Abstract
Marine mollusc production contributes to food and economic security worldwide and provides valuable ecological services, yet diseases threaten these industries and wild populations. Although the infrastructure for mollusc aquaculture health management is well characterized, its foundations are not without flaws. Use of notifiable pathogen lists can leave blind spots with regard to detection of unlisted and emerging pathogens. Increased reliance on molecular tools has come without similar attention to diagnostic validation, raising questions about assay performance, and has been accompanied by a reduced emphasis on microscopic diagnostic expertise that could weaken pathogen detection capabilities. Persistent questions concerning pathogen biology and ecology promote regulatory paralysis that impedes trade and which could weaken biosecurity by driving commerce to surreptitious channels. Solutions that might be pursued to improve shellfish aquaculture health management include the establishment of more broad-based surveillance programmes, wider training and use of general methods like histopathology to ensure alertness to emerging diseases, an increased focus on assay assessment and validation as fundamental to assay development, investment in basic research, and application of risk analyses to improve regulation. A continual sharpening of diagnostic tools and approaches and deepening of scientific knowledge is necessary to manage diseases and promote sustainable molluscan shellfish industries.
Keywords: emerging disease, aquaculture health, OIE
1. Introduction
Global aquaculture production is now a US$157 billion industry, with US$20.5 billion of this total representing the production of molluscs for food in the marine environment [1]. The largest share of this represents culture of bivalves such as oysters, clams and scallops, but gastropods such as abalone and conch are important culture products as well. Many molluscs also play key roles in ecosystem structure and function [2], and their populations are often enhanced for these reasons [3]. Mollusc mariculture spans temperate and tropical regions, but potential exists for further growth, particularly as ‘the great potential of marine bivalve aquaculture in most maritime countries in Africa and Central America remains untapped’ [1]. Marine mollusc production can be a key nutritional source as well as an economic driver in coastal areas while also providing important ecosystem services such as filtration of phytoplankton (in the case of bivalve molluscs in particular) and carbonate buffering.
Infrastructure development remains a challenge to the growth of mollusc aquaculture, even in countries with advanced aquaculture where access to waterfront can be limited. Sanitation of aquacultured shellfish is a concern, with pathogens such as noroviruses [4] and Vibrio parahaemolyticus [5–7] causing more cases of foodborne illness with warming seawater temperatures in temperate areas and increased development at the coasts [8]. Yet infectious diseases of the cultured animals themselves are perhaps the most serious threat to marine mollusc production. Severe outbreaks have wrought significant economic destruction, with some essentially causing the collapse of entire industries. For instance, epizootics of protistan parasites Haplosporidium nelsoni in the 1950s–1960s [9,10] and Perkinsus marinus in the 1980s–1990s [11,12] devastated the planting industry for oyster Crassostrea virginica in the US mid-Atlantic. An iridoviral outbreak in oyster Crassostrea angulata in southern Europe in the 1960s drove this species to commercial extinction [13]. And emergence of protistan parasites Marteilia refringens [14,15] and Bonamia ostreae [16] in Ostrea edulis in Europe in the 1960s–1970s greatly diminished populations of this oyster and the industries reliant on it. More recently, herpesviruses have affected abalone populations and related industries in Taiwan since 2003 [17] and Victoria, Australia since 2006 [18,19], and emergence of ‘microvariants' of herpesvirus OsHV-1 in Crassostrea gigas in Europe, Australia and New Zealand [20,21] threatens global production of this key aquaculture species.
Maintaining the biosecurity of international aquaculture industries requires control of these and other pathogens of concern [22]. This can include rapid detection in animals proposed for sale or transfer to prevent pathogen introduction to new areas and maintain disease-free facilities and regions, and focused management of pathogens where they do occur, including the use of pathogen-resistant broodstocks [23] to reduce the magnitude of seasonal epizootics. International frameworks for promoting effective disease management such as the World Organisation for Animal Health (OIE) [24], more powerful diagnostic methods and a deepening base of scientific knowledge are the foundation for potent control of pathogens harmful to mollusc production. There is a growing awareness of flaws in this foundation, however. For example, focusing diagnostic effort and management on some pathogens can ignore others. Additionally, the continual evolution of diagnostic tools without continual assessment prevents a firm grasp on how ‘state-of-the-art’ assays are actually performing. Finally, basic questions about pathogen biology and ecology remain unanswered. In this review, we explore these three fundamental areas of concern, which we identify as the Paradox of the List of Notifiable Diseases, the Paradox of Advanced Diagnostics and the Paradox of Uncertainty. We suggest potential solutions as we strive towards more effective control of the pathogens in significant mariculture industries for molluscs.
2. The paradox of the list of notifiable diseases
The need for better information exchange on animal diseases among countries led to the creation of the OIE (originally the Office International des Epizooties, now the World Organisation for Animal Health) in the 1920s. To facilitate this exchange of information and promote more effective disease control, a list of significant diseases was established. Besides the exchange of information, the objective of listing diseases is to support the efforts of OIE member countries in preventing the interstate spread of important diseases through transparent and consistent reporting and finally to ensure safe trade. The OIE Aquatic Animal Health Code aims to assure the sanitary safety of international trade in aquatic animals (amphibians, fish, crustaceans and molluscs) and their products in a context of expanding world trade in these species and products [25].
From nine diseases affecting mammals in 1924, the number of listed diseases increased to approximately 100 distributed in two lists (List A and List B) in the early 1980s. In 2001, a single list of animal diseases was established and replaced Lists A and B. At present, notifiable diseases include 72 terrestrial animal diseases and 28 aquatic animal diseases among which seven are diseases of molluscs. While protozoans cause most of the historically significant mollusc diseases (figure 1) and the majority of those that are notifiable to the OIE, the OIE notifiable list also includes one bacterial disease, infection of abalone with Xenohaliotis californiensis, and one viral disease, infection with abalone herpesvirus. While the total volume of worldwide abalone fisheries has declined since the 1970s, farm production and trade has increased significantly over the past few years [26]. The increase in abalone production has been accompanied by disease outbreaks including ‘withering syndrome’ due to X. californiensis and abalone viral ganglioneuritis due to the herpesvirus.
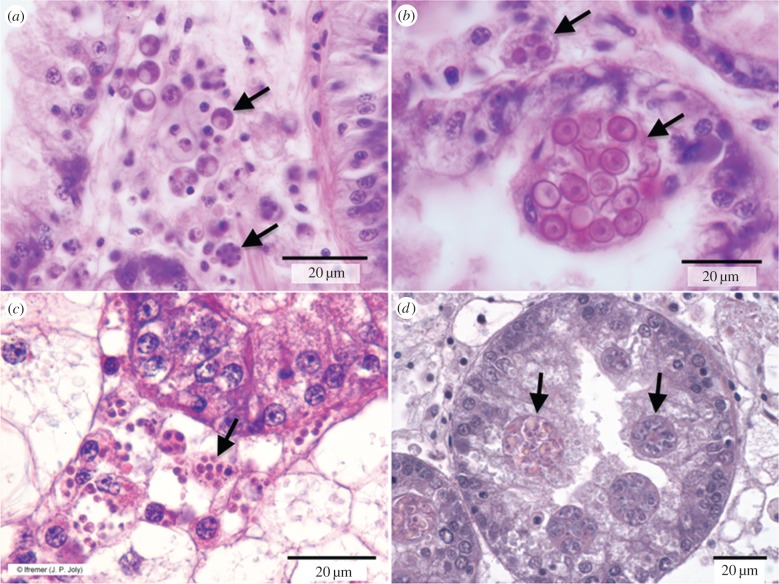
Figure 1.
Representatives of major groups of mollusc pathogens. Arrows in each panel indicate representative cell forms. (a) Perkinsus marinus infecting oyster C. virginica. (b) Haplosporidium nelsoni infecting C. virginica. (c) Bonamia ostreae infecting oyster O. edulis. (d) Marteilia refringens infecting O. edulis. All scale bars, 20 µm.
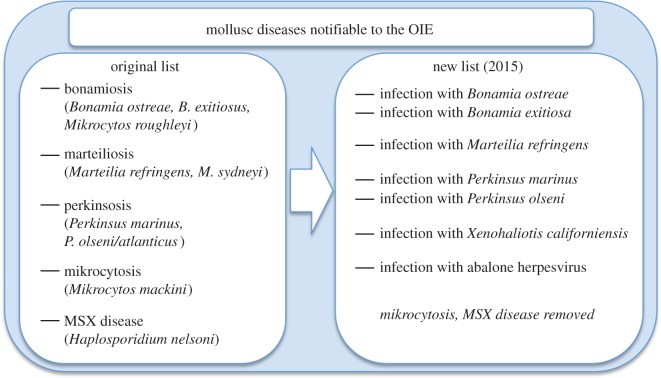
The present OIE notifiable list reflects one major shift, from the earlier disease list to the more specific pathogen list. ‘Marteiliosis', for example, is now listed as ‘Infection by Marteilia refringens' (figure 2). The reason for this is that mollusc diseases are rarely associated with clear and specific clinical signs or gross pathology. Suspicion of disease is often based on mortality reports that occur while infections are rather advanced. By listing pathogens, surveillance relies more straightforwardly on pathogen detection rather than on often nebulous clinical signs. Listing pathogens only requires a clear definition of these notifiable pathogens and effective and appropriate diagnostic tools. The shift towards listing specific pathogens improved a powerful tool for aquatic animal health management, one that focused surveillance and reporting effort on specific disease agents. The creation and use of a notifiable disease or pathogen list, however, is not as simple and straightforward as it might seem.
Figure 2.
Original and current lists of mollusc diseases notifiable to the World Organisation for Animal Health (OIE).
A listed disease must be accompanied by a clear case definition. The overriding criterion for listing a disease is its potential for international spread. Other criteria include zoonotic potential and capacity to spread to naive populations. In addition, a repeatable and robust means of detection/diagnosis is essential for the pathogen under consideration [27]. Paradoxically, a single pathogen might be viewed dissimilarly with regard to these criteria by different advisory or regulatory bodies or in different regions. Indeed, the volume of trade in molluscs, the potential for disease spread and the impact can vary widely from one country or region to another. For example, the list of mollusc diseases notifiable to the European Union (EU) displays several differences from the OIE list. The EU notifiable list includes infection with Mikrocytos mackini, a parasite infecting the oyster C. gigas along the Pacific coasts of Canada and the United States that was delisted by the OIE. The potential impact of M. mackini on oyster production in Europe, and the likelihood that the parasite could find suitable environmental conditions to complete its life cycle in several EU states, has justified continued listing of this pathogen by the EU. In contrast, infection with the host generalist Perkinsus olseni is no longer included on the EU list despite this pathogen's presence in several European countries including Portugal, Spain, Italy and France as well as its listing by the OIE [28]. Discrepancies between the EU and OIE lists could confound commerce in molluscs between EU and non-EU countries, but the potential biosecurity costs are no less a concern. While costs to biosecurity would not be obvious in the example of M. mackini, the non-listing of P. olseni within the trading network of the EU could allow potential spread of this pathogen within the EU to areas where it does not presently occur. Thus, although establishment of notifiable lists using the above criteria is helpful for disease management and control, inconsistencies in interpretation of criteria and the lists established by different bodies can complicate aquaculture commerce and potentially reduce biosecurity at regional scales.
Another paradox is that surveillance and transfer regulations are based on species susceptible to listed diseases, yet firm scientific demonstration of the host range of pathogens is elusive. The definition of susceptible species is critical for determining the scope of disease surveillance, especially in the context of the aquaculture sector and the large and growing number of diverse aquaculture species. Scientific criteria have been established for assessment of host species susceptibility, including existence of transmission pathways consistent with natural routes and clear evidence that presence of an agent in association with a putative host constitutes a genuine infection [27]. However, evidence to demonstrate susceptibility of a species is often lacking, which can keep true hosts from being identified as potential carriers of notifiable pathogens and lead to unintentional spread of these diseases. Such a situation might occur in the case of generalist pathogens such as P. olseni. Indeed, P. olseni has an extremely wide host range that includes a long list of commercially important clam, oyster and abalone species [29]. However, many other bivalve and gastropod species could be susceptible to this parasite, both within and outside its known geographical range. Most surveillance programmes are established to monitor known susceptible hosts, and transfer or import regulations are based on resulting data. This is necessary because regulations cannot be based on uncertainty alone. Exploratory surveillance of potential alternative host species, however, would provide a stronger scientific basis for conclusions concerning the host range of pathogens, and should be a research priority.
Assessment of susceptibility of host species requires clear definitions of pathogens and differentiation of pathogen strains as any change in these definitions would impact their host and geographical ranges, which would in turn have regulatory consequences. Accurate definition of taxa, however, is not simple. Despite major advances in diagnostic methodologies over the past two centuries, a consensus definition of ‘species' remains elusive [30] and evolves as our knowledge is improved. Marteilia refringens was initially defined as a protozoan infecting flat oysters, O. edulis [15]. A congeneric parasite, M. maurini, was characterized based on morphological and ultrastructural criteria in mussels Mytilus edulis and M. galloprovincialis [31,32]. Molecular analyses subsequently suggested that these parasites could be viewed as one species, but comprising two distinct genetic lineages [33]. Polymorphic sites in the ITS-1 region of the ribosomal RNA gene complex distinguish an ‘O’ genotype more often detected in oysters from an ‘M’ genotype more often detected in mussels [34]. Genotype host specificity is not strict, however, and it is common to find both types in oysters or mussels. OIE listing of infection with M. refringens as a notifiable disease currently covers both type O and type M. The evolution of parasite species definitions, i.e. synonymizing M. maurini with M. refringens, in this case has constructively expanded the list of susceptible species for the notifiable M. refringens for which regulations should apply.
Ostreid herpesvirus type 1 (OsHV-1) presents a more challenging case. OsHV-1 (figure 3) is a pathogen that infects oysters, clams and scallops [35]. Several genotypes have been identified among which one, the OsHV-1 ‘µvar’, has been associated with elevated mortality of Pacific oysters, C. gigas, since 2008. While OsHV-1 more generally is distributed around much of the world, the OsHV-1 µvar and closely related genotypes are known only from Europe, Australia and New Zealand [20,21]. The scope of the OsHV-1 pathogen definition has huge consequences not only for globally significant Pacific oyster trade but also for risk of pathogen distribution. Listing OsHV-1 generally, including all the genotypes, would allow movements of animals between OsHV-1-endemic areas regardless of the presence or absence of the virulent strain OsHV-1 µvar. The result would risk the rapid spread of OsHV-1 µvar beyond its current, limited range. On the contrary, listing OsHV-1 µvar only would permit transfer of animals from countries infected with other strains of OsHV-1 even if very similar to OsHV-1 µvar. An intermediate solution has been chosen in the EU with the listing of OsHV-1 microvariants covering genotypes with common sequence features but displaying small differences. Within the wider context of global aquaculture commerce, however, the OIE has not placed the OsHV-1 µvar on its notifiable list, allowing the pathogen to remain substantially below the radar of disease surveillance and raising the possibility of unintentional further spread.
Figure 3.
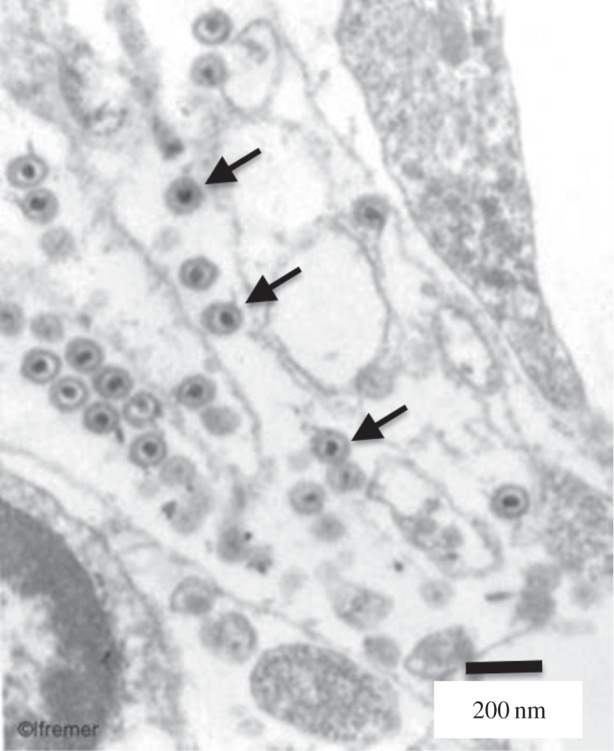
Virions of the herpesvirus OsHV-1 (examples indicated by arrows). Scale bar, 200 nm.
OIE member countries have the responsibility to report not only listed diseases but also emerging diseases. As a consequence of globalization, urbanization and global change, the number of emerging diseases is increasing and special attention is given to their detection. These diseases have a significant impact on aquatic animals resulting from (i) a change of known pathogenic agent or its spread to a new geographical area or species; or (ii) a newly recognized or suspected pathogenic agent. Different scenarios can lead to emergence of diseases but the cause is often the introduction of infected animals. The introduction of B. ostreae to Europe is a notorious example, and it is believed to have occurred via infected O. edulis seed from California [36]. Occasionally, disease emerges from the introduction of a susceptible host to an established pathogen. The Japanese carpet clam, Ruditapes philippinarum, introduced into Europe, is substantially more susceptible to infection with European endemic Vibrio tapetis (the agent of ‘brown ring disease’) compared to the native carpet clam species, Ruditapes decussatus [37]. The outbreak of disease caused by Bonamia exitiosa in experimentally cultured Crassostrea ariakensis along the US East Coast [38] is another example that was ultimately used as part of the rationale against the widespread introduction of C. ariakensis to eastern North America.
Once declared to harbour a pathogen, zones or countries may lose interest in maintaining surveillance efforts that would detect emerging pathogens whatever their source, particularly if the pathogens known to be present are unlikely to ever be eradicated. Very few countries maintain active surveillance of their mollusc populations in endemic zones. However, long-term study on the evolution of some diseases such as infection with B. ostreae in flat oyster populations in the Netherlands [39] and H. nelsoni in eastern oyster C. virginica populations along the mid-Atlantic coast of the USA [40,41] has been illuminating with regard to the evolution of host–parasite relationships, and has helped identify measures for disease management in the context of aquaculture and fisheries. Surveillance in known infected zones can also help detect abnormal situations that could be due to the emergence of new, more virulent genotypes or colonization of new host species.
The case of OsHV-1 µvar highlights an additional issue related to emerging diseases: it can be challenging to place them on notifiable lists. Listing diseases contributes to safe trade through the acquisition and exchange of information about these diseases. By facilitating regulations about the transfer of animals from infected zones to non-infected zones, listing diseases should prevent their spread. Inclusion in the OIE list requires national governments to report outbreaks promptly but may lead to trade restrictions of live animals from infected areas into areas free from the pathogen. The potential for economic losses associated with trade restrictions reduces the incentive for countries affected by an emerging pathogen to advance its listing.
Finally, dispersal of pathogens occurs despite the presence of restrictions. For example, with trade restrictions in place, infection of B. ostreae was recently reported in several bays of Ireland and the UK previously recognized as free of the parasite [42,43]. One unexpected report was the detection of the parasite in O. edulis from Limfjorden, Denmark, a zone that gained its free status regarding bonamiosis in 2004 [44]. The detection of the parasite by histology in 58% of the tested oysters raises questions regarding its introduction: was it introduced through import of infected animals or by transfer of contaminated aquaculture equipment or another vector? Similarly, a surveillance programme was implemented in several bays in Ireland and the United Kingdom regarding infection with OsHV-1 µvar, yet despite trade measures based on this surveillance programme several OsHV-1 µvar outbreaks have been reported. Some of these reports have been associated with shellfish passing through depuration centres (M Gubbins 2015, personal communication). There is presently no legislation regarding water treatment in depuration centres because there are no data on the efficacy of treatment on pathogen inactivation. While animal movements are considered as a first risk for disease introduction, other routes of disease transfer exist such as ballast waters and hull attachment, which have been incriminated in the context of the emergence of H. nelsoni in Nova Scotia, Canada and Marteilioides chungmuensis in Darwin Harbour, Australia [45,46]. Furthermore, climate change is increasingly recognized as a driving force in pattern changes of the distribution as well as activity of pathogens [47].
While animal diseases clearly gain attention by their inclusion on notifiable lists, we must acknowledge the limitation of lists as management tools and consider other or additional means including more broad-based surveillance to avoid pathogen spread and ensure the collection of robust epizootiological information.
3. The paradox of advanced diagnostics
Over the last two decades, DNA-based polymerase chain reaction (PCR) diagnostic assays have evolved from specialized tools to workhorse platforms for detection of pathogens from molluscs and other marine and aquatic organisms [48]. Quantitative polymerase chain reaction (qPCR) assays now offer advantages of quantifying infection intensities [49–52], with new tools already appearing on the horizon in the application of proteomic methods such as matrix-assisted laser desorption time-of-flight mass spectrometry [53]. We accept PCR assays as having lived up to promises of improved sensitivity and specificity relative to older methods, which in many cases were based on microscopic methods, including paraffin histopathology for haplosporidians, cytological evaluation of stained haemolymph preparations or tissue imprints for Bonamia and Marteilia species, and Ray's fluid thioglycollate method [54] for Perkinsus parasites (table 1). While traditional microscopic methods maintain their own unique value, for example for the perspective they provide on infection intensities and the severity of disease caused by pathogens that are present, molecular assays are now routinely used for detection of myriad viral, bacterial and protistan pathogens.
Table 1.
General advantages and disadvantages of diagnostic platforms broadly available at present for mollusc pathogens.
| assay | advantages | disadvantages |
|---|---|---|
| gross pathology | allows rapid preliminary assessment in systems where specific gross signs are commonly associated with infection (e.g. mikrocytosis in C. gigas, Brown Ring Disease in R. philippinarum) | few pathogens are associated with relatively unambiguous gross signs of disease |
| paraffin histopathology | provides excellent perspective on animal health; allows detection of multiple infections and emerging disease events; technology widely available | slow; requires specialized expertise; can be insensitive for detection of small pathogens (e.g. Bonamia); not suitable for viruses and bacteria |
| cytology, tissue imprints | rapid and inexpensive; useful for specific evaluation of haemolymph and haemocytes for Bonamia, of digestive gland imprints for Marteilia | not considered effective for detection of Perkinsus or Haplosporidium parasites; not useful for bacteria, viruses |
| Ray's fluid thioglycollate method | rapid and inexpensive; useful for specific evaluation of tissue samples for most Perkinsus parasites; can be quantitative | only specific to Perkinsus species |
| transmission electron microscopy | allows ultrastructural description; suitable for distinguishing parasites belonging to different genera (e.g. Bonamia from Mikrocytos) and sometimes congeneric species, identifying and characterizing viral infections | expensive, slow; requires specialized expertise and technology that is not universally available; focus on very small tissue areas could result in pathogens being missed |
| PCR, conventional | relatively rapid; required skills common in students of biology; technology widely available; can be more sensitive for detection of small pathogens that are difficult to visualize microscopically; specificity can be ‘tuned’ to be broad or narrow | provides indirect and imperfect perspective on animal health; positive results only indicate presence of pathogen DNA, not necessarily viable pathogen or actual infection; requires substantial background knowledge of the genetics of targeted (and ideally, related) pathogens |
| PCR, quantitative | same as conventional PCR, but with added advantages of pathogen quantitation, greater speed and likely sensitivity; with validation quantitation may allow stronger inferences about actual infection than are possible with conventional PCR | Same as conventional PCR; platform more expensive, less widely available than for conventional PCR and requires greater expertise for proper interpretation; copy number of target imperfectly correlated with infection intensity |
| ISH | best single method for linking a DNA sequence to pathogen observed in tissue sections; as with PCR, sensitivity can be tuned; requires histopathology but more sensitive than conventional histopathology for detection of small cryptic pathogens | (very) slow, very specialized |
| DNA sequencing | constitutes the definitive identification of pathogen DNA sequences | same as quantitative PCR; requires PCR amplification first, which is not always straightforward for detection or characterization of new pathogens |
| next-generation sequencing | allows rapid profiling of pathogen diversity | expensive, with technology not universally available; requires substantial bioinformatics expertise and resources; as for PCR, positive results not clearly indicative of actual infection |
The usefulness of PCR assays is unquestionable, especially with regard to viral pathogens such as OsHV-1 [55] that cannot readily be visualized microscopically. In situ hybridization (ISH), the detection of pathogens in histological sections using DNA probes, is a similarly valuable molecular assay but a more specialized tool used primarily for characterizing new pathogens [56,57] and linking known pathogens to new hosts [58,59]. Both PCR and ISH are valuable in elucidating life cycles. However, as with the use of notifiable lists, the application of PCR diagnostics can produce ‘blind spots' with regard to pathogen detection that may reduce, rather than enhance, biosecurity. The first of these is a by-product of the specificity of the PCR assays. PCR can only detect the pathogens it specifically targets. It thus provides a powerful means for inferring the presence or absence (and in the case of qPCR, the infection intensity) of these specific pathogens in sample materials. The problem potentially arises when PCR tools are applied to the exclusion of more general methods such as histology that provide broader perspective on pathogen presence and disease status. Applying a battery of specific PCR tests for notifiable pathogens may allow effective management of those particular pathogens, for example in transfers of mollusc seed from hatcheries or nurseries to farms where the product will be grown to market size. Yet it provides no perspective on other pathogens that may be present in a population. Lack of attention to the broader array of potential pathogens could allow spread of important new pathogens before they come to the attention of health managers. The continuing growth of mollusc aquaculture worldwide will require that PCR diagnostics play an ever more central role in quickly determining the infection status of product destined for transfer, especially when evaluations are particularly time-sensitive as in the transfer of germplasm or larvae. It is important, however, that ‘classical’ microscopic methods play their role in establishing the general health of aquaculture populations and maintaining vigilance towards detection of emerging diseases. Molecular and microscopic methods are complementary tools for managing molluscan health, but each has purposes for which it is most suitable.
A second, related issue is the relative scarcity of expertise in invertebrate pathology relative to molecular diagnostics. Despite their importance to global economies, invertebrates ‘have been ignored to some extent by veterinary medicine’ [60], and relatively few students are trained in the recognition of pathogens and pathologies of molluscs and other invertebrates. Many students, on the other hand, are educated in molecular genetics, which means that molecular diagnostics for invertebrate pathogens can be more capably applied at a broad scale than can histopathology. This can promote an overreliance on molecular diagnostic assays because these are the tools most familiar and readily employed. Although national laboratories responsible for ensuring the biosecurity of international commerce are generally expert in the full range of diagnostic methods for shellfish pathogens, smaller local and regional laboratories may have expertise primarily in molecular diagnostics, leading to overemphasis of these specific assays and a reduced effectiveness in general health screening at local and regional levels. This may ultimately have relevance at the national level for international commerce, particularly if pathogens emerging locally become widespread and threaten the security of exports. Training of the next generation of shellfish health professionals not only in molecular genetics but also in histopathology and the basic recognition of shellfish pathogens will be important for strengthening the biosecurity foundations of the field.
The lack of attention to, and support for, assessment and validation of the molecular diagnostic tools in use is a different and more serious problem. Without a proper assessment, we are unable to determine how effective these assays are. The assay development and validation pathway recommended by the OIE includes definition of the intended purpose of the assay, assay design and optimization (development pathway); followed by determination of analytical characteristics (validation pathway, Stage 1), diagnostic characteristics (Stage 2) and reproducibility (Stage 3), and then implementation (Stage 4), with subsequent monitoring and maintenance of the validation criteria [61]. Key molecular assays for mollusc pathogens have been subject to little formal assessment and very few have moved beyond Stage 1 in the assessment pathway. Only PCR assays for the Australian abalone herpesvirus [62] and oyster parasites M. mackini [63] and B. ostreae and B. exitiosa [52] are noteworthy for the degree of assessment applied. Although a long history of use does provide confidence that important tools such as the conventional PCR assays for H. nelsoni [64,65], M. refringens [34], B. ostreae [66] and P. marinus [49] perform reliably despite limited formal assessment, a ‘long history of use’ is no substitute for the formal validation that is essential to demonstrate that these molecular assays perform as intended. Lack of validation has opened the door to a proliferation of redundant assays that complicate the matter of formally determining which assays are suitable for which application. While the importance of pursuing formal validation of key assays is recognized by the mollusc health community, funding agencies ultimately need to appreciate the immense value and need of formal assay validation and commit to it, which will cost more and take more time than simple assay design.
Increasingly powerful technology should allow ever more effective diagnostics of shellfish pathogens. Despite the attractiveness and promises of advanced diagnostics, the paradox is that the effectiveness of diagnostics will remain illusory without proper consideration of which tools we use for what application and for which purpose each assay is best fit. Furthermore, developing a cadre of professionals fully trained and expert in the range of diagnostics who can conduct thorough evaluations of both new and existing tools is essential to determine which tools are most appropriate and most accurate moving forward.
4. The paradox of uncertainty in shellfish disease management
Managing molluscan shellfish health is in large part an exercise in risk management (see for example [67,68]). Specifically, the rapid growth of shellfish aquaculture described above has led to increased movements of shellfish stocks that, concomitantly, increase the risk of disease transfer [69]. It follows that there is an increasing need for more effective and efficient management of shellfish health.
Managing shellfish health must balance preventing the spread and transfer of pathogens with the desire to transfer shellfish to restore, sustain or increase shellfish populations. This desire may be derived from economic or ecologic interests, or both. Complicating this challenge, however, is a climate of uncertainty resulting from the complex interactions among hosts, pathogens and the environs in which they interact. Management is further confounded by political boundaries because they create jurisdictional limits that are often inconsistent with the ecological boundaries that shape pathogen distributions. Ultimately, balancing shellfish health management with shellfish production, trade and restoration requires a collaborative science-driven risk analysis that begins by recognizing that the risk of disease can never be eliminated.
Uncertainty often understandably invokes the precautionary principle, but application of this principle varies [70]. In extreme cases, a risk-averse strategy may invoke the precautionary principle to prohibit activities that might cause harm regardless of the magnitude or probability or any mitigating measures that could be taken to reduce risks. As a result, uncertainty can derail shellfish transfers by enacting severe restrictions that may prohibit commerce regardless of whether it actually presents a disease introduction risk. For example, in the United States, the South Carolina Department of Natural Resources (SCDNR), charged with protecting the natural resources of the state, recently issued the following ban on shellfish importations:
Based on recent information on oyster pathogens and considering the potential risk to native resources (which may be the last intact healthy populations of oysters in the world), SCDNR is declaring a moratorium on importing oyster seed into South Carolina from hatcheries located in areas of disease concern. This includes all states north of South Carolina. SCDNR already had a policy against importation of oyster seed from those states unless they were coming from a hatchery. This policy is expanded to include hatcheries, effective immediately. This moratorium will remain in effect until such time as we feel the risk has been removed. We regret any inconvenience this may cause the shellfish industry but our paramount concern must be to protect our natural resources (East Coast Shellfish Growers Association e-mail listserve, 1 April 2014).
Similarly, the Department of Marine Fisheries in the state of Massachusetts, USA, requires all importations to be certified ‘disease-free by a qualified marine pathologist’ (http://www.mass.gov/eea/agencies/dfg/dmf/laws-and-regulations/322-cmr-15-00-management-of-marine-aquaculture.html). The department has ‘maintained a ‘Zero Tolerance’ for any degree of infection (heavy or light) or prevalence (number or percentage of animals tested) in any sample from an aquaculture facility or body of water whether the shellfish are cultured or wild’. The precautionary principle states that actions should be taken to avoid or diminish potential harm [71] and these are certainly actions towards that objective, but they are extreme positions that exclude consideration of existing mitigating conditions as well as potential prevention measures, namely endemic pathogens in the recipient waters. Unlike the EU, disease zones have not been designated in the US. Such designations may help regulators move away from strict zero-tolerance policies.
When the precautionary principle is applied in the absence of a risk analysis, the effect can be stifling. In most cases, an action that poses a risk typically provides potential benefits. Risk analysis considers the magnitudes of the risk and the benefits and compares these to the impacts of not allowing the activity. Invoking the precautionary principle without any risk analysis often pits regulators against practitioners by eliminating opportunities for discourse and resolution, including taking precautionary steps that minimize risk. Additionally, although application of the precautionary principle without a risk analysis overemphasizes biosecurity at the cost of commerce, it may have the unintended consequence of driving transfers through surreptitious channels, which could have the effect of reducing biosecurity. This is thus a central paradox regarding uncertainty: it can produce risk aversion to the detriment of both trade and biosecurity.
In most cases, information exists about the level of harm that may occur and the level of risk involved with a particular action. Weighing the degree of harm along with the likelihood of harm provides a basis for assessing risk that can be used to make informed decisions while following the precautionary principle's guidance of taking action to avoid or reduce harm [72]. Managers and practitioners may ultimately disagree, but risk analysis ensures that there is a rational basis for their disagreement. Consensus may be reached through the identification of precautionary measures that significantly reduce the level of risk, e.g. limiting movement to younger animals or only animals with disease levels below those of populations in the receiving waters.
Uncertainty arises from a lack of information and/or predictability. Minimizing uncertainty provides the greatest opportunity for managing shellfish health without restricting shellfish commerce and shellfish restoration. Some uncertainty can be eliminated simply through dissemination of existing information. In other cases, uncertainty can only be addressed through the collection of new information. For example, Ulrich et al. [73] reported detecting DNA of H. nelsoni, the agent of MSX disease in C. virginica, in samples that had been collected from the Gulf of Mexico. Previously, H. nelsoni had been reported only from the Atlantic coast of North America, never in the Gulf of Mexico, so this report created uncertainty about potential movements of shellfish, even shellfish gametes, from the Atlantic to the Gulf coasts of North America. Subsequently, an extensive survey was unable to confirm the presence of H. nelsoni in the Gulf of Mexico [74]. This example points to the need for diligent surveillance. In fact, basic distribution data are incomplete for many pathogens and this can lead to controversial policies with some entities denying that a pathogen is present simply due to inadequate surveillance. Inadequate distributional data can affect both import and export of host species.
Another source of uncertainty is unresolved life cycles. Examples include haplosporidians, Marteilia spp. and QPX, the labyrinthuloid parasite of the hard clam Mercenaria mercenaria. Transmission of many pathogens is often specific to certain life-history stages. Thus, uncertainty in the life history leads to uncertainty about modes of transmission. This uncertainty can lead to caution over transfers, but in many cases (e.g. H. nelsoni) data indicate that direct transmission is not possible, and because an alternate host/vector remains unknown [41] there is little basis upon which to indicate risk. Bushek & Allen [75] highlighted the importance of strain and stock variability in virulence and susceptibility for pathogens and hosts, respectively. Little data exist to suggest this is a pervasive problem, yet it is often cited as a major source of uncertainty. Finally, there is uncertainty about how hosts and pathogens might interact in different systems under slightly or substantially different environmental conditions. A lack of understanding about environmental tolerances confounds an understanding of ecological boundaries.
Regulatory authority typically resides within political jurisdictions, but political jurisdictions often include multiple ecological boundaries that may or may not be recognized by regulatory agencies within a political jurisdiction. The problem is probably related to regulatory expediency rather than any biological or ecological rationale related to host or pathogen distributions. For example, the Delaware Bay is bisected along its length by the states of Delaware and New Jersey in the United States. Movement of oyster seed from New Jersey's portion of Delaware Bay to oyster beds on the Delaware side of the bay requires disease-free certification to ensure diseases are not moved with the seed, even though P. marinus and H. nelsoni are readily transmitted naturally from one side of the bay to the other. In such cases, political jurisdictions should work collaboratively to facilitate rational transfers. Zonal strategies may help regulators overcome political jurisdictional limitations such as this.
Knowledge of the basic biology and ecology of host–pathogen interactions as well as the movements of shellfish for trade are key to successful management but we often have an insufficient understanding of both to effectively manage shellfish diseases (for example [68]). A unique and perhaps unprecedented evaluation occurred during the 2000s along the east coast of the United States following a proposal to introduce a western Pacific oyster, C. ariakensis, to the Chesapeake Bay [71]. The proposal had three goals. First was to restore the ecological role of oysters by creating a self-recruiting population that would build reefs comparable to those that once existed extensively throughout the bay. Second was to revitalize the nearly extinct oyster fishery. Third was to develop a viable aquaculture industry through the introduction of a fast growing oyster that could reach market size in much less than a year to make oyster aquaculture viable. The proposal met much resistance and an intensive decade long study was conducted to evaluate the multitude of concerns [76]. Two primary concerns were the potential for introducing a non-native pathogen and the susceptibility of C. ariakensis to known native pathogens. Neither turned out to be major concerns, but a surprise was the discovery of a novel oyster pathogen during experiments conducted in North Carolina [38]. Ultimately, the introduction was denied, but the data collected were extremely valuable and provided a model for the level of investigation necessary to reduce uncertainty for such bold proposals.
The paradox of uncertainty in shellfish disease management is that management errors have two general outcomes: increasing the spread and severity of disease or unnecessarily limiting the production of shellfish. Finding a balance is, to some extent, a moving target that requires persistent surveillance, evaluation, innovation and adaptation.
5. Conclusion
There are multiple challenges associated with determining which shellfish pathogens most deserve surveillance and regulatory attention, and how best to detect and monitor those deemed most significant without losing capacity to detect emerging diseases. At the same time, fundamental questions of parasite distributions and ecology remain unanswered despite an ever-expanding scientific knowledge base, promoting a high level of caution among regulators. As described above, this situation creates a paradox that can have the effect of diminishing both trade and biosecurity, but actions can be taken to improve this paradox of shellfish health management.
(1) The establishment of more broad-based surveillance programmes will help define the distribution of pathogens within host populations at risk.
(2) Ensuring wider training in the use of general methods such as histopathology will help ensure alertness to emerging diseases that may otherwise go undetected until major losses occur.
(3) Demanding a focus on assay assessment and validation as fundamental to assay development will ensure that ‘state-of-the-art’ technologies perform to standards required for disease detection, surveillance and management.
(4) Providing investment in research to eliminate key knowledge gaps will increase confidence by increasing certainty across multiple aspects of management and regulation.
(5) The application of risk analyses is paramount to avoid regulatory paralysis that confronts managers called on to protect both natural and cultivated resources within their jurisdictional control.
Given the increasing production of molluscan species in aquaculture and the continued importance of capture fisheries in many areas [1], pursuing these solutions will benefit food security worldwide.
Acknowledgements
We thank the NSF RCN Award 1215977 team of investigators for inviting and encouraging this review, and Bruno Chollet and Jean Pierre Joly for providing images of M. refringens and B. ostreae, respectively, for figure 1. We appreciate the generous input from Kevin Lafferty and two anonymous reviewers whose suggestions improved the manuscript. This is Virginia Institute of Marine Science contribution number 3526.
Authors' contributions
R.B.C., I.A. and D.B. contributed equally to this publication.
Competing interests
We have no competing interests.
Funding
Partial support was provided by NSF Award 1216220 and NOAA Sea Grant Aquaculture Research Program Award NA14OAR4170093. Additional support was provided by the Virginia Institute of Marine Science Foundation A. Marshall Acuff, Sr., Memorial Endowment for oyster disease research to R.B.C., and USDA NIFA Hatch award 32109 to D.B.
References
- 1.FAO. 2015. FAO Global Aquaculture Production database updated to 2013 – Summary information. See http://www.fao.org/3/a-i4899e.pdf.
- 2.Dame R. 1996. Ecology of marine bivalves: an ecosystem approach. Boca Raton, FL: CRC Marine Science Series. [Google Scholar]
- 3.Coen LD, Brumbaugh RD, Bushek D, Grizzle R, Luckenbach MW, Posey MH, Powers SP, Tolley GS. 2007. As we see it: ecosystem services related to oyster restoration. Mar. Ecol. Prog. Ser. 341, 303–307. ( 10.3354/meps341303) [DOI] [Google Scholar]
- 4.Yu X, Cai H, Hu L, Lei R, Pan Y, Yan S, Wang Y. 2015. Molecular epidemiology of oyster-related human noroviruses: global genetic diversity and temporal-geographical distribution from 1983 to 2014. Appl. Env. Microbiol. 81, 7615–7624. ( 10.1128/AEM.01729-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newton AE, Garrett N, Stroika SG, Halpin JL, Turnsek M, Mody RK. 2014. Increase in Vibrio parahaemolyticus infections associated with consumption of Atlantic Coast shellfish-2013. MMWR Morb. Mortal. Wkly Rep. 63, 335–336. [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YS, Park KH, Chun HS, Choi C, Bahk GJ. 2015. Correlations between climatic conditions and foodborne disease. Food Res. Intl 68, 24–30. ( 10.1016/j.foodres.2014.03.023) [DOI] [Google Scholar]
- 7.Froelich BA, Noble RT. 2016. Vibrio bacteria in raw oysters: managing risks to human health. Phil. Trans. R. Soc. B 371, 20150209 ( 10.1098/rstb.2015.0209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groner ML, et al. 2016. Managing marine disease emergencies in an era of rapid change. Phil. Trans. R. Soc. B 371, 20150364 ( 10.1098/rstb.2015.0364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews JD. 1962. Oyster mortality studies in Virginia IV. MSX in James River public seed beds. Proc. Natl Shellfish. Assoc. 53, 65–84. [Google Scholar]
- 10.Haskin HH, Stauber LA, Mackin JA. 1966. Minchinia nelsoni n. sp. (Haplosporida, Haplosporidiidae): causative agent of the Delaware Bay oyster epizootic. Science 153, 1414–1416. ( 10.1126/science.153.3742.1414) [DOI] [PubMed] [Google Scholar]
- 11.Burreson EM, Andrews JD.1988. Unusual intensification of Chesapeake Bay oyster diseases during recent drought conditions. In Proc. Oceans ’88 Conf. , Baltimore, MD , 31 October–2 November 1988 , pp. 799–802. Piscataway, NJ: Institute of Electrical and Electronic Engineers. ( ) [DOI]
- 12.Ford SE. 1996. Range extension by the oyster parasite Perkinsus marinus into the northeastern United States: response to climate change? J. Shellfish Res. 15, 45–56. [Google Scholar]
- 13.Marteil L. 1968. La ‘maladie des branchies’. Int. Counc. Explor. Sea K. 5, 1–5. [Google Scholar]
- 14.Comps M. 1970. Observations sur les causes d'une mortalité anormal des huitres plates dans le basin de Marennes. Rev. Trav. Inst. Peches. Marit. 34, 317–326. [Google Scholar]
- 15.Grizel H, Comps M, Bonami JR, Cousserans F, Duthoit JL, Pennec M. 1974. Epizooty of the common oyster Ostrea edulis. Part 1. Study of the agent of digestive gland disease in Ostrea edulis (Linne). Sci. Peche 240, 7–30. [Google Scholar]
- 16.Pichot Y, Comps M, Tigé G, Grizel H, Rabouin M-A. 1980. Recherches sur Bonamia ostreae gen. n., sp. n., parasite nouveau de l'huître plate Ostrea edulis. Rev. Trav. Inst. Pêches Marit. 43, 131–140. [Google Scholar]
- 17.Chang PH, Kuo ST, Lai SH, Yang HS, Ting YY, Hsu CL, Chen HC. 2005. Herpes-like virus infection causing mortality of cultured abalone Haliotis diversicolor supertexta in Taiwan. Dis. Aquat. Org. 65, 23–27. ( 10.3354/dao065023) [DOI] [PubMed] [Google Scholar]
- 18.Hooper C, Hardy-Smith P, Handlinger J. 2007. Ganglioneuritis causing high mortalities in farmed Australian abalone (Haliotis laevigata and Haliotis rubra). Austral. Vet. J. 85, 188–193. ( 10.1111/j.1751-0813.2007.00155.x) [DOI] [PubMed] [Google Scholar]
- 19.Ellard K, Pyecroft S, Handlinger J, Andrewartha R.2009. Findings of disease investigations following the recent detection of AVG in Tasmania. In Proc. Fourth National FRDC Aquatic Animal Health Scientific Conf., Cairns, Australia, 22–24 July 2009. Deakin, Australia: Fisheries Research and Development Corporation.
- 20.Segarra A, Pepin JF, Arzul I, Morga B, Faury N, Renault T. 2010. Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 153, 92–95. ( 10.1016/j.virusres.2010.07.011) [DOI] [PubMed] [Google Scholar]
- 21.Jenkins C, et al. 2013. Identification and characterisation of an ostreid herpesvirus-1 microvariant (OsHV-1 µ-var) in Crassostrea gigas (Pacific oysters) in Australia. Dis. Aquat. Org. 105, 109–126. ( 10.3354/dao02623) [DOI] [PubMed] [Google Scholar]
- 22.Lafferty KD, Hofmann EE. 2016. Marine disease impacts, diagnosis, forecasting, management and policy. Phil. Trans. R. Soc. B 371, 20150200 ( 10.1098/rstb.2015.0200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragone Calvo LM, Calvo GW, Burreson EM. 2003. Dual disease resistance in a selectively bred eastern oyster, Crassostrea virginica, strain tested in Chesapeake Bay. Aquaculture 220, 69–87. ( 10.1016/S0044-8486(02)00399-X) [DOI] [Google Scholar]
- 24.Rodgers CJ, Carnegie RB, Chávez-Sánchez MC, Martínez-Chávez CC, Furones Nozal MD, Hine PM. 2000. Legislative and regulatory aspects of molluscan health management. J. Invertebr. Pathol. 131, 242–255. ( 10.1016/j.jip.2015.06.008) [DOI] [PubMed] [Google Scholar]
- 25.Washington S, Ababouch L. 2011. Private standards and certification in fisheries and aquaculture. FAO Fisheries Technical Paper No. 553, p. 181 Rome, Italy: FAO. [Google Scholar]
- 26.Cook PA. 2014. The worldwide abalone industry. Mod. Econ. 5, 1181–1186. ( 10.4236/me.2014.513110) [DOI] [Google Scholar]
- 27.OIE. 2015. Aquatic animal health code, eighteenth edition. Paris, France: World Organisation for Animal Health. [Google Scholar]
- 28.Arzul I, Chollet B, Michel J, Robert M, Garcia C, Joly J-P, Francois C, Miossec L. 2012. One Perkinsus species may hide another: characterization of Perkinsus species present in clam production areas of France. Parasitology 139, 1757–1771. ( 10.1017/S0031182012001047) [DOI] [PubMed] [Google Scholar]
- 29.Villalba A, Reece KS, Ordas MC, Casas SM, Figueras A. 2004. Perkinsosis in molluscs: a review. Aquat. Living Resour. 17, 411–432. ( 10.1051/alr:2004050) [DOI] [Google Scholar]
- 30.Hey J. 2001. The mind of the species problem. Trends Ecol. Evol. 16, 326–329. ( 10.1016/S0169-5347(01)02145-0) [DOI] [PubMed] [Google Scholar]
- 31.Comps M, Pichot Y, Papagianni P. 1981. Recherche sur Marteilia maurini n. sp. parasite de la moule Mytilus galloprovincialis Lmk. Rev. Trav. Inst. Pêches Marit. 45, 211–214. [Google Scholar]
- 32.Auffret M, Poder M. 1983. Recherches sur Marteilia maurini, parasite de Mytilus edulis sur les cotes de Bretagne nord. Rev. Trav. Inst. Pêches Marit. 47, 105–109. [Google Scholar]
- 33.Lopez-Flores I, De La Herran R, Garrido-Ramos MA, Navas JI, Ruiz-Rejon C, Ruiz-Rejon M. 2004. The molecular diagnosis of Marteilia refringens and differentiation between Marteilia strains infecting oysters and mussels based on the rDNA IGS sequence. Parasitology 129, 411–419. ( 10.1017/S0031182004005827) [DOI] [PubMed] [Google Scholar]
- 34.Le Roux F, Lorenzo G, Peyret P, Audemard C, Figueras A, Vivares C, Gouy M, Berthe FCJ. 2001. Molecular evidence for the existence of two species of Marteilia in Europe. J. Euk. Microbiol. 48, 449–454. ( 10.1111/j.1550-7408.2001.tb00178.x) [DOI] [PubMed] [Google Scholar]
- 35.Renault T, Novoa B. 2004. Viruses infecting bivalve molluscs. Aquat. Living Resour. 17, 397–409. ( 10.1051/alr:2004049) [DOI] [Google Scholar]
- 36.Elston RA, Farley CA, Kent ML. 1986. Occurrence and significance of bonamiasis in European flat oysters Ostrea edulis in North America. Dis. Aquat. Org. 2, 49–54. ( 10.3354/dao002049) [DOI] [Google Scholar]
- 37.Paillard C, Le Roux F, Borrego JJ. 2004. Bacterial diseases in marine bivalves, a review of recent studies: trends and evolution. Aquat. Living Resour. 17, 477–498. ( 10.1051/alr:2004054) [DOI] [Google Scholar]
- 38.Burreson EM, Stokes NA, Carnegie RB, Bishop MJ. 2004. Bonamia sp. (Haplosporidia) found in non native oysters Crassostrea ariakensis in Bogue Sound, North Carolina. J. Aquat. Animal Health 16, 1–9. ( 10.1577/H03-008.1) [DOI] [Google Scholar]
- 39.Engelsma MY, Kerkhoff S, Roozenburg I, Haenen OLM, van Gool A, Sistermans W, Wijnhoven S, Hummel H. 2010. Epidemiology of Bonamia ostreae infecting European flat oysters Ostrea edulis from Lake Grevelingen, The Netherlands. Mar. Ecol. Prog. Ser. 409, 131–142. ( 10.3354/meps08594) [DOI] [Google Scholar]
- 40.Carnegie RB, Burreson EM. 2011. Declining impact of an introduced pathogen: Haplosporidium nelsoni in the oyster Crassostrea virginica in Chesapeake Bay. Mar. Ecol. Prog. Ser. 432, 1–15. ( 10.3354/meps09221) [DOI] [Google Scholar]
- 41.Ford SE, Bushek D. 2012. Development of resistance to an introduced marine pathogen by a native host. J. Mar. Res. 70, 205–223. ( 10.1357/002224012802851922) [DOI] [Google Scholar]
- 42.Engelsma MY, Culloty SC, Lynch SA, Arzul I, Carnegie RB. 2014. Bonamia parasites: a rapidly changing perspective on a genus of important mollusc pathogens. Dis. Aquat. Org. 110, 5–23. ( 10.3354/dao02741) [DOI] [PubMed] [Google Scholar]
- 43.Laing I, Dunn P, Peeler EJ, Feist SW, Longshaw M. 2014. Epidemiology of Bonamia in the UK, 1982 to 2012. Dis. Aquat. Org. 110, 101–111. ( 10.3354/dao02647) [DOI] [PubMed] [Google Scholar]
- 44.Madsen L, Thomassen HEH.2015. First detection of Bonamia ostreae in native flat oysters from the Limfjord in Denmark. In 17th International Conference on Diseases of Fish And Shellfish: Abstract book, p. 92. Las Palmas de Gran Canaria, Spain: European Association of Fish Pathologists.
- 45.Stephenson MF, McGladdery SE, Maillet M, Veniot A, Meyer G. 2003. First reported occurrence of MSX in Canada. J. Shellfish Res. 22, 355. [Google Scholar]
- 46.Hine PM. 1996. Southern hemisphere mollusc diseases and an overview of associated risk assessment problems. Rev. Sci. Tech. OIE. 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 47.Marcogliese DJ. 2001. Implications of climate change for parasitism of animals in the aquatic environment. Can. J. Zool. 79, 1331–1352. ( 10.1139/cjz-79-8-1331) [DOI] [Google Scholar]
- 48.Burge CA, et al. 2016. Complementary approaches to diagnosing marine diseases: a union of the modern and the classic. Phil. Trans. R. Soc. B 371, 20150207 ( 10.1098/rstb.2015.0207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Audemard C, Reece KS, Burreson EM. 2004. Real-time PCR for detection and quantification of the protistan parasite Perkinsus marinus in environmental waters. Appl. Env. Microbiol. 70, 6611–6618. ( 10.1128/AEM.70.11.6611-6618.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gauthier JD, Miller CR, Wilbur AE. 2006. TaqMan® MGB real-time PCR approach to quantification of Perkinsus marinus and Perkinsus spp. in oysters. J. Shellfish Res. 25, 619–624. ( 10.2983/0730-8000(2006)25%5B619:TMRPAT%5D2.0.CO;2) [DOI] [Google Scholar]
- 51.Robert M, Garcia C, Chollet B, Lopez-Flores I, Ferrand S, François C, Joly JP, Arzul I. 2009. Molecular detection and quantification of the protozoan Bonamia ostreae in the flat oyster, Ostrea edulis. Mol. Cell. Probes 23, 264–271. ( 10.1016/j.mcp.2009.06.002) [DOI] [PubMed] [Google Scholar]
- 52.Ramilo A, Navas JI, Villalba A, Abollo E. 2013. Species-specific diagnostic assays for Bonamia ostreae and B. exitiosa in European flat oyster Ostrea edulis: conventional, real-time and multiplex PCR. Dis. Aquat. Org. 104, 149–161. ( 10.3354/dao02597) [DOI] [PubMed] [Google Scholar]
- 53.Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Inf. 16, 1614–1619. ( 10.1111/j.14690691.2010.03311) [DOI] [PubMed] [Google Scholar]
- 54.Ray SM. 1952. A culture technique for diagnosis of infections with Dermocystidium marinum Mackin, Owen and Collier in oysters. Science 116, 360–361. ( 10.1126/science.116.3014.360) [DOI] [PubMed] [Google Scholar]
- 55.Batista FM, Arzul I, Pepin JF, Ruano F, Friedman CS, Boudry P, Renault T. 2007. Detection of ostreid herpesvirus 1 DNA by PCR in bivalve molluscs: a critical review. J. Virol. Meth. 139, 1–11. ( 10.1016/j.jviromet.2006.09.005) [DOI] [PubMed] [Google Scholar]
- 56.Carnegie RB, Burreson EM, Hine PM, Stokes NA, Audemard C, Bishop MJ, Peterson CH. 2006. Bonamia perspora n. sp. (Haplosporidia), a parasite of the oyster Ostreola equestris, is the first Bonamia species known to produce spores. J. Euk. Microbiol. 53, 232–245. ( 10.1111/j.1550-7480.2006.00100.x) [DOI] [PubMed] [Google Scholar]
- 57.Stentiford GD, Bateman KS, Stokes NA, Carnegie RB. 2013. Haplosporidium littoralis sp. nov.: a crustacean pathogen within the Haplosporidia (Cercozoa, Ascetospora). Dis. Aquat. Org. 105, 243–252. ( 10.3354/dao02619) [DOI] [PubMed] [Google Scholar]
- 58.Elgharsalli R, Aloui-Bejaoui N, Salah H, Chollet B, Joly JP, Robert M, Couraleau Y, Arzul I. 2013. Characterization of the protozoan parasite Marteilia refringens infecting the dwarf oyster Ostrea stentina in Tunisia. J. Invertebr. Pathol. 112, 175–183. ( 10.1016/j.jip.2012.11.004) [DOI] [PubMed] [Google Scholar]
- 59.da Silva PM, Scardua MP, Vianna RT, Mendonca RC, Vieira CB, Dungan CF, Scott GP, Reece KS. 2014. Two Perkinsus spp. infect Crassostrea gasar oysters from cultured and wild populations of the Rio São Francisco estuary, Sergipe, northeastern Brazil. J. Invertebr. Pathol. 119, 62–71. ( 10.1016/j.jip.2014.04.005) [DOI] [PubMed] [Google Scholar]
- 60.Braun ME, Heatley JJ, Chitty J. 2006. Clinical techniques of invertebrates. Vet. Clin. Exot. Anim. Practic. 9, 205–221. ( 10.1016/j.cvex.2006.02.001) [DOI] [PubMed] [Google Scholar]
- 61.OIE. 2014. Principles and methods of validation of diagnostic assays for infectious diseases. In Manual of Diagnostic Tests for Aquatic Animals 2014, ch. 1.1.2. Paris, France: World Organisation for Animal Health. [Google Scholar]
- 62.Corbeil S, et al. 2010. Development and validation of a TaqMan PCR assay for the Australian abalone herpes-like virus. Dis. Aquat. Organ. 92, 1–10. ( 10.3354/dao02277) [DOI] [PubMed] [Google Scholar]
- 63.Polinski M, Lowe G, Meyer G, Corbeil S, Colling A, Caraguel C, Abbott CL. 2015. Molecular detection of Mikrocytos mackini in Pacific oysters using quantitative PCR. Mol. Biochem. Parasitol. 200, 19–24. ( 10.1016/j.molbiopara.2015.04.004) [DOI] [PubMed] [Google Scholar]
- 64.Stokes NA, Siddall ME, Burreson EM. 1995. Detection of Haplosporidium nelsoni (Haplosporidia, Haplosporidiidae) in oysters by PCR amplification. Dis. Aquat. Org. 23, 145–152. ( 10.3354/dao023145) [DOI] [Google Scholar]
- 65.Renault T, Stokes NA, Chollet B, Cochennec N, Berthe F, Gerard A, Burreson EM. 2000. Haplosporidiosis in the Pacific oyster Crassostrea gigas from the French Atlantic coast. Dis. Aquat. Org. 42, 207–214. ( 10.3354/dao042207) [DOI] [PubMed] [Google Scholar]
- 66.Cochennec N, Le Roux F, Berthe F, Gérard A. 2000. Detection of Bonamia ostreae based on small subunit ribosomal probe. J. Invertebr. Pathol. 76, 26–32. ( 10.1006/jipa.2000.4939) [DOI] [PubMed] [Google Scholar]
- 67.Maynard J, et al. 2016. Improving marine disease surveillance through sea temperature monitoring, outlooks and projections. Phil. Trans. R. Soc. B 371, 20150208 ( 10.1098/rstb.2015.0208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pernet F, Lupo C, Bacher C, Whittington RJ. 2016. Infectious diseases in oyster aquaculture require a new integrated approach. Phil. Trans. R. Soc. B 371, 20150213 ( 10.1098/rstb.2015.0213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lafferty KD, Porter JA, Ford SE. 2004. Are diseases increasing in the ocean? Annu. Rev. Ecol. Evol. Syst. 35, 31–54. ( 10.1146/annurev.ecolsys.35.021103.105704) [DOI] [Google Scholar]
- 70.UNESCO and COMEST (World Commission on the Ethics of Scientific Knowledge and Technology). 2005. The precautionary principle, p. 52 Paris, France: UNESCO. [Google Scholar]
- 71.NRC (National Research Council). 2004. Nonnative oysters in the Chesapeake Bay. Washington, DC: The National Academies Press. [Google Scholar]
- 72.Ben-Horin T, Lafferty KD, Bidegain G, Lenihan HS. 2016. Fishing diseased abalone to promote yield and conservation. Phil. Trans. R. Soc. B 371, 20150211 ( 10.1098/rstb.2015.0211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ulrich PN, Colton CM, Hoover CA, Gaffney PM, Marsh AG. 2007. Haplosporidium nelsoni (MSX) rDNA detected in oysters from the Gulf of Mexico and the Caribbean Sea. J. Shellfish Res. 26, 195–199. ( 10.2983/0730-8000(2007)26%5B195:HNMRDI%5D2.0.CO;2) [DOI] [Google Scholar]
- 74.Ford SE, Paterno J, Scarpa E, Stokes NA, Kim Y, Powell EN, Bushek D. 2011. Widespread survey finds no evidence of Haplosporidium nelsoni (MSX) in Gulf of Mexico oysters. Dis. Aquat. Org. 93, 251–256. ( 10.3354/dao02306) [DOI] [PubMed] [Google Scholar]
- 75.Bushek D, Allen SK. 1996. Host–parasite interactions among broadly distributed populations of the eastern oyster Crassostrea virginica and the protozoan Perkinsus marinus. Mar. Ecol. Prog. Ser. 139, 127–141. ( 10.3354/meps139127) [DOI] [Google Scholar]
- 76.U.S. Army Corps of Engineers, Norfolk District. 2009. Final programmatic environmental impact statement for oyster restoration in Chesapeake Bay including the use of native and/or nonnative oyster, p. 402 Norfolk, VA: U.S. Army Corps of Engineers. [Google Scholar]