Abstract
Sugars are the most functionally and structurally diverse molecules in the biological world. Glycan structures range from tiny single monosaccharide units to giant chains thousands of units long. Some glycans are branched, their monosaccharides linked together in many different combinations and orientations. Some exist as solitary molecules; others are conjugated to proteins and lipids and alter their collective functional properties. In addition to structural and storage roles, glycan molecules participate in and actively regulate physiological and developmental processes. Glycans also mediate cellular interactions within and between individuals. Their roles in ecology and evolution are pivotal, but not well studied because glycan biochemistry requires different methods than standard molecular biology practice. The properties of glycans are in some ways convenient, and in others challenging. Glycans vary on organismal timescales, and in direct response to physiological and ecological conditions. Their mature structures are physical records of both genetic and environmental influences during maturation. We describe the scope of natural glycan variation and discuss how studying glycans will allow researchers to further integrate the fields of ecology and evolution.
Keywords: glycans, glycoconjugates, natural variation, polymorphism, cellular ecology
1 – The Complex and Labile Forms of Glycans
No Template Required
DNA is an inert strand whose information encodes ribbons of RNA and protein that put instructions into action. For biologists, this one-to-one correspondence is convenient. It ties contemporary molecules to their history, allowing us to pinpoint the origins of molecular functions and trace their modifications through time. DNA is a template, transcribed and translated by a common set of mechanisms into just two different kinds of molecule. Because of this commonality, molecular biologists tend to use a standard set of tools to determine how information in DNA encodes the biological functions of RNA and proteins. Now that these tools exist, there is a tendency to regard template-based molecules as the most important sources of heritable variation, and other molecules as merely structural. But heritable information in the genome is also manifest at other levels; these are less bound to the barely mutable instructions in DNA, and as a consequence more sensitive to current conditions and more capable of dynamic response. A well-understood example is RNA and protein expression, which can change in response to environment and physiological condition. Less well studied are glycans – whose structures and biochemical properties have evolved to enact traits with dynamic ecology and evolution [1].
Synthesis
Glycans are sugar molecules – modular arrangements of monosaccharide units that are often attached to proteins and lipids. Glycans range in size from mono- and oligosaccharides a few nanometers long, to polysaccharide chains thousands of subunits and several micrometers in length. The monosaccharide constituents of glycans are linked together by glycosidic bonds. These can originate from several of the hydroxylated carbon atoms that form each monosaccharide ring, so glycan structures can be extensively branched. Glycans decorate the majority of extracellular proteins and many lipids (the combined molecules are called glycoconjugates) [2]. These exist primarily on the outer surface of the cell membrane, and are also secreted into extracellular spaces (Box 1). Unlike protein and RNA, glycans are not built from a template. Their synthesis is an assembly process of sequential addition, removal, and modification of sugar subunits. The form and diversity of glycan structures are guided by proteins, but also depend intrinsically on diet and physiological condition. Because the structures of mature glycans are not encoded in the genome, their evolutionary history can only be uncovered indirectly. For example, the evolution of glycan modifying proteins can reveal historical glycan changes. The gain and loss of sugar transporters, transferases, and glycosidases have a direct bearing on the suite of glycans an organism can synthesize and degrade, or use in the diet.
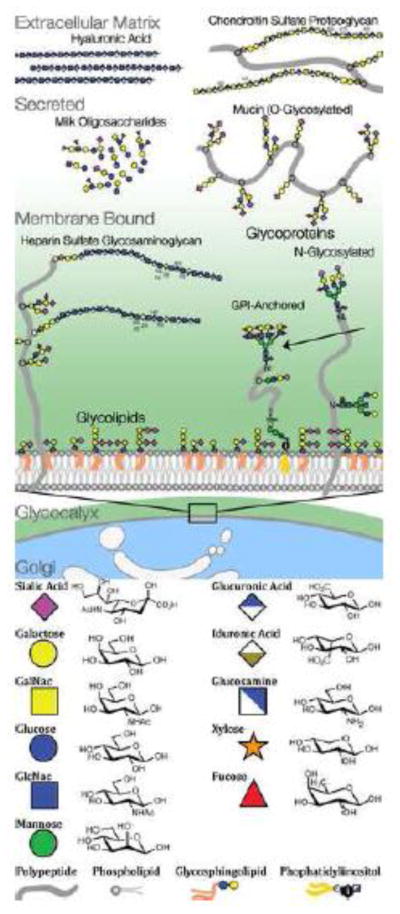
Box 1. Types of Glycans and Glycoconjugates.
Glycans are sugars, composed of monosaccharide building blocks. These basic units can be linked into chains and branched structures. Mammals use only a dozen basic monosaccharide units, but these are combined into many different types of glycan structures. Glycans can be connected with several different linkages to proteins (N- and O- glycans, glycosaminoglycans, proteoglycans) and to lipids (glycolipids, GPI-anchored proteins). Glycans can also be secreted as free oligo- or polysaccharides (hyaluronic acid, milk oligosaccharides). In eukaryotic cells, N-glycans are attached to glycoproteins in the endoplasmic reticulum, and extensively remodeled in the golgi before moving outside the cell. In prokaryotes, N- and O- glycans are built in a stepwise fashion in the cytoplasm but not extensively pruned or remodeled. Bacteria and archaea use hundreds of different monosaccharides, but these are only rarely linked into the more complex structures typical of eukaryote glycans [76].
Glycans and glycoconjugates are classified in several different ways. N-linked and O-linked glycans are categorized by their chemical attachment to a protein. Glycosaminoglycans can be recognized by their monosaccharide composition. Other glycoconjugates, like GPI-anchored proteins and glycolipids, are recognizable by the linkages and combinations of their constituent molecules. Some monosaccharides, glycans, and glycoconjugates are expressed only in particular phylogenetic lineages (mammalian glycosaminoglycans) others exist across most of cellular life (N- glycans). There are many rare glycans, monosaccharides, linkages, and modifications known to exist, and it is likely that many more remain to be discovered. We describe some of the most abundant classes of glycans.
N-linked glycans
N-glycans, are oligosaccharides, each covalently linked to a protein by the nitrogen atom of an asparagine. Potential N-linked glycosylation sites are called sequons, and have an N-X-S/T consensus amino-acid sequence. N-linked glycans are found across all three domains of life, but they are synthesized differently in eukaryotes than in eubacteria or archaea [77]. In eukaryotes, N-linked glycosylation regulates protein folding. Changes in N-glycan composition track the protein folding process in the endoplasmic reticulum. After folding, glycoproteins mature in the Golgi apparatus, where attached N-glycans are restructured into their mature forms [78]. In the golgi, N-glycans are first trimmed back to an oligomannose base, and then built into complex multiply branched forms [79]. The golgi is a multi-compartment vesicular system, and the presence or absence of glycan modifying enzymes and nucleotide sugar donors in a given golgi compartment regulates N-glycan composition. N-glycan structures vary widely in different organisms. Mammalian N-glycans are most-often terminated by sialic acids. Plant N-glycans are never terminated in sialic acids and contain modifications not seen in mammalian N-glycans. Fungi make N-glycans rich in mannose residues. Invertebrates produce hybrid N-glycans with fewer branches than vertebrates and with some of the same modifications as plants [78]. Invertebrates such as ggastropods also utilize some different monosaccharides [80]. In general multicellular organisms produce a greater abundance and variety of N-glycans than bacteria and archaea. The diversity of multicellular N-glycans may help define cellular identity and modulate cell-cell signaling during development [61].
O-linked glycans
O-Glycans are oligosaccharide chains conjugated to a protein by the oxygen atom of a serine or threonine [81]. O-glycans contain fewer branches and fewer mannose residues than N-glycans. Bacteria and archaea have diverse suites of O-glycans. A large family of enzymes synthesizes eukaryote O-glycans during protein translation. O-glycans are abundant in animal secretions such as mucus on epithelia. Mucin glycoproteins can be 80% O-glycans by mass and form protective hydrated gels. Most O-glycans are attached to a protein by a GalNac monosaccharide. Recently, O-linked GlcNac was discovered and found to interact directly with cellular metabolism and phosphorylation. O-GlcNac glycans may act as direct sensors of physiological state. Glycosaminoglycans are also attached to proteins by the oxygen atom of a serine, but they are not typically categorized as O-linked glycans.
Glycosaminoglycans
Glycosaminoglycans (GAGs ) are among the largest molecules synthesized by animals. Their synthesis begins with an O-linked xylose, which is then elongated by two galactose molecules. Many thousands of N-Acetylglucosamine and glucuronic acid disaccharides are added to this initial trisaccharide to form gigantic chains. Importantly, GAGs, undergo further modification: glucuronic acid can be epimerized into iduronic acid, and N-Acetylglucosamines can be de-N-acteylated and N-sulfated, N-acetylated or O-sulfated. These post-translational modifications generate complex chemical patterns along the length of each GAG chain. We do not yet know how to read the syntax, but it is clear that these patterns have important signaling functions. They act as binding sites for growth factors (FGFs), anti-coagulation factors (anti-thrombin), and pathogens (HIV and HSV1) [11]. GAGs are involved in a diversity of physiological and developmental processes. They interface with metabolic state via lipid particles on liver cell proteoglycans [82]. They also guide morphological development [83], and even potentially behavior via their effects on neuronal migration [84].
Glycolipid glycoconjugates
Glycolipids are oligosaccharides conjugated to lipid molecules. Gangliosides, are sialic acid containing glycosphingolpids, which are often form patterns associated with lipid rafts on the outer cell membrane. Certain cell types can have outer membrane leaflets that consist almost entirely of glycolipid instead of phospholipid [85]. The spatial orientation of glycans is important, because their binding is often multivalent, involving multiple chemical contacts in close proximity. The clustering of glycans on the cell surface by glycolipids may thus have important functional effects.
GPI-anchored glycoconjugates
GPI-anchored glycoproteins are complex molecules that combine a lipid, a glycan, and a protein. The lipid is a phosphoinositol tail, which is linked to a short oligosaccharide glycan, itself linked to a polypeptide by a phosphoethanolamine bond. The polypeptide components of most GPI-anchored proteins are further glycosylated and can contain N- and O-glycans and even glycosaminoglycans called glypicans. Interestingly, GPI anchored proteins can transfer information from one cell to another. They can be shed from one cell surface and incorporated into the membranes of other cells [86].
Rare glycans and glycoconjugates
There are many other glycan classes that are less abundant than the classes described above. Some have only a few known examples; others are expressed only in particular phylogenetic lineages. A huge unexplored diversity of glycans is likely to exist in bacteria and archea. Because they lack an ER-Golgi compartment, these taxa have evolved different ways of assembling glycans than eukaryotes. The capsids and glycoconjugates of bacteria and archaea are already known to contain a bewildering diversity of complex glycans and the study of these is in its infancy [76, 87]. Recent years have seen the discovery of many additional glycan classes including: O-xylose, O-fucose, C-Mannose, and S-linked glycans on cysteine residues [88] New classes of glycans, and new glycan functions are continually being discovered [89].
Figure.
Glycans on a mammalian cell
Privacy
Glycans are the most variable class of biological molecules, both in structural diversity and taxonomic breadth of particular molecules (Box 1) [3]. Glycans often exist at the interface of conflicts between parties with different evolutionary interests: parents and offspring, hosts and pathogens, cooperators and cheats [1]. Glycans that signal or conceal self-identity, or mediate evolutionary conflicts can change rapidly and evolve toward esoteric structures and biochemistries. Glycans also form protective structures and accumulate as stores of energy. Some storage glycans, such as glycogen and starch, can be exploited by competing organisms and must be kept sequestered inside cells. Others, like mammalian lactose, are secreted. Lactose is composed of just two simple monosaccharides, but the linkage connecting them is difficult to synthesize and equally difficult to degrade so the combined molecule can be transferred to offspring with little risk of exploitation. Structural glycans including cellulose, hemicellulose, and chitin are the most abundant biopolymers on earth. These linked monosaccharides can accumulate to such extraordinary abundances only because they are difficult to break down for energy. The space of possible glycan structures and linkages is so large that some glycans are private, in the sense that few organisms can synthesize, degrade, or bind them. Thus, the diversity of glycans allows them to be used in contexts where less diverse molecules would be vulnerable to exploitation.
Dynamics
Glycans have a remarkable degree of spatial and temporal organization, patterns that can enact changes in development and physiology. The dynamic properties of glycans influence aspects of trait development, cellular behavior, and molecular function. For example, N-linked glycans attract chaperone proteins, guiding nacent glycoproteins into their mature fold as they are translated in the endoplasmic reticulum. Later, in the golgi, N-glycan molecules are themselves remodeled into forms that alter the function of the folded glycoprotein. During synthesis, glycans can be modified by elongation, shedding, cleaving of terminal monosaccharides, and by modifications such as: N- or O-acetylation [4], deacetylation, epimerization, or N- and O sulfation [5]. Glycan patterns change with cell type and cell stage, and influence cell migration, tissue modulation, repair and regeneration [6]. Glycans also change with cell age. The Aswell receptor monitors deglycosylation as a sign that a cell or plasma glycoprotein needs recycling [7]. Glycans exist in organisms as clouds of variants whose properties integrate evolutionary and ecological information. The proteins that interact with glycans are robust to this ever-present variation, but can also evolve binding patterns that are sensitive to glycan changes that indicate development, diet, health, or environment.
Glycomics
Glycan distribution and pattern on cells and tissues was first probed with lectins – glycan binding proteins with well-defined specificities. An important challenge is to scale these studies up, to bridge the gap between lists of glycans identified by mass spectrometry, and their true biological abundance, topology, distribution and variability. New glycomic methods will allow us to look directly at the diversity and variability of glycans: their structures, locations, and their biochemistry (Box 2). Glycomics is revealing a world of new biochemical forms and functions: molecular mechanisms that underpin ecological and evolutionary processes at every spatial and temporal scale. The study of natural glycan variation is one of the most interesting new prospects in biology today.
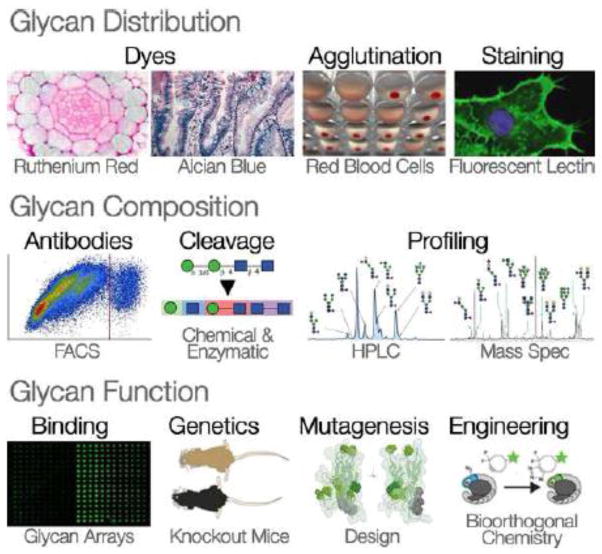
Box 2. Methods of Glycan Discovery.
Glycan distribution
Dyes
Cationic dyes stain the glycocalyx and extracellular matrix on epithelial cells because these structures have a negative charge overall. Several cationic dyes have been traditionally used to stain anionic glycans – such as Alcian Blue for mucins in epithelial secretions or hyaluronic acid in cartilage, and Ruthenium Red to stain the glycocalyx [90]. Cationic dyes do stain glycans, but they are generally not specific to particular molecules, so they don’t provide the resolution required for detailed analysis.
Agglutination
Blood cells often clump together when exposed to lectins. Plant lectins differentially agglutinate the red blood cells from different individuals and from different mammalian species. This is caused by the specificity of a multivalent lectin for polymorphic glycans on the red blood cell surface. The understanding that glycans caused blood cell agglutination allowed the development of lectin histology – the characterization of specific glycan patterns on the cell surface.
Staining
There is now a large collection of natural lectins from plants, fungi and invertebrates with a variety of binding specificities for different glycans [91]. Animal cells or tissue sections can be stained with lectins to reveal the abundance and distributions of glycans. Lectins with a particular specificity can be attached to fluorescent markers to reveal cellular structures and patterns of glycan distribution on cells. The use of glycan modifying enzymes to directly remodel cell surface glycans allows the target of a particular lectin stain to be verified. Lectin staining even permits simple analyses of the composition of glycans on the cell surface.
Glycan composition
Antibodies
The presence or absence of intact glycans on glycoconjugates can be assessed with antibodies. For glycoproteins, antibodies can be used to specifically stain western blots to identify proteins bearing particular glycans. Fluorescence activated cell sorting (FACS) can be combined with antibody or lectin staining to build sophisticated analyses of the presence and composition of glycans on cell surfaces. Anti-glycan antibodies were added to the tool kit of molecular probes for glycans and so were recombinant glycan binding proteins expressed in microbial, plant or animal cells [92].
Cleavage
A variety of chemical, biochemical and biophysical tools have been developed to characterize the structures of glycans: their monosaccharide composition, glycosidic linkage types and branching patterns [93]. Compounds such as 4-methyl umbelliferyl (4-MU), which fluoresce once a monosaccharide has been cleaved, can monitor the presence or specificity of degradation enzymes. Chemical and enzymatic methods sequentially degrade glycans or release them from their glycoconjugates. The structures of free glycans can then be inferred by chromatography and mass spectrometry.
Profiling
Glycans released from the underlying protein or lipid and are then chemically derivatized prior to separation by chromatography. Glycans are identified on an HPLC column by comparing their characteristic retention with standards. Chromatography can be coupled with mass spectrometry such as MALDI TOF (matrix assisted laser desorption/ionization time of flight) mass spectrometry to identify the individual glycan species. Even intact glycan structures, can be identified by the characteristic masses of their fragments [94].
Glycan function
Binding
Glycan arrays can assess binding specificity by presenting many glycans each with a slightly different structure. Lectin arrays can be coupled with surface plasmon resonance to directly measure the force of interaction between glycan binding proteins and glycans. In rare cases, lectins can be co-crystallized with their glycan ligand to characterize the glycan-protein interaction at the atomic level [95]. Glycan arrays require careful interpretation since binding on an array may not perfectly reflect natural binding. Recently synthetic glycans arranged in multivalent assemblages on scaffold molecules have allowed researchers to probe for glycan binding proteins in more naturalistic circumstances [96].
Genetics
Genomic comparisons of genes encoding glycan synthesis, modification, or degradation enzymes can indicate historical glycan presence and absence. Phylogenetic comparisons of particular glycan binding proteins can test whether their evolution is driven by selection. Genetic tools have uncovered the ER and Golgi enzymes that act during glycan synthesis and remodeling [97]. Genetic manipulation in model organisms can give clues to natural glycan functions in vivo. Findings from medicine inform us about about the role of glycans in disease [21]. For example, congenital diseases of glycosylation can have extraordinarily simple genetic causes, and equally simple solutions. Many are caused by the loss of a single glycan-modifying enzyme, and some can be completely reversed by supplementing the missing glycan in the diet [98].
Mutagenesis
Artificial mutations of glycan binding proteins can be used in conjunction with glycan arrays to determine which amino-acid residues promote glycan specificity, or to design new specificities. Cloning animal glycan binding proteins and their recombinant expression provides additional molecular probes for glycans. It also allows functional studies of lectin-glycan interactions in animals [99].
Engineering
Bio-orthogonal compounds are molecules that cannot be synthesized or broken down by organisms. Biorthogonal glycans combined with click chemistry artificial molecules can be used to image changes in the distribution and metabolism of natural glycans in real time [100]. Synthesis of glycans using non-natural monosaccharides also allows metabolic labeling and functional manipulation in vivo.
Challenges
Glycans are chemically diverse, so that even superficially similar classes of molecules like O- and N-glycans cannot be fully analyzed by any single set of methods. For some glycans, like GAGs, even their basic physical structures cannot yet be fully resolved. Decoding the patterns of chemical modification that occur in glycans will be difficult, and determining the function of these patterns more difficult still. The study of glycan function relies heavily on decomposition analysis and genetic manipulation of their synthesis: both tools could benefit from refinement. For example, many different glycans can have identical signatures by mass spectrometry, and it is often difficult to know which branch of a complex N-glycan contained which monosaccharide. Not all of the approaches used to characterize glycans are yet amenable to high throughput methods.
Additionally, the involvement of glycans in many biological processes can be missed because their mode of chemical interaction is different from proteins or antibodies. Glycan interactions, with each other and with the proteins that bind them, are typically multivalent – it requires several molecular contacts in close physical proximity to form a stable attachment [101]. This complicates the assessment of their function, because the molecular orientations that allow binding in natural conditions cannot always be replicated in an artificial assay. There is a critical need for binding assays that take into account multivalency, and the spatial orientations of glycans and their binding partners.
Finally, the functional analysis of glycans is potentially more complicated than that of proteins or RNA, there are many potential modifications, and it is not always possible to make them independently at each site in the molecule. It is also difficult to know which changes are realistic, in the sense that they can be achieved by mutations of a glycan synthesis protein. Genetic studies, and analysis of glycan variants produced by mutation in model organisms will yield important insights into glycan function and variation in vivo [102]. Ecological glycan functions may be restricted to certain interactions, phases of development, or to specific parts of an organism. Studying these traits will require new chemical methods of uncovering glycan variation, new statistical methods of describing it, and new theory to describe the patterns of glycan variation expected in natural circumstances.
Standards
The structural diversity of glycans and glycoconjugates has been an impediment to their systematic analysis. This is an underdeveloped area of glycobiology that deserves research effort. The first step is simply cataloging the variation of glycans, glycoconjugates and glycan-modifying enzymes as it exists in nature. There is a rapidly growing repository of glycan active enzymes: CAZy (http://www.cazy.org/). There are also databases that attempt to record the diversity of glycan structures themselves: (http://www.glycome-db.org/), (http://expasy.org/resources/search/keywords:glycomics), (http://www.functionalglycomics.org/glycomics/molecule/jsp/carbohydrate/carbMoleculeHome.jsp). Resources for glycobiology methods are available from several organizations (http://glycosciences.de./), (http://www.functionalglycomics.org/static/consortium/resources.shtml). The open glycoworkbench is a resource for interpreting and annotating mass spectrometry data (http://www.glycoworkbench.org/). Glycopedia is a general reference for glycoscience related information (http://www.glycopedia.eu/).
Glycobiologists should aim to develop standards of analysis that allow comparison between studies, to the extent that this will be possible for such diverse molecules. Journals and funding agencies should mandate the deposition of glycan data into public databases, where it can be accessed and subject to synthetic analysis.
Figure.
Methods of studying glycan distribution, composition, and function
2 – Cellular Ecology
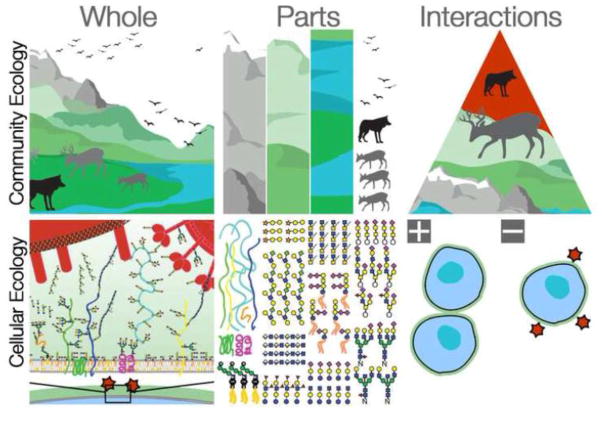
Classically, ecologists have studied the interactions of organisms with each other and with their environment. But ecological interactions are not limited to just one spatial scale. Cells also have ecology, with interactions as nuanced and interesting as those at larger scales (Fig. 1). But there is one very important difference between community and cellular ecology – the mechanistic complexity of the traits involved. Many of the interesting phenomena in community ecology result from patterns of organismal response, which can be developmental, physiological, or in animals neural. The responses are organismal, so the mechanisms are often too complex to be traced down to a particular molecular cause. The behavioral responses of cellular ecology are equally sophisticated in their scope, but vastly simpler mechanistically. Recognizable ecological interactions can be caused by very simple, tractable, chemical interactions [8].
Figure 1. Community and Cellular Ecology.
Classically, ecological studies consider interactions at the organismal level and above. The interacting parts are whole organisms and their biotic and abiotic environments. The variability in these interactions results from organismal behaviors and physiologies whose mechanistic bases are complex. Cellular ecology is concerned with the interaction of cells, whether within or between individuals. As with traditional ecology, the first task is to survey the parts of the ecosystem. Glycomic methods now permit the enumeration of parts lists for a given interaction, and often these interacting parts are glycans. Unraveling the molecular details and ecological consequences of these mechanistically simple interactions will be the domain of cellular ecology.
The distinction is not important for a pure ecologist, but for an evolutionary ecologist mechanistically simple systems are key because their history can be more easily traced. Glycans are ecological traits. As a consequence their structures are condition dependent and their historical variation is not as easy to trace as a template based molecule. But glycans are a middle ground – their evolutionary history can be inferred from the evolution of their synthetic enzymes. Thus, the application of phylogenetic tools to glycans with recognizable cellular ecology is a bridge that links the fields of ecology and evolution. Figures 2, 3, and 4 present case studies from primates. These examples were developed as extensions of human medical studies, but there will be countless other examples. Cellular ecology pervades biology. Glycomic methods will allow researchers to enumerate the parts of each system, and from there cellular ecologists can begin to unravel the interactions.
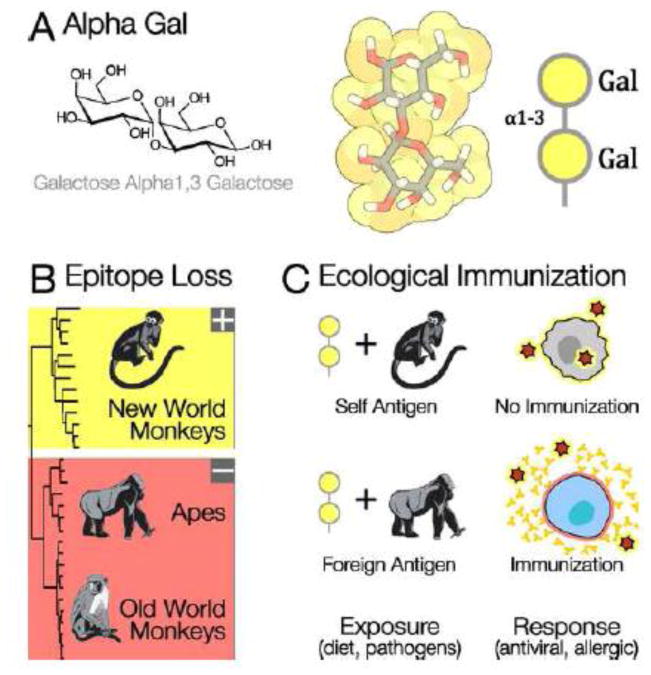
Figure 2. Alpha Gal: A Recently Evolved, Highly Reactive, Foreign Antigen.
A) Alpha gal, a common structure on animal glycans, is a terminal galactose alpha 1,3 linked to an underlying galactose. B) 1,3galactosyltransferase ( 1,3GT), the enzyme that synthesizes alpha gal, was inactivated at the common ancestor of Old World Primates [63]. Apes, humans, and Old World monkeys cannot synthesize the terminal alpha gal linkage. Most mammals, including the New World primates, do synthesize the alpha gal epitope. C) Individuals can be immunized against foreign glycans by exposure to them on pathogens, or by their presence in the diet. The loss of alpha gal may have made Old World primates resistant to infection, because anti alpha gal antibodies target this foreign epitope on enveloped viruses coming from other mammalian species [64]. Anti alpha gal antibodies can cause severe allergic reactions in humans that eat red meat after immunization by a tick bite [65]. Alpha gal antibodies also prevent organ transplantation from other mammal species – pig organs are rejected partly because they are rich in alpha gal [66]. Anti alpha gal antibodies have a potential medical applications if they could be directed against pathogens or cancerous cells [67].
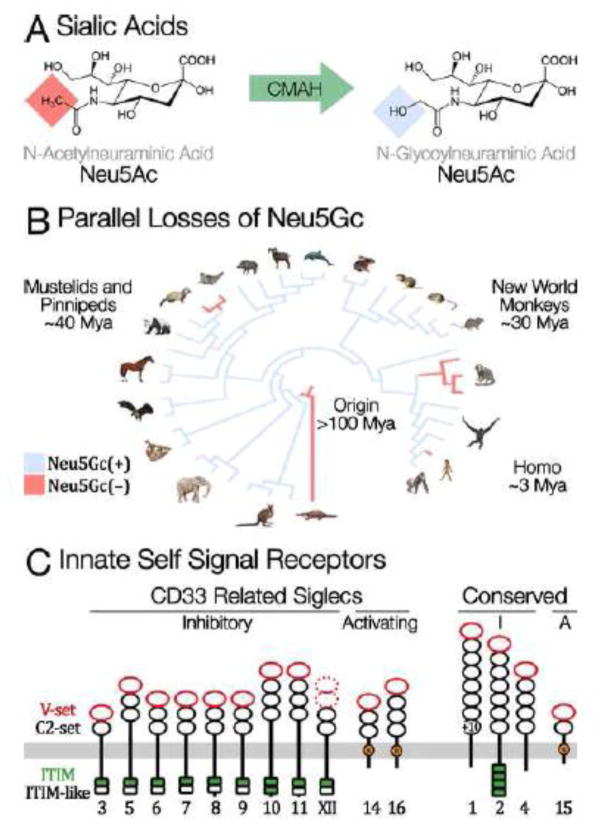
Figure 3. Sialic Acids: Parallel Evolution of Innate Self-Signals.
A) Sialic acids are nine-carbon monosaccharides found at the terminal tips of N-glycans, on glycolipids, and as long chains of polysialic acid. N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) are two of the most common forms: each a family of molecules with various modifications of the canonical monosaccharide. Neu5Gc is synthesized from Neu5Ac by the CMAH protein (cytidine monophosphate-N-acetylneuraminic acid hydroxylase), which hydroxylates a terminal methyl group. Neu5Gc is a private glycan in the sense that no pathogen has yet been found to synthesize this monosaccharide. B) Humans cannot synthesize Neu5Gc, because human CMAH was inactivated over two million years ago [68]. The inactivating mutation fixed rapidly after originating, which suggests that the loss could have been adaptive – driven by pathogen avoidance, sexual conflict, or a combination of the two [69]. Independent losses of CMAH have recently been found in New World Primates [62] and in Mustelids [70]. C) Sialic acids are bound by a family of Siglec receptor proteins expressed on most immune cells [71]. Binding generally inhibits inflammation, thus sialic acids are innate markers of self. Many pathogens bind Siglecs to exploit these immunosuppressive effects, and in response several immune-activating Siglecs have evolved. Other pathogens use Neu5Gc itself as a receptor on host cells. Several human pathogens have modified their receptor affinity to recognize Neu5Ac following the loss of Neu5Gc in humans.
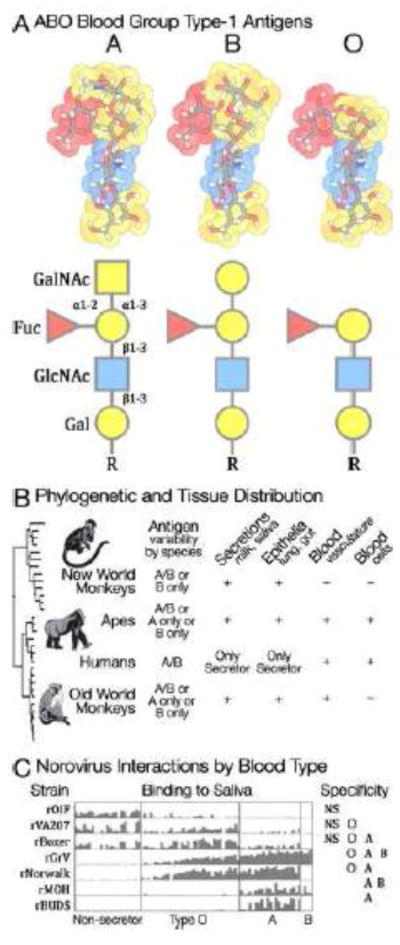
Figure 4. ABO Blood Group: Pathogens and Polymorphism.
A) The first molecular polymorphism discovered in humans was the ABO blood group [72]. Blood types were discovered because certain lectins agglutinate the red blood cells of some individuals and not others. These lectins bind glycans that are polymorphic within human populations. The glycans that define A, B, and O blood types are created by the activity of a GalNAc/Gal transferase. An enzyme that transfers N-Acetylgalactosamine results in blood type A; one that transfers Galactose results in type B. An inactive enzyme does not modify the underlying glycan and results in type O. B) The ABO polymorphism originated at the common ancestor of primates [73]. Various patterns of ABO polymorphism are found among primate species. Additionally, a fucosyl transferase (FUT2) causes variation in blood group antigen expression in different tissues. Individual humans are either secretors (ABO glycans are found on epithelia and secretions) or non-secretors (ABO glycans are found only in the blood and vasculature). Other primates have various different secretion patterns. C) Many pathogens exploit specific blood type glycans for host recognition and attachment including noroviruses and rotaviruses [74]. Anti-AB antibodies may help maintain ABO polymorphisms by targeting incoming enveloped viruses bearing mismatched epitopes [75].
Parts
Prokaryotes
All living cells are coated by a glycocalyx, a complex layer of protein and lipid glycoconjugates with protective and signaling functions (Box 1). Prokaryote glycans act as defensive barriers, provide support, store resources, and can also act as signaling molecules [9] [10]. Eubacteria and Archeae use over 700 different monosaccharides to produce their bacterial capsid polysaccharides and archaeal cell envelopes. Unicellular organisms use the glycocalyx to shield themselves from environmental influence, and these extracellular glycoconjugates also mediate cellular interactions such as niche construction and the formation of biofilms.
Eukaryotes
In Metazoans the glycocalyx must additionally hold information about cell lineage identity, and mediate and cell-cell communication during development and cell migration. Animal glycans are built from only about a dozen different monosaccharides, in many different linkages. The extracellular matrix (ECM) of multicellular animals is rich in glycosaminoglycans, long secreted polymers such as hyaluronan, or proteoglycans such as heparan sulfate, chondroitin sulfate, and dermatan sulfate [11]. In plants, cell wall polysaccharides are key structural molecules [12]. Plants make complex glycans consisting of many more individual monosaccharide types than found in animals.
Interactions
Within Individuals
Glycans act at all life history stages, their composition and patterns on cell surfaces changing during the course of development. Sperm-egg interactions of invertebrates and vertebrates require glycan mediated binding [13–15]. Developing tissues can be identified by glycans called Stage Specific Embryonic Antigens (SSEAs)[16]. Different surfaces of the same cell can even bear different glycans. For example, heparan sulfate proteoglycans give polarity to fly germ line cells [17, 18]. Glycans pattern the extracellular matrix. Their structural and informational roles allow signaling gradients to form: a critical requirement of proper development [19, 20].
Physiological and dietary changes also influence glycan synthesis [21]. The abundant O-GlcNAc modification (of transcription factors, kinases, histones and other proteins) directly connects nutrient levels with glycosylation, gene regulation, and epigenetic marks on histone tails [22]. Glycan receptors, such as siglecs and other lectins, are expressed on immune cells. These bind self-glycan patterns and regulate the innate immune response [23]. Other glycan receptors induce inflammatory responses when they bind fragments of large structural glycans like cellulose or hyaluronan: these fragments presumably indicate a lysed cell, a pathogen intrusion, or some other danger [24, 25].
Glycans are often used in more than one context within an organism. The asialoglycoprotein (Ashwell) receptor binds aging platelets, RBCs and glycoproteins so they can be removed from circulation [26] [7]. This same receptor mitigates bacterial infection [27]. Polysialic acids mediate neuronal development, and are also expressed by all three germ layer precursors [28, 29]. Galectins form intercellular lattices by cross-linking N-glycans terminating in galactose [30], and also aid in cytoplasmic pathogen defense [31]. Selectins on leukocytes, platelets and endothelia mediate both leukocyte trafficking [32] and blastocyst implantation [33]. Within individuals, glycans take on a diversity of roles, and the synthesis and regulation of individual glycan molecules is tailored to suit each particular circumstance.
Between Individuals
Glycans are also a nexus for interactions between individuals: in symbiosis and in pathogenesis. Animals recruit microbes to expand the range of glycans available to them in the diet. Gut microbe genomes are typically enriched with polysaccharide utilizing loci that allow symbionts to digest glycans otherwise unavailable to the host [34]. Animals do not have cellulose-digesting enzymes, but many (termites, ruminants, koalas, leaf eating monkeys) have evolved symbioses with microbes that can digest cellulose for energy [35]. In mammals [36], the establishment of a specific gut microbiota (and with it proper immune maturation) is aided by oligosaccharides secreted in mother’s milk [37]. Some symbiotic microbes evade host immunity by molecular mimicry: camouflaging their surfaces by synthesizing glycans that resemble the host [38]. Other symbionts incorporate glycans synthesized by the host to decorate their surfaces and increase host tolerance [39]. Oligosaccharides such as polysaccharide A, secreted by the mammalian gut symbiont Bacteroides fragilis, can directly induce tolerance by interacting with T regulatory cells in the gut [40].
The potential for glycans to induce immune tolerance can also be exploited, and this conflict results in in evolutionary arms races between hosts and their pathogens. Some pathogenic bacteria attach to host cells using adhesins with high specificity for host glycans (E.coli, P. falciparum). Other pathogens secrete toxins that target host glycans (S. Typhi and Clostridium tetani), or modify host glycans into appropriate ligands for binding (V. cholerae sialidase) [41]. Viruses also use host glycans for attachment (Paramyxoviruses for example) [3, 42]. In response, hosts have evolved lectins: including high mannose receptor, DC-SIGN, dectin and intelectin-1, which are expressed on a variety of immune cells. These innate immune receptors allow plants and animals to rapidly detect foreign antigens [24], and some innate lectins like RegIII even have direct bactericidal properties [43].
Hosts also use glycans to defend themselves against pathogens. Defensive mucins are secreted on most animal epithelia. These heavily O-glycosylated glycoproteins resist proteolytic degradation and form hydrated gels, forming a physical barrier against incoming microbes [44]. Mucins can also act as decoys, mimicking the structures of cell surface receptors. Many viruses express sialidases (neuramindases) so that they can bypass sialic acid rich mucin decoys. Bacteria are also infected by viruses, and bacteriophages are known to target glycans on the surface of their bacterial hosts [45]. Most viruses do not synthesize their own glycans, but viral proteins can be glycosylated by the host cell, allowing them to evade host immunity (HIV1 GP120)[46] and/or inhibition (Filovirus mucin-like glycoprotein)[47]. Parasites with complex host cycles are glyco-engineers, negotiating and manipulating the different glycan environments in each of their vertebrate, insect and mollusk hosts [48]. Glycans mediate aspects of ecology that cause them to evolve rapidly. The tools developed by ecologists and evolutionary biologists to study diverse and rapidly evolving traits will help us better understand the extraordinary diversity of glycans.
3 – Ecology and Evolution: Tools to Understand Glycans
Ecology and evolutionary biology study the distribution of biological variation in space and in time. Each field has developed statistical methods for quantifying variation, and each uses null models to predict the patterns of variation expected under specific processes [49–51]. We are only now gaining the technology to observe most natural glycan variants. Applying existing methods from ecology and evolution will expand our understanding of the determinants and consequences of glycan variation. The first step is developing biochemical tools and statistical metrics to quantify glycan variation in nature: as it responds to ecology, and as it evolves. Soon, system-level models will predict the patterns of glycan variation expected by specific genetic or regulatory changes. Similarly, appropriate null ecological or evolutionary models could be used as means of inferring the processes that shape glycan variation in space and time. The features of these models will be specific to glycans, but it is certain that they will resemble, and likely extend, statistical methods of analysis that already exist in ecology and evolution.
Quantifying glycan variation
One of the main uses of statistics in ecology and evolution is to separate phenotypic variation into components, so that differences caused by genetics and environment can be isolated and studied independent of measurement error [49, 52]. Ecologists have developed many different metrics to quantify patterns of variation in space, and these are flexible enough that most can be applied directly to glycan variation. Statistics from ecology can be used to quantify glycan distribution and abundance, to compare diversity and patterns of association, or to study community structure and dynamics [51]. Experimental methods developed by ecologists also translate directly to glycobiology, and are thus already widely used [53, 54]. Evolutionary methods are less straightforward to apply. Many rely on estimates of history, which are difficult to infer because traces of historical change are only partially evident in the structure of non-template based molecules. Quantitative genetic methods that draw inference solely from phenotypic variation are directly applicable, although the discrete nature of some glycan variation may violate the assumption that phenotypic variation arises by mutations of tiny effect [55]. However, with methods to quantify natural glycan variation, it should be possible to dissect genetic and environmental effects, estimate heritability, response to selection, rate of evolution, and other metrics applicable to quantitative trait evolution [52, 56].
Predicting glycan variation
Many ecological and evolutionary processes occur on timescales that are too long to observe directly. We must infer the action of a given process by observing its outcomes in nature and comparing them to an expectation. Ecology and evolution use models to precisely specify assumptions that describe a process, and to predict which outcomes we should observe if that process is operating. For example, models of molecular evolution allow us to generate null predictions of evolution in the absence of selection and thus to separate patterns expected by neutral and adaptive evolution [50]. Glycans are often found in phenotypes that mediate evolutionary conflicts, so they are likely to evolve rapidly. There is a genuine need for theory that can provide null expectations for the evolution of glycan phenotypes. This will be perhaps the most difficult synthesis of glycobiology and evolution because it requires substantial innovation in both fields. In particular, theorists should attempt to develop metrics that can infer deviations from neutral glycan distribution in time and space so that glycobiologists can develop methods to measure glycan variation appropriately.
4 – Glycans: Tools to Understand Ecology and Evolution
Sources of glycan variation
Evolutionary biology is now reconciling the modern synthesis of selection and genetics with the reality that phenotypic traits are not entirely genetically determined [57]. Development, diet, environment, and many other non-genetic processes connect information encoded in genes and the phenotypes that are realized in nature. Non-genetic processes could even create additional sources of heritable variation on which selection can act, or impose constraints on evolution that are not obvious from studying genetic variation alone [58]. For example changes in nutrient availability can directly alter available UDP-GlcNAc levels, which can change epigenetic marks on histone tails and alter the regulation of gene expression [59]. These mechanisms directly link environment and gene regulation, and could underpin phenotypically plastic traits where environmental cues create regulatory changes that cause the development of appropriate phenotypes.
Attempts at a broader evolutionary synthesis face the difficulty of uncovering processes that connect genotype and phenotype [60]. Glycans are useful in this respect, because their structures are physical records of the genetic and environmental processes that created them. Synthesis proteins encoded directly in the genome control some synthesis processes, and others depend intrinsically on organismal context. Sugars integrate both kinds of information, and thus will offer a unique opportunity to study the roles of non-genetic information in evolution. Additionally, the spectrum of phenotypic variation created by mutations of glycan modifying proteins varies enormously. Some synthesis steps have specific effects temporally and spatially; others have broad organismal effects. For example, genetic manipulation of N-glycan synthesis in mammals shows that the early steps of glycan synthesis are more conserved than the late steps [61]. As systems-level information accumulates, it may be possible to examine these outcomes and look at relations between pleiotropy, evolutionary rate, and patterns of gene family diversification.
Interpreting glycan variation
Ecology has yet to undergo a full integration with molecular biology, in part because the ecological relevance of most molecular traits is not yet easily recognizable [8]. Most current molecular ecology studies uncover history using molecular markers but do not elucidate molecular mechanism. Here too, studying glycans will offer new opportunities. Glycans are variable on ecological timescales and the interactions mediated by glycans occur at small spatial scales. Glycans are involved in traits whose ecological importance is recognizable, so glycans will allow experimental tests of ecological theory that take molecular mechanisms of interaction explicitly into account [1]. Glycan phenotypes will also aid on the ground conservation efforts because their variants are sensors of organismal condition as it responds to local ecology. Some preserve aspects of each individual’s ecological history such as dietary changes or health; others are indicative of disease susceptibility [62].
5 – Glycan Phenotypes and the Synthesis of Ecology and Evolution
Almost every biological process involves sugars. Their ubiquity reflects an unmatched capacity for structural and functional variability. Glycans can respond to demands that change on physiological, developmental, ecological, and evolutionary timescales. As such, they are often recruited as mechanisms to enact dynamic traits. Glycans are molecules that record the outcome of entire systems of protein interaction – their non-template based synthesis integrates environmental and genetic information. The specificity of glycosylation (position, timing, and variability) is a physical representation of how tightly the synthesis process is regulated, and the effects of dysregulation. This mode of phenotype production will allow progress on fronts in ecology and evolution that are difficult to study with proteins or RNA, because the structures, distribution and variability of glycans can vary on organismal timescales. The variability of glycan molecules is caused both by information in DNA and by the context of their synthesis. Mature glycans represent both factors, and so they are the phenotypic records that we need to recognize the ecological relevance of molecular traits.
It has been argued that the disciplines and methods of ecology and evolution still do not fully interface [57]. Difficulties in uniting scientific fields arise either because the information required for synthesis is difficult to collect, or because the importance of existing information is difficult to recognize. Studying glycans can help bridge these divides. Glycans are tractable molecular traits whose genetic and environmental determinants can be recognized and whose ecological relevance can be understood. The non-template based synthesis of glycans creates many technical and theoretical challenges. Glycans do not vary in ways that are fully compatible with existing experimental methods, statistical methods, or theory. But new glycomic methods are emerging that will collect information on natural glycan variation at full biological resolution and scale. Quantifying and predicting the synthesis, regulation, and function of natural glycan variation will allow us to study aspects of ecology and evolution that are not accessible to studies of proteins and RNA alone. Because glycans vary on organismal timescales, they promise an opportunity to further integrate ecology and evolution themselves: two closely aligned but still separate fields of biology.
Significance.
This article introduces methods of discovering glycans and isolating their biological functions. Glycans have dynamic patterns of evolution, and can change in direct response to ecological conditions. We highlight some of the early results of this expansive new field and discuss the many exciting intersections of glycobiology with ecology and evolution.
Acknowledgments
We thank HS Reynoso, A Lizcano, J Okerblom, and TK Altheide for discussion. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation, Grant CSD081 (to P.G.) and by the National Institutes of Health, Grant R01 GM095882 (to P.G.)
Footnotes
Conflict of Interest
The authors declare that there are no competing interests, financial or otherwise.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Springer SA, Gagneux P. Glycan evolution in response to collaboration, conflict, and constraint. J Biol Chem. 2013;288:6904–6911. doi: 10.1074/jbc.R112.424523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marth JD. A unified vision of the building blocks of life. Nat Cell Biol. 2008;10:1015–1016. doi: 10.1038/ncb0908-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop JR, Gagneux P. Evolution of carbohydrate antigens--microbial forces shaping host glycomes? Glycobiology. 2007;17:23R–34R. doi: 10.1093/glycob/cwm005. [DOI] [PubMed] [Google Scholar]
- 4.Kasaai MR. Various methods for determination of the degree of N-acetylation of chitin and chitosan: a review. J Agric Food Chem. 2009;57:1667–1676. doi: 10.1021/jf803001m. [DOI] [PubMed] [Google Scholar]
- 5.Honke K, Taniguchi N. Sulfotransferases and sulfated oligosaccharides. Med Res Rev. 2002;22:637–654. doi: 10.1002/med.10020. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu T, Muramatsu H. Carbohydrate antigens expressed on stem cells and early embryonic cells. Glycoconj J. 2004;21:41–45. doi: 10.1023/B:GLYC.0000043746.77504.28. [DOI] [PubMed] [Google Scholar]
- 7.Yang WH, Aziz PV, Heithoff DM, Mahan MJ, Smith JW, Marth JD. An intrinsic mechanism of secreted protein aging and turnover. Proc Natl Acad Sci U S A. 2015;112:13657–13662. doi: 10.1073/pnas.1515464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Springer SA, Crespi BJ, Swanson WJ. Beyond the phenotypic gambit: molecular behavioural ecology and the evolution of genetic architecture. Mol Ecol. 2011;20:2240–2257. doi: 10.1111/j.1365-294X.2011.05116.x. [DOI] [PubMed] [Google Scholar]
- 9.Ridley BL, O’Neill MA, Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry. 2001;57:929–967. doi: 10.1016/s0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 10.Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 11.Esko JD, Kimata K, Lindahl U. Proteoglycans and Sulfated Glycosaminoglycans. Essentials of Glycobiology. 2009 [PubMed] [Google Scholar]
- 12.Sorensen I, Domozych D, Willats WG. How have plant cell walls evolved? Plant Physiol. 2010;153:366–372. doi: 10.1104/pp.110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biermann CH, Marks JA, Vilela-Silva AC, Castro MO, Mourao PA. Carbohydrate-based species recognition in sea urchin fertilization: another avenue for speciation? Evol Dev. 2004;6:353–361. doi: 10.1111/j.1525-142X.2004.04043.x. [DOI] [PubMed] [Google Scholar]
- 14.Pang PC, Chiu PC, Lee CL, Chang LY, Panico M, Morris HR, Haslam SM, Khoo KH, Clark GF, Yeung WS, Dell A. Human sperm binding is mediated by the sialyl-Lewis(x) oligosaccharide on the zona pellucida. Science. 2011;333:1761–1764. doi: 10.1126/science.1207438. [DOI] [PubMed] [Google Scholar]
- 15.Tecle E, Gagneux P. Sughar Coated Sperm: Unravelling the function of the mammalian sperm glycocalyx. Molecular Reproduction and Development. 2015;82:635–650. doi: 10.1002/mrd.22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hennen E, Czopka T, Faissner A. Structurally distinct LewisX glycans distinguish subpopulations of neural stem/progenitor cells. J Biol Chem. 2011;286:16321–16331. doi: 10.1074/jbc.M110.201095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirouse V, Christoforou CP, Fritsch C, St Johnston D, Ray RP. Dystroglycan and perlecan provide a basal cue required for epithelial polarity during energetic stress. Dev Cell. 2009;16:83–92. doi: 10.1016/j.devcel.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Ikonen E, Simons K. Protein and lipid sorting from the trans-Golgi network to the plasma membrane in polarized cells. Semin Cell Dev Biol. 1998;9:503–509. doi: 10.1006/scdb.1998.0258. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo I, Kimura-Yoshida C. Extracellular distribution of diffusible growth factors controlled by heparan sulfate proteoglycans during mammalian embryogenesis. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenderson BA, Stamenkovic I, Aruffo A. Localization of hyaluronan in mouse embryos during implantation, gastrulation and organogenesis. Differentiation. 1993;54:85–98. doi: 10.1111/j.1432-0436.1993.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 21.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Hart GW. Three Decades of Research on O-GlcNAcylation - A Major Nutrient Sensor That Regulates Signaling, Transcription and Cellular Metabolism. Front Endocrinol (Lausanne) 2014;5:183. doi: 10.3389/fendo.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varki A. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology. 2011;21:1121–1124. doi: 10.1093/glycob/cwr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 25.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashwell G, Harford J. Carbohydrate-specific receptors of the liver. Annu Rev Biochem. 1982;51:531–554. doi: 10.1146/annurev.bi.51.070182.002531. [DOI] [PubMed] [Google Scholar]
- 27.Grewal PK, Uchiyama S, Ditto D, Varki N, Le DT, Nizet V, Marth JD. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat Med. 2008;14:648–655. doi: 10.1038/nm1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnaar RL, Gerardy-Schahn R, Hildebrandt H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol Rev. 2014;94:461–518. doi: 10.1152/physrev.00033.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lackie PM, Zuber C, Roth J. Polysialic acid of the neural cell adhesion molecule (N-CAM) is widely expressed during organogenesis in mesodermal and endodermal derivatives. Differentiation. 1994;57:119–131. doi: 10.1046/j.1432-0436.1994.5720119.x. [DOI] [PubMed] [Google Scholar]
- 30.Compagno D, Jaworski FM, Gentilini L, Contrufo G, Gonzalez Perez I, Elola MT, Pregi N, Rabinovich GA, Laderach DJ. Galectins: major signaling modulators inside and outside the cell. Curr Mol Med. 2014;14:630–651. doi: 10.2174/1566524014666140603101953. [DOI] [PubMed] [Google Scholar]
- 31.Chen HY, I, Weng C, Hong MH, Liu FT. Galectins as bacterial sensors in the host innate response. Curr Opin Microbiol. 2014;17:75–81. doi: 10.1016/j.mib.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 32.McEver RP, Moore KL, Cummings RD. Leukocyte trafficking mediated by selectin-carbohydrate interactions. J Biol Chem. 1995;270:11025–11028. doi: 10.1074/jbc.270.19.11025. [DOI] [PubMed] [Google Scholar]
- 33.Genbacev OD, Prakobphol A, Foulk RA, Krtolica AR, Ilic D, Singer MS, Yang ZQ, Kiessling LL, Rosen SD, Fisher SJ. Trophoblast L-selectin-mediated adhesion at the maternal-fetal interface. Science. 2003;299:405–408. doi: 10.1126/science.1079546. [DOI] [PubMed] [Google Scholar]
- 34.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 35.Smant G, Stokkermans JP, Yan Y, de Boer JM, Baum TJ, Wang X, Hussey RS, Gommers FJ, Henrissat B, Davis EL, Helder J, Schots A, Bakker J. Endogenous cellulases in animals: isolation of beta-1, 4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc Natl Acad Sci U S A. 1998;95:4906–4911. doi: 10.1073/pnas.95.9.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 37.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ilg K, Yavuz E, Maffioli C, Priem B, Aebi M. Glycomimicry: display of the GM3 sugar epitope on Escherichia coli and Salmonella enterica sv Typhimurium. Glycobiology. 2010;20:1289–1297. doi: 10.1093/glycob/cwq091. [DOI] [PubMed] [Google Scholar]
- 39.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surana NK, Kasper DL. The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunol Rev. 2012;245:13–26. doi: 10.1111/j.1600-065X.2011.01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hooper LV, Gordon JI. Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology. 2001;11:1R–10R. doi: 10.1093/glycob/11.2.1r. [DOI] [PubMed] [Google Scholar]
- 42.Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- 43.Lehotzky RE, Partch CL, Mukherjee S, Cash HL, Goldman WE, Gardner KH, Hooper LV. Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc Natl Acad Sci U S A. 2010;107:7722–7727. doi: 10.1073/pnas.0909449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen M, Zhang XQ, Senaati HP, Chen HW, Varki NM, Schooley RT, Gagneux P. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J. 2013;10:321. doi: 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindberg AA. Bacteriophage receptors. Annu Rev Microbiol. 1973;27:205–241. doi: 10.1146/annurev.mi.27.100173.001225. [DOI] [PubMed] [Google Scholar]
- 46.Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 47.Cook JD, Lee JE. The secret life of viral entry glycoproteins: moonlighting in immune evasion. PLoS Pathog. 2013;9:e1003258. doi: 10.1371/journal.ppat.1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smit CH, van Diepen A, Nguyen DL, Wuhrer M, Hoffmann KF, Deelder AM, Hokke CH. Glycomic Analysis of Life Stages of the Human Parasite Schistosoma mansoni Reveals Developmental Expression Profiles of Functional and Antigenic Glycan Motifs. Mol Cell Proteomics. 2015;14:1750–1769. doi: 10.1074/mcp.M115.048280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.King RA. Statictical Ecology. Annual Reviews of Statistics and its Application. 2014;1:401–426. [Google Scholar]
- 50.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng Q, Sukumaran J, Wu S, Rodrigo A. Neutral Models of Microbiome Evolution. PLoS Comput Biol. 2015;11:e1004365. doi: 10.1371/journal.pcbi.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barton NH, Keightley PD. Understanding quantitative genetic variation. Nat Rev Genet. 2002;3:11–21. doi: 10.1038/nrg700. [DOI] [PubMed] [Google Scholar]
- 53.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013;110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grafen A. Natural Selection, Kin Selection and Group Selection. In: Krebs JR, Davies NB, editors. Behavioural Ecology an Evolutionary Approach. Blackwell Scientific Publications; Oxford: 1984. pp. 62–84. [Google Scholar]
- 56.Hendry AP, Kinnison MT. An introduction to microevolution: rate, pattern, process. Genetica. 2001;112–113:1–8. [PubMed] [Google Scholar]
- 57.Pigliucci M. An extended synthesis for evolutionary biology. Ann N Y Acad Sci. 2009;1168:218–228. doi: 10.1111/j.1749-6632.2009.04578.x. [DOI] [PubMed] [Google Scholar]
- 58.Day T, Bonduriansky R. A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am Nat. 2011;178:E18–E36. doi: 10.1086/660911. [DOI] [PubMed] [Google Scholar]
- 59.Hardiville S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014;20:208–213. doi: 10.1016/j.cmet.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrett RD, Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nat Rev Genet. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- 61.Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 62.Springer SA, Diaz SL, Gagneux P. Parallel evolution of a self-signal: humans and new world monkeys independently lost the cell surface sugar Neu5Gc. Immunogenetics. 2014;66:671–674. doi: 10.1007/s00251-014-0795-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galili U, Shohet SB, Kobrin E, Stults CL, Macher BA. Man, apes, and Old World monkeys differ from other mammals in the expression of alpha-galactosyl epitopes on nucleated cells. J Biol Chem. 1988;263:17755–17762. [PubMed] [Google Scholar]
- 64.Takeuchi Y, Porter CD, Strahan KM, Preece AF, Gustafsson K, Cosset FL, Weiss RA, Collins MK. Sensitization of cells and retroviruses to human serum by (alpha 1–3) galactosyltransferase. Nature. 1996;379:85–88. doi: 10.1038/379085a0. [DOI] [PubMed] [Google Scholar]
- 65.Wolver SE, Sun DR, Commins SP, Schwartz LB. A peculiar cause of anaphylaxis: no more steak? The journey to discovery of a newly recognized allergy to galactose-alpha-1,3-galactose found in mammalian meat. J Gen Intern Med. 2013;28:322–325. doi: 10.1007/s11606-012-2144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper DK. Identification of alpha Gal as the major target for human anti-pig antibodies. Xenotransplantation. 2009;16:47–49. doi: 10.1111/j.1399-3089.2009.00513.x. [DOI] [PubMed] [Google Scholar]
- 67.Kristian SA, Hwang JH, Hall B, Leire E, Iacomini J, Old R, Galili U, Roberts C, Mullis KB, Westby M, Nizet V. Retargeting pre-existing human antibodies to a bacterial pathogen with an alpha-Gal conjugated aptamer. J Mol Med (Berl) 2015;93:619–631. doi: 10.1007/s00109-015-1280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chou HH, Hayakawa T, Diaz S, Krings M, Indriati E, Leakey M, Paabo S, Satta Y, Takahata N, Varki A. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci U S A. 2002;99:11736–11741. doi: 10.1073/pnas.182257399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghaderi D, Springer SA, Ma F, Cohen M, Secrest P, Taylor RE, Varki A, Gagneux P. Sexual selection by female immunity against paternal antigens can fix loss of function alleles. Proc Natl Acad Sci U S A. 2011;108:17743–17748. doi: 10.1073/pnas.1102302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ng PS, Bohm R, Hartley-Tassell LE, Steen JA, Wang H, Lukowski SW, Hawthorne PL, Trezise AE, Coloe PJ, Grimmond SM, Haselhorst T, von Itzstein M, Paton AW, Paton JC, Jennings MP. Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nat Commun. 2014;5:5750. doi: 10.1038/ncomms6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwarz F, Fong JJ, Varki A. Human-specific evolutionary changes in the biology of siglecs. Adv Exp Med Biol. 2015;842:1–16. doi: 10.1007/978-3-319-11280-0_1. [DOI] [PubMed] [Google Scholar]
- 72.Aymard JP. Karl Landsteiner (1868–1943) and the discovery of blood groups. Transfus Clin Biol. 2012;19:244–248. doi: 10.1016/j.tracli.2012.08.127. [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto F, Cid E, Yamamoto M, Saitou N, Bertranpetit J, Blancher A. An integrative evolution theory of histo-blood group ABO and related genes. Sci Rep. 2014;4:6601. doi: 10.1038/srep06601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marionneau S, Ruvoen N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology. 2002;122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neil SJ, McKnight A, Gustafsson K, Weiss RA. HIV-1 incorporates ABO histo-blood group antigens that sensitize virions to complement-mediated inactivation. Blood. 2005;105:4693–4699. doi: 10.1182/blood-2004-11-4267. [DOI] [PubMed] [Google Scholar]
- 76.Adibekian A, Stallforth P, Hecht M, Werz DB, Gagneux P, Seeberger PH. Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chemical Science. 2:337–344. [Google Scholar]
- 77.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 78.Stanley P, Schachter H, Taniguchi N. N-Glycans. Essentials of Glycobiology 2009 [Google Scholar]
- 79.Hang I, Lin CW, Grant OC, Fleurkens S, Villiger TK, Soos M, Morbidelli M, Woods RJ, Gauss R, Aebi M. Analysis of site-specific N-glycan remodelling in the ER and the Golgi. Glycobiology. 2015 doi: 10.1093/glycob/cwv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stepan H, Staudacher E. Optimization of monosaccharide determination using anthranilic acid and 1-phenyl-3-methyl-5-pyrazolone for gastropod analysis. Anal Biochem. 2011;418:24–29. doi: 10.1016/j.ab.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brockhausen I, Schachter H, Stanley P. O-GalNAc Glycans. Essentials of Glycobiology 2009 [Google Scholar]
- 82.Stanford KI, Bishop JR, Foley EM, Gonzales JC, Niesman IR, Witztum JL, Esko JD. Syndecan-1 is the primary heparan sulfate proteoglycan mediating hepatic clearance of triglyceride-rich lipoproteins in mice. J Clin Invest. 2009;119:3236–3245. doi: 10.1172/JCI38251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crawford BE, Garner OB, Bishop JR, Zhang DY, Bush KT, Nigam SK, Esko JD. Loss of the heparan sulfate sulfotransferase, Ndst1, in mammary epithelial cells selectively blocks lobuloalveolar development in mice. PLoS One. 2010;5:e10691. doi: 10.1371/journal.pone.0010691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miller GM, Hsieh-Wilson LC. Sugar-Dependent Modulation of Neuronal Development, Regeneration, and Plasticity by Chondroitin Sulfate Proteoglycans. Exp Neurol. 2015 doi: 10.1016/j.expneurol.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schnaar RL, Suzuki A, Stanley P. Glycosphingolipids. Essentials of Glycobiology 2009 [Google Scholar]
- 86.Ferguson MAJ, Kinoshita T, Hart GW. Glycosylphosphatidylinositol Anchors. Essentials of Glycobiology 2009 [Google Scholar]
- 87.Schaffer C, Graninger M, Messner P. Prokaryotic glycosylation. Proteomics. 2001;1:248–261. doi: 10.1002/1615-9861(200102)1:2<248::AID-PROT248>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 88.Freeze HH, Haltiwanger RS. Other Classes of ER/Golgi-derived Glycans. Essentials of Glycobiology 2009 [Google Scholar]
- 89.Defaus S, Gupta P, Andreu D, Gutierrez-Gallego R. Mammalian protein glycosylation--structure versus function. Analyst. 2014;139:2944–2967. doi: 10.1039/c3an02245e. [DOI] [PubMed] [Google Scholar]
- 90.Ito S. Form and function of the glycocalyx on free cell surfaces. Philos Trans R Soc Lond B Biol Sci. 1974;268:55–66. doi: 10.1098/rstb.1974.0015. [DOI] [PubMed] [Google Scholar]
- 91.Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology. 2004;14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 92.O’Reilly MK, Paulson JC. Multivalent ligands for siglecs. Methods Enzymol. 2010;478:343–363. doi: 10.1016/S0076-6879(10)78017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 94.Tissot B, North SJ, Ceroni A, Pang PC, Panico M, Rosati F, Capone A, Haslam SM, Dell A, Morris HR. Glycoproteomics: past, present and future. FEBS Lett. 2009;583:1728–1735. doi: 10.1016/j.febslet.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moothoo DN, Naismith JH. A general method for co-crystallization of concanavalin A with carbohydrates. Acta Crystallogr D Biol Crystallogr. 1999;55:353–355. doi: 10.1107/S0907444998008919. [DOI] [PubMed] [Google Scholar]
- 96.Huang ML, Cohen M, Fisher CJ, Schooley RT, Gagneux P, Godula K. Determination of receptor specificities for whole influenza viruses using multivalent glycan arrays. Chem Commun (Camb) 2015;51:5326–5329. doi: 10.1039/c4cc08613a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Orr SL, Le D, Long JM, Sobieszczuk P, Ma B, Tian H, Fang X, Paulson JC, Marth JD, Varki N. A phenotype survey of 36 mutant mouse strains with gene-targeted defects in glycosyltransferases or glycan-binding proteins. Glycobiology. 2013;23:363–380. doi: 10.1093/glycob/cws150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Freeze HH, Eklund EA, Ng BG, Patterson MC. Neurological Aspects of Human Glycosylation Disorders. Annu Rev Neurosci. 2015;38:105–125. doi: 10.1146/annurev-neuro-071714-034019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 100.Chang PV, Prescher JA, Hangauer MJ, Bertozzi CR. Imaging cell surface glycans with bioorthogonal chemical reporters. J Am Chem Soc. 2007;129:8400–8401. doi: 10.1021/ja070238o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cohen M, Varki A. Modulation of glycan recognition by clustered saccharide patches. Int Rev Cell Mol Biol. 2014;308:75–125. doi: 10.1016/B978-0-12-800097-7.00003-8. [DOI] [PubMed] [Google Scholar]
- 102.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]