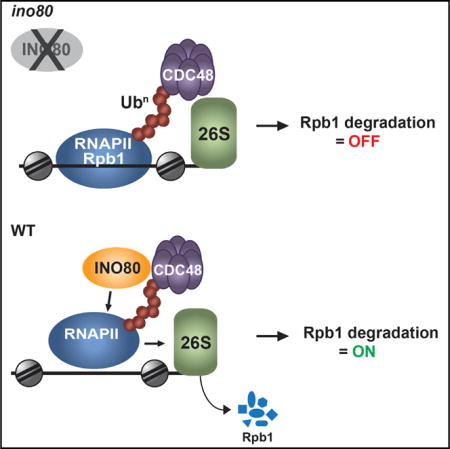
SUMMARY
Stalling of RNA Polymerase II (RNAPII) on chromatin during transcriptional stress results in polyubiquitination and degradation of the largest subunit of RNAPII, Rpb1, by the ubiquitin proteasome system (UPS). Here, we report that the ATP-dependent chromatin remodeling complex INO80 is required for turnover of chromatin-bound RNAPII in yeast. INO80 interacts physically and functionally with Cdc48/p97/VCP, a component of UPS required for degradation of RNAPII. Cells lacking INO80 are defective in Rpb1 degradation and accumulate tightly bound ubiquitinated Rpb1 on chromatin. INO80 forms a ternary complex with RNAPII and Cdc48 and targets Rpb1 primed for degradation. The function of INO80 in RNAPII turnover is required for cell growth and survival during genotoxic stress. Our results identify INO80 as a bona fide component of the proteolytic pathway for RNAPII degradation and suggest that INO80 nucleosome remodeling activity promotes the dissociation of ubiquitinated Rpb1 from chromatin to protect the integrity of the genome.
Graphical abstract
INTRODUCTION
Transcriptional elongation by RNA Polymerase II (RNAPII) is a discontinuous process. Backtracking of RNAPII or hindrance from chromatin structure, DNA damage, or other DNA metabolic processes during elongation can cause RNAPII to stall or arrest irreversibly (Svejstrup, 2007). RNAPII can be an obstacle to DNA replication and DNA damage repair machineries, posing a severe threat to cell viability (Daulny and Tansey, 2009; Helmrich et al., 2013). Polyubiquitination and degradation of RNAPII by the ubiquitin proteasome system (UPS) is a mechanism known to prevent transcriptional interference and resolve stalled polymerases on DNA (Wilson et al., 2013).
Proteolysis of RNAPII is an evolutionarily conserved, tightly regulated, multistep pathway (Wilson et al., 2013). In budding yeast, it involves mono- and polyubiquitination of Rpb1 by the E3 ligases Rsp5 and Cul3, respectively (Huibregtse et al., 1997; Ribar et al., 2007). Ubiquitination of RNAPII is inhibited by phosphorylation of serine 5 at the C-terminal domain of Rpb1, thereby restricting degradation of RNAPII by the 26S proteasome to the elongating complex (Somesh et al., 2005). The 26S proteasome associates with transcribing genes (Auld et al., 2006), supporting the idea that proteolysis of stalled RNAPII takes place on chromatin. How stalled RNAPII is released from its site of arrest for proteasomal degradation is a largely unresolved question. A recent study in yeast proposed the involvement of the protein segregase Cdc48 in this process (Verma et al., 2011). Cdc48/p97/VCP is an evolutionarily conserved essential AAA+ ATPase with a well-established role in dissociating ubiquitinated substrates from protein complexes, aggregates, or membranes (Jentsch and Rumpf, 2007; Meyer et al., 2012). Cdc48 function is regulated by its binding to adaptor proteins of the UBX family of ubiquitin receptors (Schuberth and Buchberger, 2008). Cdc48 and its adaptor proteins Ubx4 and Ubx5 are required for the turnover of chromatin-bound ubiquitinated RNAPII under UV-induced DNA damage conditions (Verma et al., 2011). While Deshaies and colleagues envisioned a role of Cdc48 in the dissociation of ubiquitinated Rpb1 from chromatin-bound Pol II holoenzyme, the molecular mechanism for the release of stalled RNAPII from chromatin remains unknown.
Chromatin is a compacted, yet highly dynamic nucleoprotein structure. The SWI/SNF family of ATP-dependent chromatin remodeling enzymes plays an important role in regulating chromatin architecture. The SWI/SNF-like enzymes are DNA translocases, which use the energy of ATP hydrolysis to move, eject, or restructure nucleosomes, leading to profound changes in chromosome organization (Saha et al., 2006). The current model of function posits that nucleosome remodeling enzymes control spatiotemporal accessibility of DNA to regulatory factors (Bartholomew, 2014; Clapier and Cairns, 2009). INO80 is an evolutionarily conserved ATP-dependent chromatin remodeling complex (Conaway and Conaway, 2009) that controls genome-wide organization of the chromatin landscape (Papamichos-Chronakis et al., 2011; Yen et al., 2012). INO80 mediates nucleosome sliding (Udugama et al., 2011; Yen et al., 2012) and nucleosome turnover (Yen et al., 2013) and facilitates H2A.Z/H2B dimer eviction (Papamichos-Chronakis et al., 2011). INO80 has been directly implicated in a wide variety of DNA metabolic processes, including transcription, DNA replication, DNA-damage repair, and chromosome segregation across species (Conaway and Conaway, 2009). However, how INO80 function regulates nuclear processes remains largely unknown. Here, we report that in Saccharomyces cerevisiae, INO80 functions in the ubiquitin-proteasome system for RNAPII proteolysis. We show that INO80 promotes degradation of Rpb1 upon DNA damage conditions. INO80 physically interacts with Cdc48 and associates with Rpb1 in the presence of Cdc48, targeting Rpb1 primed for degradation. We demonstrate that INO80 destabilizes nucleosomes and disrupts the contacts between ubiquitinated Rpb1 and chromatin, suggesting that INO80 is required for the release of poly-ubiquitinated RNAPII arrested on chromatin. Our data provide evidence that INO80 promotes cell viability and suppresses genomic instability by chromatin clearance of RNAPII, revealing a link between chromatin regulation and nuclear protein turnover.
RESULTS
INO80 Physically Interacts with the CDC48 Complex
To gain insight into the role of INO80 in DNA metabolism, we conducted a proteomic screen for protein-interacting partners of the Ino80 ATPase. We performed mass spectrometry analysis on calmodulin pull downs from exponentially growing yeast cells expressing Ino80-TAP. Surprisingly, GO analysis and protein network analysis by GENEMANIA (http://genemania.org/) of the MS results revealed a statistically significant enrichment in “protein catabolic process” and “protein complex disassembly” pathways, with false discovery rate < 10−5, with several INO80 interactors involved in the ubiquitin signaling and ubiquitin-proteosomal degradation pathways (Figure S1A). Among the hits, we found the AAA-ATPase Cdc48 and its cofactors Ubx4, Ubx6, and Ubx7, which form a nuclear complex with Cdc48 (Decottignies et al., 2004) (Figure S1A). TAP co-immunoprecipitation experiments confirmed the interaction between Ino80-TAP and Cdc48 (Figure 1A). In a reciprocal GFP-IP, Cdc48-GFP associated with both Ino80 and the Arp5 subunit of INO80 (Figure 1B). Arp5 also associated with the GFP-tagged Cdc48 cofactors Ubx4, Ubx5, and Ubx7 in a GFP-IP assay (Figure 1C). These results indicate that INO80 and CDC48 complexes associate in vivo.
Figure 1. INO80 Physically Associates with the CDC48 Complex.
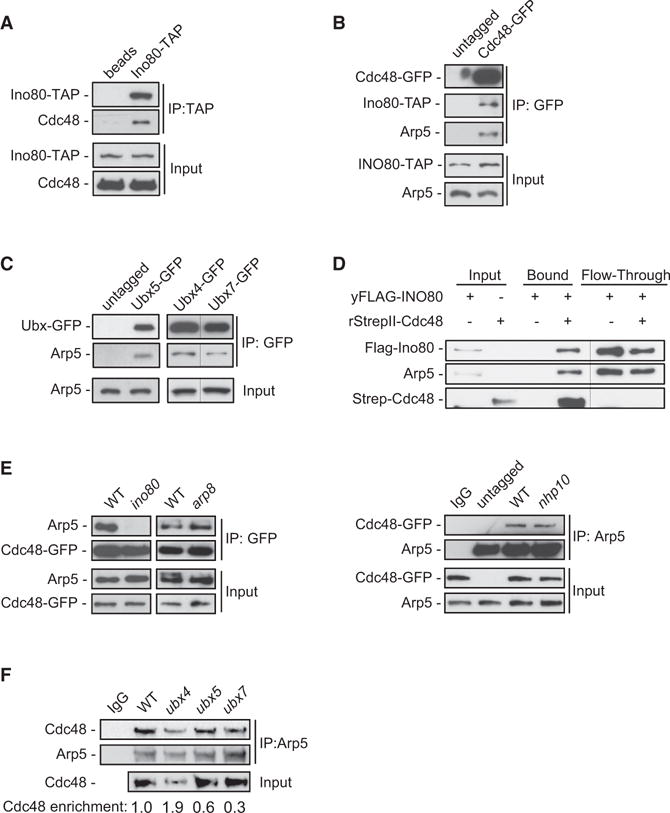
(A) Lysates from cells expressing Ino80-TAP were subjected to mock-IP (beads) or IP against TAP and immunoblotted for TAP and Cdc48.
(B) Lysates from cells co-expressing Ino80-TAP and Cdc48 either untagged or tagged with GFP were subjected to GFP-IP. Inputs and IP samples were immunoblotted for GFP and Arp5.
(C) Lysates from cells expressing Ubx5, Ubx4, or Ubx7 either untagged (control lane) or tagged with GFP were subjected to GFP-IP and immunoblotted for Arp5 and GFP. Lanes for Ubx4 and Ubx7 pull downs were cropped from the same blot and displayed side by side for clarity.
(D) Purified yeast FLAG-INO80 complex was incubated with purified recombinant StrepII-Cdc48 tethered to Strep-Tactin beads. Input, bound, and flow-through samples were immunoblotted for Strep, Flag, and Arp5. Flow-through lanes were cropped from a different exposure of the same blot and displayed side by side with input and bound samples for clarity.
(E) Lysates from cells expressing untagged or GFP-tagged Cdc48 in the indicated strains were subjected to GFP-IP (left) or IP against Arp5 (right) and immunoblotted for Arp5 and GFP.
(F) Lysates from cells from the indicated strains were subjected to mock-IP (IgG) or IP against Arp5 and immunoblotted for Arp5 and Cdc48. Values reflect the enrichment of Cdc48 relative to Arp5 in the IP, after normalization against the amount of Cdc48 in the input. The value in WT was set arbitrarily to 1.0. The WT input is common for IgG and WT samples. This experiment was performed in parallel with the experiment in Figure 5E.
See also Figure S1.
Next, we tested whether INO80 interacts directly with Cdc48 in an in vitro assay. INO80 was FLAG-purified to homogeneity from yeast cells expressing Flag-Ino80 (Figure S1B) and incubated with recombinant StrepII-Cdc48 (Figure S1C). Both Flag-Ino80 and Arp5 were pulled down by Strep-Cdc48 (Figure 1D). This result indicates a direct physical interaction between Cdc48 and INO80.
The INO80 complex contains several distinct multi-protein modules, which are brought together through interactions of specific subunits of the complex with the Ino80 ATPase (Tosi et al., 2013). Notably, the specific subunits Arp8 and Arp5 control the chromatin remodeling activity of INO80 (Shen et al., 2003), while the high-affinity binding of Nhp10 to nucleosomes and distorted DNA has been proposed to target INO80 on specific DNA sites (Ray and Grove, 2012; Tosi et al., 2013). Co-IP analysis revealed that binding of Cdc48 to Arp5 was abolished in cells lacking Ino80, demonstrating that the interaction of Cdc48 with Arp5 requires the catalytic Ino80 subunit and intact INO80 complex (Figure 1E). The association of Cdc48 with Arp5 remained unchanged in arp8 and nhp10 mutants compared to wild-type (WT) cells (Figure 1E). Therefore, the chromatin-related ARP5, ARP8, and NHP10 modules are dispensable for the binding of Cdc48 to INO80.
We further tested, by a co-IP against Arp5, whether Ubx4, Ubx5, or Ubx7 may regulate the interaction between INO80 and CDC48. Arp5 interacted stronger with Cdc48 in the ubx4 mutant, while we also observed that Cdc48 protein levels were decreased compared to WT (Figure 1F). The interaction between Cdc48 and Arp5 was moderately reduced in the ubx5 mutant and strongly decreased in ubx7 (Figure 1F). This result suggests that Ubx5 and Ubx7 regulate the interaction between INO80 and CDC48. Collectively, these data provide evidence for CDC48 as an interacting partner of INO80.
Functional Interactions between INO80 and CDC48 Promote Cell Viability, Resistance to DNA Damage, and Efficient DNA Replication
To test whether INO80 is functionally related to CDC48, we analyzed the genetic interactions between components of the two complexes. We deleted ARP8 in the temperature-sensitive cdc48-3 mutant strain, since disruption of INO80 results in lethality in the W303 strain background of cdc48-3. Strikingly, while both arp8 and cdc48-3 mutants do not show substantial growth defects at 25°C and 30°C, the arp8,cdc48-3 double-mutant strain demonstrated synthetic sickness at 25°C and synthetic lethality at 30°C (Figure 2A). These gross synthetic phenotypes suggest that INO80 and CDC48 function in parallel pathways to promote cell viability.
Figure 2. Functional Interactions between INO80 and Cdc48Ubx Complexes.
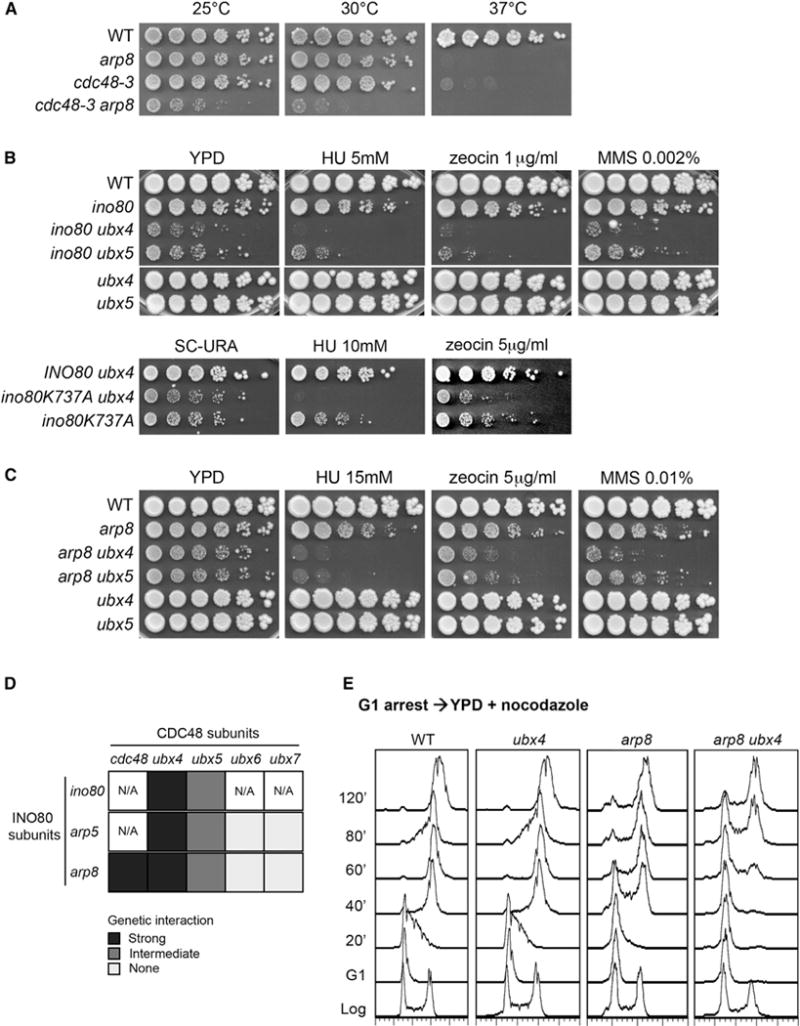
(A) 5-fold serial dilution of cells from the indicated strains were plated onto YPD and incubated at the indicated temperatures for 3 days.
(B) Upper panel: serial dilution of cells from the indicated strains were plated onto YPD or YPD containing the indicated concentrations of hydoxyurea (HU), zeocin, or methyl methanesulfonate (MMS) and incubated at 30°C for 4 days. All strains were grown in the same plate. Lower panel: serial dilutions of ino80 and ino80,ubx4 cells expressing a WT INO80 allele or an ATPase-defective allele of INO80 (ino80-K277A) from the pRS416 plasmid were plated onto SC-URA medium containing HU or zeocin at the indicated concentrations and incubated at 30°C for 3–4 days.
(C) 5-fold serial dilutions of cells from the indicated strains were plated onto YPD or YPD containing the indicated concentrations of HU, zeocin, or MMS and incubated at 30°C for 4 days.
(D) Table summarizing the genetic interactions between different INO80 and Cdc48 co-factors, as shown in (A)–(C) and Figure S2A–S2C. The genetic interactions are classified according to their strength, as following: strong (dark gray), intermediate (gray), and none (pale gray). N/A corresponds to genetic interactions that have not been tested.
(E) Cells from the indicated strains were synchronized in G1 phase with α-factor and subsequently released into YPD containing nocodazole. Cell samples were collected at the indicated times and analyzed for DNA content by flow cytometry analysis. Microscopy analysis showed similar bud emergence kinetics for arp8 and arp8,ubx4 strains (not shown).
See also Figure S2.
We evaluated whether the ATPase activity of Ino80 participates in the functional interactions with CDC48 under normal conditions and under genotoxic stress induced by the presence of the S-phase-dependent, DNA damage-inducing alkylating agent methyl methane sulfonate (MMS) (Tercero et al., 2003), the replicative stress-inducing ribonucleotide reductase inhibitor hydroxyurea (HU) (Allen et al., 1994), and the radiomimetic agent zeocin, which induces chromosome breaks independently of DNA replication (Ramotar and Wang, 2003). Cell growth or viability was not affected by disruption of the UBX factors in any of the conditions tested (Figures 2B, 2C, and S2A). Inactivation of Ino80 by deleting INO80 or expressing an ATPase-dead INO80 allele (ino80K737A) in ubx4 resulted in growth synthetic defect in normal conditions and severe synthetic lethality in all drugs tested (Figure 2B). The ino80,ubx5 strain also exhibited synergistic defects, albeit to a lesser extent (Figure 2B). These synthetic genetic interactions further support the view that INO80 and CDC48 act in parallel pathways.
Genetic interactions were not observed when either ARP5 or ARP8 was disrupted in the ubx6 and ubx7 mutants (Figures S2B and S2C). However, slow growth and synthetic lethality in genotoxic stress conditions were observed in deletion of either ARP5 or ARP8 when combined with ubx4,ubx5 single or ubx4,ubx5 double mutants (Figures 2C, S2B, and S2C). These results indicate specific functional relationships between INO80 and CDC48 and implicate the chromatin function of INO80 in the functional crosstalk with CDC48 (Figure 2D).
We further evaluated the functional interaction between INO80 and CDC48Ubx4 in DNA replication by flow cytometry analysis. The ubx4,arp8 cells required more than double the time to complete DNA replication compared to arp8 and three times more than WT and ubx4 cells (Figure 2E). This result suggests that INO80 and CDC48 promote DNA replication in a synergistic manner. No phosphorylation of the DNA damage marker gH2AX (H2AS129Phos) or hyperphosphorylation of the checkpoint protein Rad53 was observed in the ubx4,arp8 strain under normal conditions (Figure S2D). Therefore, the synthetic delay in S phase progression of the ubx4,arp8 strain is due to neither persistent DNA damage (Rogakou et al., 1998) nor permanent activation of the S phase checkpoint (Pellicioli et al., 1999). Taken together, these data indicate that the function of INO80 in cell homeostasis and in viability under genotoxic stress are related to CDC48-associated pathways.
INO80 Promotes Degradation of Rpb1
Cdc48, together with Ubx4 and Ubx5, facilitates degradation of Rpb1 (Verma et al., 2011). The genetic interactions of INO80 with CDC48Ubx4,Ubx5 in DNA damage prompted us to investigate a potential role for INO80 in the degradation of Rpb1 under genomic instability-inducing conditions. We arrested WT, ino80, and ino80K737A mutant cells in G1 and subsequently released into S phase in the presence of MMS. Cycloheximide was added to inhibit protein synthesis after cells had entered S phase, in order to avoid blockage at the G1/S transition. Rpb1 abundance decreased in WT cells over time, indicating degradation of Rpb1 (Figure 3A). However, ino80 and ino80K737A cells maintained high levels of Rpb1 protein, supporting a role for INO80 in RNAPII degradation upon S-phase-dependent DNA damage (Figure 3A).
Figure 3. INO80 Controls Degradation of RNAPII.
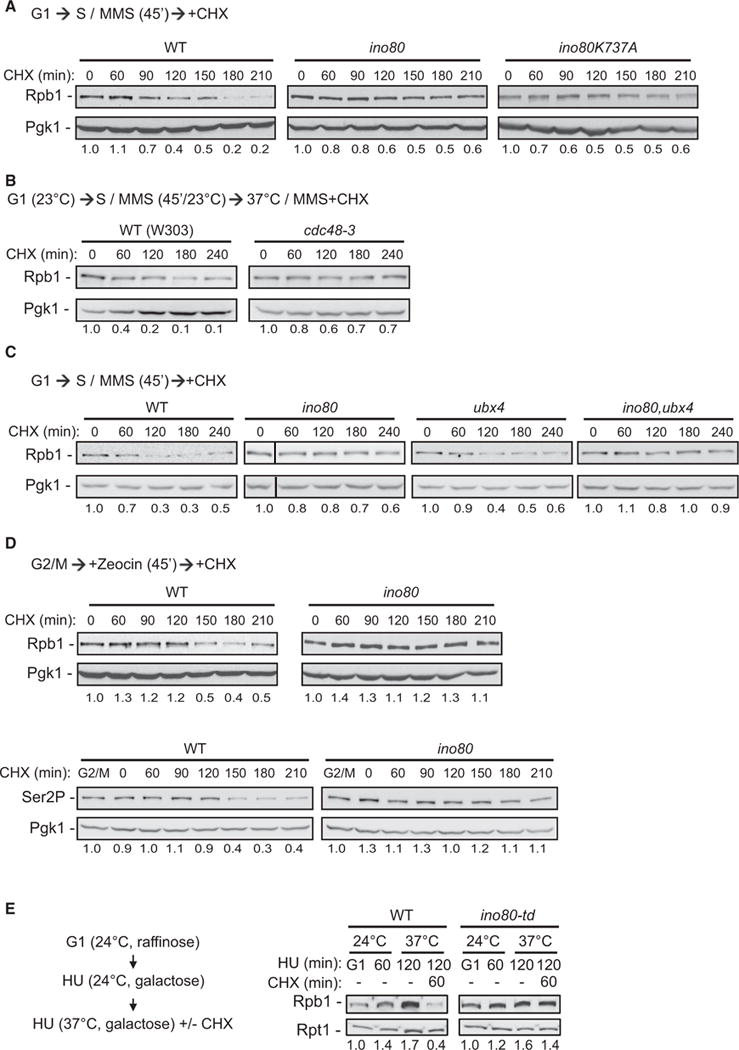
(A) Rpb1 abundance in WT and ino80 cells synchronized in G1 with a-factor and subsequently released into YPD containing 0.06% MMS. Cycloheximide (CHX) was added 45 min after release. Protein samples were acid-extracted at the indicated time points after cycloheximide addition and analyzed by immunoblotting against Rpb1. Pgk1 serves as loading control. Values reflect the amount of Rpb1 in the specific conditions relative to the starting time point after normalization against the respective loading control. The first point at each strain was set arbitrarily to 1.0.
(B) Rpb1 abundance in WT and cdc48-3 cells synchronized in G1 and released into YPD containing 0.1% MMS at 23°C for 45 min. Cells were subsequently shifted to 37°C in medium containing 0.1% MMS and cycloheximide. Protein samples were collected at the indicated times after temperature shift. Analysis and quantifications are as in (A).
(C) Rpb1 abundance in cells from the indicated strains synchronized in G1 and released into YPD containing 0.1% MMS. Cycloheximide treatment, sample analysis, and quantifications are as in (A). The immunoblot of the 0 min time point sample for ino80 was cropped from the same exposure of the same blot and displayed next to the 60 min samples for clarity. For original image, see Figure S3B.
(D) Rpb1 abundance in WT and ino80 cells synchronized at G2/M with nocodazole and treated with 100 μg/ml zeocin. Cycloheximide was added to the medium 45 min after addition of zeocin. Protein samples were collected at the indicated times after cycloheximide addition and analyzed by immunoblotting against Rpb1 (upper panel) and serine 2 phosphorylated form of Rpb1 (Ser2P, lower panel). Values are as in (A).
(E) Left panel: schematic representation for conditional degradation of Ino80 in HU. In brief, wild-type and ino80-td cells carrying a galactose-inducible UBR1 gene were grown in raffinose medium and arrested in G1 at 24°C. Cells were subsequently released at the permissive temperature in medium containing 100 mM HU. After 30 min, galactose was added in the medium for 30 min to induce expression of UBR1. Cells were subsequently shifted to 37°C to induce degradation of Ino80, and cycloheximide was added in the medium. Right panel: WT and ino80-td cells, grown as described in the left panel, were collected at the indicated times in HU and protein extracts were prepared and analyzed as in (A).
See also Figure S3.
Rpb1 degradation was defective in cdc48-3 and ubx4,ubx5 mutants upon MMS treatment (Figures 3B and S3A). Deletion of INO80 in the ubx4 mutant strain resulted in a greater defect in Rpb1 degradation (Figures 3C and S3B). These results implicate INO80 and CDC48 in RNAPII degradation in MMS and point toward discrete functions for INO80 and CDC48 in the RNAPII proteolytic pathway.
The release of cells into S phase in the presence of HU led to a strong turnover in Rpb1 protein levels in WT but not in ino80 (Figure S3C). In contrast, we observed normal degradation of Rpb1 in UV-irradiated ino80 mutant cells (Figure S3D), indicating a role for INO80 under specific genotoxic conditions.
G2/M arrested ino80 cells treated with zeocin also exhibited severe defects in Rpb1 degradation, indicating that the role of INO80 in degradation of RNAPII is not exclusively coupled to S phase (Figure 3D). The levels of elongating RNAPII phosphorylated at serine 2 (Rpb1S2P) were decreased in WT but remained largely unchanged in ino80 (Figure 3D). This data points to a role for INO80 in the UPS-dependent inhibition of transcriptional elongation during the DNA damage response (Pankotai et al., 2012; Somesh et al., 2005).
To exclude long-term effects of INO80 deletion in the RNAPII proteolytic pathway, we monitored Rpb1 degradation in a yeast strain that allows for rapid, inducible degradation of Ino80 (ino80-td) at 37°C (Jónsson et al., 2004) (Figure S3E). WT and ino80-td cells were arrested in G1 at 24°C and subsequently released at permissive temperature in medium containing HU (Figure 3E) for sufficient time to activate the intra-S phase checkpoint (Figure S3E). When cells were shifted to 37°C, Rpb1 was efficiently degraded in the WT cells, but not in the ino80-td strain (Figure 3E). These data directly implicate INO80 in RNAPII degradation.
The Function of INO80 in Cell Growth and Maintenance of Genome Stability Is Coupled to Ubiquitin-Dependent Proteolysis of RNAPII
We tested for genetic interactions between INO80 and RNAPII mutants deficient for ubiquitin-mediated proteolysis. Rpb1 degradation is mediated by ubiquitination at lysines K330 and K695 (Somesh et al., 2007). While single K330 or K695 mutations confer no sickness or sensitivity to the genotoxic stress conditions (Somesh et al., 2007) (Figure 4), the double-mutant rpb1K330R,K695R is inviable, demonstrating the importance of RNAPII ubiquitination in cell homeostasis (Somesh et al., 2007). Strikingly, deletion of ARP8 in either rpb1K330R or rpb1K695R mutants resulted in extreme synthetic sickness in normal conditions and hypersensitivity in HU, zeocin, and MMS (Figure 4). Therefore, a functional relationship between INO80 and RNAPII proteolysis is critical for cell viability in normal and genome instability-inducing conditions.
Figure 4. The Function of INO80 in Cell Growth and Resistance to Genotoxic Stress Is Coupled to RNAPII Proteolysis.
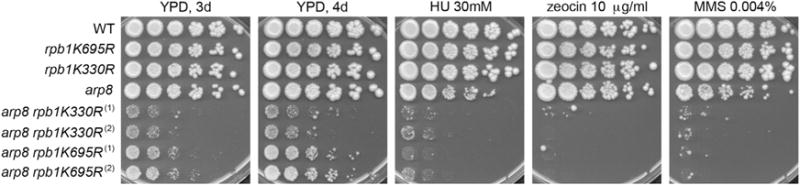
Serial dilutions of cells from the indicated strains were plated onto YPD or YPD containing the indicated concentrations of HU, zeocin, or MMS. Pictures of the YPD plates were taken after 3 or 4 days of incubation at 30°C, as indicated. Pictures of the plates containing drugs were taken after 4 days of incubation. Numbering indicates different isolates.
Concurrent Interaction of INO80 with RNAPII and Cdc48
We sought to understand how INO80 is integrated into the proteolytic pathway for RNAPII. Interestingly, Rpb1 was identified in our proteomic screen and in a co-IP assay for Ino80-TAP (data not shown and Figure 5A). Both Ino80 and Arp5 associated with Rpb1 in a reciprocal co-IP against Rpb1-GFP, confirming the interaction between INO80 and RNAPII (Figure 5B). Furthermore, Rpb1S2P co-precipitated with Arp5 in a co-IP assay (Figure 5C). This result implies that INO80 is in contact with elongating RNAPII.
Figure 5. INO80 and Cdc48 Associate Simultaneously with Rpb1.
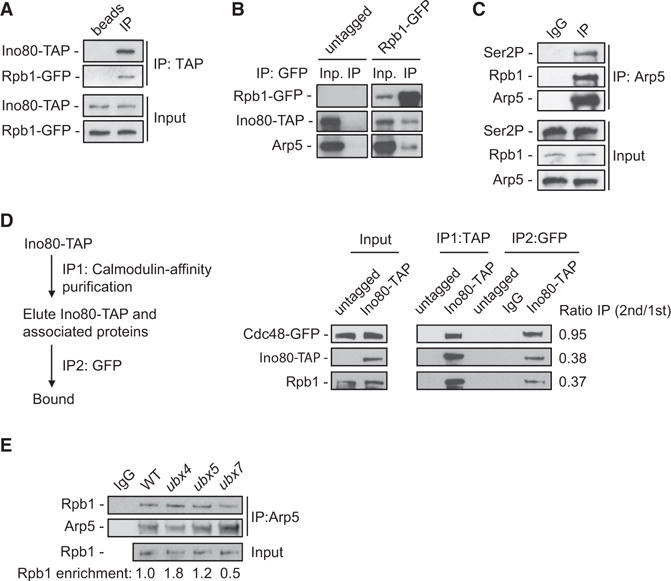
(A) Lysates from cells co-expressing Ino80-TAP and Rpb1-GFP were subjected to mock-IP or IP against TAP and immunoblotted for GFP and TAP.
(B) Lysates from cells co-expressing Ino80-TAP and Rpb1 untagged or tagged with GFP were subjected to GFP-IP and immunoblotted for GFP, TAP, and Arp5.
(C) Lysates from WT cells were subjected to mock-IP or IP against Arp5 and immunoblotted for Rpb1, Rpb1S2P, and Arp5.
(D) Tandem immunoprecipitation assay was performed on log-phase cells co-expressing either untagged Ino80 (control) or Ino80-TAP with Cdc48-GFP. Calmodulin affinity pull down for Ino80-TAP was conducted in the first step (IP1), followed by a second pull down against GFP (IP2). Samples were immunoblotted for TAP, Rpb1, and GFP. Values reflect the relative enrichment of the corresponding protein in the second IP, over the amount of the respective protein in the first IP.
(E) Lysates from cells from the indicated strains were subjected to IP against Arp5 and immunoblotted for Arp5 and Rpb1. The WT input corresponds to both IgG and WT IPs. This experiment was performed from the same cell extracts as in Figure 1F. Values are as in Figure 1F.
See also Figure S4.
To determine whether INO80 binds simultaneously to CDC48 and RNAPII, we developed an in vivo tandem pull-down assay, first for Ino80-TAP and subsequently for Cdc48-GFP (Figure 5D, scheme). Both Cdc48 and Rpb1 co-purified with Ino80-TAP (Figure 5D). Almost all of Cdc48-GFP from the INO80-TAP pull down was recovered in the GFP IP, demonstrating the efficiency of our assay (Figure 5D). Furthermore, approximately 40% of both Ino80 and Rpb1 released from the first IP were detected in the second pull down for Cdc48, suggesting concomitant interaction of INO80 with CDC48 and RNAPII (Figure 5D). In a different tandem pull-down assay for Ino80-TAP followed by IP against Rpb1, we also observed that a fraction of Cdc48 was recovered in the second IP (Figure S4A). These results strongly suggest that INO80, CDC48, and RNAPII engage into a ternary complex formation.
To understand how the concurrent association of INO80 with RNAPII and CDC48 is regulated, we investigated whether Ino80 and Ubx co-factors of Cdc48 participate in the association of RNAPII with CDC48 or INO80, respectively. No significant change in the interaction between Cdc48 and Rpb1 was observed in the absence of Ubx4, Ubx5, or Ubx7 cofactors (Figure S4B). GFP-IP demonstrated that the interaction of Cdc48-GFP with Rpb1 is not altered in the absence of INO80, ruling out the possibility that INO80 mediates the recruitment of Cdc48 to Rpb1 (Figure S4C). Binding of Rpb1 to Arp5 was not altered in the ubx5 strain, while it was increased in the ubx4 strain (Figure 5E). In contrast, cells lacking Ubx7 demonstrated a reduced association of Rpb1 with Arp5 (Figure 5E). This result indicates a role for Ubx7 in promoting the interaction between INO80 and RNAPII.
The Interaction between INO80 and RNAPII Is Regulated by the Ubiquitin-Proteasome System
The observation that Ubx7 regulates the Arp5-Rpb1 interaction raises the possibility that ubiquitination may be important for the association of INO80 with RNAPII. Notably, the binding of Rpb1 to Arp5 was reproducibly decreased in the E3 ubiquitin ligase rsp5-1 mutant strain (Figure 6A, top panel), which is defective in ubiquitination of Rpb1 (Huibregtse et al., 1997). Furthermore, higher migratory species of Rpb1, indicative of ubiquitinated Rpb1, were observed in the Arp5 pull down in WT, but not in rsp5-1 cells (Figure 6A, bottom panel). This result indicates that Rsp5 promotes the interaction of INO80 with Rpb1.
Figure 6. INO80 Interacts with Rpb1 in the Context of the UPS.
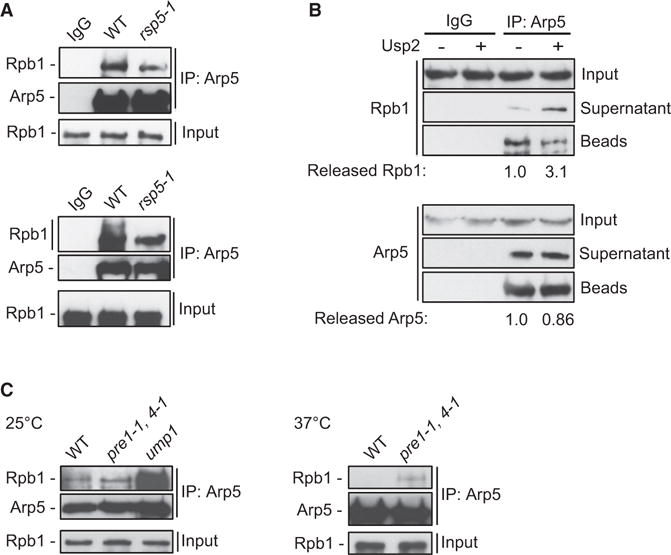
(A) Lysates from WT and rsp5-1 cells grown at 25°C and shifted to 37°C for 2 hr were subjected to mock-IP with IgG (WT) or IP against Arp5 and analyzed by immunoblot against Rpb1 and Arp5. Upper and lower panels come from two independent experiments.
(B) Lysates from WT cells were subjected to mock-IP with IgG or IP against Arp5. The bead-bound samples were subjected to deubiquitination assay in the presence or absence of recombinant Usp2. Soluble and bead-bound fractions were isolated and analyzed as in (A). Arp5 in supernatant reflects the dissociation rates of the immunoprecipitated samples in the deubiquitination reaction conditions and serves as quality control. Values reflect the ratio of the amount in the supernatant over the total amount (supernatant + beads) for the respective protein. Values of mock reactions were set arbitrarily to 1.0. SD was 8%.
(C) Cell lysates from the indicated strains grown at 25°C or shifted to 37°C for 2 hr were subjected to IP against Arp5 and analyzed as in (A).
To directly test whether ubiquitination is necessary for the association between INO80 and RNAPII, Arp5 co-IP from WT cells was subjected to in vitro deubiquitination (DUB) by incubating the pull-down sample with recombinant ubiquitin-specific protease Usp2, followed by separation of the supernatant from the beads. We found a 3-fold increase of the amount of Rpb1 released from Arp5 in the soluble fraction of the reaction containing Usp2 (Figure 6B). This result demonstrates that either deubiquitination by Usp2 promotes dissociation of Rpb1 from INO80 or that Usp2 competes for binding with UbRpb1 and thereby reduces its interaction with Arp5. Both scenarios further underline the importance of ubiquitin in the interaction of INO80 with RNAPII.
Polyubiquitinated Rpb1 accumulates in cells with compromised proteasome function, which cannot efficiently degrade Rpb1 (Beaudenon et al., 1999). Deletion of the 20S maturation factor Ump1 dramatically increased the amount of Rpb1 bound to Arp5 (Figure 6C). Similarly, in the conditional pre1-1, pre4-1ts double-mutant strain for the assembly subunits of the 20S proteolytic core Pre1 and Pre4, the interaction between Rpb1 and Arp5 was strongly increased at the restrictive temperature of 37°C (Figure 6C). These results indicate that INO80 interacts with RNAPII primed for degradation, identifying INO80 as a bona fide component of the proteolytic pathway for RNAPII degradation.
Tight Binding of RNAPII to Chromatin in the Absence of INO80
We next sought to delineate the role of INO80 in RNAPII proteolysis. A chromatin fractionation assay revealed that both Cdc48 and the Rpt1 subunit of the 19S regulatory particle of the 26S proteasome associated normally with chromatin in ino80 cells in either normal or DNA damage-inducing conditions (Figure S5A). This result excludes the possibility that impaired degradation of Rpb1 in ino80 is due to defective recruitment of either Cdc48 or the proteasome to chromatin.
We asked whether INO80 regulates the binding of ubiquitinated RNAPII to chromatin. Yeast cells expressing (His)6-tagged Ub were subjected to chromatin fractionation, followed by a NiNTA pull down under denaturing conditions to enrich for ubiquitinated proteins, including poly-ubiquitinated species of Rpb1 (henceforth UbRpb1, Figure S5B). The rpb1K330R mutant exhibited a significant decrease in the total and the chromatin-bound UbRpb1 (Figure S5C), indicating binding of ubiquitinated RNAPII to chromatin. Interestingly, UbRpb1 was highly enriched in the chromatin fraction of ino80 and ino80K737A cells when compared to WT (Figure 7A). Quantitative analysis indicated an increase of UbRpb1 on chromatin in the absence of INO80 by greater than 3-fold (Figure 7B). This result indicates that INO80 prevents accumulation of ubiquitinated RNAPII onto chromatin.
Figure 7. Tight Association of UbRpb1 with Chromatin and Increased Nucleosome Stability in the Absence of INO80.
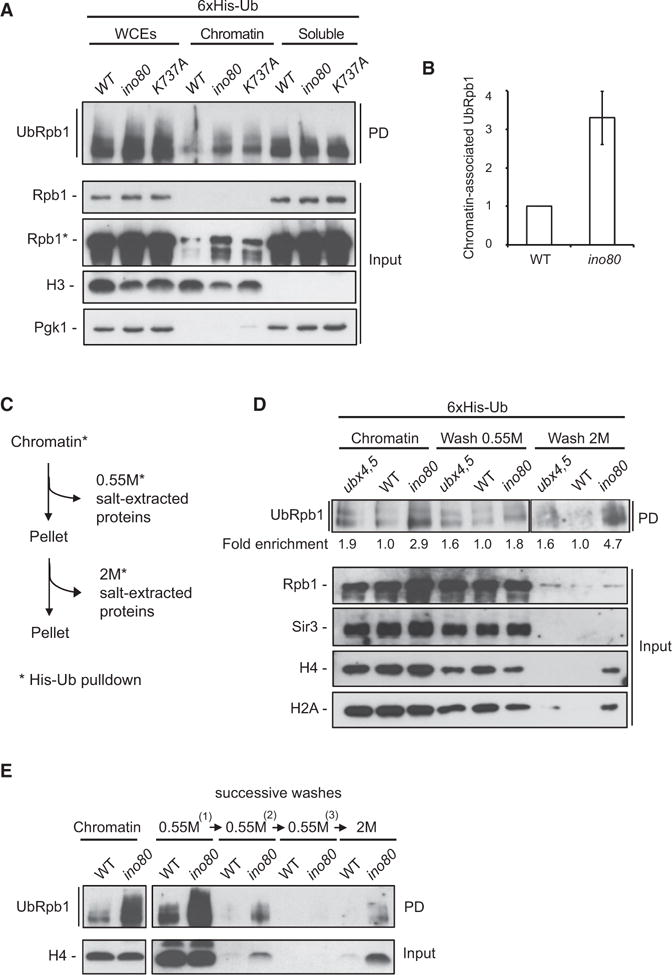
(A) Log-phase WT, ino80, and ATPase-dead ino80-K737A cells expressing 6xHis-tagged Ub were fractionated into chromatin and soluble fractions. NiNTA pull down under denaturing conditions was conducted in all fractions. Immunoblot analysis was performed against Rpb1 on the His pull-down samples (PD) and against Rpb1, histone H3, and Pgk1 on the total proteins samples (Input). Asterisk (*) denotes long time exposure.
(B) Quantification reflects the amount of chromatin-associated ubiquitinated Rpb1 relative to histone H3 in WT and ino80 null cells. The amount of ubiquitinated Rpb1 in the WT strain was set arbitrarily to 1.0. The values represent the means from five independent experiments, with error bars reflecting SD.
(C) Schematic representation of the sequential salt extraction assay for chromatin-associated proteins as described in Experimental Procedures.
(D) Chromatin from WT, ubx4,ubx5, and ino80 cells expressing (His)6-tagged Ub was isolated by chromatin fractionation and subjected to salt extraction as described in (C) followed by NiNTA pull down under denaturing conditions. Immunoblot analysis was performed against Rpb1 on the His pull-down samples (PD) and against Rpb1, Sir3, H4, and H2A on the total proteins samples (Input). Values reflect the enrichment of UbRpb1 in the respective mutant strain, after normalization against the amount of UbRpb1 in WT in the same fraction. The amount of UbRpb1 in WT for each condition was set arbitrarily to 1.0. Lanes corresponding to chromatin, wash 0.55 M, and wash 2 M were cropped from different exposures of the same blot.
(E) Chromatin from WT and ino80 cells expressing (His)6-tagged Ub was isolated by chromatin fractionation and subjected to salt extraction as follows: chromatin was successively washed three times with 0.55 M NaCl, followed by a final wash at 2 M. The different fractions were treated as in (A).
See also Figure S5.
To test whether INO80 deletion leads to stronger binding of UbRpb1 to chromatin, we developed an assay to evaluate the in vivo binding capacity of UbRpb1 to chromatin (Figure 7C). Chromatin from yeast cells expressing His-Ub was isolated and washed with 0.55 M and subsequently with 2 M NaCl solutions. The wash fractions were collected and subjected to denatured NiNTA pull down. Immunoblot analysis of the wash fractions for proteins released from DNA under increased concentrations of NaCl provides a mean to evaluate how strongly proteins, including ubiquitinated Rpb1, associate with chromatin. Treatment with 0.55 M NaCl released most of UbRpb1 from the chromatin of WT cells (Figure 7D). Interestingly, a long exposure revealed the presence of a small amount of UbRpb1 in the WT 2 M NaCl wash fraction (Figure S5D). This suggests that a population of UbRpb1 binds to chromatin tightly in normal conditions. In agreement with a previous report (Verma et al., 2011), higher levels of UbRpb1 bound to chromatin in the ubx4,ubx5 strain compared to WT (Figure 7D). However, the amount of UbRpb1 in ubx4,ubx5 relative to WT remained essentially the same in all fractions, indicating that CDC48Ubx4,Ubx5 does not regulate the binding capacity of UbRpb1 to DNA. In contrast, the amount of UbRpb1 detected in the 2 M NaCl wash in ino80 relative to WT was increased when compared to the relative amount of UbRpb1 found in the total chromatin of ino80 (Figure 7D). This result indicates that in the absence of INO80 a greater population of UbRpb1 binds tightly to chromatin.
It is possible that the increased amount of UbRpb1 on ino80 chromatin requires extensive washes, rather than higher ionic conditions, in order to be released. We therefore modified our assay, washing chromatin three times with 0.55 M NaCl before proceeding with the final 2 M NaCl wash. Under these conditions, UbRpb1 from WT cells was undetectable at the third 0.55 M NaCl wash fraction but could be readily detected in the 2 M NaCl fraction (Figures 7E and S5E). Importantly, the amount of UbRpb1 in the 2 M NaCl wash fraction of the ino80 mutant (Figure 7E) was substantially higher compared to WT. This result provides further evidence that UbRpb1 tightly associates with DNA in cells lacking INO80.
The persistent binding of UbRpb1 to DNA in the absence of INO80 prompted us to evaluate the binding of other chromatin bound proteins, including histones, under the same conditions. In all strains, the heterochromatin protein Sir3 was completely released from the chromatin fraction upon 0.55 M NaCl wash (Figure 7D), demonstrating that stronger binding of proteins to chromatin is not a general effect of INO80 loss. Histones H4 and H2A were released from WT chromatin at 0.55 M NaCl, as expected (Piñeiro et al., 1991), and the same was observed in ubx4,ubx5 (Figure 7D). In contrast, a substantial portion of histones H4 and H2A was detected in the 2.0 M NaCl wash fraction of the ino80 mutant (Figure 7D). This result shows that histones are more resistant to dissociation from DNA in the absence of INO80. Likewise, a 2 M NaCl wash was required for the release of histone H3 from DNA in the ino80 mutant (Figure S5F) and for the release of histone H4 from ino80 chromatin, which had been previously washed three times with 0.55 M NaCl (Figure 7E). In conclusion, loss of INO80 leads to strong binding of nucleosomal histones to DNA, correlating with the tighter association of UbRpb1 with chromatin observed in the same strain.
DISCUSSION
Here, we provide evidence that the ATP-dependent chromatin remodeling complex INO80 is directly implicated in degradation of stalled RNAPII. Cells lacking INO80 exhibit aberrant accumulation of polyubiquitinated Rpb1 on chromosomal DNA. In the absence of INO80, UbRNAPII binds tightly to chromatin and is impervious to degradation. INO80 interacts with RNAPII in the context of the UPS. Our analyses show that the DNA-dependent ATPase activity of the INO80 complex is required for the function of INO80 in RNAPII proteolysis. We therefore propose that clearance of stalled, ubiquitinated RNAPII from chromatin occurs via an INO80-mediated chromatin remodeling mechanism.
Integration of INO80 in the Ubiquitin-Mediated RNAPII Degradation Pathway
Our data suggest that INO80 is a component of the ubiquitin-proteasome system for RNAPII proteolysis. First, INO80 simultaneously interacts with RNAPII and Cdc48, a factor known to promote proteolysis of Rpb1 (Verma et al., 2011). Second, both Ubx7 and the E3 ligase Rsp5 promote the interaction between INO80 and RNAPII. Third, the INO80-RNAPII interaction is mediated by ubiquitin and enhanced upon disruption of the 26S proteasome. These observations posit INO80 downstream of Rpb1 ubiquitination and upstream of RNAPII degradation by the proteasome. The strong synthetic negative genetic interactions between INO80 and CDC48 mutants and the fact that simultaneous deletion of INO80 and UBX4 exacerbates the defect in degradation of Rpb1 raise the possibility of crosstalk between INO80 and CDC48 functions. It is therefore plausible that INO80 and CDC48 may converge on stalled RNAPII and act in concert in order to facilitate its efficient degradation. The role of the interaction between INO80 and CDC48 in RNAPII turnover is not clear yet. Nevertheless, our discovery of a physical and functional interplay between INO80 and CDC48 complexes in targeting RNAPII provides a molecular framework within which to investigate the mechanistic underpinnings of their role in the RNAPII degradation pathway.
Evidence for a Chromatin-Related Mechanism for RNAPII Extraction
The UPS pathway for RNAPII proteolysis is an extensively studied, multi-step process. Cdc48 has been implicated in the UV-induced degradation of Rpb1 (Verma et al., 2011). We find that INO80 is required for Rpb1 degradation in several different DNA damage-inducing conditions, but not upon UV irradiation. This is in agreement with the weak sensitivity of ino80 cells in UV damage (Sarkar et al., 2010). While the reasons underlying the specificity for INO80 requirement in RNAPII degradation under different conditions are not clear, these data indicate that global chromatin aberrations caused by ino80 mutations are not a de facto obstacle for RNAPII degradation.
Our study reveals that Ino80 facilitates the release of stalled RNAPII from DNA. Remarkably, we find that INO80, but not Cdc48Ubx4/Ubx5, promotes loosening the contact between UbRNAPII and chromatin. Moreover, deletion of INO80 does not result in accumulation of the 26S proteasome on chromatin, as has been reported for Cdc48 mutants (Verma et al., 2011). These results point toward a distinction of function between the chromatin remodeling and protein segregase activities of INO80 and CDC48, respectively.
The Swi/Snf-like ATPase factor Rad26 and its human homolog Cockayne syndrome protein B (CSB) have been shown to be involved in RNAPII processing during transcription-coupled nucleotide excision repair (TC-NER). Rad26/CSB stimulates lesion bypass by RNAPII, averting ubiquitination and proteolysis of RNAPII (Anindya et al., 2007; Charlet-Berguerand et al., 2006; Selby and Sancar, 1997; Woudstra et al., 2002). In contrast, absence of INO80 leads to increased ubiquitination and defective degradation of RNAPII, suggesting that INO80 targets terminally arrested RNAPII. Thus, it is interesting to speculate that INO80 and Rad26/CSB might work in two independent and compensatory chromatin-related pathways that control the fate of RNAPII upon DNA damage.
How could INO80 facilitate dissociation of RNAPII from chromatin? In a fashion similar to that of the prokaryotic ATP-dependent DNA translocase protein Mfd (Selby and Sancar, 1993), the DNA translocase activity of INO80 may disrupt the association of RNAPII to DNA. In eukaryotes, elongating RNAPII frequently stalls and can terminally arrest inside nucleosomes (Bintu et al., 2012; Bondarenko et al., 2006; Churchman and Weissman, 2011). Therefore, it is conceivable that release of arrested RNAPII from nucleosomal DNA may require the chromatin remodeling activity of INO80. The requirement of the ATPase activity of Ino80 in Rpb1 degradation and for preventing accumulation of UbRNAPII on chromatin is in line with this model. In addition, our genetic analysis implicates the chromatin-related Arp8 module of INO80 in RNAPII proteolysis. Our observation that INO80 promotes loosening of the histone-DNA contacts is in agreement with such a mechanistic role for INO80. In such a scenario, we envisage that the remodeling activity of INO80 could potentially expose the active site at the clamp of RNAPII, which is protected by the nucleosome (Chang et al., 2014). Such a nucleosome remodeling action could thus allow access to termination or other specialized RNAPII-release factors, unlocking RNAPII from DNA. Irrespective of the precise potential mechanism of action, our data reveal that INO80 plays a key role in orchestrating the extraction of RNAPII from chromatin.
Chromatin Remodeling at the Interface of Nuclear Proteostasis and DNA Metabolism: Implication for Genome Stability and Disease
Transcriptional interference with DNA replication and repair is strongly associated with chromosomal recombination and mutagenesis, leading to increased genomic instability and high cancer occurrence (Aguilera and García-Muse, 2013; Haffner et al., 2011; Helmrich et al., 2013). Our data establish chromatin clearance of UbRNAPII by INO80 as an essential step in the RNAPII proteolytic pathway, critical for cell growth, in promoting DNA replication and for prevention of genomic instability. It is likely that INO80 function alleviates transcriptional stress from sites of transcriptional interference with DNA replication and repair.
The metazoan INO80 complex contains the deubiquitinating enzyme Uch37 (UCH-L5), (Jin et al., 2005; Yao et al., 2008). This is strong evidence for a putative functional link between the metazoan INO80 and the UPS. A recent report has associated INO80, along with Uch37, the proteasome, and the RNAPII machinery, with progression of Alzheimer’s disease in humans (Kikuchi et al., 2013). Furthermore, two recent studies have provided individual evidence for Ino80 and the nuclear UPS in maintaining pluripotency and self-renewal of embryonic stem cells and reprogramming of somatic cells into induced pluripotent stem cells (Buckley et al., 2012; Wang et al., 2014). Whether the role of INO80 in development and disease is linked to the UPS is unknown. Our work opens exciting avenues for the investigation of the role of the metazoan INO80 in the nuclear UPS.
EXPERIMENTAL PROCEDURES
Yeast Strains
Strains used in this study are listed in Table S1. Gene deletions and other standard procedures were performed as described (Longtine et al., 1998).
Biochemical Techniques
Ino80-TAP was purified as in Sinha et al. (2009). Benzonase-treated yeast cell extracts from Ino80-TAP or untagged strains were incubated with Calmodulin Sepharose beads, and pull downs were analyzed by MS. The experiment was conducted twice. Proteins recovered in the mock pull down or not recovered in the biological replicates were removed from the Ino80 interactors’ list. Immunoprecipitation against GFP-tagged proteins was conducted using GFP antibody or GFP-TRAP beads. Chromatin fractionation was performed as described in Papamichos-Chronakis et al. (2011). His-ubiquitin pull downs in denaturing conditions were conducted as described in Becuwe et al. (2012). The deubiquitination assay was conducted in the presence or absence of purified Usp2 (Enzo Life Sciences). The soluble and bead bound fractions were collected separately and treated with SDS sample buffer. For the tandem immunoprecipitation experiments, yeast cell lysates were first subjected to pull down with calmodulin beads. The bound proteins were eluted, and a second pull down was performed on the eluate using either anti-GFP or anti-Rpb1 antibody. In vitro pull-down experiments were conducted incubating purified S. cerevisiae Flag-Ino80 complex with recombinant purified StrepII-Cdc48 tethered to Strep-Tactin beads.
All biochemical experiments were reproduced at least twice. Images were acquired by radiography film and the ImageQuant LAS 4000 mini Imager (GE Healthcare). Quantification of non-saturated images was performed using ImageJ software.
Cell Biology Assays
Cell spotting, cell-cycle arrest, and FACS analysis were performed as described previously (Papamichos-Chronakis and Peterson, 2008).
Supplementary Material
Highlights.
INO80 interacts with the protein segregase Cdc48
INO80 physically associates with RNAPII in the context of the UPS
INO80 is required for degradation of RNAPII
INO80 promotes dissociation of ubiquitinated Rpb1 from chromatin
Acknowledgments
This work has received support by ATIP-Avenir (INSERM), Marie Curie CIG (FP7 EU), Schlumberger Foundation for Education and Research (FSER), Epigenesys NoE. La Ligue Contre le Cancer (A.L.), Association “le Cancer du sein, Parlons en” (S.T.), and under the program «Investissements d’Avenir» launched by the French Government and implemented by ANR with the references ANR-10-LABX-0044_DEEP and ANR-10-IDEX-0001-02 PSL. We thank R. Deshaies and A. Peyroche for yeast strains, K. Labib and A. Taddei for antibodies, and S. Léon for plasmids and protocols. We thank B.F. Pugh, K. Yen, R. Margueron, L. Prendergast, and members of the Papamichos team for helpful discussions and critical reading of the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.10.028.
AUTHOR CONTRIBUTIONS
M.P.-C. conceived the project. F.P. carried out the INO80-TAP screen. F.D. and D.L. performed the mass spectrometry analysis. S.B. and B.B. purified INO80-Flag. A.L., S.T., and M.P.-C. designed the experiments, analyzed and interpret data, and wrote the manuscript. A.L. conducted experiments for Figures 1, 2, 4, 5, and 7. S.T. conducted experiments for Figures 1, 5, and 6. M.P.-C. conducted experiments for Figure 3.
References
- Aguilera A, García-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- Anindya R, Aygün O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol Cell. 2006;21:861–871. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha C, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Léon S. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J Cell Biol. 2012;196:247–259. doi: 10.1083/jcb.201109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal elements that control the topography of the barrier to transcription. Cell. 2012;151:738–749. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Ujvári A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Kulaeva OI, Shaytan AK, Kibanov M, Kuznedelov K, Severinov KV, Kirpichnikov MP, Clark DJ, Studitsky VM. Analysis of the mechanism of nucleosome survival during transcription. Nucleic Acids Res. 2014;42:1619–1627. doi: 10.1093/nar/gkt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet-Berguerand N, Feuerhahn S, Kong SE, Ziserman H, Conaway JW, Conaway R, Egly JM. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Daulny A, Tansey WP. Damage control: DNA repair, transcription, and the ubiquitin-proteasome system. DNA Repair (Amst) 2009;8:444–448. doi: 10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Decottignies A, Evain A, Ghislain M. Binding of Cdc48p to a ubiquitin-related UBX domain from novel yeast proteins involved in intracellular proteolysis and sporulation. Yeast. 2004;21:127–139. doi: 10.1002/yea.1071. [DOI] [PubMed] [Google Scholar]
- Haffner MC, De Marzo AM, Meeker AK, Nelson WG, Yegnasubramanian S. Transcription-induced DNA double strand breaks: both oncogenic force and potential therapeutic target? Clin Cancer Res. 2011;17:3858–3864. doi: 10.1158/1078-0432.CCR-10-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Nudler E, Tora L. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol. 2013;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutiérrez JL, Coleman MK, Workman JL, Mushegian A, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- Jónsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Kikuchi M, Ogishima S, Miyamoto T, Miyashita A, Kuwano R, Nakaya J, Tanaka H. Identification of unstable network modules reveals disease modules associated with the progression of Alzheimer’s disease. PLoS ONE. 2013;8:e76162. doi: 10.1371/journal.pone.0076162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro M, Puerta C, Palacián E. Yeast nucleosomal particles: structural and transcriptional properties. Biochemistry. 1991;30:5805–5810. doi: 10.1021/bi00237a025. [DOI] [PubMed] [Google Scholar]
- Ramotar D, Wang H. Protective mechanisms against the antitumor agent bleomycin: lessons from Saccharomyces cerevisiae. Curr Genet. 2003;43:213–224. doi: 10.1007/s00294-003-0396-1. [DOI] [PubMed] [Google Scholar]
- Ray S, Grove A. Interaction of Saccharomyces cerevisiae HMO2 domains with distorted DNA. Biochemistry. 2012;51:1825–1835. doi: 10.1021/bi201700h. [DOI] [PubMed] [Google Scholar]
- Ribar B, Prakash L, Prakash S. ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells. Mol Cell Biol. 2007;27:3211–3216. doi: 10.1128/MCB.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Kiely R, McHugh PJ. The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. J Cell Biol. 2010;191:1061–1068. doi: 10.1083/jcb.201006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell. 2009;138:1109–1121. doi: 10.1016/j.cell.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somesh BP, Reid J, Liu WF, Søgaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Somesh BP, Sigurdsson S, Saeki H, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell. 2007;129:57–68. doi: 10.1016/j.cell.2007.01.046. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci. 2007;32:165–171. doi: 10.1016/j.tibs.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner KP. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–1219. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Udugama M, Sabri A, Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Miao YL, Zhou B, Han L, Fargo DC, et al. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14:575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Harreman M, Svejstrup JQ. Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim Biophys Acta. 2013;1829:151–157. doi: 10.1016/j.bbagrm.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Woudstra EC, Gilbert C, Fellows J, Jansen L, Brouwer J, Erdjument-Bromage H, Tempst P, Svejstrup JQ. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415:929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013;154:1246–1256. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


