Abstract
Elevated concentrations of albumin in the urine, albuminuria, are a hallmark of diabetic kidney disease and are associated with an increased risk for end-stage renal disease and cardiovascular events. To gain insight into the pathophysiological mechanisms underlying albuminuria, we conducted meta-analyses of genome-wide association studies and independent replication in up to 5,825 individuals of European ancestry with diabetes and up to 46,061 without diabetes, followed by functional studies. Known associations of variants in CUBN, encoding cubilin, with the urinary albumin-to-creatinine ratio (UACR) were confirmed in the overall sample (P = 2.4 × 10−10). Gene-by-diabetes interactions were detected and confirmed for variants in HS6ST1 and near RAB38/CTSC. Single nucleotide polymorphisms at these loci demonstrated a genetic effect on UACR in individuals with but not without diabetes. The change in the average UACR per minor allele was 21% for HS6ST1 (P = 6.3 × 10–7) and 13% for RAB38/CTSC (P = 5.8 × 10−7). Experiments using streptozotocin-induced diabetic Rab38 knockout and control rats showed higher urinary albumin concentrations and reduced amounts of megalin and cubilin at the proximal tubule cell surface in Rab38 knockout versus control rats. Relative expression of RAB38 was higher in tubuli of patients with diabetic kidney disease compared with control subjects. The loci identified here confirm known pathways and highlight novel pathways influencing albuminuria.
Introduction
Urinary albumin and serum creatinine are two biomarkers recommended for the routine assessment of chronic kidney disease (CKD) (1). Even at physiological rates of glomerular filtration, small elevations in urinary albumin concentrations are associated with an increased risk for CKD progression, end-stage renal disease (ESRD), cardiovascular events, and cardiovascular and all-cause mortality (2–4). Patients with diabetes are at a particularly high risk for CKD and its sequelae: the prevalence of CKD among individuals with diabetes is >40% compared with ∼10% in the general U.S. adult population (5), and the presence of CKD is an important contributor to the excess mortality in diabetes (6). The appearance of significant amounts of albumin in the urine (albuminuria) is a hallmark of diabetic kidney disease (DKD), the incidence of which continues to rise along with type 2 diabetes worldwide (7). Residual diabetes-related microvascular risk represents an important challenge even in treated individuals (8), and DKD remains the leading cause of ESRD. No new effective treatments for DKD have been approved in more than two decades (9), highlighting the importance to better understand its underlying mechanisms.
Using genome-wide association study (GWAS) meta-analysis in general population cohorts, we previously identified a missense single nucleotide polymorphism (SNP) in the gene encoding cubilin (CUBN) in association with the urinary albumin-to-creatinine ratio (UACR) (10). CUBN is currently the only genome-wide significant locus for UACR. However, this variant explains only a small fraction of the previously reported heritability of albuminuria, ranging from 0.2 to 0.46 in the general population and in those with diabetes (11–13), suggesting that additional genetic variants remain to be found. Here we report the results of a GWAS meta-analysis of albuminuria traits in the general population performed in almost twice the sample size of our previous study (10), with a special focus on those with diabetes, replication in additional independent individuals, and follow-up investigations in human tissues and a genetically modified animal model of diabetes.
Research Design and Methods
Study Populations
Our study was based on 30 discovery and replication studies mostly from the general population, with the exception of Action in Diabetes and Vascular Disease: Preterax and Diamicron MR Controlled Evaluation (ADVANCE) and Genetics of Diabetic Nephropathy (GENDIAN), which enrolled exclusively individuals with type 2 diabetes, totaling 67,452 participants of European ancestry across the different analyses (up to 7,787 with diabetes in discovery and replication). The study characteristics, including the distribution of albuminuria and diabetes, are reported in Supplementary Table 1. Study protocols were approved by each local institutional review board or ethics committee, and all human participants gave written informed consent.
Phenotype Definitions and Analytical Strategy
The measurement of urinary albumin and creatinine in each study is reported in Supplementary Table 2. Urinary albumin values below the detection limit of the used assays were set to the lower limit of detection. Rather than using urinary albumin, the UACR was calculated as urinary albumin/urinary creatinine (mg/g) to account for differences in urine concentration. Microalbuminuria (MA) was defined as UACR >25 mg/g in women and >17 mg/g in men (10). Diabetes was defined as fasting glucose ≥126 mg/dL, nonfasting glucose ≥200 mg/dL, or treatment for diabetes, or by self-report if this information was not available. Across studies, we evaluated two traits, UACR and MA, and performed four GWAS meta-analyses: MA and UACR in the overall sample, as well as UACR—a continuous trait with higher statistical power—separately among those with and without diabetes. Diabetes-stratified genome-wide association analyses of MA were not performed due to limited sample size. Detailed information on the design, genotyping, imputation, and data management of each study is provided in Supplementary Tables 2 and 3.
Discovery Meta-Analysis, Replication, and Power
Stringent quality control of the genetic data was performed at the individual study level and again at the meta-analysis level using state-of-the-art methods. Missing genotypes were imputed using the HapMap reference panels in 19 studies and the 1000 Genomes reference panels in two studies. Details of genotyping, imputation software, reference panels, and quality filters in each study are reported in Supplementary Table 3.
All GWAS were performed following a standardized analysis protocol. In each study, the natural logarithm of UACR was taken. Subsequently, sex-specific residuals were obtained from linear regression models of ln (UACR) on age and study-specific covariates, including study center and genetic principal components, to adjust for possible population stratification, if applicable. The continuous sex-specific residuals were then combined and used as the dependent variable that was regressed on imputed allelic dosages for each SNP in the GWAS.
Before the meta-analyses, all study-specific GWAS summary files underwent quality control using the GWAtoolbox (14). Genomic-control (GC) (15) correction was applied when the GC factor was >1. Inverse variance weighted fixed-effects meta-analyses were conducted using METAL (16). The I2 statistic was used to evaluate between-study heterogeneity (17). All meta-analyses were performed in duplicate by two independent researchers.
After meta-analysis, SNPs with average minor allele frequency (MAF) <0.01 were excluded, and another GC correction was applied. There were 2,191,945 SNPs with average MAF >0.05 and present in >50% of the studies, which were then clustered based on correlation (linkage disequilibrium pruning using r2 ≤ 0.2) with the respective index SNP (the SNP with the lowest P value) within windows of ± 1 MB to identify independent SNPs with suggestive association (P < 10−5) in one or more of the four analyses.
Replication testing was then performed for signals that were genome-wide significant (P < 5 × 10−8) in any analysis or that showed suggestive association among those with diabetes, motivated by the clinical importance of DKD and the stronger association of the known and validated CUBN variant on UACR among those with diabetes (18). Replication was defined as a one-sided P value <0.05 in the meta-analysis of independent replication studies. Five of the nine studies that contributed to replication used imputed dosage, and four studies performed replication genotyping of the index SNPs. A meta-analysis of the replication results was performed. The double GC-corrected results from the discovery meta-analysis and the results of the nine replication studies were meta-analyzed to obtain the overall statistical significance. Unless stated otherwise, all reported P values are two-sided.
Assuming that associated SNPs explain a respective 0.6% and 0.5% of the UACR variance in diabetes (Table 1), there was 95% and 91% power, respectively, to replicate the seven suggestive loci from the discovery stage in an additional 1,800 samples with a one-sided P value of <0.05.
Table 1.
Replicated SNP associations with UACR in individuals with diabetes
| Sample size (n) | Effect on log(UACR) mg/g | SE | P value | I2 % | |
|---|---|---|---|---|---|
| rs649529, RAB38 | |||||
| Discovery | 5,825 | −0.15 | 0.03 | 9.3 × 10−6 | 0 |
| Replication | 1,962 | −0.12 | 0.05 | 0.02 | 0 |
| Combined | 7,787 | −0.14 | 0.03 | 5.8 × 10−7 | 0 |
| rs13427836, HS6ST1 | |||||
| Discovery | 5,509 | 0.20 | 0.04 | 6.1 × 10−6 | 10 |
| Replication | 1,890 | 0.16 | 0.07 | 0.03 | 58 |
| Combined | 7,399 | 0.19 | 0.04 | 6.3 × 10−7 | 30 |
For both variants, the effect of each additional copy of the minor allele (T) on UACR was modeled in an additive fashion. I2 is provided as a measure of heterogeneity across studies. Imputation quality ranged from 0.41 to 1.0 for rs649529 and from 0.44 to 1.0 for rs13427836. The variants were directly genotyped in four of the replication studies, with a call rate ranging from 0.98 to 1.0 for rs649529 and from 0.99 to 1.0 for rs13427836. The estimated proportion of explained variance in UACR among those with diabetes is 0.6% for rs649529 and 0.5% for rs13427836, using the formula 2 × MAF × (1-MAF) × effect2/var(log[UACR]), based on the combined effect estimates from Table 1 and the phenotypic variance in the large population-based ARIC Study.
Additional Analyses to Characterize Novel Loci
Replicated SNPs were further evaluated even in the absence of genome-wide significance because, in addition to the significant replication P value, the low heterogeneity across cohorts and the biological plausibility of the RAB38 locus further increased confidence in the findings. The SNPs were evaluated in the Diabetes Control and Complications Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications (EDIC) study for association with a primary clinical end point defined as time from the DCCT baseline until time to persistent MA or a secondary end point of time to incident albumin excretion rate >300 mg/24 h or ESRD (10). Time to outcome development or censoring was determined as the number of visit years from the DCCT baseline up to and including the 12th year of EDIC follow-up. Subjects with persistent MA at DCCT baseline and DCCT year 1 were excluded from analyses of that outcome.
Epigenomic map analyses were performed, as described previously (19), by using data from human kidney and kidney proximal tubule epithelial cells that can be accessed at Gene Expression Omnibus (GSE49637).
Genetic associations with additional renal function traits, estimated glomerular filtration rate (eGFR), and CKD, were evaluated based on results from GWAS meta-analysis of the corresponding traits within the CKDGen Consortium (C.P., personal communication).
Gene Expression Analyses in Human Tissues
Quantification of transcript abundance in microdissected fractions of human glomeruli and tubuli from surgical nephrectomies, living allograft donors, and portions of diagnostic kidney biopsies (20) was performed using RNA sequencing. Tissue from different renal compartments was separated using microdissection, homogenized, and stored at −80°C. Total RNA of human proximal tubule fractions (n = 256) and glomerular cells (n = 48) was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA quality was assessed with the Agilent Bioanalyzer 2100, and RNA preparations exhibiting RNA integrity number scores >7 were used for cDNA synthesis (library preparation at DNA Sequencing Core at UT Southwestern Medical Center). In short, 1 μg total RNA was used to isolate poly A purified mRNA using the Illumina TruSeq RNA Preparation Kit. Single-end 100 bp sequencing was performed, and the annotated RNA counts (fastq) were calculated by CASAVA 1.8.2 (Illumina). Reads were mapped to the reference genome (National Center for Biotechnology Information build 37, hg19) using Spliced Transcripts Alignment to a Reference (STAR). Reads per kilobase of transcript per million mapped for HS6ST1 and RAB38 were compared between glomerular and tubular fractions using a two-sided t test.
Comparison of candidate gene expression between case subjects with DKD proven by biopsy specimen and healthy control subjects was based on publicly available microarray data from human microdissected glomeruli and tubuli (Gene Expression Omnibus, GSE 30122) (20). Raw data were analyzed using the R package “Affy” version 1.44.0, and expression levels were normalized using robust multiarray average. Transcript abundance between patients and control subjects was compared using two-sided t tests; statistical significance was defined as P < 8.3 × 10−3 (α = 0.05 corrected for six comparisons).
Studies of Rab38 in Rats
To better understand the association of RAB38 with albuminuria in diabetes, we studied genetically modified rat models of diabetes. Eight Rab38 knockout (KO) rats on a fawn-hooded hypertensive (FHH) background, seven rats transgenic for the wild-type Brown Norway rat Rab38 allele, and seven congenic rats were generated and raised as described previously (21–23). Rab38 KO rats did not express the protein (22). These references also describe the recording of blood pressure and the measurement of glucose and albuminuria. Diabetes was induced by administering streptozotocin (STZ) (STZ; 50 mg/kg i.p.; Sigma-Aldrich, St. Louis, MO) to 9-week-old male rats.
Paraffin blocks of rat kidney samples were sectioned (6-μm thickness) with a Leica RM2255 rotary microtome (Thermo-Fisher Scientific, Waltham, MA) on Superfrost Plus glass slides (12-550-15, Thermo-Fisher Scientific). Before staining, slides were deparaffinized in changes of CitriSolv (22-143-975, Thermo-Fisher Scientific) and 70% isopropanol. Antigen retrieval was accomplished by incubating in sodium citrate buffer (1.8% 0.1 mol/L citric acid, 8.2% 0.1 mol/L sodium citrate, in distillated water, pH 6.0) in a rice cooker for 30 min. Slides were blocked with PBS blocking buffer (1% BSA, 0.2% nonfat dry milk in PBS) for 30 min and stained with primary antibodies specific for megalin or cubilin diluted in blocking buffer overnight at 4°C. Sheep anti-megalin and rabbit anti-cubilin were provided by Dr. P. Verroust, INSERM, Paris, France. After two washes in 0.1% Tween 20 (v/v in PBS), slides were incubated with corresponding fluorophore-conjugated secondary antibodies (Invitrogen), diluted in blocking buffer at room temperature for 1 h, and counterstained with 10 μmol/L Hoechst 33342 (Molecular Probes-Invitrogen, H1399). Slides were subsequently mounted in Prolong Gold Antifade reagent (Invitrogen), and images were acquired with a Leica SP5 confocal laser scanning microscope (Center for Microscopy and Image Analysis, University of Zurich) equipped with a Leica APO ×63 NA 1.4 oil immersion objective.
All experiments were performed in compliance with National Institutes of Health Guide for Care and Use of Laboratory Animals, and all used protocols were approved by the local institutional animal care and use committee.
Results
Discovery of Genomic Loci Associated With Albuminuria Traits
The discovery GWAS meta-analyses for the four traits included up to 20 studies and up to 54,450 individuals per trait. The median UACR in the 20 individual studies that contributed to the UACR meta-analysis ranged from 2.5 to 15.6 mg/g. Across all studies, the mean proportion of women was 53%, and the median of average age was 57 years. The prevalence of diabetes in the population-based studies ranged from 1 to 14% (Supplementary Table 1).
There was no evidence of systematic biases influencing the genome-wide association results, as indicated by low genomic control parameters (Supplementary Fig. 1). Only SNPs in the previously identified CUBN locus showed genome-wide significant association with both UACR (P = 2.4 × 10−10, Supplementary Table 4 and Supplementary Fig. 2) and MA (P = 1.3 × 10−10, Supplementary Table 5 and Supplementary Fig. 2). The effect of the minor C allele of the index SNP rs10795433 on logarithmic UACR values was fourfold larger among 5,825 individuals with diabetes (0.19 log[mg/g], P = 2.0 × 10−5) compared with 46,061 individuals without diabetes (0.045 log[mg/g], P = 8.7 × 10−6; P = 8.2 × 10−4 for difference). This corresponds, for each additional C allele, to a 5% higher geometric mean of UACR (exp[0.045]) in individuals without diabetes compared with 21% higher average UACR in those with diabetes (exp[0.19]).
Suggestive associations were identified for all four analyses (Supplementary Tables 4–7 and regional association plots in Supplementary Fig. 3). Among the clinically important group of individuals with diabetes, seven genomic loci contained one or more SNPs showing a suggestive association with UACR. These were exclusively identified in the meta-analysis of individuals with diabetes and mapped into or near HS6ST1, CNTN4, KBTBD8, TFAP2B/PKHD1, CHN2, WDR11/FGFR2, and RAB38/CTSC (Supplementary Table 7). Following our analytical strategy, we selected the index SNP in each of these seven regions for follow-up among up to 1,962 independent individuals with diabetes.
The Supplementary PDF document contains the QQ and Manhattan plots of all GWAS meta-analyses, the regional association plots, tables with cohort descriptions, and association results of SNPs at P < 10−5. Results from discovery GWAS meta-analysis are publicly available at http://fox.nhlbi.nih.gov/CKDGen/.
Replication Analyses Implicate RAB38/CTSC and HS6ST1 as Novel Loci for UACR in Diabetes
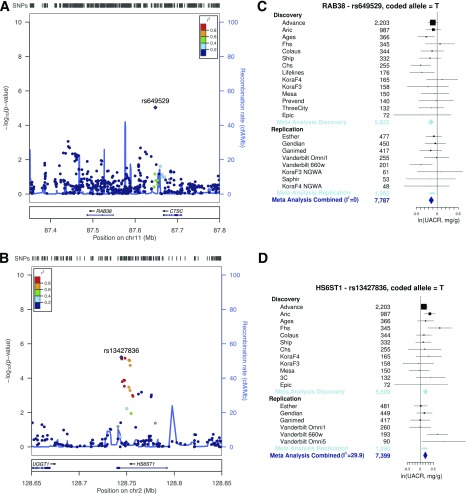
The replication analyses included nine studies and up to 1,962 individuals with diabetes. The median UACR across replication studies ranged from 3.8 to 14.5 mg/g. The mean proportion of women was 49%, and the median of average age was 55 years. For the seven SNPs tested for replication (Supplementary Table 8), we assessed whether the one-sided P value was <0.05 in the combined replication studies (see research design and methods). This was the case for two SNPs: intergenic rs649529 upstream of RAB38/downstream of CTSC on chromosome 11q14 (Fig. 1A) and the intronic variant rs13427836 in HS6ST1 on chromosome 2q21 (Fig. 1B). As illustrated in Fig. 1C, each additional copy of the minor T allele of rs649529 at RAB38/CTSC was consistently associated with lower UACR among the 5,825 individuals in the discovery and 1,962 in the replication cohorts (combined P = 5.8 × 10−7) (Table 1), with no evidence of heterogeneity across cohorts (I2 = 0%). This effect corresponded to 13% lower geometric mean of UACR per copy of the T allele. Similarly, rs13427836 in HS6ST1 showed consistent effects across cohorts (combined P = 6.3 × 10−7) (Table 1), with each copy of the T allele associated with an ∼21% higher mean UACR but moderate heterogeneity (I2 = 29.9%) (Fig. 1D).
Figure 1.
Overview of associated genomic loci at RAB38/CTSC and HS6ST1 and consistent association with albuminuria in diabetes across the contributing studies. A: Regional association plot of the RAB38/CTSC locus on chromosome 11. B: The T allele at rs649529 is associated with lower UACR across discovery and replication studies. C: Regional association plot of the HS6ST1 locus on chromosome 10. D: The T allele of intronic rs13427836 is associated with higher UACR across discovery and replication studies. The solid squares indicate the mean difference and are proportional to the weights used in the meta-analysis. The solid vertical line indicates no effect. The diamond indicates the weighted mean difference, and the lateral tips of the diamond indicate the associated CIs. The horizontal lines represent the 95% CI.
The association of rs649529 near RAB38/CTSC and rs13427836 in HS6ST1 was not found in individuals without diabetes (P = 0.64 and P = 0.38, respectively) (Table 2). Differences in the association with UACR among those with and without diabetes were significant (t test for difference: P = 6.9 × 10−6 for rs649529 and P = 1.7 × 10−5 for rs13427836). Effects for the index variant in CUBN are provided for comparison. We also evaluated the association of the replicated SNPs with MA in the setting of diabetes. Information was obtained from a subset of studies with sufficiently high numbers of individuals with diabetes and MA (n = 2,552; Atherosclerosis Risk in Communities Study [ARIC], Cardiovascular Health Study [CHS], Cohorte Lausannoise [CoLaus], European Prospective Investigation into Cancer and Nutrition [EPIC], Framingham Heart Study [FHS], Kooperative Gesundheitsforschung in der Region Augsburg [KORA] F3, KORAF4, and Study of Health in Pomerania [SHIP]). Across cohorts, the odds ratio for MA for each copy of the minor allele was 0.84 for rs649529 near RAB38 (P = 0.019) and 1.39 for rs13427836 in HS6ST1 (P = 7.8 × 10−4), consistent with the direction of the SNP effects on UACR.
Table 2.
Replicated SNP associations with additional kidney function and diabetes-related traits
| Trait |
n |
Effect (OR) |
SE |
P value |
P difference* |
|---|---|---|---|---|---|
| rs649529, RAB38/CTSC | |||||
| UACR, diabetes; log(mg/g) | 7,787 | −0.14 | 0.03 | 5.8 × 10−7 | 6.9 × 10−6 |
| UACR, no diabetes; log(mg/g) | 45,094 | −0.004 | 0.008 | 0.64 | |
| eGFRcrea, diabetes; log(mL/min/1.73 m2) | 11,527 | 0.003 | 0.004 | 0.46 | 2.8 × 10−1 |
| eGFRcrea, no diabetes; log(mL/min/1.73 m2) | 118,427 | −0.001 | 0.001 | 0.59 | |
| CKD (eGFR <60 mL/min/1.73 m2) | 118,114 | (1.01) | 0.02 | 0.57 | |
| Type 2 diabetes | 63,390 | (1.02) | 0.02 | 0.32 | |
| Fasting glucose (mmol/L) | 46,186 | 0.003 | 0.006 | 0.65 | |
| HbA1c (%) | 46,368 | 0.004 | 0.004 | 0.31 | |
| rs13427836, HS6ST1 | |||||
| UACR, diabetes; log(mg/g) | 7,399 | 0.19 | 0.04 | 6.3 × 10−7 | 1.7 × 10−5 |
| UACR, no diabetes; log(mg/g) | 34,830 | 0.010 | 0.012 | 0.38 | |
| eGFRcrea, diabetes; log(mL/min/1.73 m2) | 11,092 | 0.008 | 0.006 | 0.13 | 1.3 × 10−1 |
| eGFRcrea, no diabetes; log(mL/min/1.73 m2) | 114,247 | 0.000 | 0.001 | 0.94 | |
| CKD (eGFR <60 mL/min/1.73m2) | 113,612 | (0.97) | 0.02 | 0.23 | |
| Type 2 diabetes | 63,390 | (1.00) | 0.03 | 0.94 | |
| Fasting glucose (mmol/L) | 46,186 | −0.005 | 0.004 | 0.22 | |
| HbA1c (%) | 46,368 | 0.003 | 0.005 | 0.61 | |
| rs10795433, CUBN† | |||||
| UACR, diabetes; log(mg/g) | 5,825 | 0.19 | 0.04 | 2.0 × 10−5 | 8.2 × 10−4 |
| UACR, no diabetes; log(mg/g) | 46,061 | 0.045 | 0.01 | 8.7 × 10−6 | |
| eGFRcrea, diabetes; log(mL/min/1.73 m2) | 11,522 | 0.007 | 0.005 | 0.18 | 1.9 × 10−1 |
| eGFRcrea, no diabetes; log(mL/min/1.73 m2) | 118,299 | 0.0007 | 0.001 | 0.61 | |
| CKD (eGFR <60 mL/min/1.73 m2) | 118,121 | (1.04) | 0.02 | 0.08 | |
| Type 2 diabetes | 63,390 | (1.00) | 0.03 | 0.88 | |
| Fasting glucose (mmol/L) | 46,186 | −0.003 | 0.005 | 0.52 | |
| HbA1c (%) | 46,368 | −0.002 | 0.005 | 0.73 | |
DM, diabetes mellitus; eGFRcrea, GFR estimated by serum creatinine levels; OR, odds ratio.
Effects represent the change in trait associated with each additional copy of the minor allele for each of the SNPs. For continuous traits, units are provided; the effect for binary outcomes, shown in parentheses, represents the OR. Except for UACR in diabetes, estimates refer to the discovery samples of the respective trait and to the published resources for the glycemic traits. Fasting glucose and HbA1c were evaluated among individuals free of diabetes. For the kidney traits, P values and SEs are corrected using genomic control.
*P value for difference from a two-sample t test: t = (effectDM − effectnonDM)/(SEDM2 + SEnonDM2)0.5 which, for large sample sizes is distributed as a normal (0,1). The correlation between effectDM and effectnonDM is assumed to be 0.
Associations with type 2 diabetes were tested using the publicly available summary statistics dataset from the DIAGRAM (DIAbetes Genetics Replication And Meta-analysis) Consortium (12,171 case subjects and 56,862 control subjects) (40). Associations with fasting glucose and plasma hemoglobin A1c concentrations were evaluated using the publicly available results from the MAGIC Consortium (www.magicinvestigators.org) (41,42).
†UACR effect estimates for UACR in diabetes for CUBN are provided from the discovery stage.
Characterization of Genetic Effects by Markers of Kidney Function and Diabetes
We next investigated whether the gene-by-environment interaction was also observed for the eGFR, another measure of kidney function, and/or diabetes or glycemic traits. No statistically significant associations were found between rs649529, rs13427836, rs10795433, and eGFR in those with or without diabetes (Table 2) and no associations were found with CKD. There were also no statistically significant associations of these variants with type 2 diabetes, fasting blood glucose, or plasma hemoglobin A1c concentrations (Table 2), indicating that the observed associations pertain to albuminuria in the setting of diabetes rather than to diabetes or impaired glucose metabolism per se. A comprehensive search in the National Human Genome Research Institute GWAS Catalog (24) did not reveal any significant associations between the two validated SNPs or their proxies with other diseases or traits.
Variant Evaluation
Using publicly available data of genetic effects on gene expression (25), we found an association in cis between rs649529 and transcript levels of RAB38 (P = 5.4 × 10−6) and the neighboring CTSC (P = 7.6 × 10−7), consistent with a regulatory effect of this variant in whole blood. Corresponding data for kidney-specific tissues are currently not available, but we used epigenetic maps generated from human adult kidney tissue (19) (see research design and methods) to further examine the regulatory potential of index SNPs. The intronic index SNP in HS6ST1 and several proxies mapped into enhancer regions. Similarly, the CUBN index variant rs10795433 mapped into an intronic enhancer region. The region in which the index variant at RAB38/CTSC is located was annotated as not mapped/repressed in these cells, thus preventing further examination. All proxies in strong linkage disequilibrium with these three index SNPs (r2 > 0.6, 1000G v5 reference panel) (26) were intronic (CUBN and HS6ST1) or intergenic (RAB38/CTSC).
Clinical Characterization Including Gene Expression of Replicated Loci
To evaluate target tissues within the kidney, we characterized the identified loci using data for tissue-specific gene expression. Clinical characterization was conducted using data from patients with DKD and healthy control subjects (27) and a prospective study of individuals with type 1 diabetes (28).
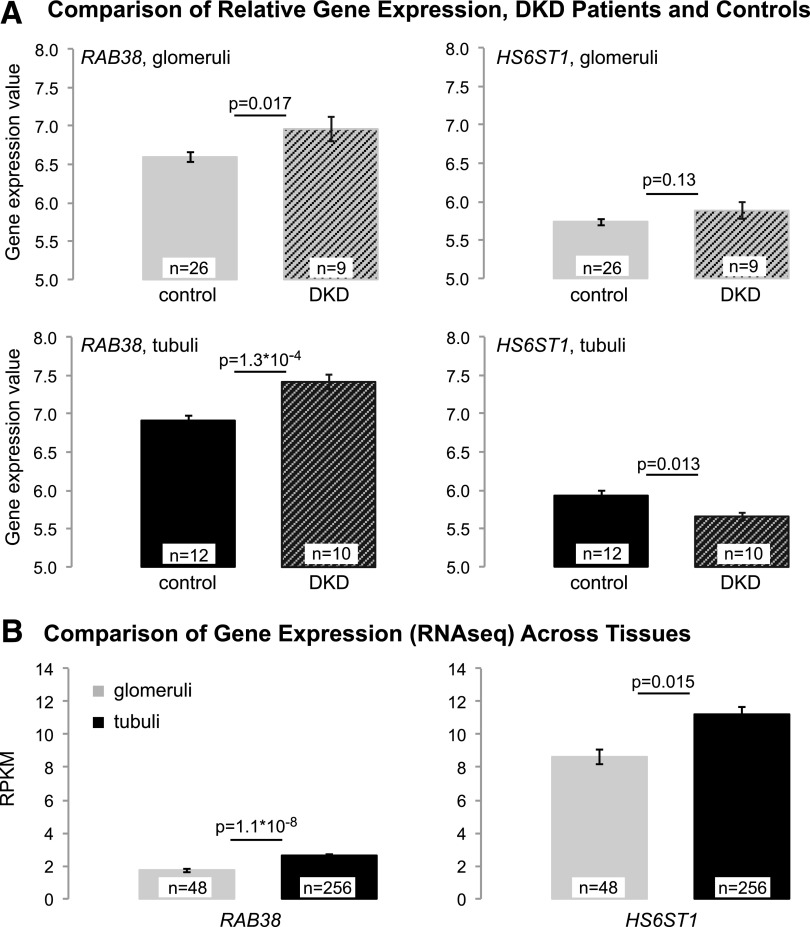
We used publicly available data (27) to compare relative expression of RAB38, CTSC, and HS6ST1 between patients with DKD confirmed by biopsy specimen and healthy control subjects (see research design and methods). After multiple testing correction, only RAB38 expression levels were significantly different, with higher expression in tubuli of DKD patients compared with control subjects (P = 1.3 × 10−4) (Fig. 2A). We also used RNA sequencing data from microdissected human kidney samples to quantify RAB38 and HS6ST1 expression in human glomeruli and tubuli. HS6ST1 showed higher expression levels than RAB38, and both genes showed higher expression in tubuli than in glomeruli (Fig. 2B). The difference between tubular and glomerular expression was more pronounced for RAB38 (P = 1.1 × 10−8) than for HS6ST1 (P = 0.015).
Figure 2.
RAB38 and HS6ST1 expression across kidney tissues. A: Comparison of RAB38 and HS6ST1 expression (microarray) in tubuli and glomeruli of patients with DKD and control subjects shows significantly higher RAB38 expression in tubuli of DKD patients than in tubuli of control subjects (significance threshold 0.05/6 = 8.3 × 10−3 for investigating RAB38, CTSC, and HS6ST1 in tubuli and glomeruli). CTSC expression was not significantly different between DKD case subjects and control subjects in tubuli (P = 0.11) or glomeruli (P = 0.03). Expression levels are shown as robust multiarray average–processed gene intensity values. B: RAB38 and HS6ST1 transcript abundance quantified from RNA sequencing (RNAseq) is detected at high levels in human tubuli but also in glomerular cells. Transcripts were quantified by reads per kilobase of transcript per million mapped (RPKM). The error bars in A and B correspond to the SEM.
To investigate whether the effect of the replicated SNPs extended to kidney disease progression in the setting of type 1 diabetes, the SNPs were tested for association with incident MA (268 case subjects, primary end point) and a combined end point of time to macroalbuminuria or ESRD (133 case subjects, secondary end point) among up to 1,304 participants with type 1 diabetes in the DCCT/EDIC study (28). Neither SNP showed a significant association (Supplementary Table 9).
Diabetic Rab38 KO Rats Show Increased Urinary Albumin Excretion
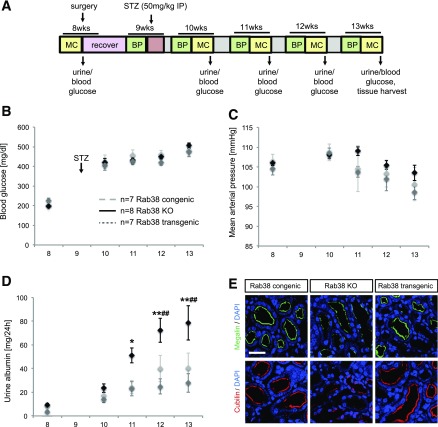
We aimed to further substantiate our findings by obtaining experimental support. We focused on the examination of RAB38 because it was the gene implicated by higher gene expression in tubuli of DKD patients compared with control subjects and because previous studies of Rab38 KO and transgenic rats have confirmed its role in albuminuria in FHH rats and highlighted a role in tubular albumin reuptake (21,22). We thus examined these animals in the setting of diabetes as outlined in Fig. 3A. Injection of STZ in 9-week-old rats successfully induced diabetes in all strains (Fig. 3B). Blood glucose rose from normal values before injection of STZ (congenic, 205 ± 3 mg/dL; transgenic, 227 ± 11 mg/dL; KO, 198 ± 7 mg/dL) to high values that indicate severe hyperglycemia 1 week after STZ (congenic, 422 ± 35 mg/dL; transgenic, 406 ± 27 mg/dL; KO, 420 ± 21 mg/dL). Blood glucose levels remained high at age 11, 12, and 13 weeks and showed no significant differences among strains (Fig. 3B). There were no significant differences in mean arterial blood pressure among congenic, transgenic, and KO animals freely moving around the cage. All animal strains showed a tendency toward decreased blood pressure 3–4 weeks after injection of STZ (Fig. 3C).
Figure 3.
Comparison of Rab38 congenic, transgenic, and KO rats after induction of diabetes. A: Experimental setup and timeline. BP, blood pressure measurement via radio telemetry; MC, metabolic cage. B: Comparison of blood glucose concentrations. C: Comparison of mean arterial pressure. D: Comparison of urinary albumin concentrations. *P < 0.05, **P < 0.01 KO vs. transgenic, ##P < 0.01 KO vs. congenic. E: Expression of endocytic markers. Immunofluorescence staining for megalin (green, top panel) and cubilin (red, bottom panel) in kidneys from all three rat strains. Nuclei counterstained with DAPI (blue). Scale bar, 50 μm. Data are presented as mean ± SEM. The results for blood pressure measurement, urinary albumin excretion, and blood glucose were analyzed by two-way ANOVA, followed by the Tukey post hoc test.
As illustrated in Fig. 3D, Rab38 KO animals showed a progressive increase in urinary albumin excretion that became statistically significant 2 weeks after injection of STZ. At 4 weeks after injection, Rab38 KO animals had an albumin excretion of 79 ± 14 mg/day, whereas albumin excretion was only 28 ± 8 mg/day in transgenic (P < 0.01) and 41 ± 13 mg/day in congenic animals (P < 0.01). These data indicate that diabetic rats without Rab38 are more susceptible to the development of albuminuria than congenic and transgenic animals with functional Rab38 despite a similar degree of hyperglycemia in all animals. Kidney sections obtained from a subset of animals showed a higher average glomerulosclerosis score (2.9 ± 0.3) compared with congenic (2.2 ± 0.1) and transgenic (2.2 ± 0.1) rats (P < 0.05) (Supplementary Fig. 4), but differences were subtler than those observed for urinary albumin excretion.
To further clarify how loss of Rab38 may lead to albuminuria, we performed immunohistochemistry staining of megalin and cubilin, known to mediate albumin reuptake in the proximal tubulus, in kidney sections of all three animal strains. Cubilin and megalin were markedly reduced at the luminal membrane of proximal tubular cells in Rab38 KO rats compared with congenic and transgenic control animals (Fig. 3E), consistent with a role of Rab38 in regulating the abundance of cubilin and megalin at the cell surface. In contrast, there was no significant difference in the number of structures positive for the lysosomal marker LAMP1 among the three strains.
Discussion
In this GWAS discovery meta-analysis of 2,191,945 SNPs in up to 54,450 participants of 20 studies, we replicated the association of the previously identified CUBN locus and UACR as well as MA at genome-wide significance and identified several suggestive signals among individuals with diabetes. Two of these loci, RAB38/CTSC and HS6ST1, showed evidence of independent replication, and RAB38 was further supported by functional studies in a rat model. Our findings point to mechanisms in renal handling of albumin that associate with albuminuria in humans in the setting of diabetes. They thus represent examples of gene-by-diabetes interactions resulting in a complex trait that manifests when environmental exposure and genetic susceptibility variants occur together (29).
Not all individuals with diabetes develop DKD, suggesting that neither the presence of hyperglycemia nor genetic variants alone are sufficient to elicit the renal damage that typically manifests itself as albuminuria in diabetes. Our observations therefore raise the question of how the diabetic environment may result in the manifestation of genetic effects on albuminuria. The lack of association between both genetic variants and type 2 diabetes or specific glycemic measures in humans indicates that their effects occur without influencing diabetes per se. This notion is further substantiated by the fact that diabetic Rab38 KO rats showed higher urinary albumin concentrations compared with controls despite the presence of similar blood glucose concentrations.
A difference between our observations in humans and rats is that the effect of genetic variation near RAB38 on albuminuria was only found in humans with but not without diabetes, whereas Rab38 KO rats without diabetes also progress to albuminuria (22). A potential explanation is that KO rats represent a null mutation, allowing for the genetic component to take full effect without needing further aggravation by environmental factors. Conversely, many human susceptibility variants of complex traits do not result in a complete loss of function but instead are of a regulatory nature. The effect of such variants may become apparent only upon an environmental challenge, such that genetically determined alterations in renal albumin handling could manifest themselves in the setting of hyperglycemia and/or diabetes due to a number of mechanisms that secondarily affect albumin reabsorption, including an increased load of filtered albumin due to hyperfiltration or impairment of the glomerular filter. Along these lines, our observation of significantly higher RAB38 transcript abundance in tubuli of DKD patients than control subjects may indicate an adaption of the tubular machinery for albumin reabsorption in this setting. Moreover, genetics effects of the index SNP at the CUBN locus on albuminuria were four times as large in individuals with diabetes compared with those without diabetes, supporting alterations of tubular albumin handling in the setting of diabetes.
RAB38 encodes a member of the small Rab GTPase protein family that regulates intracellular vesicle trafficking between organelles and is important in exo-, endo-, and transcytosis (30). Expression of Rab38 at the mRNA and protein level was observed in proximal tubule cells of wild-type rats (31). FHH rats, a natural Rab38-null mutation, show increased urinary albumin excretion without changes in glomerular permeability (21). In these animals, the expression of a Brown Norway Rab38 transgene led to phenotypic rescue, and knockdown of Rab38 in a proximal tubule cell system significantly decreased albumin endocytosis (22). Together, these observations support an important role for the small Rab GTPase RAB38 in the reabsorption of filtered albumin.
Impaired RAB38 function may lead to increased albumin excretion via different mechanisms: altered intracellular vesicle transport may affect albumin reabsorption or recycling of reabsorbed albumin back to the plasma membrane (32). Alternatively, altered RAB38 function may affect the delivery of proteins required, such as cubilin or megalin, for albumin endocytosis, the mechanism underlying albuminuria in Dent disease (33). Our experimental data showing reduced abundance of cubilin and megalin in Rab38 KO but not in control rats are consistent with the latter hypothesis. Finally, that impaired RAB38 function may directly cause glomerular damage, in turn leading to increased concentrations of urinary albumin, is also conceivable.
Although the combined evidence from Rab38 KO rats along with the gene expression and GWAS data strongly implicate RAB38 as the gene underlying albuminuria in humans, the intergenic index SNP mapped upstream of RAB38 and downstream of CTSC and was associated with transcript levels of both genes in whole blood. We can therefore not exclude the possibility that CTSC may be the causal gene underlying the observed associations or that it contributes to the phenotype in addition to RAB38. CTSC encodes for a lysosomal cysteine protease. Rare mutations in the gene cause autosomal-recessive Papillon-Lefèvre syndrome. No renal abnormalities have been reported in affected patients (34), Ctsc KO mice do not show kidney abnormalities (35), and the gene has not been linked to albuminuria or kidney disease.
The other genomic locus associated with albuminuria in diabetes contains HS6ST1, encoding the enzyme heparan sulfate (HS) 6-O-sulfotransferase that catalyzes the 6-O-sulfation of HS and heparin (36,37). HS are anionic side chains of HS proteoglycans, which are components of basement membranes, extracellular matrix, and cell surfaces. Several studies have reported that inactivation or removal of HS leads to proteinuria, and biopsy specimens from patients with diabetes revealed changes in HS sulfation patterns compared with control subjects (38). Thus, a genetic variant altering the activity or abundance of the enzyme may lead to altered albuminuria. The underlying mechanisms could be manifold, because HS has been reported to affect not only glomerular filtration but also growth factor signaling, composition, and functions of the glomerular basement membrane as well as functions at the endothelial surface layer (38) and the proximal tubule (39).
Strengths of our study include its large sample size, specific examination of individuals with diabetes, and consistent effects across a variety of studies underscoring the relevance of our findings at the general population level. In addition, we performed careful characterization of a replicated finding through in vivo experiments in RAB38 KO and control rats that cannot, however, elucidate the exact mechanism by which genetic variation at this locus influences albuminuria in humans.
Limitations include the fact that the replicated SNPs did not achieve genome-wide significance, necessitating future confirmation in even larger studies, and that we could not assess allele-specific gene expression in human kidney tissues. We focused on European ancestry study participants and mostly on individuals with type 2 diabetes. Future studies should therefore examine these associations among individuals of additional ancestries and in well-powered studies of patients with type 1 diabetes. Although results were combined after study-specific analyses, biological variation in UACR and different urine collection and storage methods may have resulted in increased variation and thus reduced statistical power to reveal significant associations.
Additional studies are required to determine the causal variants and the exact underlying molecular mechanism by which genetic variation at RAB38/CTSC and HS6ST1 associates with albuminuria in humans. An elucidation of the underlying mechanisms and the contributions and differences of albuminuria of glomerular and tubular origin may improve our understanding of proteinuric kidney diseases in general but may be especially relevant to DKD, the most common cause of ESRD.
Supplementary Material
Article Information
Acknowledgments and Funding. 3C Study: The work was made possible by the generous participation of the control subjects, the patients, and their families. The authors thank Dr. Anne Boland, Centre National de Génotypage, for her technical help in preparing the DNA samples for analyses. This work was supported by the National Foundation for Alzheimer’s disease and related disorders, the Institut Pasteur de Lille, and the Centre National de Génotypage. The Three-City (3C) Study was performed as part of a collaboration between INSERM, the Victor Segalen Bordeaux II University, and Sanofi-Synthélabo. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study was also funded by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, MGEN, Institut de la Longévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Aquitaine and Bourgogne Regional Councils, Fondation de France, and the joint French Ministry of Research/INSERM “Cohortes et collections de données biologiques” programme. Lille Génopôle received an unconditional grant from Eisai.
ADVANCE Study: The genetic epidemiological work was funded by Prognomix, Inc., and by grants from Genome Quebec and Canadian Institutes of Health Research (CIHR). The clinical study was managed by The George Institute for Global Health (University of Sydney, Sydney, New South Wales, Australia) with grants received from Les Laboratoires Servier, France, and from the National Health and Medical Research Council of Australia. The genotyping was performed at the genomic platform of Centre de Recherche du Centre hospitalier de l'Université de Montréal (CRCHUM). The authors acknowledge the technical help of Carole Long and Mounsif Haloui and the bioinformatic analyses performed by Gilles Godefroid, François-Christophe Blanchet-Marois, and François Harvey (CRCHUM, Université de Montréal, Montreal, Quebec, Canada). Also acknowledged are the members of the genetic substudy of ADVANCE, Stephen Harrap (Department of Physiology, University of Melbourne, Melbourne, Victoria, Australia) and Michel Marre (Diabétologie, Endocrinologie, Nutrition at Hôpital Bichat, Paris, France). M.W. did not participate in animal experiments.
AGES Study: The study has been funded by National Institutes of Health (NIH) contracts N01-AG-1-2100 and HHSN271201200022C, the National Institute on Aging Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). The study is approved by the Icelandic National Bioethics Committee, VSN: 00-063. The researchers are indebted to the participants for their willingness to participate in the study.
Amish Study: The authors thank the Amish research volunteers for their long-standing partnership in research and the research staff at the Amish Research Clinic for their hard work and dedication. The study is supported by grants and contracts from the NIH, including R01-AG-18728 (Amish Longevity Study), R01-HL-088119 (Amish Calcification Study), U01-GM-074518-04 (Pharmacogenomics of Anti-Platelet Intervention [PAPI] Study), U01-HL0-72515-06 (Heredity and Phenotype Intervention [HAPI] Study), U01-HL0-84756, and K12-RR0-23250 (University of Maryland Multidisciplinary Clinical Research Career Development Program), the University of Maryland General Clinical Research Center, grant M01-RR-16500, the Baltimore Veterans Administration Medical Center Geriatric Research Education and Clinical Center, and the Paul Beeson Physician Faculty Scholars in Aging Research Program. Partial funding was also provided by the Mid-Atlantic Nutrition Obesity Research Center (P30-DK0-72488).
ARIC Study: The ARIC is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01-HL0-87641, R01-HL-9367 and R01-HL0-86694, National Human Genome Research Institute contract U01-HG0-04402, and NIH contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by grant number UL1-RR0-25005, a component of the NIH and NIH Roadmap for Medical Research. This work as well as Y.L. and A.K. were supported by the Emmy Noether Programme of the German Research Foundation (KO 3598/2-1 to A.K.).
Baltimore Longitudinal Study of Aging (BLSA): BLSA was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
CHS: The CHS research reported in this article was supported by National Heart, Lung, and Blood Institute contracts N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01-HC-15103, N01-HC-55222, N01-HC-75150, N01-HC-45133, and grant numbers U01-HL0-80295 and R01-HL0-87652, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01-RR-00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK-063491 to the Southern California Diabetes Endocrinology Research Center.
CoLaus Study: The CoLaus study received financial contributions from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (33CSCO-122661).
CROATIA-Split Study: The CROATIA-Split study was supported through grants from the Medical Research Council U.K., and Ministry of Science, Education and Sport of the Republic of Croatia (No. 108-1080315-0302). The authors acknowledge the invaluable contributions of the recruitment team from the Croatian Centre for Global Health, University of Split, the administrative teams in Croatia and Edinburgh, and the people of Split. The SNP genotyping for the Split cohort was performed by AROS Applied Biotechnology, Aarhus, Denmark.
EPIC-Norfolk Study: The EPIC-Norfolk Study is supported by program grants from the Medical Research Council, and Cancer Research U.K. The authors acknowledge the contribution of the staff and participants of the EPIC-Norfolk Study.
ESTHER Study: The authors thank all the individuals who took part in the ESTHER (Epidemiological investigations of the chances of preventing, recognizing early and optimally treating chronic diseases in an elderly population) study and all the researchers, clinicians, technicians, and administrative staff who have enabled this work to be carried out. This work was supported in part by the Baden-Württemberg State Ministry of Science, Research and Arts, by the German Federal Ministry of Education and Research (grant number 01ET0717), and by the CHANCES (Consortium on Health and Ageing: Network of Cohorts in Europe and the United States) project funded in the Seventh Framework Programme of the Directorate-General for Research and Innovation in the European Commission (grant number 242244).
Fenland Study: The Fenland study is funded by the Medical Research Council (MC_U106179471) and Wellcome Trust. The authors are grateful to all the volunteers for their time and help and to the general practitioners and practice staff for assistance with recruitment. The authors thank the Fenland Study Investigators, Fenland Study Co-ordination team, and the Epidemiology Field, Data and Laboratory teams.
Framingham Heart Study: This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart, Lung, and Blood Institute of the NIH and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung and Blood Institute Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc. for genotyping services (Contract No. N02-HL-6-4278). A portion of this research used the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Boston University School of Medicine Department of Medicine and Boston Medical Center.
GENDIAN Study: The support of the physicians, the patients, and the staff of the Diabetes Zentrum Mergentheim (Head: Prof. Dr. Thomas Haak), the diabetes outpatient clinic Dr. Nusser – Dr. Kreisel, the dialysis centers KfH Amberg, KfH Bayreuth, KfH Deggendorf, KfH Donauwörth, KfH Freising, KfH Freyung, KfH Fürth, KfH Hof, KfH Ingolstadt, KfH Kelheim, KfH München Elsenheimerstraße, KfH München-Schwabing, KfH Neumarkt, KfH Neusäß, KfH Oberschleißheim, KfH Passau, KfH Plauen, KfH Regensburg Günzstraße, KfH Regensburg Caritas-Krankenhaus, KfH Straubing, KfH Sulzbach-Rosenberg, KfH Weiden, Dialysezentrum Augsburg Dr. Kirschner, Dialysezentrum Bad Alexandersbad, KfH Bamberg, Dialysezentrum Emmering, Dialysezentrum Klinikum Landshut, Dialysezentrum Landshut, Dialysezentrum Pfarrkirchen, Dialysezentrum Schwandorf, Dr. Angela Götz, the medical doctoral student Johanna Christ, and the study nurse Ingrid Lugauer. The expert technical assistance of Claudia Strohmeier is gratefully acknowledged. Phenotyping was funded by the Dr. Robert Pfleger-Stiftung (C.A.B.), the MSD Stipend Diabetes (C.A.B.), and the University Hospital of Regensburg (intramural grant ReForM A to Dr. Angela Götz, ReForM C to C.A.B.). Genome-wide genotyping was funded by the KfH Stiftung Präventivmedizin e.V. (C.A.B., Dr. Jens Brüning), the Else Kröner-Fresenius-Stiftung (2012_A147 to C.A.B. and I.M.H.), and the University Hospital Regensburg (C.A.B.). Data analysis was funded by the Else Kröner-Fresenius Stiftung (2012_A147 to I.M.H. and C.A.B. and P48/08//A11/08 to C.A.B. and B.K.K.). GENDIAN Study Group: M.G., I.M.H., B.K.K., M.R., Michael Broll, Alexander Lammert, Jens Brüning, M.O., Klaus Stark, Claudia Strohmeier, Simone Neumeier, Sarah Hufnagel, Petra Jackermeier, Emilia Ruff, Johanna Christ, Peter Nürnberg, Thomas Haak, and C.A.B.
INCIPE Study: The INCIPE (Initiative on Nephropathy, of relevance to public health, which is Chronic, possibly in its Initial stages, and carries a Potential risk of major clinical End-points) study was cosponsored by Fondazione Cassa di Risparmio di Verona, Azienda Ospedaliera di Verona, and University of Verona, Italy. N.S.’s research is supported by the Wellcome Trust (grant codes WT098051 and WT091310), the European Union Framework Program 7 (EPIGENESYS grant code 257082 and BLUEPRINT grant code HEALTH-F5-2011-282510).
KORA-F3/KORA-F4 Study: The genetic epidemiological work was funded by the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ, and by the Else Kröner-Fresenius-Stiftung (P48/08//A11/08 to C.A.B., B.K.K. and 2012_A147 to C.A.B. and I.M.H.). The kidney parameter measurements in F3 were funded by the Else Kröner-Fresenius-Stiftung (C.A.B., B.K.K.) and the Regensburg University Medical Center, Germany, and in F4 by the University of Ulm, Germany (W.K.). De novo genotyping as well as partly genome-wide genotyping costs in F3 and F4 were funded by the Else Kröner-Fresenius-Stiftung (C.A.B., B.K.K.). The KORA research platform and the MONICA (Multinational MONItoring of Trends and Determinants in CArdiovascular Disease) Augsburg studies were initiated and financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, by the German Federal Ministry of Education and Research and by the State of Bavaria. Genotyping was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum München. The LINUX platform for computation were funded by the University of Regensburg for the Department of Epidemiology and Preventive Medicine at the Regensburg University Medical Center.
LifeLines Study: The LifeLines Cohort Study and generation and management of GWAS genotype data for the LifeLines Cohort Study is supported by the Netherlands Organization of Scientific Research NWO grant 175.010.2007.006, the Economic Structure Enhancing Fund (FES) of the Dutch government, the Ministry of Economic Affairs, the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the Northern Netherlands Collaboration of Provinces (SNN), the Province of Groningen, University Medical Center Groningen, the University of Groningen, Dutch Kidney Foundation, and Dutch Diabetes Research Foundation. LifeLines is a multidisciplinary prospective population-based cohort study examining in a unique three-generation design the health and health-related behaviors of 165,000 individuals living in the Northeast region of the Netherlands. It uses a broad range of investigative procedures in assessing the biomedical, sociodemographic, behavioral, physical, and psychological factors that contribute to the health and disease of the general population, with a special focus on multimorbidity and complex genetics.
MESA (The Multi-Ethnic Study of Atherosclerosis) Study: University of Washington (N01-HC-95159), Regents of the University of California (N01-HC-95160), Columbia University (N01-HC-95161), Johns Hopkins University (N01-HC-95162, N01-HC-95168), University of Minnesota (N01-HC-95163), Northwestern University (N01-HC-95164), Wake Forest University (N01-HC-95165), University of Vermont (N01-HC-95166), New England Medical Center (N01-HC-95167), Harbor-UCLA Research and Education Institute (N01-HC-95169), Cedars-Sinai Medical Center (R01-HL-071205), University of Virginia (subcontract to R01-HL-071205).
MICROS (Microisolates in South Tyrol) Study: The authors are grateful to all participants. The authors thank the primary care practitioners Raffaela Stocker, Stefan Waldner, Toni Pizzecco, Josef Plangger, Ugo Marcadent, and the personnel of the Hospital of Silandro Department of Laboratory Medicine for their participation and collaboration in the research project. The authors thank Dr. Peter Riegler (Hemodialysis Unit, Hospital of Merano) for the important discussions. In South Tyrol, the study was supported by the Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bolzano, the South Tyrolean Sparkasse Foundation, and the European Union Framework Program 6 EUROSPAN project (contract No. LSHG-CT-2006-018947).
PREVEND Study: PREVEND (Prevention of Renal and Vascular End-stage Disease) genetics is supported by the Dutch Kidney Foundation grant E033; the EU project grant GENECURE (FP-6 LSHM CT 2006 037697); the NIH grant 2R01-LM0-10098; The Netherlands Organisation for Health Research and Development NWO-Groot grant 175.010.2007.006, NWO VENI grant 916.761.70, ZonMw grant 90.700.441; and the Dutch Inter University Cardiology Institute Netherlands (ICIN).
SAPHIR Study: The SAPHIR (Salzburg Atherosclerosis Prevention Program in Subjects at High Individual Risk) study was partially supported by a grant from the Kamillo Eisner Stiftung to B.P. and by grants from the Genomics of Lipid-associated Disorders (GOLD) of the “Austrian Genome Research Programme GEN-AU” to F.K.
SHIP/SHIP-Trend/GANI_MED Study: SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research grants 01ZZ9603, 01ZZ0103, and 01ZZ0403; the Ministry of Cultural Affairs, as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania, and the network “Greifswald Approach to Individualized Medicine (GANI_MED)” funded by the Federal Ministry of Education and Research grant 03IS2061A. Genome-wide data have been supported by the Federal Ministry of Education and Research grant 03ZIK012 and a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the “Center of Knowledge Interchange” program of the Siemens AG, the German Centre for Diabetes Research (DZD), and the Caché Campus program of the InterSystems GmbH. The SHIP authors are grateful to Mario Stanke for the opportunity to use his Server Cluster for the SNP imputation as well as to Holger Prokisch and Thomas Meitinger (Helmholtz Zentrum München) for the genotyping of the SHIP-Trend cohort.
SKIPOGH (Swiss Kidney Project on Genes in Hypertension) Study: This research was funded by grant 33CM30-124087 from the Swiss National Science Foundation. The study also received support from Lausanne University Hospital, Geneva University Hospital, and Bern University Hospital, Switzerland. M.B. received support from the Swiss School of Public Health Plus (SSPH+).
Vanderbilt Study: Genotyping was supported by National Institute of General Medical Sciences (NIGMS) RC2-GM092318 (Omni1 and Omni5 array). This work was supported by the Electronic Medical Records and Genomics (eMERGE) Network, initiated and funded by the National Human Genome Research Institute, with additional funding from the NIGMS, through U01-HG04603 (660 array).
Susztak Laboratory: Work in the laboratory of K.S. was supported by NIH R01-DK0-76077 and DK0-87635.
Jacobs Laboratory: Work in the H.J.J. and J.L. laboratories was supported by 2R01-069321 to H.J.J.
Devuyst Laboratory: This work was supported by the European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 305608 (EURenOmics) and the Swiss National Science Foundation project grant 310030_146490.
Additional Data Resources: Data on glycemic traits have been contributed by MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium) investigators and have been downloaded from www.magicinvestigators.org.
Duality of Interest. C.He. received honoraria from Novartis. J.Ch., J.T., and P.H. received research grants and honoraria from Servier. M.W. had consultancies with Amgen and Novartis and received support from Sanofi. K.S. received research support from Boehringer Ingelheim and was on the advisory board of Abbvie. No other potential conflicts of interest relevant to this article were reported.
The sponsors had no role in the study design, analyses, drafting of the manuscript, or the decision to publish.
Author Contributions. A.L., B.S., B.K.K., B.P., C.He., C.Me., C.G., G.G., G.E., H.E.W., H.V., J.C.D., L.J.L., M.J.H., M.N., N.J.W., P.P.P., P.S.W., R.R., R.B., R.J.C., T.B.H., V.G., A.D.P., J.La., J.T., P.H., H.J.J., C.S.F., and A.K. designed this study. A.K.D., A.R.S., B.K., B.Po., B.K.K., B.Pa., B.D.M., C.Ha., C.He., C.Me., C.G., D.A., D.S., E.B., F.K., G.B.E., G.W., G.N., G.G., G.E., H.E.W., H.G., H.V., H.B., H.K., I.M.L., I.R., J.L.H., J.S.B., J.H.Z., J.Ch., J.Co., K.B., L.J.L., L.F., L.K., M.Wo., M.J.H., M.Wa., M.P., M.B., N.J.W., N.E., O.P., P.v.d.H., P.P.P., P.V., P.S.W., R.T.G., R.R., R.K., S.C., S.B., T.B.H., T.C., T.I., T.T., U.L., U.V., V.G., W.K., I.H.d.B., J.La., P.H., C.S.F., C.P., and A.K. were involved in the study management. A.R.S., A.L., B.K.K., B.Pa., C.He., C.Me., C.G., G.G., G.E., H.E.W., H.W., H.V., J.L.H., J.Co., M.N., L.K., N.J.W., O.P., P.v.d.H., P.P.P., P.S.W., R.J.C., R.T.G., S.S., V.G., O.D., P.H., and C.S.F. recruited the subjects. A.Te., A.Ti., R.S., M.G., N.C.Y., M.L., Y.L., V.M., B.O.T., A.V.S., B.Pa., F.K., J.Co., L.K., M.J.H., M.R., S.C., T.A., A.D.P., J.La., K.E., J.T., P.H., C.S.F., C.P., and A.K. interpreted the results. A.Te., A.Ti., M.G., N.C.Y., M.L., Y.-A.K., M.J.H., I.M.H., J.La., K.E., K.S., J.T., P.H., H.J.J., C.A.B., C.P., and A.K. drafted the manuscript. A.Te., A.Ti., R.S., M.G., N.C.Y., A.Y.C., M.L., Y.L., V.M., Y.-A.K., D.T., M.-H.C., Q.Y., M.C.F., M.O., L.T.H., B.O.T., C.F., A.V.S., B.K., C.Ha., C.M.S., C.Mü., G.L., J.Lu., M.M., M.J.H., M.R., N.V., R.J.C., R.K., S.-J.H., S.E.R., T.A., Z.K., J.R.O., A.P., I.M.H., C.A.B., A.D.P., K.E., K.S., C.P., and A.K. developed statistical methods and performed the analyses. A.R.S., A.M.Z., B.D.M., C.Ha., C.L., E.B., G.H., H.G., M.H., M.Wa., M.R., N.S., O.P., R.J.F.L., S.C., T.Z., T.I., and U.V. performed the genotyping. M.G., Y.L., V.M., Y.-A.K., D.T., C.M.S., C.Mü., G.M., J.C.L., J.Lu., N.V., P.v.d.H., V.C., J.R.O., I.M.H., K.S., and C.A.B. conducted the bioinformatics analyses. N.C.Y., A.L., A.M.Z., M.J.H., O.D., J.La., and H.J.J. did the animal work or provided functional data. All authors critically reviewed the manuscript. A.Te., C.S.F., C.P., and A.K. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Findings from this study were presented at the 51st Congress of the European Renal Association (ERA)-European Diabetes and Transplant Association (EDTA), Amsterdam, the Netherlands, 31 May–3 June 2014, and at the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Investigator Meeting, Jackson, MS, 1–2 July 2015. An abstract and platform presentation of this work was presented at the American Society of Human Genetics (ASHG) 2015 Annual Meeting, Baltimore, MD, 6–10 October 2015.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-1313/-/DC1.
H.J.J., C.A.B., C.S.F., C.P., and A.K. provided joint oversight of this work.
References
- 1.Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group. KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Kidney Int Suppl 2013;3:1–150
- 2.Gansevoort RT, Matsushita K, van der Velde M, et al. Lower estimated GFR and higher albuminuria are associated with adverse kidney outcomes. A collaborative meta-analysis of general and high-risk population cohorts. Kidney Int 2011;80:93–104 [DOI] [PMC free article] [PubMed]
- 3.Astor BC, Matsushita K, Gansevoort RT, et al. Lower estimated glomerular filtration rate and higher albuminuria are associated with mortality and end-stage renal disease. A collaborative meta-analysis of kidney disease population cohorts. Kidney Int 2011;79:1331–1340 [DOI] [PMC free article] [PubMed]
- 4.Van der Velde M, Matsushita K, Coresh J, et al. Lower estimated glomerular filtration rate and higher albuminuria are associated with all-cause and cardiovascular mortality. A collaborative meta-analysis of high-risk population cohorts. Kidney Int 2011;79:1341–1352 [DOI] [PubMed]
- 5.Plantinga LC, Crews DC, Coresh J, et al. Prevalence of chronic kidney disease in US adults with undiagnosed diabetes or prediabetes. Clin J Am Soc Nephrol 2010;5:673–682 [DOI] [PMC free article] [PubMed]
- 6.Afkarian M, Sachs MC, Kestenbaum B, et al. Kidney disease and increased mortality risk in type 2 diabetes. J Am Soc Nephrol 2013;24:302–308 [DOI] [PMC free article] [PubMed]
- 7.Tuttle KR, Bakris GL, Bilous RW, et al. Diabetic kidney disease: a report from an ADA Consensus Conference. Am J Kidney Dis 2014;64:510–533 [DOI] [PubMed]
- 8.Fioretto P, Dodson PM, Ziegler D, Rosenson RS. Residual microvascular risk in diabetes: unmet needs and future directions. Nat Rev Endocrinol 2010;6:19–25 [DOI] [PubMed]
- 9.Himmelfarb J, Tuttle KR. New therapies for diabetic kidney disease. N Engl J Med 2013;369:2549–2550 [DOI] [PubMed]
- 10.Böger CA, Chen MH, Tin A, et al. CUBN is a gene locus for albuminuria. J Am Soc Nephrol 2011;22:555–570 [DOI] [PMC free article] [PubMed]
- 11.Forsblom CM, Kanninen T, Lehtovirta M, Saloranta C, Groop LC. Heritability of albumin excretion rate in families of patients with type II diabetes. Diabetologia 1999;42:1359–1366 [DOI] [PubMed]
- 12.Fox CS, Yang Q, Guo CY, et al. Genome-wide linkage analysis to urinary microalbuminuria in a community-based sample: the Framingham Heart Study. Kidney Int 2005;67:70–74 [DOI] [PubMed]
- 13.Langefeld CD, Beck SR, Bowden DW, Rich SS, Wagenknecht LE, Freedman BI. Heritability of GFR and albuminuria in Caucasians with type 2 diabetes mellitus. Am J Kidney Dis 2004;43:796–800 [DOI] [PubMed]
- 14.Fuchsberger C, Taliun D, Pramstaller PP, Pattaro C; CKDGen Consortium . GWAtoolbox: an R package for fast quality control and handling of genome-wide association studies meta-analysis data. Bioinformatics 2012;28:444–445 [DOI] [PubMed] [Google Scholar]
- 15.Devlin B, Roeder K. Genomic control for association studies. Biometrics 1999;55:997–1004 [DOI] [PubMed] [Google Scholar]
- 16.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–560 [DOI] [PMC free article] [PubMed]
- 18.Böger CA, Gorski M, Li M, et al.; CKDGen Consortium . Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD. PLoS Genet 2011;7:e1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko YA, Mohtat D, Suzuki M, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol 2013;14:R108 [DOI] [PMC free article] [PubMed]
- 20.Woroniecka KI, Park ASD, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011;60:2354–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rangel-Filho A, Sharma M, Datta YH, et al. RF-2 gene modulates proteinuria and albuminuria independently of changes in glomerular permeability in the fawn-hooded hypertensive rat. J Am Soc Nephrol 2005;16:852–856 [DOI] [PubMed]
- 22.Rangel-Filho A, Lazar J, Moreno C, Geurts A, Jacob HJ. Rab38 modulates proteinuria in model of hypertension-associated renal disease. J Am Soc Nephrol 2013;24:283–292 [DOI] [PMC free article] [PubMed]
- 23.Katter K, Geurts AM, Hoffmann O, et al. Transposon-mediated transgenesis, transgenic rescue, and tissue-specific gene expression in rodents and rabbits. FASEB J 2013;27:930–941 [DOI] [PMC free article] [PubMed]
- 24.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001–D1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013;45:1238–1243 [DOI] [PMC free article] [PubMed]
- 26.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 2014;31:1334–1336 [DOI] [PMC free article] [PubMed]
- 27.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011;60:2354–2369 [DOI] [PMC free article] [PubMed]
- 28.Effect of intensive therapy on the development and progression of diabetic nephropathy in the Diabetes Control and Complications Trial. The Diabetes Control and Complications (DCCT) Research Group. Kidney Int 1995;47:1703–1720 [DOI] [PubMed]
- 29.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest 2008;118:1590–1605 [DOI] [PMC free article] [PubMed]
- 30.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009;10:513–525 [DOI] [PubMed]
- 31.Osanai K, Takahashi K, Nakamura K, et al. Expression and characterization of Rab38, a new member of the Rab small G protein family. Biol Chem 2005;386:143–153 [DOI] [PubMed]
- 32.Bultema JJ, Di Pietro SM. Cell type-specific Rab32 and Rab38 cooperate with the ubiquitous lysosome biogenesis machinery to synthesize specialized lysosome-related organelles. Small GTPases 2013;4:16–21 [DOI] [PMC free article] [PubMed]
- 33.Christensen EI, Devuyst O, Dom G, et al. Loss of chloride channel ClC-5 impairs endocytosis by defective trafficking of megalin and cubilin in kidney proximal tubules. Proc Natl Acad Sci U S A 2003;100:8472–8477 [DOI] [PMC free article] [PubMed]
- 34.Toomes C, James J, Wood AJ, et al. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet 1999;23:421–424 [DOI] [PubMed]
- 35.Pham CTN, Ivanovich JL, Raptis SZ, Zehnbauer B, Ley TJ. Papillon-Lefèvre syndrome: correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J Immunol 2004;173:7277–7281 [DOI] [PubMed]
- 36.Habuchi H, Kobayashi M, Kimata K. Molecular characterization and expression of heparan-sulfate 6-sulfotransferase. Complete cDNA cloning in human and partial cloning in Chinese hamster ovary cells. J Biol Chem 1998;273:9208–9213 [DOI] [PubMed]
- 37.Habuchi H, Tanaka M, Habuchi O, et al. The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J Biol Chem 2000;275:2859–2868 [DOI] [PubMed]
- 38.Kolset SO, Reinholt FP, Jenssen T. Diabetic nephropathy and extracellular matrix. J Histochem Cytochem 2012;60:976–986 [DOI] [PMC free article] [PubMed]
- 39.Masola V, Gambaro G, Tibaldi E, Onisto M, Abaterusso C, Lupo A. Regulation of heparanase by albumin and advanced glycation end products in proximal tubular cells. Biochim Biophys Acta 2011;1813:1475–1482 [DOI] [PubMed]
- 40.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed]
- 41.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed]
- 42.Soranzo N, Sanna S, Wheeler E, et al. Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes 2010;59:3229–3239 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.