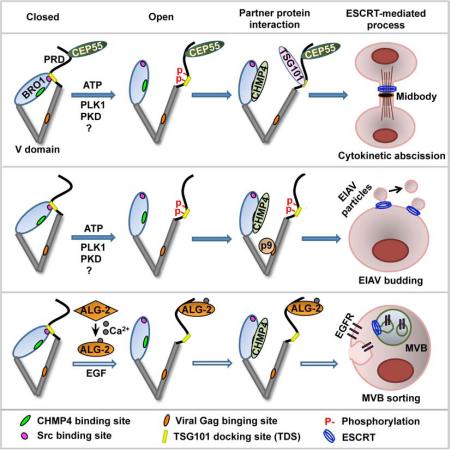
SUMMARY
The modular adaptor protein ALIX is a key player in multiple ESCRT-III-mediated membrane remodeling processes. ALIX is normally present in a closed conformation due to an intramolecular interaction that renders ALIX unable to perform its ESCRT functions. Here we demonstrate that M phase-specific phosphorylation of the intramolecular interaction site within the proline-rich domain (PRD) of ALIX transforms cytosolic ALIX from closed to open conformation. Defining the role of this mechanism of ALIX regulation in three classical ESCRT-mediated processes revealed that phosphorylation of the intramolecular interaction site in the PRD is required for ALIX to function in cytokinetic abscission and retroviral budding but not in MVB sorting of activated EGFR. Thus phosphorylation of the intramolecular interaction site in the PRD is one of the major mechanisms that activates the ESCRT function of ALIX.
Graphical Abstract
INTRODUCTION
The endosome sorting complexes required for transport (ESCRT) proteins constitute an evolutionary conserved membrane remodeling system that buds membranes and severs membrane necks in the opposite topology of endocytosis (Henne et al., 2011; Henne et al., 2013; Hurley, 2015). The best-characterized functions of ESCRT proteins are the multivesicular body (MVB) sorting of internalized membrane receptors (Babst et al., 2002a; Babst et al., 2002b; Katzmann et al., 2001), the budding of enveloped retroviruses from infected cells (Strack et al., 2003; von Schwedler et al., 2003), and the membrane scission at the end of cytokinesis (Carlton and Martin-Serrano, 2007; Morita et al., 2007). The central event in ESCRT-mediated membrane remodeling is the assembly of a highly oligomerized ESCRT-III complex from monomeric proteins (McCullough et al., 2015; Teis et al., 2008), followed by the timely disassembly of this complex by the ATPase Vps4 (Caillat et al., 2015; Yang et al., 2015). Upstream of the core event, ALIX (ALG-2 interacting protein X), in parallel with the ESCRT-I-ESCRT-II complex, binds both a membrane remodeling cargo and the ESCRT-III component CHMP4 to initiate ESCRT-III assembly at the right time and in the right location (Carlson and Hurley, 2012; Pashkova et al., 2013).
Structural studies of ALIX revealed that ALIX consists of an N-terminal banana-shaped Bro1 domain, a middle V letter-shaped domain, and an intrinsically disordered C-terminal proline-rich domain (PRD) (Fisher et al., 2007). The Bro1 domain contains a three-dimensional docking site for CHMP4 (hydrophobic Patch 1) and a linear docking site for Src (hydrophobic Patch 2) (Kim et al., 2005; McCullough et al., 2008). The V domain contains a three dimensional hydrophobic pocket around F676 (the F676 pocket), which is the docking site for retroviral Gag proteins (Lee et al., 2007; Zhai et al., 2008). The PRD contains two linear docking sites (717-720 and 852-855) for the ESCRT-I component TSG101 and an ALIX multimerization sequence (Carlton et al., 2008). Using monoclonal antibodies that recognize different epitopes in ALIX, we discovered that cytosolic ALIX is normally present in a closed conformation that is unable to interact with CHMP4, Src or viral GAG (Zhou et al., 2009; Zhou et al., 2008). This is achieved through an intramolecular interaction between the Src docking site in the Bro1 domain and the first TSG101 docking site (TDS) in the PRD (Zhou et al., 2010). These findings predict that relieving the intramolecular interaction of ALIX is critical for multiple ESCRT-III mediated processes.
In MVB sorting of activated EGFR, calcium-dependent ALG-2 interaction with cytosolic ALIX relieves its intramolecular interaction and promotes its association with the membrane through interaction with membrane-bound CHMP4 (Sun et al., 2015b). ALG-2 knockdown inhibited ALIX-supported MVB sorting and degradation of activated EGFR, but did not affect ESCRT-mediated cytokinetic abscission or retroviral budding (Sun et al., 2015a). These findings identify ALG-2 as a critical regulator of ALIX in calcium-dependent ESCRT processes and predict that a different mechanism activates ALIX for it to function in calcium-independent ESCRT-mediated processes.
The Xenopus ortholog of ALIX, Xp95, is phosphorylated both at the conserved tyrosine residue (Y318) within the Patch 2 of the Bro1 domain (Che et al., 1999) and at multiple sites within the N-terminal half of the PRD (nPRD) during Xenopus oocyte maturation (Dejournett et al., 2007). Since these phosphorylations also occur in ALIX (Dejournett et al., 2007; Schmidt et al., 2005), phosphorylation of one or both of the intramolecular interaction sites may transform ALIX from closed to open conformation. In this study, we demonstrated that mitotic phosphorylation of cytosolic ALIX at the intramolecular interaction site within the nPRD transforms ALIX from closed to open conformation. We also showed that this activating phosphorylation of ALIX is required for ALIX to function in cytokinetic abscission and equine infectious anemia virus (EIAV) budding but not in MVB sorting of activated EGFR.
RESULTS
Cytosolic ALIX changes from closed to open conformation during M phase induction
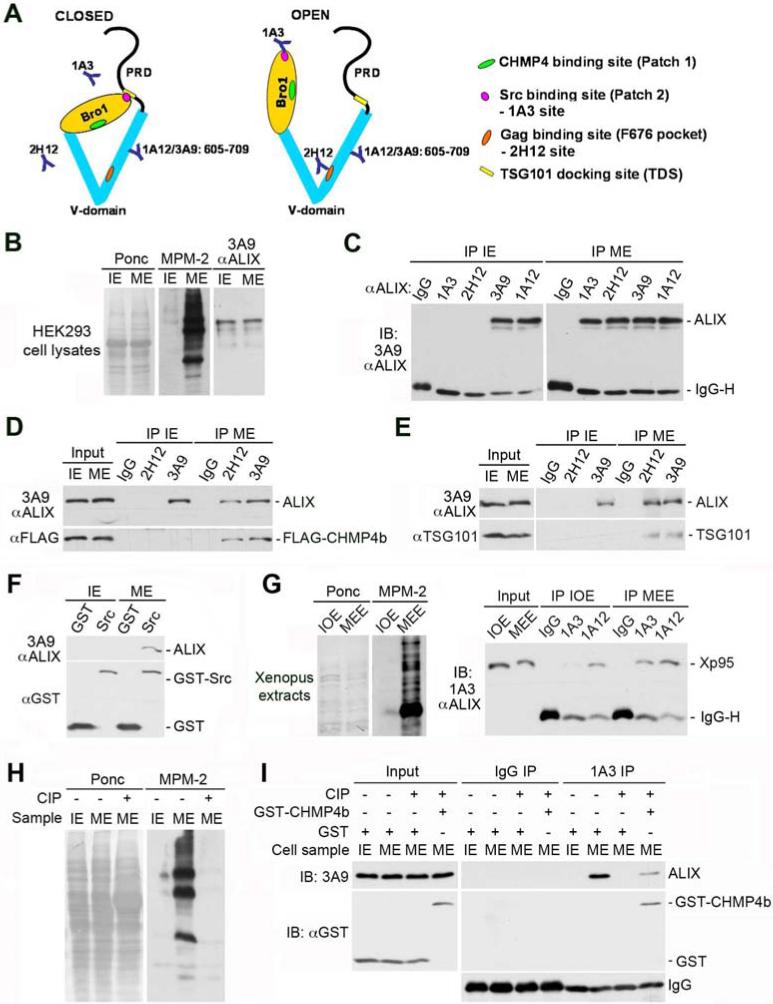
As illustrated in Fig. 1A, the anti-ALIX antibody 1A3 recognizes the Patch2/Src docking site in the Bro1 domain, and the anti-ALIX antibody 2H12 recognizes the F676 pocket/the viral GAG docking site in the V domain. Because the intramolecular interaction of ALIX renders these two partner protein docking sites inaccessible, 1A3 and 2H12 only immunoprecipitate opened ALIX. In contrast, the anti-ALIX antibodies 3A9 and 1A12 recognize conformation-insensitive epitopes within the V domain and thus immunoprecipitate both opened and closed ALIX (Zhou et al., 2010). We used immunoprecipitation (IP) with 1A3 or 2H12 to determine whether cytosolic ALIX undergoes a conformational change when HEK293 cells proceed from interphase (I) to mitosis (M).
Figure 1. Cytosolic ALIX changes from closed to open conformation during M phase induction.
(A) Schematic illustration of the closed and open conformations of ALIX. (B) IB of IE and ME with MPM-2 and 3A9 after Ponceau S (Ponc) staining. (C-E) IP of IE and ME with the indicated antibodies, followed by IB with the indicated antibodies. (F) Incubation of IE and ME with GST or GST-Src, followed by IB of bound proteins with the indicated antibodies. (G) IB of IOE or MEE with MPM-2, and IP of IOE or MEE with the indicated antibodies, followed by IB with 1A3. (H) IB of IE, ME and CIP-treated ME with MPM-2. (I) ME was first incubated with GST or GST-CHMP4b at 4°C for 2 h, and then treated with CIP. IE, ME and the two samples of differently treated ME were immunoprecipitated with the indicated antibodies, followed by IB with the indicated antibodies. See also Figure S1.
Asynchronously growing cells (>95% in I, called I cells) or mitotically arrested cells (>80% in M, called M cells) were extracted with a detergent-free buffer to isolate cytosolic proteins without affecting the intramolecular interaction of ALIX (Zhou et al., 2009; Zhou et al., 2008). ATP and the PP1/PP2A inhibitor microcystin were added to the extraction buffer for M cells to stabilize the M phase status of the extracts. Immunoblotting (IB) of I cell extracts (IE) or M cell extracts (ME) with the mitotic phosphoprotein monoclonal antibody 2 (MPM-2) (Wu et al., 2010) demonstrated a dramatically increased level of protein phosphorylation in M cells. ALIX IB showed that levels of ALIX are similar between I and M cells (Fig. 1B). IP results showed that 1A12 and 3A9 immunoprecipitated ALIX from both IE and ME, whereas 1A3 and 2H12 immunoprecipitated ALIX only from ME (Fig. 1C), indicating a conformational change of ALIX during mitotic entry. To determine whether the difference in ALIX conformation between IE and ME was due to different buffer conditions, we prepared both IE and ME in the presence of ATP and microcystin, and performed parallel IP with 2H12 and 3A9. Under the same buffer condition, 2H12 immunoprecipitated a trace amount of ALIX from IE but a high level of ALIX from ME (Fig. S1A). The trace amount of 2H12-immunoprecipitable ALIX in IE could be from a small percentage of mitotic cells present in I cells. Together, these results indicate that ALIX changes from closed to open conformation during mitotic entry.
To characterize opened ALIX in M cells, we examined ALIX interaction with ectopically expressed CHMP4b, endogenous TSG101 and GST-Src. Both FLAG-CHMP4b and TSG101 coimmunoprecipitated with ALIX in ME but not in IE (Fig. 1D, 1E). GST-Src also specifically pulled down ALIX from ME (Fig. 1F). These results indicate that the opened cytosolic ALIX in mitotic cells is able to interact with multiple partner proteins.
We previously showed that relieving the intramolecular interaction of cytosolic ALIX promotes CHMP4-mediated ALIX association with the membrane in I cells (Sun et al., 2015a; Zhou et al., 2010). To determine whether this also occurs in M cells, we fractionated the post nuclear supernatant (PNS) of crude cell lysates from I or M cells by membrane flotation centrifugation. The distribution of ALIX and CHMP4b between membrane and soluble protein fractions was similar between I and M cells (Fig. S1C), suggesting that membrane-bound CHMP4 in M cells cannot interact with newly opened cytosolic ALIX.
To determine whether Xp95 undergoes a conformational change during oocyte maturation, we immunoprecipitated Xp95 from interphase-arrested Xenopus oocyte extracts (IOE) or M phase-arrested Xenopus egg extracts (MEE), which were also prepared in the absence of detergent, with 1A3 and 1A12. While 1A12 immunoprecipitated Xp95 from both IOE and MEE, 1A3 specifically immunoprecipitated Xp95 from MEE (Fig. 1G), indicating that the M phase-associated conformational change of cytosolic ALIX is a conserved phenomenon that applies to both mitotic and meiotic cycles.
To determine the role of protein phosphorylation in the conformational change of ALIX during mitotic entry, we treated ME with calf intestinal alkaline phosphatase (CIP) and determined the effect on ALIX conformation. CIP treatment both eliminated the MPM-2 reactivity (Fig. 1H) and reversed the 1A3 immunoprecipitability of ALIX (Fig. 1I and Fig. S1B), indicating that protein phosphorylation plays a critical role in the conformational change of ALIX during mitotic entry. Interestingly, if GST-CHMP4b or myc-TSG101 was added to ME before the CIP treatment, the CIP-induced reversal of the 1A3 immunoprecipitability became partial (Fig. 1I and Fig. S1B), indicating that the acquired intermolecular interaction of ALIX decreases the probability of reforming the intramolecular interaction of ALIX after dephosphorylation of mitotic phosphoproteins.
MEE treatment of ALIXnPRD inhibits its interaction with ALIXBro1
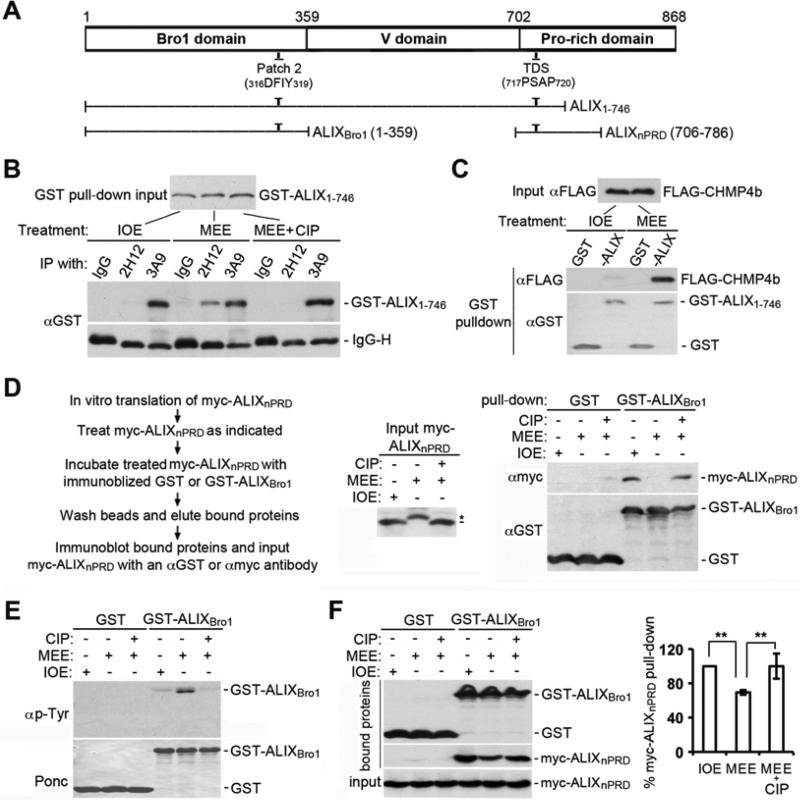
To define the protein phosphorylation that relieves the intramolecular interaction of ALIX in mitotic cells, we first incubated GST tagged ALIX1-746, which contains both of the intramolecular interaction sites (Fig. 2A) (Zhou et al., 2010), with IOE, MEE, or MEE plus CIP, and immunoprecipitated the end products with 3A9 or 2H12. 3A9 immunoprecipitated high levels of GST-ALIX1-746 under all three conditions, whereas 2H12 immunoprecipitated a readily detectable level of GST-ALIX1-746 only after its treatment with MEE (Fig. 2B). That the MEE induces an open conformation of GST-ALIX1-746 was further confirmed by GST-ALIX1-746 pull-down of FLAG-CHMP4b (Fig. 2C).
Figure 2. MEE treatment of ALIXnPRD inhibits its interaction with ALIXBro1.
(A) Schematic illustration of the regions of ALIX that comprise the three structural domains and the ALIX fragments used in this study. (B) GST-ALIX1-746 was phosphorylated with IOE, MEE, or MEE plus CIP, and end products were immunoprecipitated with the indicated antibodies, followed by IB with α-GST. (C) GST or GST-ALIX1-746 was phosphorylated with IOE or MEE, and then absorbed onto GSH beads. The washed beads were used to pull down FLAG-CHMP4b in IE. IB of input and bound proteins with the indicated antibodies. (D) Left: experimental flowchart. Middle: IB of input myc-ALIXnPRD; the short line and the asterisk indicate the start and shift positions of myc-ALIXnPRD, respectively. Right: IB of bound proteins. (E) Phosphorylation of GST or GST-ALIXBro1 with IOE, MEE, or MEE plus CIP, followed by IB of washed proteins with αphosphotyrosine (p-Tyr). (F) Washed GST or GST-ALIXBro1 proteins in (E) were tested for interaction with myc-ALIXnPRD by GST pull-down (n = 3 ± SD).
To identify the ALIX domain through which MEE treatment relieves the intramolecular interaction of ALIX, we produced GST-ALIXBro1 and myc-ALIXnPRD (Fig. 2A) and verified their interaction. We then incubated the in vitro transcription-linked-translation (TNT) product of myc-ALIXnPRD with IOE, MEE, or MEE plus CIP as diagrammed (Fig. 2D, left) and determined the effect on myc-ALIXnPRD interaction with GST-ALIXBro1. Treating myc-ALIXnPRD with MEE but not with IOE or MEE plus CIP induced a gel mobility shift (Fig. 2D, middle). Accordingly, GST-ALIXBro1 pulled down myc-ALIXnPRD treated with IOE or MEE plus CIP, but not with MEE (Fig. 2D, right). We also incubated GST-ALIXBro1 with IOE, MEE, or MEE plus CIP, examined the tyrosine phosphorylation of the washed substrate and determined its interaction with myc-ALIXnPRD. Treating GST-ALIXBro1 with MEE but not with IOE or MEE plus CIP generated immunoreactivity to anti-phosphotyrosine antibodies (Fig. 2E). However, the MEE treatment only moderately reduced the ability of GST-ALIXBro1 to interact with myc-ALIXnPRD (Fig. 2F). Together, these results indicate that the MEE treatment relieves the intramolecular interaction of ALIX mainly through targeting the nPRD.
The S718-S721 phosphorylation inhibits the ALIXnPRD interaction with ALIXBro1
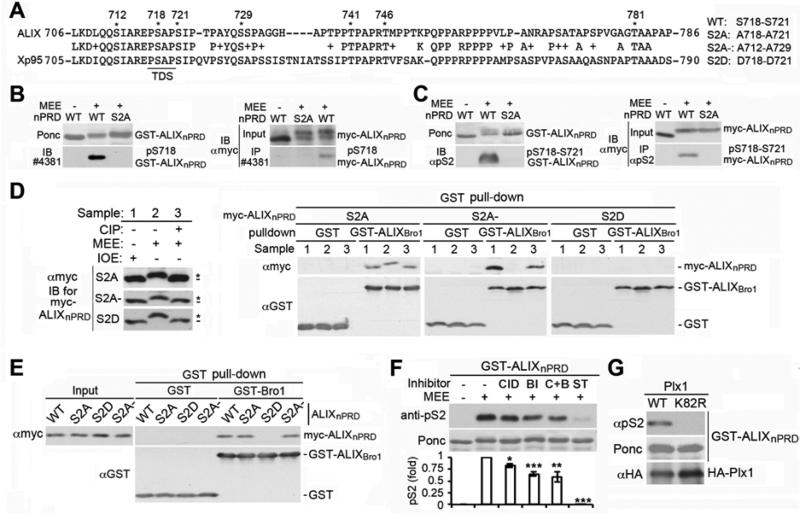
The nPRD of Xp95 is phosphorylated at multiple sites in mature oocytes (Dejournett et al., 2007). Sequence alignment of the nPRD of ALIX with that of Xp95 showed that the nPRD contains seven conserved S/T residues, two of which (S718 and S721 in ALIX) localize at or near the TDS at 717-720 (Fig. 3A). We thus determined whether MEE phosphorylates the S718 and S721 residues in ALIXnPRD.
Figure 3. The S718-S721 phosphorylation inhibits the ALIXnPRD interaction with ALIXBro1.
(A) Sequence alignment of the ALIX nPRD with the Xp95 nPRD, and indication of the conserved S/T sites with asterisks. (B&C) Left: IB of mock treated or MEE phosphorylated GST-ALIXnPRD with #4381 (B) or αpS2 (C). Right: dIP of mock treated or MEE phosphorylated myc-ALIXnPRD with #4381 (B) or αpS2 (C), followed by IB with αmyc. (D) GST or GST-ALIXBro1 pull-down of the indicated forms of myc-ALIXnPRD after their treatment with IOE, MEE, or MEE plus CIP, followed by IB with the indicated antibodies. (E) Pull-down of the indicated forms of myc-ALIXnPRD with GST or GST-ALIXBro1, followed by IB with the indicated antibodies. (F) GST-ALIXnPRD was mock treated or phosphorylated at 22°C for 1 h with MEE in the presence or absence of CID755673 (CID), BI-2536 (BI), both inhibitors (C+B) or staurosporine (ST). After the substrate was immobilized onto GSH beads and washed, bound proteins were probed with αpS2 to determine the level of the S718-S721 phosphorylation (n = 3 ± SD). (G) Phosphorylation of GST-ALIXnPRD with HA-Plx1, followed by IB of the substrate with αpS2.
The Phospho-(Ser/Thr) PKD Substrate Antibody #4381 from CST detects peptides containing a phospho-S/T residue with arginine at the −3 position and leucine at the −5 position, preferring proline at the −1 position. Since the S718 context meets all of these criteria and #4381 preferentially detected a polypeptide about the size of ALIX in ME (Fig. S2A), we used #4381 to determine whether MEE phosphorylates the S718 residue in ALIXnPRD. MEE treatment of the wild type (WT) but not the S718A-S721A (S2A) mutant form GST-ALIXnPRD or myc-ALIXnPRD generated reactivity to #4381, as determined by IB or IP (Fig. 3B). We also generated rabbit polyclonal antibodies against a synthetic ALIX peptide that is phosphorylated at both S718 and S721, and observed that the antibodies, named the anti-pS2 antibody (αpS2), also preferentially recognized a polypeptide about the size of ALIX in ME (Fig. S2B). As observed with #4381, αpS2 specifically recognized WT but not S2A GST-ALIXnPRD or myc-ALIXnPRD upon MEE treatment (Fig. 3C). The recognition was sensitive to CIP treatment (Fig. S2C). Incubation of WT but not S2A GSTALIXnPRD with progesterone-matured oocyte extracts (MOE), freshly prepared in the absence of microcystin, also generated reactivity to αpS2 in a CIP-sensitive manner (Figs. S2D-F). Collectively, these results indicate that MEE phosphorylates S718-S721 in ALIXnPRD.
To determine whether the phosphorylation of ALIXnPRD at S718-S721 is required for MEE-induced inhibition of the ALIXnPRD interaction with ALIXBro1, we produced a phosphomimetic form of myc-ALIXnPRD on S718-S721 (S718D-S721D, S2D) and a control phosphodefective form of myc-ALIXnPRD on S712-S729 (S712A-S729A, S2A-). We then incubated S2A, S2A- or S2D myc-ALIXnPRD with IOE, MEE, or MEE plus CIP, and characterized their interaction with GST-ALIXBro1. None of the double mutations eliminated the gel mobility shift of myc-ALIXnPRD in MEE (Fig. 3D, left), consistent with previous results (Dejournett et al., 2007). However, while S2A myc-ALIXnPRD interacted with GST-ALIXBro1 under all three conditions, S2D myc-ALIXnPRD did not do so under any of the three conditions. Only S2A- myc-ALIXnPRD interacted with GST-ALIXBro1 upon incubation with IOE or MEE plus CIP but not with MEE (Fig. 3D, right). These results indicate that the S718-S721 phosphorylation is required for MEE-induced inhibition of the ALIXnPRD interaction with ALIXBro1.
The S718-S721 phosphorylation may directly inhibit the ALIXnPRD interaction with ALIXBro1 through phosphorylation-produced negative charges or by generating a docking site for a cofactor that prevents the ALIXnPRD interaction with ALIXBro1. To distinguish between these two possibilities, we determined the interaction of different mutant forms of myc-ALIXnPRD with GST-ALIXBro1 in the absence of Xenopus extracts. GST-ALIXBro1 pulled down WT, S2A and S2A- myc-ALIXnPRD but not S2D myc-ALIXnPRD (Fig. 3E), favoring the direct inhibition of the ALIXnPRD interaction with ALIXBro1 by the S718-S721 phosphorylation.
The S718 context fits with the phosphorylation consensus sequences for PKD and PLK1. Since both PKD and PLK1 are activated in mitotic cells (Golsteyn et al., 1995; Kienzle et al., 2013), we determined whether PKD and/or PLK1 are the major kinases in MEE that phosphorylate S718-S721. GST-ALIXnPRD was phosphorylated with MEE in the presence or absence of the PKD inhibitor CID755673 and/or the PLK1 inhibitor BI-2536 or the pan kinase inhibitor staurosporine, and the substrate washed was probed with αpS2. The pan kinase inhibitor dramatically inhibited the phosphorylation of GST-ALIXnPRD. In contrast, the PKD and PLK1 inhibitors reduced the phosphorylation to 83% and 65% of the control level, respectively, and their combined use reduced the phosphorylation to 59% of the control level (Fig. 3F). We also phosphorylated GST-ALIXnPRD with purified Xenopus PLK1 (Plx1). WT but not a catalytically inactive (K82R) Plx1 generated immunoreactivity to αpS2 (Fig. 3G). These results indicate that PKD and PLK1 are among the protein kinases in MEE that phosphorylate S718-S721 in ALIXnPRD.
The S718-S721 phosphorylation relieves the intramolecular interaction of ALIX
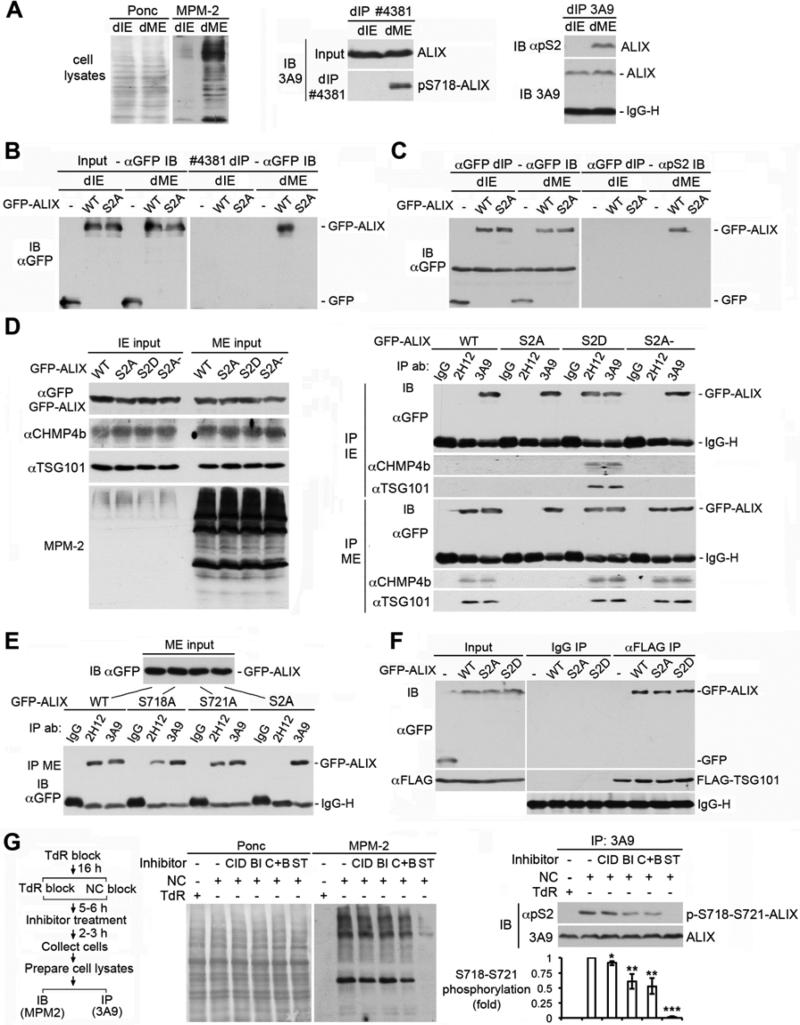
To determine whether ALIX is phosphorylated at S718-S721 in M cells, we determined the recognition of ALIX from I and M cells by #4381 and αpS2. Denatured IE or ME (dIE or dME) were prepared with 1% SDS to preserve the phosphorylation status, and denaturing immunoprecipitation (dIP) was performed after neutralization of SDS with NP-40. Both phosphospecific antibodies differentially recognized ALIX from M cells (Fig. 4A). When WT or S2A GFP-ALIX was expressed in HEK293 cells, both #4381 (Fig. 4B) and αpS2 (Fig. 4C) recognized WT GFP-ALIX but not S2A GFP-ALIX in ME, confirming that the antibody recognition was due to the S718-S721 phosphorylation.
Figure 4. The S718-S721 phosphorylation relieves the intramolecular interaction of ALIX in mitotic cells.
(A) Left: IB of dIE and dME with MPM2. Middle: dIP with #4381, followed by IB with 3A9. Right: dIP with 3A9, followed by IB with 3A9 and αpS2. (B) dIP of IE and ME from cells ectopically expressing WT or S2A GFP-ALIX with #4381, followed by IB with αGFP. (C) dIP of the IE and ME samples in (B) with αGFP, followed by IB with αGFP and αpS2. (D) Left: IB of IE and ME from cells ectopically expressing indicated forms of GFP-ALIX with the indicated antibodies. Right: IP of the IE or ME samples with IgG or the indicated α-ALIX, followed by IB with the indicated antibodies. (E) IP of ME from cells ectopically expressing the indicated forms of GFP-ALIX with IgG or the indicated α-ALIX, followed by IB of input proteins and immunocomplexes with αGFP. (F) IP of IE from cells ectopically coexpressing FLAG-TSG101 and GFP or the indicated forms of GFP-ALIX with αFLAG in the presence of 1% Triton X-100, followed by IB of input proteins and immunocomplexes with the indicated antibodies. (G) Left: experimental flowchart for cell treatments. Middle: IB of IE and ME from collected cells with MPM2. Right: determination of the S718-S721 phosphorylation by dIP with 3A9, followed by IB with 3A9 and αpS2 (n = 3 ± SD). See also Figure S2.
To determine the effect of S718-S721 phosphorylation on the intramolecular interaction of ALIX, we ectopically expressed WT, S2A, S2D or S2A- GFP-ALIX in HEK293 cells and probed their conformation by IP with 2H12. 2H12 immunoprecipitated WT and S2A- GFP-ALIX from ME but not IE, S2A GFP-ALIX from neither IE nor ME, but S2D GFP-ALIX from both IE and ME. IB of the immunocomplexes with αCHMP4b and αTSG101 showed that in all cases, the 2H12 immunoprecipitability correlated GFP-ALIX interaction with CHMP4b and TSG101 (Fig. 4D). These results indicate that the S718-S721 phosphorylation is responsible for relieving the intramolecular interaction of cytosolic ALIX in M cells.
To determine whether phosphorylation of both the S718 and S721 residues is required for relieving the intramolecular interaction of ALIX, we expressed the S718A and S721A single mutant forms of GFP-ALIX, and probed their conformation in M cells by IP with 2H12. While 2H12 did not immunoprecipitate S2A GFP-ALIX, it immunoprecipitated the S718A and S721A single mutant forms of GFP-ALIX at ~40% and ~70% of the level of WT GFP-ALIX, respectively (Fig. 4E). These results indicate that phosphorylation of both the S718 and S721 residues is required for most efficiently relieving the intramolecular interaction of ALIX.
The 717-720 motif is also critically involved in ALIX interaction with TSG101 (Carlton et al., 2008). Since ALIX differentially interacted with TSG101 in mitotic cells (Fig. 1E), we speculated that the phosphorylation of this dual protein interaction site specifically regulates the intramolecular interaction of ALIX without affecting ALIX interaction with TSG101. To test this possibility, we co-expressed FLAG-TSG101 with WT, S2A or S2D GFP-ALIX in HEK293 cells and determined FLAG-TSG101 interaction with each form of GFP-ALIX in IE in the presence of 1% Triton X-100, which disrupts the intramolecular interaction of ALIX (Zhou et al., 2008). FLAG-TSG101 interacted with both mutant forms of GFP-ALIX as efficiently as with WT GFP-ALIX (Fig. 4F), supporting our hypothesis.
To determine the role of PKD and/or PLK1 in the S718-S721 phosphorylation in M cells, we accumulated cells in mitosis in the presence or absence of kinase inhibitors and probed the ALIX phosphorylation with αpS2. As anticipated, the pan kinase inhibitor inhibited both mitotic entry and the S718-S721 phosphorylation. In contrast, the PKD inhibitor slightly inhibited the ALIX phosphorylation without affecting mitotic entry; the PLK1 inhibitor reduced the ALIX phosphorylation to 75% of the control level without affecting mitotic entry; and the combined use of the two inhibitors reduced the ALIX phosphorylation to 61% of the control level without affecting mitotic entry (Fig. 4G). These results indicate that PLK1 and likely also PKD contribute to the S718-S721 phosphorylation in M cells.
The S718-S721 phosphorylation is required for ALIX to function in cytokinetic abscission
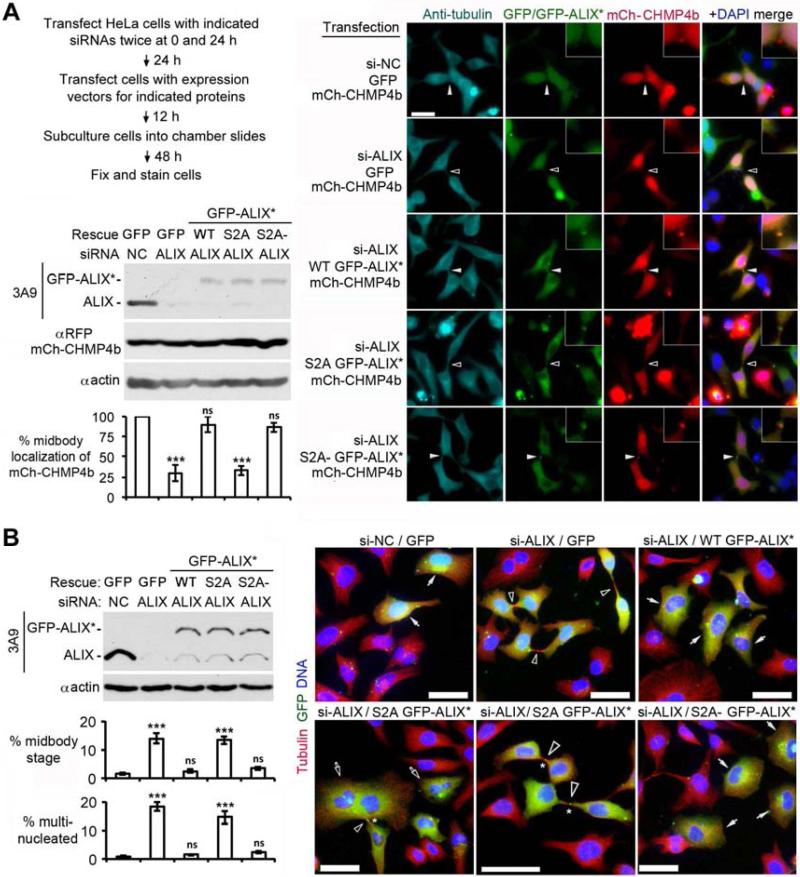
Both ALIX and TSG101 interact with cep55 at the midbody and promote ESCRT-III assembly through recruiting CHMP4 (Carlton and Martin-Serrano, 2007; Morita et al., 2007). Since the cep55 interaction site in ALIX localizes at 801-806 (Carlton et al., 2008), which is outside the region required for the intramolecular interaction of ALIX (Zhou et al., 2010), the S718-S721 phosphorylation is unlikely to be required for ALIX localization at the midbody but may be required for ALIX to recruit CHMP4 to the midbody. To test these predictions, we first examined the effect of ALIX knockdown on the midbody localization of mCherry (mCh)-tagged CHMP4b or TSG101 in HeLa cells. Consistent with the current understanding, ALIX knockdown inhibited the midbody localization of mCh-CHMP4b in >70% of the cells examined but had little effect on the midbody localization of mCh-TSG101 (Fig. S3A and Fig. S3B). We then compared the abilities of different forms of siRNA-insensitive GFP-ALIX (GFP-ALIX*) to rescue the defect of the midbody localization of mCh-CHMP4b. All forms of GFP-ALIX* examined were able to localize at the midbody. However, while WT and S2A- GFP-ALIX* restored the midbody localization of mCh-CHMP4b to near control levels, S2A GFP-ALIX* did not (Fig. 5A). These results demonstrate that the S718-S721 phosphorylation is required for ALIX to recruit CHMP4 to the midbody.
Figure 5. The S718-S721 phosphorylation is required for ALIX to function in cytokinetic abscission.
(A) HeLa cells were transfected with the indicated agents and cultured as diagrammed, followed by IB of cell lysates with the indicated antibodies. Fixed cells were stained with αtubulin and αGFP, and GFP-mCherry double positive cells were scored for the midbody localization of mCh-CHMP4b (n = 3 ± SD). Solid and hollow arrowheads in the representative images indicate the presence or absence of mCh-CHMP4b at the midbody, respectively. Enlargements (3×) depict the midbody area; scale bar: 50 μm. (B) HeLa cells were transfected with the indicated agents and cultured as done in (A), followed by IB of cell lysates with the indicated antibodies. Fixed cells were stained with αtubulin and αGFP, and GFP positive cells were scored for midbody-stage or multinucleation (n = 3 ± SD). Solid and hollow arrows in representative images indicate GFP positive mononucleated and multinucleated cells, respectively; hollow arrowheads indicate midbodies between GFP positive cells; and the asterisks indicate the midbody localization of GFP-ALIX*. Scale bar: 50 μm. See also Figure S3.
The above results predicted that the S718-S721 phosphorylation is required for ALIX to function in cytokinetic abscission. To test this, we compared the abilities of WT, S2A and S2A- GFP-ALIX* to rescue the defect of ALIX knockdown cells in cytokinetic abscission. Consistent with previous observations (Carlton and Martin-Serrano, 2007; Morita et al., 2007), ALIX knockdown increased the percentages of midbody stage and multinucleated cells from <2% to ~14% and ~17%, respectively (Fig. S3C). While the expression of WT or S2A- GFP-ALIX* reduced the percentage of midbody stage or multinucleated cells to near control levels, the expression of S2A GFP-ALIX* did not rescue the cytokinetic abscission defect despite its ability to localize to the midbody (Fig. 5B). The expression of S2A GFP-ALIX* in control knockdown cells did not generate any phenotypes in cytokinetic abscission (data not shown). The intracellular bridge in S2A GFP-ALIX* expressing cells appeared largely normal without the secondary ingression. These results supported our prediction.
Previous studies indicated that ~50% of ALIX knockdown cells had anti-tubulin staining persisting through the Flemming body, whereas ~10% of control cells had such aberrant midbodies (Carlton et al., 2008). Under our experimental conditions, however, only 3% and 12% of control and ALIX knockdown cells had aberrant midbodies, respectively (Fig. S3D). The discrepancy of our observation with that in previous studies could be due to different culture conditions, which may affect the rate of conversion of midbody-stage cells to multinucleated cells.
The S718-S721 phosphorylation is required for ALIX to function in EIAV budding
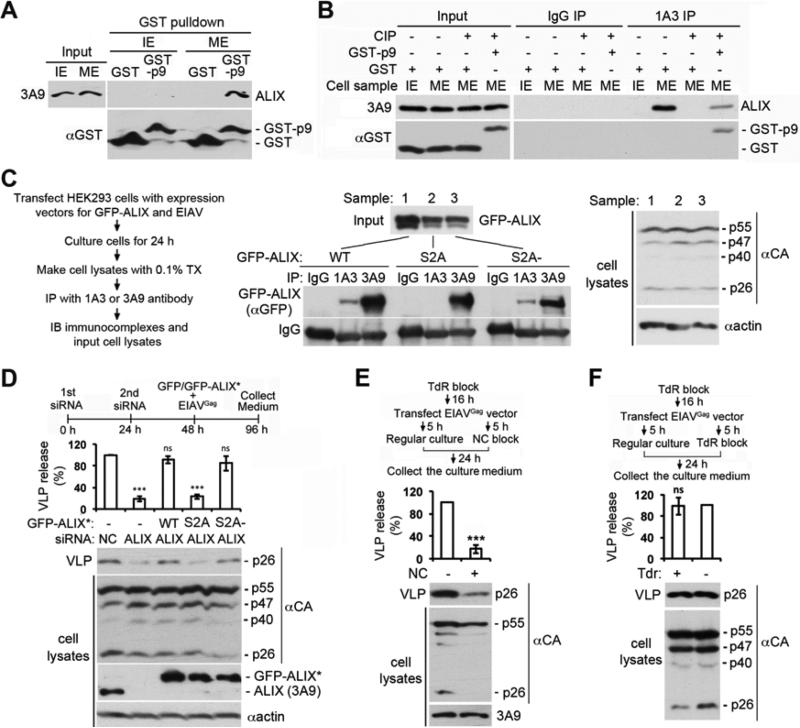
EIAV budding requires ALIX interaction with both CHMP4b and the GAG protein p9 (Strack et al., 2003; von Schwedler et al., 2003). Since relieving the intramolecular interaction of ALIX is required for both events (Zhou et al., 2010), EIAV budding is an ideal model system for defining the role of the S718-S721 phosphorylation in retroviral budding. GST-p9 pulled down ALIX from ME but not IE (Fig. 6A), indicating that the phosphorylation-induced open conformation of ALIX supports ALIX interaction with p9. IP with 1A3 showed that the prior incubation of ME with GST-p9 partially sustained the open conformation of ALIX after ME dephosphorylation (Fig. 6B). GST-p9 also sustained the open conformation of GFP-ALIX when GFP-ALIX was first phosphorylated by MEE and then dephosphorylated by IOE (Fig. S4). These results indicate that the p9 interaction with phosphorylated ALIX helps sustain the open conformation of ALIX after ALIX dephosphorylation.
Figure 6. The S718-S721 phosphorylation is required for ALIX to function in EIAV budding.
(A) Incubation of GST or GST-p9 with IE or ME, followed by IB of input and bound proteins with the indicated antibodies. (B) ME was first incubated with GST or GST-p9 at 4°C for 2 h, and then treated with CIP. IE, ME, and the two samples of differently treated ME were immunoprecipitated with IgG or 1A3, followed by IB with the indicated antibodies. (C) Left: experimental flowchart. Middle: IP of cell lysates with the indicated antibodies, followed by IB with αGFP. Right: IB of cell lysates with the indicated antibodies. (D-F) HEK293 cells were processed as diagrammed. IB of VLPs and cell lysates with the indicated antibodies. Relative levels of the VLP production were determined (n = 3 ± SD). See also Figure S4.
To determine the role of the S718-S721 phosphorylation in EIAV budding, we first coexpressed WT, S2A or S2A- GFP-ALIX with an infection-defective EIAV in HEK293 cells and examined their conformation by IP with 1A3. 1A3 immunoprecipitated a readily detectable level of WT or S2A- GFP-ALIX but not S2A GFP-ALIX (Fig. 6C), indicating that the S718-S721 phosphorylation is required for EIAV expressing cells to generate a significant pool of opened ALIX. We then determined the ability of different forms of GFP-ALIX* to rescue the defect of ALIX knockdown cells in EIAV budding. While WT and S2A-GFP-ALIX rescued the defect, S2A GFP-ALIX did not (Fig. 6D), indicating that the S718-S721 phosphorylation is required for ALIX to function in EIAV budding.
To determine whether the S718-S721 phosphorylation that supports EIAV budding occurs in I or M cells, we determined the effect of mitotic or S phase arrest on EIAV budding. Arresting cells in mitosis greatly inhibited EIAV budding (Fig. 6E), whereas arresting cells in S phase had no effect (Fig. 6F), indicating that the S718-S721 phosphorylation that supports EIAV budding occurs in interphase cells.
The S718-S721 phosphorylation does not affect the function of ALIX in MVB sorting of activated EGFR
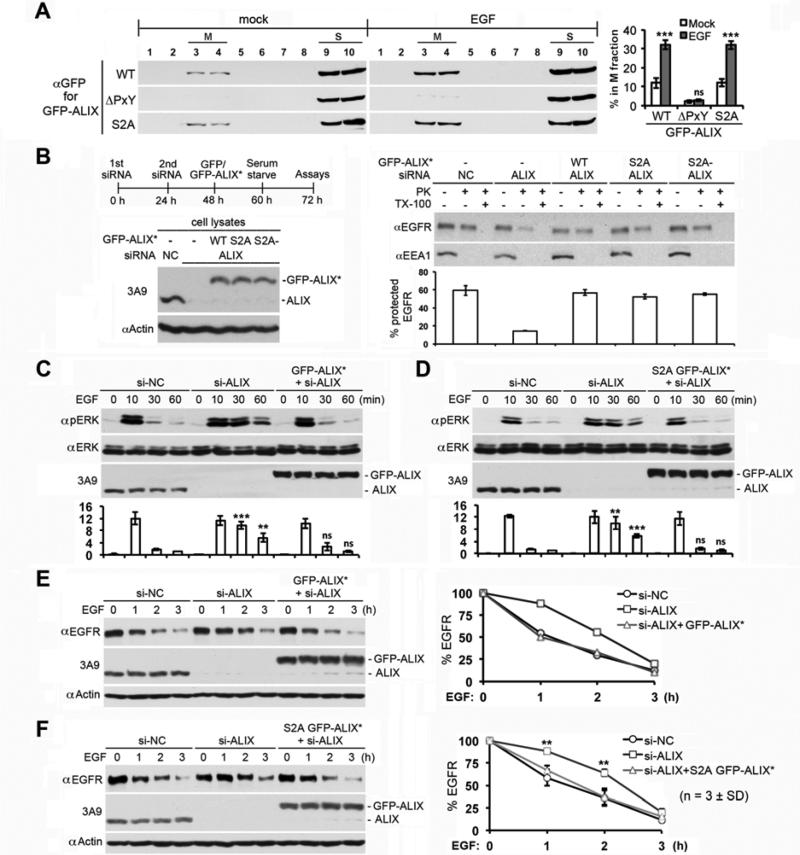
Our previous studies demonstrated that calcium-dependent ALG-2 interaction with ALIX relieves the intramolecular interaction of ALIX in EGF stimulated cells and promotes MVB sorting of activated EGFR (Sun et al., 2015a). To determine whether the S718-S721 phosphorylation may cooperate with the ALG-2-dependent mechanism to fully activate the MVB sorting function of ALIX, we first determined the effect of the S2A mutation on ALIX association with the membrane, which indicates ALIX interaction with membrane-bound CHMP4. ALIX with deletion of the ALG-2 binding site (ΔPxY), which inhibits CHMP4-dependent ALIX association with the membrane, was used as a control (Sun et al., 2015a). As previously observed, EGF stimulation increased the membrane associated WT GFP-ALIX by ~3 fold, and ΔPxY GFP-ALIX barely associated with the membrane irrespective of EGF stimulation. However, S2A GFP-ALIX behaved similarly as WT GFP-ALIX did (Fig. 7A and Fig. S5A), indicating that the S718-S721 phosphorylation does not affect CHMP4-dependent ALIX association with the membrane.
Figure 7. The S718-S721 phosphorylation does not affect the function of ALIX in MVB sorting of activated EGFR.
(A) HEK293 cells ectopically expressing indicated forms of GFP-ALIX were mock-treated or stimulated with EGF for 1 h, and the average percentage of each GFP-ALIX in the M fraction was determined by membrane flotation centrifugation of the PNS (n = 3 ± SD). (B-F) Left: HEK293 cells were processed as diagrammed, and cell lysates were immunoblotted with the indicated antibodies. Right: cells were stimulated with EGF for 30 min, and assayed for MVB sorting of activated EGFR by the proteinase K protection assay (n = 2 ± data range). (C&D) Cells were stimulated with EGF for indicated minutes, and cell lysates were immunoblotted with the indicated antibodies. The relative levels of p-ERK were determined and normalized against the level of si-NC cells at 60 min (n = 3 ± SD). (E&F) Cells were stimulated with EGF for indicated hours, and cell lysates were immunoblotted with the indicated antibodies. The percentages of remaining EGFR at different time points were determined. See also Figure S5.
We then determined the ability of S2A GFP-ALIX to rescue the defect of ALIX knockdown cells in MVB sorting of activated EGFR by the proteinase K protection assay used in our previous studies (Sun et al., 2015a; Sun et al., 2015b). Consistent with our previous results, ALIX knockdown reduced the percentage of protected EGFR from ~60% to ~15%. Importantly, both S2A and S2A- GFP-ALIX* rescued the inhibitory effect of ALIX knockdown as efficiently as WT GFP-ALIX* did (Fig. 7B, right). These results indicate that the S718-S721 phosphorylation does not affect MVB sorting of activated EGFR.
Further, we determined the ability of S2A GFP-ALIX to rescue the defect of ALIX knockdown cells in the timely silencing of activated EGFR. As previously observed, ALIX knockdown prevented the quick and dramatic inactivation of ERK1/2 after 10 min and sustained the ERK1/2 activation at 80% and 50% of the peak level at 30 min and 60 min, respectively. While the effect was rescued by the expression of WT GFP-ALIX* (Fig. 7C), it was not rescued by the expression of ΔPxY GFP-ALIX* (Fig. S5B). However, S2A GFP-ALIX* rescued the sustaining effect of ALIX knockdown on ERK1/2 activation (Fig. 7D), indicating that the S718-S721 phosphorylation does not affect the timely silencing of activated EGFR.
Finally, we determined the ability of S2A GFP-ALIX to reverse the retardation of ALIX knockdown cells in EGFR degradation under EGF continuous stimulation conditions. Consistent with our previous results (Sun et al., 2015b), ALIX knockdown retarded the 50% EGFR degradation from 1 h to 2 h, and the effect was rescued by the expression of WT GFP-ALIX* (Fig. 7E) but not by ΔPxY GFP-ALIX* (Fig. S5C). Importantly, the expression of S2A GFP-ALIX* rescued the retardation effect of ALIX knockdown on EGFR degradation (Fig. 7F), indicating that the S718-S721 phosphorylation does not affect the timely degradation of activated EGFR.
Collectively, these results indicate that the S718-S721 phosphorylation is not involved in generating opened ALIX that supports endolysosomal trafficking of activated EGFR.
DISCUSSION
Mitosis is a special time in the cell cycle when a great portion of protein kinases are simultaneously activated (Daub et al., 2008) and thousands of proteins are robustly phosphorylated (Dephoure et al., 2008; Olsen et al., 2010). Although none of the known ESCRT-mediated processes occurs in mitosis, mitotic cells provide a unique platform to determine the role of protein phosphorylation in the regulation of ALIX conformation. Using this platform, we discovered that the S718-S721 phosphorylation transforms ALIX from closed to open conformation and that multiple protein kinases, including PLK1 and likely also PKD, catalyze the S718-S721 phosphorylation. Following this lead, we demonstrated that the S718-S721 phosphorylation is required for ALIX to function in cytokinetic abscission and EIAV budding, but does not affect the function of ALIX in MVB sorting of activated EGFR. These findings identify the phosphorylation of the intramolecular interaction site within the nPRD as one of the major mechanisms that activates the ESCRT function of ALIX.
Since cells undergoing cytokinetic abscission have already biochemically entered interphase (Gershony et al., 2014), there seems to be a time lag between the activating phosphorylation of ALIX and the function of ALIX in cytokinetic abscission. However, accumulating evidence indicates that multiple activated mitotic kinases and phosphoproteins remain at the midbody much longer than the duration of mitosis. For example, the midbody was specifically recognized by the mitotic phosphoprotein monoclonal antibody MPM-2 (Davis et al., 1983; Vandre et al., 1986), by antibodies that recognize activated MEK, ERK and RSK (Willard and Crouch, 2001), by antibodies that recognize aurora B kinase (Crosio et al., 2002), and by antibodies that recognize citron kinase and its activator RhoA (Madaule et al., 1998). #4381 also stained the midbody area after the robust and widespread mitotic staining disappeared (unpublished results). Therefore, it is possible that some of the mitotic kinases that catalyze the S718-S721 phosphorylation remain active at the midbody. Once transformed into open conformation by the activating phosphorylation, the midbody-localized ALIX recruits CHMP4 and interacts with TSG101. These acquired intermolecular interactions of ALIX in turn inhibit reformation of the intramolecular interaction of ALIX even after ALIX dephosphorylation. We have yet to identify the ALIX activating kinases at the midbody. Although our data indicate that PLK1 is one of the kinases in mitotic cells that catalyzes the activating phosphorylation of ALIX, PLK1 negatively regulates cep55 recruitment to the midbody (Bastos and Barr, 2010), disfavoring this possibility.
EIAV budding neither occurs in M cells nor requires cells to first pass through mitosis. Thus it is somewhat surprising that the S718-S721 phosphorylation is required for ALIX to function in EIAV budding. For this to occur, one potential scenario is that EIAV infection induces the activation of a protein kinase that catalyzes the activating phosphorylation of ALIX. However, when we probed the ALIX phosphorylation in EIAV expressing cells with the phosphospecific antibodies, we did not detect positive signals (unpublished results), suggesting that either EIAV expression did not induce the activating phosphorylation of ALIX, or the induction level was too low to be detected. In any event, it seems improbable that EIAV expressing cells contain a level of the activating phosphorylation of ALIX that can independently explain the ALIX activation required for EIAV budding. Another potential scenario is that EIAV expressing cells contain a low background or induced level of the S718-S721 phosphorylation, which cooperates with a viral factor to generate a significant pool of activated ALIX. Since EIAV expressing cells have high levels of p9 expression, and p9 interaction with phosphorylated ALIX maintains the open conformation of ALIX after ALIX dephosphorylation, it is possible that the combined effects of low levels of ALIX phosphorylation and high levels of p9 expression generate a significant pool of activated ALIX that supports EIAV budding.
EGF-induced downregulation of EGFR is a commonly used model system to study ESCRT-mediated MVB sorting and lysosome targeting of ubiquitinated membrane receptors in vertebrate cells. Our previous studies demonstrated that calcium-dependent ALG-2 interaction with ALIX generates opened ALIX that supports MVB sorting, silencing and degradation of activated EGFR (Sun et al., 2015a). Since EGF-induced signal transduction activates multiple protein kinases, it is possible that the activating phosphorylation of ALIX cooperates with the ALG-2 dependent mechanism to maximally activate the MVB sorting function of ALIX in EGF-stimulated cells. Results in this study disprove this hypothesis.
The MVB sorting of membrane receptors, the retroviral budding, and the cytokinetic abscission are three classical ESCRT-mediated processes that critically involve ALIX function (Bissig and Gruenberg, 2014; McCullough et al., 2013). Although ALG-2 expression is required for ALIX to function in MVB sorting of activated EGFR, it is not important for ALIX to function in EIAV budding or cytokinetic abscission (Sun et al., 2015a). These results are in sharp contrast to the roles of the S718-S721 phosphorylation in the three classical ESCRT-mediated processes. Thus it seems that the two identified mechanisms of ALIX activation are independently involved in distinct ESCRT-mediated membrane remodeling processes. Since only ALG-2 interaction with ALIX requires calcium, it is possible that ALIX activation by ALG-2 is primarily involved in calcium-dependent ESCRT-mediated processes, whereas the activating phosphorylation of ALIX is primarily involved in calcium-independent ESCRT-mediated processes.
EXPERIMENTAL PROCEDURES
Cell culture, transfection and cell synchronization
Culture, transfection and EGF stimulation of HEK293 or HeLa cells were performed as previously described (Sun et al., 2015b). HEK293 cells were synchronized in mitosis through a single thymidine (TdR) (Sigma) block, followed by nocodazole (NC) (Sigma) block (Wu et al., 2014). HEK293 cells were arrested in S phase by a single TdR block. The Plk1 inhibitor BI-2536 (Axon Medchem), PKD inhibitor CID755673 (BioVision Inc.), and pan-kinase inhibitor staurosporine (LC Laboratories) were dissolved in dimethyl sulfoxide (DMSO) and added to culture medium to reach a final concentration of 100 nM, 3 μM, and 50 nM, respectively. The siRNAs, mammalian expression vectors and PCR primers used in this study are summarized in Tables S1, S2 and S3, respectively.
Protein extraction, immunoblotting, immunoprecipitation, and immunostaining
Preparation of crude cell lysates for IB were performed as we previously described (Sun et al., 2015a). Relative signals on immunoblots were quantified by analyzing scanned images with NIH ImageJ version 1.41o. To prepare cytosolic proteins for IP or GST pull-down, pelleted cells were sonicated with 10 volumes of extraction buffer (EB), consisting of 80 mM beta-glycerophosphate, 20 mM EGTA, 15 mM MgCl2, 150 mM NaCl, 1 mM DTT, and proteinase inhibitor cocktail (Sigma), pH 7.4. For extraction of M cells and occasionally also I cells, EB was freshly supplemented with 1 μM microcystin (Sigma) and 1 mM ATP. Cell lysates were cleared by centrifugation at 16,000 g for 10 min at 4°C. CIP (New England Biolabs) was added to ME/MEE at a final concentration of 1 unit/ g substrate proteins. Samples were immunoprecipitated with the indicated antibodies, and immunocomplexes were washed five times with EB. To prepare cell lysates for dIP, pelleted cells were re-suspended with 10 volumes of denaturing buffer consisting of 50 mM Tris-HCl (pH 7.5), 1% SDS and 5 mM DTT, and sonicated. After samples were boiled for 5 min and cleared by centrifugation at 16,000 g for 5 min, they were diluted 10 fold with an SDS-neutralizing IP buffer consisting of 50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM DTT (Tansey, 2007) and proteinase inhibitor cocktail (Sigma). Samples were immunoprecipitated with the indicated antibodies, and immunocomplexes were washed five times with the IP buffer. Production of αpS2 and immunostaining are described in Supplemental Experimental Procedures.
In vitro phosphorylation of ALIX fragments with Xenopus extracts and GST pull-down
MEE and IOE were prepared as previously described (Wu et al., 2010). In vitro transcription and linked translation was performed by using TNT Quick Coupled Transcription/Translation System (Promega). GST and GST-tagged proteins were produced and purified using our standard procedures (Che et al., 1997). Phosphorylation reaction included one volume of substrate proteins and three volumes of IOE or MEE. The reaction was performed at 22°C for 2 h unless otherwise indicated, and terminated by adding SDS-PAGE sample buffer. BI-2536, CID755673, and staurosporine were added to MEE at 4°C 15 min prior to the phosphorylation reaction to reach a final concentration of 2 μM, 5 μM and 5 μM, respectively. GST tagged proteins were absorbed onto glutathione (GSH) beads (GenScript) at 4°C for 2 h. After GSH beads were washed five times with EB, proteins remaining on the beads were eluted with SDS-PAGE sample buffer for IB.
EIAV VLP release assay, membrane flotation centrifugation and proteinase K protection assay
Assays of VLP release from HEK293 cells transfected with the pEV53B EIAVGag vector,fractionation of the PNS of crude lysates of HEK293 cells by membrane flotation centrifugation, and measurement of MVB sorting of activated EGFR by the proteinase K protection assay were performed exactly as we previously described (Sun et al., 2015b).
Statistical analysis
Statistical analyses were performed using Student's t-test.
Supplementary Material
Highlights.
Cytosolic ALIX changes from closed to open conformation during M phase induction
Phosphorylation of the S718-S721 residues produces an open conformation of ALIX
The S718-S721 phosphorylation allows ALIX to function in cytokinetic abscission
The S718-S721 phosphorylation is required for ALIX to support EIAV budding
ACKNOWLEDGEMENTS
We thank Drs. Masatoshi Maki (Nagoya, Japan), Wesley I. Sundquist (Salt Lake City, UT), William G. Dunphy (Pasadena, CA), and Robert Mealey (Pullman, WA) for generously sharing their reagents. This work was supported by NHARP grant 01878, and an IRG grant awarded to J. Kuang, and grants from NIH (P50 CA140388 and CA174798) and Cancer Prevention and Research Institute of Texas (RP150179 and RP150282) awarded to SH Lin. DNA sequencing was performed by the DNA Analysis Facility of The University of Texas Anderson Cancer Center, which is supported by the NIH/NCI grant P30CA016672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.S. designed and performed most of the experiments and participated in manuscript writing; L.S. generated αpS2; X.Z. performed some of the pilot experiments; C.W. and R.W. contributed critical reagents and participated in manuscript preparation; S.L. participated in data analyses and manuscript writing; and J.K. directed the project and wrote the manuscript.
CONFLICT OF INTERESTS
Authors declare no conflict of interests.
REFERENCES
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002a;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002b;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Bastos RN, Barr FA. Plk1 negatively regulates Cep55 recruitment to the midbody to ensure orderly abscission. J. Cell Biol. 2010;191:751–760. doi: 10.1083/jcb.201008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig C, Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 2014;24:19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Caillat C, Macheboeuf P, Wu Y, McCarthy AA, Boeri-Erba E, Effantin G, Gottlinger HG, Weissenhorn W, Renesto P. Asymmetric ring structure of Vps4 required for ESCRT-III disassembly. Nature Commun. 2015;6:8781. doi: 10.1038/ncomms9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Hurley JH. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc Natl Acad Sci U S A. 2012;109:16928–16933. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci U S A. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Che S, El-Hodiri HM, Wu CF, Nelman-Gonzalez M, Weil MM, Etkin LD, Clark RB, Kuang J. Identification and cloning of xp95, a putative signal transduction protein in Xenopus oocytes. J. Biol. Chem. 1999;274:5522–5531. doi: 10.1074/jbc.274.9.5522. [DOI] [PubMed] [Google Scholar]
- Che S, Weil MM, Nelman-Gonzalez M, Ashorn CL, Kuang J. MPM-2 epitope sequence is not sufficient for recognition and phosphorylation by ME kinase-H. FEBS Lett. 1997;413:417–423. doi: 10.1016/s0014-5793(97)00948-4. [DOI] [PubMed] [Google Scholar]
- Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Davis FM, Tsao TY, Fowler SK, Rao PN. Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA. 1983;80:2926–2930. doi: 10.1073/pnas.80.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejournett RE, Kobayashi R, Pan S, Wu C, Etkin LD, Clark RB, Bogler O, Kuang J. Phosphorylation of the proline-rich domain of Xp95 modulates Xp95 interaction with partner proteins. Biochem. J. 2007;401:521–531. doi: 10.1042/BJ20061287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Gershony O, Pe'er T, Noach-Hirsh M, Elia N, Tzur A. Cytokinetic abscission is an acute G1 event. Cell Cycle. 2014;13:3436–3441. doi: 10.4161/15384101.2014.956486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn RM, Mundt KE, Fry AM, Nigg EA. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell. Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kienzle C, Eisler SA, Villeneuve J, Brummer T, Olayioye MA, Hausser A. PKD controls mitotic Golgi complex fragmentation through a Raf-MEK1 pathway. Mol. Biol. Cell. 2013;24:222–233. doi: 10.1091/mbc.E12-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 2015;350:1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci U S A. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Sunshine AB, Rich M, Dunham MJ, Yu L, Piper RC. The yeast Alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev. Cell. 2013;25:520–533. doi: 10.1016/j.devcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH, Dikic I, Bogler O. Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. J. Biol. Chem. 2005;280:3414–3425. doi: 10.1074/jbc.M409839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Sun S, Zhou X, Corvera J, Gallick GE, Lin SH, Kuang J. ALG-2 activates the MVB sorting function of ALIX through relieving its intramolecular interaction. Cell Discovery. 2015a;1:15018. doi: 10.1038/celldisc.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhou X, Zhang W, Gallick GE, Kuang J. Unravelling the pivotal role of Alix in MVB sorting and silencing of the activated EGFR. Biochem. J. 2015b;466:475–487. doi: 10.1042/BJ20141156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey WP. Denaturing protein immunoprecipitation from Mammalian cells. CSH protocols. 20072007 doi: 10.1101/pdb.prot4619. pdb prot4619. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Vandre DD, Davis FM, Rao PN, Borisy GG. Distribution of cytoskeletal proteins sharing a conserved phosphorylated epitope. Eur. J. Cell Biol. 1986;41:72–81. [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Willard FS, Crouch MF. MEK, ERK, and p90RSK are present on mitotic tubulin in Swiss 3T3 cells: a role for the MAP kinase pathway in regulating mitotic exit. Cell. Signal. 2001;13:653–664. doi: 10.1016/s0898-6568(01)00185-1. [DOI] [PubMed] [Google Scholar]
- Wu CF, Liu S, Lee YC, Wang R, Sun S, Yin F, Bornmann WG, Yu-Lee LY, Gallick GE, Zhang W, et al. RSK promotes G2/M transition through activating phosphorylation of Cdc25A and Cdc25B. Oncogene. 2014;33:2385–2394. doi: 10.1038/onc.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CF, Wang R, Liang Q, Liang J, Li W, Jung SY, Qin J, Lin SH, Kuang J. Dissecting the M phase-specific phosphorylation of serine-proline or threonine-proline motifs. Mol. Biol. Cell. 2010;21:1470–1481. doi: 10.1091/mbc.E09-06-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Stjepanovic G, Shen Q, Martin A, Hurley JH. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 2015;22:492–498. doi: 10.1038/nsmb.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- Zhou X, Pan S, Sun L, Corvera J, Lee YC, Lin SH, Kuang J. The CHMP4b and Src docking sites in the Bro1 domain are autoinhibited in the native state of Alix. Biochem. J. 2009;418:277–284. doi: 10.1042/BJ20081388. [DOI] [PubMed] [Google Scholar]
- Zhou X, Pan S, Sun L, Corvera J, Lin SH, Kuang J. The HIV-1 p6/EIAV p9 docking site in Alix is autoinhibited as revealed by a conformation-sensitive anti-Alix monoclonal antibody. Biochem. J. 2008;414:215–220. doi: 10.1042/BJ20080642. [DOI] [PubMed] [Google Scholar]
- Zhou X, Si J, Corvera J, Gallick GE, Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem. J. 2010;432:525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.