Abstract
The role of the Hippo signaling pathway in cranial neural crest (CNC) development is poorly understood. We used the Wnt1Cre and Wnt1Cre2SOR drivers to conditionally ablate both Yap and Taz in the CNC of mice. When using either Cre driver, Yap and Taz deficiency in the CNC resulted in enlarged, hemorrhaging branchial arch blood vessels and hydrocephalus. However, Wnt1Cre2SOR mutants had an open cranial neural tube phenotype that was not evident in Wnt1Cre mutants. In O9-1 CNC cells, the loss of Yap impaired smooth muscle cell differentiation. RNA-sequencing data indicated that Yap and Taz regulate genes encoding Fox transcription factors, specifically Foxc1. Proliferation was reduced in the branchial arch mesenchyme of Yap and Taz CNC conditional knockout (CKO) embryos. Moreover, Yap and Taz CKO embryos had cerebellar aplasia similar to Dandy-Walker spectrum malformations observed in human patients and mouse embryos with mutations in Foxc1. In embryos and O9-1 cells deficient for Yap and Taz, Foxc1 expression was significantly reduced. Analysis of Foxc1 regulatory regions revealed a conserved recognition element for the Yap and Taz DNA binding co-factor Tead. ChIP-PCR experiments supported the conclusion that Foxc1 is directly regulated by the Yap-Tead complex. Our findings uncover important roles for Yap and Taz in CNC diversification and development.
KEY WORDS: Yap and Taz, Cranial neural crest, Craniofacial development
Summary: Conditional ablation of Yap and Taz in the cranial neural crest reveals how these proteins regulate Fox gene expression and, in turn, cranial neural crest diversification and development.
INTRODUCTION
The neural crest (NC) is a migratory, multipotent cell population that originates in the embryonic dorsal neural tube. Developmental defects in NC formation result in numerous human congenital anomalies. Based on the site of origin, NC cells are divided into cranial, cardiac and trunk populations, each of which has its own unique developmental potential. The cranial neural crest (CNC) ultimately diversifies into multiple cell types, including neuronal, glial, cartilage, bone and smooth muscle cells (Santagati and Rijli, 2003). Mutations in the genes required for CNC development are often associated with the pathophysiology of human congenital malformations (Acloque et al., 2009; Cordero et al., 2011), highlighting the clinical importance of understanding the molecular mechanisms governing CNC development. Much effort has been expended interrogating the gene regulatory networks underlying NC development (Sauka-Spengler and Bronner-Fraser, 2008). Major signaling pathways, including Wnt, Fgf, Bmp and Notch signaling, have been shown to play important roles in regulating NC induction, proliferation and migration (Sela-Donenfeld and Kalcheim, 1999; Garcia-Castro et al., 2002; Coles et al., 2004; Glavic et al., 2004; Carmona-Fontaine et al., 2008). However, the function of Hippo signaling in CNC development remains poorly understood.
Hippo signaling is an essential pathway that regulates organ size through the modulation of cell proliferation (Heallen et al., 2011). The key components of the Hippo signaling pathway are evolutionarily conserved. In mice, Mst1/2 (orthologous to Drosophila melanogaster Hpo) and Salv (orthologous to human WW45) form a complex that phosphorylates the kinases Lats1 and Lats2 (orthologous to Drosophila Warts). Lats1/2, in turn, phosphorylate the most downstream Hippo signaling components Yap (also known as Yap1 – Mouse Genome Informatics) and Taz, thus promoting their binding to 14-3-3 proteins and inhibiting them from shuttling into the nucleus. In the absence of the repressive activity from Hippo signaling, Yap and Taz localize to the nucleus and partner with transcription factors, such as transcriptional enhancer activator (TEA) domain (TEAD) family members, to promote gene programs favoring proliferation. Recently, familial studies have shown that heterozygous nonsense mutations in YAP1 are associated with variable phenotypes in the affected families, including orofacial clefting and intellectual disability (Williamson et al., 2014). However, the mechanisms underlying these phenotypic alterations remain unclear. In this study, we investigated the function of Yap and Taz in the CNC. Using two Wnt1 Cre drivers, we uncover important functions of Yap and Taz in CNC proliferation and subsequent differentiation.
RESULTS
Yap and Taz deletion in CNC-derived cells results in embryonic lethality
To determine the function of Yap and Taz in the CNC, we generated compound Yap and Taz conditional mutants by using conditional null alleles and the Wnt1Cre and Wnt1Cre2SOR drivers. We collected embryos at multiple developmental stages to analyze the morphogenesis of several CNC-derived structures. Importantly, the Wnt1Cre driver (Chai et al., 2000) has been shown to induce ectopic expression of Wnt1, leading to defects in midbrain development, whereas the Wnt1Cre2SOR driver does not have these issues (Lewis et al., 2013).
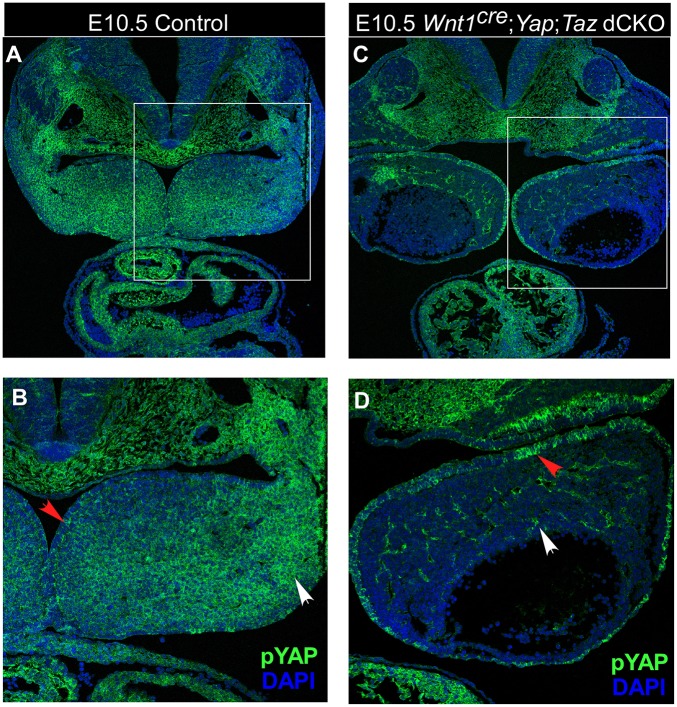
We first evaluated embryos in which Yap and Taz were deleted using the Wnt1Cre driver. In control embryos, phosphorylated Yap (pYAP) – a readout for Hippo signaling activity – was present in CNC-derived cells, such as mandibular mesenchymal cells (Fig. 1A,B). However, in Wnt1Cre; Yap; Taz double conditional knockout (dCKO) embryos, the level of pYAP was dramatically reduced in CNC-derived cells, but unchanged in non-CNC-derived cells such as endothelium (Fig. 1C,D). These data indicated that the Wnt1Cre driver efficiently inactivated Yap and Taz in CNC-derived cells. Among the different mutant genotypes, both Wnt1Cre; Yapf/f; Tazf/+ and Wnt1Cre; Yap; Taz dCKO were embryonic lethal at embryonic day (E) 10.5 (Table S1). Mutant embryos with the reciprocal genotype of Wnt1Cre; Yapf/+; Tazf/f displayed lethality over a wide range of developmental time points, from E14.5 to postnatal week 8 (Table S1).
Fig. 1.
Efficient deletion of Yap and Taz in CNC-derived cells. Hippo signaling activity, indicated by the level of phosphorylated Yap (pYAP) in control embryos (A,B) and in Wnt1Cre; Yapf/f; Tazf/f embryos (C,D). pYAP levels were reduced in CNC-derived cells (white arrows) but not in non-CNC derived cells (red arrows). Hippo-active cells (green) were stained with pYAP; nuclei (blue) were stained with DAPI.
In addition, we used the Wnt1Cre2SOR driver (Lewis et al., 2013) to inactivate Yap and Taz in CNC. Similar to Wnt1Cre embryos, lethality was observed at E10.5 in both Wnt1Cre2SOR; Yapf/f; Tazf/+ and Wnt1Cre2SOR; Yap; Taz dCKO embryos, whereas Wnt1Cre2SOR; Yapf/+; Tazf/f embryos survived until E15.5, the latest developmental stage examined in this study (Table S2). Together, our findings indicate that embryos with Yap and Taz compound loss of function or Yap deletion with Taz haploinsufficiency in the CNC exhibit early embryonic lethality, whereas embryos with Taz deletion and Yap haploinsufficiency in the CNC survive until later developmental stages.
Yap and Taz deletion in Wnt1Cre and Wnt1Cre2SOR embryos results in similar vascular defects, but distinct neural tube phenotypes
No obvious craniofacial morphologic defects were observed in Yap and Taz compound mutants at E9.5. At E10.5, neither control embryos, including Yap and Taz compound heterozygous embryos and embryos without Wnt1Cre (Fig. S1A-C), nor Wnt1Cre; Yapf/+; Tazf/f embryos (Fig. S1D-F) displayed any obvious craniofacial defects. The Wnt1Cre; Yapf/f; Tazf/+ (Fig. S1G-I) and Wnt1Cre; Yap; Taz dCKO (Fig. S1J-L,P-R) embryos survived until E10.5 and showed disrupted craniofacial structures, including enlarged blood vessels in the branchial arch and hemorrhage in the forebrain and mandible at E10.5.
Similar to Wnt1Cre; Yap; Taz mutant embryos, lethality at E10.5 was observed in Wnt1Cre2SOR; Yap; Taz dCKO (Fig. 2A-D) and Wnt1Cre2SOR; Yapf/f; Tazf/+ (Fig. 2M-P) embryos. The E10.5 mutant embryos exhibited disrupted craniofacial structures, including blood vessel enlargement and hemorrhage, which are phenotypes we observed in Wnt1Cre compound mutant embryos. These early CNC phenotypes were not observed in Wnt1Cre2SOR; Yapf/+; Tazf/f embryos that have one functional copy of Yap or in controls (Fig. 2E-L). Wnt1Cre2SOR compound mutant embryos lacked any obvious morphologic defects at E9.5. Notably, both E10.5 Wnt1Cre2SOR; Yapf/f; Tazf/+ (Fig. 2Pa) and Wnt1Cre2SOR; Yap; Taz dCKO (Fig. 2D,R) embryos had neural tube defects, which is one of the most common human birth defects. At E10.5, all (14/14) Wnt1Cre2SOR; Yap; Taz dCKO embryos displayed open anterior neural tubes (Fig. 2D,R), and most (7/11) Wnt1Cre2SOR; Yapf/f; Tazf/+ embryos presented with less severe, but still abnormal, anterior neural tube morphology (Fig. 2Pa). Neural tube defects were not observed in any of the Yap; Taz compound mutant embryos generated using Wnt1Cre (Fig. S1).
Fig. 2.
Vascular defects, hemorrhage and neural tube defects following Yap and Taz inactivation using the Wnt1Cre2SOR Cre driver. Both Wnt1Cre2SOR; Yap; Taz dCKO (A-D) and Wnt1Cre2SOR; Yapf/f; Tazf/+ (M-P) embryos show lethality at E10.5 with severe vascular defects, hemorrhage and defects in neural tube closure, whereas control embryos (I-L) and Wnt1Cre2SOR; Yapf/+; Tazf/f embryos (E-H) show no obvious defects. (Ma-Pa) Zoom images of branchial arch vessel defects, hemorrhage and neural tube defects of Wnt1Cre2SOR; Yapf/f; Tazf/+ embryos. Three-dimensional imaging using optical projection tomography microscopy indicates that the neural tube of control embryos closed normally (Q), whereas Wnt1Cre2SOR; Yap; Taz dCKO embryos have a defect in neural tube closure (R).
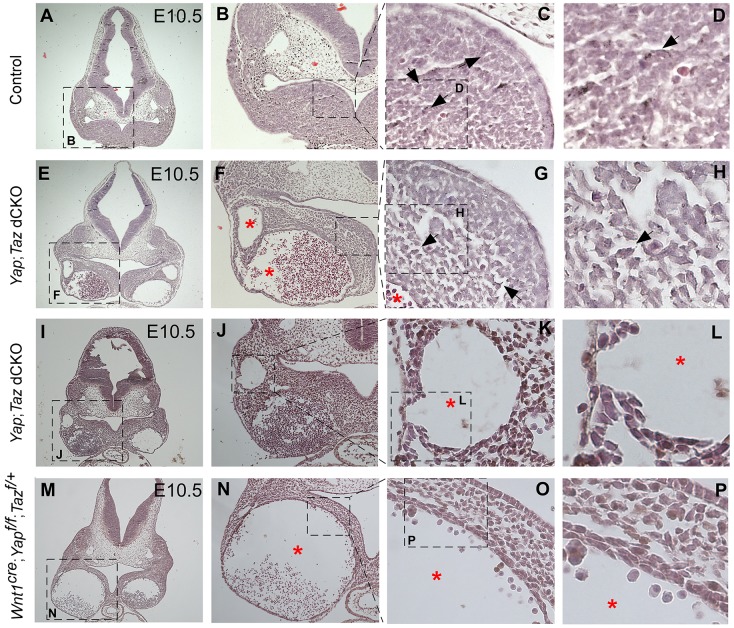
Histological analysis indicated that, compared with control embryos (Fig. 3A-D, Fig. S2A-C), both Wnt1Cre; Yap; Taz dCKO (Fig. 3E-L, Fig. S2D-F) and Wnt1Cre; Yapf/f; Tazf/+ (Fig. 3M-P, Fig. S2G-I) embryos had enlarged blood vessels. Moreover, the mandibular mesenchyme was disorganized in Wnt1Cre; Yap; Taz dCKO (Fig. 3G,H) and Wnt1Cre; Yapf/f; Tazf/+ (Fig. 3O,P) embryos. The sparse mesenchyme in the mandible suggests that there are fewer pericytes to be recruited from the surrounding mesenchyme because of the lack of Yap and Taz in CNC-derived tissues; thus, deficient support from the blood vessels surrounding cells probably caused hemorrhage in mutants.
Fig. 3.
Histological analysis showing the disruption of mandibular structure in Yap and Taz-deficient embryos. Coronal sections stained with hematoxylin and eosin showing that, in contrast to control embryos (A-D), both Wnt1Cre; Yap; Taz dCKO (E-L) and Wnt1Cre; Yapf/f; Tazf/+ (M-P) embryos have disorganized, sparse mesenchyme (black arrows) and enlarged vessels (red stars) in the mandible. Boxed areas are shown at higher magnification in panels to the right, as labeled.
Yap and Taz deletion causes severe neural tube vessel regression
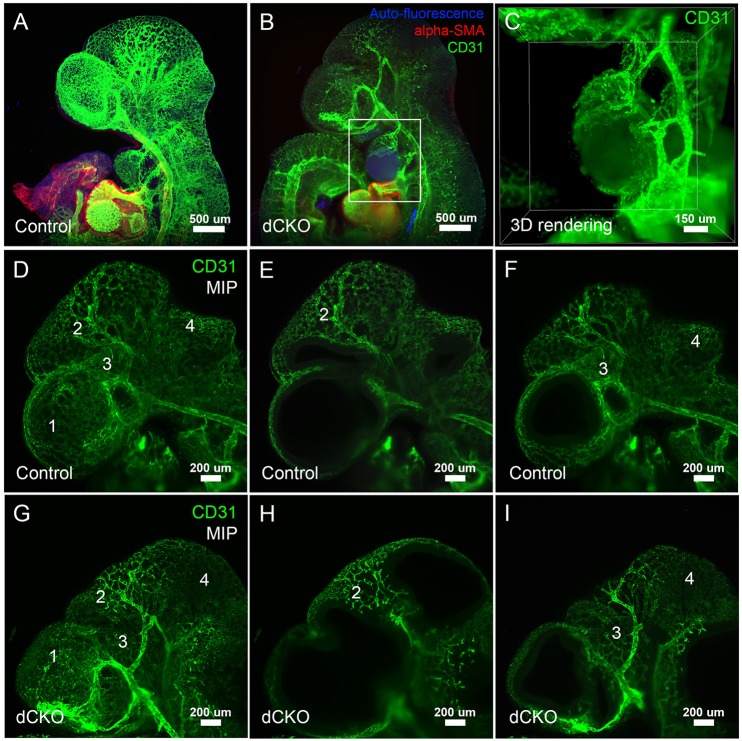
To more closely examine the hemangioma phenotype in the forebrain, branchial arch and mandibular regions in Wnt1Cre; Yap; Taz dCKO embryos, we performed whole-mount immunofluorescence staining for CD31 (Pecam1, endothelial cell marker) and smooth muscle actin (SMA, a smooth muscle cell marker), followed by imaging with Lightsheet microscopy. In contrast to control embryos (Fig. 4A), the hemangiomas present in the branchial arch of Wnt1Cre; Yap; Taz dCKO embryos are encapsulated by CD31-positive endothelium (Fig. 4B). In addition, the results of three-dimensional rendering and maximum intensity projection of the branchial arch in a Yap; Taz dCKO embryo further supported the finding that the vessels are wrapped around the hemangioma in the branchial arch of the Yap; Taz dCKO embryo (Fig. 4C). Similar endothelial-lined hemangiomas were also identified in the forebrain and mandibular regions in other dCKO embryos. Images of CD31 immunostaining also revealed abnormal vessel regression throughout the brain of Wnt1Cre; Yap; Taz dCKO embryos. By comparing control embryos (Fig. 4D-F) with Wnt1Cre; Yapf/f; Tazf/f embryos (Fig. 4G-I), we detected several regions where vessel regression and/or disorganization was present in Wnt1Cre; Yapf/f; Tazf/f embryos (regions labeled 1-4 in Fig. 4G-I), demonstrating that expression of both Yap and Taz within the CNC is required for normal vascularization of the early brain and mandibular region.
Fig. 4.
Severe vessel defects caused by deletion of Yap and Taz. Whole-mount CD31 immunofluorescence staining in a control (A) and a Wnt1Cre; Yap; Taz dCKO mutant (B) reveals vessel defects and endothelial-lined hemangiomas in the forebrain and mandible in Wnt1Cre; Yap; Taz dCKO mutants (B). (C) The boxed area in B at a higher magnification, focusing on the endothelial-lined hemangiomas in the branchial arch in the Yap; Taz dCKO embryo. Whereas control embryos have normal vessel development in brain (D-F), Wnt1Cre; Yap; Taz dCKO mutants have vessel regression and disorganization (different regions are labeled 1-4) (G-I). Endothelial cells are stained with CD31 antibody (green), smooth muscle cells are stained with SMA antibody (red) and auto fluorescence is blue.
RNA sequencing reveals genes regulated by the Hippo pathway in the CNC
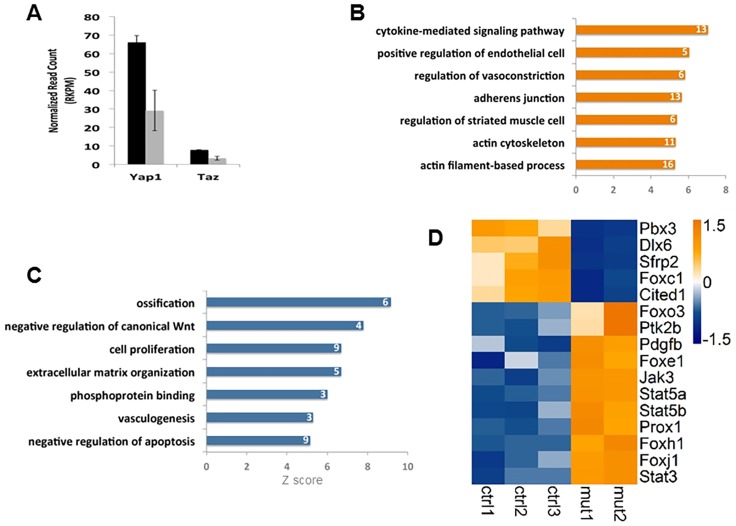
Because the mandibular phenotype in Yap; Taz dCKO embryos appeared to be consistent in both CNC-specific Cre lines, we focused our attention on the molecular mechanism underlying the enlarged cranial and facial vessels. We performed RNA-sequencing (RNA-Seq) analysis by using mandibular RNA isolated from E10.5 Wnt1Cre; Yap; Taz dCKO and control embryos. RNA-Seq analysis of the mandibular tissue from Wnt1Cre; Yap; Taz dCKO embryos revealed efficient ablation of Yap and Taz (Fig. 5A). Among the differentially expressed genes, 355 were upregulated and 77 were downregulated (Fig. S3A). We noted that a large percentage of the differentially expressed genes (11%) were DNA binding factors (Fig. S3B).
Fig. 5.
The regulation of multiple signals by the Hippo pathway in CNC-derived cells. RNA-Seq analysis was performed by using mandibular tissues from E10.5 control embryos and Wnt1Cre Taz; Yap dCKO mutants. (A) RNA-Seq analysis indicates that Yap and Taz expression levels (indicated by reads) decreased in Wnt1Cre; Taz; Yap dCKO embryos compared with control embryos. (B) Gene ontology analysis shows genes that are upregulated in Wnt1Cre; Taz; Yap dCKO embryos compared with control embryos, which includes genes that regulate adherens junctions, vasoconstriction and the cytoskeleton. (C) Genes that are downregulated in Wnt1Cre; Taz; Yap dCKO embryos compared with control embryos, which includes genes that regulate cell proliferation and vasculogenesis and that negatively regulate canonical Wnt. Numbers in bars in B and C indicate gene number of each gene ontology term. (D) Heat map of RNA-Seq data shows that, compared with controls, Wnt1Cre; Taz; Yap dCKO mutants downregulate expression of Foxc1 and upregulate expression of Foxe1, Prox1, Pdgfb and Jak-Stat genes, including Jak3, Ptk2b, Stat3, Stat5a and Stat5b.
Gene ontology analysis indicated that genes upregulated in Wnt1Cre; Yap; Taz dCKO embryos are involved in regulating adherens junction formation, vasoconstriction, cytoskeleton and positive regulation of endothelial cell migration (Fig. 5B). Genes downregulated in Wnt1Cre; Yap; Taz dCKO embryos primarily function in cell proliferation, extracellular matrix organization and vasculogenesis (Fig. 5C).
Among the downregulated genes, we identified those that negatively regulate the canonical Wnt signaling pathway, including the secreted Frizzled-related protein (SFRP) Sfrp2, a Wnt inhibitor that directly binds Wnt ligands (Ladher et al., 2000). Notably, Wnt1Cre; Yap; Taz dCKO embryos had reduced Foxc1 expression (Fig. 5D). Foxc1 has been implicated in ocular and cerebellar malformations in human patients, as well as in vascular malformations in mice (Kume et al., 2001; Kume, 2009; Delahaye et al., 2012; Haldipur et al., 2014).
Among the transcripts upregulated in Wnt1Cre; Yap; Taz dCKO embryos, we identified genes encoding components of the Jak-Stat cascade, including Jak3, Ptk2b, Stat3, Stat5a and Stat5b (Fig. 5D). Interestingly, Jak-Stat signaling controls organ size in Drosophila, much like the Hippo pathway. In vertebrates, the Jak-Stat pathway is involved in growth hormone signaling, although it is less clear what the predicted consequence of increased Jak-Stat levels would be during development. We also found that mRNA expression of prospero homeobox protein 1 (Prox1) is upregulated in Wnt1Cre; Yap; Taz dCKO embryos. Prox1 encodes a transcription factor that is a master regulator of lymphatic endothelial cell specification and identity, and is essential for normal vascular development (Wigle and Oliver, 1999; Wigle et al., 2002; Johnson et al., 2008). Another upregulated vascular mitogen was platelet-derived growth factor b (Pdgfb), which promotes the recruitment and proliferation of vascular cells (Yancopoulos et al., 2000). Pdgfb deficiency leads to reduced numbers of microvascular-associated pericytes in mouse embryos (Lindahl et al., 1997). Wnt1Cre; Yap; Taz dCKO embryos also had upregulated mRNA expression of forkhead box E1 (Foxe1), which has been implicated as a causative gene in human orofacial clefting (Moreno et al., 2009).
Yap and Taz regulate proliferation and apoptosis in the CNC
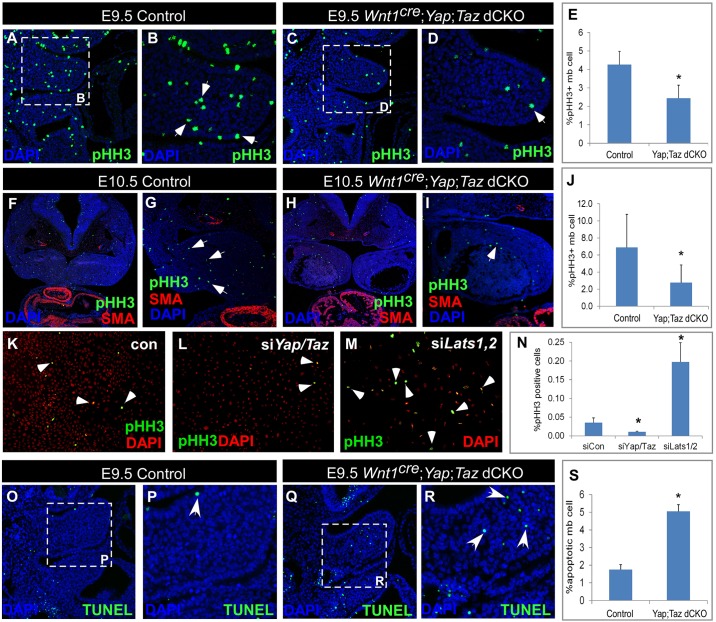
As indicated by our RNA-Seq data analysis (Fig. 5C), cell proliferation genes were downregulated in Wnt1Cre; Yap; Taz dCKO embryos compared with control embryos. To evaluate differences in proliferation in vivo, we performed phospho-histone H3 (pHH3) immunofluorescence staining on sections from both E9.5 and E10.5 Yap; Taz dCKO and control embryos. The percentage of proliferating, pHH3-positive cells was lower in the mandible of Wnt1Cre; Yap; Taz dCKO embryos (E9.5, Fig. 6C,D; E10.5, Fig. 6H,I) than in that of control embryos (E9.5, Fig. 6A,B; E10.5, Fig. 6F,G). Furthermore, quantification of pHH3-positive cells indicated that the rate of proliferation was significantly lower in Wnt1Cre; Yap; Taz dCKO embryos than in control embryos at both E9.5 and E10.5 (Fig. 6E,J).
Fig. 6.
Regulation of proliferation and apoptosis in CNC cells by the Hippo pathway. (A-J) Compared with control embryos, Wnt1Cre; Yap; Taz dCKO embryos have a significantly diminished percentage of pHH3-positive proliferating cells in the mandible at E9.5 and E10.5. Proliferating cells are stained with pHH3 antibody (green), smooth muscle cells with SMA antibody (red) and nuclei with DAPI (blue). *P<0.05. (K-N) Compared with O9-1 cells transfected with control (con) siRNA, O9-1 cells transfected with Yap and Taz siRNA have significantly reduced proliferation, and O9-1 cells transfected with Lats1 and Lats2 siRNA have significantly increased proliferation (*P<0.05). Proliferating cells are stained with pHH3 antibody (green), nuclei with DAPI (red). Arrows indicate proliferating cells. (O-S) Compared with control embryos, Wnt1Cre; Yap; Taz dCKO embryos have a significantly increased percentage of apoptotic cells in the mandible at E9.5 (*P<0.05). Apoptotic cells are stained with TUNEL (green), nuclei with DAPI (blue). All error bars represent s.e.m. Arrows indicate apoptotic cells.
To further evaluate the role of the Hippo pathway in CNC cells, we used the O9-1 cell line for in vitro analyses. The O9-1 cell line is a stable, multipotent, mesenchymal CNC cell line, originally derived from Wnt1Cre; R26R-green fluorescent protein (GFP)-expressing cells and can differentiate into multiple CNC derivatives, including osteoblasts, chondrocytes, smooth muscle cells and glial cells (Ishii et al., 2012). To examine cell proliferation in response to altered Hippo signaling, we used siRNA-mediated knockdown to reduce expression of Yap and Taz, as well as Lats1 and Lats2, in O9-1 cells, and performed pHH3 immunofluorescence staining to assess cell proliferation. The percentage of pHH3-positive cells was decreased in cells treated with siRNA against Yap and Taz (Fig. 6L), but was increased in cells treated with siRNAs targeting Lats1 and Lats2 (Fig. 6M) compared with cells treated with control siRNA (Fig. 6K). Based on cell counting, the proliferation rate was significantly reduced in Yap and Taz-knockdown cells but significantly increased in Lats1 and Lats2-knockdown cells when compared with the control (Fig. 6N).
Although cell proliferation was reduced in the mandible of Wnt1Cre; Yap; Taz dCKO embryos, these mutant embryos showed no obvious defects in neural tube morphogenesis. We also evaluated cell proliferation in Wnt1Cre2SOR; Yap; Taz dCKO embryos, which did display neural tube closure defects (Fig. 2R). However, no obvious difference was detected in cell proliferation within the neural tube between Wnt1Cre2SOR; Yap; Taz dCKO and control embryos (Fig. S4). Moreover, when we performed immunofluorescence studies to evaluate the expression of the actin-severing protein cofilin 1 (CFL1), an important factor for neural tube closure, we found no obvious difference between Wnt1Cre2SOR; Yap; Taz dCKO and control embryos (Fig. S5).
Our RNA-Seq data indicated that the expression levels of negative regulators of apoptosis were lower in Wnt1Cre; Yap; Taz dCKO embryos than in control embryos (Fig. 5C). To evaluate cell apoptosis in vivo, we performed terminal deoxynucleotidyl transferase (dUTP) nick end labeling (TUNEL) analysis in both E9.5 Wnt1Cre; Yap; Taz dCKO mutant embryos and control embryos. Our TUNEL data indicated that cell apoptosis was significantly increased in E9.5 Wnt1Cre; Yap; Taz dCKO embryos compared with control embryos (Fig. 6O-S).
Yap and Taz promote smooth muscle differentiation
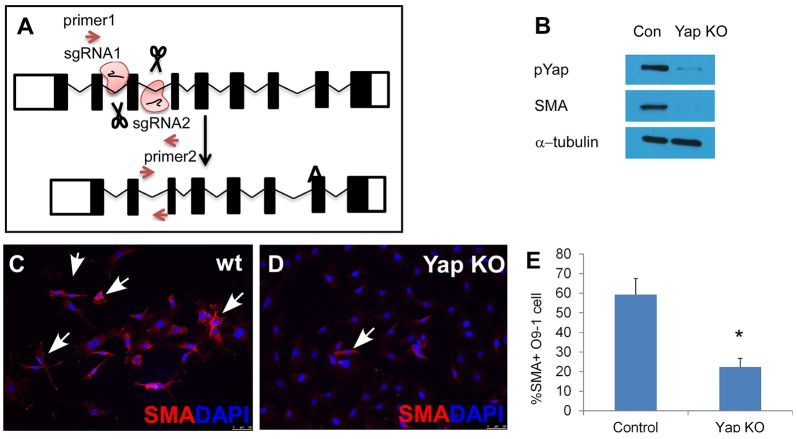
Our RNA-Seq data indicated that genes involved in vasculogenesis are downregulated in Wnt1Cre; Yap; Taz dCKO embryos when compared with control embryos (Fig. 5C). Moreover, Lightsheet microscopy revealed severe vessel defects in Wnt1Cre; Yap; Taz dCKO embryos than in control embryos (Fig. 4). A recent study indicated that Yap and Taz deficiency gives rise to smooth muscle differentiation defects partially derived from the CNC (Manderfield et al., 2015). Given the similar vessel defects in Wnt1Cre; Yapf/f; Tazf/+ embryos and Wnt1Cre; Yap; Taz dCKO embryos, we hypothesized that Yap alone plays a crucial role in the regulation of smooth muscle differentiation. Accordingly, we created a Yap-null O9-1 cell line (Yap KO O9-1) by removing exon 3 of Yap using CRISPR/Cas9-mediated genome editing (strategy is shown in Fig. 7A; details in Materials and Methods). Compared with wild-type O9-1 cells, Yap KO O9-1 cells had diminished SMA protein expression and Yap activity (Fig. 7B). SMA immunofluorescence indicated that under differentiation conditions (see Materials and Methods), wild-type O9-1 cells gave rise to SMA-positive smooth muscle cells, but the ability of Yap KO O9-1 cells to generate SMA-positive cells was significantly diminished, supporting the idea that Yap plays the predominant function is SMC differentiation, perhaps because it is more highly expressed in CNC (Fig. 7C-E).
Fig. 7.
Yap is required for smooth muscle differentiation. (A) Strategy for establishing a Yap-knockout (KO) O9-1 cell line by using CRISPR/Cas9 system. Specifically, exon 3 was deleted from Yap. (B) Western blot data shows diminished SMA and pYap expression in Yap KO O9-1 cells compared with wild-type (wt) O9-1 cells. (C) Under differentiation conditions, most wild-type O9-1 cells give rise to smooth muscle cells. (D) Yap KO O9-1 cells did not give rise to smooth muscle cells. Arrows indicate SMA-positive cells. Smooth muscle cells are stained with SMA antibody (red), nuclei with DAPI (blue). (E) Cell counting data show that the percentage of SMA-positive cells is significantly reduced in Yap KO O9-1 cells compared with wild-type O9-1 cells (*P<0.01). Error bars represent s.e.m.
Hydrocephalus in Yapf/+; Tazf/f CKO embryos
Although Wnt1Cre; Yapf/+; Tazf/f and Wnt1Cre2SOR; Yapf/+; Tazf/f embryos did not show any obvious defects before E10.5 (Fig. 2, Fig. S1), they developed hydrocephalus at later stages (Fig. S6E-G, Fig. S7D-F). Hydrocephalus is mainly characterized by the abnormal widening of brain spaces caused by the excessive accumulation of cerebrospinal fluid, which places harmful pressure on the surrounding tissues of the brain. Fig. S6E-G shows a representative example of an E12.5 Wnt1Cre; Yapf/+; Tazf/f embryo that had severe hydrocephalus in the hindbrain. In addition, in the sagittal sections of control embryos (Fig. S6D,d), the cerebellum structure is clearly visible, whereas it is missing in the Wnt1Cre; Yapf/+; Tazf/f embryos (Fig. S6H,h). The hydrocephalus phenotype was detected as early as E11 in Wnt1Cre; Yapf/+; Tazf/f embryos, but it was not observed in either Wnt1Cre; Yapf/f; Tazf/+ or Wnt1Cre; Yap; Taz dCKO embryos, most likely because of their lethality at E10.5. Moreover, Wnt1Cre; Yapf/f embryos at E11 also had severe hydrocephalus (Fig. S8M-P) when compared with controls (Fig. S8I-L). Wnt1Cre2SOR; Yapf/+; Tazf/f embryos at E12.5 presented more severe hydrocephalus (Fig. S7D-F) than that in Wnt1Cre; Yapf/+; Tazf/f embryos in the hindbrain region and forebrain region. Notably, our RNA-Seq data (Fig. 5) suggested that genes required for forebrain and hindbrain morphogenesis, such as Foxc1, were regulated by Taz and Yap. Foxc1 loss of function also leads to hydrocephalus in mice (Kume et al., 1998).
Yap and Taz regulate Fox genes
Our RNA-Seq data indicated that Yap and Taz modulated the expression of multiple members of the winged-helix/forkhead box (Fox) transcription factor family, including the upregulation of Foxe1, Foxh1, Foxj1 and Foxo3, as well as the downregulation of Foxc1 (Fig. 5D). We focused on Foxc1 in part because its expression was reduced in Wnt1Cre; Yap; Taz dCKO embryos and Yap and Taz are thought to be transcriptional activators. Indeed, Yap ChIP-Seq data showed that genome-wide Yap peaks are enriched in active chromatin regions, as defined by H3K27Ac chromatin marks (Morikawa et al., 2015). We reasoned that downregulated genes were more likely to be direct Yap/Taz target genes. Moreover, Foxc1 is the gene mutated in the congenital hydrocephalus mouse mutant (Kume et al., 1998).
Immunohistochemistry data indicated that Foxc1 is normally expressed in the majority of the epithelial and mesenchymal cells in the mandible (sagittal view in Fig. 8Bb; coronal view in Fig. S9Bb), yet Foxc1 expression was missing or reduced in the majority of mesenchymal cells and unchanged in the mandibular epithelial cells in Wnt1Cre; Yap; Taz dCKO embryos (sagittal view in Fig. 8Db; coronal view in Fig. S9Db). On the basis of cell counting, the number of Foxc1-positive cells was significantly lower in the Wnt1Cre; Yap; Taz dCKO mandible than in the control mandible (P<0.01, Fig. 8E). Furthermore, Foxc1 immunofluorescence staining indicated that Foxc1 expression was significantly lower in Yap knockout; Taz knockdown O9-1 cells (Fig. 8G) than in wild-type O9-1 cells (Fig. 8F) (P<0.05, Fig. 8H). Western blot analysis of Foxc1 further indicated that Foxc1 expression was decreased in response to the decrease in Yap and Taz expression levels (Fig. 8I).
Fig. 8.
Regulation of Foxc1 by Yap and Taz. Immunohistochemical staining of Foxc1 in sagittal sections of the mandible show that the majority of cells in control embryos express Foxc1 (A-Bb), whereas the expression of Foxc1 is diminished in Wnt1Cre; Yap; Taz dCKO embryos (C-Db). Boxed areas are shown at higher magnification in panels to the right, as labeled. (E) Cell counting data show that the percentage of cells positive for Foxc1 expression in the mandible is significantly reduced in Wnt1Cre; Yap; Taz dCKO embryos compared with control embryos (*P<0.01). (F-H) Immunohistochemical staining of Foxc1 in O9-1 cells shows significantly decreased Foxc1 expression in Yap knockout (KO); Taz knockdown (KD) O9-1 cells compared with wild-type O9-1 cells (*P<0.05). Arrows indicate Foxc1-positive cells. (I) Western blot analysis of Foxc1 in O9-1 cells shows decreased Foxc1 expression in response to decreased Yap and Taz expression level. (J) Conserved Tead binding site located in the upstream region of FOXC1. Peaks in ATAC-seq data (accession no. GSE70751) indicate chromatin accessibility (Prescott et al., 2015). (K) In vivo real-time PCR using ChIP DNA indicates that Foxc1 is bound by the Yap-Tead complex in embryonic facial tissue. P<0.05. All error bars represent s.e.m.
Importantly, we identified a conserved binding site for Tead, the Yap and Taz cofactor, in Foxc1 (Fig. 8J). Craniofacial cis-regulatory landscapes were recently studied by deep-sequencing of transposase-accessible chromatin (ATAC-seq) in human and chimpanzee cranial neural crest cells (Prescott et al., 2015). Bioinformatic analysis of these data sets (Gene Expression Omnibus, accession no. GSE70751) revealed increased chromatin accessibility in a region located upstream of the 5′ UTR of FOXC1. Significantly, this putative enhancer region contained a potential TEAD DNA-binding element (Fig. 8J). The TEAD binding element was confirmed by chromatin immunoprecipitation (ChIP) PCR in murine embryonic facial tissue using an anti-Yap antibody, indicating that a Yap-Tead complex directly binds to Foxc1 chromatin during embryonic facial morphogenesis (Fig. 8K).
DISCUSSION
During normal development, regeneration and cancer progression, Hippo signaling inhibits proliferation while promoting apoptosis. Here, we show that, during craniofacial development, Yap and Taz, the final downstream effectors of Hippo signaling, regulate multiple events in CNC diversification including vasculogenesis, smooth muscle differentiation, cerebellar development, and neural tube closure. Furthermore, our data indicate that Foxc1 is an important downstream target of Yap and Taz. Significantly, Foxc1 loss of function in mouse embryos phenocopies several of the morphological defects present in Yap and Taz mutant embryos.
Overlapping functions for Yap and Taz during craniofacial development
Using either a Wnt1Cre or Wnt1Cre2SOR driver, Yap and Taz conditional CNC mutants presented with disrupted craniofacial vascular development and hemorrhage. We also observed hydrocephalus in Yapf/+; Tazf/f mutants established by using both Cre drivers, at later embryonic stages. It is most likely that Yap and Taz have redundant functions when they are co-expressed, but at earlier developmental stages, Taz expression is lower than that of Yap. Interestingly, we did observe a phenotypic difference in embryos when we deleted Yap and Taz using the two Cre drivers. Deleting Yap and Taz with the Wnt1Cre2SOR driver resulted in anterior neural tube closure defects at E10.5, whereas Wnt1Cre; Yapf/f; Tazf/+ and Wnt1Cre; Yap; Taz dCKO embryos showed no neural tube closure defects. Wnt1Cre directs ectopic Wnt1 activity in the midbrain, whereas Wnt1Cre2SOR has normal Wnt1 activity in the midbrain (Lewis et al., 2013). Because Wnt signaling is known to stabilize Yap and Taz (Azzolin et al., 2014), our data suggest that the phenotypic differences observed between mutants obtained by using Wnt1Cre and Wnt1Cre2SOR might be caused by elevated Wnt signaling from the Wnt1Cre transgene (Chai et al., 2000).
Yap and Taz regulate vascular development
The diminished expression of CD31 in E10.5 Yap and Taz mutants indicated that Yap and Taz have an essential role in craniofacial vascular development. We found that the multipotent O9-1 NC cells treated with siRNAs targeting Yap and Taz were defective in their ability to differentiate into smooth muscle cells. Moreover, our RNA-Seq data revealed that genes functioning in adherens junction formation, endothelial cell migration, vasoconstriction and the cytoskeleton were differentially expressed in Yap and Taz mutants, suggesting that other developmental events also contribute to defective vascular development.
We noted that genes encoding the Jak-Stat cascade components, including Jak3, Ptk2b, Stat3, Stat5a and Stat5b, were upregulated in Wnt1Cre; Yap; Taz dCKO mutants. A previous study reported that the activation of Stat3 promotes the apoptosis of vascular smooth muscle cells by triggering mitochondria-mediated cell death receptors and cell death pathways (Bai et al., 2008). Further work is required to investigate the role of increased Jak-Stat pathway components during craniofacial development in an in vivo setting.
Transcripts of Prox1, a regulator of lymphatic development, were also elevated in Yap and Taz mutant embryos. Specification of lymphatic endothelial cells is an essential event in vascular development (Wigle and Oliver, 1999; Wigle et al., 2002; Johnson et al., 2008); thus, elevated Prox1 levels may conceivably disrupt normal vascular specification. It will be interesting to determine in future experiments whether increased Prox1 expression leads to fate switching to a predominantly lymphatic endothelial cell phenotype.
We found that Pdgfb is also upregulated in Yap and Taz mutant embryos. PDGF signaling has been implicated both directly and indirectly in the regulation of vasculogenesis and angiogenesis. Given the crucial role of Pdgfb in regulating the recruitment and proliferation of vascular cells (Yancopoulos et al., 2000), the defective vascular development observed in Yap and Taz mutant embryos might be related to increased Pdgfb expression.
The Hippo pathway regulates CNC proliferation and apoptosis
We showed that proliferation was significantly reduced in the mandibular mesenchyme in Yap and Taz mutant embryos. This diminished proliferation is consistent with our observation of reduced cell density surrounding the enlarged vessels in Yap and Taz mutant embryos. Furthermore, a recent study showed that NC migration and fate specification were spared in Wnt1Cre; Yap; Taz embryos at E10.5, suggesting that Yap and Taz regulate proliferation in the post migratory CNC (Manderfield et al., 2014). In Wnt1Cre2SOR; Yap; Taz embryos, we also found that proliferation was intact in the neural tube, despite the open neural tube phenotype, indicating that other mechanisms might account for the failure of neural tube closure.
Consistent with our in vivo observations, we found that the siRNA-mediated knockdown of Yap and Taz significantly reduced the proliferation of multipotent NC O9-1 cells, further suggesting that Yap and Taz are essential for NC cell proliferation. We also showed that the knockdown of Lats1 and Lats2 significantly increased the proliferation of O9-1 cells, which is consistent with the idea that Yap and Taz function as Hippo pathway effectors to regulate NC proliferation.
In addition, apoptosis was significantly increased in the mandibular mesenchyme in Yap and Taz mutant embryos. Both decreased proliferation and increased apoptosis were observed in E9.5 Yap and Taz mutant embryos in the absence of any detectable hemorrhage, indicating that the reduced cell proliferation and sparse cell density observed in E10.5 Yap and Taz mutant embryos were caused by a direct, primary effect of Yap and Taz, rather than a secondary effect due to the hemorrhaging.
Regulation of Fox genes by Yap and Taz and their potential roles in human disease
Fox transcription factors are key regulators of embryogenesis and control fundamental biological processes, including cell proliferation, fate determination, differentiation and growth (Kaufmann and Knochel, 1996; Kume et al., 1998, 2001; Kidson et al., 1999; Tuteja and Kaestner, 2007a,b; Benayoun et al., 2011; Haldipur et al., 2014). Notably, Fox genes are evolutionarily and functionally conserved across multiple species, including mice and humans, making animal models invaluable tools for understanding the mechanisms underlying human diseases caused by Fox genes.
Expression of Foxc1 was downregulated at the mRNA and protein level in Yap and Taz mutant embryos. Foxc1 has been implicated in several different human disorders including the most common cerebellar malformation, Dandy-Walker malformation (DWM), Axenfeld-Rieger syndrome (ARS), 6p25 deletion syndrome and iridogoniodysgenesis (Mears et al., 1998; Nishimura et al., 1998; Maclean et al., 2005; Aldinger et al., 2009; Delahaye et al., 2012). In addition to the function of Foxc1 in different diseases, it also has crucial roles in the development of multiple organs and tissues, including cerebellar, skull, ocular and cardiovascular development (Kume, 2009). Notably, compound Foxc1 and Foxc2-knockout mouse embryo mutants have craniofacial abnormalities similar to those of Yap; Taz mutants, such as enlarged blood vessels, sparse mesenchyme and an open neural tube (Kume et al., 2001).
Patients with mutations in Foxc1 display a variant of the Dandy-Walker malformation, including cerebellar vermis hypoplasia (CVH). Notably, Foxc1 mutant mice also have an enlarged fourth ventricle roof plate. Similar to Foxc1 mutant mice, Yap; Taz dCKO mice also display cerebellar hypoplasia with an enlarged fourth ventricle roof plate. In addition, Foxc1-knockout mice are reported to have hydrocephalus (Kume et al., 1998), a severe and common lethal human birth defect that was also observed in mice with Yap inactivation and Taz heterozygosity in NC cells. Collectively, these findings further support the hypothesis that Foxc1 is a downstream effector of Yap and Taz.
MATERIALS AND METHODS
Mouse alleles and transgenic lines
All animal experiments in this study were approved by the Baylor College of Medicine Institutional Review Board. The Wnt1Cre, Wnt1Cre2SOR, Yapflox/+ and Tazflox/+ mouse lines and alleles used in this study have been described previously (Chai et al., 2000; Xin et al., 2011, 2013; Lewis et al., 2013).
Histology and hematoxylin and eosin staining
All embryos were dissected in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde (PFA) overnight at 4°C. For H&E staining, the fixed embryos were dehydrated with an ethanol series (70% to 100%) and xylene and then embedded in paraffin. These tissues were subsequently cut into 7 μm sections and stained with H&E, as previously described (Lu et al., 1999).
O9-1 cell culture and siRNA knockdown
The O9-1 cells were cultured under undifferentiating conditions by following a previously published protocol (Ladher et al., 2000). The culture medium used consisted of Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, 0.1 mM MEM nonessential amino acids, 1 mM sodium pyruvate, 55 µM β-mercaptoethanol, 100 units/ml penicillin/streptomycin and 2 mM L-glutamine. Importantly, the medium was conditioned with growth-inhibited STO (mouse embryonic fibroblast cell line) feeder cells overnight, filtered (0.22 µm pore size) and further supplemented with 25 ng/ml basic fibroblast growth factor (R&D Systems, 233-FB) and 1000 U leukemia inhibitory factor (Millipore, ESG1106). The conditions used for smooth muscle differentiation were described previously (Ishii et al., 2012). For the siRNA knockdown experiments in O9-1 cells, siRNA SMARTpools targeting Yap, Taz and Hippo kinases Lats1 and Lats2 were purchased from Dharmacon and the transfections were performed by following a typical RNAiMAX transfection procedure (Thermo Fisher Scientific).
Yap exon 3 deletion using CRISPR/Cas9
To make a Yap-null O9-1 cell line, exon 3 of Yap was deleted using CRISPR/Cas9 genome editing. Two sgRNAs flanking exon 3 were identified using the sgRNA design tool (http://www.dna20.com). Four sgRNA oligonucleotides with overhanging BbsI restriction sites were synthesized by IDT (Coralville, IA): 5′-CACCGtggattacgtgggtatgtt-3′ (sgRNA1-forward), 5′-AAACaacatacccacgtaatccac-3′ (sgRNA1-reverse), 5′-CACCGagatggtctaatgtagtga-3′ (sgRNA2-forward) and 5′-AAACtcactacattagaccatctC-3′ (sgRNA2-reverse). The paired oligonucleotides were annealed and ligated into a pSpCas9(bb)-2a-GFP vector (Addgene, plasmid #48138). To create cell lines with a deletion of Yap exon 3, we transfected pSpCas9-GFP-YAP-sgRNA1 and pSpCas9-GFP-YAP-sgRNA2 into O9-1 cells using Lipofectamine 2000 (Thermo Fisher Scientific). Twenty-four hours after the transfection, the cells underwent fluorescence-activated cell sorting (FACS) with GFP/7-AAD. Viable, single cells (7-AAD−;GFP+) were seeded into 96-well plates with culture medium. After 4-5 days, single colonies could be observed in some of the wells; wells that had more than one colony were excluded from further analyses. Single colonies were passaged into 12-well plates and incubated for 2-4 days to expand the clones. When the cells were more than 80% confluent, they were dissociated with trypsin and half of the sample was processed for DNA extraction while the other half was used to make a freezer stock culture. To detect the Yap exon 3 deletion, we designed PCR primers flanking the sgRNA recognition sites: 5′-AAAACAGTCTCCACTACCCCTT-3′ (forward) and 5′-GGCCATCATAGATCCTGGACG-3′ (reverse). Clones harboring a homozygous deletion for Yap exon 3 were retained and used for experimental analyses.
3D embryo imaging with optical projection tomography microscopy
Optical projection tomography microscopy was used to determine embryonic craniofacial morphology. Specimens were embedded in 1% agarose (Amresco LLC, Solon, OH) and dehydrated in 25%, 50%, 75% and 100% ethanol (Sigma-Aldrich) for 2 h each. Samples were then stored in fresh 100% ethanol overnight before clearing. The dehydrated samples were then completely cleared in BABB solution, which contained one part benzyl alcohol (Thermo Fisher Scientific) and two parts benzyl benzoate (Acros Organics, Waltham, MA). Samples were then imaged on a custom-built OPT system (BCM-OPT) (Singh et al., 2015) modified from a previous version (Wong et al., 2013). The BCM-OPT microscope consisted of an Optem Zoom 125C lens (Qioptiq, Waltham, MA) and an auxiliary objective lens attached to a Retiga 4000DC FAST 1394 CCD camera (QImaging, Surrey, BC) with a 1.38× TV tube (Qioptiq). Autofluorescence images of the embryos were acquired with an X-Cite illumination light source (Exfo, Quebec, QE) and both a 425/26 nm BrightLine bandpass excitation filter and a 520/20 nm emission filter (Semrock, Rochester, NY). The embryos were rotated 360°, with each view taken with a 0.3° step size. Acquired images were reconstructed using NRecon Reconstruction software and 3D rendering was performed by using CTVox software (Bruker Corporation, Camarillo, CA).
Immunofluorescence and immunohistochemistry
After the embryos were fixed overnight in 4% PFA and dehydrated, they were embedded in paraffin, cut into 7 µm sections, and collected on Superfrost Plus slides (Thermo Fisher Scientific). The sections were rehydrated using xylene and a graded ethanol series to a final concentration of 70% and then processed for antigen retrieval. For immunofluorescence analysis, the antigens were retrieved by incubating the slides in citrate buffer (10 mM) for 2 min in a microwave oven. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) at a 1:500 dilution. Primary antibodies against phospho-Yap (Ser127) (Cell Signaling, 4911), phospho-histone H3 (Cell Signaling, 9701) and Foxc1 (Abcam, ab5079) were used at a 1:200 dilution. A broad HRP-conjugated secondary antibody (Invitrogen) was used according to the manufacturer's guidelines. An anti-SMA-Cy3 antibody (Sigma-Aldrich, Clone 1A4) was used at a 1:200 dilution. Staining was then visualized by using a TSA Plus Fluorescence System (PerkinElmer) and a Zeiss LSM 510 confocal microscope.
For immunohistochemistry analysis, antigens were retrieved by boiling the sections in ethylenediaminetetraacetic acid for 15 min and then tissues were permeabilized by incubating sections with 0.5% Triton X-100 for 15 min. Next, the sections were treated with 0.3% hydrogen peroxide for 15 min and blocked with 10% donkey serum for 1 h. The sections were incubated with primary antibody against Foxc1 (Abcam, ab5079) at a 1:200 dilution overnight at 4°C. The next day, the sections were incubated with HRP-conjugated anti-goat IgG antibody (1:200 dilution) for 1 h and then developed with DAB substrate (Vector Laboratories) for 2 min and 30 s. Sections were subsequently washed with water, stained with hematoxylin for 3 min and washed with water again for 5 min. Then, they were dehydrated with an ethanol series to a final concentration of 100% ethanol, fixed in xylene and mounted.
Whole-mount immunohistochemistry
E10.5 embryos were processed as described (Wythe et al., 2013). Briefly, the embryos were fixed in 4% PFA overnight, serially dehydrated to absolute methanol, bleached in 5% hydrogen peroxide/95% methanol overnight, and rehydrated to PBS-Tween (0.1%). The embryos were blocked for 2 h (PBS/5% goat serum/0.5% Triton X-100), then incubated with an anti-CD31 antibody (BD Biosciences, MEC13.3, 1:200 dilution) in blocking solution for 3 days. The embryos were then washed and incubated with biotin-conjugated goat anti-rat IgG secondary antibody (Vector Laboratories, 1:250 dilution) for 2 days. The signal was amplified with Alexa Fluor 488 Tyramide (Thermo Fisher Scientific, T20948). The embryos were then washed in blocking solution and incubated with anti-SMA-Cy3 antibody (Sigma-Aldrich, Clone 1A4, 1:200 dilution) for 3 days at 4°C. After washing, the embryos were cleared in ScaleA2 for 2 days, ScaleB4 for 2 days, then embedded in 1% agarose in ddH2O and cleared for another 2 days in ScaleA2. The embryos were then imaged on a Lightsheet Z.1 (Zeiss) using a 5× air lens (NA=0.16) in ScaleA2. All images were obtained using the same laser power and exposure time. Serial images were aligned using Zen software (Zeiss) and 3D rendering was performed with Imaris software (Bitplane).
RNA-Seq and data analysis
Mandibular tissues from Wnt1cre, Yap and Taz dCKO mutant and control E10.5 embryos were dissected in diethylpyrocarbonate-treated PBS. Total RNA extracted from these tissues was treated with RNase-free DNase I (Ambion) for 30 min at 37°C. Then, the poly(A) RNA was purified with the MicroPoly(A) Purist Kit according to the manufacturer's instructions (Thermo Fisher Scientific). The purified RNA samples were processed for cDNA synthesis and library preparation. cDNA sequencing using the Illumina platform and read mapping were performed as previously described (Nagalakshmi et al., 2008). Datasets were processed for gene ontology analysis, which provided different terms that represent gene properties including cellular components, molecular function and biological processes. The data were deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE69311). More information is available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ejajwkiynvevjuj&acc=GSE69311.
Chromatin immunoprecipitation
Embryonic orofacial tissues from wild-type mice at stages E10.5-E11.5 were dissected for Yap ChIP analysis, as previously described (Wang et al., 2013; Morikawa et al., 2015). Normal mouse IgG was used as a replacement control for the anti-Yap ChIP assay to show nonspecific immunoprecipitation of the chromatin. The primers used to amplify the TEAD regulatory element in the 5′ upstream region of the Foxc1 genomic sequence were 5′-CCTTGGCATCTCTCAGAAAGTC-3′ (sense) and 5′-TAGTCCTATCCCAGTGAGCATC-3′ (antisense).
Real-time PCR
For real-time RT-PCR analysis, total RNA was isolated from the mandibles of E10.5 embryos or O9-1 cells by using the RNeasy Micro Kit (Qiagen) and processed for cDNA synthesis using Super Script II Reverse Transcriptase (Invitrogen). For the real-time ChIP-PCR analysis, immunoprecipitated DNA and input DNA were used as templates. All real-time thermal cycling was performed with the StepOne Real-time PCR System (Thermo Fisher Scientific). SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) was used for real-time thermal cycling. All error bars represent s.e.m.
Cell counts and data analyses
For embryonic cell counting, all embryos used were at the same embryonic stage. The embryos were embedded carefully to maintain the same orientation and were sectioned at identical thickness (5 µm). Sections were carefully chosen to represent the matching regions between control and mutant embryos. Our analyses included at least three embryos for each genotype. For O9-1 cell counting, each experiment was performed in duplicate and all experiments were repeated three times or more. ImageJ software (National Institutes of Health, Bethesda, MD) was used to perform the cell counting and over 200 cells were counted for each experiment. The percentage of proliferating cells was determined by counting pHH3-positive cells and dividing that number by the number of DAPI-positive cells. The percentage of apoptotic cells was determined by counting the number of TUNEL-positive cells. All counting data are represented in graphs as the mean±s.e.m. The two-tailed Student's t-test was used to determine statistical significance and P<0.05 was considered statistically significant.
Acknowledgements
We thank Dr Nicole Stancel and Heather Leibrecht for editorial assistance in the preparation of the manuscript. We also thank Matthew C. Hill for Tead binding site analysis and Shichu Chen for cell counting.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
J.W. and J.F.M. designed research; J.W., Y.X., C.-W.H., I.M.M-T., Y.B. and J.D.W. performed research; J.W., Y.X., C.-W.H., I.M.M-T., M.Z., J.D.W., M.E.D. and J.F.M. analyzed data; M.I., R.E.M. and E.N.O provided reagents; J.W., C.-W.H., J.D.W. and J.F.M. wrote the manuscript.
Funding
This work was supported by American Heart Association (AHA) National Center Scientist Development Grant (SDG) 14SDG19840000 (to J.W.), 2014 Lawrence Research Award from the Rolanette and Berdon Lawrence Bone Disease Program of Texas (to J.W.). AHA National Center [12060353SDG to J.D.W.], National Institutes of Health [2T32GM088129-06 to I.M.M.-T.; NIH R01HL118761 and R01DE023177 to J.F.M.] and the Vivian L. Smith foundation (to J.F.M.). J.F.M. was supported by Transatlantic Network of Excellence Award LeDucq Foundation Transatlantic Networks of Excellence in Cardiovascular Research 14CVD01: ‘Defining the genomic topology of atrial fibrillation’. Also supported by Baylor College of Medicine IDDRC grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development and the Mouse Phenotyping Core at Baylor College of Medicine with funding from the NIH [U54 HG006348]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.126920/-/DC1
References
- Acloque H., Adams M. S., Fishwick K., Bronner-Fraser M. and Nieto M. A. (2009). Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J. Clin. Invest. 119, 1438-1449. 10.1172/JCI38019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldinger K. A., Lehmann O. J., Hudgins L., Chizhikov V. V., Bassuk A. G., Ades L. C., Krantz I. D., Dobyns W. B. and Millen K. J. (2009). FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy-Walker malformation. Nat. Genet. 41, 1037-1042. 10.1038/ng.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., Bresolin S., Frasson C., Basso G., Guzzardo V. et al. (2014). YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 158, 157-170. 10.1016/j.cell.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Bai Y., Ahmad U., Wang Y., Li J. H., Choy J. C., Kim R. W., Kirkiles-Smith N., Maher S. E., Karras J. G., Bennett C. F. et al. (2008). Interferon-gamma induces X-linked inhibitor of apoptosis-associated factor-1 and Noxa expression and potentiates human vascular smooth muscle cell apoptosis by STAT3 activation. J. Biol. Chem. 283, 6832-6842. 10.1074/jbc.M706021200 [DOI] [PubMed] [Google Scholar]
- Benayoun B. A., Caburet S. and Veitia R. A. (2011). Forkhead transcription factors: key players in health and disease. Trends Genet. 27, 224-232. 10.1016/j.tig.2011.03.003 [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C., Matthews H. K., Kuriyama S., Moreno M., Dunn G. A., Parsons M., Stern C. D. and Mayor R. (2008). Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456, 957-961. 10.1038/nature07441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Jiang X., Ito Y., Bringas P. Jr, Han J., Rowitch D. H., Soriano P., McMahon A. P. and Sucov H. M. (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, 1671-1679. [DOI] [PubMed] [Google Scholar]
- Coles E., Christiansen J., Economou A., Bronner-Fraser M. and Wilkinson D. G. (2004). A vertebrate crossveinless 2 homologue modulates BMP activity and neural crest cell migration. Development 131, 5309-5317. 10.1242/dev.01419 [DOI] [PubMed] [Google Scholar]
- Cordero D. R., Brugmann S., Chu Y., Bajpai R., Jame M. and Helms J. A. (2011). Cranial neural crest cells on the move: their roles in craniofacial development. Am. J. Med. Genet. A 155, 270-279. 10.1002/ajmg.a.33702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahaye A., Khung-Savatovsky S., Aboura A., Guimiot F., Drunat S., Alessandri J.-L., Gerard M., Bitoun P., Boumendil J., Robin S. et al. (2012). Pre- and postnatal phenotype of 6p25 deletions involving the FOXC1 gene. Am. J. Med. Genet. A 158A, 2430-2438. 10.1002/ajmg.a.35548 [DOI] [PubMed] [Google Scholar]
- Garcia-Castro M. I., Marcelle C. and Bronner-Fraser M. (2002). Ectodermal Wnt function as a neural crest inducer. Science 297, 848-851. [DOI] [PubMed] [Google Scholar]
- Glavic A., Silva F., Aybar M. J., Bastidas F. and Mayor R. (2004). Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development 131, 347-359. 10.1242/dev.00945 [DOI] [PubMed] [Google Scholar]
- Haldipur P., Gillies G. S., Janson O. K., Chizhikov V. V., Mithal D. S., Miller R. J. and Millen K. J. (2014). Foxc1 dependent mesenchymal signalling drives embryonic cerebellar growth. Elife 3, e03962 10.7554/eLife.03962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R. L. and Martin J. F. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458-461. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M., Arias A. C., Liu L., Chen Y.-B., Bronner M. E. and Maxson R. E. (2012). A stable cranial neural crest cell line from mouse. Stem Cells Dev. 21, 3069-3080. 10.1089/scd.2012.0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. C., Dillard M. E., Baluk P., McDonald D. M., Harvey N. L., Frase S. L. and Oliver G. (2008). Lymphatic endothelial cell identity is reversible and its maintenance requires Prox1 activity. Genes Dev. 22, 3282-3291. 10.1101/gad.1727208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E. and Knochel W. (1996). Five years on the wings of fork head. Mech. Dev. 57, 3-20. 10.1016/0925-4773(96)00539-4 [DOI] [PubMed] [Google Scholar]
- Kidson S. H., Kume T., Deng K., Winfrey V. and Hogan B. L. M. (1999). The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev. Biol. 211, 306-322. 10.1006/dbio.1999.9314 [DOI] [PubMed] [Google Scholar]
- Kume T. (2009). The cooperative roles of Foxc1 and Foxc2 in cardiovascular development. Adv. Exp. Med. Biol. 665, 63-77. 10.1007/978-1-4419-1599-3_5 [DOI] [PubMed] [Google Scholar]
- Kume T., Deng K.-Y., Winfrey V., Gould D. B., Walter M. A. and Hogan B. L. M. (1998). The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell 93, 985-996. 10.1016/S0092-8674(00)81204-0 [DOI] [PubMed] [Google Scholar]
- Kume T., Jiang H., Topczewska J. M. and Hogan B. L. M. (2001). The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 15, 2470-2482. 10.1101/gad.907301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladher R. K., Church V. L., Allen S., Robson L., Abdelfattah A., Brown N. A., Hattersley G., Rosen V., Luyten F. P., Dale L. et al. (2000). Cloning and expression of the Wnt antagonists Sfrp-2 and Frzb during chick development. Dev. Biol. 218, 183-198. 10.1006/dbio.1999.9586 [DOI] [PubMed] [Google Scholar]
- Lewis A. E., Vasudevan H. N., O'Neill A. K., Soriano P. and Bush J. O. (2013). The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev. Biol. 379, 229-234. 10.1016/j.ydbio.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl P., Johansson B. R., Leveen P. and Betsholtz C. (1997). Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277, 242-245. 10.1126/science.277.5323.242 [DOI] [PubMed] [Google Scholar]
- Lu M.-F., Pressman C., Dyer R., Johnson R. L. and Martin J. F. (1999). Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 401, 276-278. 10.1038/45797 [DOI] [PubMed] [Google Scholar]
- Maclean K., Smith J., St Heaps L., Chia N., Williams R., Peters G. B., Onikul E., McCrossin T., Lehmann O. J. and Ades L. C. (2005). Axenfeld-Rieger malformation and distinctive facial features: clues to a recognizable 6p25 microdeletion syndrome. Am. J. Med. Genet. A 132A, 381-385. 10.1002/ajmg.a.30274 [DOI] [PubMed] [Google Scholar]
- Manderfield L. J., Engleka K. A., Aghajanian H., Gupta M., Yang S., Li L., Baggs J. E., Hogenesch J. B., Olson E. N. and Epstein J. A. (2014). Pax3 and hippo signaling coordinate melanocyte gene expression in neural crest. Cell Rep. 9, 1885-1895. 10.1016/j.celrep.2014.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manderfield L. J., Aghajanian H., Engleka K. A., Lim L. Y., Liu F., Jain R., Li L., Olson E. N. and Epstein J. A. (2015). Hippo signaling is required for Notch-dependent smooth muscle differentiation of neural crest. Development 142, 2962-2971. 10.1242/dev.125807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears A. J., Jordan T., Mirzayans F., Dubois S., Kume T., Parlee M., Ritch R., Koop B., Kuo W.-L., Collins C. et al. (1998). Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am. J. Hum. Genet. 63, 1316-1328. 10.1086/302109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno L. M., Mansilla M. A., Bullard S. A., Cooper M. E., Busch T. D., Machida J., Johnson M. K., Brauer D., Krahn K., Daack-Hirsch S. et al. (2009). FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum. Mol. Genet. 18, 4879-4896. 10.1093/hmg/ddp444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y., Zhang M., Heallen T., Leach J., Tao G., Xiao Y., Bai Y., Li W., Willerson J. T. and Martin J. F. (2015). Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci. Signal. 8, ra41 10.1126/scisignal.2005781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M. and Snyder M. (2008). The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344-1349. 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura D. Y., Swiderski R. E., Alward W. L. M., Searby C. C., Patil S. R., Bennet S. R., Kanis A. B., Gastier J. M., Stone E. M. and Sheffield V. C. (1998). The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat. Genet. 19, 140-147. 10.1038/493 [DOI] [PubMed] [Google Scholar]
- Prescott S. L., Srinivasan R., Marchetto M. C., Grishina I., Narvaiza I., Selleri L., Gage F. H., Swigut T. and Wysocka J. (2015). Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163, 68-83. 10.1016/j.cell.2015.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati F. and Rijli F. M. (2003). Cranial neural crest and the building of the vertebrate head. Nat. Rev. Neurosci. 4, 806-818. 10.1038/nrn1221 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T. and Bronner-Fraser M. (2008). A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557-568. 10.1038/nrm2428 [DOI] [PubMed] [Google Scholar]
- Sela-Donenfeld D. and Kalcheim C. (1999). Regulation of the onset of neural crest migration by coordinated activity of BMP4 and Noggin in the dorsal neural tube. Development 126, 4749-4762. [DOI] [PubMed] [Google Scholar]
- Singh M., Nair A., Vadakkan T., Piazza V., Udan R., Frazier M. V., Janecek T., Dickinson M. E. and Larin K. V. (2015). Comparison of optical projection tomography and optical coherence tomography for assessment of murine embryonic development. SPIE Proc. 9334 10.1117/12.2078319 [DOI] [Google Scholar]
- Tuteja G. and Kaestner K. H. (2007a). SnapShot: forkhead transcription factors II. Cell 131, 192 10.1016/j.cell.2007.09.016 [DOI] [PubMed] [Google Scholar]
- Tuteja G. and Kaestner K. H. (2007b). SnapShot: forkhead transcription factors I. Cell 130, 1160.e1-1160.e2. 10.1016/j.cell.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Wang J., Bai Y., Li H., Greene S. B., Klysik E., Yu W., Schwartz R. J., Williams T. J. and Martin J. F. (2013). MicroRNA-17-92, a direct Ap-2alpha transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genet. 9, e1003785 10.1371/journal.pgen.1003785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle J. T. and Oliver G. (1999). Prox1 function is required for the development of the murine lymphatic system. Cell 98, 769-778. 10.1016/S0092-8674(00)81511-1 [DOI] [PubMed] [Google Scholar]
- Wigle J. T., Harvey N., Detmar M., Lagutina I., Grosveld G., Gunn M. D., Jackson D. G. and Oliver G. (2002). An essential role for Prox1 in the induction of the lymphatic endothelial cell phenotype. EMBO J. 21, 1505-1513. 10.1093/emboj/21.7.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson K. A., Rainger J., Floyd J. A. B., Ansari M., Meynert A., Aldridge K. V., Rainger J. K., Anderson C. A., Moore A. T., Hurles M. E. et al. (2014). Heterozygous loss-of-function mutations in YAP1 cause both isolated and syndromic optic fissure closure defects. Am. J. Hum. Genet. 94, 295-302. 10.1016/j.ajhg.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. D., Dazai J., Walls J. R., Gale N. W. and Henkelman R. M. (2013). Design and implementation of a custom built optical projection tomography system. PLoS ONE 8, e73491 10.1371/journal.pone.0073491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wythe J. D., Dang L. T. H., Devine W. P., Boudreau E., Artap S. T., He D., Schachterle W., Stainier D. Y. R., Oettgen P., Black B. L. et al. (2013). ETS factors regulate Vegf-dependent arterial specification. Dev. Cell 26, 45-58. 10.1016/j.devcel.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Kim Y., Sutherland L. B., Qi X., McAnally J., Schwartz R. J., Richardson J. A., Bassel-Duby R. and Olson E. N. (2011). Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 4, ra70 10.1126/scisignal.2002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Kim Y., Sutherland L. B., Murakami M., Qi X., McAnally J., Porrello E. R., Mahmoud A. I., Tan W., Shelton J. M. et al. (2013). Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 110, 13839-13844. 10.1073/pnas.1313192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G. D., Davis S., Gale N. W., Rudge J. S., Wiegand S. J. and Holash J. (2000). progress Vascular-specific growth factors and blood vessel formation. Nature 407, 242-248. 10.1038/35025215 [DOI] [PubMed] [Google Scholar]