Abstract
Despite significant efforts in the past decade towards complete mapping of the human proteome, 3564 proteins (neXtProt, 09-2014) are still “missing proteins”. Over one-third of these missing proteins are annotated as membrane proteins, owing to their relatively challenging accessibility with standard shotgun proteomics. Using non-small cell lung cancer (NSCLC) as a model study, we aim to mine missing proteins from disease-associated membrane proteome, which may be still largely under-represented. To increase identification coverage, we employed Hp-RP StageTip pre-fractionation of membrane-enriched samples from 11 NSCLC cell lines. Analysis of membrane samples from 20 pairs of tumor and adjacent normal lung tissue were incorporated to include physiologically expressed membrane proteins. Using multiple search engines (X!Tandem, Comet and Mascot) and stringent evaluation of FDR (MAYU and PeptideShaker), we identified 7702 proteins (66% membrane proteins) and 178 missing proteins (74 membrane proteins) with PSM-, peptide-, and protein-level FDR of 1%. Through multiple reaction monitoring (MRM) using synthetic peptides, we provided additional evidences for 8 missing proteins including 7 with transmembrane helix domains (TMH). This study demonstrates that mining missing proteins focused on cancer membrane sub-proteome can greatly contribute to map the whole human proteome. All data were deposited into ProteomeXchange with the identifier PXD002224.
Keywords: Missing Proteins, Hp-RP StageTip, Membrane Proteins, MRM, Lung Cancer
Introduction
The completion of the human genome project which decoded more than 20,000 protein-coding genes has inspired enthusiastic efforts towards complete mapping of the human proteome in order to understand the human biology. Mass spectrometry has become a promising tool for large-scale profiling of the proteome particularly when coupled to advances in biological sample preparation, and bioinformatics algorithms. Recently, mass spectrometry-based draft map of the human proteome provided by two independent groups1,2 followed by antibody-based tissue mapping by the Human Proteome Atlas (HPA) group3 marked a huge progress with a claim of identifying and characterizing over 90% of the human proteome. After the first human proteome draft maps, based on the neXtProt database (09-2014 release), there are still 3564 proteins with no or inadequate evidence of translation, and considered as “missing proteins”.4 Missing proteins are those predicted to be encoded from the gene but with no available protein expression evidence from mass spectral detection, antibody-capture, 3D structures (X-Ray or NMR) or Edman sequencing.5 The current list of coding genes for 3564 missing proteins includes 2647 genes having transcript expression evidence, 214 genes inferred from homologous proteins in related species, 87 genes hypothesized from gene models and 616 “dubious” or “uncertain” genes.4,5
The expression of some proteins may vary in different tissues or cell types which may contribute to the difficulty of detecting these proteins with common proteomic workflows. Some of these missing proteins may be expressed only in rarely available samples, tissue or cell types like the brain, nasal epithelium, skeletal muscle and testis.5 Guruceaga et al. also studied gene expression profiles using over 3400 public microarray experiments and showed the importance of prioritizing normal tissues such as testis, brain, and skeletal muscle, as well as cancer samples of ovary, lung, kidney, breast, uterus, prostate, and lymph node.6 Some missing proteins are also likely to be expressed only under certain stimulus or stress.5 Recently, some mass spectrometry (MS) evidences were reported evidences to identify missing proteins from human brain tissues,7 lung tissues and cell lines,8 colorectal cancer samples,9 and hepatocellular carcinoma samples10, showing the significance of clinical samples in detecting missing proteins. In addition, the distinct proteome profiles deciphered by the draft map of the human proteome1 also revealed that missing proteins may even be expressed only during development in embryo or fetal tissues, with over 700 proteins having ten-fold increase in expression level compared to the adult counterparts.1
Another critical factor that contributes to the lack of protein-level evidence for missing proteins is related to physico-chemical characteristic of the proteins. Some missing protein sequences are unlikely to yield detectable peptides with the commonly employed tryptic digestion method, while others are composed only of few amino acid residues producing less number of observable peptides for MS analysis.11 Among the different structural features, 34% of the current missing proteins in neXtProt are annotated membrane proteins with some composed of multiple hydrophobic TMH domains. Chang et al. found that hydrophobicity and protein abundance greatly influence the detectability of a protein.12 Beck et al. were able to identify more than 10000 proteins from a single cell type of human osteosarcoma cell line (U2OS), however they observed that around 33% of the mRNAs corresponding to the unidentified proteins encodes for transmembrane proteins.13 The difficulty to detect membrane proteins consisting of TMH domains even with advanced MS platforms is due to the high concentration of detergents usually required for solubilization, resistance to enzymatic cleavage due to inaccessible sequences, and their inherent low abundance.5,14,15 Muraoka et al. have reported 851 missing membrane proteins in the membrane fraction of breast cancer tissues, providing a significant contribution to the efforts of completely mapping the whole human proteome.16 Taken together, membrane sub-proteome in human cancer samples may be a source for mining missing protein.
The technical limitations of current shotgun proteomics approach also add as a deterring factor in missing protein identification, especially in extremely complex biological samples. Recent studies have emphasized the evaluation of the analytic strategies, including sample pre-fractionation and improvement of MS analytic approach for large-scale protein identification in complex samples.17-20 Iwasaki and Ishihama highlighted the importance of further advances in sample pre-fractionation and sensitive mass spectrometry detection to address the wide dynamic range and huge complexity of the proteome.18 Peptide fractionation is a vital approach for enhancing the proteome coverage by separating co-eluting peptides for more efficient mass spectrometry analysis.19,20 Kim et al. also performed pre-fractionation by separating peptides into 96 fractions followed by concatenation to 24 fractions in generating a draft map of the human proteome.1
MRM has been underlined by C-HPP to be a promising targeted technique for validating the expression of missing protein coding genes due to its high sensitivity (low-attomolar), broad dynamic range (up to five orders of magnitude), and reproducibility better than the common data dependent acquisition (DDA) mode.17,21 Chen et al. used MRM detection method to confirm the expression of 57 targeted missing proteins in normal human liver tissue samples from 185 MRM assays,22 while Segura et al. performed MRM analysis in multiple laboratories for the detection of recombinant forms of 24 missing proteins.23
In this study, we hypothesized that deep membrane sub-proteomic profiling in human cancer cells and tissues can be an efficient strategy for the identification of missing proteins, even for a single cancer type. To provide higher sensitivity, high-pH reverse phase stop-and-go extraction tip (Hp-RP StageTip) fractionation followed by detection with high-resolution MS was applied for more comprehensive analysis. Hp-RP StageTip fractionation allowed increased recovery of the hydrophobic peptides and enhanced the identification of membrane proteins.24 For confident identification of missing proteins, multiple search engines and two false discovery rate (FDR) estimation approaches (PeptideShaker and MAYU), as well as unique peptide filtering were employed. In addition to 11 NSCLC cell lines, the patient-to-patient heterogeneity from 20 pairs of tumor and adjacent normal tissues of NSCLC patients were also utilized to increase the coverage of the membrane proteome of lung cancer. Under strict criteria of 1% FDR at the peptide-to-spectrum match (PSM-), peptide- and protein-level with peptides having 7 or more amino acid residues and at least one unique peptide for each protein, the in-depth membrane proteome profiling documented 7702 proteins from which 5121 (66%) were annotated to be membrane proteins. This provided mass spectral evidence for 178 missing proteins, among which 139 (78%) already possessed transcript-level protein evidence and 74 (41%) were annotated to be membrane proteins. Using synthetic reference peptides and MRM acquisition, we were able to further validate the expression of 8 selected missing proteins in Hp-RP StageTip fractionated membrane-enriched samples. Seven of these validated missing proteins were membrane proteins with TMH domains and confirmed in multiple cell lines.
Experimental Section
Materials and Reagents
Triethylammonium bicarbonate (TEABC), methyl methanethiosulfonate (MMTS), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), trifluoroacetic acid (TFA), 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES), magnesium chloride (MgCl2), potassium chloride (KCl), Hydrochloric acid (HCl), HPLC grade acetonitrile (ACN), and sodium chloride (NaCl) were purchased from Sigma-Aldrich (St. Louis, MO). Urea was purchased from USB Corporation (Cleveland, OH USA). Protease inhibitor cocktail tablet was obtained from Roche Diagnostics (Mannheim, Germany). Sodium dodecyl sulfate (SDS), sucrose, and ethylenediaminetetraacetic acid (EDTA) were obtained from Merck (Darmstadt, Germany). The bicinchoninic acid (BCA) assay reagent kit was obtained from Pierce (Rockford, IL). Formic acid (FA) was purchased from Riedel de Haen (Seelze, Germany). Tris (hydroxymethyl)aminomethane (Tris) was purchased from PlusOne (GE Healthcare, Orsay, France). C8 membrane was purchased from 3M Empore (St. Paul, MN). 3 μm and 5 μm C18-AQ beads were purchased from Dr. Maisch-GmbH (Ammerbuch, Germany). Synthetic peptides (95% purity) were purchased from Abomics Co. Ltd. (New Taipei City, Taiwan)
Cell Culture, Lysis and Tissue Collection
The human primary lung cancer cell lines: A549, CL100, CL141, CL152, CL25, CL83, CL97, H1975, H3255, PC9 and PC9/gef (Table S1) were obtained from Dr. Pan-Chyr Yang, National Taiwan University Hospital at Taipei, Taiwan and grown in RPMI 1640 medium. The cell cultures were supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic at 37°C in 5% CO2. The cells were lysed using a hypotonic buffer (10 mM HEPES, pH 7.5, 1.5 mM MgCl2, 10 mM KCl) with protease inhibitor cocktail (100:1, sample/protease inhibitor, v/v). The cells were homogenized using 50 strokes of a Dounce homogenizer.
Clinical tissue samples were obtained from National Taiwan University Hospital at Taipei, Taiwan in accordance with approved human subject guidelines authorized by Medical Ethics and Human Clinical Trial Committee at National Taiwan University Hospital. Following surgery, the tumor and adjacent normal tissues were collected in separate tubes, kept on dry ice for 30 mins. during transportation, and stored at -80 °C before further processing. Adjacent normal tissues were obtained from the distal edge of the resection ≥ 10 cm from the tumor. In this study, a total of 20 pairs of tumor and adjacent normal tissue were collected and analyzed from individual patients. All lung cancer patients had histologically been confirmed by pathologists. The detailed clinical information was shown in Table S2.
Membrane Protein Extraction and Digestion of NSCLC Cell Lines
The data of 11 NSCLC cell lines were taken from our previous study,24 and then re-analyzed in this work. Briefly, membrane proteins were isolated through a two-step centrifugation followed by gel-assisted digestion described previously.25 First, the nuclei and other heavy cell debris were separated by centrifugation at 3000 × g for 10 mins. at 4 °C. Then the supernatant was mixed with 1.8 M sucrose to a final concentration of 0.25 M and was centrifuged for 1 h. at 13000 × g at 4 °C to pellet out the remaining membrane proteins. The pellet was then washed with 1 mL of 0.1 M Na2CO3 (pH 11.5) for 1 h., and recovered through centrifugation at 13000 rpm for 1 h. at 4 °C. Membrane proteins were then suspended in the digestion buffer (6 M urea, 5 mM EDTA, 2% SDS and 0.1 M TEABC) and then sonicated at 4 °C for 15 mins. Disulfide bonds were cleaved through incubation with 5 mM TCEP at 37 °C for 30 mins. and alkylated with 2 mM MMTS at room temperature for 30 mins. The membrane proteins were then embedded into the polyacrylamide gel directly formed in the sample tube. The gel was cut into small pieces and then washed several times with 25 mM TEABC and 25 mM TEABC in 50% ACN, then further dehydrated by adding 100% ACN. Trypsin in 25 mM TEABC was incubated with the membrane proteins (protein:trypsin = 10:1 g/g) for 16 h. at 37 °C. Tryptic peptides were extracted from the gel by sequential washing with 25 mM TEABC, 0.1% TFA, 0.1% TFA in 50% ACN and 100% ACN. The amount of peptide produced was determined using the BCA protein assay.
Tissue Membrane Protein Extraction and Digestion
Frozen tissues were thawed rapidly at 37 °C, cut into small pieces, weighed and then washed by 0.9% NaCl to remove blood. The pre-cleaned tissues were homogenized in STM buffer solution (5mL/g tissue, 0.25 M sucrose, 10 mM Tris-HCl, and 1 mM MgCl2) with protease inhibitor mixture (100:1, sample/protease inhibitor, v/v) using mechanical homogenizer (Precellys®24, Bertin Technologies) in 2.0 mL standard tubes (STURDY TUBE®) containing ceramic zirconium oxide beads (5 beads of 2.8 mm diameter and 10 beads of 1.4 mm diameter). The tissue samples were homogenized at 6500 rpm three times for 15 seconds pausing for 5 mins. in between each homogenization steps, and then tissue debris was removed by centrifugation (260 × g) for 5 mins. at 4 °C. The supernatant was centrifuged at 1500 × g for 10 mins. at 4 °C to pellet the nucleus, and then the obtained supernatant was centrifuged again at 13000 rpm for 1 h. at 4 °C to precipitate the crude membrane pellet. The pellet was washed in 1 mL of 0.1 M Na2CO3 overnight at 4 °C and re-collected by centrifugation at 13000 rpm for 1 h. at 4 °C. Digestion of the tissue membrane samples was performed using the gel-assisted digestion method described above.
Hp-RP StageTip Fractionation
Hp-RP StageTips were prepared as described from the protocol of Rappsilber et al.26 Briefly, 1.25 mg of 5 μm C18-AQ beads suspended in 100 mM ammonium formate (NH4HCO2, pH 10) in 50% ACN were packed into the Gilson 200-μL tip with a C8 membrane frit by centrifugation at 1500 × g for 2 mins. After sufficient washing and conditioning, membrane peptides reconstituted in the loading solution (200 mM NH4HCO2, pH 10) were bound to the StageTips through centrifugation. The membrane peptides were eluted from the tip with increasing concentration of ACN to separate the peptides into 6 fractions. Detailed procedure of the Hp-RP StageTip fractionation was previously described.24
LC-MS/MS Analysis
Fractionated membrane peptides were analyzed using Synapt G1 High Definition Mass Spectrometer (HDMS, Waters Corp., UK) and TripleTOF 5600 System (AB SCIEX Concord, ON Canada). For LC-MS/MS analysis through Synapt G1 HDMS, the peptides reconstituted in buffer A (0.1% FA in H2O) were injected into a 2 cm × 180 μm capillary trap column and separated by 75 μm × 25 cm nanoACQUITY 1.7 μm BEH C18 column using nanoACQUITY Ultra Performance LCTM (Waters Corporation, Milford, MA, USA). For the analysis using TripleTOF 5600 System, peptide samples were injected into a 100 μm × 150 mm self-packed 3 μm C18-AQ column in a nanoACQUITY Ultra Performance LCTM. The bound peptides were eluted with a gradient of 0-80% buffer B (0.1% FA in ACN) for 120 mins., operated in ESI-positive V mode. The LC gradient for each Hp-RP StageTip fractions is previously described.24 Data acquisition for Synapt G1 HDMS was done by DDA mode to automatically switch between a full MS scan (400-1600 m/z, 0.6 s.) and six MS/MS scans (100-1990 m/z, 0.6 s. for each scan) on the six most intense ions present. Data from 5600 TripleTOF System were obtained through the same acquisition mode by selecting 15 most intense precursor peaks and performing 15 MS/MS (100-1800 m/z, 200 ms. for each scan) for each full MS scan (300-1600 m/z, 200 ms.). The mass spectrometry proteomics data have been deposited into the ProteomeXchange Consortium27 via the PRIDE partner repository with the dataset identifier PXD002224.
Database Search for Peptide and Protein Identification
The acquired MS/MS spectra were searched against UniProtKB/Swiss-Prot human database (2014_05 release, 20,264 entries) appended with reversed decoy sequences using Mascot28 (Matrix Science; version 2.3.02) with p-value < 0.05, and against UniProtKB/Swiss-Prot human database (2014_11 release, 20,193 entries) with reversed sequences added for target-decoy analysis using X!Tandem29, and Comet30 search engines. For database searches using X!Tandem and Comet, the .wiff raw files from 5600 TripleTOF system were first converted into mzML format by the MS Data Converter (AB SCIEX version 1.3 beta) using “centroid” option, and the resulting mzML files were further converted into mzXML format by the msconvert.exe from ProteoWizard (version 3.0.4462)31,32 package using default parameters. Raw files from Synapt G1 HDMS were converted directly into mzXML format using msconvert.exe. For database searching, a maximum of 2 missed cleavages were allowed for trypsin digestion with variable modifications of deamidation (Asn and Gln), oxidation (Met) and carbamidomethylation (Cys). In the Mascot search, a parent ion tolerance of 20.0 ppm and fragment ion tolerance of 0.1 Da for TripleTOF 5600 runs were used, and similar parameters were applied in both X!Tandem and Comet searches. For Synapt G1 HDMS runs 0.1 Da tolerance for both parent and fragment ion were used in the three search engines. Additional X!Tandem parameters include using top 160 peaks for matching, and default modifications: pyroglutamate from Gln and Glu and N-terminal acetylation. For Comet search, high resolution binning was used (0.02 tolerance and 0.0 offset).
To ensure high confidence in identification results, we further evaluated the FDR at PSM-, peptide- and protein-level. Search results from Mascot were imported into the PeptideShaker version 0.38.433 for peptide and protein inference. PSMs, distinct peptides and protein groups were validated at a 1% FDR estimated using the decoy hit distribution. Output files from X!Tandem and Comet were processed by PeptideProphet34 using the parameters “-OpdEAP -PPM”. The PepXML files from PeptideProphet were further combined using iProphet35 via the trans proteomic pipeline (TPP)36. The resulting protein and peptide identification lists were then determined by MAYU37 (version 1.07) based on 1% FDR. The parameters used for MAYU analysis are the following: “-I 2 -P pepFDR=0.01:t -G 0.1 -H 200 -PprotFeat –PmFDR”. Only proteins with protein FDR of 1% and with matched peptides passing the FDR threshold of 1% in both the peptide- and PSM-level, having at least one unique peptide were considered as identified. In addition, only peptides with at least 7 amino acid residues were accepted as confident identification.
Cellular Localization, Family/Domain, and Missing Protein Annotations
Membrane protein annotation was based on UniProtKB (http://www.uniprot.org/)38, neXtProt (http://www.nextprot.org/)39, and The Human Protein Atlas (HPA) (http://www.proteinatlas.org/)40. Missing protein annotation, protein existence evidence, and chromosome assignments were based on neXtProt (2014-09 release). TMH distribution was obtained from UniProtKB (2015_05 release). Family or domain annotation was performed by InterPro (v50.0)41. All the peptides were also cross-referenced to the peptide entries of the Peptide Atlas (http://www.peptideatlas.org/)42 repository (Human 2015-03).
MRM Method Development, Optimization and Acquisition
In order to further confirm the identification of some missing proteins, 14 synthetic peptide sequences of 11 missing proteins were purchased from Abomics Co. Ltd. (New Taipei City, Taiwan) with high purity of 95% and used to develop MRM method. The unique proteotypic peptides selected from the discovery mode used for the MRM fulfilled the following criteria: no cite susceptible to modification, no missed cleavage, and with the suitable length of 8 amino acids or more. The 14 peptides were divided into 5 groups (F1, F2, F3, F4 and F5/F6) based on the Hp-RP StageTip fractions in which they were detected in the DDA mode (Table S3). Five to eight most abundant MRM transitions for each peptide were selected having two or more unique ion signatures based from SRMCollider (v1.4).43 A total of 13 to 31 transitions were monitored for each Hp-RP StageTip fraction groups, with a maximum 10 transition analyzed simultaneously to allow a dwell time of 100 ms. or more.
All MRM acquisitions were performed using QTRAP5500 System (AB SCIEX Concord, ON Canada), and the peptide samples were injected in a 100 μm × 150 mm self-packed 3 μm C18-AQ column in a nanoACQUITY Ultra Performance LCTM. The same LC gradient optimized for each Hp-RP StageTip fraction as described previously24 were also used for the 14 synthetic peptides. The MS instrument was operated in positive mode with the following parameters: ion spray voltage of 2500 V, curtain gas at 25 psi, nebulizer gas at 20 psi, unit resolution (0.7 Da full-width-at-half-maximum) for both Q1 and Q3 quadrupoles, interface temperature at 150 °C, and scan mass range of m/z > 300 - 1250. The scheduled MRM was performed with 5-min retention time window and instrument cycle time of 1.5 s.
In MRM modes, collision energies (CE) and declustering potential (DP) were optimized. Using Skyline (version 3.1), the default CE used for the QTRAP 5500 instrument were calculated according to the formulas CE = 0.036 × (precursor m/z) + 8.857 and CE = 0.0544 × (precursor m/z) -2.4099, for doubly and triply charged precursor ions, respectively. For each parent ion, 11 different CE values (default CE ± 2V, 5 steps) were measured to obtain the optimized CE. The Skyline default DP for QTRAP 5500 calculated according to the formula DP = 0.0729 × (precursor m/z) + 31.117 and used for all peptides ranging from 62.2 V to 102.2 V. Further optimization did not generate noticeable increment in in the peak area. All MRM data analyses were performed using Skyline software.44
Results and Discussion
Large Scale Profiling of Membrane Proteome in Lung Cancer
In this study, lung cancer was chosen as the model for deep profiling of the membrane proteome. Lung cancer is one of the most frequently diagnosed type of cancer and the leading cause of cancer-related death worldwide.45 The overall survival rate of lung cancer is below 15% with over 85% of all cases comprising NSCLC.46 Although certain tyrosine kinase inhibitors (TKI) targeting epidermal growth factor receptor (EGFR) with constitutively activated mutation were developed and currently used in clinical trial, high percentage of the patients eventually develop resistance to treatment.47 Towards comprehensive profiling, 11 primary NSCLC cell lines and 20 pairs of tumor and adjacent normal tissue samples from NSCLC patients were used for in-depth membrane sub-proteome identification. These NSCLC cell lines harbor different EGFR mutation status and various sensitivity to TKIs, including CL141, CL152, CL83 and A549 cell lines with wild-type EGFR; PC9, PC9IR, CL25, and CL100 cell lines with exon 19 deletion of EGFR; H3255 cell line with a point mutation of L858R; and cell lines CL97 and H1975 with double EGFR point mutation of G719/T790M and L858R/T790M, respectively. Compared to cell lines, the tissue sample reflects a heterogeneous population of cell types possessing distinct molecular phenotype with tissue-specific function and protein expression in vivo. The tissue samples collected for this study also vary in EGFR mutation status consisting of wild type (n = 12 pairs), exon 19 deletion (n = 2 pairs), and a point mutation of L858R (n = 6 pairs), and were obtained from patients in different lung cancer stages (10 patients in the early stage-1, 4 patients in stage-2, 5 patients in stage-3, and one with an unknown stage).
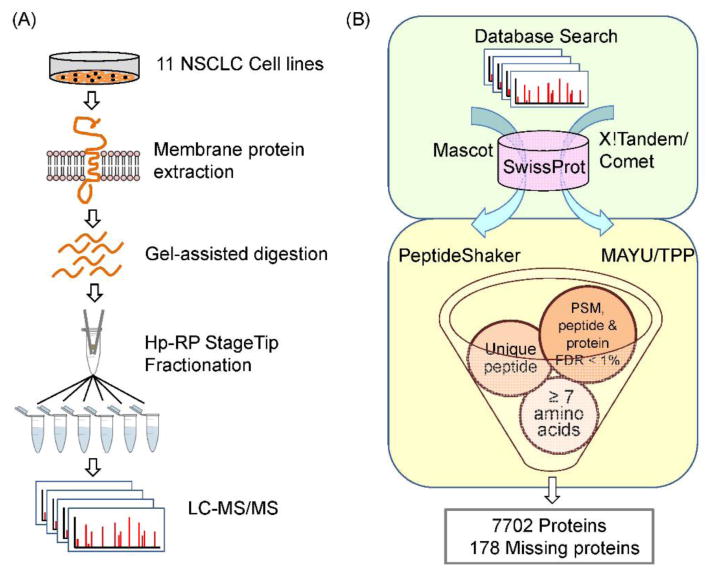
As illustrated in Figure 1A, membrane proteins extracted from the 11 NSCLC cell lines were digested using the gel-assisted digestion method, which allowed better solubilization of the hydrophobic proteins through the use of detergent and easy removal of the detergent before MS/MS analysis.25 To increase the coverage of membrane proteins, we utilized the C18-based Hp-RP StageTip pre-fractionation for sensitive and efficient separation of the hydrophobic peptides.24 The Hp-RP StageTip fractions were then analyzed using Q-TOF MS instruments performing duplicate runs for each fraction. Due to the limited amount of clinical tissue samples available and further reduction after membrane protein enrichment, one shot LC-MS/MS analysis was performed for the tissue membrane samples without Hp-RP StageTip fractionation of the peptides.
Figure 1. Workflow for missing protein identification.
A) Analytical workflow for increased coverage of membrane proteins. B) Database searching and bioinformatics screening for high confident identification.
Shotgun proteomics heavily relies on statistical computation which can result in protein inference problems, particularly in large-scale proteomic experiments.48 In order to alleviate this problem, the use of multiple search engines has been found to increase confidence and provide better identification coverage.49 In accordance with this, protein identification was achieved by combining results from three commonly used search engines: X!Tandem,29 Comet,30,50 and Mascot50 software. The FDR at the PSM-, peptide- and protein-level in this reported large-scale data set were evaluated through MAYU37 for search results obtained from X!Tandem and Comet, while PeptideShaker was used for the Mascot output (Figure 1B). Strict filtering criteria were implemented to ensure the high confidence of the identification results. The identified proteins are all within the FDR threshold of 1% in the PSM-, peptide-, and protein-level to minimize the occurrence of false positive identification. Furthermore, all the identified proteins are inferred from peptides with 7 or more amino acid residues consisting of at least one uniquely matched to the particular protein sequence. This was achieved through peptide screening using our in-house database of unique tryptic peptides derived from in silico tryptic digestion of the whole human proteome. In this overall workflow, we aim to achieve sensitive and comprehensive proteome profiling through our analytical techniques, while the deep bioinformatics analyses provided high confident identification of the lung cancer proteome and missing proteins. For NSCLC cell lines, about 1500 - 4500 proteins were identified from each cell line, accounting a total of 6820 proteins (Table S4 and S5). For each tissue membrane sample, about 800 - 2300 proteins were identified comprising a total of 4406 proteins (Table S4 and S5).
Mining Missing Proteins from Identified Membrane Proteome of Lung Cancer Samples
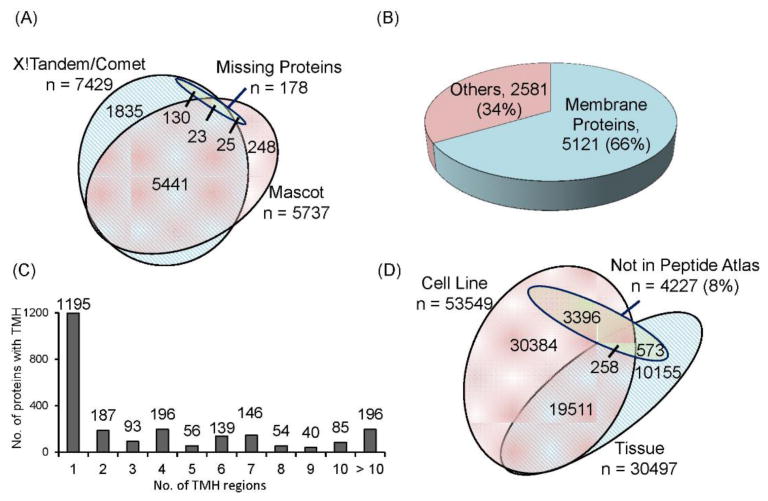
From the combined MS analysis result of 11 lung cancer cell lines and 20 pairs of tumor and adjacent normal tissue samples, a total 64277 non-redundant peptides corresponding to 7702 proteins with 1% FDR at the PSM-, peptide- and protein-level were identified (Table S6). Integrating the search results from Mascot and X!Tandem/Comet search engines, 5464 of all 7702 proteins (71%) were commonly identified, indicating high confidence of these proteins (Figure 2A). Membrane protein annotation was performed by using three databases: UniProtKB, HPA and neXtProt, wherein proteins were considered membrane protein if found in at least one of the databases. Based on these databases, 5121 (66%) out the 7702 proteins were annotated to be membrane proteins (Figure 2B). We further analyzed the structural features of these annotated membrane proteins, wherein 2387 were found to contain TMH domains (Figure 2C) with almost half found to possess multipass TMH (consisting of 2 or more TMH domains) (Table S7). The result revealed that our strategy provides high efficiency for increasing the recovery of the hydrophobic peptides and enhancing the identification of membrane proteins. The detailed evaluation of higher recovery of Hp-RP StaheTip for hydrophobic peptide of membrane proteins in comparison with the commonly employed pre-fractionation techniques, including strong-cation exchange (SCX) and strong-anion exchange (SAX) StageTip was demonstrated using HeLa cell lines.24
Figure 2. The 7702 overall proteins and 178 missing proteins identified with PSM-, peptide-, and protein-level FDR of 1%.
(A) Comparison of proteins identified through Mascot and X!Tandem/Comet search engines. The 178 missing proteins were also indicated. (B) 5121 membrane proteins were annotated from the overall identification. (C) Almost 50% of the 2387 transmembrane proteins have multiple TMH regions. (D) 4227 identified peptides were not yet observed in the Peptide Atlas.
Comparison with the Peptide Atlas repository42 (Human 2015-03, 1025698 distinct peptides) revealed 60050 (92%) identified peptides in this study were already present in the Peptide Atlas from the collective data of different sample types. On the other hand, our dataset provided previously un-reported mass spectral evidences for 4227 additional peptides from 2791 protein groups. Among these proteins, 1917 were annotated as membrane proteins, from which 66% were found to consist of TMH domains, with the highest consisting of up to 36 TMH regions (Figure 2D and Table S8). Among the 214 unique peptides of the 178 missing proteins, it was noted that 19 peptides corresponding to 9 missing proteins have been found to be deposited in the Peptide Atlas. Among the 9 missing proteins, 3 have additional unique peptides (Table S9).
Among these confident proteins identified in lung cancer cell lines and tissue samples, 178 proteins were annotated to be missing proteins by neXtProt (09-2014 release). In addition to 12 missing proteins commonly identified in both samples, 144 missing proteins were only found in the cell lines and 21 only identified in the tissue samples (Figure S1A). Among the 178 missing proteins, 52 were identified with multiple PSMs for unique peptides with the highest of 31 matched spectra, and 28 missing proteins were found in multiple cell lines or tissue specimens (Table 1 and see details in Table S9). The identification of single PSM for the remaining missing proteins still revealed the challenges of multi-peptide identification of missing proteins by the current technology.
Table 1.
The 52 missing proteins with multiple identified unique peptides, PSMs (only from unique peptides) or cell line/tissue sources.
| Protein Accession # | Gene Name | Chromosome | Evidence | Membrane (TMH) | No. of unique peptides | No. of Cell line/tissue | No. of PSMs |
|---|---|---|---|---|---|---|---|
| Q5T601 | GPR110 | 6 | PE2 | Yes (7TMH) | 4 | 3 | 12 |
| O60330 | PCDHGA12 | 5 | PE2 | Yes (1TMH) | 3 | 2 | 7 |
| Q14773 | ICAM4 | 19 | PE2 | Yes (1TMH) | 3 | 7 | 23 |
| Q8N0V5 | GCNT2 | 6 | PE2 | Yes (1TMH) | 3 | 2 | 8 |
| Q9P2D7 | DNAH1 | 3 | PE2 | No | 3 | 6 | 10 |
| Q9UMS5 | PHTF1 | 1 | PE2 | Yes | 3 | 4 | 11 |
| A6NE01 | FAM186A | 12 | PE2 | No | 2 | 4 | 5 |
| Q5T2N8 | ATAD3C | 1 | PE2 | No | 2 | 2 | 3 |
| Q6UWH6 | TEX261 | 2 | PE2 | Yes (5TMH) | 2 | 3 | 22 |
| Q8N5D6 | GBGT1 | 9 | PE2 | Yes (1TMH) | 2 | 1 | 7 |
| Q9Y5E3 | PCDHB6 | 5 | PE2 | Yes (1TMH) | 2 | 1 | 5 |
| A6NIZ1 | - | 5 | PE2 | Yes | 1 | 2 | 2 |
| P09131 | SLC10A3 | X | PE2 | Yes (8TMH) | 1 | 3 | 5 |
| P25089 | FPR3 | 19 | PE2 | Yes (7TMH) | 1 | 1 | 2 |
| P59826 | BPIFB3 | 20 | PE2 | No | 1 | 4 | 4 |
| Q08AG5 | ZNF844 | 19 | PE2 | No | 1 | 2 | 2 |
| Q13360 | ZNF177 | 19 | PE2 | No | 1 | 1 | 2 |
| Q14588 | ZNF234 | 19 | PE2 | No | 1 | 3 | 6 |
| Q53R12 | TM4SF20 | 2 | PE2 | Yes (4TMH) | 1 | 1 | 3 |
| Q5JT25 | RAB41 | X | PE2 | No | 1 | 5 | 7 |
| Q69YG0 | TMEM42 | 3 | PE2 | Yes (4TMH) | 1 | 1 | 3 |
| Q6IF42 | OR2A2 | 7 | PE2 | Yes (7TMH) | 1 | 1 | 5 |
| Q6NVV3 | NIPAL1 | 4 | PE2 | Yes (9TMH) | 1 | 1 | 4 |
| Q6PGQ1 | DRICH1 | 22 | PE2 | No | 1 | 1 | 2 |
| Q6ZVZ8 | ASB18 | 2 | PE2 | Yes | 1 | 4 | 7 |
| Q7Z5H4 | VN1R5 | 1 | PE2 | Yes (7TMH) | 1 | 4 | 6 |
| Q8IZC6 | COL27A1 | 9 | PE2 | No | 1 | 2 | 2 |
| Q8IZD6 | SLC22A15 | 1 | PE2 | Yes (12TMH) | 1 | 3 | 7 |
| Q8N239 | KLHL34 | X | PE2 | No | 1 | 5 | 31 |
| Q8N8Q1 | CYB561D1 | 1 | PE2 | Yes | 1 | 2 | 3 |
| Q8TDW4 | ST7L | 1 | PE2 | Yes (2TMH) | 1 | 2 | 4 |
| Q96JL9 | ZNF333 | 19 | PE2 | Yes | 1 | 3 | 4 |
| Q96SR6 | ZNF382 | 19 | PE2 | No | 1 | 1 | 5 |
| Q9BWX1 | PHF7 | 3 | PE2 | No | 1 | 3 | 6 |
| Q9NUH8 | TMEM14B | 6 | PE2 | Yes (4TMH) | 1 | 6 | 27 |
| Q9NZP0 | OR6C3 | 12 | PE2 | Yes (7TMH) | 1 | 1 | 2 |
| Q9Y5R2 | MMP24 | 20 | PE2 | Yes (1TMH) | 1 | 1 | 2 |
| H7BZ55 | - | 2 | PE5 | No | 14 | 2 | 25 |
| B5MCN3 | SEC14L6 | 22 | PE5 | Yes | 2 | 9 | 11 |
| P0C7P4 | UQCRFS1P1 | 22 | PE5 | Yes | 2 | 10 | 16 |
| Q58FF7 | HSP90AB3P | 4 | PE5 | No | 2 | 3 | 4 |
| Q58FG1 | HSP90AA4P | 4 | PE5 | No | 2 | 3 | 6 |
| Q8N7Z5 | ANKRD31 | 5 | PE5 | No | 2 | 13 | 20 |
| P54792 | DVL1P1 | 22 | PE5 | No | 1 | 1 | 2 |
| Q5JNZ5 | RPS26P11 | X | PE5 | No | 1 | 1 | 2 |
| Q6ZS52 | - | 6 | PE5 | No | 1 | 2 | 2 |
| Q86SG4 | HMGN2P46 | 15 | PE5 | No | 1 | 2 | 4 |
| Q8TAF5 | FLVCR1-AS1 | 1 | PE5 | No | 1 | 4 | 9 |
| Q92928 | RAB1C | 9 | PE5 | Yes | 1 | 2 | 2 |
| Q9BYX7 | POTEKP | 2 | PE5 | No | 1 | 4 | 6 |
| Q9BZK3 | NACAP1 | 8 | PE5 | No | 1 | 5 | 7 |
| Q9UKY3 | CES1P1 | 16 | PE5 | No | 1 | 17 | 19 |
The existence of mRNA level expression is indicative of the high probability that the corresponding genes are coding for the proteins. As expected, 77% (139) of the identified missing proteins already have transcript level evidence (PE2) while fewer numbers were identified in other categories of protein existence evidence level: 5 missing proteins were among those inferred from homologous species (PE3) and 3 were predicted from gene models (PE4). Moreover, 31 missing proteins belonging to the “uncertain” (PE5) evidence level were also identified with some detected in multiple tissue or cell line samples, or inferred from multiple spectra (Table 1 and Figure S1B). For example, H7BZ55 (no given gene name) belongs to the PE5 category, however, the identification of as many as 14 unique peptides from two tissue and cell line sources (tissue # 162 and H3255) show confidence evidence of its expression in lung cancer samples. UQCRFS1P1 (P0C7P4), annotated as a membrane protein, was also confidently identified with two unique peptides in 4 cell lines (CL141, CL97, PC9, and Cl52) and 6 tissue samples (tissue # 171, 184, 262, 269, 275, and 299). We also performed manual inspection of the 17 missing protein in the PE5 category with single PSMs. Based on the criteria of continues y-and b-fragment ions and high signal-to-noise ratio, 10 proteins have medium to high quality spectra (Supplementary Figure S2). Two proteins, PPP1R2P9 and Q8NDZ9 (no given name), were removed due to bad quality of MS/MS spectra.
The single amino acid variations (SAAVs) have been found to link with in disease progression.51 The presence of SAAVs also presents challenge for unambiguous peptide identification and protein inference for large-scale high throughput mass spectrometry datasets. In order to check the possibility of mis-identification of a protein despite a correct peptide identification,52,53 all the 216 unique peptides of the missing proteins were analyzed against in silico trypsin digested human proteome (UniProtKB/Swiss-Prot human, 2014-09, 20,188 sequences) for a match upon I/L, N/D or Q/K amino acids substitutions. Upon I/L substitution, our analysis revealed that the ILVAIMK peptide annotated for OR1M1 may likely match to the tryptic peptide LIVALMK from Annexin A5 (ANXA5). In addition, we also found TUBA4B protein identified by a peptide QIFHPEQLITGK, which also likely maps to other tubulin family member (alpha -1B/4A/3C/3E/1A/8//1C) with a peptide QLFHPEQLITGK. These two peptides of OR1M1 and UBA4B were removed from the total list of the peptides of the missing proteins. However, TUBA4B protein was retained since it was identified by additional unique peptide resulting in overall confident identification of 214 unique peptides corresponding to the 178 missing proteins.
Annotation of Membrane Protein and Family/Domain of the Missing Proteins
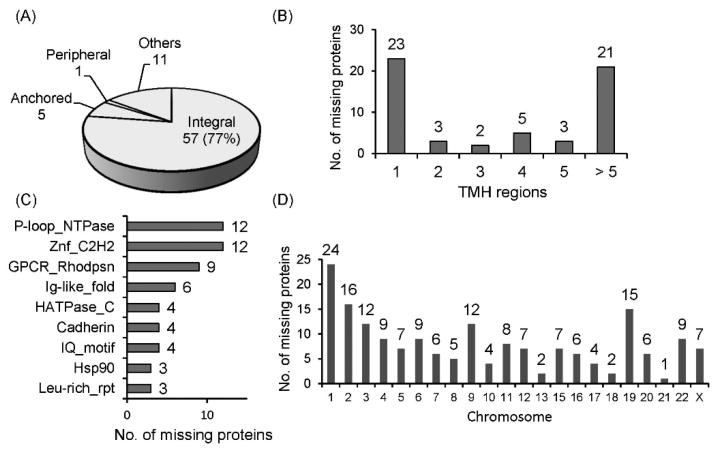
We further looked into the structural features and potential function of these missing proteins. Out of 178 missing proteins reported in this study, 74 were annotated to be membrane proteins, among which 57 (77%) consist of TMH domain (Figure 3A). Furthermore, 34 (60%) of the transmembrane missing proteins contains 2 or more TMH domains, which are usually difficult to be detected due to the highly hydrophobic regions encompassing the membrane (Figure 3B). In order to further decipher the potential function of these missing proteins, information from the InterPro database (v50.0)41 was extracted to annotate the domain and protein family. As shown in Figure 3C, many missing proteins belong to protein families such as the P-loop containing NTPase family, Znf C2H2 domains, multipass transmembrane GPCR, and Ig-like proteins. These families possessed several key biological functions including nucleotide binding, receptors, gene transcription and translation as well as cell adhesion. Further functional studies of the identified missing proteins may elucidate important functions. In particular, 9 new GPCRs were identified, which are among the membrane protein class with 7 TMH domains and have been known to mediate many crucial cellular responses, and thus possess important therapeutic target characteristics.54 The missing proteins were also found to be distributed in all chromosomes except in chromosome Y, with the highest number of 24 proteins in chromosome 1 (Figure 3D and Table S9).
Figure 3. Membrane, TMH, function or domain, and chromosome annotation of the identified missing proteins.
(A) The proportion of the different membrane proteins types for 74 missing membrane proteins, showing 77% as integral membrane proteins. (B) The number of TMH domains in each of the 57 missing integral membrane proteins with 60% having multiple TMH regions. (C) Protein function or domain annotation. (D) Chromosome distribution of the 178 missing proteins. P-loop_NTPase = P-loop containing nucleoside triphosphate hydrolase, Znf_C2H2 = Zinc finger C2H2 domains, GPCR_Rhodpsn = G protein-coupled receptors rhodopsin-like, Ig-like_fold = Immunoglobulin-like fold, HATPase_C = Histidine kinase-like ATPase C-terminal domain, Hsp90 = Heat shock protein 90, Leu-rich_rpt = Leucine-rich repeats.
Validation of Missing Proteins by MRM
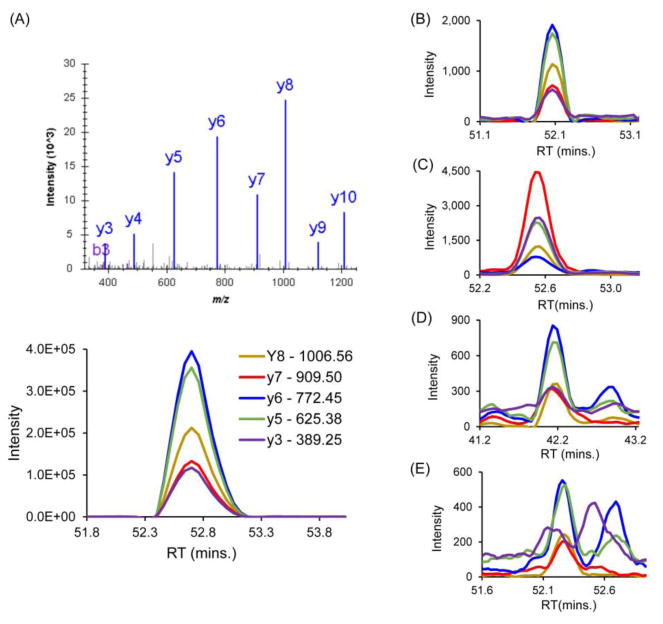
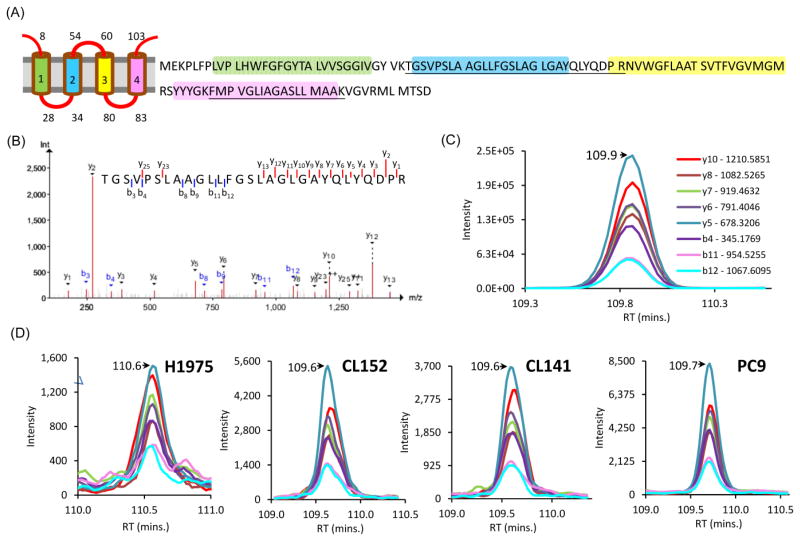
In order to confirm the protein expression of some missing proteins, we selected 11 missing proteins for validation by MRM MS including 8 annotated as membrane proteins: SLC10A3 (8TMH), GPR110 (7TMH), TEX261 (5TMH), TM4SF20 (4TMH), TMEM14B (4TMH), ST7L (2TMH), GCNT2 (1TMH), and PCDHB6 (1TMH) and 3 consisting of unique peptides suitable for MRM acquisition (BPIFB3, CACTIN-AS1, and TTC16) (Table S3). A total of 14 unique peptides from these proteins were used for validating their protein-level expression in different NSCLC cell lines. All the acquired MRM peaks from the membrane-enriched samples were verified by using peak features like peak shape, signal-to-noise ratio, relative fragment intensities and co-elution of fragment ions.17,55 MRM peaks and DDA spectra of synthetic peptides served as a reference to compare the similarities of the peak patterns. Figure 4 illustrates the criteria used to select and confirm that the sample MRM peaks were correctly matched to the reference peaks. Fragment ions in each synthetic reference peptides were chosen from the acquired DDA and MRM spectra (Figure 4A). By comparison of the acquired MRM peaks with the reference MRM, Figure 4B shows one example that passed the criteria for a confirmed validation; the sample spectra showed similar relative intensities, peak shape, co-elution and retention time as compared to the MRM peak pattern from the reference peptide. However, sample MRM peaks in Figure 4C differ in relative fragment ion intensities particularly for y7 ion. Figure 4D shows another example of false MRM peak due to difference in the retention time. The peaks for the fragment ions in Figure 4E are not co-eluting and has low signal-to-noise ratio, which are also indicative of a poor match with the reference MRM spectra, and cannot be used for validation of the missing protein expression. Using this approach, 10 peptides corresponding to 8 missing proteins were validated, among which 7 are transmembrane proteins. As shown in Table 2, most of these missing proteins were validated in multiple cell lines providing numerous MRM assays for proof of existence (see detail in Figure S3). The DDA identification result of the 10 validated peptides were also provided in supplementary figure (Figure S4). For example, TMEM14B is a small missing membrane protein composed of 114 amino acid residues with predicted 4 TMH domains (Figure 5A). It is noted that 74% of the whole protein sequence are predicted to have TMH. Two peptides, 88FMPVGLIAGASLLMAAK105 (845.480 Da, 2+) and 33TGSVPSLAAGLLFGSLAGLGAYQLYQDPR61 (974.085, 3+) of TMEM14B were identified in six types of NSCLC cell lines by the DDA method. Figure 5B shows the representative MS/MS spectra of the peptide sequence unique to TMEM14B (Figure 5B). The two peptides identified were found to be both located in the TMH region of the proteins, with one uniquely matched to the protein. By the comparison with the reference MRM profiles from synthetic peptide (Figure 5C), the MRM validation in Hp-RP StageTip fractionated peptides from H1975, CL152, CL100, and PC9 cell lines further confirmed the expression of the missing protein at the protein-level (Figure 5D). Without pre- Hp-RP StageTip fractionation, TMEM14B was still validated in membrane digest samples although the intensities of the obtained peaks were relatively lower compared to that of the fractionated samples (Figure S5). Previous studies have shown that identification of low-molecular weight proteins pose many challenges due to the low number of observable peptides for MS/MS detection.11,15 Taking into account the added challenges of recovering hydrophobic peptides, our approach demonstrated the capability to identify and validate even small hydrophobic membrane proteins.
Figure 4. Representative MRM spectra showing the peak feature criteria to validate the expression of missing proteins.
(A) Reference MS/MS spectrum in DDA mode (upper) and MRM spectra of different transitions from the synthetic peptide. In comparison with the reference MRM peaks, sample peak (B) has similar peak features confirming validation. The relative intensities of the co-eluting fragment ions and the retention time are the same as in the reference peak. Sample peak (C) has different relative intensities of fragments compared to the reference peak, sample peak (D) widely varies in retention time relative to the reference peak, while sample peak (E) has low signal-to-noise ratio with fragment peaks not co-eluting. Sample peaks (C), (D), and (E) represent MRM results that does not match with the reference peptide spectrum, and therefore cannot be used for confirming the existence of the missing protein.
Table 2.
Summary of 10 peptides corresponding to 8 missing proteins validated with MRM approach in lung cancer cell lines membrane sample.
| Gene Name | Membrane(TMH) | Peptide Sequence (z) | Cell line where validated |
|---|---|---|---|
| SLC10A3 | Yes (8TMH) | VTSLDTEVLTIK (+2) | CL152, CL141, PC9, H3255, CL100 |
| TMEM14B | Yes(4TMH) | TGSVPSLAAGLLFGSLAGLGAYQLYQDPR (+3) | CL152, CL141, PC9, H3255, H1975, CL100 |
| TEX261 | Yes(5TMH) | LGILVVFSFIK (+2) | CL152, CL141, PC9, H3255, H1975, CL100 |
| GCNT2 | Yes (1TMH) | YVHQELLNHK (+3) | CL152, CL141, PC9, H3255 |
| TTC16 | no | LQEFDGAVEDFLK (+2) | CL152, CL100, PC9 |
| PCDHB6 | Yes (1TMH) | SLDYEALQSFEFR (+2) | CL152, CL100, PC9, CL141 |
| YTISSNPHFHVLTR (+3) | CL152, PC9, CL141 | ||
| GPR110 | Yes (7TMH) | VLIGSDQFQR (+2) | H3255, CL152 |
| IQGFESVQVTQFR (+2) | H3255 | ||
| ST7L | Yes (2TMH) | GLSTAEINAVEAIHR (+3) | CL100, PC9 |
Figure 5. Mass spectral evidences from DDA discovery and MRM Validation of Transmembrane protein 14B (TMEM14B).
(A) TMEM14B composed of 114 amino acid residues possesses 4 TMH domains. The two peptides identified (underlined) from this protein are located in the TMH region. (B) DDA spectral evidence of the identified unique peptide (TGSVPSLAAGLLFGSLAGLGAYQLYQDPR). (C) Reference MRM peak from the synthetic peptide. (D) MRM peaks showing similar peak features as the reference peak for the peptide found in H1975, CL152, CL100, and PC9.
Conclusion
Membrane sub-proteome analysis has been recognized to be useful for drug discovery in clinical applications, although the challenges in membrane protein solubilization and peptide fractionation techniques still require further improvement. Through efficient peptide pre-fractionation of membrane samples from lung cancer cell lines and human tissue specimens, this sub-proteome has been shown by this study to be a good mining resource for many missing proteins. Deep bioinformatics analysis of mass spectral data for identification, FDR estimation and unique peptide filtering provided highly confident evidences of translation of missing proteins. Targeted MRM-based approach has been applied to confirm the expression of 8 of 11 selected missing proteins. For biological prospective, the evidences for the presence of missing membrane proteins in lung cancer may suggest their potential function in lung tumorigenesis. On the technical front, we expect that combining efficient membrane proteomic profiling, multiple search engines and FDR estimations followed by MRM validation could be a feasible approach to identify missing membrane proteins. Application of this platform to other disease types may facilitate the search of the remaining missing proteins.
Supplementary Material
Figure S1. Comparison of highly-confident missing proteins identified between the lung cancer cell line and lung tissue samples (with PSM-, peptide-, and protein-level FDR of 1%) and protein existence evidence of the identified missing proteins.
Figure S2. Sample DDA spectra of PE5 proteins with single hit spectrum.
Figure S3. Sample MRM spectral evidences of the 10 validated peptides corresponding to the 8 validated missing proteins.
Figure S4. Sample DDA spectra for the 8 validated missing proteins.
Figure S5. Comparison of validation results between Hp-RP StageTip fractionated peptides and peptides without fractionation prior to MRM analysis.
Table S2. Clinical information of the lung tissue samples.
Table S3. List of missing proteins selected for MRM validation.
Table S4. List of all identified proteins searched against Swiss-Prot (2014_06) appended with decoy through Mascot (version 2.3.02) and processed by PeptideShaker.
Table S5. List of all identified proteins searched against Swiss-Prot (2014_11) appended with decoy through X!Tandem and Comet, and processed by PeptideProphet, iProphet, and MAYU.
Table S6. Overall 7702 proteins Identified with PSM, peptide and protein FDR = 1%, having at least one unique peptide and peptides with > 6 amino acid length.
Table S7. Information about the membrane annotation, HPA reliability index and number of TMH regions for the overall 7702 proteins.
Table S8. Comparison of the identified peptides with the Peptide Atlas repository. The frequency of observation for each peptide was also specified.
Table S9. The 178 missing proteins identified with PSM, peptide and protein FDR = 1%, having at least one unique peptide and peptides with > 6 amino acid length. This material is available free of charge via the Internet at http://pubs.acs.org
Acknowledgments
This work was supported by Academia Sinica and the Ministry of Science and Technology in Taiwan, and the Department of Science and Technology (DOST) in the Philippines. Dr. Alexey I. Nesvizhskii and Mr. Chih-Chang Tsou were supported by US National Institutes of Health grant 5R01GM94231 (to A.I.N.).
Abbreviations
- Hp-RP
High-pH Reverse phase chromatography
- StageTip
stop-and-go extraction tip
- NSCLC
non-small cell lung cancer
- MRM
multiple reaction monitoring
- EGFR
epidermal growth factor receptor
- TKI
tyrosine kinase inhibitors
- TMH
transmembrane helices
- PSM
peptide-spectrum match
- FDR
false discovery rate
- HPA
Human Protein Atlas
- TPP
Trans Proteomic Pipeline
- DDA
Data Dependent Acquisition
References
- 1.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LD, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TS, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 3.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue-based map of the human proteome. Science (New York, N Y) 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 4.Gaudet P, Michel PA, Zahn-Zabal M, Cusin I, Duek PD, Evalet O, Gateau A, Gleizes A, Pereira M, Teixeira D, Zhang Y, Lane L, Bairoch A. The neXtProt knowledgebase on human proteins: current status. Nucleic acids research. 2015;43:D764–770. doi: 10.1093/nar/gku1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane L, Bairoch A, Beavis RC, Deutsch EW, Gaudet P, Lundberg E, Omenn GS. Metrics for the human proteome project 2013–2014 and strategies for finding missing proteins. J Proteome Res. 2014;13:15–20. doi: 10.1021/pr401144x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guruceaga E, Sanchez Del Pino MM, Corrales FJ, Segura V. Prediction of a missing protein expression map in the context of the human proteome project. J Proteome Res. 2015;14:1350–1360. doi: 10.1021/pr500850u. [DOI] [PubMed] [Google Scholar]
- 7.Martins-de-Souza D, Carvalho PC, Schmitt A, Junqueira M, Nogueira FC, Turck CW, Domont GB. Deciphering the human brain proteome: characterization of the anterior temporal lobe and corpus callosum as part of the Chromosome 15-centric Human Proteome Project. J Proteome Res. 2014;13:147–157. doi: 10.1021/pr4009157. [DOI] [PubMed] [Google Scholar]
- 8.Ahn JM, Kim MS, Kim YI, Jeong SK, Lee HJ, Lee SH, Paik YK, Pandey A, Cho JY. Proteogenomic analysis of human chromosome 9-encoded genes from human samples and lung cancer tissues. J Proteome Res. 2014;13:137–146. doi: 10.1021/pr400792p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, Tomonaga T. Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the Chromosome-centric Human Proteome Project. J Proteome Res. 2013;12:2414–2421. doi: 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C, Li N, Zhai L, Xu S, Liu X, Cui Y, Ma J, Han M, Jiang J, Yang C, Fan F, Li L, Qin P, Yu Q, Chang C, Su N, Zheng J, Zhang T, Wen B, Zhou R, Lin L, Lin Z, Zhou B, Zhang Y, Yan G, Liu Y, Yang P, Guo K, Gu W, Chen Y, Zhang G, He QY, Wu S, Wang T, Shen H, Wang Q, Zhu Y, He F, Xu P. Systematic analysis of missing proteins provides clues to help define all of the protein-coding genes on human chromosome 1. J Proteome Res. 2014;13:114–125. doi: 10.1021/pr400900j. [DOI] [PubMed] [Google Scholar]
- 11.Landry CR, Zhong X, Nielly-Thibault L, Roucou X. Found in translation: functions and evolution of a recently discovered alternative proteome. Current opinion in structural biology. 2015;32:74–80. doi: 10.1016/j.sbi.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Chang C, Li L, Zhang C, Wu S, Guo K, Zi J, Chen Z, Jiang J, Ma J, Yu Q, Fan F, Qin P, Han M, Su N, Chen T, Wang K, Zhai L, Zhang T, Ying W, Xu Z, Zhang Y, Liu Y, Liu X, Zhong F, Shen H, Wang Q, Hou G, Zhao H, Li G, Liu S, Gu W, Wang G, Wang T, Zhang G, Qian X, Li N, He QY, Lin L, Yang P, Zhu Y, He F, Xu P. Systematic analyses of the transcriptome, translatome, and proteome provide a global view and potential strategy for the C-HPP. J Proteome Res. 2014;13:38–49. doi: 10.1021/pr4009018. [DOI] [PubMed] [Google Scholar]
- 13.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Molecular systems biology. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichacker LA, Granvogl B, Mirus O, Müller BC, Miess C, Schleiff E. Hiding behind hydrophobicity: transmembrane segments in mass spectrometry. J Biol Chem. 2004;279:50915–50922. doi: 10.1074/jbc.M405875200. [DOI] [PubMed] [Google Scholar]
- 15.Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, Vazquez J, Valencia A, Tress ML. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Human molecular genetics. 2014;23:5866–5878. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muraoka S, Kume H, Adachi J, Shiromizu T, Watanabe S, Masuda T, Ishihama Y, Tomonaga T. In-depth membrane proteomic study of breast cancer tissues for the generation of a chromosome-based protein list. J Proteome Res. 2013;12:208–213. doi: 10.1021/pr300824m. [DOI] [PubMed] [Google Scholar]
- 17.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki M, Ishihama Y. Challenges facing complete human proteome analysis. Chromatography. 2014;35:73–80. [Google Scholar]
- 19.Di Palma S, Hennrich ML, Heck AJR, Mohammed S. Recent advances in peptide separation by multidimensional liquid chromatography for proteome analysis. J Proteomics. 2012;75:3791–3813. doi: 10.1016/j.jprot.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 20.Chen EI, Hewel J, Felding-Habermann B, Yates JR., 3rd Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT) Molecular & cellular proteomics : MCP. 2006;5:53–56. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Paik YK, Omenn GS, Uhlen M, Hanash S, Marko-Varga G, Aebersold R, Bairoch A, Yamamoto T, Legrain P, Lee HJ, Na K, Jeong SK, He F, Binz PA, Nishimura T, Keown P, Baker MS, Yoo JS, Garin J, Archakov A, Bergeron J, Salekdeh GH, Hancock WS. Standard guidelines for the chromosome-centric human proteome project. J Proteome Res. 2012;11:2005–2013. doi: 10.1021/pr200824a. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Liu X, Zheng W, Zhang L, Yao J, Yang P. Screening of missing proteins in the human liver proteome by improved MRM-approach-based targeted proteomics. J Proteome Res. 2014;13:1969–1978. doi: 10.1021/pr4010986. [DOI] [PubMed] [Google Scholar]
- 23.Segura V, Medina-Aunon JA, Mora MI, Martinez-Bartolome S, Abian J, Aloria K, Antunez O, Arizmendi JM, Azkargorta M, Barcelo-Batllori S, Beaskoetxea J, Bech-Serra JJ, Blanco F, Monteiro MB, Caceres D, Canals F, Carrascal M, Casal JI, Clemente F, Colome N, Dasilva N, Diaz P, Elortza F, Fernandez-Puente P, Fuentes M, Gallardo O, Gharbi SI, Gil C, Gonzalez-Tejedo C, Hernaez ML, Lombardia M, Lopez-Lucendo M, Marcilla M, Mato JM, Mendes M, Oliveira E, Orera I, Pascual-Montano A, Prieto G, Ruiz-Romero C, Sanchez del Pino MM, Tabas-Madrid D, Valero ML, Vialas V, Villanueva J, Albar JP, Corrales FJ. Surfing transcriptomic landscapes. A step beyond the annotation of chromosome 16 proteome. J Proteome Res. 2014;13:158–172. doi: 10.1021/pr400721r. [DOI] [PubMed] [Google Scholar]
- 24.Dimayacyac-Esleta BRT, Tsai CF, Kitata RB, Lin PY, Choong WK, Weng SH, Yang PC, Arco SD, Sung TY, Chen YJ. Rapid high-pH reverse phase StageTip for sensitive small-scale membrane proteomic profiling. Manuscript Submitted. doi: 10.1021/acs.analchem.5b03639. [DOI] [PubMed] [Google Scholar]
- 25.Han CL, Chien CW, Chen WC, Chen YR, Wu CP, Li H, Chen YJ. A multiplexed quantitative strategy for membrane proteomics: opportunities for mining therapeutic targets for autosomal dominant polycystic kidney disease. Molecular & cellular proteomics : MCP. 2008;7:1983–1997. doi: 10.1074/mcp.M800068-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 27.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Elecrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics (Oxford, England) 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 30.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 31.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak MY, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics (Oxford, England) 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaudel M, Burkhart JM, Zahedi RP, Oveland E, Berven FS, Sickmann A, Martens L, Barsnes H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat Biotechnol. 2015;33:22–24. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]
- 34.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 35.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Molecular & cellular proteomics : MCP. 2011;10:M111. doi: 10.1074/mcp.M111.007690. 007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Molecular & cellular proteomics : MCP. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Consortium U. The Universal Protein Resource (UniProt) Nucleic acids research. 2007;35:D193–197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lane L, Argoud-Puy G, Britan A, Cusin I, Duek PD, Evalet O, Gateau A, Gaudet P, Gleizes A, Masselot A, Zwahlen C, Bairoch A. neXtProt: a knowledge platform for human proteins. Nucleic acids research. 2012;40:D76–D83. doi: 10.1093/nar/gkr1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pontén F, Schwenk JM, Asplund A, Edqvist PHD. The Human Protein Atlas as a proteomic resource for biomarker discovery. J Intern Med. 2011;270:428–446. doi: 10.1111/j.1365-2796.2011.02427.x. [DOI] [PubMed] [Google Scholar]
- 41.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, Copley R, Courcelle E, Durbin R, Falquet L, Fleischmann W, Gouzy J, Griffith-Jones S, Haft D, Hermjakob H, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Orchard S, Pagni M, Peyruc D, Ponting CP, Servant F, Sigrist CJ. InterPro: an integrated documentation resource for protein families, domains and functional sites. Briefings Bioinf. 2002;3:225–235. doi: 10.1093/bib/3.3.225. [DOI] [PubMed] [Google Scholar]
- 42.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rost H, Malmstrom L, Aebersold R. A computational tool to detect and avoid redundancy in selected reaction monitoring. Molecular & cellular proteomics : MCP. 2012;11:540–549. doi: 10.1074/mcp.M111.013045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics (Oxford, England) 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 46.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annual review of pathology. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 47.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nature reviews Clinical oncology. 2014;11:473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 48.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Molecular & cellular proteomics : MCP. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Shteynberg D, Nesvizhskii AI, Moritz RL, Deutsch EW. Combining results of multiple search engines in proteomics. Molecular & cellular proteomics : MCP. 2013;12:2383–2393. doi: 10.1074/mcp.R113.027797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Song C, Wang F, Cheng K, Wei X, Bian Y, Wang K, Tan Y, Wang H, Ye M, Zou H. Large-Scale Quantification of Single Amino-Acid Variations by a Variation-Associated Database Search Strategy. Journal of Proteome Research. 2014;13:241–248. doi: 10.1021/pr400544j. [DOI] [PubMed] [Google Scholar]
- 52.Deutsch EW, Sun Z, Campbell D, Kusebauch U, Chu CS, Mendoza L, Shteynberg D, Omenn GS, Moritz RL. The State of the Human Proteome in 2014/2015 as viewed through PeptideAtlas: enhancing accuracy and coverage through the AtlasProphet. J Proteome Res. 2015 doi: 10.1021/acs.jproteome.5b00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nat Meth. 2014;11:1114–1125. doi: 10.1038/nmeth.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular systems biology. 2008;4:222–222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of highly-confident missing proteins identified between the lung cancer cell line and lung tissue samples (with PSM-, peptide-, and protein-level FDR of 1%) and protein existence evidence of the identified missing proteins.
Figure S2. Sample DDA spectra of PE5 proteins with single hit spectrum.
Figure S3. Sample MRM spectral evidences of the 10 validated peptides corresponding to the 8 validated missing proteins.
Figure S4. Sample DDA spectra for the 8 validated missing proteins.
Figure S5. Comparison of validation results between Hp-RP StageTip fractionated peptides and peptides without fractionation prior to MRM analysis.
Table S2. Clinical information of the lung tissue samples.
Table S3. List of missing proteins selected for MRM validation.
Table S4. List of all identified proteins searched against Swiss-Prot (2014_06) appended with decoy through Mascot (version 2.3.02) and processed by PeptideShaker.
Table S5. List of all identified proteins searched against Swiss-Prot (2014_11) appended with decoy through X!Tandem and Comet, and processed by PeptideProphet, iProphet, and MAYU.
Table S6. Overall 7702 proteins Identified with PSM, peptide and protein FDR = 1%, having at least one unique peptide and peptides with > 6 amino acid length.
Table S7. Information about the membrane annotation, HPA reliability index and number of TMH regions for the overall 7702 proteins.
Table S8. Comparison of the identified peptides with the Peptide Atlas repository. The frequency of observation for each peptide was also specified.
Table S9. The 178 missing proteins identified with PSM, peptide and protein FDR = 1%, having at least one unique peptide and peptides with > 6 amino acid length. This material is available free of charge via the Internet at http://pubs.acs.org