Significance
Although the pathogenicity of autoantibodies (autoAbs) targeting the desmosomal cadherins desmoglein (Dsg) 3 and 1 is well established in pemphigus vulgaris (PV), the disease relevance of other autoAbs remains an area of intense research. By using disease-specific protein microarrays to characterize the autoAb response in PV, we identified Dsg and non-Dsg antigens as primary targets of patient autoAbs. Additionally, we found that healthy first- or second-degree relatives of patients with PV, specifically those expressing HLA DRB1*0402 or DQB1*0503 (PV susceptibility alleles) exhibit patterns of autoreactivity similar to that of patients with PV. These findings suggest a potential role for non-Dsg autoAbs in disease pathogenesis and implicate HLA as a primary driver of autoAb specificity in PV.
Keywords: pemphigus, autoantibody, protein microarray
Abstract
Patients with pemphigus vulgaris (PV) harbor antibodies reactive against self-antigens expressed at the surface of keratinocytes, primarily desmoglein (Dsg) 3 and, to a lesser extent, Dsg1. Conventionally, only antibodies targeting these molecules have been thought to contribute to disease pathogenesis. This notion has been challenged by a growing pool of evidence that suggests that antibodies toward additional targets may play a role in disease. The aims of this study were to (i) establish high-throughput protein microarray technology as a method to investigate traditional and putative autoantibodies (autoAbs) in PV and (ii) use multiplexed protein array technology to define the scope and specificity of the autoAb response in PV. Our analysis demonstrated significant IgG reactivity in patients with PV toward the muscarinic acetylcholine receptor subtypes 3, 4, and 5 as well as thyroid peroxidase. Furthermore, we found that healthy first- and second-degree relatives of patients with PV express autoAbs toward desmoglein and non-Dsg targets. Our analysis also identified genetic elements, particularly HLA, as key drivers of autoAb expression. Finally, we show that patients with PV exhibit significantly reduced IgM reactivity toward disease-associated antigens relative to controls. The use of protein microarrays to profile the autoAb response in PV advanced the current understanding of disease and provided insight into the complex relationship between genetics and disease development.
Pemphigus vulgaris (PV) is a blistering autoimmune skin disease characterized by the presence of autoantibodies (autoAbs) directed against keratinocyte surface antigens (1, 2). Although early immunofluorescence studies demonstrated the presence of autoAbs in patient sera that bound to the surface of keratinocytes, a direct role of autoAbs in disease pathogenesis was not established until purified patient IgG (PVIgG) was shown to elicit blister formation upon passive transfer in mice (3). The main targets of these autoAbs were identified as Desmoglein (Dsg) 3 and 1, cadherin proteins that constitute key components of desmosomes, protein complexes responsible for maintaining cell–cell adhesion (4–7). Later experiments in which patient sera depleted of anti-Dsg3 Abs failed to produce blisters when passively transferred to mice (8) seemingly cemented the notion that these autoAbs alone were responsible for blister formation. As a result, subsequent research in the field has focused primarily on autoAbs directed against Dsgs.
The assertion that anti-Dsg autoAbs alone are pathogenic was first challenged when PVIgG, lacking any anti-Dsg1 autoAbs, produced blisters when passively transferred into Dsg3-null mice (9). Additionally, several studies have shown that anti-Dsg Ab titers do not necessarily correlate with disease activity, and a subset of patients with PV do not harbor any detectable anti-Dsg Abs (10–14). The presence of pathogenic autoAbs directed against non-Dsg targets could account for these findings. Early studies established the presence of non-Dsg autoAbs in PV sera by showing that PVIgG depleted of anti-Dsg3 Abs recognized a number of non-Dsg antigens (15), and subsequent work identified several specific non-Dsg proteins as targets of autoAbs in PV (9, 16–20).
Currently, the scope and specificity of non-Dsg autoAbs in PV has not been fully examined, and the genetic factors underlying autoAb generation, including the impact of HLA allele expression on PV autoAb repertoires, are not well understood. Without a detailed understanding of the specificity of the autoimmune response, broad-scale immunosuppression, whose side effects alone can be severe or even life-threatening, remains the mainstay treatment of PV. Characterization of the scope and specificity of autoAbs in PV represents a potentially pivotal step toward a better understanding of disease mechanisms in pemphigus, facilitating the development of more specific and safe treatments.
In recent years, the development of protein microarrays has enabled high-throughput analysis and improved sensitivity in the detection of autoAbs compared with conventional methods such as ELISA (21, 22). Array technology has been successfully used to assess autoAb responses in several autoimmune diseases, including systemic lupus erythematous, polymyositis, rheumatoid arthritis, and multiple sclerosis (23–25). In the present study, we used protein array technology to experimentally examine the scope and specificity of autoAbs in PV. Candidate antigens were identified by a thorough review of the literature (5, 9, 26–34). The selected proteins were printed onto glass slides and used to probe the serum of patients with pemphigus, healthy first- or second-degree relatives of patients with PV, and unrelated control subjects. By using this methodology, we clearly demonstrated increased autoAb reactivity in patients with active disease toward four non-Dsg antigenic targets in addition to Dsg3. We also found that, compared with the unrelated control group, related control subjects express greater IgG autoAb reactivity toward a subset of the antigens recognized by the patient autoAbs. Importantly, we found that autoAb reactivity in related control subjects is tightly associated with the expression of either of two HLA alleles known to be highly associated with PV susceptibility, DRB1*0402 and DQB1*0503. Surprisingly, in contrast to the IgG response, IgM reactivity toward Dsg and non-Dsg antigens was found to be depressed in patients with active pemphigus relative to unrelated control subjects.
Materials and Methods
Patient Recruitment and Serum Samples.
Patients with PV and control subjects were recruited at meetings of the International Pemphigus and Pemphigoid Foundation as well as the outpatient clinics of the departments of dermatology at Weill Medical College of Cornell University [institutional review board (IRB) 0998–398] and Michigan State University (IRB 05-1034). Diagnosis of PV was based on established clinical and histopathological criteria. Patients and controls donated blood after signing an informed consent document. Demographic information as well as information regarding duration, morphology and current treatment was obtained at the time of blood draw. Serum was isolated from peripheral blood immediately and stored at −80 °C until further use. A total of 40 active pemphigus patients [39 PV and 1 pemphigus foliaceous (PF); disease activity defined as presence of nontransient lesions, i.e., lesions lasting more than 1 wk, according to published disease definitions (35)] and 40 control patients (20 unrelated subjects and 20 first- or second-degree relatives of patients with PV) with no history of pemphigus or other autoimmune diseases were included in this study. Of the active patients, 26 were female and 14 male, with ages ranging from 21 to 76 y (mean age, 53.2 ± 15.8 y). More detailed demographic information of patients is provided in Table S1.
Table S1.
Demographic information of patient and control groups
| Group | Age (y) | Sex | Ethnicity |
| PVA | 52.0 ± 15.8 | 26 F/14 M | 17 Ashkenazi, 15 Caucasian, 2 Asian, 6 other |
| UCR | 32.4 ± 13.9 | 10 F/10 M | 1 Ashkenazi, 7 Caucasian, 10 Asian, 2 other |
| RCR | 50.8 ± 15.1 | 13 F/7 M | 9 Ashkenazi, 7 Caucasian, 0 Asian, 4 other |
PVA, active PV; RCR, related controls; UCR, unrelated controls. Demographic information for all subjects was obtained at the time of enrollment. Age shown as mean ± SD. The majority of subjects stated their ethnicity as Ashkenazi, Caucasian, or Asian. Patients who identified their ethnicity as African American, African, Hispanic, or “other” are listed as “other”.
Autoantigen Microarray Design and Printing.
Antigens were selected based on a comprehensive survey of literature aimed at identifying potentially disease relevant targets of autoAbs in PV (5, 9, 26–34). Fifteen PV-associated autoantigens were identified and printed on the array: Dsg1–4, muscarinic acetylcholine receptor subtypes M1 and M3–M5 (mAChR1 and -3–5), desmocollins 2 and 3 (Dsc2 and -3), E-cadherin (E-cad), Fc receptor-ε 1 (FcεR1), pemphaxin (ANXA9), plakoglobin (PKG), and thyroid peroxidase (TPO). The bullous pemphigoid-associated antigen BP230 (no known reactivity with pemphigus sera) and an HIV-peptide (corresponding to a highly conserved region in the gp41protein with no known reactivity in HIV negative individuals) were included as negative biological controls. Tetanus toxoid (Ttox) was included as a positive biological control antigen. All antigens used were purified recombinant proteins produced in various expression systems and were plated in their native, nondenatured state (Table S2). Technical controls consisted of human IgA, IgM, IgG1–4, normal BSA, and biotinylated BSA. A TECAN scanner was used to spot the proteins onto nitrocellulose-coated glass slides. Preliminary experiments determined that the optimal printing concentration was 0.2 mg/mL for all printed proteins. All antigens were printed in triplicate. A representative image of a slide and array template is shown in Fig. 1A.
Table S2.
List of antigens printed on protein microarray
| Antigen | Supplier | Catalogue no. |
| Dsg1 | Abnova | H00001828-P01 |
| Dsg2 | R & D Systems | 947-DM |
| Dsg3 | R & D Systems | 1720-DM |
| Dsg4 | Abnova | H00147409-Q01 |
| Dsc2 | Novus Biologicals | H00001824-Q01 |
| Dsc3 | Novus Biologicals | H00001825-Q01 |
| mAChR1 | Perkin-Elmer | RBHM1M |
| mAChR3 | Perkin-Elmer | RBHM3M |
| mAChR4 | Perkin-Elmer | RBHM4M |
| mAChR5 | Perkin-Elmer | RBHM5M |
| E-cad | Spring Bioscience | P3464 |
| PKG | Novus Biologicals | H00003728-P01 |
| ANXA9 | US Biological | A2296-69A |
| FcER1 | Novus Biologicals | H00002205-P01 |
| TPO | GenWay | 10–663-45310 |
| Ttox | ANASPEC | 61518 |
| BP230 | Novus Biologicals | H00000667-Q01 |
| HIV-1 gp41 | Polimun Scientific | AB001 |
Fig. 1.
Slide template and verification of protein printing. (A) Each microarray slide (Left) consists of 48 identical arrays (zoomed-in image of one representative array is shown at Top Right), each printed with 48 antigen triplicates. (B) Arrays were probed with select antibodies directed against various printed antigens to verify proper printing and demonstrate the specificity of the array (a representative selection of antigen features visualized with their respective fluorochrome-labeled antibody is shown).
Hybridization and Scanning of Autoantigen Microarrays.
Before the experiment, the microarray slides were brought to room temperature for 30 min. and washed four times in PBS solution/Tween 0.05%. After drying by centrifugation for 1 min at 150 × g at 23 °C, the slides were incubated in blocking buffer (1% BSA in PBS/Tween 0.05%) for 1 h at room temperature under gentle rocking. Six microliters of serum diluted to 1:150 was added to each array and incubated at room temperature for 1 h. After an additional washing step, slides were incubated in biotinylated anti-human IgG or IgM antibody at 1 µg/mL at room temperature for 1 h. Slides were then washed and dried as outlined earlier. Streptavidin–phycoerythrin was added to each array and incubated for 1 h at room temperature, followed by another washing and drying step. Finally, the slides were scanned by using a TECAN scanner (Tecan Group) at 532 nm to generate TIFF images for analysis.
Determination of Anti-Dsg3 and -1 Serum Levels.
A commercially available 96-well Dsg1 and Dsg3 ELISA kit (MESACUP Dsg1 and Dsg3 ELISA Test System; MBL International) was used to categorize serum antibody levels of anti-Dsg1 and anti-Dsg3 antibodies of all patient and control samples. Briefly, patient serum diluted 1:100 was added to wells precoated with the ectodomains of Dsg1 or Dsg3. AutoAbs were detected by addition of a secondary HRP-labeled antibody and color development by tetramethylbenzidine substrate per manufacturer’s instructions. OD values were assessed under a spectrophotometer (Molecular Devices) at 450 nm. To compare samples from different ELISA plates, optical densities were adjusted relative to a positive and negative control sample in each plate and are reported as index values [(OD of tested serum − OD negative control)/(OD of positive control − OD of negative control) × 100]. Patients were defined as positive for either of the anti-Dsg Abs if the ELISA index value exceeded 20.0 U/mL.
HLA Typing.
HLA typing of PV patients and related and unrelated controls was performed at the Rogosin Institute (New York) and the Tissue Typing Laboratory at Michigan State University by amplification with specific primers (36). Patients and controls were defined as HLA-positive (HLA+) if they possessed at least one of the PV-associated HLA class II alleles DRB*0402 or DQB*0503 (37) or HLA-negative (HLA−) if expressing neither allele.
Data Analysis.
The image of each array was analyzed using Genepix Pro-5.0 software to generate a Gene Pix Results file. The net fluorescence intensities (spot fluorescence intensity minus background intensity) were determined for every printed spot of every triplicate on every array. The mean net fluorescence intensity (MFI) for each triplicate was taken, and each value was normalized to the fluorescence intensity of the appropriate negative controls. Significance analysis of microarrays (SAM) was then applied to detect statistically significant differences [defined as q-value < 0.05 and false discovery rate (FDR) < 0.05] in array reactivity between active patient subgroups and healthy controls (38). Unbiased hierarchal cluster analysis was performed by using Cluster (version 3.0). Heat maps and node trees were generated by using TreeView (version 1.1.6) (39). Dot plots were prepared with KaleidaGraph software suite (version 4.5; Synergy Software).
Results
Development and Validation of Protein Microarrays.
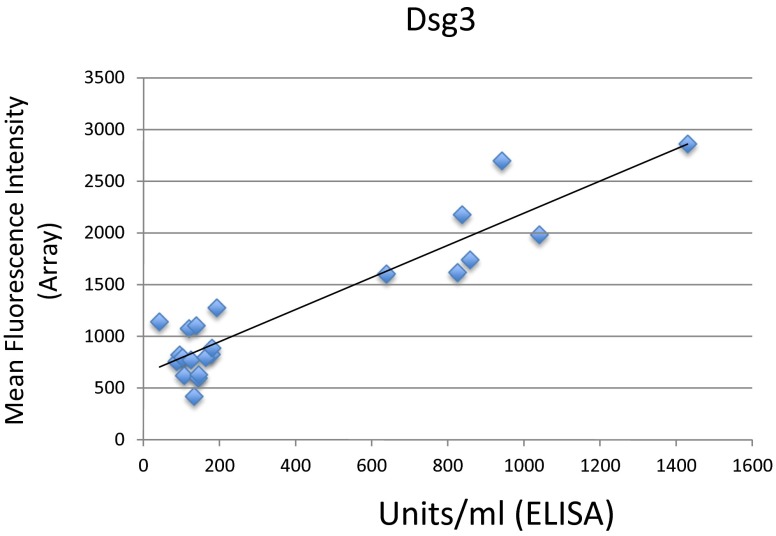
Fifteen protein antigens with potential relevance to PV (5, 20, 27–35) were printed on glass slides in triplicate (details provided in Materials and Methods). To establish and validate our methodology, individual arrays were probed with commercially available antibodies generated against specific printed antigens to demonstrate the specificity of Abs to the plated proteins and to ensure that the proteins were antigenically intact (Fig. 1B). Comparison of array values for Dsg3 reactivity in patients with active pemphigus to anti-Dsg3 ELISA index values obtained in separate experiments showed that array values for anti-Dsg3 were highly correlated to ELISA index values (Fig. S1). Together, these results demonstrate proper array manufacturing and functioning.
Fig. S1.
Reactivity toward Dsg3 measured by protein microarray is highly correlated to anti-Dsg3 ELISA values. Commercially available anti-Dsg3 ELISA kits were used to determine the level of anti-Dsg3 antibodies in the active PV patient group shown by ELISA to be positive for anti-Dsg3 autoAbs (n = 23) and compared with values obtained my protein microarray. Microarray reactivity toward Dsg3 was positively correlated to values obtained by ELISA (R = 0.93).
Patients with Active PV Exhibit Greater IgG Reactivity to Dsg and Non-Dsg Targets than Healthy, Unrelated Controls.
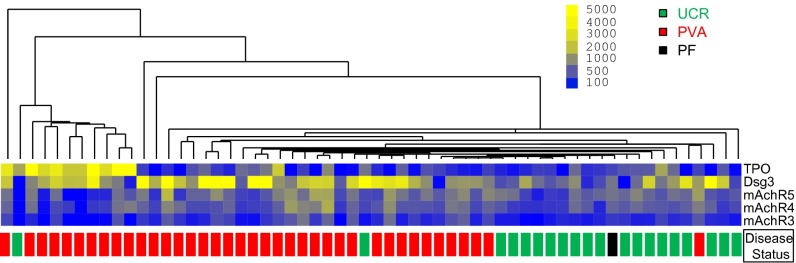
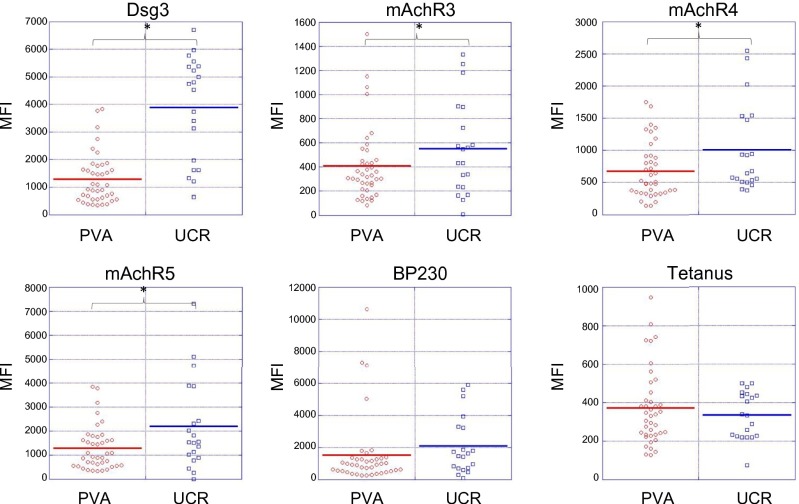
We first assessed serum reactivities to the panel of 15 autoantigens in active pemphigus patients (PV, n = 39; PF, n = 1) compared with healthy unrelated controls with no personal history of autoimmune disease or family history of PV (n = 20). Five antigen features were identified by SAM to have statistically significant increases in IgG autoAb reactivity in active pemphigus patients vs. healthy controls: Dsg3, mAChR3, mAchR4, mAchR5, and TPO (q-value < 0.05, FDR < 0.05). No significant differences in IgG reactivity were found for the positive and negative control antigens (Ttox and BP230, respectively). Unbiased hierarchical cluster analysis of array values based on these five autoantigen reactivities demonstrated an almost complete separation of active pemphigus patients from unrelated controls (Fig. 2). Assessment of treatment status revealed that therapy did not drive the clustering of patients with PV (Fig. S2). Of the 40 pemphigus patients tested, only two clustered within the unrelated control group and vice versa, indicating that, in addition to reactivity toward Dsg3, reactivity toward the non-Dsg targets mAChR3, mAchR4, mAchR5, and TPO distinguishes patients from healthy controls lacking self or family history of autoimmune disease. One of the two patient samples clustering with the controls was the only patient with PF included in the study, indicating that this patient with PF displays a distinct autoAb profile from PV.
Fig. 2.
Patients with active PV exhibit increased IgG autoAb reactivity against Dsg- and non-Dsg antigens. Sera from active patients (PVA, n = 39) and unrelated controls (UCR, n = 20) were probed for autoAbs directed against plated antigens. SAM identified five antigens (mAChR3, mAChR4, mAChR5, Dsg3, TPO) that showed significantly greater reactivity (q < 0.05) in active PV patients compared with unrelated control subjects. These five antigens were used in an unbiased clustering approach to reveal similarities/differences in autoAb profiles between subjects. Active PV, red boxes; unrelated controls, green boxes; PF, black box.
Fig. S2.
Description of therapy status of active patients and its association with autoAb expression. Information regarding any current treatment was obtained at the time of blood draw. The therapy status of each patient was classified as minimal (marked “M”), more than minimal (marked “>M”), or off therapy based on published guidelines and definitions (35). Addition of the therapy status of each patient to the cluster analysis performed in Fig. 3 reveals that therapy does not underlie patient clustering.
Healthy Controls Related to PV Patients Exhibit autoAb Specificities Similar to Active PV Patients and Distinct from Unrelated Controls.
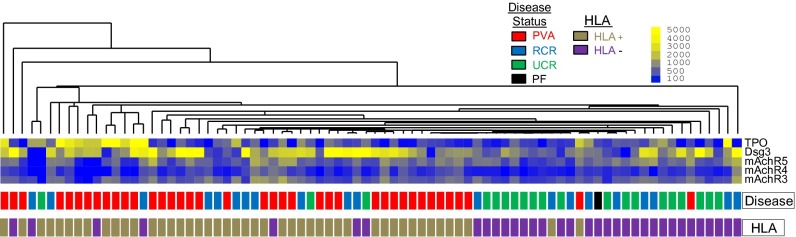
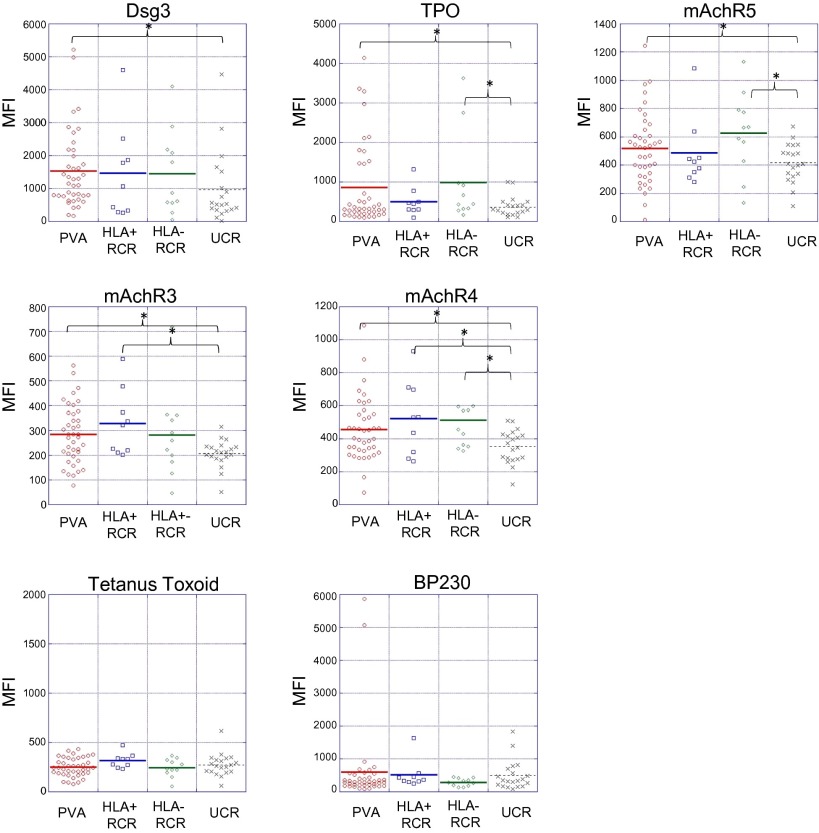
The presence of Abs directed at self-antigens has traditionally been viewed as a consequence of a break in immune tolerance that ultimately contributes to the development of autoimmune pathologies. Recent studies demonstrating the presence of abundant autoAbs in healthy individuals have challenged this view, suggesting that these “natural” autoAbs may play a role in immune homeostasis (40–46). The detection of Abs directed against Dsg1 and -3 in healthy controls related to patients with PV (47, 48) suggests that shared genetic (and possibly environmental) factors may contribute to the development of autoreactivity, if not autoimmune disease. To investigate further, in the next analysis, we included control subjects who were first- or second-degree blood relatives of patients with PV (n = 20). Compared with all (unfractionated) controls (related, n = 20 plus unrelated, n = 20; total n = 40), patients with active pemphigus had significantly increased reactivity only to Dsg3. In addition, the distinction between patients and controls based on the five autoantigens described in the previous analysis (in which only unrelated control subjects were used) was not as clear. Approximately half of the related controls clustered with the active patient group, whereas the remainder clustered with unrelated controls (Fig. 3). Directly comparing related with unrelated control subjects, we found increased reactivities to the same autoantigens as in the comparison of active pemphigus patients vs. healthy unrelated controls: significant for mAChR4 and TPO at q < 0.05, FDR < 0.05; mAChR3 and -5 and Dsg3 were found to be significant at lower stringencies (q < 0.05, FDR = 0.15). These data indicate that related controls recognize autoantigens similar to those targeted by patients with PV and distinct from those of unrelated control subjects.
Fig. 3.
Clustering of related control subjects with patients with PV and unrelated control subjects is mainly driven by HLA expression. Sera from patients with active PV, unrelated control subjects, and related control subjects was probed by using protein microarray. Based on the reactivity data toward the five antigens that were up-regulated in active PV patients vs. unrelated control subjects, all three groups were clustered by using an unbiased hierarchal clustering algorithm. Subsequently, HLA information was overlaid onto the clustered groups of patients and control subjects to assess the influence of HLA genetics on the expression of potentially disease-relevant autoAbs. HLA+ indicates expression of the PV-associated HLA alleles DRB1*0402 and/or DQB1*0503 (gold boxes); HLA− represents the absence of either allele (purple boxes). PVA (active PV), red boxes; PF, black box; unrelated controls, green boxes; RCR (related control subjects), blue boxes.
HLA Expression Underlies AutoAb Specificity in Healthy Subjects Related to Patients with PV.
Given the tight association of certain HLA alleles with PV, we sought to determine the extent to which HLA expression affects autoAb specificity. Subjects carrying one or both of the two PV-associated HLA alleles (DRB1*0402 and DQB1*0503) were classified as HLA+, whereas subjects carrying neither allele were classified as HLA−. HLA association data were then overlaid on the unbiased hierarchical cluster generated with data from patients with PV, unrelated control subjects, and related control subjects (Fig. 3 and Table S3). Our data suggest that HLA expression may impact the autoAb profiles of healthy controls. Of the 11 related controls who clustered with the patients with PV, 8 were HLA+, i.e., typed as DRB1*0402 and/or DQB1*0503. Conversely, of the 9 related controls that clustered separately from patients with, 8 were HLA−. In addition, both HLA+ unrelated control subjects clustered with patients with PV (Fig. 3). No significant differences in autoAb reactivity patterns were identified between HLA+ (n = 9) and HLA− (n = 11) related control groups. However, these groups demonstrated somewhat different reactivities compared with the unrelated control group: HLA−-related controls showed increased reactivity toward mAchR4, mAchR5, and TPO, whereas HLA+-related controls had significantly increased reactivity toward mAchR3 and 4 (Fig. 4). A direct comparison of HLA+ vs. HLA− active patients was omitted because of the small number (n = 5) of HLA− patients.
Table S3.
Description of HLA status and cluster analysis of all controls
| Control ID | PV-associated HLA alleles | HLA status | Cluster | |
| DRB*0402 | DQB*0503 | |||
| Related controls | ||||
| PVCR 103 | x | HLA+ | Active patients | |
| PVCR 106 | x | HLA+ | Active patients | |
| PVCR 119 | x | HLA+ | Active patients | |
| PVCR 120 | x | HLA+ | Active patients | |
| PVCR 122 | x | HLA+ | Active patients | |
| PVCR 131 | x | HLA+ | Active patients | |
| PVCR 157 | x | HLA+ | Active patients | |
| PVCR 159 | x | x | HLA+ | Active patients |
| PVCR 185 | x | HLA+ | Healthy controls | |
| PVCR 160 | HLA− | Active patients | ||
| PVCR 105 | HLA− | Active patients | ||
| PVCR 133 | HLA− | Active patients | ||
| PVCR 130 | HLA− | Healthy controls | ||
| PVCR 132 | HLA− | Healthy controls | ||
| PVCR 117 | HLA− | Healthy controls | ||
| PVCR 155 | HLA− | Healthy controls | ||
| PVCR 156 | HLA− | Healthy controls | ||
| PVCR 104 | HLA− | Healthy controls | ||
| PVCR 186 | HLA− | Healthy controls | ||
| PVCR 187 | HLA− | Healthy controls | ||
| Unrelated controls | ||||
| PVCR 168 | x | HLA+ | Active patients | |
| PVCR 195 | x | HLA+ | Active patients | |
| PVCR 166 | HLA− | Active patients | ||
| PVCR 112 | HLA− | Healthy controls | ||
| PVCR 114 | HLA− | Healthy controls | ||
| PVCR 115 | HLA− | Healthy controls | ||
| PVCR 135 | HLA− | Healthy controls | ||
| PVCR 158 | HLA− | Healthy controls | ||
| PVCR 110 | HLA− | Healthy controls | ||
| PVCR 169 | HLA− | Healthy controls | ||
| PVCR 171 | HLA− | Healthy controls | ||
| PVCR 172 | HLA− | Healthy controls | ||
| PVCR 173 | HLA− | Healthy controls | ||
| PVCR 174 | HLA− | Healthy controls | ||
| PVCR 176 | HLA− | Healthy controls | ||
| PVCR 177 | HLA− | Healthy controls | ||
| PVCR 178 | HLA− | Healthy controls | ||
| PVCR 179 | HLA− | Healthy controls | ||
| PVCR 192 | HLA− | Healthy controls | ||
| PVCR 193 | HLA− | Healthy controls | ||
An “x” denotes expression of a given HLA PV associated HLA allele (DRB1*0402 or DQB1*0503). HLA+ indicates expression of at least one of the PV associated HLA alleles and HLA− indicates the absence of either allele. The “cluster” column refers to the cluster analysis described in Fig. 4 and denotes if a given patient control was seen to associate with the “active patient” cluster or the “healthy control” group.
Fig. 4.
Related control subjects exhibit increased levels of reactivity toward the same antigens recognized by patients with active PV. Microarray analysis of autoAb reactivity in related control subjects reveals an increase in autoAb reactivity toward a subset of the antigens recognized by patients with active PV. Presence or absence of a disease-associated HLA allele in related control subjects underlies unique autoAb reactivities (asterisk indicates differences in array reactivity determined to be significant by SAM; q < 0.05). No significant differences in reactivity toward Ttox or BP230 were observed between the groups. Units shown reflect mean fluorescence of antigen triplicates normalized to the appropriate negative control. HLA+ indicates expression of the PV-associated HLA alleles DRB1*0402 and/or DQB1*0503, HLA− indicates the absence of either allele. PVA, active PV; UCR, unrelated control subjects; RCR, related control subjects.
IgM Reactivity Levels Are Significantly Higher in Healthy Controls than in Patients with Active PV.
Previous studies concerning autoAbs in PV have focused almost solely on those of the IgG subtype. To expand our understanding of the role of other autoreactive immunoglobulin subtypes in PV, we probed patient sera for IgM autoAbs using our protein microarray. Unexpectedly, compared with patients with active PV, unrelated control subjects were seen to express significantly increased IgM reactivity toward Dsg3 and mAchR3, -4, and -5 (q < 0.05; FDR < 0.05; Fig. 5). With the exception of TPO, these represent all of the antigens that were targeted by IgG autoAbs in the patient population.
Fig. 5.
Unrelated control subjects exhibit increased IgM autoreactivity against disease-related antigens compared with patients with active PV. Patient and unrelated control sera were probed to determine the IgM subtype-specific response to disease-relevant antigens. SAM identified increased IgM reactivity directed against Dsg3, mAChR3, mAChR4, and mAChR5 in unrelated control subjects compared with patients with active PV (asterisk indicates differences in array reactivity determined to be significant by SAM; q < 0.05). With the exception of TPO, the antigens targeted by IgM autoAbs in controls were the same as those targeted by IgG autoAbs in patients with active PV. BP230 and Ttox (negative biological controls) did not show significant differences in IgM reactivity between patients and unrelated control subjects. PVA, active PV; UCR, unrelated controls.
Discussion
AutoAbs directed against the desmosomal cadherins Dsg3 and Dsg1 have classically been considered the principal mediators of disease pathogenesis in PV. As a result, the monitoring of disease activity and effect of clinical interventions have centered on studying these autoAbs. However, as more targets of autoAbs in PV have been reported, there has been a growing pool of evidence suggesting autoAbs directed against non-Dsg antigens may also play a role in disease (9–20). Current knowledge of non-Dsg autoAbs in PV is segmented and discontinuous, with individual studies focusing on non-Dsg targets individually, providing little insight into their disease relevance as biomarkers or drivers of disease. Although non-Dsg autoAbs have been identified in patients, little is known about their prevalence and even less is known concerning any potential effect they may have on disease expression. Understanding the broader scope and specificity of the autoAb response in PV has implications regarding mechanisms of blister formation and may uncover biomarkers capable of more accurately monitoring disease activity and predicting disease progression or response to therapy.
Protein microarrays allow for the rapid assessment of antibodies toward a large number of targets simultaneously, and, not surprisingly, have been successfully used to characterize the autoAb response of several autoimmune diseases (22, 23, 49–52). Here, we used this technology in an attempt to address a number of gaps that exist in our current understanding of PV. Probing the scope and specificity of the autoAb response in PV with our custom-designed, disease-specific protein microarray enabled us to assimilate decades of disparate studies into a more cohesive understanding of autoAbs in PV.
Early studies aimed at evaluating autoAbs in PV were almost all confined to the study of a single antigen, and only a relatively small number of sera were probed in each study. Our methodology exhibits two main advantages compared with previous work: (i) the capability to rapidly assess larger patient populations, resulting in more accurate estimates of autoAb prevalence; and (ii) simultaneous examination of multiple autoAb specificities to allow for an enhanced characterization of the broader autoAb milieu in patients. These advantages greatly increase the likelihood of uncovering novel disease-relevant antigenic targets. Our array studies reveal an increased autoAb reactivity in patients compared with control subjects toward a specific subset of antigens: Dsg3, mAchR3, mAchR4, mAchR5, and TPO. To our knowledge, this finding represents the first time that extensive profiles of autoAbs, beyond only anti-Dsg Abs, have been described in patients with PV, and represents a critical step in identifying potentially functionally significant non-Dsg autoAbs.
Increased autoAb reactivity toward muscarinic acetylcholine receptors suggests that perturbation of cholinergic signaling could represent an alternate pathway contributing to blister formation in PV. Cholinergic signaling plays a vital role in cell adhesion in the epidermis: muscarinic antagonists can disrupt cellular adhesion in vitro and muscarinic agonists can strengthen cell–cell adhesion (53). Additionally, it has been shown that treatment with cholinergic agonists can inhibit the formation of blisters in neonatal mice after the passive transfer of PVIgG (54). Although these findings support a functional role for these autoAbs in PV, future work is required to determine what effects, if any, patient autoAbs have on cholinergic signaling in the epidermis.
We also found TPO, an enzyme expressed primarily on thyroid tissue that is responsible for the organification of iodine, to be a target of PV autoAbs. Interestingly, TPO is not thought to be expressed in keratinocytes at the protein or mRNA level (55). However, anti-TPO Abs have been associated with dysfunctions in the epidermis, mainly in chronic autoimmune urticaria, although a direct pathogenic role has yet to be established (56). IgG purified from patients with Graves disease, an autoimmune thyroid condition, has been shown to effect BrdU incorporation and cAMP accumulation in primary cultures of human keratinocytes (55), further suggesting a potential effect of anti-TPO Abs in cutaneous processes. Alternatively, it is also possible that Abs directed against TPO may cross react with an as yet unknown keratinocyte associated antigen. Additional studies are needed to explore if, and how, Abs directed against TPO may play a role in PV.
The existence of functionally relevant non-Dsg autoAbs could shed light on some of the many questions currently surrounding the pathogenesis of blister formation in PV. In some cases, non-Dsg autoAbs may be sufficient to induce the formation of blisters, which would explain the presence of disease in the subset of patients who do not express anti-Dsg3 or anti-Dsg1 Abs. Non-Dsg autoAbs could also work in concert with anti-Dsg autoAbs, augmenting the ability of pathogenic Abs to cause blistering. A certain combination of anti-Dsg and non-Dsg autoAbs might be necessary to cross the threshold of pathogenicity, tipping the scale toward self-recognition that distinguishes disease remission and activity. This theory may help to explain the lack of a tight correlation between anti-Dsg Ab titers and lesional activity. Moreover, non-Dsg autoAbs may contribute to autoAb profiles that underlie the clinical variability of disease morphologies that exists across patients. Identification of functionally significant autoAb could open the door for monitoring multiple autoAb levels to more accurately assess and classify disease activity, evaluate the efficacy of therapy, and potentially predict disease expression and prognosis.
Findings from this study also shed light on the influence of genetics on autoAb generation. Genetic factors are known to strongly contribute to a number of autoimmune diseases, and it is likely that individuals related to patients with autoimmune disease share some of these genetic elements. However, the large majority do not develop disease. Examination of the related healthy control population may help to identify key elements or disturbances in immune function that are lacking in this group, but required for transition to a disease state in patients. Our demonstration of anti-Dsg autoAbs in these individuals is in line with the findings of other groups who have shown that relatives of patients with pemphigus can exhibit positive Dsg3 titers and positive immunofluorescence results (47, 48). However, to our knowledge, we are the first to demonstrate that healthy relatives of patients with PV also possess autoreactivity toward additional non-Dsg antigens that are also targeted by patients.
Most interestingly, when grouped by the presence or absence of the PV-associated HLA alleles DRB1*0402 and/or DQB1*0503, the majority of related control subjects who harbored these alleles exhibited autoAb profiles more similar to that of active pemphigus patients, whereas relatives who were negative for the risk alleles had autoAb profiles more similar to unrelated control subjects. In fact, even unrelated control subjects who expressed the PV-associated HLA risk alleles clustered with active patients based on autoAb reactivity. These data indicate that the expression of HLA DRB1*0402 or DQB1*0503 may be key to the generation of autoAbs against particular sets of autoantigens. However, even though HLA+ and HLA− related control subjects cluster separately, our data show that both groups possess comparable levels of autoreactivity. One explanation for this may be that, because of the higher sensitivity of the protein microarray, the levels of autoAbs expressed by patients and related control subjects are above the linear range of detection, resulting in a plateau of signal and a subsequent inability to identify the full amplitude of patient responses. Alternatively, these similarities in autoAb reactivity may emphasize the role of other shared genetic and/or environmental in driving the autoAb response. Regardless, these findings suggest that the presence of autoAbs directed against disease relevant targets in healthy control subjects is not sufficient by itself to lead to clinical pathology.
Several other lines of investigation from our laboratory support the impact of PV-associated HLA alleles on autoimmunity. Healthy control subjects expressing the DRB*0402 or DQB*0503 susceptibility allele (HLA-matched controls) have altered expression of a number of cytokines that are also dysregulated in patients with pemphigus (57). We have also shown that the peripheral blood cells of HLA-matched control subjects exhibit similar patterns of gene expression as patients with PV (58). Together, these data implicate HLA as a central driver in the breakdown of self-tolerance and the specificity of the autoimmune response. It remains to be determined how genetically predisposed individuals who have activated certain autoimmune pathways remain disease free. Relevant to this point, in recent work, we found that HLA-matched healthy control subjects, in addition to sharing some transcriptional alterations with patients with pemphigus, also express a set of genes that are down-regulated compared with patients and control subjects who do not carry the PV-associated risk alleles (58), suggesting the hypothesis that expression of these genes in susceptible individuals may function to protect this group from disease development, despite their genetic inclination to disease. Collectively, these data demonstrate the profound influence of HLA on disease activity and shed light on the complex regulations within the immune system.
Examination of IgG and IgM autoAb responses identified another unique aspect of the autoAb response in PV. In contrast to IgG reactivities, control subjects surprisingly exhibited a significant elevation of IgM reactivity toward mAChR3, mAChR4, mAChR5, and Dsg3, again indicating that disease-free individuals can harbor Abs that recognize self-antigens. The presence of Abs capable of recognizing self-antigens in unrelated healthy patients suggest that at least a significant subset of healthy individuals possess B-cell receptors capable of recognizing self-antigens, but there are other mechanisms in place that prevent the activation of a full autoimmune response with functional consequences. Because IgM isotype Abs represent a more acute B-cell response and prolonged exposure toward antigens is known to drive the isotype switch from IgM to IgG, the cause of elevated IgM autoAbs in healthy control subjects compared with patients with active pemphigus may result from proper functioning of peripheral tolerance mechanisms that inhibit the activation and isotype switch of self-reactive B cells. It may also be that the lower levels of IgM reactivity seen in patients with active pemphigus results from the depletion of IgM-expressing B cells as they switch the isotype of their Abs from IgM to IgG.
The current paradigm of PV pathogenesis is beginning to shift from the limited lens of anti-Dsg autoAbs to a more complex and intricate view that incorporates a more comprehensive understanding of disease relevant autoAbs. Foremost, our study illuminates the existence of autoAbs that are directed at non-Dsg antigens in a significant proportion of patients with PV. We also demonstrate the utility of protein microarrays, not only for rapidly assessing the autoAb response in PV, but also as an instrument that is capable of dissecting the complex relationships between genetics and the expression of autoAbs. Our data highlight HLA as the central driver of the autoAb response to Dsg and non-Dsg targets. However, the increased autoreactivity observed in the HLA− related control subjects in Fig. 4 supports the notion that shared genetic and/or environmental factors, beyond that of HLA, may influence autoAb specificity. Direct comparison of HLA+ and HLA− related control subjects did not reveal any significant differences in autoreactivity, but further analyses in larger groups will be required to determine the impact of non-HLA genetic factors.
Identification of the full set of PV autoAbs across patients can be expected to facilitate the development of a more targeted and specific therapeutic approach that is at once more comprehensive and individualized. It remains to be seen if these specificities hold the potential to serve as actionable biomarkers that will allow clinicians to more accurately predict, classify, monitor, and treat disease subgroups. Finally, future studies must be aimed at assessing the functional effects of autoAbs directed at non-Dsg targets to determine the mechanisms by which they may contribute, alone or in concert with anti-Dsg autoAbs, to disease development.
Acknowledgments
The authors thank Birendra Kumar Sinha for continued guidance and support.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525448113/-/DCSupplemental.
References
- 1.Beutner EH, Lever WF, Witebsky E, Jordon R, Chertock B. Autoantibodies in pemphigus vulgaris: Response to an intercellular substance of epidermis. JAMA. 1965;192:682–688. doi: 10.1001/jama.1965.03080210026006. [DOI] [PubMed] [Google Scholar]
- 2.Anderson HJ, Newcomer VD, Landau JW, Rosenthal LH. Pemphigus and other diseases. Results of indirect intercellular immunofluorescence. Arch Dermatol. 1970;101(5):538–546. [PubMed] [Google Scholar]
- 3.Peterson LL, Wuepper KD. Isolation and purification of a pemphigus vulgaris antigen from human epidermis. J Clin Invest. 1984;73(4):1113–1120. doi: 10.1172/JCI111297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley JR, Yaar M, Hawley-Nelson P, Katz SI. Pemphigus antibodies identify a cell surface glycoprotein synthesized by human and mouse keratinocytes. J Clin Invest. 1982;70(2):281–288. doi: 10.1172/JCI110615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amagai M, Klaus-Kovtun V, Stanley JR. Autoantibodies against a novel epithelial cadherin in pemphigus vulgaris, a disease of cell adhesion. Cell. 1991;67(5):869–877. doi: 10.1016/0092-8674(91)90360-b. [DOI] [PubMed] [Google Scholar]
- 6.Stanley JR, Koulu L, Thivolet C. Distinction between epidermal antigens binding pemphigus vulgaris and pemphigus foliaceus autoantibodies. J Clin Invest. 1984;74(2):313–320. doi: 10.1172/JCI111426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanley JR, Koulu L, Klaus-Kovtun V, Steinberg MS. A monoclonal antibody to the desmosomal glycoprotein desmoglein I binds the same polypeptide as human autoantibodies in pemphigus foliaceus. J Immunol. 1986;136(4):1227–1230. [PubMed] [Google Scholar]
- 8.Amagai M, Hashimoto T, Shimizu N, Nishikawa T. Absorption of pathogenic autoantibodies by the extracellular domain of pemphigus vulgaris antigen (Dsg3) produced by baculovirus. J Clin Invest. 1994;94(1):59–67. doi: 10.1172/JCI117349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vu TN, et al. The pathophysiological significance of nondesmoglein targets of pemphigus autoimmunity. Development of antibodies against keratinocyte cholinergic receptors in patients with pemphigus vulgaris and pemphigus foliaceus. PV Arch Dermatol. 1998;134(8):971–980. doi: 10.1001/archderm.134.8.971. [DOI] [PubMed] [Google Scholar]
- 10.Abasq C, et al. ELISA testing of anti-desmoglein 1 and 3 antibodies in the management of pemphigus. Arch Dermatol. 2009;145(5):529–535. doi: 10.1001/archdermatol.2009.9. [DOI] [PubMed] [Google Scholar]
- 11.Akman A, Uzun S, Alpsoy E. Immunopathologic features of pemphigus in the east Mediterranean region of Turkey: A prospective study. Skinmed. 2010;8(1):12–16. [PubMed] [Google Scholar]
- 12.Arin MJ, Engert A, Krieg T, Hunzelmann N. Anti-CD20 monoclonal antibody (rituximab) in the treatment of pemphigus. Br J Dermatol. 2005;153(3):620–625. doi: 10.1111/j.1365-2133.2005.06651.x. [DOI] [PubMed] [Google Scholar]
- 13.Kwon EJ, Yamagami J, Nishikawa T, Amagai M. Anti-desmoglein IgG autoantibodies in patients with pemphigus in remission. J Eur Acad Dermatol Venereol. 2008;22(9):1070–1075. doi: 10.1111/j.1468-3083.2008.02715.x. [DOI] [PubMed] [Google Scholar]
- 14.Belloni-Fortina A, et al. Detection of autoantibodies against recombinant desmoglein 1 and 3 molecules in patients with pemphigus vulgaris: Correlation with disease extent at the time of diagnosis and during follow-up. Clin Dev Immunol. 2009;2009:187864. doi: 10.1155/2009/187864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen VT, Ndoye A, Shultz LD, Pittelkow MR, Grando SA. Antibodies against keratinocyte antigens other than desmogleins 1 and 3 can induce pemphigus vulgaris-like lesions. J Clin Invest. 2000;106(12):1467–1479. doi: 10.1172/JCI10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Den Z, Koch PJ. Loss of desmocollin 3 in mice leads to epidermal blistering. J Cell Sci. 2008;121(pt 17):2844–2849. doi: 10.1242/jcs.031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amagai M, Kàrpàti S, Klaus-Kovtun V, Udey MC, Stanley JR. Extracellular domain of pemphigus vulgaris antigen (desmoglein 3) mediates weak homophilic adhesion. J Invest Dermatol. 1994;103(4):609–615. doi: 10.1111/1523-1747.ep12397292. [DOI] [PubMed] [Google Scholar]
- 18.Green KJ, Simpson CL. Desmosomes: New perspectives on a classic. J Invest Dermatol. 2007;127(11):2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- 19.Desai BV, Harmon RM, Green KJ. Desmosomes at a glance. J Cell Sci. 2009;122(pt 24):4401–4407. doi: 10.1242/jcs.037457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chernyavsky AI, Arredondo J, Kitajima Y, Sato-Nagai M, Grando SA. Desmoglein versus non-desmoglein signaling in pemphigus acantholysis: Characterization of novel signaling pathways downstream of pemphigus vulgaris antigens. J Biol Chem. 2007;282(18):13804–13812. doi: 10.1074/jbc.M611365200. [DOI] [PubMed] [Google Scholar]
- 21.Tozzoli R. The diagnostic role of autoantibodies in the prediction of organ-specific autoimmune diseases. Clin Chem Lab Med. 2008;46(5):577–587. doi: 10.1515/cclm.2008.138. [DOI] [PubMed] [Google Scholar]
- 22.Robinson WH, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8(3):295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 23.Hueber W, Utz PJ, Steinman L, Robinson WH. Autoantibody profiling for the study and treatment of autoimmune disease. Arthritis Res. 2002;4(5):290–295. doi: 10.1186/ar426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fathman CG, Soares L, Chan SM, Utz PJ. An array of possibilities for the study of autoimmunity. Nature. 2005;435(7042):605–611. doi: 10.1038/nature03726. [DOI] [PubMed] [Google Scholar]
- 25.Plebani M, Pittoni M, Celadin M, Bernardi D, Mion MM. Recent advances in diagnostic technologies for autoimmune diseases. Autoimmun Rev. 2009;8(3):238–243. doi: 10.1016/j.autrev.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen VT, Ndoye A, Grando SA. Pemphigus vulgaris antibody identifies pemphaxin. A novel keratinocyte annexin-like molecule binding acetylcholine. J BiolChem. 2000;275(38):29466–29476. doi: 10.1074/jbc.M003174200. [DOI] [PubMed] [Google Scholar]
- 27.Emery DJ, et al. Pemphigus foliaceus and pemphigus vulgaris autoantibodies react with the extracellular domain of desmoglein-1. J Invest Dermatol. 1995;104(3):323–328. doi: 10.1111/1523-1747.ep12665364. [DOI] [PubMed] [Google Scholar]
- 28.Evangelista F, Dasher DA, Diaz LA, Prisayanh PS, Li N. E-cadherin is an additional immunological target for pemphigus autoantibodies. J Invest Dermatol. 2008;128(7):1710–1718. doi: 10.1038/sj.jid.5701260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiebiger E, Hammerschmid F, Stingl G, Maurer D. Anti-FcepsilonRIalpha autoantibodies in autoimmune-mediated disorders. Identification of a structure-function relationship. J Clin Invest. 1998;101(1):243–251. doi: 10.1172/JCI511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwatsuki K, et al. Internalization of constitutive desmogleins with the subsequent induction of desmoglein 2 in pemphigus lesions. Br J Dermatol. 1999;140(1):35–43. doi: 10.1046/j.1365-2133.1999.02604.x. [DOI] [PubMed] [Google Scholar]
- 31.Kljuic A, et al. Desmoglein 4 in hair follicle differentiation and epidermal adhesion: Evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell. 2003;113(2):249–260. doi: 10.1016/s0092-8674(03)00273-3. [DOI] [PubMed] [Google Scholar]
- 32.Spindler V, et al. Desmocollin 3-mediated binding is crucial for keratinocyte cohesion and is impaired in pemphigus. J BiolChem. 2009;284(44):30556–30564. doi: 10.1074/jbc.M109.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korman NJ, Eyre RW, Klaus-Kovtun V, Stanley JR. Demonstration of an adhering-junction molecule (plakoglobin) in the autoantigens of pemphigus foliaceus and pemphigus vulgaris. N Engl J Med. 1989;321(10):631–635. doi: 10.1056/NEJM198909073211002. [DOI] [PubMed] [Google Scholar]
- 34.Pitoia F, et al. Prevalence of thyroid autoimmunity in patients with pemphigus vulgaris. Medicina (B Aires) 2005;65(4):307–310. [PubMed] [Google Scholar]
- 35.Murrell DF, et al. Consensus statement on definitions of disease, end points, and therapeutic response for pemphigus. J Am Acad Dermatol. 2008;58(6):1043–1046. doi: 10.1016/j.jaad.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: An alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992;39(5):225–235. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee E, et al. Disease relevant HLA class II alleles isolated by genotypic, haplotypic, and sequence analysis in North American Caucasians with pemphigus vulgaris. Hum Immunol. 2006;67(1-2):125–139. doi: 10.1016/j.humimm.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen IR. The cognitive paradigm and the immunological homunculus. Immunol Today. 1992;13(12):490–494. doi: 10.1016/0167-5699(92)90024-2. [DOI] [PubMed] [Google Scholar]
- 41.Nobrega A, et al. Global analysis of antibody repertoires. II. Evidence for specificity, self-selection and the immunological “homunculus” of antibodies in normal serum. Eur J Immunol. 1993;23(11):2851–2859. doi: 10.1002/eji.1830231119. [DOI] [PubMed] [Google Scholar]
- 42.Poletaev AB. The immunological homunculus (immunculus) in normal state and pathology. Biochemistry (Mosc) 2002;67(5):600–608. doi: 10.1023/a:1015514732179. [DOI] [PubMed] [Google Scholar]
- 43.Quintana FJ, Getz G, Hed G, Domany E, Cohen IR. Cluster analysis of human autoantibody reactivities in health and in type 1 diabetes mellitus: A bio-informatic approach to immune complexity. J Autoimmun. 2003;21(1):65–75. doi: 10.1016/s0896-8411(03)00064-7. [DOI] [PubMed] [Google Scholar]
- 44.Merbl Y, Zucker-Toledano M, Quintana FJ, Cohen IR. Newborn humans manifest autoantibodies to defined self molecules detected by antigen microarray informatics. J Clin Invest. 2007;117(3):712–718. doi: 10.1172/JCI29943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madi A, Bransburg-Zabary S, Kenett DY, Ben-Jacob E, Cohen IR. The natural autoantibody repertoire in newborns and adults: a current overview. Adv Exp Med Biol. 2012;750:198–212. doi: 10.1007/978-1-4614-3461-0_15. [DOI] [PubMed] [Google Scholar]
- 46.Nagele EP, et al. Natural IgG autoantibodies are abundant and ubiquitous in human sera, and their number is influenced by age, gender, and disease. PLoS One. 2013;8(4):e60726. doi: 10.1371/journal.pone.0060726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kavala M, et al. Detection of pemphigus autoantibodies in healthy relatives of Turkish patients with pemphigus. Indian J Dermatol Venereol Leprol. 2007;73(4):240–242. doi: 10.4103/0378-6323.32889. [DOI] [PubMed] [Google Scholar]
- 48.Torzecka JD, et al. Circulating pemphigus autoantibodies in healthy relatives of pemphigus patients: Coincidental phenomenon with a risk of disease development? Arch Dermatol Res. 2007;299(5-6):239–243. doi: 10.1007/s00403-007-0760-y. [DOI] [PubMed] [Google Scholar]
- 49.von Mühlen CA, Tan EM. Autoantibodies in the diagnosis of systemic rheumatic diseases. Semin Arthritis Rheum. 1995;24(5):323–358. doi: 10.1016/s0049-0172(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 50.Büssow K, et al. A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res. 1998;26(21):5007–5008. doi: 10.1093/nar/26.21.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2(2):RESEARCH0004. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pietropaolo M, Eisenbarth GS. Autoantibodies in human diabetes. Curr Dir Autoimmun. 2001;4:252–282. doi: 10.1159/000060541. [DOI] [PubMed] [Google Scholar]
- 53.Grando SA. Muscarinic receptor agonists and antagonists: Effects on keratinocyte functions. Handb Exp Pharmacol. 2012;208:429–450. doi: 10.1007/978-3-642-23274-9_18. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen VT, et al. Pemphigus vulgaris acantholysis ameliorated by cholinergic agonists. Arch Dermatol. 2004;140(3):327–334. doi: 10.1001/archderm.140.3.327. [DOI] [PubMed] [Google Scholar]
- 55.Cianfarani F, et al. TSH receptor and thyroid-specific gene expression in human skin. J Invest Dermatol. 2010;130(1):93–101. doi: 10.1038/jid.2009.180. [DOI] [PubMed] [Google Scholar]
- 56.Nuzzo V, Tauchmanova L, Colasanti P, Zuccoli A, Colao A. Idiopathic chronic urticaria and thyroid autoimmunity: Experience of a single center. Dermatoendocrinol. 2011;3(4):255–258. doi: 10.4161/derm.3.4.17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo C, Seiffert-Sinha K, Sinha AA. 2014. Differential cytokine profiles offer multi-dimensional view on immune pathways involved in Pemphigus vulgaris. J Invest Dermatol 134(suppl 1):S17 (abstr 95)
- 58.Dey-Rao R, Seiffert-Sinha K, Sinha AA. Genome-wide expression analysis suggests unique disease-promoting and disease-preventing signatures in Pemphigus vulgaris. Genes Immun. 2013;14(8):487–499. doi: 10.1038/gene.2013.44. [DOI] [PubMed] [Google Scholar]