Abstract
REV-ERBα and REV-ERBβ nuclear receptors regulate several physiological processes, including circadian rhythm and metabolism. A previous study reported the REV-ERBα gene to be co-overexpressed with ERBB2 in breast cancer cell lines. Surprisingly, we found that several tumor types, including a number of breast cancer cell lines, predominantly express the REV-ERBβ variant. This pattern was independent of ERBB2 and ER status, and opposite to that of non-cancer mammary epithelial HMEC cells, in which REV-ERBα was the major variant. Consistent with this molecular profile, REV-ERB target genes in both circadian and metabolic pathways were derepressed upon silencing of REV-ERBβ, but not REV-ERBα. Strikingly, we found that REV-ERBβ is a determinant of sensitivity to chloroquine, a clinically relevant lysosomotropic agent that suppresses autophagy. The cytoprotective function of REV-ERBβ appears to operate downstream of autophagy blockade. Through compound screening, we identified ARN5187, a novel lysosomotropic REV-ERBβ ligand with a dual inhibitory activity toward REV-ERB-mediated transcriptional regulation and autophagy. Remarkably, although ARN5187 and chloroquine share similar lysosomotropic potency and have a similar effect on autophagy inhibition, ARN5187 is significantly more cytotoxic. Collectively, our results reveal that dual inhibition of REV-ERBβ and autophagy is an effective strategy for eliciting cytotoxicity in cancer cells. Furthermore, our discovery of a novel inhibitor compound of both REV-ERB and autophagy may provide a scaffold for the discovery of new multifunctional anticancer agents.
Introduction
REV-ERBα (NR1D1) and its variant REV-ERBβ (NR1D2) proteins belong to the nuclear receptor superfamily, which is composed of large number of ligand-activated transcription factors. REV-ERB proteins lack a transcriptional activation domain and repress target genes bearing REV-ERB Responsive Elements within their promoter.1, 2, 3 REV-ERBα regulates several processes, including circadian rhythm and metabolism.2,4 In mice, REV-ERBα reportedly confers robustness to the oscillatory clock.4 However, recent studies revealed that the REV-ERBα and REV-ERBβ variants compensate for one another in the repression of common target genes, indicating a more prominent function of REV-ERB proteins in circadian regulation.5,6
Disruption of the circadian clock is associated with a variety of human pathologies, including cancer.7, 8, 9, 10 Accordingly, the expression of several clock genes is perturbed in many tumors.11, 12, 13 Aberrant clock gene expression in tumors likely has a causal role in tumor development and survival. For instance, the incidence of breast cancer is higher among women who predominantly work nightshifts.9
REV-ERBα possesses a prosurvival function in ERBB2-positive breast cancer cells,14 which is of interest as REV-ERB proteins are proven druggable targets.15,16 In fact, pharmacological screening identified the compound SR6452 (GSK4112) as a REV-ERB synthetic agonist.17 Further studies identified several SR6452 analogs with optimized REV-ERB agonistic activity.16 The only REV-ERB antagonist identified to date is the compound SR8278.18 Although SR8278 is a useful probe for cellular REV-ERB activity, the pharmacokinetic properties of this compound limit its pharmacological uses,18 underscoring the need for additional REV-ERB inhibitors.
To determine whether pharmacological modulation of REV-ERB proteins may be a viable anticancer strategy, we investigated the contribution of both REV-ERB variants to the survival of ERBB2-positive cancer cells by both molecular genetic and pharmacological approaches.
Results
REV-ERBβ is the major variant in cancer cells
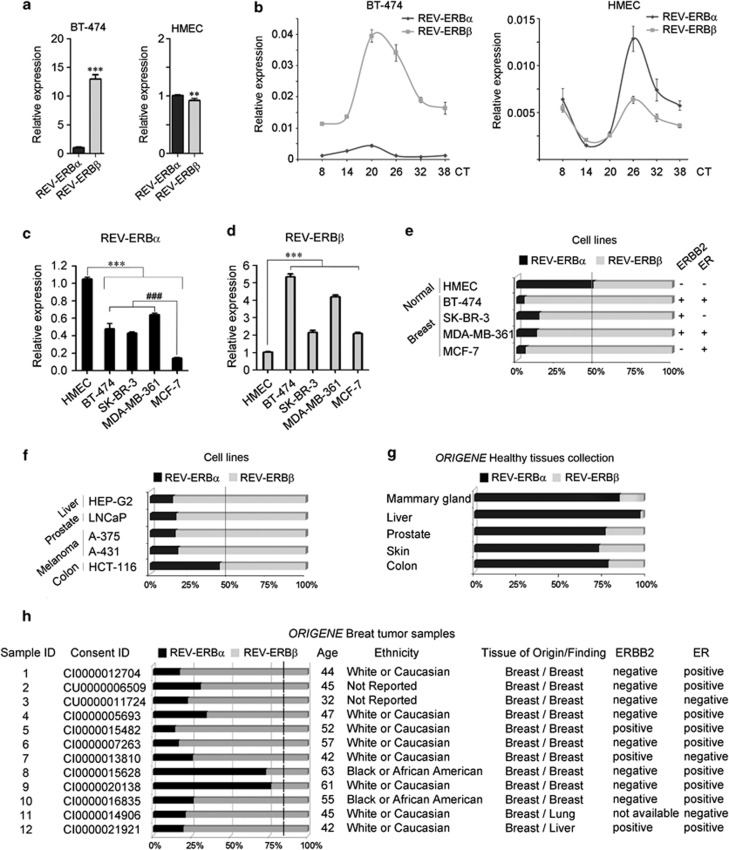
Because REV-ERBα and REV-ERBβ have redundant functions, we first determined the relative contribution of each variant to REV-ERB-dependent gene expression and to the survival of breast cancer cells. We initially analyzed their transcript levels in BT-474 breast cancer cells, in which the REV-ERBα gene has been reported to be co-overexpressed with ERBB2.19 Contrary to our expectations, the REV-ERBβ transcript was considerably more abundant than REV-ERBα (Figure 1a), and represented more than the 95% of total REV-ERB mRNA (Figure 1e). Notably, a different molecular phenotype was observed in non-cancer primary human mammary epithelial cells HMEC, in which levels of each variant were equally represented (Figures 1a and e).
Figure 1.
REV-ERBβ is the prominent variant in various cancer cells. (a) REV-ERBα and REV-ERBβ relative expression in breast cancer BT-474 and primary human mammary epithelial HMEC cells was determined by quantitative reverse transcriptase–PCR (qRT–PCR). GAPDH expression was used as normalizer and the REV-ERBα relative expression was set to 1. Shown as mean±s.e.m., n=6. **P<0.01 and ***P<0.001, β versus α variant. (b) BT-474 and HMEC cells were circadian synchronized by dexamethasone (DEX) treatment for 2 h and the relative expression of REV-ERBα and REV-ERBβ was evaluated at the indicated post-treatment time points (circadian time=CT). CT8 has been chosen as the first point for the analysis to avoid the interference of the DEX response in the early time of synchronization.20 (c and d) The expression of REV-ERBα (c) and REV-ERBβ (d) in BT-474, SK-BR-3, MDA-MB-361 and MCF-7 breast cancer cells relative to their levels in normal HMEC cells was determined by qRT–PCR. GAPDH expression was used as the normalizer and the REV-ERB variant relative expression in HMEC cells was set to 1. Shown as mean±s.e.m., n=6. ***P<0.001, cancer versus normal HMEC cells. ###P<0.001, ERBB2-negative MCF-7 versus ERBB2-positive BT-474, SK-BR-3 and MDA-MB-361 cells. (e) Relative abundance of REV-ERBα and REV-ERBβ in non-cancer human mammary epithelial HMEC cells and in breast cancer cell lines with a different ERBB2 and ER status was evaluated by qRT–PCR. Presented as mean of the percentage of each isoform expression contribution to the total REV-ERBs expression (n=6, s.e.m.<3%). The ERBB2 and ER status of each cell line is indicated at the right. (f) Relative abundance of REV-ERBα and REV-ERBβ in liver, prostate, melanoma and colon cancer cell lines. (g) Relative abundance of REV-ERBα and REV-ERBβ in the OriGene Healthy tissues cDNA collection, which contains cDNA of various tissues pooled from multiple healthy individuals of different ethnicity to avoid detection of individual differences. (h) Relative abundance of REV-ERBα and REV-ERBβ in OriGene breast tumor cDNA samples. Information on age, ethnicity, tissue of origin/finding, ERBB2 and ER status are reported for each sample. Dashed line refers to the REV-ERBα percentage in normal tissue.
REV-ERB gene expression is under the control of the circadian clock; we thus investigated whether this regulation also applied in cells in which REV-ERBβ is overexpressed. Accordingly, we analyzed the circadian expression profile of REV-ERBα and REV-ERBβ in BT-474 and HMEC cells synchronized by dexamethasone treatment.20 REV-ERB transcripts showed a similar oscillatory pattern in both BT-474 and HMECs (Figure 1b). In HMEC, levels of both transcripts peaked at CT26, where the relative REV-ERBα expression became higher than REV-ERBβ. In marked contrast, REV-ERBβ expression was higher than that of REV-ERBα at all timepoints in BT-474 cells (Figure 1b). Furthermore, the peak of both REV-ERB transcripts occurred earlier (CT20) when compared with the HMEC circadian profile (Figure 1b). Overall, our results indicate that REV-ERBβ is the more abundant variant in both non-synchronized and synchronized BT-474 cells. BT-474 belong to the Luminal B subtype of breast carcinomas, which are generally ERBB2 and ER positive.21 We thus investigated whether REV-ERBα and REV-ERBβ expression was dependent on ERBB2 and/or ER. The levels of the two REV-ERB transcripts were analyzed in BT-474, SK-BR-3, MDA-MB-361 and MCF-7 breast cancer cells relative to their levels in HMECs (Figures 1c and d). As reported,19 REV-ERBα levels were higher in ERBB2-positive (BT-474, SK-BR-3, MDA-MB-361) than in ERBB-2 negative (MCF-7) cells (Figure 1c). However, REV-ERBα levels were significantly lower in all the breast cancer cell lines versus normal HMECs. In marked contrast, REV-ERBβ transcript levels were significantly higher in all breast cancer cell lines analyzed when compared with normal epithelial cells (Figure 1d). REV-ERBβ overexpression in cancer cells was also confirmed at the protein level (Supplementary Figure S1a). Because relative copy number (Q) analysis22 indicated that the REV-ERBβ locus is not amplified in cancer cells, REV-ERBβ overexpression likely depends on a different transcriptional regulation in cancer versus normal cells (Supplementary Figures S1b and c). Finally, we observed that REV-ERBβ preponderance was independent of both ERBB2 and ER expression (Figure 1e).
We next addressed whether REV-ERBβ overrepresentation was a general feature of transformed cells, and found that REV-ERBβ was the most highly expressed variant in liver (HEP-G2), prostate (LNCaP) and melanoma (A-375 and A-431) cancer cells (Figure 1f). In contrast, a colon carcinoma cell line (HCT-116) expressed comparable levels of each variant (Figure 1f). The REV-ERBβ/REV-ERBα ratio was also higher in immortalized/transformed embryonic kidney HEK-293 cells, in which REV-ERBβ mRNA accounted for >90% of total REV-ERB transcripts (data not shown).
To evaluate whether this REV-ERBβ abundance was a feature of cancer cell lines, or rather was related to a human tissue-specific expression of this variant, we determined the REV-ERBα/REV-ERBβ ratio from the OriGene (Rockville, MD, USA) Healthy tissues cDNA collection. This panel contains pooled cDNA from tissues of multiple healthy individuals of different ethnicity. Remarkably, this analysis showed that REV-ERBα is more abundant than REV-ERBβ in various normal human tissues (Figure 1g and Supplementary Figure S2).
Conversely, similar analysis in breast, colon, liver and prostate cancer tissue cDNA acquired from OriGene, revealed that REV-ERBβ was highly expressed compared to the REV-ERBα variant in the majority of tumor samples (10/12 breast tumors, 10/12 colon tumors, 12/12 liver tumors, 12/12 prostate tumors, REV-ERBβ ⩾65% total REV-ERB; Figure 1h and Supplementary Figure S3). Consistent with our observations in breast cancer cell lines, this analysis also showed that preferential overexpression of REV-ERBβ is not related to the ERBB2 or ER status of tumors (Figure 1h).
REV-ERBβ overrepresentation corresponds to its dominant role in REV-ERB-mediated transcriptional regulation
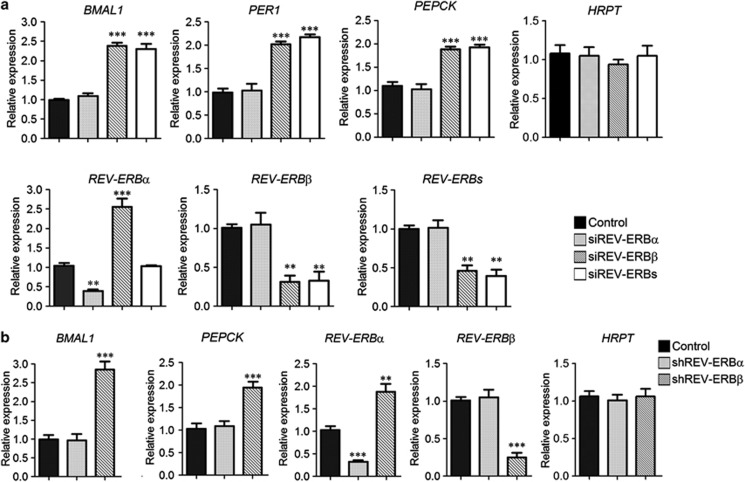
Studies in rodents have shown that REV-ERBα and REV-ERBβ variants compensate for one another to repress a number of common targets, including genes of circadian and metabolic pathways.5,6 We therefore examined whether the REV-ERBβ preponderance in cancer cell lines corresponds with a predominant functional role of this variant in REV-ERB-regulated gene expression. Thus, we knocked down expression of REV-ERBα, REV-ERBβ or both REV-ERB variants in BT-474 cells. Strikingly, REV-ERBβ silencing significantly enhanced the expression of circadian and metabolic genes, whereas REV-ERBα silencing had no such effect (Figure 2a). This result is consistent with a major dominant regulatory role of REV-ERBβ in repressing REV-ERB target genes. Consistent with the well-established feedback loop in the circadian signaling network,23 we observed an upregulation of the REV-ERBα gene following knockdown of REV-ERBβ. Indeed, the derepression of the circadian transcriptional activator, BMAL1, following REV-ERBβ depletion may account for the transcriptional upregulation of REV-ERBα, which is a BMAL1 target gene. However, as the basal level of REV-ERBα transcription is extremely low in BT-474 cells, the increase in REV-ERBα was insufficient to compensate for loss of REV-ERBβ, and therefore did not re-establish repression (Figure 2a).
Figure 2.
REV-ERBβ overrepresentation corresponds to a preponderant functional role in REV-ERB-mediated transcriptional regulation. (a) The expression of the circadian (BMAL1, PER1) and metabolic (PEPCK) REV-ERB-regulated genes was analyzed in BT-474 cells 72 h after transfection with pooled siRNA sequences against REV-ERBα (siREV-ERBα), REV-ERBβ (siREV-ERBβ) and both REV-ERBα and REV-ERBβ (siREV-ERBs), with a non-targeting pool as a negative control (Control). Relative expression was determined by quantitative reverse transcriptase–PCR (qRT–PCR) using GAPDH for normalization. HRPT expression is reported as representative of a REV-ERB-independent gene. The effect of REV-ERBs silencing on the two nuclear receptor variants was also evaluated. Levels of total REV-ERB transcripts (REV-ERBα plus REV-ERBβ) is indicated as REV-ERBs. Shown as mean±s.e.m., n=3. **P<0.01 and ***P<0.001, siRNA target sequences versus control. (b) BT-474 cells were transfected with vectors co-expressing a GFP protein with shRNA sequences against a non-targeting (Control), REV-ERBα (shREV-ERBα), REV-ERBβ (shREV-ERBβ) genes. Forty eight hours post-transfection, GFP-positive cells were sorted by fluorescence-activated cell sorting and processed for qRT–PCR analysis to evaluate the expression of REV-ERB-regulated genes. The relative expression was determined using GAPDH for normalization. HRPT expression is reported as representative of a REV-ERB-independent gene. Data are shown as mean±s.e.m., n=3. **P<0.01 and ***P<0.001, shRNA samples versus control.
To rule out potential off-target effects of our small interfering RNA (siRNA) experiments, we performed an alternative silencing method (small hairpin RNA (shRNA)) that utilized non-overlapping sequences. Supporting a predominant role for REV-ERBβ, the expression of the REV-ERB-regulated targets BMAL1 and PEPCK significantly increased in shRNA REV-ERBβ cells, whereas negligible changes were observed in shRNA REV-ERBα cells (Figure 2b). Similar results were obtained in MCF-7 and HEK-293 cells, which predominantly expressed the REV-ERBβ variant (Supplementary Figure S4).
Our hypothesis is that the lack of target gene upregulation upon REV-ERBα knockdown is related to its low abundance relative to REV-ERBβ consistent with this (and in contrast to cancer cells) we observed significantly increased expression of REV-ERB-dependent targets in REV-ERBα-silenced HMECs, which expressed similar levels of both REV-ERB variants (Supplementary Figure S5). In addition, and fully confirming the redundant function of the two REV-ERB proteins in transcriptional regulation, the depletion of both REV-ERB transcripts further enhanced the expression of the REV-ERB targets in HMEC cells (Supplementary Figure S5).
Overall, our results indicate that REV-ERBβ overrepresentation in cancer cells underlies a major functional role of this variant in the regulation of REV-ERB target genes.
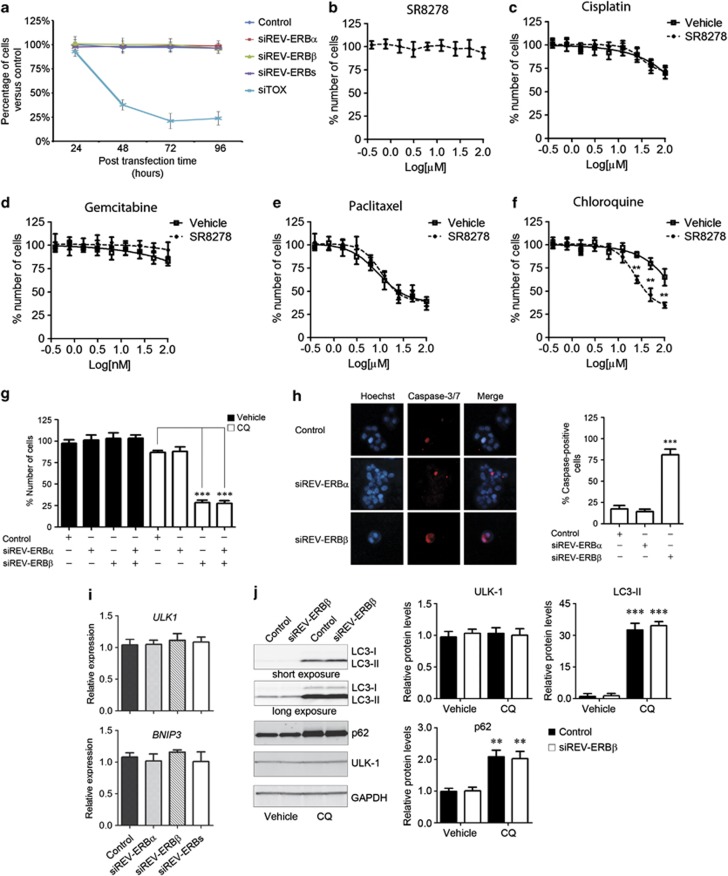
We next assessed the contribution of REV-ERBα in REV-ERBβ silencing to BT-474 viability. Unexpectedly, although REV-ERBα has been reported to promote survival of these cells,14 we did not observe a reduction of cell viability following knockdown of either REV-ERBα or REV-ERBβ (Figure 3a and Supplementary Figure S6).
Figure 3.
REV-ERBβ silencing enhances the cytotoxicity of the lysosomotropic autophagy inhibitor chloroquine. (a) BT-474 cells were transfected with pooled siRNA sequences against REV-ERBα (siREV-ERBα), REV-ERBβ (siREV-ERBβ), both REV-ERBα and REV-ERBβ (siREV-ERBs), a non-targeting pool (Control) and a pool of cytotoxic siRNA sequences (siTOX). The percentage of cells versus the Control transfection was evaluated at the indicated post-transfection times. (b) Concentration response plots of REV-ERB antagonist SR8278 cytotoxicity in BT-474 cells after 72 h treatment. Values of cells treated with vehicle were set to 100% of number of cells. Compound concentration is reported as log[μM]. Data expressed as mean±s.e.m., n=6. (c–f) Concentration response plots of Cisplatin (c),Gemcitabine (d), Paclitaxel (Taxol) (e) and Chloroquine (f) cytotoxicity in BT-474 cells in presence (dash line) or absence (solid line) of 10 μM SR8278 for 72 h. Values of cells treated with vehicle were set to 100% of number of cells. Compound concentration is reported as log[μM]. Data expressed as mean±s.e.m., n=6. **P<0.01, SR8278 versus vehicle. (g) BT-474 cells were reverse transfected with pooled siRNA sequences against REV-ERBα (siREV-ERBα), REV-ERBβ (siREV-ERBβ) and both REV-ERBα and REV-ERBβ genes or a non-targeting pool as a negative control (Control). One day post transfection, cells were treated with vehicle or chloroquine (CQ) for 48 h and the relative number of cells was evaluated by CyQUANT assay. Results are shown as mean±s.e.m. of the percentage number of cells normalized setting a control sample to 100%. ***P<0.001 CQ-treated siREV-ERBβ, and siREV-ERBα plus siREV-ERBβ versus Control samples. (h) Caspase-3 and -7 induction in BT-474 cells treated as in g was evaluated with the fluorescent SR-DEVD-FMK compound (Invitrogen). Hoechst 33342 (Hoechst) was used to stain cell nuclei. Count of the percentage of caspase-positive cells is given at the right. Shown as mean±s.e.m., n=3. ***P<0.001, siREV-ERBβ versus control. (i) The expression of the autophagy-related genes ULK-1 and BNIP3 was analyzed in BT-474 cells 72 h after transfection with pooled siRNA sequences against REV-ERBα (siREV-ERBα), REV-ERBβ (siREV-ERBβ) and both REV-ERBα and REV-ERBβ (siREV-ERBs), with a non-targeting pool as a negative control (Control). Relative expression was determined by quantitative reverse transcriptase–PCR using GAPDH for normalization. Data are shown as mean±s.e.m., n=3. (j) BT-474 cells were transfected with pooled siRNA sequences against REV-ERBβ (siREV-ERBβ) or a non-targeting pool as a negative control (Control) and then treated with Vehicle (water) or 50 μM CQ. The levels of LC3, p62, ULK-1 and GAPDH proteins were analyzed by Immunoblot analysis with specific antibodies. Densitometry analysis of protein signals is reported as relative protein levels normalized by GAPDH. Vehicle-treated Control sample value was set to 1. Shown as mean±s.e.m., n=3. **P<0.01 and ***P<0.001, CQ versus vehicle.
REV-ERBβ inhibition enhances the cytotoxicity of the lysosomotropic autophagy inhibitor, chloroquine
Because derepression of the clock machinery can sensitize cancer cells to an apoptotic stimulus,24 and REV-ERB proteins are clock-repressor factors, we decided to test whether REV-ERBα and REV-ERBβ inhibition enhanced the cytotoxicity of current anticancer drugs. As ERBB2-positive BT-474 cells are particularly resistant to cisplatin, gemcitabine, paclitaxel (Taxol) and chloroquine,25,26 we decided to study the cytotoxicity of these drugs in the presence or absence of the REV-ERB inhibitor compound SR827818 (Supplementary Figure S7).
Pharmacological inhibition of REV-ERB by SR8278 did not affect BT-474 cell viability (Figure 3b). Notably, although co-treatment with the REV-ERB antagonist had negligible effect in cisplatin-, gemcitabine- and paclitaxel-treated cells (Figures 3c–e), SR8278 significantly enhanced the cytotoxic activity of the lysosomotropic autophagy inhibitor drug, chloroquine (Figure 3f). To confirm that this was due to specific inhibition of REV-ERB, we evaluated chloroquine cytotoxicity in REV-ERBα and REV-ERBβ-silenced BT-474 cells. Indeed, a subtoxic dose of chloroquine markedly reduced the number of REV-ERBβ and REV-ERBα/β-interfered cells (Figure 3g). Conversely, the knockdown of REV-ERBα did not sensitize cells to chloroquine (Figure 3g). Consistent with increased apoptosis, the percentage of caspase-positive cells was significantly higher in REV-ERBβ-silenced cells upon chloroquine treatment (Figure 3h). This analysis also confirmed a specific cytoprotective action of the REV-ERBβ variant, since the percentage of caspase-positive cells did not significantly differ between REV-ERBα-silenced and control cells (Figure 3h).
Overall, our data indicate that chemical or genetic inhibition of REV-ERBβ significantly compromises viability in the context of a CQ-mediated autophagy blockade. Consistent with this, knockdown of the essential autophagy gene ATG527 enhanced the toxicity of SR8278, further demonstrating that REV-ERB inhibition induces apoptosis when autophagy is genetically inhibited (Supplementary Figure S8).
In mouse muscle, REV-ERB regulates the transcription of autophagy-related genes, such as ULK-1 and BNIP3.28,29 However, REV-ERB silencing did not affect the transcription of these genes in BT-474 cells (Figure 3i). To further analyze whether REV-ERBβ silencing affects autophagy in both basal and chloroquine-stimulated conditions, we analyzed autophagy-related proteins by immunoblot analysis in REV-ERBβ knockdown cells in the presence or absence of chloroquine.
Consistent with the expression analysis, ULK-1 protein levels were not affected by REV-ERBβ silencing (Figure 3j). REV-ERBβ knockdown also had no effect on the levels of the autophagy marker LC3 in untreated cells. Consistent with this, pharmacological REV-ERB inhibition did not affect LC3 levels (Supplementary Figure S9a). As expected, autophagy blockade upon chloroquine treatment increased the levels of the phosphatidylethanolamine-conjugated LC3-II form and the p62/SQSTM1 protein, the degradation of which is mediated by the autophagolysosome30 (Figure 3j). A similar level of LC3-II and p62 induction was observed in both REV-ERBβ-interfered and control cells (Figure 3j). Ruling out possible side effects related to prolonged chloroquine treatment, a similar induction of LC3-II protein in REV-ERBβ-silenced and control cells was also observed using a shorter incubation time (2 h) and lower concentration of drug (25 μM; Supplementary Figure S9b).
Overall, our data indicate that REV-ERBβ acts as a cytoprotective factor downstream of a blockade of autophagy; the precise molecular mechanism(s) underlying this prosurvival function remain to be determined.
Identification of a novel lysosomotropic compound with a dual inhibitory activity toward REV-ERB and autophagy
Although the molecular basis of the cytoprotective REV-ERBβ function requires more investigation, our results indicate that REV-ERBβ inhibition enhances the cytotoxicity of chloroquine, a lysosomotropic compound with relevant anticancer applicative uses.31,32
A subset of lysosomotropic compounds share certain physicochemical properties, in that they possess a ClogP >2 and a basic pKa between 6.5 and 11.33 Because several reported REV-ERB synthetic ligands have a ClogP >3 and contain a protonable amino group,16 we speculated that some novel REV-ERB inhibitor compounds might also possess lysosomotropic properties.
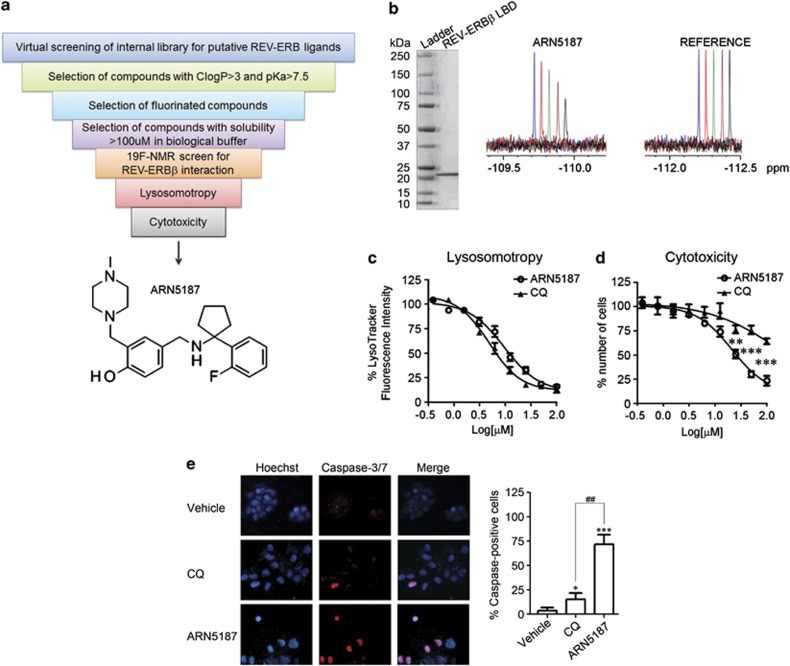
Thus, we performed an in silico screen of a diverse and non-redundant set of approximately 15 000 molecules present in our internal chemical collection (Figure 4a). An energy minimum conformation of the REV-ERB synthetic ligand SR6452 was used to generate Atomic Property Field (APF) potentials,34 setting the presence of at least two protonable amino group as a constriction. The members of the library were thus ranked according to the generated APF score values. Of the 200 top-ranking compounds, 25% possessed a ClogP >3 and a basic pKa >7 and contained at least one fluorine atom, which facilitates sensitive fluorine nuclear magnetic resonance (NMR)-based screening (FNBS).35 The 31 most soluble molecules (solubility >100 μM) were thus tested by FNBS in the presence of the purified LBD of REV-ERBβ (Supplementary Figure S10).
Figure 4.
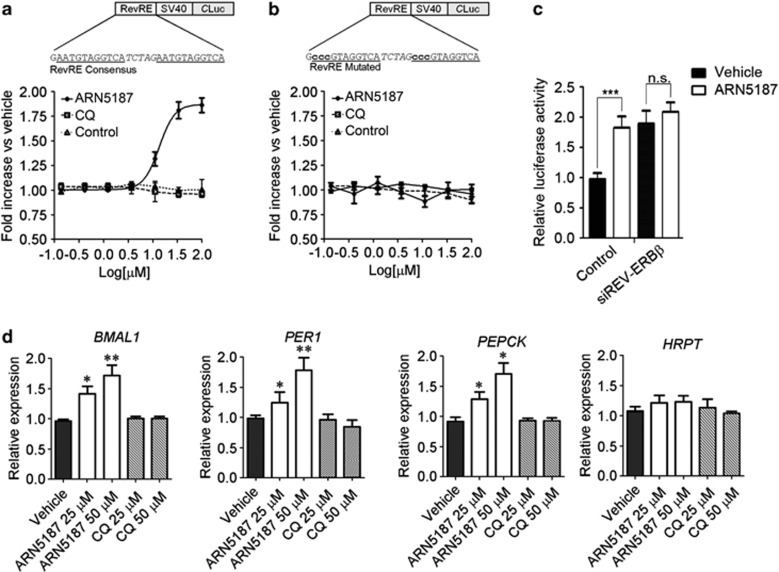
Identification of a novel lysosomotropic REV-ERB ligand. (a) Schematic representation of the work-flow used to identify ARN5187. (b) Representative of 19F-NMR spectra of ARN5187 in the presence of different concentration of a purified REV-ERBβ LBD (left). ARN5187 interaction with the protein resulted in line broadening effects producing a progressive decrease of 19F-NMR signal as function of increasing protein amounts. Excluding artifacts deriving from protein aggregation, no effect was observed for a Reference compound (blue=no protein; red, green, violet, and black=1, 2, 4, and 8 μM, respectively). (c) Concentration response plots of ARN5187 (open circles) and chloroquine (CQ; triangles) lysosomotropism evaluated by LysoTracker fluorescence. Values of cells treated with vehicle were set to 100% of fluorescence intensity. Data expressed as mean±s.e.m., n=6. EC50 values for lysosomotropy responses of ARN5187 and CQ were 9.5±1.2 and 4.8±1.9 μM, respectively. (d) Concentration response plots of ARN5187 (open circles) and CQ (triangles) cytotoxicity in BT-474 cells treated 48 h. Values of cells treated with vehicle were set to 100% of number of cells. Data expressed as mean±s.e.m., n=6. EC50 values for cytotoxic responses of ARN5187 and CQ were 23.5±7.3 and >100 μM, respectively. **P<0.01 and ***P<0.001, ARN5187 versus CQ. (e) Caspase-3 and -7 induction in BT-474 cells treated 48 h with vehicle, ARN5187 and chloroquine (CQ) was evaluated with the fluorescent SR-DEVD-FMK compound (Invitrogen). Hoechst 33342 (Hoechst) was used to stain cell nuclei. Count of the percentage of caspase-positive cells is given at the right. Shown as mean±s.e.m., n=3. **P<0.05 and ***P<0.001, compounds versus vehicle. ##P<0.01, ARN5187 versus CQ.
This lead to the identification of ARN5187, a novel REV-ERBβ ligand (Figure 4a). Indicating a direct interaction of ARN5187 with the LBD of REV-ERBβ, the incubation of ARN5187 in the presence of increasing receptor concentration caused line broadening and a progressive decrease of signal on the 19F-NMR spectra (Figure 4b).
We then compared the lysosomotropic and cytotoxic effects of ARN5187 and chloroquine in BT-474 cells. Although ARN5187 displayed a lysosomotropic potency comparable to chloroquine (Figure 4c), it was significantly more cytotoxic (Figures 4d and e). We thus investigated whether ARN5187 cytotoxicity was due to combined inhibition of REV-ERB activity and autophagy.
ARN5187 relieves REV-ERB-mediated clock transcriptional repression
We evaluate the effect of ARN5187 on REV-ERB transcriptional repression activity, using a RevRE reporter assay. A 24-h treatment only modestly reduced cell number at the highest tested ARN5187 dose in both BT-474 and HEK-293 cells (Supplementary Figure S10d). We thus adopted this time point for the analysis in order to avoid the confounding effects of compound cytotoxicity.
Consistent with its ability to relieve REV-ERB-mediated transcriptional repression, ARN5187 activated the RevRE reporter in a concentration-dependent manner, whereas no change was observed with the control compound (Figure 5a). Supporting the specificity of our assay for REV-ERB-mediated transcription, ARN5187 did not alter the activity of a reporter bearing a mutated RevRE that is not recognized by the REV-ERB DNA binding domain1 (Figure 5b). Analogous to other synthetic REV-ERB effectors,15 ARN5187 was not preferentially selective for either REV-ERB variant (Supplementary Figure S11).
Figure 5.
ARN5187 relieves REV-ERB-mediated transcriptional repression. (a) Co-transfection assay in ARN5187- and Chloroquine (CQ)-treated HEK-293 cells with REV-ERBβ and a luciferase REV-ERB-responsive reporter driven by two repetition of a RevRE consensus. An inactive compound from the FNBS screen was used as a negative ‘Reference' control. Schematic representation of the reporters is shown at the top. Data expressed as fold increase of luciferase activity versus vehicle (DMSO-d6). Shown as mean±s.e.m., n=9. EC50 for ARN5187 antagonism versus REV-ERBβ was 15±3.2 μM. (b) Co-transfection assay in ARN5187- and CQ-treated HEK-293 cells with REV-ERBβ and a REV-ERB-unresponsive reporter bearing a mutated RevRE that is not recognized by REV-ERB DNA binding domain. Shown as mean±s.e.m., n=6. (c) HEK-293 cells were co-transfected with a luciferase REV-ERB-responsive reporter and vectors containing a non-targeting shRNA sequence (Control) or a sequence against REV-ERBβ (shREV-ERBβ). Twenty-four hours post transfection, cells were treated with vehicle (DMSO-d6) or 50 μM ARN5187 and the normalized luciferase expression was evaluated. Shown as mean±s.e.m., n=6. ***P<0.001 ARN5187-treated versus vehicle-treated Control cells. (d) Expression of REV-ERB-regulated BMAL1, PER1 and PEPCK genes in breast cancer BT-474 cells after treatment with two doses of ARN5187 or CQ. The HPRT gene was used as representative of a REV-ERB independent gene. Shown as relative expression normalized by GAPDH. The value in vehicle sample was set to 1. Reported as mean±s.e.m., n=6. *P<0.05; **P<0.01 ARN5187 versus vehicle.
Notably, chloroquine had negligible effects on RevRE reporter activity (Figure 5a), suggesting that the effect of ARN5187 on REV-ERB transcriptional repression was not related to its lysosomotropic properties.
To further demonstrate the REV-ERBβ inhibitory activity of ARN5187, we tested its effect on reporter expression in REV-ERBβ-knockdown cells. Indeed, ARN5187 treatment increased luciferase activity in the control, but not in REV-ERBβ-knockdown cells (Figure 5c).
We then examined ARN5187 effects on the expression of endogenous REV-ERB target genes in BT-474 cells. Consistent with lysosomotropic-independent inhibition of REV-ERB activity, ARN5187 significantly enhanced the expression of BMAL1, PER1 and PEPCK in a dose-dependent manner, whereas chloroquine treatment had no effect (Figure 5d). Collectively, these results reveal a lysosomotropic-independent REV-ERB antagonistic activity of ARN5187.
ARN5187 is a novel autophagy inhibitor
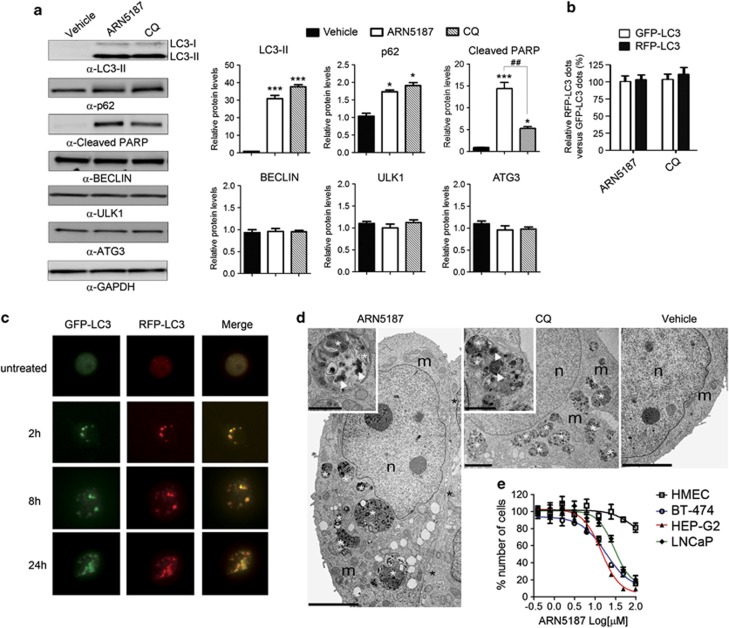
We next tested the ability of ARN5187 to inhibit autophagy and observed a robust increase in LC3-II levels after treatment with equimolar concentrations of ARN5187 or chloroquine (Figure 6a). Furthermore, levels of the autophagy-associated protein p62 significantly increased in either ARN5187- or chloroquine-treated cells. Although both compounds produced a comparable increase of LC3-II and p62, cleaved poly ADP-ribose polymerase (PARP) levels were significantly elevated in ARN5187-treated cells (Figure 6a), consistent with its higher cytotoxicity.
Figure 6.
ARN5187 is a novel autophagy inhibitor. (a) Immunoblot analysis of protein samples from BT-474 cells treated 24 h with Vehicle, 50 μM ARN5187 and 50 μM chloroquine (CQ) with the indicated antibodies. Cross-hybridization signal corresponding to LC3-I form is also indicated. Densitometry analysis of protein signals is reported as relative protein levels normalized by GAPDH. Vehicle sample value was set to 1. Shown as mean±s.e.m., n=6. *P<0.05; **P<0.01; ***P<0.001 compounds versus vehicle. ##P<0.01 ARN5187 versus CQ. (b) BT-474 cells transduced with a GFP/RFP/LC3 chimera protein were treated with Vehicle, 50 μM ARN5187 and 50 μM CQ. After 24 h, the number of GFP- and RFP-positive dots per cell was quantified under a fluorescent microscope. Shown as mean relative RFP-positive dots versus GFP-positive dots±s.e.m., n=6. (c) BT-474 cells transduced with a GFP/RFP/LC3 chimera protein were treated 50 μM ARN5187 and the GFP- and RFP-positive dots was visualized under a fluorescent microscope 2, 8 and 24 h after treatment. Merges images showing co-localization of red and green dots is shown. Fluorescent signals in untreated cells is also shown. (d) Transmission electron microscopy images of BT-474 cells treated 24 h with 50 μM ARN5187, 50 μM CQ and Vehicle. Left, low magnification of a cell exposed to ARN5187. The cytoplasm contains numerous Autophagic Vacuole derivatives (AVds) of different sizes (white asterisks). The black asterisks mark a portion of another cell in close contact. Inset: high magnification of a single AVd. Note the electron-dense materials (arrowheads) and the membrane remnants (white asterisks). Center, representative of a cell exposed to CQ. Numerous AVds are present inside the cell cytoplasm (white asterisks). Inset: high magnification of a single AVd containing electron dense material (arrowheads) and membrane remnants (white asterisks). Right, low magnification of a BT-474 cell exposed to Vehicle (DMSO-d6). m, mitochondria, n, nucleus. Scale bars are 5 μm for ARN5187- and Vehicle-treated cell; 2 μm for CQ-treated cell; 1 μm for insets. (e) Concentration response plots of ARN5187 cytotoxicity evaluated in normal primary human mammary epithelial cells (black), and the breast (BT-474, blue), liver (HEP-G2, red) and prostate (LNCaP, green) cancer cell lines. Values of cells treated with vehicle were set to 100% of number of cells. Data expressed as mean±s.e.m., n=6. EC50 values for BT-474, HEP-G2, LNCaP and HMEC were 23.5±7.3, 14.4±3.4, 29.8±10.9 and >100, respectively.
Owing to the complex transcriptional regulation of the p62 gene,30 we cannot exclude an autophagy-independent increase of p62. Thus, the elevated p62 levels in ARN5187-treated cells do not definitively demonstrate an autophagy inhibitory activity of this compound. Nevertheless, the levels of proteins involved in the initiation (ULK1), nucleation (BECLIN) and elongation (ATG3) phases of autophagy were unaffected by either ARN5187 or chloroquine treatment (Figure 6a). To avoid confounding side effects related to the prolonged treatment, we repeated the experiment with 25 μM of ARN5187 and chloroquine, and analyzed the levels of LC3-II and p62 after 2 and 4 h. Consistent with our previous analysis, ARN5187 and CQ similarly increased the levels of LC3-II after 2 h (Supplementary Figure S12). LC3-II levels further increased at 4 h. Although neither ARN5187 nor CQ significantly increased p62 levels at 2 h, both compounds generated a similar induction of p62 after 4 h (Supplementary Figure S12).
These data support the hypothesis that ARN5187 blocks autophagy at a late stage. To further evaluate this possibility, we exploited the fluorescence properties of an acid-sensitive green fluorescent protein (GFP) that can be quenched upon acidification in the mature autophagolysosome.36 BT-474 cells were transduced with a chimeric protein in which an acid-sensitive GFP and an acid-insensitive red fluorescent protein (RFP) were fused to LC3. As expected from an autophagy blockade at the late stage, the number of GFP/RFP fluorescent dots increased proportionally in ARN5187- or chloroquine- treated cells compared with vehicle (Figure 6b).
Because autophagy is a very dynamic process, the analysis of a single time point is not sufficient to rule out the possibility that ARN5187 also affected autophagy formation and maturation. We therefore analyzed the appearance of GFP- and RFP-positive dots at different time points in ARN5187-treated cells. Consistent with the idea that ARN5187 blocks autophagy by disrupting the lysosomal function and preventing autophagolysosome final maturation, we observed a similar accumulation of both GFP- and RFP-positive dots over time (Figure 6c).
To further test this hypothesis, we used transmission electron microscopy to detect the presence of autophagic vesicles and to distinguish early autophagic compartments (autophagosomes) from late autophagic structures (autophagolysosomes).30 This revealed a robust increase in the number of autophagic vacuoles in cells treated with ARN5187 or chloroquine compared with vehicle (Figure 6d and Supplementary Figure S13). Clearly indicating an inhibition of late-stage autophagy, the majority of autophagic vacuoles were autophagolysosomes, as revealed by the ultrastructure of their cytoplasmic content, including membrane stacks and vesicles, which appeared electron dense, altered and partially disintegrated (Figure 6d and Supplementary Figure S13).
Collectively, our results indicate that ARN5187 is a novel dual autophagy/REV-ERB inhibitor. Considering that the inhibition of autophagy has very low cytotoxicity in several non-cancer cells, including HMEC,37 and that REV-ERBβ seems to act as a cytoprotective factor downstream of autophagy blockade, we compared the cytotoxicity of ARN5187 in HMEC and BT-474 cells. In addition, to extend our observations to non-breast cancer cells, we also assessed the toxicity of ARN5187 in prostate (LNCaP) and liver (HEP-G2) cells. We found that the compound had negligible effects on HMEC viability, while it was equally cytotoxic in all the cancer cell lines (Figure 6e).
Discussion
Although circadian regulation is altered in several tumor types, whether pharmacological targeting of circadian regulators is a viable anticancer strategy remains to be determined. In contrast, the efficacy of autophagy inhibition in cancer therapy has been well documented.31,38,39 Here, we provide the first indication that dual inhibition of the circadian regulator REV-ERBβ and the autophagy process may be a novel anticancer therapeutic approach.
In cancer cells, we found that in marked contrast to normal epithelial cells, REV-ERBβ is the predominantly expressed REV-ERB variant (Figure 1 and Supplementary Figure S1). Our knockdown experiments are also the first to indicate that REV-ERBβ overrepresentation corresponds to a principal functional role of this variant in the transcriptional regulation of REV-ERB target genes in cancer cells (Figure 2 and Supplementary Figure S4). We observed that the alpha variant is the prominent REV-ERB form in several tissue types (Figure 1g and Supplementary Figure S2), consistent with a previous report.40 Conversely, REV-ERBβ levels were higher in the majority of tumor samples examined (Figure 1h and Supplementary Figure S3).
Although REV-ERBβ does not seem to contribute to cancer cell viability per se, our data indicate that it has a protective function when the autophagy flux of cancer cells is compromised (Figure 3 and Supplementary Figure S8). We did not observe effects of REV-ERBβ silencing on autophagy markers under basal or chloroquine-treated conditions (Figures 3i and j, and Supplementary Figure S9b). This suggests that the cytoprotective function of REV-ERBβ likely operates downstream of the chloroquine-mediated blockade of autophagy. Further investigation will help to clarify the molecular mechanism behind this novel REV-ERBβ cytoprotective role.
We also identified a compound, ARN5187, with dual inhibitory activity toward both REV-ERB and autophagy. ARN5187 is apparently a novel autophagy inhibitor that disrupts lysosomal function, blocks the autophagy process at the late stage, and reduces cancer cell viability (Figure 6 and Supplementary Figure S12). In addition, ARN5187 inhibits REV-ERB mediated transcriptional regulation (Figure 5). ARN5187 was more cytotoxic than the clinically relevant lysosomotropic autophagy inhibitor, chloroquine (Figures 4d and e). Our data support the view that the stronger apoptotic induction of ARN5187 compared with chloroquine is mainly due to the dual inhibitory activity of this compound with respect to both autophagy and REV-ERB function.
We suggest that this multifunctional REV-ERB and autophagy inhibitor provides a scaffold for the discovery of new anticancer agents. Further examination of the efficacy and pharmacokinetic/pharmacodynamic studies of ARN5187 in vivo will provide useful information on the potential of ARN5187 as an anticancer agent.
Materials and methods
Cell lines
BT-474 (HTB-20), MDA-MB-316 (HT-B27), SK-BR-3 (HTB-30), HEK-293 (CRL-1573), A-375 (CRL-1619), and A-431 (CRL-1555) cells were acquired from the American Type Culture Collection and the National Collection of Type Cultures (ATCC). MCF-7, HEP-G2 and HCT-116 cells were kindly provided by Istituto di Ricerche Farmacologiche ‘Mario Negri', Milan, Italy. BT-474 cells were grown in Dulbecoo's Modified Eagle's Medium High Glucose (4.5 g/l D-Glucose) containing 4 mM L-glutamine, 10% fetal bovine serum (FBS) and 0.25 mM sodium pyruvate. HEK-293 and A-375 cells were grown in Dulbecoo's Modified Eagle's Medium High Glucose containing 4 mM L-glutamine, 10% FBS and 0.5 mM sodium pyruvate. MCF-7, H-1299, HEP-G2 and HCT-116 cells were grown in RPMI-1640 containing 4 mM L-glutamine and 10% FBS. A-431 cells were grown in Eagle's minimum essential medium containing 10% FBS and non-essential amino acids. SK-BR-3 cells were grown in McCoy's medium containing 10% FBS.
Reagents
Chloroquine (>98%), SR8278 (>98%), vorinostat (>98%), dichloromethane, diethyl ether (Et2O), methanol (MeOH), 2 M HCl solution in Et2O and sodium triacetoxyborohydride [NaBH(OAc)3] were acquired from Sigma-Aldrich, Milan, Italy. For the initial screening, ARN5187 (CAS n. 1287451-26-6) was acquired from Asinex (Moscow, Russia). ARN5187 was re-synthesized as hydrochloride salt with purity >99.5%.
Synthesis of 4-[[[1-(2-Fluorophenyl)cyclopentyl]-amino]methyl]-2-[(4-methylpiperazin-1-yl)methyl]phenol-trihydrochloride (ARN5187)
ARN5187 was resynthesized by reductive amination between 1-(2-fluorophenyl)cyclopentanamine (1.0 equiv.) and 4-hydroxy-3-[(4-methylpiperazin-1-yl)methyl]benzaldehyde (1.0 equiv.), which were obtained by chemical synthesis according to the procedures described elsewhere.41 The free base (60.0 mg, 0.15 mmol) was dissolved in dichloromethane (1.5 ml) and 2.0 M HCl solution in Et2O (1.5 ml, 3.0 mmol, 20.0 equiv.) was added observing the formation of precipitate. Excess solvent was removed under reduced pressure and the resulting solid was triturated with Et2O, then dissolved in a solution of CH3CN/H2O (1:1) and lyophilized. Representative ultra performance liquid chromatography/mass spectrometry and NMR spectra of ARN5187 are reported in Supplementary Figures S14 and S15.
Computational methods
Calculations were performed using ICM v.3.7 (San Diego, CA, USA). Molecular structures were prepared assigning three-dimensional coordinates to each compound. Each molecule was assigned the right bond orders, stereochemistry, hydrogen atoms and protonation states at pH 7.4. Merck molecular force field atom types and charges were used. The dihedral space of the template was sampled by means of the ICM stochastic optimizer. Thoroughness and vicinity parameters were set equal to 1.0 and 15.0, respectively. Fifty conformations were retained and subjected to fully flexible energy minimization. An energy minimum conformation of SR6452 was used to generate APF potential maps as described by.34 Grids covered the template plus a 5 Å margin. Each molecule from the library was iteratively sampled in the APF grid complement and its APF energy calculated.
Purification of REV-ERBβ LBD
Human REV-ERBβ sequence coding for the LBD from aa377 to aa579 was cloned in the pMAL-c5x vector (Biolabs, Hitchin, UK). The resulting MBP fusion protein was expressed in BL21 cells and purified by affinity chromatography as described in Supplementary Figure S10.
NMR experiments
NMR spectra were recorded at 298 K with a Bruker FT NMR Avance III 600 MHz spectrometer (Milan, Italy), equipped with a 5 mm CryoProbeQCI-1H/19F–13C/15N–D quadruple resonance with a shielded Z-gradient coil. Compounds and protein were tested in 50 mM Tris-HCl, pH 7.5, 138 mM NaCl, 1 mM DTT and 8% D2O for the lock signal. R2 filter experiments were recorded as described by Dalvit et al.42 with the Carr-Purcell-Meiboom-Gill scheme with a time interval of 47 ms between the 180° pulses and with different total lengths. The spectra were acquired with proton decoupling using the Waltz-16 composite pulse sequence with a spectral width of 80 p.p.m., a 5-s relaxation delay, and 256 scans. Chemical shifts were referenced to the CFCl3 signal in water.
REV-ERB luciferase assay
Two repetitions of the RevRE consensus (5′-AGA ATG TAG GTC ATC TAG AAT GTA GGT CA-3′) or a mutated version (5′-AGC CCG TAG GTC ATC TAG CCC GTA GGT CA-3′) that is no longer able to bind REV-ERB were cloned in the pSV-40Cluc vector (Biolabs). For the assay, HEK-293 cells were co-transfected with the reporters and a plasmid expressing a REV-ERBα or REV-ERBβ. The following day, cells were treated with the compounds and after 24 h luciferase activities were measured according to the manufacturer's instructions. A vector with an SV40 promoter-driven Gaussia luciferase was used for normalization.
Relative copy number analysis
Relative copy number (Q) of ERBB2 vs GAPDH and REV-ERBβ vs GAPDH in BT-474 and HMEC cells was calculated as described elsewhere.22 Primers used for the analysis are reported in Supplementary Table S1.
Quantitative reverse transcriptase–PCR
RNA sample preparation and relative transcript expression levels were assessed as described previously.43 GAPDH transcript was used for normalization. Primer sequences are listed in Supplementary Table S1. For human tissue expression experiments, cDNA from OriGene Technologies Inc. was analyzed.
Immunoblot analysis
Protein samples were extracted in RIPA buffer as described previously.43 Antibodies and immunoblot conditions are listed in Supplementary Table S2.
Transmission electron microscopy
For transmission electron microscopy studies, cells were grown on glass coverslips. After treatment with ARN5187, Chloroquine or vehicle (DMSO-d6), cells were processed as described elsewhere.44 transmission electron microscopy images were collected by a FEI Tecnai G2 F20 equipped with a field-emission gun operating at 80 kV of acceleration voltage and recorded with a four Mp-Gatan-BM-UltraScan-Charge-Coupled Device camera.
Lysosomotropy and cytotoxicity assays
Cells were seeded in 96-well plates at 2000 cells/100 μl/well in culture medium and incubated overnight. For lysosomotropy assays, 24 h post-treatment cells lysosomal staining was measured as described previously.33
For cytotoxicity assays, the percentage of cell number was evaluated with the CyQUANT kit (Invitrogen, Carlsbad, CA, USA). This method was adopted because it is independent of cellular metabolic activity, which may be affected by both REV-ERB and autophagy inhibition.
Caspase-3 and -7 activity induction was evaluated with the Image-iT LIVE Red Caspase-3 and -7 Detection Kit (Invitrogen, Life Technologies).
siRNA and shRNA
Sequence information for siRNAs and shRNAs used in the silencing experiments are listed in Supplementary Table S3. DharmaFECT 1 (Thermo Fisher Scientific, Waltham, MA, USA) and Fugene (Roche Italia, Monza, Italy) transfection reagents were used for siRNA and shRNA experiments, respectively. Knockdown was verified by immunoblot analysis (Supplementary Figure S16).
Statistical analysis
Log(inhibitor)-versus-response curves, one-way analysis of variance with Dunnett's post test and two-tailed t-test were performed using GraphPad-Prism Software (San Diego, CA, USA).
Acknowledgments
We thank Professor Daniele Piomelli for helpful discussions and for revising the manuscript. We thank Dr Gennaro Colella for MCF-7, HEP-G2 and HCT-116 cell lines, and Dr Natalia Realini for providing cDNA from H-1299, HEP-G2, A-375, A-431 and HCT-116 cells. We also thank Professor Mariapia Abbracchio, Dr Ana Guijarro, Dr Andrea Contestabile and Dr Francesco Niola for discussions. Dr L Ercolani is supported by an interdepartmental post-doctoral fellowship from the Istituto Italiano di Tecnologia (IIT).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
GB, ET, RS and BG are co-inventors in a patent that includes ARN5187 owned by Istituto Italiano di Tecnologia (IIT). The remaining authors declare no conflict of interest.
Supplementary Material
References
- 1Harding HP, Lazar MA. The orphan receptor Rev-ErbA alpha activates transcription via a novel response element. Mol Cell Biol 1993; 13: 3113–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Duez H, Staels B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J Appl Physiol 2009; 107: 1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 2007; 14: 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Teboul M, Grechez-Cassiau A, Guillaumond F, Delaunay F. How nuclear receptors tell time. J Appl Physiol 2009; 107: 1965–1971. [DOI] [PubMed] [Google Scholar]
- 5Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 2012; 485: 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F et al. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev 2012; 26: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci 2003; 4: 649–661. [DOI] [PubMed] [Google Scholar]
- 8Rossetti S, Esposito J, Corlazzoli F, Gregorski A, Sacchi N. Entrainment of breast (cancer) epithelial cells detects distinct circadian oscillation patterns for clock and hormone receptor genes. Cell Cycle 2012; 11: 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer 2009; 9: 886–896. [DOI] [PubMed] [Google Scholar]
- 10Engelen E, Janssens RC, Yagita K, Smits VA, van der Horst GT, Tamanini F. Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PLoS ONE 2013; 8: e56623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer 2003; 3: 350–361. [DOI] [PubMed] [Google Scholar]
- 12Kelleher FC, Rao A, Maguire A. Circadian molecular clocks and cancer. Cancer Lett 2013; 342: 9–18. [DOI] [PubMed] [Google Scholar]
- 13Savvidis C, Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med 2012; 18: 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res 2010; 70: 1783–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012; 485: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Trump RP, Bresciani S, Cooper AW, Tellam JP, Wojno J, Blaikley J et al. Optimized chemical probes for REV-ERBalpha. J Med Chem 2013; 56: 4729–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Kumar N, Solt LA, Wang Y, Rogers PM, Bhattacharyya G, Kamenecka TM et al. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology 2010; 151: 3015–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Kojetin D, Wang Y, Kamenecka TM, Burris TP. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem Biol 2011; 6: 131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Bertucci F, Borie N, Ginestier C, Groulet A, Charafe-Jauffret E, Adelaide J et al. Identification and validation of an ERBB2 gene expression signature in breast cancers. Oncogene 2004; 23: 2564–2575. [DOI] [PubMed] [Google Scholar]
- 20Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2000; 289: 2344–2347. [DOI] [PubMed] [Google Scholar]
- 21Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res 2011; 13: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Konigshoff M, Wilhelm J, Bohle RM, Pingoud A, Hahn M. HER-2/neu gene copy number quantified by real-time PCR: comparison of gene amplification, heterozygosity, and immunohistochemical status in breast cancer tissue. Clin Chem 2003; 49: 219–229. [DOI] [PubMed] [Google Scholar]
- 23Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int J Biochem Cell Biol 2009; 41: 81–86. [DOI] [PubMed] [Google Scholar]
- 24Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci USA 2009; 106: 2841–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Hastak K, Alli E, Ford JM. Synergistic chemosensitivity of triple-negative breast cancer cell lines to poly(ADP-Ribose) polymerase inhibition, gemcitabine, and cisplatin. Cancer Res 2010; 70: 7970–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Carew JS, Espitia CM, Esquivel JA 2nd, Mahalingam D, Kelly KR, Reddy G et al. Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem 2011; 286: 6602–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer 2007; 7: 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med 2013; 19: 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15: 406–416. [DOI] [PubMed] [Google Scholar]
- 30Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Carew JS, Kelly KR, Nawrocki ST. Autophagy as a target for cancer therapy: new developments. Cancer Manag Res 2012; 4: 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E et al. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell 2009; 16: 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Nadanaciva S, Lu S, Gebhard DF, Jessen BA, Pennie WD, Will Y. A high content screening assay for identifying lysosomotropic compounds. Toxicol In Vitro 2011; 25: 715–723. [DOI] [PubMed] [Google Scholar]
- 34Totrov M. Atomic property fields: generalized 3D pharmacophoric potential for automated ligand superposition, pharmacophore elucidation and 3D QSAR. Chem Biol Drug Des 2008; 71: 15–27. [DOI] [PubMed] [Google Scholar]
- 35Vulpetti A, Dalvit C. Fluorine local environment: from screening to drug design. Drug Discov Today 2012; 17: 890–897. [DOI] [PubMed] [Google Scholar]
- 36Zhou C, Zhong W, Zhou J, Sheng F, Fang Z, Wei Y et al. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy 2012; 8: 1215–1226. [DOI] [PubMed] [Google Scholar]
- 37Hu C, Solomon VR, Ulibarri G, Lee H. The efficacy and selectivity of tumor cell killing by Akt inhibitors are substantially increased by chloroquine. Bioorg Med Chem 2008; 16: 7888–7893. [DOI] [PubMed] [Google Scholar]
- 38Checinska A, Soengas MS. The gluttonous side of malignant melanoma: basic and clinical implications of macroautophagy. Pigment Cell Melanoma Res 2011; 24: 1116–1132. [DOI] [PubMed] [Google Scholar]
- 39White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Nishimura M, Naito S, Yokoi T. Tissue-specific mRNA expression profiles of human nuclear receptor subfamilies. Drug Metab Pharmacokinet 2004; 19: 135–149. [DOI] [PubMed] [Google Scholar]
- 41Tagad HD, Hamada Y, Nguyen JT, Hidaka K, Hamada T, Sohma Y et al. Structure-guided design and synthesis of P1' position 1-phenylcycloalkylamine-derived pentapeptidic BACE1 inhibitors. Bioorg Med Chem 2011; 19: 5238–5246. [DOI] [PubMed] [Google Scholar]
- 42Dalvit C, Flocco M, Veronesi M, Stockman BJ. Fluorine-NMR competition binding experiments for high-throughput screening of large compound mixtures. Comb Chem High Throughput Screen 2002; 5: 605–611. [DOI] [PubMed] [Google Scholar]
- 43Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab 2010; 12: 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Marotta R, Falqui A, Curcio A, Quarta A, Pellegrino T. Immunocytochemistry, electron tomography, and energy dispersive X-ray spectroscopy (EDXS) on cryosections of human cancer cells doped with stimuli responsive polymeric nanogels loaded with iron oxide nanoparticles. Methods Mol Biol 2013; 1025: 179–198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.