Abstract
Nosocomial infectious outbreaks caused by multidrug-resistant Acinetobacter baumannii have emerged as a serious threat to human health. Phosphoproteomics of pathogenic bacteria has been used to identify the mechanisms of bacterial virulence and antimicrobial resistance. In this study, we used a shotgun strategy combined with high-accuracy mass spectrometry to analyze the phosphoproteomics of the imipenem-susceptible strain SK17-S and -resistant strain SK17-R. We identified 410 phosphosites on 248 unique phosphoproteins in SK17-S and 285 phosphosites on 211 unique phosphoproteins in SK17-R. The distributions of the Ser/Thr/Tyr/Asp/His phosphosites in SK17-S and SK17-R were 47.0%/27.6%/12.4%/8.0%/4.9% versus 41.4%/29.5%/17.5%/6.7%/4.9%, respectively. The Ser-90 phosphosite, located on the catalytic motif S88VS90K of the AmpC β-lactamase, was first identified in SK17-S. Based on site-directed mutagenesis, the nonphosphorylatable mutant S90A was found to be more resistant to imipenem, whereas the phosphorylation-simulated mutant S90D was sensitive to imipenem. Additionally, the S90A mutant protein exhibited higher β-lactamase activity and conferred greater bacterial protection against imipenem in SK17-S compared with the wild-type. In sum, our results revealed that in A. baumannii, Ser-90 phosphorylation of AmpC negatively regulates both β-lactamase activity and the ability to counteract the antibiotic effects of imipenem. These findings highlight the impact of phosphorylation-mediated regulation in antibiotic-resistant bacteria on future drug design and new therapies.
Members of the genus Acinetobacter are nonmotile Gram-negative bacteria, many of which cause severe, life-threatening infections and hospital outbreaks (1). Although Acinetobacter baumannii is regarded as an opportunistic pathogen with low virulence, this species infects the soft tissues, bone, bloodstream, and urinary tract and is an important cause of pneumonia and meningitis in immune-compromised patients (2). Crude mortalities because of nosocomial pneumonia and bloodstream infections caused by A. baumannii ranged from 30–75% and 25–54%, respectively (3–5). In intensive care units (ICU), outbreaks of infection caused by multidrug-resistant A. baumannii strains exhibit a crude mortality rate as high as 91.7% (4, 5). The poor outcome in patients with invasive multidrug-resistant A. baumannii infection highlights the urgent need for new therapeutic agents and vaccines to reduce the associated morbidity and mortality.
The survival of A. baumannii is enhanced by its ability to acquire foreign genes, thus increasing the number of vulnerable hosts, producing biofilms, and displaying an open pan-genome (6, 7). These abilities enable A. baumannii to persist in nosocomial environments and to survive even under antibiotic treatment. Numerous studies have reported the emergence of A. baumannii clinical isolates that are resistant to multiple antimicrobials such as carbapenems, colistin, sulbactam, and tigecycline, thus reducing the number of effective therapeutic options (8, 9). In epidemiological studies, the incidence rate of carbapenem-resistant A. baumannii in countries such as Australia, Brazil, Singapore, Canada, South Korea, Taiwan, and Thailand is in the range of 47–80% (10). A study showed that 11% of nosocomial isolates of A. baumannii were carbapenem-resistant; resulting in a morbidity and mortality rate of 52% as compared with a rate of 19% of patients infected with carbapenem-sensitive isolates (4, 11–13). Among the many carbapenem derivatives, imipenem initially was highly effective in the treatment of patients with A. baumannii infections; however, imipenem resistance has been confirmed in 53.7% of Acinetobacter nosocomial infections since the early 1990s (4, 14, 15). The most common pathways leading to carbapenem resistance are associated with the loss of outer membrane porins, overexpression of efflux pumps, and overproduction of Ambler class B metallo-β-lactamases, class D oxacillinases, and AmpC cephalosporinase (16–18). In the case of Acinetobacter-derived cephalosporinase (ADC)1, the key upstream insertion sequence (IS) element, ISAba1, provides promoter sequences that confer bacterial resistance to broad-spectrum cephalosporins (3, 19, 20). In a study of Pseudomonas aeruginosa, the overproduction of AmpC β-lactamase exhibited weak carbapenem-hydrolyzing activity and thus contributed to carbapenem resistance in porin-deficient isolates (21). Although the study suggested a link between AmpC β-lactamase and carbapenem resistance, the regulatory mechanisms remain unclear.
Kinase-induced protein phosphorylation and phosphatase-induced protein dephosphorylation are crucial for signal transduction in both prokaryotic and eukaryotic species (22–26). Hence, bacterial phosphoproteomic analysis is a promising and accurate tool to study biological networks, including the mechanisms of antibiotic resistance. In a recent comparative phosphoproteomic study of A. baumannii ATCC17978 and the multidrug-resistant clinical isolate A. baumannii Abh12O-A2, the relationship between phosphoproteins and antibiotic resistance remained unclear because of the lack of biological confirmation (27). In this study, we used two clinical isolates of A. baumannii to establish comparative phosphoproteomic maps and to conduct biological validation to explore the mechanisms of imipenem resistance (28). Phosphoproteomic analysis of A. baumannii SK17 clinical strains was carried out using a shotgun strategy combined with phosphopeptides enrichment techniques and high-performance mass spectrometry, and thus the identified phosphosites were verified by site-directed mutagenesis (23, 29–31). Our findings clearly show that AmpC β-lactamase activity is regulated by phosphorylation and is involved in imipenem resistance.
EXPERIMENTAL PROCEDURES
Bacterial Strains Growth Conditions and Protein Extraction
The two clinical isolates of A. baumannii used in this study, SK17-S and SK17-R, were originally isolated from the same male patient at a hospital in southern Taiwan (28). Strain SK17-S, which was isolated first, was susceptible to imipenem, whereas strain SK17-R was resistant to imipenem (28). Accordingly, SK17-R harbors plasmid-borne ISAba1-blaOXA-82, a blaOXA-51-like gene with the upstream insertion sequence ISAba1 (GenBank; GQ352402.1), which is widely found in A. baumannii isolates (28, 32).
Acinetobacter baumannii SK17-S was maintained on lysogeny broth (LB) agar plates (BioShop, Burlington, Canada), whereas strain SK17-R on LB agar plates containing imipenem (4 μg/ml; USP, Rockville, MD). Protein extracts were prepared by inoculating a single colony of A. baumannii SK17-S into 5 ml of LB medium, and strain SK17-R into 5 ml LB medium containing imipenem (4 μg/ml). Both cultures were grown at 37 °C with vigorous shaking for 24 h (OD600 nm = 2.0). The cultures were then transferred at a dilution of 1:100 into flasks containing LB medium with or without imipenem (4 μg/ml). After ∼6 h (OD600 nm = 0.8), the cells were pelleted at 3000 × g for 15 min at 4 °C, washed twice with PBS buffer (pH 7.4), resuspended in fresh lysis buffer (25 mm ammonium bicarbonate, PhosSTOP phosphatase inhibitor mixture tablets (Roche, Basel, Switzerland), 6 m urea, and 2 m thiourea, pH 8.0), and disrupted by sonication on ice. Cellular debris was removed by centrifugation at 12,000 × g for 30 min at 4 °C. The supernatant was recovered, and the protein concentrations were determined using the Bradford assay (Bio-Rad, Hercules, CA).
In-solution Protein Digestion and Phosphopeptide Enrichment
Ten milligrams of crude protein was digested using an in-solution method for phosphoproteomic analysis. The protein extracts were reduced using 10 mm dithiothreitol (Sigma, St. Louis, MO) at 37 °C for 1 h and then alkylated with 55 mm iodoacetamide (Sigma) at room temperature in the dark for 1 h. The alkylated proteins were diluted by 4-fold with 25 mm ammonium bicarbonate buffer (pH 8.5) and then incubated overnight at 37 °C with trypsin (Promega, Mannheim, Germany) at a dilution of 1:50 (w/w). The tryptic peptides were centrifuged for 10 min at 6000 × g. The supernatants were desalted using SDB-XC StageTips with SDB-XCEmpore disc membranes (3 M Bioanalytical Technologies, St. Paul, MN) (33), eluted in buffer containing 0.1% trifluoroacetic acid (TFA)/80% acetonitrile (ACN), and then dried in a SpeedVac concentrator (Thermo Electron, Milford, MA) to remove any partial salts of the ammonium bicarbonate. The digested peptides were stored at −20 °C until phosphopeptide enrichment.
Phosphopeptides were enriched using hydroxy-acid-modified metal-oxide chromatography (HAMMOC) with 0.5 mg TiO2 beads (GL Sciences, Tokyo, Japan) packed into 10 μl of C8-StageTips (34). The custom-made HAMMOC tips were washed with solution A (0.1% TFA, 80% ACN), after which solution B (solution A containing lactic acid (300 mg/ml)) was added as a selectivity enhancer to equilibrate the tips. Each tip contained 100 μg of dry digested SK17-S or SK17-R peptides that had been redissolved in solution A and diluted with an equal volume of solution B before loading. Solutions A and B were used to wash the tips and to remove the nonspecific binding of peptides. Sequential elution to obtain pure phosphopeptides was carried out using 0.5 and 5% piperidine (WAKO, Osaka, Japan). The eluted phosphopeptides were acidified in 20% phosphoric acid (WAKO) to pH 2.5, desalted, concentrated as described above, and used in nano-scale liquid chromatography with tandem MS (nanoLC-MS/MS) analysis.
NanoLC-MS/MS Analysis
An online nanoLC-MS/MS LTQ-Orbitrap Velos (Thermo Electron, Bremen, Germany) equipped with a PicoView nanospray interface (New Objective, Woburn, MA) and coupled with a nanoAcquity system (Waters, Milford, MA) were used for sample analysis throughout this study. Enriched phosphopeptides were loaded onto a 75-μm × 250-mm nanoACQUITY UPLC BEH130 column packed with C18 resin (Waters). Elution was carried out at a flow rate of 300 nl/min over a linear gradient of 5–30% ACN in 0.1% formic acid (FA), followed by a sharp increase to 90% ACN and holding at 95% ACN. Instrument control was achieved using Tune 2.6.0 and Xcalibur 2.1 and the HPLC column effluent was directly electrosprayed into the mass spectrometer.
The MS scan range was m/z 300–2000, and the multistage activation (MSA)-MS/MS top20 method was acquired using the Orbitrap analyzer. A survey scan was performed after accumulation to a target value of 5 × 106 ions in the linear ion trap with the resolution set to 60,000 at m/z 400. Peptide ions with charge states ≥2 were selectively isolated to a target value of 5,000 and fragmented in the high pressure linear ion trap by MSA with normalized collision energy of 35%. Neutral loss masses were 97.98, 48.99, and 32.66 Da (single-, double-, and triple-charged phosphopeptides), and the ion selection threshold for MS/MS was 500 counts. The maximum allowed ion accumulation times were 500 ms for full scans and 100 ms for MSA-MS/MS measurements in the LTQ. An activation of q = 0.25 and an activation time of 10 ms were used. For all experiments, standard MS conditions were as follows: spray voltage, 2.0 kV; heated capillary temperature, 200 °C; no sheath and auxiliary gas flow; predictive automatic gain control enabled, and an S-lens RF level of 69%.
Data Validation and Classification
The combined database used in this study was generated from the DNA sequencing results of strains SK17-S and SK17-R. Detailed information regarding DNA sequencing and database generation are provided in the supporting information. Raw files were analyzed using the default settings of MaxQuant versions 1.4.1.2 and 1.3.0.5 (http://www.maxquant.org/) combined with the standard MaxQuant contaminants database and searched against the A. baumannii SK17 database. The search criteria applied for phosphopeptide and phosphosite identification were as follows: trypsin digestion and two missed cleavages were allowed; carbamido-methylation of cysteine (+57.0214 Da) was the fixed modification; methionine oxidation (+15.9949 Da), Ser/Thr/Tyr/Asp/His phosphorylation, and protein N-terminal acetylation were variable modifications; each peptide had a minimum of seven amino acids; and the mass tolerance window was 10 ppm for the parent ion and 0.6 Da for the fragment ions. The false discovery rate (FDR) of the peptides, protein groups, and modification sites was set to 1% for the MS/MS spectra automatically processed by MaxQuant for statistical validation and quantification. The minimum Maxquant score for phosphorylation sites was 25. The acceptance criteria of the localization probabilities of all Ser/Thr/Tyr/Asp/His phosphosites were applied, including only peptides with a localization probability ≥75% (calculated based on the post-translational modification (PTM) score) (35). The assembled contigs in fasta format were uploaded to the RAST server (http://rast.nmpdr.org/) to identify the protein encoding, rRNA, and tRNA genes to assign gene functions and thereby classify biological functions (36). The Uniprot and Protein Information Resource databases were used in parallel as complementary methods to further classify each identified protein.
Homology Modeling
A homology modeling approach was used to model SK17 AmpC and to investigate the importance of each conserved Ser residue (Ser-81, Ser-88, and Ser-90) identified in this study. The refined 1.2Å resolution crystal structure of ADC-1, the extended-spectrum class C β-lactamase Acinetobacter-derived cephalosporinase (Protein Data bank (PDB): 4net), was used to construct the active site of SK17 AmpC, based on the 99% sequence identity between these two enzymes (37). The bioinformatics tools PDB, and Discovery Studio 4.0 (Accelrys, San Diego, CA) were used to build a homology model using default protocols. The structure file was visualized using the PyMol molecular graphics system (http://www.pymol.org).
Construction of AmpC Expression Plasmids and Site-Directed Mutagenesis
The ampC of A. baumannii SK17 was optimized for expression in the multiple antimicrobials-susceptible strain A. baumannii 290 (Ab290) isolated from Taipei Veterans General Hospital in Taiwan in 1999 (38–40). The conserved forward primer sequence of ISAba1, located upstream of ampC to facilitate efficient production of the enzyme, and the conserved reverse primer of ampC were used in this study (32). The PCR product was cloned between the XbaI and XhoI sites of pYMAb2 and transformed into chemically competent Escherichia coli JM109 cells (39). The primers for ISAba1-ampC wild-type (WT) and the site-directed mutants (S81A, S81D, S88A, S88D, S90A, S90D, S88A/S90A, S88A/S90D, S88D/S90A, S88D/S90D, S81A/S90A, S81A/S90D, S81D/S90A, and S81D/S90D) were synthesized based on the criteria of site-directed ligation-independent mutagenesis (SLIM) (supplemental Table S7) (41). The PCR products of randomly selected isolates were verified by sequencing (Genomics, New Taipei City, Taiwan). For the expression and purification of the WT and mutant forms of AmpC, the respective plasmids were first amplified from E. coli JM109 and then electro-transformed into Ab290 cells.
Electrotransformation of Plasmid-borne ISAba1-AmpC
The recombinant shuttle vector pYMAb2, which contains the kanamycin resistance determinant and the entire ISAba1-ampC sequence (WT or mutants), or the control vector (pYMAb2 only), was extracted from donor E. coli JM109 cells using a Plasmid Mini Kit (Viogene-BioTek, New Taipei City, Taiwan). These recombinant pYMAb2 plasmids were transformed into the kanamycin-susceptible strain Ab290 by electroporation using a gene pulser electroporator (Bio-Rad) and 2-mm electrode gap cuvettes (7, 28, 39). Briefly, electrocompetent cells were prepared as follows: overnight cultures of strain Ab290 were diluted 1:100 and grown to the exponential phase (OD600 nm = 0.5–0.7). The cells were collected at 4 °C by centrifugation, washed with ice-cold 10% glycerol in double-distilled water (ddH2O) three times, and resuspended in the same buffer. Electroporation was conducted at 2.0 kV with the pulse controller at a parallel resistance of 200 Ω and a capacity of 25 μF (42). The electroporated cell mixture was suspended in 2 ml of LB broth and incubated at 37 °C for 1 h with shaking. The cells were then plated on kanamycin agar plates (25 μg/ml) and incubated overnight at 37 °C, and the kanamycin-resistant transformants were selected. The ampC of the recombinant plasmids was verified by colony PCR (38, 43).
Purification of AmpC and Mutant Proteins
Colonies of strain Ab290 containing the distinct plasmids were grown overnight at 37 °C in 100 ml of LB medium containing kanamycin (25 μg/ml). Overnight cultures of the transformants were diluted 1:100 into 1000 ml of fresh medium and grown for 6 h (OD600 nm = 0.8–1.0). The cells were collected at 4 °C by centrifugation at 6000 × g for 25 min at 4 °C, re-suspended in cold lysis buffer (20 mm Tris-HCl pH 7.5, 500 mm NaCl, 20 mm imidazole), and then disrupted using a French-press. Cell debris was removed by centrifugation at 8000 × g for 25 min at 4 °C; the clear supernatant fluid was filtered through 0.45-μm pore size filters (Millipore, Bedford, MA). The hexahistidine-tagged proteins from strain Ab290 carrying wild-type AmpC (AmpC-WT) or mutants (S90A and S90D) were purified from a Ni2+-NTA column (Sepharose 6 Fast Flow resin, GE Healthcare, Piscataway, NJ; Econo-Pac column, Bio-Rad), and eluted in 200 mm imidazole (44). Fractions containing the desired protein were identified by 12.5% SDS-PAGE, based on the presence of a 43 kDa band, and then concentrated using an Amicon Ultra-15 filter (Millipore).
β-Lactamase Assay
To correlate the AmpC phosphorylation status with β-lactamase activity, the purified hexahistidine-tagged AmpCs (WT, S90A, and S90D) were treated prior to the assays with alkaline phosphatase (Thermo Scientific, Pittsburgh, PA) or with reaction buffer only for 1 h at 37 °C (45, 46). The β-lactamase activities of phosphatase-untreated (control) and phosphatase-treated (dephosphorylated) recombinant AmpCs were then analyzed using nitrocefin (BioVision, Milpitas, CA) as the substrate. AmpC activity of the purified enzymes was determined spectrophotometrically at 500 nm at room temperature by measuring the rate of hydrolysis (milliOD/min). The first 10 min of the linear enzymatic data were collected, and the hydrolytic activity in each reaction was measured at a final substrate (nitrocefin) concentration of 100 μm and a final enzyme concentration of 0.1 nm (47, 48). All kinetic assays were carried out in PBS buffer (pH 7.4) in a microplate reader Paradigm (Molecular Devices, Sunnyvale, CA). The kinetic values were calculated and plotted using GrahPad PRISM 5.0 (GraphPad Software, La Jolla, CA).
Antibiotic Resistance Profiles and Identification
The differential susceptibilities of the clinical strains (SK17-S and SK17-R) and Ab290 transformants (pYMAb2 only, AmpC-WT, S81A, S81D, S88A, S88D, S90A, S90D, S88A/S90A, S88A/S90D, S88D/S90A, S88D/S90D, S81A/S90A, S81A/S90D, S81D/S90A, and S81D/S90D) to imipenem (5 or 10 μg) and ceftazidime (20 or 30 μg) (Sigma) were compared in a standard disc diffusion assay (49). Ceftazidime, a third-generation cephalosporin, can serve as a substrate of AmpC. A previous study showed that ceftazidime resistance in A. baumannii can arise as a consequence of increased expression of chromosomal ampC (50). Here the assay was carried out as follows: overnight cultures of A. baumannii clinical strains and transformants were spread evenly on Mueller-Hinton (M-H) agar plates (BD Biosciences, Sparks, MD). The antibiotic resistance of each strain was determined using discs containing imipenem (5 or 10 μg) and ceftazidime (20 or 30 μg). After incubating the plates at 37 °C for 24 h, the inhibition zone diameters were measured to the nearest millimeter.
Neutralization of Imipenem by Purified AmpCs
Because the strain SK17-S is imipenem-susceptible, it was used as the test strain in disc diffusion assays to determine the different imipenem-susceptibility profiles conferred by AmpC-WT, S90A, and S90D (51, 52). A fixed amount (5 μg) of imipenem was mixed with various amounts (4–7.5 μg) of recombinant AmpCs in a final volume of 10 μl. These mixtures were incubated for 1 h at 37 °C and then loaded onto the discs. Blank discs with imipenem (5 μg), purified AmpC-WT protein, or mutant proteins (S90A and S90D) (7.5 μg) alone served as the controls. Each disc was placed on MH agar plates containing an overnight culture of SK17-S. Following incubation of the plates at 37 °C for 24 h, the differential susceptibility profile of SK17-S to imipenem was determined by measuring the inhibition zones as described previously (53).
RESULTS
Comparison of A. baumannii SK17-S and SK17-R Phosphoproteomes
Based on the whole shotgun genome sequences of SK17-S and SK17-R, the two strains are highly similar except that the strain SK17-R harbors a plasmid containing the ISAba1-blaOXA-82 gene (28, 32). The high genetic similarities between SK17-S and SK17-R suggest that they are suitable for investigating imipenem resistance. Phosphoproteomic analyses of SK17 were conducted using the shotgun LC-MS/MS approach to obtain large-scale data sets from biological triplicate experiments for each strain. Using the robust HAMMOC enrichment method and high mass accuracy LTQ-Orbitrap Velos, we identified 248 unique phosphoproteins with 351 phosphopeptides from SK17-S (supplemental Table S1), as well as 211 unique phosphoproteins with 240 phosphopeptides from SK17-R (supplemental Table S2) (34, 44, 54). Only 70 phosphoproteins overlapped in these two strains; 178 and 141 uniquely identified phosphoproteins were found in SK17-S and SK17-R, respectively (Fig. 1 and supplemental Table S3). That there were so many more uniquely identified phosphoprotiens than overlapping suggested the basis for the differential phosphorylation-dependent regulation of imipenem resistance between SK17-S and SK17-R. Therefore, these phosphoproteins are potential candidates responsible for imipenem resistance for further validation.
Fig. 1.
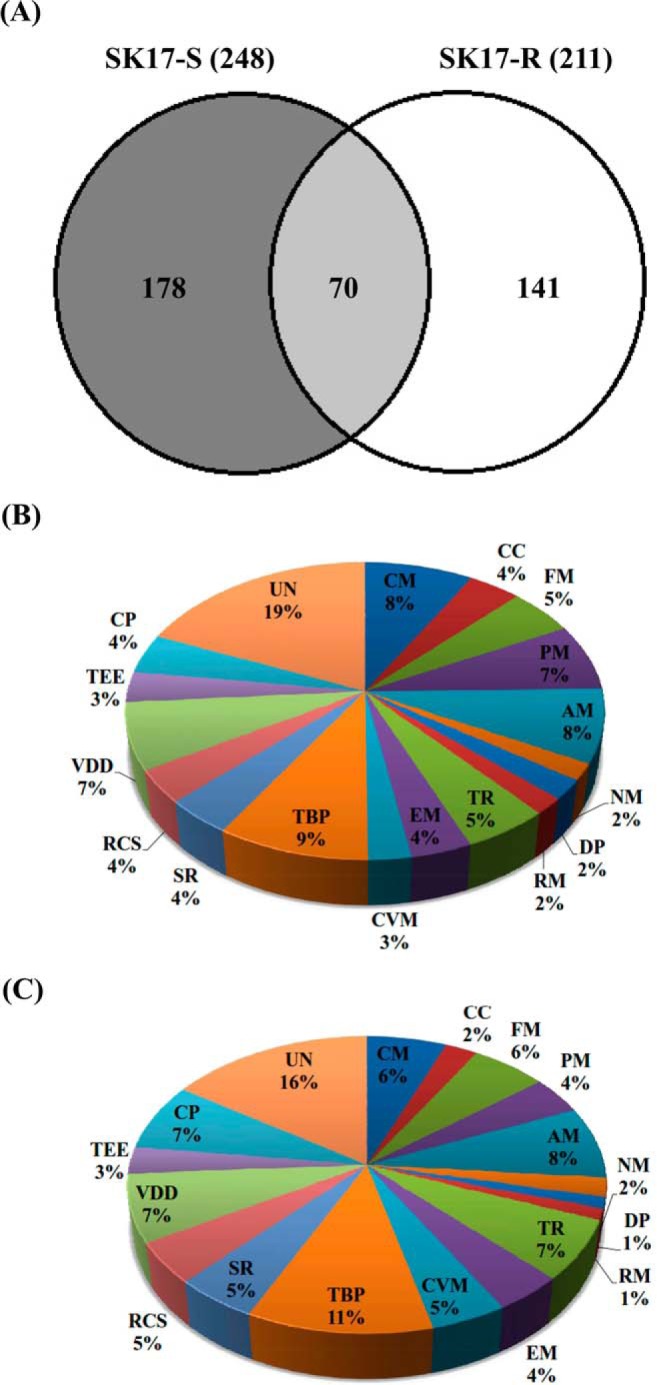
Classification of the identified phosphoproteins in A. baumannii strain SK17. A, Venn diagrams are used to show the proportion of identified phosphoproteins shared by A. baumannii strains SK17-S and SK17-R. The identified phosphoproteins are grouped by biological function in SK17-S (B) and SK17-R (C). Abbreviations: CM, Carbohydrate metabolism; CC, Cell wall and capsule; FM, Fatty acids lipids and isoprenoids metabolism; PM, Protein metabolism; AM, Amino acid metabolism; NM, Nucleosides and nucleotides metabolism; DP, DNA binding proteins; RM, RNA metabolism; TR, Transcription; EM, Energy metabolism; CVM, Cofactors and vitamins metabolism; TBP, Transport and binding proteins; SR, Stress response; RCS, Regulation and cell signaling; VDD, Virulence disease and defense; TEE, Transposable and extrachromosomal elements; CP, Cellular processes; and UN, Unknown.
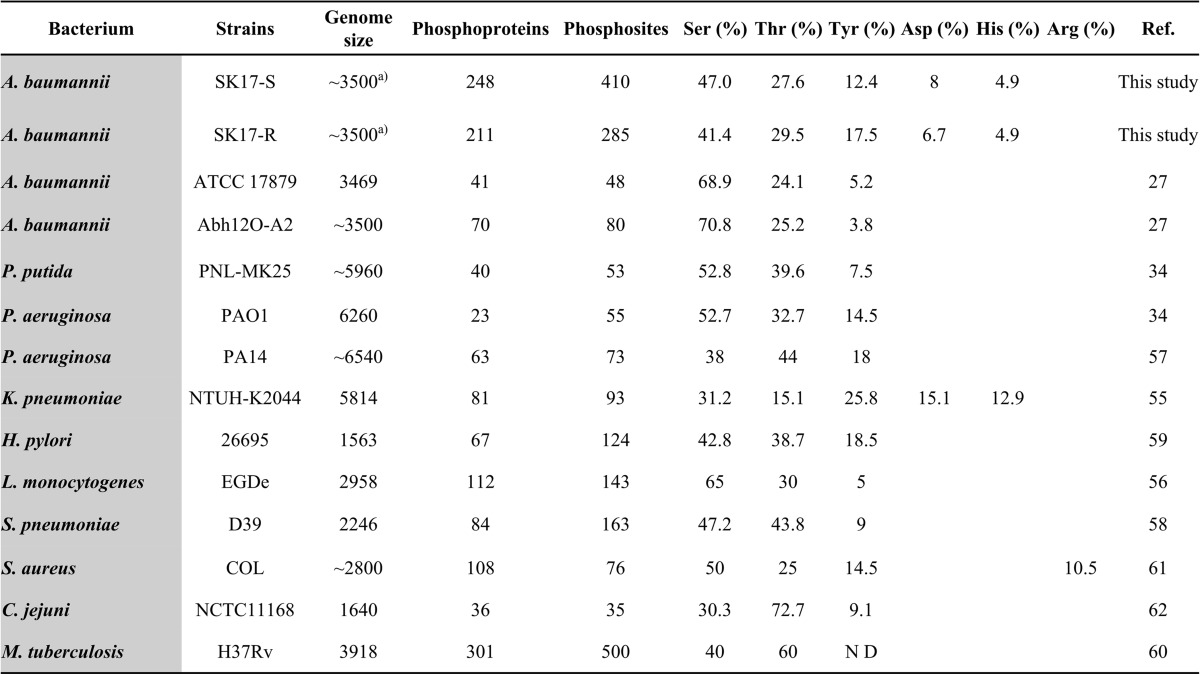
Of the 410 phosphosites identified in SK17-S, 193 (47.0%) were Ser, 113 (27.6%) Thr, 51 (12.4%) Tyr, 33 (8.0%) Asp, and 20 (4.9%) His (Table I). Of the 285 phosphosites in SK17-R, 118 (41.4%) were Ser, 84 (29.5%) Thr, 50 (17.5%) Tyr, 19 (6.7%) Asp, and 14 (4.9%) His (Table I). These phosphopeptides were calculated using the PTM score and were confirmed manually (class I, p > 0.75). The annotated MS/MS spectra of the phosphopeptides identified in SK17-S and SK17-R are shown in supplemental Fig. S1 and S2, respectively. Notably, the extra-plasmid ISAba1-blaOXA-82 (GenBank: GQ352402.1) of SK17-R was included in our database but none of the phosphopeptides encoded by the plasmid were observed in this study. Thus, the relationship between the plasmid ISAba1-blaOXA-82 encoded proteins and imipenem resistance remains unclear and requires further analysis.
Table I. Comparison of the A. baumannii strain SK17 phosphoproteome with other pathogenic bacterial phosphoproteomes.
a Incomplete sequence, ND: not detectable.
According to site-specific phosphoproteomic analysis of pathogenic bacteria, we found that the numbers of phosphoproteins/phosphosites detected in A. baumannii SK17-S and SK17-R were higher than in other pathogenic bacteria (Table I). Nevertheless, our results were comparable to those reported for Mycobacterium tuberculosis which has a similar genome size (Table I) (27, 34, 55–62). Accordingly, the phosphoproteins identified from SK17-S accounted for 8.1% all open reading frames encoded in the genome of A. baumannii SK17 compared with 7.5% of M. tuberculosis (56, 63). In the phosphoproteomic results of four strains of A. baumannii (ATCC17978, Abh12O-A2, SK17-S, and SK17-R), we found eight overlapping phosphopeptides from the 18 corresponding phosphoproteins (supplemental Table S4). Based on our results for the imipenem-resistant strain SK17-R and the previous results for multidrug-resistant strain Abh12O-A2, with both strains being resistant to carbapenem-type antibiotics, we sought to improve the understanding of phosphorylation-dependent antibiotic resistance to carbapenems in A. baumannii (27).
Classification of the Identified Phosphoproteins
To gain insight into the biological functions of the phosphoproteins identified in SK17-S and SK17-R, the phosphoproteins were classified into 18 groups based on their Gene Ontology (GO) assignments (Fig. 1B, 1C, supplemental Table S1 and S2). Approximately 80% of the identified phosphoproteins showed clear functional tendencies (Figs. 1B and 1C). Subcellular localization analysis indicated that most of the phosphoproteins could be assigned to specific locations, whereas only 23 and 17% of the phosphoproteins in SK17-S and SK17-R, respectively, were annotated as unknown (supplemental Fig. S3). Most identified phosphoproteins were assigned to the cytoplasm, and the phosphoproteins in the two strains showed similar distributions for subcellular localization (supplemental Fig. S3).
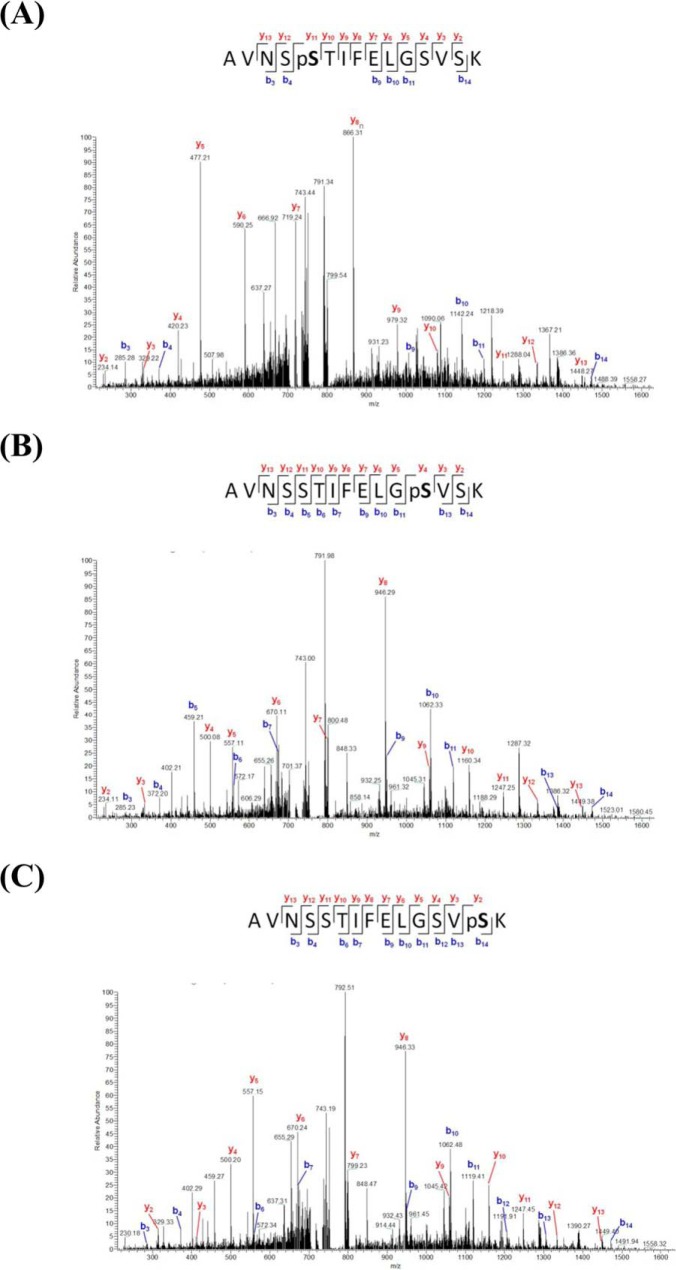
To determine the correlation between the identified phosphoproteins and antibiotic resistance, we focused on two functional categories: transport and binding proteins (TBP), and virulence, disease and defense (VDD). Among the identified phosphoproteins from SK17-S and SK17-R are 25 (9%) and 27 (11%), respectively, that are related to TBP, including numerous factors associated with antibiotic pump efflux systems in bacterial antibiotic resistance (supplemental Table S5) (64). Similarly, phosphoproteins classified in the VDD category contributed to pathogenesis and antibiotics resistance, although the phosphoproteins were present in the same percentages (7%) in both strains (supplemental Table S6) (18). Interestingly, one of the identified phosphoproteins, a class C β-lactamase (470.191.peg.910; AmpC, cephalosporinase), was previously shown to be responsible for the resistance to multiple β-lactam antibiotics (65). Eight phosphopeptides of AmpC β-lactamase were identified in SK17-S but only two phosphopeptides were identified in SK17-R, indicating that the low level of AmpC phosphorylation contributed to imipenem resistance in SK17-R (supplemental Table S6). The multiple phosphorylated AmpCs in SK17-S contained three phosphosites, Ser-81, Ser-88, and Ser-90, located in the active-site phosphopeptide AVNSS81TIFELGS88VS90K91 (Fig. 2).
Fig. 2.
MS/MS spectra of the serine-phosphorylated peptides of AmpC β-lactamase. Rich backbone fragmentation spectra of the active-site phosphopeptides (AVNSS81TIFELGS88VS90K91) carry Ser phosphosites on Ser-81 (A), Ser-88 (B), and Ser-90 (C). The m/z values and relative intensities of the measured fragment signals are shown. The b- and y-ion series are almost complete and labeled in blue and red, respectively.
Effects of AmpC Phosphorylation on Antibiotics Susceptibility
Overexpression of β-lactamases (AmpC, TEM, VEB, PER, CTX-M, SHV, OXA, IMP, and VIM) is a common mechanism in the resistance to β-lactam antibiotics (66, 67). To verify the structure-function relationships of the identified phosphosites, a homology model of AmpC based on ADC-1 (with 99% sequence identity) was generated (Fig. 3A and supplemental Fig. S4) (37). According to the model, Ser-88 and Ser-90 are located in the catalytic motif S88VS90K91 of typical β-lactamases (domain 2), whereas Ser-81 and Thr-151 are positioned relatively close to the protein surface, based on their locations in domain 1 and the P2-loop domain, respectively (Fig. 3A) (37, 68). The hydrolytic reaction catalyzed by AmpC β-lactamase includes two steps, acylation and deacylation (69, 70). First, Lys-91, aligned as Lys-67 in E. coli, may act as a general base and contribute to the deprotonation of Ser-88. The deprotonated Ser-88, aligned as Ser-64, attacks the β-lactam ring carbon of the substrate and forms a covalent acyl-enzyme complex in the acylation step (69, 70). Second, catalytic water reacts with the covalent linkage in the acyl-enzyme complex leading to release of the hydrolyzed product in the deacylation step (69, 70). Additionally, although residues Ser-88 and Lys-91 have been thoroughly examined, the function of the highly conserved Ser-90 remains unknown (51, 71, 72).
Fig. 3.
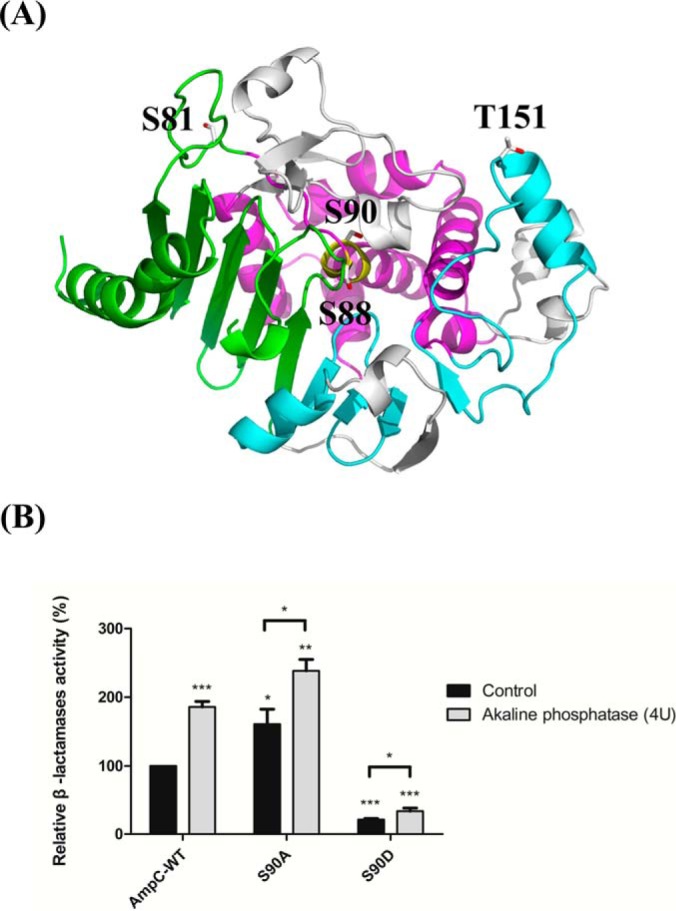
The homology model of AmpC and β-lactamase assays of recombinant AmpCs. The model was generated using the A. baumannii ADC-1 (PDB entry 4net) as the template and the identified phosphosites were structurally mapped. A, The cartoon representation of the overall structure shows the predicted relative positions of each phosphosite (white sticks). The phosphosites Ser-88 and Ser-90 are both located in the motif S88VS90K91 (yellow) of domain 2 (magenta). Ser-81 and Thr-151 are located close to the surface of domain 1 (green) and in the P2-loop (cyan), respectively. B, The relative β-lactamase activity of recombinant AmpCs (WT, S90A, and S90D) in the presence or absence of alkaline phosphatase. The data are presented as the means with error bars based on experiments carried out in triplicate. The values were all normalized to that of phosphatase-untreated AmpC-WT. Asterisks (*) indicate statistical significance (p < 0.05).
To validate the importance of all three serine phosphosites (Ser-81, Ser-88, and Ser-90) in the AmpC, transformants carrying plasmids of AmpC-WT or site-specific mutants (S81A, S81D, S88A, S88D, S90A, S90D, S81AS90A, S81AS90D, S81DS90A, S81DS90D, S88AS90A, S88AS90D, S88DS90A, and S88DS90D) were examined using the disc diffusion method with imipenem (10 μg) (Table II) (9, 50, 68). After normalization of the results with those obtained from the control strain containing the unmodified vector, the AmpC-WT, S81A, S81D, and S90A transformants showed moderate sensitivity (46.4–57.3%) (Table II). The findings that strains S81D and S81A both retained their resistance to imipenem rule out a correlation between Ser-81 phosphorylation and imipenem resistance (Table II). In contrast, the S90D strain was much more sensitive to imipenem than was the S90A strain (84.3% versus 50.2%) (Table II). According to the results of the disc diffusion assay with 5 μg imipenem, the slightly imipenem-sensitive (40.6%) transformants, S81A and S81A/S90A, exhibited a clear role for nonphosphorylatable AmpC in imipenem resistance (supplemental Table S8). Consistent with these results, analysis of double mutants containing Ser-81 and Ser-90 (S81A/S90A, S81D/S90A, S81A/S90D, and S81D/S90D) showed that mutants containing the S90A mutation were more resistant to imipenem than those containing the S90D mutation (supplemental Table S8).
Table II. Antibiotic susceptibility of A. baumannii strain SK17 and transformants.
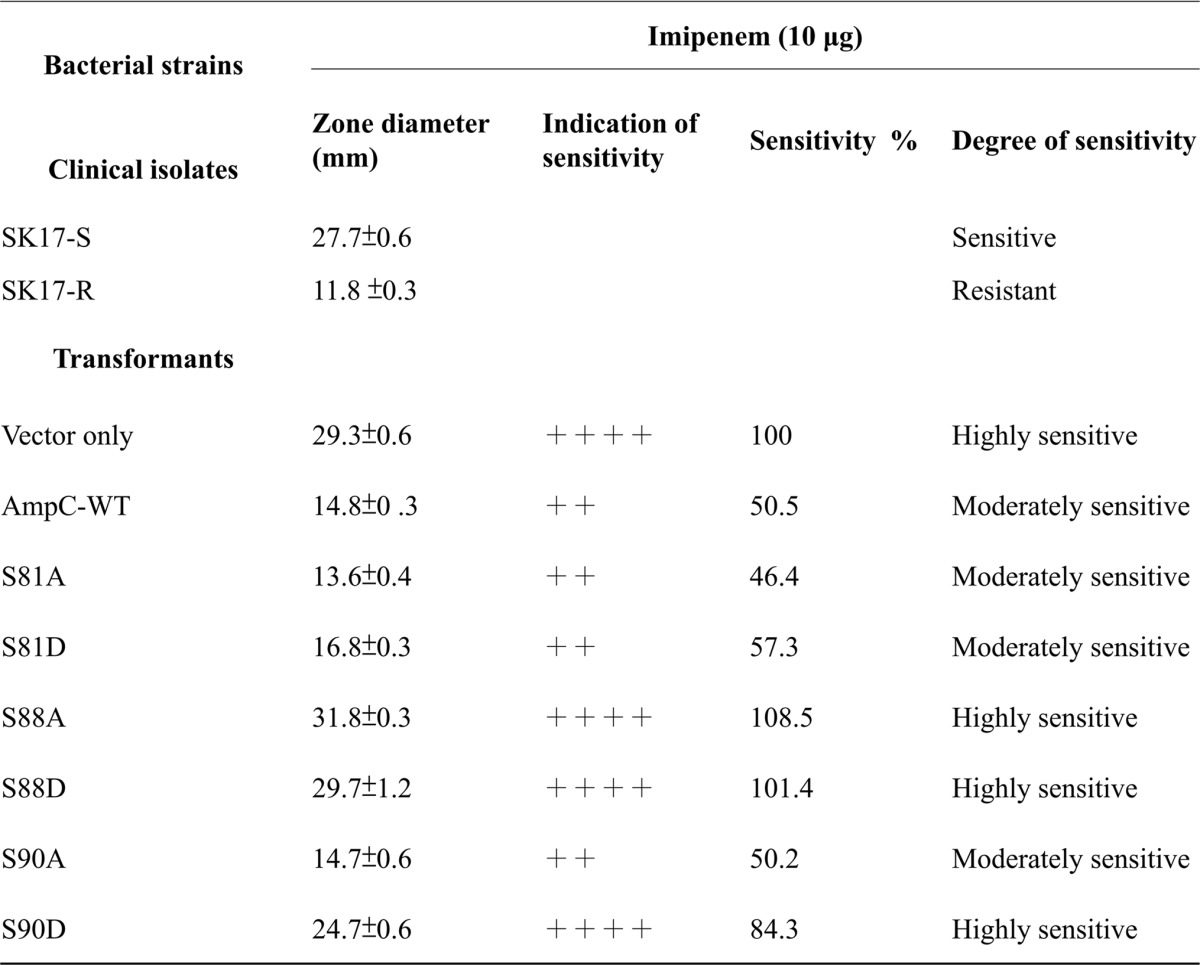
Resistant (absence of zone around discs) -, slightly sensitive (sensitivity 21–40%) +, moderately sensitive (sensitivity 41–60%) ++, quite sensitive (sensitivity 61–80%) +++, highly sensitive (sensitivity 81–100%) ++++, all the results of sensitivity in this table were normalized with the diameter values of vector only.
We also investigated whether phosphorylation of AmpC cephalosporinase can regulate the ability to hydrolyze ceftazidime (a third-generation cephalosporin), because ceftazidime-resistant A. baumannii has increased enormously in the past decade (50). Interestingly, most of the strains, including SK17-S, SK17-R, S81A, S81D, S90A, and S90D, were resistant to ceftazidime, indicating the clear regulatory role of Ser-90 phosphorylation of AmpC in imipenem-specific resistance (supplemental Table S8). Among the analysis of double mutants containing Ser-81 and Ser-90, only the S81D/S90D strain was sensitive to both imipenem and ceftazidime (supplemental Table S8). Single- or double-mutagenesis screening of the catalytic residue Ser-88 (S88A, S88D, S88AS90A, S88AS90D, S88DS90A, and S88DS90D) yielded both imipenem- and ceftazidime-sensitive phenotypes; thus, Ser-88 phosphorylation is likely not involved in antibiotic resistance (supplemental Table S9). Taken together, our findings indicate that Ser-90, but not Ser-81, plays a regulatory role in imipenem resistance through phosphorylation/de-phosphorylation (Table II and supplemental Table S8). Because of the location of Ser-90 in the catalytic motif S88VS90K91, its phosphorylation may be involved in regulating AmpC β-lactamase activity. Thus, we conducted kinetic analysis of β-lactamase activity using purified recombinant AmpCs (WT, S90A, and S90D).
Effect of AmpC Ser-90 Phosphorylation on β-Lactamase Activity and the Ability to Neutralize Imipenem
To gain insight into the regulation of AmpC phosphorylation and enzyme activity, phosphatase was used to remove covalently bound phosphate groups from recombinant AmpCs (46). Kinetic analysis showed that the S90A mutant protein exhibited greater β-lactamase activity with an ∼twofold higher kcat/Km value compared with AmpC-WT, suggesting that Ser-90-dephosphorylated AmpC increased enzyme activity (Fig. 3B and supplemental Table S10). Interestingly, similarly enhanced activity was observed for the phosphatase-treated AmpC-WT and S90A mutant proteins, indicating that Ser-90 phosphorylation of AmpC negatively regulated β-lactamase activity (Fig. 3B). This conclusion is supported by the fact that we were unable to obtain kinetic values for the mimic phosphorylated S90D mutant protein, as its β-lactamase activity was dramatically reduced (Fig. 3B and supplemental Table S10). Additionally, the difference in enzyme activities of the phosphatase-treated and phosphatase-untreated S90D proteins was relatively low, suggesting that the Ser-90 phosphosite is crucial for the phosphorylation network of global AmpC (Fig. 3B). Based on the circular dichroism spectra, all AmpC mutants showed similar components in their secondary structure compared with AmpC-WT (supplemental Fig. S5).
Imipenem acts as an inhibitor of AmpC β-lactamase, and the covalent bond between the catalytic serine residue and imipenem molecule has been determined by x-ray crystallography (51, 71, 72). Therefore, formation of an AmpC-imipenem covalent complex inhibits both AmpC enzyme activity and the efficacy of imipenem against bacteria. Given the proximity of Ser-90 to the imipenem-bonding site that includes Ser-88, we proposed that Ser-90 phosphorylation of AmpC may influence covalent bond formation. To better understand the efficacy of imipenem for recombinant AmpCs in vivo, neutralization assays were carried out. The results showed that the imipenem-induced clear zones could be neutralized by 5.5 μg and 7.0 μg of the S90A and AmpC-WT proteins, respectively (Fig. 4 and supplemental Fig. S6). In agreement with the results of the kinetic assay, the neutralization of imipenem was achieved with a lower dose of S90A protein than of AmpC-WT because of the higher enzyme activity of the S90A protein (Fig. 4 and Supplemental Fig. S6). Therefore, overexpression of the S90A protein exhibited greater protection of strain SK17-S against imipenem compared with AmpC-WT. For the discs containing S90D protein, the sensitivity of strain SK17-S to imipenem was similar to that of the control (imipenem only), demonstrating that Ser-90-phosphorylated AmpC failed to show neutralizing activity against imipenem (Fig. 4 and supplemental Fig. S6). Additionally, the predicted interactions between the four AmpCs (WT, phosphorylated-Ser-90, S90A, and S90D) and imipenem complexes from the docking experiments supported our finding that the S90D protein may be unable to bond covalently to imipenem (supplemental Figs. S7–S10). In summary, the phosphorylation/dephosphorylation of Ser-90 of AmpC has large and specific influences on not only imipenem susceptibility in vivo but also β-lactamase activity in vitro. Moreover, Ser-90-dephosphorylated AmpC regulated imipenem neutralization, conferring protection to the imipenem-susceptible strain SK17-S.
Fig. 4.
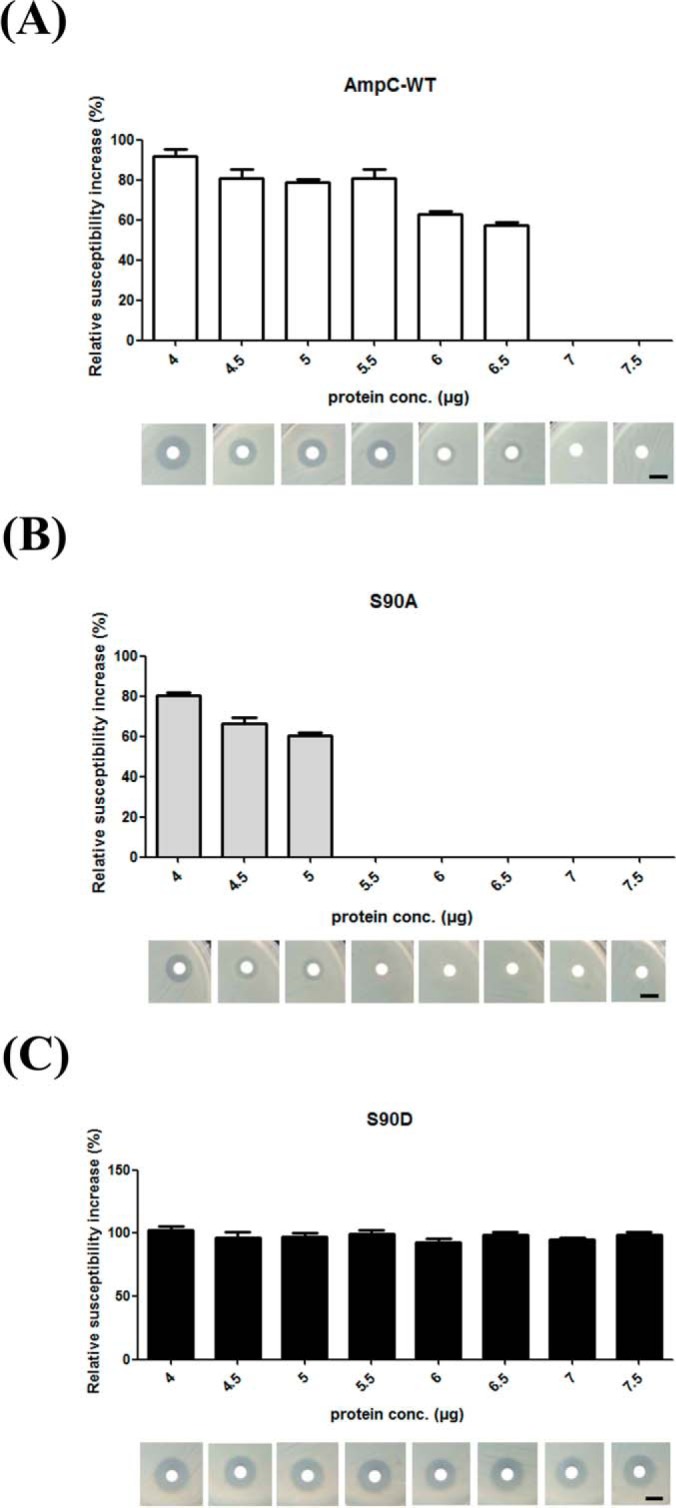
Neutralization of imipenem by recombinant AmpCs. The relative susceptibility of A. baumannii SK17-S to imipenem in the presence of recombinant AmpCs (WT, S90A, and S90D) were identified by the imipenem neutralization assay. The discs contained increasing amounts (4–7.5 μg) of recombinant proteins and imipenem (5 μg). Imipenem susceptibility was determined by measuring the diameters of the bacterial growth inhibition zones. All values were normalized to the mean value of the imipenem-only disc. Scale bars correspond to 10 mm.
DISCUSSION
The virulence of the Acinetobacter species is based on their adherence, colonization, and invasion of human epithelial cells, but the mechanisms of the virulent factors and antibiotic resistance remain unclear (5). Although AmpC β-lactamase is known to mediate the resistance to multiple β-lactam antibiotics, the mechanisms underlying its regulation are poorly understood (16). In this study, we used a shotgun approach to conduct comparative phosphoproteomic analysis of imipenem-susceptible and imipenem-resistant strains of A. baumannii SK17, as well as site-directed mutagenesis to determine whether AmpC phosphorylation is involved in regulating β-lactamase activity and imipenem resistance.
Phosphoproteomic Comparison of A. baumannii Strains
Because of the biased nature of data-dependent acquisition (DDA) used in conventional shotgun proteomic studies; low-abundance peptides are unlikely to be sequenced in a complex mixture of peptides. Therefore, our findings may represent only high-abundance phosphoproteins because of the highly dynamic process of phosphorylation in living cells and the settings used in the LC-MS/MS method in DDA mode. Western blot analysis of lysates from SK17-S and SK17-R revealed high levels of phosphorylation on Ser/Thr/Tyr residues (Supplemental Fig. S11B, S11C, and S11D). Interestingly, Ser and Tyr showed different phosphorylation levels in SK17-S and SK17-R (supplemental Fig. S11A, S11B, and S11D). A total of 178 and 141 phosphoproteins were uniquely identified in SK17-S and SK17-R, respectively, but only 70 phosphoproteins overlapped between the two strains (Fig. 1A and supplemental Table S3). The observation of a higher number of unique phosphoproteins between SK17-S and SK17-R compared with the number of overlapping phosphoproteins is consistent with the different phosphorylation patterns observed in Western blot analysis. Although our results identified relatively high abundant phosphoproteins that may be linked to imipenem resistance, further biological validations are required to enhance the significance of our findings. Based on the Western blotting and phosphoproteomic analyses results, our study contributes to understanding of the potential phosphoproteins involved in imipenem resistance mechanisms.
Recently, a study comparing A. baumannii ATCC17978 and Abh12O-A2 pointed out potential virulence-related phosphoproteins in this emerging pathogen (27). Our results revealed many of the same phosphoproteins whose biological functions may be correlated to antibiotic resistance, such as pathogenesis (e.g. protein tyrosine kinase), virulence (e.g. KdpD, KdpE), drug resistance (e.g. ABC-type multidrug transport system), and stress response protein (e.g. superoxide dismutase) (see supplemental Table S11) (13, 27). Hence, the overlapping phosphoproteins and phosphopeptides identified from the four A. baumannii strains may represent the most reliable candidates involved in carbapenem resistance (supplemental Table S4).
Some of these phosphoproteins were previously linked to imipenem resistance in A. baumannii, including AmpC, Cpn60 chaperonin (GroEL), ATP synthase, OmpA (73), AdeT, RND-type efflux system transporters (74–76), and penicillin-binding protein (73, 77–79) (supplemental Table S11). In particular, the phosphosites of AmpG (88, 89) and LysR (90, 91), which are transcriptional regulators involved in AmpC induction, were also identified in this study (supplemental Table S11). The RND multidrug efflux transporter-acriflavin resistance protein is a component of the efflux pump acriflavine resistance proteins A/B-Tolerance to colicins (AcrAB-TolC) (supplemental Table S11). The multidrug AcrAB-TolC efflux system was found to be involved in imipenem resistance in Enterobacter aerogenes (80–82).
Furthermore, we identified several functional phosphoproteins related to bacterial pathogenesis, such as those involved in toxicity (phospholipase C (6, 18, 83)) and adherence to host epithelial cells (TonB-dependent receptor (84, 85)) (supplemental Table S11). Phosphoproteins involved in polysaccharide synthesis/biofilm development (PgaA and PgaB (86, 87)) and the siderophore-mediated iron acquisition system (2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28) (enterobactin) siderophore (6, 18)) are also potential antimicrobial targets that are worth investigating in future studies (supplemental Table S11).
Phosphorylation of AmpC at Ser-90 Regulates Imipenem Susceptibility and β-Lactamase Activity
Previously, it has been reported that phosphorylation-dependent regulation of the β-lactamase modulated its enzyme activity and secretion. In Myxococcus Xanthus, the transmembrane protein kinase Pkn2 can phosphorylate threonine residues of class A TEM-type β-lactamase, thus controlling its enzyme activity and localization (66, 67). The Ser-88 phosphosite of AmpC was identified in a phosphoproteome study of E. coli; however, the regulatory networks of phosphorylation-mediated AmpC in bacterial signal transduction remain unclear (27, 54). Previous studies only revealed a connection between overexpression of AmpC and β-lactam-resistant pathogens (21, 92). Our results may provide insight into the phosphorylation-dependent regulation of AmpC in imipenem-resistant A. baumannii.
The identification of two phosphorylated serine residues (Ser-88 and Ser-90) in the catalytic motif S88VS90K91 of AmpC suggested that these residues regulate enzyme activity and substrate recognition (Fig. 3A). Thus, strains carrying a substitution at the catalytic Ser-88 were sensitive to both imipenem and ceftazidime, which agrees with the results of studies showing that the mutation and phosphorylation of Ser-88 significantly impaired AmpC enzyme function (supplemental Table S9) (71, 93). Moreover, our antibiotic susceptibility results for mutants S90A and S90D revealed that the regulation of imipenem resistance was mediated by AmpC Ser-90 phosphorylation (Table II and supplemental Table S8). The S90A mutant showed both enhanced β-lactamase activity and the ability to neutralize imipenem; in contrast, the S90D mutant caused the imipenem-sensitive phenotype because of the inability to neutralize imipenem (Fig. 3B, 4, supplemental Table S10, and supplemental Fig. S6). The inability of the S90D mutant to neutralize imipenem suggests that Ser-90-phosphorylated AmpC interferes with the AmpC-imipenem interaction (51). Here, we provided the first evidence highlighting the significance of phosphorylation-mediated regulation of AmpC with respect to the imipenem resistance of A. baumannii.
Phosphorylation of AmpC at Ser-90 may Modulate the Interaction between AmpC and Imipenem from Docking Calculations
Numerous studies of prokaryotic and eukaryotic enzymes have shown that conserved phosphosites stabilize the phosphate group in the catalytic site to modulate conformational changes during substrate binding, thereby achieving phosphorylation-mediated signal transduction (94–96). As a covalent inhibitor of class C β-lactamases, imipenem may initially noncovalently bind to AmpC and then form a nonhydrolyzable covalent intermediate in the acylation step, thus inactivating enzymatic function (51). Based on the substrate-assisted catalysis model, imipenem lacking the equivalent nitrogen on its 6(7)α substituent is trapped in the acyl complex state without the deacylation step (51, 69, 70). Ser-90 phosphorylation may perturb the initial noncovalent binding step indirectly when AmpC encounters imipenem by altering the charge state or shape of the binding pocket (51, 97).
Our docking calculations of four AmpCs (WT, phosphorylated-Ser-90, S90A, and S90D) in complex with imipenem revealed their pocket shapes and the relative distances between imipenem and the catalytic residues in the motif S88VS90K91 (supplemental Fig. S7 and S8). Lys-91, which plays an electrostatic role, may contribute to the deprotonation of Ser-88Oγ to attack the β-lactam ring carbon of imipenem during the acylation step (51, 69). Next, Lys-91Nζ forms hydrogen bonds with Ser-88Oγ and the carbonyl oxygen O-7 of imipenem (Imi O-7) in the nonhydrolyzable AmpC-imipenem complex (51, 69). From the minimized pockets of AmpC-WT and the S90A mutant, the pose of imipenem suggests that the orientations are favorable for Lys-91Nζ to form hydrogen bonds with Imi O-7, and Ser-88Oγ to attack the β-lactam ring carbon of imipenem (supplemental Fig. S7A, S7C, S8A, S8C, and S9B). Additionally, the Lys-91Nζ in the models of AmpC-WT and the S90A mutant are close to Ser-88Oγ, with distances of 3.4Å and 2.7Å, respectively, suggesting that Lys-91Nζ facilitates the deprotonation of Ser-88 (supplemental Fig. S8A and S8C). However, the pose of imipenem in the pockets of the phosphorylated-Ser-90 and S90D mutant may reflect the loss of hydrogen bonding between Lys-91Nζ and Imi O-7, as well as the inability of Ser-88Oγ to attack the β-lactam ring carbon of imipenem (supplemental Fig. S8B, S8D; S9A, and S9C). In the phosphorylated-Ser-90 model, rotation of Lys-91Nζ toward the phosphate group of phosphorylated-Ser-90 suggested that Lys-91Nζ loses the ability to act as a general base for Ser-88Oγ as well as its hydrogen bonding interaction with Imi O-7 (supplemental Fig. S8B and S9A). Moreover, the S90D mutant model showed the predicted structural impediment around the pocket and the σ loop, between the B8 and B9 strands from domain 1, in the active site (supplemental Fig. S7D, S9C, and S10). Taken together, both the phosphorylated-Ser-90 and S90D mutant models revealed an unfavorable pocket shape or orientation of Lys-91Nζ for acylation, which agrees with the results of high imipenem sensitivity and the inability of neutralizing imipenem from the mutant S90D (Table II and Fig. 4). Our docking results revealed that Ser-90 phosphorylation of AmpC may modulate the pocket shape or the orientation of Lys-91Nζ, which may indirectly interfere with the interaction between AmpC and imipenem.
Protein phosphorylation plays an important role in the regulation of a wide variety of cellular functions in both prokaryotic and eukaryotic cells. Bacterial protein phosphorylation is required for the biosynthesis of capsular polysaccharides, biofilm development, virulence, and antibiotic resistance, particularly through two-component signaling systems (44, 55, 98, 99). Thus, it has been demonstrated that His and Asp phosphorylation regulate the functions of pathogenesis, such as adherence, motility, enhancement of toxicity, quorum sensing, capsule formation, and drug resistance (100). Recently, in cancer drug development studies, it has been suggested that changes in phosphorylation stoichiometry are helpful for characterizing the kinase-dependent pathway in gefitinib-resistant lung cancer cells (101). Our study showed that Ser-90 phosphorylation of AmpC may be part of the kinase/phosphatase machinery conferring imipenem resistance, β-lactamase activity, and the ability to neutralize imipenem in A. baumannii. Thus, upstream cognate kinases/phosphatases involved in the phosphorylation-mediated regulation of AmpC β-lactamase activity may serve as targets for the design of effective antibacterial agents (102, 103). Therefore, drugs designed to interfere with bacterial protein phosphorylation cascades may provide an alternative way for eliminating pathogenic bacteria in the post antibiotic-resistant era.
Supplementary Material
Acknowledgments
We thank Yi-Tzu Lee MD, PhD (Institute of Clinical Medicine, School of Medicine, National Yang-Ming University, Taipei, Taiwan) for the clinical isolates A. baumannii SK17, and Dr. Suh-Yuen Liang (Core Facilities for Protein Structural Analysis, Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan) for the annotated database (supplemental Table S12). Additional help and advice in validating the kinetic assays were kindly provided by Dr. Meng-Ru Ho (IBC Biophysical Instrumentation Laboratory, Academia Sinica) and Dr. Hsien-Ya Lin (Department of Chemistry, National Taiwan University). The proteomic MS data have been deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (104) with the data set identifier PXD002033.
Footnotes
Author contributions: J. Lai and S.W. designed research; J. Lai, J.Y., and J.C. performed research; T.C., S.T., S.L., C.C., and S.W. contributed new reagents or analytic tools; J. Lai and J. Liao analyzed data; J. Lai wrote the paper; W.W., J. Liao, and S.W. guided the first author to solve the exp. problems.
* This work was financially supported by Ministry of Science and Technology (NSC 101-2923-B-001-005-MY3).
 This article contains supplemental Fig. S1 to S11 and Tables S1 to S12.
This article contains supplemental Fig. S1 to S11 and Tables S1 to S12.
1 The abbreviations used are:
- ADC
- Acinetobacter-derived cephalosporinase SK17-S, A. baumannii SK17-S
- SK17-R
- A. baumannii SK17-R
- TiO2
- titanium dioxide
- TFA
- trifluoroacetic acid
- ACN
- acetonitrile
- HAMMOC
- hydroxy-acid-modified metal-oxide chromatography
- FA
- formic acid
- CD
- circular dichroism
- MDRAB
- multidrug resistant A. baumannii
- IR-MDRAB
- imipenem-resistant multidrug-resistant A. baumannii
- ICU
- intensive care units.
REFERENCES
- 1.Imperi F., Antunes L. C., Blom J., Villa L., Iacono M., Visca P., and Carattoli A. (2011) The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 63, 1068–1074 [DOI] [PubMed] [Google Scholar]
- 2.Levin A. S., Levy C. E., Manrique A. E., Medeiros E. A., and Costa S. F. (2003) Severe nosocomial infections with imipenem-resistant Acinetobacter baumannii treated with ampicillin/sulbactam. Int. J. Antimicrob. Agents 21, 58–62 [DOI] [PubMed] [Google Scholar]
- 3.Manchanda V., Sanchaita S., and Singh N. (2010) Multidrug resistant Acinetobacter. J. Global Infect. Dis. 2, 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Anazi K. A., and Al-Jasser A. M. (2014) Infections caused by Acinetobacter baumannii in recipients of hematopoietic stem cell transplantation. Front. Oncol. 4, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim U. J., Kim H. K., An J. H., Cho S. K., Park K. H., and Jang H. C. (2014) Update on the Epidemiology, Treatment, and outcomes of carbapenem -resistant Acinetobacter infections. Chonnam. Med. J. 50, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerqueira G. M., and Peleg A. Y. (2011) Insights into Acinetobacter baumannii pathogenicity. IUBMB life 63, 1055–1060 [DOI] [PubMed] [Google Scholar]
- 7.Smith M. G., Gianoulis T. A., Pukatzki S., Mekalanos J. J., Ornston L. N., Gerstein M., and Snyder M. (2007) New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21, 601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang-Yu C., Kuo S. C., Liu C. Y., Luo B. S., Huang L. J., Lee Y. T., Chen C. P., Chen T. L., and Fung C. P. (2011) Difference in imipenem, meropenem, sulbactam, and colistin nonsusceptibility trends among three phenotypically undifferentiated Acinetobacter baumannii complex in a medical center in Taiwan, 1997–2007. J. Microbiol. Immunol. Infect. 44, 358–363 [DOI] [PubMed] [Google Scholar]
- 9.Poirel L., Bonnin R. A., and Nordmann P. (2011) Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB life 63, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 10.Tang S. S., Apisarnthanarak A., and Hsu L. Y. (2014) Mechanisms of beta-lactam antimicrobial resistance and epidemiology of major community- and healthcare-associated multidrug-resistant bacteria. Adv. Drug. Deliv. Rev. 78C, 3–13 [DOI] [PubMed] [Google Scholar]
- 11.Huang S. T., Chiang M. C., Kuo S. C., Lee Y. T., Chiang T. H., Yang S. P., Ti Y., Chen T. L., and Fung C. P. (2012) Risk factors and clinical outcomes of patients with carbapenem-resistant Acinetobacter baumannii bacteremia. J. Microbiol. Immunol. Infect. 45, 356–362 [DOI] [PubMed] [Google Scholar]
- 12.Karaoglan I., Zer Y., Bosnak V. K., Mete A. O., and Namiduru M. (2013) In vitro synergistic activity of colistin with tigecycline or beta-lactam antibiotic /beta-lactamase inhibitor combinations against carbapenem-resistant Acinetobacter baumannii. J. Int. Med. Res. 41, 1830–1837 [DOI] [PubMed] [Google Scholar]
- 13.Tiwari V., and Tiwari M. (2014) Quantitative proteomics to study carbapenem resistance in Acinetobacter baumannii. Front Microbiol. 5, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baran G., Erbay A., Bodur H., Onguru P., Akinci E., Balaban N., and Cevik M. A. (2008) Risk factors for nosocomial imipenem-resistant Acinetobacter baumannii infections. Int. J. Infect. Dis. 12, 16–21 [DOI] [PubMed] [Google Scholar]
- 15.Su C. H., Wang J. T., Hsiung C. A., Chien L. J., Chi C. L., Yu H. T., Chang F. Y., and Chang S. C. (2012) Increase of carbapenem-resistant Acinetobacter baumannii infection in acute care hospitals in Taiwan: association with hospital antimicrobial usage. PloS one 7, e37788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corvec S., Caroff N., Espaze E., Giraudeau C., Drugeon H., and Reynaud A. (2003) AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52, 629–635 [DOI] [PubMed] [Google Scholar]
- 17.Gordon N. C., and Wareham D. W. (2010) Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Ag. 35, 219–226 [DOI] [PubMed] [Google Scholar]
- 18.McConnell M. J., Actis L., and Pachon J. (2013) Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 37, 130–155 [DOI] [PubMed] [Google Scholar]
- 19.Hujer K. M., Hamza N. S., Hujer A. M., Perez F., Helfand M. S., Bethel C. R., Thomson J. M., Anderson V. E., Barlow M., Rice L. B., Tenover F. C., and Bonomo R. A. (2005) Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: defining a unique family of class C enzymes. Antimicrob. Agents Chemother. 49, 2941–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel L., and Nordmann P. (2006) Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12, 826–836 [DOI] [PubMed] [Google Scholar]
- 21.Quale J., Bratu S., Landman D., and Heddurshetti R. (2003) Molecular epidemiology and mechanisms of carbapenem resistance in Acinetobacter baumannii endemic in New York City. Clin. Infect. Dis. 37, 214–220 [DOI] [PubMed] [Google Scholar]
- 22.Cousin C., Derouiche A., Shi L., Pagot Y., Poncet S., and Mijakovic I. (2013) Protein-serine/threonine/tyrosine kinases in bacterial signaling and regulation. FEMS Microbiol. Lett. 346, 11–19 [DOI] [PubMed] [Google Scholar]
- 23.Imamura H., Wakabayashi M., and Ishihama Y. (2012) Analytical strategies for shotgun phosphoproteomics: status and prospects. Sem. Cell. Dev. Biol. 23, 836–842 [DOI] [PubMed] [Google Scholar]
- 24.Kobir A., Shi L., Boskovic A., Grangeasse C., Franjevic D., and Mijakovic I. (2011) Protein phosphorylation in bacterial signal transduction. Biochim. Biophys. Acta 1810, 989–994 [DOI] [PubMed] [Google Scholar]
- 25.Pinto S. M., Nirujogi R. S., Rojas P. L., Patil A. H., Manda S. S., Subbannayya Y., Roa J. C., Chatterjee A., Prasad T. S., and Pandey A. (2015) Quantitative phosphoproteomic analysis of IL-33-mediated signaling. Proteomics 15, 532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong J., Martinez M., Sengupta S., Lee A., Wu X., Chaerkady R., Chatterjee A., O'Meally R. N., Cole R. N., Pandey A., and Zachara N. E. (2015) Quantitative phosphoproteomics reveals crosstalk between phosphorylation and O-GlcNAc in the DNA damage response pathway. Proteomics 15, 591–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soares N. C., Spat P., Mendez J. A., Nakedi K., Aranda J., and Bou G. (2014) Ser/Thr/Tyr phosphoproteome characterization of Acinetobacter baumannii: comparison between a reference strain and a highly invasive multidrug-resistant clinical isolate. J. Proteomic 102, 113–124 [DOI] [PubMed] [Google Scholar]
- 28.Chen T. L., Lee Y. T., Kuo S. C., Hsueh P. R., Chang F. Y., Siu L. K., Ko W. C., and Fung C. P. (2010) Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents Chemother. 54, 4575–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mijakovic I., and Macek B. (2012) Impact of phosphoproteomics on studies of bacterial physiology. FEMS Microbiol. Rev. 36, 877–892 [DOI] [PubMed] [Google Scholar]
- 30.Wang F., Song C., Cheng K., Jiang X., Ye M., and Zou H. (2011) Perspectives of comprehensive phosphoproteome analysis using shotgun strategy. Anal. Chem. 83, 8078–8085 [DOI] [PubMed] [Google Scholar]
- 31.Yang M. K., Qiao Z. X., Zhang W. Y., Xiong Q., Zhang J., Li T., Ge F., and Zhao J. D. (2013) Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res. 12, 1909–1923 [DOI] [PubMed] [Google Scholar]
- 32.Lee Y. T., Kuo S. C., Chiang M. C., Yang S. P., Chen C. P., Chen T. L., and Fung C. P. (2012) Emergence of carbapenem-resistant non-baumannii species of Acinetobacter harboring a blaOXA-51-like gene that is intrinsic to A. baumannii. Antimicrob. Agents Chemother. 56, 1124–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 34.Ravichandran A., Sugiyama N., Tomita M., Swarup S., and Ishihama Y. (2009) Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics 9, 2764–2775 [DOI] [PubMed] [Google Scholar]
- 35.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 36.Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., Formsma K., Gerdes S., Glass E. M., Kubal M., Meyer F., Olsen G. J., Olson R., Osterman A. L., Overbeek R. A., McNeil L. K., Paarmann D., Paczian T., Parrello B., Pusch G. D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., and Zagnitko O. (2008) The RAST Server: rapid annotations using subsystems technology. BMC genomics 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharya M., Toth M., Antunes N. T., Smith C. A., and Vakulenko S. B. (2014) Structure of the extended-spectrum class C beta-lactamase ADC-1 from Acinetobacter baumannii. Acta Crystallogr. D 70, 760–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen T. L., Wu R. C., Shaio M. F., Fung C. P., and Cho W. L. (2008) Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother. 52, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo S. C., Yang S. P., Lee Y. T., Chuang H. C., Chen C. P., Chang C. L., Chen T. L., Lu P. L., Hsueh P. R., and Fung C. P. (2013) Dissemination of imipenem-resistant Acinetobacter baumannii with new plasmid-borne blaOXA-72 in Taiwan. BMC Infect. Dis. 13, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao Y. T., Kuo S. C., Lee Y. T., Chen C. P., Lin S. W., Shen L. J., Fung C. P., Cho W. L., and Chen T. L. (2014) Sheltering effect and indirect pathogenesis of carbapenem-resistant Acinetobacter baumannii in polymicrobial infection. Antimicrob. Agents Chemother. 58, 3983–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu J., March P. E., Lee R., and Tillett D. (2004) Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mammeri H., Poirel L., and Nordmann P. (2007) Extension of the hydrolysis spectrum of AmpC beta-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. J. Antimicrob. Chemother. 60, 490–494 [DOI] [PubMed] [Google Scholar]
- 43.Chen T. L., Chang W. C., Kuo S. C., Lee Y. T., Chen C. P., Siu L. K., Cho W. L., and Fung C. P. (2010) Contribution of a plasmid-borne blaOXA-58 gene with its hybrid promoter provided by IS1006 and an ISAba3-like element to beta-lactam resistance in Acinetobacter genomic species 13TU. Antimicrob. Agents Chemother. 54, 3107–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu W. L., Liao J. H., Lin G. H., Lin M. H., Chang Y. C., Liang S. Y., Yang F. L., Khoo K. H., and Wu S. H. (2013) Phosphoproteomic analysis reveals the effects of PilF phosphorylation on type IV pilus and biofilm formation in Thermus thermophilus HB27. Mol. Cell. Proteomics 12, 2701–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu C. W., Lin M. H., Huang H. C., Ku W. C., Yi T. H., Tsai C. F., Chen Y. J., Sugiyama N., Ishihama Y., Juan H. F., and Wu S. H. (2012) Phosphoproteomic analysis of Rhodopseudomonas palustris reveals the role of pyruvate phosphate dikinase phosphorylation in lipid production. J. Proteome Res. 11, 5362–5375 [DOI] [PubMed] [Google Scholar]
- 46.Ishihama Y., Wei F. Y., Aoshima K., Sato T., Kuromitsu J., and Oda Y. (2007) Enhancement of the efficiency of phosphoproteomic identification by removing phosphates after phosphopeptide enrichment. J. Proteome Res. 6, 1139–1144 [DOI] [PubMed] [Google Scholar]
- 47.Minond D., Saldanha S. A., Subramaniam P., Spaargaren M., Spicer T., Fotsing J. R., Weide T., Fokin V. V., Sharpless K. B., Galleni M., Bebrone C., Lassaux P., and Hodder P. (2009) Inhibitors of VIM-2 by screening pharmacologically active and click-chemistry compound libraries. Bioorg. Med. Chem. 17, 5027–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsang M. W., and Leung Y. C. (2007) Overexpression of the recombinant Enterobacter cloacae P99 AmpC beta-lactamase and its mutants based on a phi105 prophage system in Bacillus subtilis. Protein Express. Purif. 55, 75–83 [DOI] [PubMed] [Google Scholar]
- 49.Kim S. H., and Wei C. I. (2007) Expression of AmpC beta-lactamase in Enterobacter cloacae isolated from retail ground beef, cattle farm and processing facilities. J. Appl. Microbiol. 103, 400–408 [DOI] [PubMed] [Google Scholar]
- 50.Hamidian M., and Hall R. M. (2014) Tn6168, a transposon carrying an ISAba1-activated ampC gene and conferring cephalosporin resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 69, 77–80 [DOI] [PubMed] [Google Scholar]
- 51.Beadle B. M., and Shoichet B. K. (2002) Structural basis for imipenem inhibition of class C beta-lactamases. Antimicrob. Agents Chemother. 46, 3978–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galleni M., Amicosante G., and Frere J. M. (1988) A survey of the kinetic parameters of class C beta-lactamases. Cephalosporins and other beta-lactam compounds. Biochem. J. 255, 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziervogel B. K., and Roux B. (2013) The binding of antibiotics in OmpF porin. Structure 21, 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., and Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 55.Lin M. H., Hsu T. L., Lin S. Y., Pan Y. J., Jan J. T., Wang J. T., Khoo K. H., and Wu S. H. (2009) Phosphoproteomics of Klebsiella pneumoniae NTUH-K2044 reveals a tight link between tyrosine phosphorylation and virulence. Mol. Cell. Proteomics 8, 2613–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misra S. K., Milohanic E., Ake F., Mijakovic I., Deutscher J., Monnet V., and Henry C. (2011) Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 11, 4155–4165 [DOI] [PubMed] [Google Scholar]
- 57.Ouidir T., Jarnier F., Cosette P., Jouenne T., and Hardouin J. (2014) Potential of liquid-isoelectric-focusing protein fractionation to improve phosphoprotein characterization of Pseudomonas aeruginosa PA14. Anal. Bioanal. Chem. 406, 6297–6309 [DOI] [PubMed] [Google Scholar]
- 58.Sun X., Ge F., Xiao C. L., Yin X. F., Ge R., Zhang L. H., and He Q. Y. (2010) Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J. Proteome Res. 9, 275–282 [DOI] [PubMed] [Google Scholar]
- 59.Ge R., Sun X., Xiao C., Yin X., Shan W., Chen Z., and He Q. Y. (2011) Phosphoproteome analysis of the pathogenic bacterium Helicobacter pylori reveals over-representation of tyrosine phosphorylation and multiply phosphorylated proteins. Proteomics 11, 1449–1461 [DOI] [PubMed] [Google Scholar]
- 60.Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C. M., Bemis G., Church G. M., Steen H., and Husson R. N. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U.S.A. 107, 7521–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basell K., Otto A., Junker S., Zuhlke D., Rappen G. M., Schmidt S., Hentschker C., Macek B., Ohlsen K., Hecker M., and Becher D. (2014) The phosphoproteome and its physiological dynamics in Staphylococcus aureus. Int. Med. Microbiol. 304, 121–132 [DOI] [PubMed] [Google Scholar]
- 62.Voisin S., Watson D. C., Tessier L., Ding W., Foote S., Bhatia S., Kelly J. F., and Young N. M. (2007) The cytoplasmic phosphoproteome of the Gram-negative bacterium Campylobacter jejuni: evidence for modification by unidentified protein kinases. Proteomics 7, 4338–4348 [DOI] [PubMed] [Google Scholar]
- 63.Takahata Y., Inoue M., Kim K., Iio Y., Miyamoto M., Masui R., Ishihama Y., and Kuramitsu S. (2012) Close proximity of phosphorylation sites to ligand in the phosphoproteome of the extreme thermophile Thermus thermophilus HB8. Proteomics 12, 1414–1430 [DOI] [PubMed] [Google Scholar]
- 64.Alvarez-Ortega C., Olivares J., and Martinez J. L. (2013) RND multidrug efflux pumps: what are they good for? Front Microbiol. 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ambler R. P. (1980) The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 289, 321–331 [DOI] [PubMed] [Google Scholar]
- 66.Bonomo R. A., and Szabo D. (2006) Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43, S49–56 [DOI] [PubMed] [Google Scholar]
- 67.Udo H., Munoz-Dorado J., Inouye M., and Inouye S. (1995) Myxococcus xanthus, a gram-negative bacterium, contains a transmembrane protein serine/threonine kinase that blocks the secretion of beta-lactamase by phosphorylation. Genes dev. 9, 972–983 [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez-Martinez J. M., Poirel L., and Nordmann P. (2010) Genetic and functional variability of AmpC-type beta-lactamases from Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4930–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y., McReynolds A., and Shoichet B. K. (2009) Re-examining the role of Lys67 in class C beta-lactamase catalysis. Protein Sci. 18, 662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Y., Minasov G., Roth T. A., Prati F., and Shoichet B. K. (2006) The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution. J. Am. Chem. Soc. 128, 2970–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beadle B. M., and Shoichet B. K. (2002) Structural bases of stability–function tradeoffs in enzymes. J. Mol. Biol. 321, 285–296 [DOI] [PubMed] [Google Scholar]
- 72.Thomas V. L., McReynolds A. C., and Shoichet B. K. (2010) Structural bases for stability-function tradeoffs in antibiotic resistance. J. Mol. Biol. 396, 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee H. Y., Chen C. L., Wang S. B., Su L. H., Chen S. H., Liu S. Y., Wu T. L., Lin T. Y., and Chiu C. H. (2011) Imipenem heteroresistance induced by imipenem in multidrug-resistant Acinetobacter baumannii: mechanism and clinical implications. Int. J. Antimicrob. Ag. 37, 302–308 [DOI] [PubMed] [Google Scholar]
- 74.Fernandez L., and Hancock R. E. (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hou P. F., Chen X. Y., Yan G. F., Wang Y. P., and Ying C. M. (2012) Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy 58, 152–158 [DOI] [PubMed] [Google Scholar]
- 76.Srinivasan V. B., Rajamohan G., Pancholi P., Marcon M., and Gebreyes W. A. (2011) Molecular cloning and functional characterization of two novel membrane fusion proteins in conferring antimicrobial resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 66, 499–504 [DOI] [PubMed] [Google Scholar]
- 77.Russo T. A., MacDonald U., Beanan J. M., Olson R., MacDonald I. J., Sauberan S. L., Luke N. R., Schultz L. W., and Umland T. C. (2009) Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J. Infect. Dis. 199, 513–521 [DOI] [PubMed] [Google Scholar]
- 78.Cayo R., Rodriguez M. C., Espinal P., Fernandez-Cuenca F., Ocampo-Sosa A. A., Pascual A., Ayala J. A., Vila J., and Martinez-Martinez L. (2011) Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 5907–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gehrlein M., Leying H., Cullmann W., Wendt S., and Opferkuch W. (1991) Imipenem resistance in Acinetobacter baumanii is due to altered penicillin-binding proteins. Chemotherapy 37, 405–412 [DOI] [PubMed] [Google Scholar]
- 80.Bornet C., Chollet R., Mallea M., Chevalier J., Davin-Regli A., Pages J. M., and Bollet C. (2003) Imipenem and expression of multidrug efflux pump in Enterobacter aerogenes. Biochem. Bioph. Res. Commun. 301, 985–990 [DOI] [PubMed] [Google Scholar]
- 81.Fischer N., and Kandt C. (2013) Porter domain opening and closing motions in the multi-drug efflux transporter AcrB. Biochim. Biophys. Acta 1828, 632–641 [DOI] [PubMed] [Google Scholar]
- 82.Pradel E., and Pages J. M. (2002) The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob. Agents and Chemother. 46, 2640–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Antunes L. C., Imperi F., Carattoli A., and Visca P. (2011) Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PloS one 6, e22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smani Y., McConnell M. J., and Pachon J. (2012) Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PloS one 7, e33073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Devos S., Van Oudenhove L., Stremersch S., Van Putte W., De Rycke R., Van Driessche G., Vitse J., Raemdonck K., and Devreese B. (2015) The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in Stenotrophomonas maltophilia. Front. Microbiol. 6, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Choi A. H., Slamti L., Avci F. Y., Pier G. B., and Maira-Litran T. (2009) The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1–6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191, 5953–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abbott I., Cerqueira G. M., Bhuiyan S., and Peleg A. Y. (2013) Carbapenem resistance in Acinetobacter baumannii: laboratory challenges, mechanistic insights and therapeutic strategies. Expert. Rev. Anti. Infect. Ther. 11, 395–409 [DOI] [PubMed] [Google Scholar]
- 88.Woodhams K. L., Chan J. M., Lenz J. D., Hackett K. T., and Dillard J. P. (2013) Peptidoglycan fragment release from Neisseria meningitidis. Infect. Immun. 81, 3490–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang T. C., Chen T. F., Tsai J. J., and Hu R. M. (2014) NagZ is required for beta-lactamase expression and full pathogenicity in Xanthomonas campestris pv. campestris str. 17. Res. Microbiol. 165, 612–619 [DOI] [PubMed] [Google Scholar]
- 90.Kong K. F., Aguila A., Schneper L., and Mathee K. (2010) Pseudomonas aeruginosa beta-lactamase induction requires two permeases, AmpG and AmpP. BMC Microbiol. 10, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Everett M., Walsh T., Guay G., and Bennett P. (1995) GcvA, a LysR-type transcriptional regulator protein, activates expression of the cloned Citrobacter Freundii Ampc beta-lactamase gene in Escherichia Coli cross-talk between DNA-binding proteins. Microbiology 141, 419–430 [DOI] [PubMed] [Google Scholar]
- 92.Kuo H. Y., Chang K. C., Kuo J. W., Yueh H. W., and Liou M. L. (2012) Imipenem: a potent inducer of multidrug resistance in Acinetobacter baumannii. Int. J. Antimicrob. Ag. 39, 33–38 [DOI] [PubMed] [Google Scholar]
- 93.Trehan I., Beadle B. M., and Shoichet B. K. (2001) Inhibition of AmpC beta-lactamase through a destabilizing interaction in the active site. Biochemistry 40, 7992–7999 [DOI] [PubMed] [Google Scholar]
- 94.Birck C., Mourey L., Gouet P., Fabry B., Schumacher J., Rousseau P., Kahn D., and Samama J. P. (1999) Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure 7, 1505–1515 [DOI] [PubMed] [Google Scholar]
- 95.Ciarimboli G., Koepsell H., Iordanova M., Gorboulev V., Durner B., Lang D., Edemir B., Schroter R., Van Le T., and Schlatter E. (2005) Individual PKC-phosphorylation sites in organic cation transporter 1 determine substrate selectivity and transport regulation. J. Am. Soc. Nephrol. 16, 1562–1570 [DOI] [PubMed] [Google Scholar]
- 96.Mehrens T., Lelleck S., Cetinkaya I., Knollmann M., Hohage H., Gorboulev V., Boknik P., Koepsell H., and Schlatter E. (2000) The affinity of the organic cation transporter rOCT1 is increased by protein kinase C-dependent phosphorylation. J. Am. Soc. Nephrol. 11, 1216–1224 [DOI] [PubMed] [Google Scholar]
- 97.Singh J., Petter R. C., Baillie T. A., and Whitty A. (2011) The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 [DOI] [PubMed] [Google Scholar]
- 98.Sun F., Ding Y., Ji Q., Liang Z., Deng X., Wong C. C., Yi C., Zhang L., Xie S., Alvarez S., Hicks L. M., Luo C., Jiang H., Lan L., and He C. (2012) Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 15461–15466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaspy I., Rotem E., Weiss N., Ronin I., Balaban N. Q., and Glaser G. (2013) HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 4, 3001. [DOI] [PubMed] [Google Scholar]
- 100.Mijakovic I. (2010) Protein phosphorylation in bacteria. Febs J. 277, 20–21 [Google Scholar]
- 101.Tsai C. F., Wang Y. T., Yen H. Y., Tsou C. C., Ku W. C., Lin P. Y., Chen H. Y., Nesvizhskii A. I., Ishihama Y., and Chen Y. J. (2015) Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat. Commun. 6, 6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hirakawa H., Nishino K., Yamada J., Hirata T., and Yamaguchi A. (2003) Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52, 576–582 [DOI] [PubMed] [Google Scholar]
- 103.Jacoby G. A. (2009) AmpC beta-lactamases. Clin. Microbiol. Rev. 22, 161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.