α–β insert; PS1BH, presensor 1 β-hairpin. MCM protein also carries a nucleic acid-binding domain at the N-terminus (NA-binding domain). (C) Structure models of ATPase domain of DnaB and MCM. Phyre2 models of the ATPase domain are shown (Dte DnaB residues 176–456; Nmo MCM residues 260–696). Dte DnaB (red) has inteins at insertion point a and b, with the T + 1 residues at the intein insertion sites highlighted as gray spheres.
Another large group of intein-containing proteins is DNA/RNA helicases many of which are involved in DNA repair. Among bacterial intein-containing ATP-dependent DNA helicases are UvrD, a helicase involved in nucleotide excision repair, RecG, a helicase that catalyzes branch migration and resolves Holliday junction intermediates (fig. 4A, table 1), and the UvrD-like helicase RecD, a member of the RecBCD complex which is essential for the repair of double-strand breaks in Escherichia coli and many other bacteria (supplementary table S5, Supplementary Material online) (Morita et al. 2010; Lenhart et al. 2012). In archaea, the Ski2-like DNA helicase, thought to be involved in DNA repair and recombination, was found to carry inteins in some species (fig. 4B, table 2, and supplementary table S6, Supplementary Material online) (Buttner et al. 2007). Other replication and repair proteins containing inteins include bacterial DNA repair photolyase SplB, primary repair DNA polymerase I that also has a role in replication, and error-prone repair DNA polymerase II. In archaea, DNA ligase and chromosome condensation and segregation the structural maintenance of chromosomes ATPase all contain inteins (fig. 4, tables 1 and 2, supplementary tables S5 and S6, Supplementary Material online) (Long and Faguy 2004).
Inteins are common in ribonucleotide reductases (RNR) in both bacteria and archaea, with 211 and 43 identified inteins, respectively (fig. 4, tables 1 and 2, supplementary tables S1, S2, S5, and S6, Supplementary Material online). Although RNR is not a part of the replisome, RNR plays a crucial role in determining the rate of DNA synthesis following replication initiation, by catalyzing the rate-limiting step in dNTP production (Herrick and Sclavi 2007). Another relatively large group of inteins was found in the archaeal ATP synthase (Swithers et al. 2013).
Inteins in Replicative Helicases DnaB and MCM
For an in-depth analysis of intein clustering, we first examined intein distribution in replicative helicases, the most common protein sinks for inteins among both bacteria and archaea. DnaB and MCM are ring-shaped hexameric enzymes that move along one strand of a DNA duplex and catalyze the displacement of the complementary strand in a reaction that is coupled to ATP hydrolysis. Although DnaB and MCM (and some other helicases) share the NTPase fold, they are only distantly related to each other and belong to different divisions of the P-loop ATPases (Gorbalenya and Koonin 1989). DnaB belongs to a family which is related to the RecA-like ATPases (Leipe et al. 2000), whereas the MCM proteins are members of the AAA+ superfamily of ATPases (Neuwald et al. 1999).
DnaB helicase is a member of superfamily SF4 that also includes G40P helicase of Bacillus subtilis SSP1 bacteriophage and T7 gp4 helicase-primase of E. coli T7 bacteriophage (Ilyina et al. 1992; Sawaya et al. 1999; Patel and Picha 2000). All members from this superfamily possess 5′ → 3′ helicase activity (Patel and Picha 2000). We found intein-containing DnaB among bacteria from ten clades. Inteins are widely distributed in DnaB proteins from Actinobacteria (202 inteins) and Cyanobacteria (60 inteins). Only a few species were found to have DnaB inteins among representatives from the other clades (fig. 5A, table 1, and supplementary tables S1 and S5, Supplementary Material online). Three distinct insertion sites were identified in DnaB helicase, which were designated a–c based on the conventional intein insertion site classification scheme (Perler et al. 1997; Perler 2002). All three insertion points are located in the ATPase domain in highly conserved motifs (fig. 5B and C). Sites a and c are at the C-terminal end of the domain only two amino acid residues apart. These sites are in motif H4, which is responsible for DNA binding (Ilyina et al. 1992; Sawaya et al. 1999). Insertion point b is in the ATP-binding P-loop (fig. 5B and C). A structure model of DnaB exteins from actinobacteria Demetria terragena highlights the spatial proximity of insertion sites b and a (fig. 5C). There is a number of DnaB helicases from Actinobacteria and Cyanobacteria that carry b and a insertions simultaneously.
In contrast to DnaB, MCM belongs to superfamily SF6, and has opposing 3′ → 5′ helicase activity (Chong et al. 2000; Patel and Picha 2000). MCM proteins are conserved not only throughout archaea but also in eukaryotes, and are essential for DNA replication initiation and progression (Grainge et al. 2003; Pacek and Walter 2004). Numerous intein insertions were found among Halobacteria (76 inteins) and Thermococci (19 inteins), with a few inteins in other clades (fig. 5A, table 2, and supplementary tables S2 and S6, Supplementary Material online). All identified MCM inteins were located in highly conserved motifs of the ATPase domain (sites a–e; fig. 5B and C). The insertion point a is in the P-loop at the analogous site as insertion point b of the DnaB inteins (fig. 5B and C). Insertion point b of the MCM inteins is between the P-loop and Walker B motif of the ATPase domain in close proximity to the MCM-specific β-α-β insert (H2I), which is believed to be essential for coupling ATP hydrolysis and DNA unwinding activity (Jenkinson and Chong 2006). Insertion points e and d are around the Walker B motif; and point c is found right before the presensor 1 β-hairpin (PS1BH), which is required for DNA duplex unwinding (Brewster et al. 2008). Both H2I and PS1BH are structurally and functionally critical elements of MCM helicases (Jenkinson and Chong 2006; Brewster et al. 2008). The structure model for MCM exteins from the haloarchaeon Natronomonas moolapensis shows the intein insertion sites (a, c, and d) clustered near catalytic amino acid residues (fig. 5C).
Inteins in Clamp Loaders and Replicative Polymerases
We performed a similar analysis for clamp loader subunits belonging to the subset of accessory replicative proteins which are orthologous in all extant organisms (Leipe et al. 2000). Inteins were found in both bacterial clamp loader DNA polymerase III γ subunit (PolIIIγ) and archaeal replication factor C small subunit (RFC-S) (fig. 4, tables 1 and 2) (Kaneko et al. 1995; Bult et al. 1996). Comparative analysis showed the independent acquisition of inteins by bacterial and archaeal clamp loaders. For PolIIIγ, inteins were found only in representatives from Cyanobacteria (15 inteins) occupying a single insertion point (insertion point a), immediately after the highly conserved Walker B motif that coordinates the magnesium ion required for ATP hydrolysis (fig. 6A–C, table 1) (Walker et al. 2000). Interestingly, 13 out of 15 species possessing PolIIIγ inteins also had an intein insertion in their replicative DNA-dependent DNA polymerase catalytic subunit PolIIIα (DNA polymerase III subunit alpha) (see detailed analysis below). In contrast, inteins were found widely distributed in the ATPase domain among RFC-S proteins from diverse archaea belonging to six clades, primarily in Thermococci (13 inteins), Methanococci (7 inteins), and Halobacteria (12 inteins; also Soucy et al. 2014), at four distinct insertion points (a–d) (fig. 6A–C and table 2). None of these overlapped with the one present in Cyanobacteria indicating independent acquisition. Thus, in spite of the orthologous nature of the ATPase domains from bacterial and archaeal clamp loaders, inteins and their insertion points from the same proteins are not orthologous and likely were acquired independently.
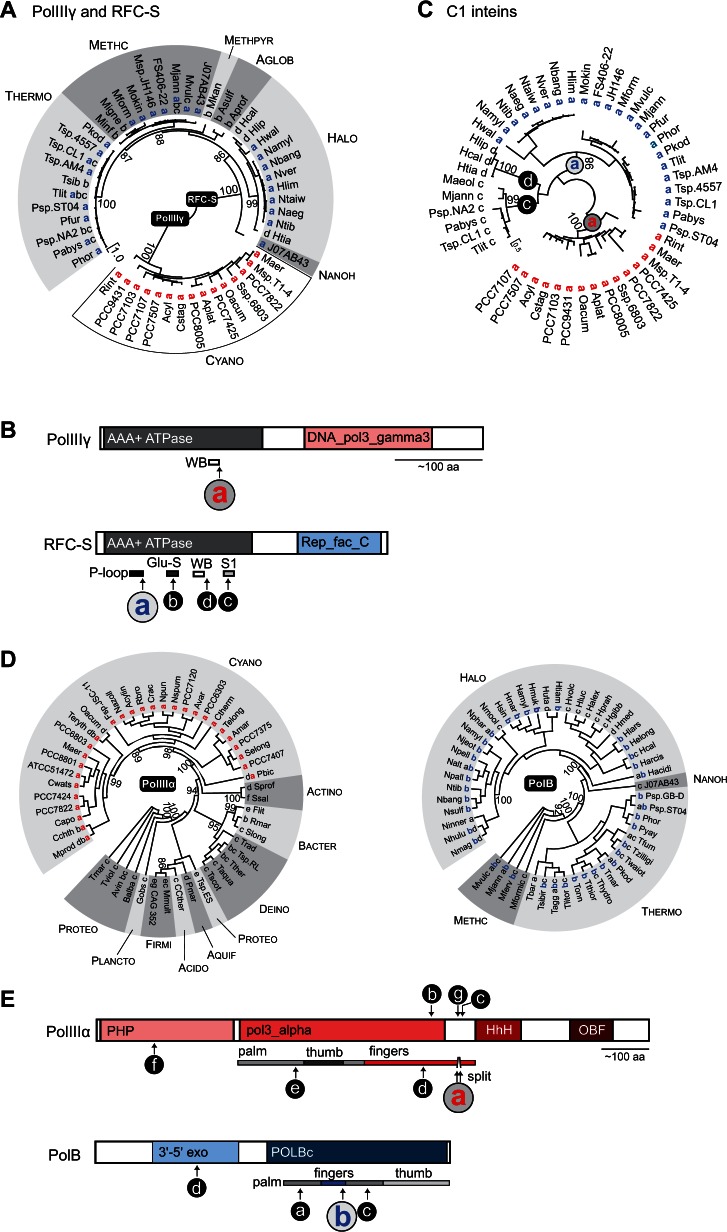
Fig. 6.
Inteins in bacterial and archaeal clamp loaders and DNA polymerases cluster in functional domains. (A) Distribution of inteins in PolIIIγ and RFC-S. Phylogenetic tree for ATPase domain of the clamp loader proteins from both bacteria and archaea was reconstructed based on the amino acid sequences using the ML algorithm with WAG model. Statistical support for the tree was evaluated with SH-aLRT; however, only values for critical nodes, which were higher than 85%, are shown. The intein insertion point(s) a–d and abbreviated species names are shown next to branches. Letters for insertion points in PolIIIγ and RFC-S do not correspond to each other (see B and C). The tree with full-length species names is available in supplementary figure S9, Supplementary Material online; the trees reconstructed based on the extended data sets including intein-containing and intein-less proteins are available in supplementary figures S10 and S11, Supplementary Material online. PolIIIγ inteins were found only in Cyanobacteria (Cyano). Archaeal clades as follows: Thermo, Thermococci; Methc, Methanococci; Methpyr, Methanopyri; Nanoh, Nanohaloarchaeota; Halo, Halobacteria; Aglob, Archaeolglobi. (B) Intein insertion points. Intein locations are shown along PolIIIγ and RFC-S relative to structural and functional domains. The ATPase domain (AAA+ ATPase, black) has a single intein insertion in PolIIIγ (site a shown in red) and multiple intein insertion points in RFC-S (sites a–d). The insertion point a in PolIIIγ is located in highly conserved Walker B motif (WB). The most common insertion point a in RFC-S (blue) is located in P-loop. Other motifs shown for RFC-S are: Glu-S, glutamine switch; S1, sensor one. PolIIIγ and RFC-S proteins have additional domains specific for respective proteins: DNA_pol3_gamma3 domain (pink) is found only in PolIIIγ, whereas Rep_fac_C domain (light blue) is present only in RFC-S proteins. (C) Phylogenetic analysis of the C1 inteins from PolIIIγ and RFC-S. Phylogenetic tree was reconstructed based on the intein splicing domain amino acid sequences using the ML algorithm with WAG model. Statistical support for the tree was evaluated with SH-aLRT; however, only values for critical nodes, which were higher than 85%, are shown. Only inteins with cysteine as the first amino acid residue (C1 inteins) were used, which included all inteins identified in PolIIIγ (insertion point a, red), and inteins from insertion points a (blue), c, and d from RFC-S. The intein insertion point(s) a, c, d and abbreviated species names are shown next to branches. The intein insertion point(s) are also indicated in the nodes. The tree with full-length species names is available in supplementary figure S12, Supplementary Material online. (D) Distribution of inteins in PolIIIα and PolB. Phylogenetic trees for bacterial replicative DNA polymerase PolIIIα and archaeal PolB were reconstructed based on the extein amino acid sequences using the ML algorithm with WAG model. Statistical support was evaluated with SH-aLRT; however, only values for critical nodes, which were higher than 85%, are shown. The intein insertion point(s) a–f and abbreviated species names are shown next to branches. Letters for insertion points in PolIIIα and PolB do not correspond to each other (see B). The full-length trees with full-length species names are available in supplementary figures S13 and S14, Supplementary Material online. Although PolIIIα and PolB are functionally equivalent counterparts in bacteria and archaea, these proteins are not related. Bacterial clades as follows: Cyano, Cyanobacteria; Actino, Actinobacteria; Bacter, Bacteroidetes; Deino, Deionococcus–Thermus; Acido, Acidobacteria; Plancto, Planctomycetes; Proteo, Proteobacteria; Aquif, Aquificae; and Firmi, Firmicutes. Archaeal clades as follows: Halo, Halobacteria; Nanoh, Nanohaloarchaeota; Methc, Methanococci; Thermo, Thermococci. (E) Intein insertion points. Intein locations are shown along PolIIIα and PolB relative to structural and functional domains. The critical catalytic domains have multiple intein insertion points in both PolIIIα (pol3_alpha) and PolB (POLBc). Additional insertion points were found in bacterial PHP (polymerase and histidinol hhosphatase domain) for PolIIIα and in archaeal 3′–5′ exo (3′–5′ exonuclease domain of archaeal family-B DNA polymerases) for PolB. Polymerase structural domains are shown on the bottom. PolIIIα inteins from insertion point a are split. Additional abbreviations: HhH, helix-hairpin-helix DNA-binding domain; OBF, (oligonucleotide/oligosaccharide binding)-fold.
Although the majority of PolIIIγ and RFC-S inteins have Cys as the first amino acid residue (C1 inteins), RFC-S inteins at the insertion point b start with either Ser or Ala (S1/A1 inteins). The initial intein residue has implications for splicing mechanism. C1 inteins belong to class 1 based on the sequence similarity and splicing mechanism, whereas S1/A1 inteins might belong to class 2 which employs a somewhat different protein splicing pathway (Southworth et al. 2000). Although PolIIIγ and RFC-S inteins seem to have been acquired independently, their protein splicing domains show similarity, and C1 inteins likely share a common ancestor. Thus, we performed a phylogenetic analysis of all C1 inteins combined. As seen from the resulting phylogenetic tree reconstructed based on the amino acid sequences of the splicing domain, inteins cluster together based on their exteins (either PolIIIγ or RFC-S) indicating vertical transmission of inteins within each cluster (fig. 6C and supplementary fig. S8, Supplementary Material online). Interestingly, RFC-S C1 inteins form distinct groups based on their insertion point, emphasizing an independent origin of the inteins not only between PolIIIγ and RFC-S, but within RFC-S as well (fig. 6C). These results reinforce the notion that despite the orthologous nature of the ATPase domains from bacterial and archaeal clamp loaders, inteins and their insertion points from the same proteins are not orthologous and inteins likely were acquired independently.
Replicative DNA-dependent DNA polymerases provide another striking example of proteins that are functionally, but not evolutionarily related between bacteria and archaea. Yet, both bacterial and archaeal replicative DNA polymerases contain multiple intein insertions, mostly in the same polymerization domain of the two disparate proteins (fig. 6D and E). Bacterial replicative PolIIIα belongs to the family C polymerases, which share no sequence similarity with other polymerases and exhibit a fold that is strikingly different from those of archaeal and eukaryotic replicative polymerases, suggesting an independent origin of the DNA replication machinery (Braithwaite and Ito 1993; Filee et al. 2002; Lamers et al. 2006). PolIIIα inteins were found primarily in Cyanobacteria (89 inteins), with 27 in the other clades shown (fig. 6D). The intein hot spot is in the “finger” domain with several distinct insertion points, a–d and g, present in a sequence stretch of 75 amino acid residues (fig. 6E). Fingers are involved in the correct positioning of the template and 3′-end of the primer in the active site as well as distinguishing between ribo- and deoxyribonucleoside triphosphates. The PolIIIα intein insertion point a is the most common and widely distributed among Cyanobacteria. All known inteins at this insertion point are split, representing the largest known group of split inteins (Dassa et al. 2009). The primary replicative DNA polymerase in archaea is from the family B (PolB). PolB inteins are widespread among Halobacteria (37 inteins), Thermococci (20 inteins), and Methanococci (5 inteins) (fig. 6D). There are four distinct insertion points for inteins in PolB, a–d, of which a–c are located in the polymerase catalytic domain (POLBc; fig. 6E), once again in the polymerase fingers. These observations reinforce the notion that intein clustering is related to protein function, regardless of evolutionary origin.
Discussion
This study shows that inteins are concentrated in proteins of the DNA replication fork, particularly ATPase domains, even in proteins that are nonorthologous but functionally similar in bacteria and archaea. These observations focus our hypothesis that some inteins are maintained because of functional adaptation. The biased intein distribution toward replisome components is puzzling. The principal proteins of the bacterial replication machinery are unrelated or only distantly related to the functionally equivalent components from archaea (Edgell and Doolittle 1997; Leipe et al. 1999). Among nonorthologous proteins performing equivalent function are replicative helicases and the replicative DNA polymerases (Braithwaite and Ito 1993; Edgell and Doolittle 1997; Leipe et al. 2000; Filee et al. 2002; Lamers et al. 2006). In contrast, the core components of transcription and translation are highly conserved in all domains of life (Edgell and Doolittle 1997; Leipe et al. 1999), and they contain few inteins. Moreover, many of the intein-containing replication proteins are functionally equivalent but are structurally highly divergent or even unrelated, indicating that intein retention in these nonorthologous replication proteins likely relates to host protein function (fig. 4). We speculate that the inteins sense specific environmental cues and act as panic buttons to reversibly halt splicing and thereby DNA synthesis during stress.
The rapidly growing sequence databases for bacteria and archaea gave us the opportunity for comprehensive intein mining. By screening genomes from a wide range of organisms for the presence of inteins, our large data set confirmed the sporadic nature of intein distribution. In adapting functional genomics approaches to intein discovery, we found that COGs are suitable for functional annotation and further categorization of the inteins based on their exteins. This classification system is a natural framework for comparative genomics of prokaryotes, including phage, within an evolutionary context (Tatusov et al. 1997, 2003). Functional annotation of newly sequenced genomes and genome-wide evolutionary analyses are the most important applications of COGs, but have found use in functional genomics of particular proteins, protein families, or large groups of proteins of interest. The COG analysis clearly demonstrated in a fully quantifiable manner that functional categories for replication, repair, recombination, and nucleotide transport and metabolism are disproportionally enriched among intein-containing proteins. We propose the use of COGs, supplemented with other relevant information, as a basis for intein classification in the future. Using COGs will help to put experimental data sets in a context of comparative and functional genomics.
To further elucidate the forces that shaped intein distribution, we performed a detailed analysis of the inteins present in three pairs of replication proteins: nonorthologous replicative helicases DnaB and MCM; orthologous clamp loaders PolIIIγ and RFC-S; and replicative DNA polymerases PolIIIα and PolB, which are functionally analogous but evolutionary unrelated (Braithwaite and Ito 1993; Leipe et al. 1999; Filee et al. 2002). Our data support a concentration of inteins not only in proteins with related roles but also within similar functional domains. This is the case even for bacterial and archaeal inteins that were presumably acquired independently by functionally identical replication genes of different evolutionary origin. For example, the ATPase domains carry inteins in nonorthologous replicative helicases DnaB/MCM and orthologous clamp loaders PolIIIγ/RFC-S. However, protein-specific domains rarely have inteins; for example, inteins were found in ATPase domains but not in the DNA_pol3_gamma3 domain specific for PolIIIγ, nor in the Rep_fac_C found only in RFC-S (figs. 5B and 6B). Intein insertions are present in the highly conserved P-loop in three of these ATPases, DnaB/MCM, and RFC-S. Moreover, there are many other examples of ATPases with inteins in their P-loop (tables 1–3 and supplementary tables S5–S7, Supplementary Material online) (Novikova et al. 2014). A different example of intein clustering in functional domains is in the polymerization domain of unrelated replicative polymerases. Interestingly, although inteins are usually present in conserved protein motifs involved in the active site and often near amino acid residues important for substrate and ligand binding, they are rarely inserted at catalytic residues. A number of inteins is located in structurally dynamic regulatory parts of proteins such as DNA-binding motif H4 of DnaB ATPase and the distal part of the fingers of the PolIIIα catalytic domain (fig. 6B and E) (Sawaya et al. 1999; Wing et al. 2008). Regardless, the theme of intein concentration in proteins and domains of like function prevails.
The current models for intein dynamics do not fully explain the observed seemingly selective distribution of inteins. The earliest model suggested that inteins are found in highly conserved proteins and conserved protein domains and motifs because the imprecise removal of the intein would lead to incapacitation of the host protein (Pietrokovski 2001). However, there is no specific mechanism for intein removal from the coding region and loss of the intein likely occurs through global processes such as gene conversion, horizontal gene transfer or large-scale rearrangements. Moreover, it was suggested that most of the examples of intein loss represent cases of being outcompeted by intein-free alleles (Fullmer et al. 2014; Soucy et al. 2014). Thus, while still valid, the original model does not alone explain the presence of inteins in conserved sequences and does not address other aspects of intein dynamics.
Some other models are focused on the dynamics of the intein HEN rather than on the protein splicing domain as inteins rely on HENs for their propagation (Gimble and Thorner 1992; Burt and Koufopanou 2004; Gogarten and Hilario 2006). HENs recognize and cleave specific DNA sequences 14–40 bp in length, the homing site for intein acquisition (Guhan and Muniyappa 2003). Importantly, HENs possess the unique ability to minimize nonspecific cleavage, while maintaining the potential to cleave closely related variants of the homing site (Chevalier et al. 2003; Posey et al. 2004). The simple model for intein persistence includes invasion of the intein-free site through HEN-mediated gene conversion, spreading of the intein in the population, fixation, HEN decay in the absence of selective pressure, loss of the intein, and, finally, reinvasion of the empty target site (Burt and Koufopanou 2004; Gogarten and Hilario 2006). This homing cycle model, however, was shown to have multiple limitations when applied to natural populations. For example, there are significant differences in gene transfer frequencies and efficiency between sexual and asexual species which will affect HEN persistence. Another significant limitation of this model is the inability to explain the simultaneous presence of intein-containing and intein-free alleles in a population. The modeling, however, demonstrated that intein-containing and intein-free alleles can potentially coexist long-term with no benefit to the host (Gogarten and Hilario 2006; Yahara et al. 2009; Barzel, Obolski, et al. 2011). Other complex models might be proposed to explain the occurrence of inteins at specific sites, like the attractiveness of the structure of a protein that would allow proper folding of itself and the intein. It is possible that various factors contribute to intein persistence in different proteins. One such factor, which seems to be often overlooked, is the possibility that inteins, or subpopulations of inteins, are in functionally important proteins and are beneficial to the host.
We argue that intein association with particular domains (such as the P-loop), proteins (such as ATPases), and functions (such as replication) indicates selective retention and likely the importance of inteins in these particular positions. The observation that inteins occupy specific proteins, even in nonorthologous functional counterparts in bacteria and archaea, suggests that some inteins are not simply selfish passengers, but are likely to perform yet unknown roles. The benefit from the intein presence can be subtle or reveal itself only under certain environmental conditions, which are not commonly encountered by host cells. Environmental stress might explain the sporadic distribution of inteins among closely related species. If there is no strong selective pressure for intein retention in a particular subpopulation and the benefit from keeping the intein is low under prevailing conditions, the intein will likely be outcompeted by intein-free alleles (Gogarten and Hilario 2006).
There are multiple examples of conditional protein splicing by engineered and native inteins indicating that splicing can be locked or triggered by a particular condition, such as a change in redox state, temperature or pH, as has been recently reviewed (Novikova et al. 2014; Shah and Muir 2014; Topilina and Mills 2014). The existence of inteins modulated in a stimulus-dependent manner points to the possibility that some inteins may adapt to their intracellular niche and become posttranslational regulatory elements. Indeed some inteins can act as sensors, and through intein chemistry can inhibit splicing under stressful environmental conditions, such as redox modulation (Callahan et al. 2011), oxidative and nitrosative stress (Topilina, Green, et al. 2015) or cold shock (Topilina, Novikova, et al. 2015). It remains to be shown that inteins are sensitive to those stimuli in their native host.
The preponderance of inteins in ATPase domains, where approximately 70% of inteins in archaea and bacteria are in proteins that bind ATP, is worthy of consideration. One possibility is that ATPases represent ancient structures to which ancestral intein folding adapted (Caetano-Anolles et al. 2007). Additionally, many of these ATPases are in proteins involved in replication, a process that must be slowed as an organism mounts a stress response. Inhibition of DNA synthesis would not only avoid replication toxicity but also would spare ATP required in the stress response. A similar ATP-sparing argument could be made for inteins in recombination functions like RecA in bacteria and RadA in archaea. We have shown that splicing is inhibited in a hyperthermophilic RadA from Pyrococcus horikoshii by formation of 3D interactions between the intein and exteins at low temperatures (Topilina, Novikova, et al. 2015). Such splicing inhibition during cold shock would spare ATP, which is a key substrate in metabolic and energy conservation (Macario et al. 1999; Lopez-Garcia and Forterre 2000). Increasing ATP concentration would maintain biochemical processes required to survive stressful conditions. Thus, we suggest that splicing inhibition would provide an immediate arrest of RadA function to spare ATP for use in the cold-shock response. In different stress responses, the intein panic button would inhibit replication and recombination functions and stop detrimental ATP consumption. Conversely, recovery from stress could be immediate, with protein splicing providing a functional premade protein, to allow the organism to rapidly re-enter a normal growth state.
Idiosyncratic intein clustering in genes involved in DNA replication, recombination and repair, and the metabolism of nucleotides might also be explained in part by the intein mobility pathway. In order to mobilize, inteins must cleave the host DNA, and they require repair, and DNA is more accessible for cleavage and recombination during replication. Inteins may therefore be favored in proteins which are expressed during replication. There are many other factors that might affect intein distribution among species. Our data set shows that there are certain bacterial clades with relatively high levels of inteins in comparison with others (fig. 1). This might be attributed to the specific ecology of the species in a given clade or their genome organization and dynamics. For example, among cyanobacteria, only one out of five species did not have any inteins in their genomes. All cyanobacteria are free-living, usually forming very large populations, which might explain such a prevalence of intein-containing species in this clade. On the other hand, in spirochetes only two species (out of 74 sequences available) have inteins. Spirochetes are highly specialized bacteria, many of which are pathogenic, with small isolated populations and streamlined genomes, possibly accounting for only a few intein-containing species.
The intrinsic nature of mobile elements is to explore new emerging niches in genomes and, ultimately, break species boundaries. In terms of intein spread, viruses and bacteriophage represent ideal “vectors” for horizontal transfer (Hambly and Suttle 2005; Nagasaki et al. 2005; Filee et al. 2007). They can carry intein sequences across cell boundaries as part of their own genes or transfer cellular genes they acquired as a result of rampant recombination (Filee et al. 2007). Many bacteriophage carry bacteria-like genes in their replication modules including genes for helicases DnaB and MCM as well as DNA polymerases (Weigel and Seitz 2006) which also might explain the biased distribution of inteins. Our analysis also showed that substantial numbers of inteins from bacterial (∼4%) and archaeal (∼5%) genomes are associated with the mobilome (such as prophages) and secretion system-related proteins, indicating potential pathways for horizontal transfer of inteins (see table 3). There are remarkable examples of the viral and bacteriophage intein-bearing genomes (Lazarevic 2001; Amitai et al. 2004; Ogata et al. 2005; Allen et al. 2011; Bigot et al. 2013; Dwivedi et al. 2013; Fouts et al. 2013). We propose that delivery of inteins can be straightforward and quasirandom. However, fixation in a population after transfer and further retention of inteins depend on many factors including population structure, gene flow and, importantly, pressure under environmental extremes. These factors cumulatively account for the profiles of inteins we observe in extant organisms.
Materials and Methods
Data Mining
The sequences used in the present study were obtained from the Protein and Genome databases at the NCBI (www.ncbi.nlm.nih.gov, last accessed December 11, 2015). Only fully sequenced, assembled, and annotated microbial genomes from NCBI RefSeq microbial genome collection were taken into consideration (Tatusova et al. 2015). By using only genomes that were added to the RefSeq genome collection we ensure the analysis of sequences that pass annotation quality control during RefSeq genome processing and annotation (Tatusova et al. 2015). As the first step, the word search query was used of the following composition: “(‘Phylum’[Organism]) AND intein [All Fields].” Phylum was iterated between the bacterial and archaeal phyla. The primary sequence data obtained for precursor as a whole and intein(s) separately were additionally analyzed using the following resources. The presence of conserved protein domains and motifs was verified using Conserved Domain Database (CDD; http://www.ncbi.nlm.nih.gov/cdd) and Conserved Domain Search Service (CD Search; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) which are available at NCBI. Additionally, protein domains and motifs were identified using InterPro protein analysis tool. Other resources heavily used in the present study are: InBase—intein database (http://tools.neb.com/inbase/; Perler 2002); European Nucleotide Archive (ENA: http://www.ebi.ac.uk/ena/); Genome at NCBI (http://www.ncbi.nlm.nih.gov/genome/); Basic Local Alignment Search Tool (BLAST) programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi); “The All-Species Living Tree” Project (http://www.arb-silva.de/projects/living-tree/). After comparative analysis of the sequences and a second review, the list of intein-containing proteins was formed.
Sequence Analysis
All protein alignments were performed by MUSCLE3.8 (Edgar 2004). Phylogenetic analysis was performed using Maximum-Likelihood (ML) method in PhyML 3.0 (Guindon et al. 2010). Statistical support for ML trees was evaluated with approximate likelihood-ratio test (aLRT). Specifically, a nonparametric version of the aLRT (Shimodaira–Hasegawa aLRT, SH-aLRT) was used (Anisimova and Gascuel 2006; Anisimova et al. 2011). Substitution models were selected using ProtTest 3 (Darriba et al. 2011). COG annotation was performed using COG database (http://www.ncbi.nlm.nih.gov/COG/). The InterPro database was used in GO enrichment analysis (http://www.ebi.ac.uk/interpro/). GO annotation results were plotted using a web-based tool, WEGO (Ye et al. 2006). The randomized sampling of the protein sequences was performed using custom BioPerl script available by request.
Structure Modeling
The extein sequences for DnaB of D. terragena and MCM of N. moolapensis were submitted for modeling to Phyre2 servers using intensive mode (Kelley et al. 2015). Structure models came back with high levels of confidence for both proteins, with 436 residues (95%) of Dte DnaB and 670 residues (96%) of Nmo MCM modeled at greater than 90% accuracy. Models were manipulated in PyMOL (v1.7.2), and the +1 residues of the relevant intein insertion sites were highlighted.
Supplementary Material
Supplementary figures S1–S17 and tables S1–S7 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/)
Acknowledgments
The authors acknowledge Rebecca McCarthy and Dorie Smith for help with the manuscript. They thank Dr David Shub for introducing them to the “panic button” concept and Dr Barry Stoddard for useful discussions. This work was supported by the National Institutes of Health (NIH) grants GM39422 and GM44844 to M.B.
References
- Allen MJ, Lanzén A, Bratbak G. 2011. Characterisation of the coccolithovirus intein. Mar Genomics. 4:1–7. [DOI] [PubMed] [Google Scholar]
- Amitai G, Dassa B, Pietrokovski S. 2004. Protein splicing of inteins with atypical glutamine and aspartate C-terminal residues. J Biol Chem. 279:3121–3131. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 55:539–552. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. 2011. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 60:685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzel A, Naor A, Privman E, Kupiec M, Gophna U. 2011. Homing endonucleases residing within inteins: evolutionary puzzles awaiting genetic solutions. Biochem Soc Trans. 39:169–173. [DOI] [PubMed] [Google Scholar]
- Barzel A, Obolski U, Gogarten JP, Kupiec M, Hadany L. 2011. Home and away—the evolutionary dynamics of homing endonucleases. BMC Evol Biol. 11:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Roberts RJ. 1997. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 25:3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot Y, Piégu B, Casteret S, Gavory F, Bideshi DK, Federici BA. 2013. Characteristics of inteins in invertebrate iridoviruses and factors controlling insertion in their viral hosts. Mol Phylogenet Evol. 67:246–254. [DOI] [PubMed] [Google Scholar]
- Braithwaite DK, Ito J. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, Chen XS. 2008. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci U S A. 105:20191–20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058–1073. [DOI] [PubMed] [Google Scholar]
- Burt A, Koufopanou V. 2004. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 14:609–615. [DOI] [PubMed] [Google Scholar]
- Butler MI, Gray J, Goodwin TJ, Poulter RT. 2006. The distribution and evolutionary history of the PRP8 intein. BMC Evol Biol. 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner K, Nehring S, Hopfner KP. 2007. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 14:647–652. [DOI] [PubMed] [Google Scholar]
- Caetano-Anolles G, Kim HS, Mittenthal JE. 2007. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Natl Acad Sci U S A. 104:9358–9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BP, Topilina NI, Stanger MJ, Van Roey P, Belfort M. 2011. Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat Struct Mol Biol. 18:630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier B, Turmel M, Lemieux C, Monnat RJ, Jr, Stoddard BL. 2003. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J Mol Biol. 329:253–269. [DOI] [PubMed] [Google Scholar]
- Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. 2000. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci U S A. 97:1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa B, London N, Stoddard BL, Schueler-Furman O, Pietrokovski S. 2009. Fractured genes: a novel genomic arrangement involving new split inteins and a new homing endonuclease family. Nucleic Acids Res. 37:2560–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE, Penny SL. 1999. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 48:236–244. [DOI] [PubMed] [Google Scholar]
- Dwivedi B, Xue B, Lundin D, Edwards RA, Breitbart M. 2013. A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol Biol. 13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgell DR, Doolittle WF. 1997. Archaea and the origin(s) of DNA replication proteins. Cell 89:995–998. [DOI] [PubMed] [Google Scholar]
- Filee J, Forterre P, Sen-Lin T, Laurent J. 2002. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol. 54:763–773. [DOI] [PubMed] [Google Scholar]
- Filee J, Siguier P, Chandler M. 2007. I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 23:10–15. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gadelle D. 2009. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 37:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE, Klumpp J, Bishop-Lilly KA, Rajavel M, Willner KM, Butani A, Henry M, Biswas B, Li M, Albert MJ, et al. 2013. Whole genome sequencing and comparative genomic analyses of two Vibrio cholerae O139 Bengal-specific Podoviruses to other N4-like phages reveal extensive genetic diversity. Virol J. 10:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullmer MS, Soucy SM, Swithers KS, Makkay AM, Wheeler R, Ventosa A, Gogarten JP, Papke RT. 2014. Population and genomic analysis of the genus Halorubrum. Front Microbiol. 5:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43:D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble FS, Thorner J. 1992. Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature 357:301–306. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Hilario E. 2006. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol Biol. 6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. 2002. Inteins: structure, function, and evolution. Annu Rev Microbiol. 56:263–287. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 17:8413–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainge I, Scaife S, Wigley DB. 2003. Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids Res. 31:4888–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhan N, Muniyappa K. 2003. Mycobacterium tuberculosis RecA intein, a LAGLIDADG homing endonuclease, displays Mn(2+) and DNA-dependent ATPase activity. Nucleic Acids Res. 31:4184–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Hambly E, Suttle CA. 2005. The viriosphere, diversity, and genetic exchange within phage communities. Curr Opin Microbiol. 8:444–450. [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32:D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick J, Sclavi B. 2007. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol. 63:22–34. [DOI] [PubMed] [Google Scholar]
- Hirata R, Ohsumk Y, Nakano A, Kawasaki H, Suzuki K, Anraku Y. 1990. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 265:6726–6733. [PubMed] [Google Scholar]
- Ilyina TV, Gorbalenya AE, Koonin EV. 1992. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J Mol Evol. 34:351–357. [DOI] [PubMed] [Google Scholar]
- Jacquier A, Dujon B. 1985. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41:383–394. [DOI] [PubMed] [Google Scholar]
- Jenkinson ER, Chong JP. 2006. Minichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc Natl Acad Sci U S A. 103:7613–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane PM, Yamashiro CT, Wolczyk DF, Neff N, Goebl M, Stevens TH. 1990. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H(+)-adenosine triphosphatase. Science 250:651–657. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. 1995. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 2:153–166. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koufopanou V, Goddard MR, Burt A. 2002. Adaptation for horizontal transfer in a homing endonuclease. Mol Biol Evol. 19:239–246. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Georgescu RE, Lee SG, O’Donnell M, Kuriyan J. 2006. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell 126:881–892. doi: 10.1016/j.cell.2006.07.028 [DOI] [PubMed] [Google Scholar]
- Lazarevic V. 2001. Ribonucleotide reductase genes of Bacillus prophages: a refuge to introns and intein coding sequences. Nucleic Acids Res. 29:3212–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Aravind L, Grishin NV, Koonin EV. 2000. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 10:5–16. [PubMed] [Google Scholar]
- Leipe DD, Aravind L, Koonin EV. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. 2012. DNA repair and genome maintenance in Bacillus subtilis. Microbiol Mol Biol Rev. 76:530–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ. 2000. Protein-splicing intein: genetic mobility, origin, and evolution. Annu Rev Genet. 34:61–76. [DOI] [PubMed] [Google Scholar]
- Long SW, Faguy DM. 2004. Anucleate and titan cell phenotypes caused by insertional inactivation of the structural maintenance of chromosomes (smc) gene in the archaeon Methanococcus voltae. Mol Microbiol. 52:1567–1577. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Forterre P. 2000. DNA topology and the thermal stress response, a tale from mesophiles and hyperthermophiles. Bioessays 22:738–746. [DOI] [PubMed] [Google Scholar]
- Luo J, Hall BD. 2007. A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol. 64:101–112. [DOI] [PubMed] [Google Scholar]
- Macario AJ, Lange M, Ahring BK, Conway de Macario E. 1999. Stress genes and proteins in the archaea. Microbiol Mol Biol Rev. 63:923–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Nakane S, Shimada A, Inoue M, Iino H, Wakamatsu T, Fukui K, Nakagawa N, Masui R, Kuramitsu S. 2010. Molecular mechanisms of the whole DNA repair system: a comparison of bacterial and eukaryotic systems. J Nucleic Acids. 2010:179594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K, Shirai Y, Tomaru Y, Nishida K, Pietrokovski S. 2005. Algal viruses with distinct intraspecies host specificities include identical intein elements. Appl Environ Microbiol. 71:3599–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27–43. [PubMed] [Google Scholar]
- Novikova O, Topilina N, Belfort M. 2014. Enigmatic distribution, evolution, and function of inteins. J Biol Chem. 289:14490–14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Raoult D, Claverie JM. 2005. A new example of viral intein in Mimivirus. Virol J. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Walter JC. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. Embo J. 23:3667–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MM, Belisle M, Belfort M. 1999. Intron homing with limited exon homology. Illegitimate double-strand-break repair in intron acquisition by phage t4. Genetics 153:1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Picha KM. 2000. Structure and function of hexameric helicases. Annu Rev Biochem. 69:651–697. [DOI] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, et al. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182. [DOI] [PubMed] [Google Scholar]
- Perler FB. 2002. InBase: the Intein Database. Nucleic Acids Res. 30:383–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perler FB, Olsen GJ, Adam E. 1997. Compilation and analysis of intein sequences. Nucleic Acids Res. 25:1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrokovski S. 2001. Intein spread and extinction in evolution. Trends Genet. 17:465–472. [DOI] [PubMed] [Google Scholar]
- Posey KL, Koufopanou V, Burt A, Gimble FS. 2004. Evolution of divergent DNA recognition specificities in VDE homing endonucleases from two yeast species. Nucleic Acids Res. 32:3947–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter RT, Goodwin TJ, Butler MI. 2007. The nuclear-encoded inteins of fungi. Fungal Genet Biol. 44:153–179. [DOI] [PubMed] [Google Scholar]
- Saleh L, Perler FB. 2006. Protein splicing in cis and in trans. Chem Rec. 6:183–193. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. 1999. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell 99:167–177. [DOI] [PubMed] [Google Scholar]
- Shah NH, Muir TW. 2014. Inteins: nature’s gift to protein chemists. Chem Sci. 5:446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy SM, Fullmer MS, Papke RT, Gogarten JP. 2014. Inteins as indicators of gene flow in the halobacteria. Front Microbiol. 5:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth MW, Benner J, Perler FB. 2000. An alternative protein splicing mechanism for inteins lacking an N-terminal nucleophile. Embo J. 19:5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swithers KS, Senejani AG, Fournier GP, Gogarten JP. 2009. Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements. BMC Evol Biol. 9:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swithers KS, Soucy SM, Lasek-Nesselquist E, Lapierre P, Gogarten JP. 2013. Distribution and evolution of the mobile vma-1b intein. Mol Biol Evol. 30:2676–2687. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. [DOI] [PubMed] [Google Scholar]
- Tatusova T, Ciufo S, Federhen S, Fedorov B, McVeigh R, O’Neill K, Tolstoy I, Zaslavsky L. 2015. Update on RefSeq microbial genomes resources. Nucleic Acids Res. 43:D599–D605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilina NI, Green CM, Jayachandrana P, Kelley DS, Stanger MJ, Piazza CL, Nayak S, Belforta M., 2015. The SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stress. Proc Natl Acad Sci U S A. 112:10348–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilina NI, Mills KV. 2014. Recent advances in in vivo applications of intein-mediated protein splicing. Mob DNA. 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilina NI, Novikova O, Stanger M, Banavali NK, Belfort M. 2015. Post-translational environmental switch of RadA activity by extein-intein interactions in protein splicing. Nucleic Acids Res. 43:6631–6648. doi:10.1093/nar/gkv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Gagnon MC, O’Kelly CJ, Otis C, Lemieux C. 2009. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol. 26:631–648. [DOI] [PubMed] [Google Scholar]
- Walker JR, Hervas C, Ross JD, Blinkova A, Walbridge MJ, Pumarega EJ, Park MO, Neely HR. 2000. Escherichia coli DNA polymerase III tau- and gamma-subunit conserved residues required for activity in vivo and in vitro. J Bacteriol. 182:6106–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu XQ. 1997. Identification of an unusual intein in chloroplast ClpP protease of Chlamydomonas eugametos. J Biol Chem. 272:11869–11873. [DOI] [PubMed] [Google Scholar]
- Weigel C, Seitz H. 2006. Bacteriophage replication modules. FEMS Microbiol Rev. 30:321–381. [DOI] [PubMed] [Google Scholar]
- Wing RA, Bailey S, Steitz TA. 2008. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III alpha-subunit. J Mol Biol. 382:859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara K, Fukuyo M, Sasaki A, Kobayashi I. 2009. Evolutionary maintenance of selfish homing endonuclease genes in the absence of horizontal transfer. Proc Natl Acad Sci U S A. 106:18861–18866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glockner FO, Rossello-Mora R. 2008. The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 31:241–250. [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, et al. 2006. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34:W293–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.