Abstract
Large studies in humans and animals have demonstrated a clear association of an adverse intrauterine environment with an increased risk of cardiovascular disease later in life. Yet mechanisms remain largely elusive. The present study tested the hypothesis that gestational hypoxia leads to promoter hypermethylation and epigenetic repression of the glucocorticoid receptor (GR) gene in the developing heart, resulting in increased heart susceptibility to ischemia and reperfusion injury in offspring. Hypoxic treatment of pregnant rats from day 15 to 21 of gestation resulted in a significant decrease of GR exon 14, 15, 16, and 17 transcripts, leading to down-regulation of GR mRNA and protein in the fetal heart. Functional cAMP-response elements (CREs) at −4408 and −3896 and Sp1 binding sites at −3425 and −3034 were identified at GR untranslated exon 1 promoters. Hypoxia significantly increased CpG methylation at the CREs and Sp1 binding sites and decreased transcription factor binding to GR exon 1 promoter, accounting for the repression of the GR gene in the developing heart. Of importance, treatment of newborn pups with 5-aza-2’-deoxycytidine reversed hypoxia-induced promoter methylation, restored GR expression and prevented hypoxia-mediated increase in ischemia and reperfusion injury of the heart in offspring. The findings demonstrate a novel mechanism of epigenetic repression of the GR gene in fetal stress-mediated programming of ischemic-sensitive phenotype in the heart.
Keywords: Fetal programming, hypoxia, glucocorticoid receptor, DNA methylation
1. Introduction
Evidence from both human and animal studies indicates a clear association of an adverse intrauterine environment with an increased risk of cardiovascular diseases in later in life [1-4]. Gestational hypoxia is a common complication during pregnancy and contributes significantly to developmental malformations in the developing fetus and cardiovascular disease later in life [5]. It has been shown that exposure to long-term high altitude hypoxia results in decreases in cardiac contractile proteins activity and cardiac output in fetal sheep hearts [6]. Due to enhanced metabolic demand, the developing heart is especially susceptible to prolonged hypoxia. Insufficient oxygen in utero results in myocardial thinning and ventricle dilation, as well as delayed fetal heart maturation [7]. Adult male rat hearts exposed to antenatal hypoxia showed increased susceptibility to ischemia and reperfusion (I/R) injury with an increase in myocardial infarction and a decrease in post-ischemic recovery [8]. Indeed, cardiomyocyte hypertrophy and myocardial hypoplasia are common in fetal hearts subjected to chronic hypoxia [7, 9]. Underlying the programming of heart structure and development is the programming of gene expression and cell function by hypoxia. Expression of pivotal genes, including the PKCε, HSP70 and eNOS has been shown to be changed by fetal hypoxia in the developing heart [8, 10, 11]. However, hypoxia-induced fetal programming of heart development is believed to involve complex mechanisms that remain elusive.
Glucocorticoids are primary stress hormones, and the glucocorticoid receptor (GR) is the major mediator for glucocorticoid (GC) action. GR signaling plays essential roles in regulating the expression of a wide range of genes in response to stress, changes in metabolism, inflammation, etc. Particularly, substantial changes in gene expression occur with the endogenous GC surge and activation of GR during late pregnancy in mammals [12]. In the heart, the GR plays pivotal roles in normal development and function. Activation of the GR in cardiomyocytes leads to cardiomyocyte hypertrophy and cell survival, accompanied by a remarkable change in the gene transcriptome [13]. Mice with cardiomyocyte-specific deletion of the GR die prematurely and aberrant regulation of a large cohort of genes can be detected before the onset of pathology [14]. This line of evidence strongly suggests that intact GR is critical for heart development as well as for function throughout life [15, 16]. The GR gene (NR3C1) structure is highly conserved, with both human and rodent NR3C1 genes containing multiple 5’ untranslated regions (5’UTR), which fine-tune tissue-specific expression of the GR. Among the multiple alternate first exons, exon 11 to 13 are located in the distal region of the gene, about −30 Kb upstream of exon 2 and exon 14 to 111 are located in the proximal region of the gene, about -5 Kb upstream of exon 2. Transcription of each GR first exon is believed to be controlled by its own promoter, located directly upstream of each exon 1 [17, 18]. Multiple transcription factor binding sites with CpGs have been predicted or demonstrated at the GR promoter, suggesting a possible involvement of DNA methylation as an epigenetic mechanism in the regulation of GR expression [18-20]. Epigenetic regulation at selective-GR promoters has been related to fetal programming in organs including the brain and liver [21-23]. Our previous findings revealed a significant decrease of GR protein abundance in rat hearts exposed to maternal hypoxia and this reduction of GR is sustained in the adult offspring [8]. However, the molecular mechanisms controlling the regulation of cardiac GR expression remain unknown.
In the present study, we investigated the effect of gestational hypoxia on epigenetic programming of the GR gene at transcriptional level in the developing heart. Our results indicate that the hypoxia-induced reduction of GR expression in the heart resulted from the selective reduction of transcription of exons 14, 15, 16, and 17 mRNA. Several functional transcription factor binding sites, including cAMP-response elements (CREs) at promoter 14 and 15 and Sp1 binding sites at 16 and 17, were identified in the GR promoter, and hypoxia-induced hypermethylation at the GR promoter led to decreased binding of the associated transcription factors. Of importance, inhibition of DNA methylation by 5-aza-2’-deoxycytidine in the developing heart reversed hypoxia-induced promoter hypermethylation, restored GR expression and recovered the hypoxia-mediated increase in ischemia and reperfusion injury of the heart in offspring.
2. Materials and methods
2.1. Experimental animals
Time-dated pregnant Sprague–Dawley rats were purchased from Charles River Laboratories (Portage, MI). The rats were randomly divided into two groups: normoxic control and hypoxic treatment of 10.5% O2 from day 15 to 21 of gestation, as described previously [11]. Neonatal rats were treated with saline control or 5-aza-2′-deoxycytidine (1 mg/kg/day) on postnatal day 1 and day 3 via intraperitoneal injection, and were allowed to grow to 4 weeks old. 5-Aza-2′-deoxycytidine is a DNA methyltransferase inhibitor that incorporates only into DNA as opposed to 5-azacytidine that incorporates into both DNA and RNA. 5-Aza-2′-deoxycytidine has been widely used to inhibit promoter methylation and rescue gene expression [8, 11, 24, 25]. Hearts were studied in gestational day 21 (E21) fetuses and 4 week old offspring. To isolate hearts, rats were anaesthetized with isoflurane (5% for induction, 2% for maintenance) in oxygen (2 L/min for induction, 1 L/min for maintenance) and hearts were removed. The adequacy of anesthesia was determined by the loss of a pedal withdrawal reflex and any other reaction from the animal in response to pinching the toe, tail, or ear of the animal. Previous [8] and preliminary studies revealed that there were no sex-dependent effects of hypoxia on GR expression in the heart. Thus, fetuses and offspring of mixed male and female were studied. All procedures and protocols were approved by the Institutional Animal Care and Use Committee of Loma Linda University and followed the guidelines by the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.2. Western blot
Fetal hearts and left ventricles of offspring hearts were homogenized in RIPA lysis buffer (Life Technologies, Carlsbad, CA) supplemented with Halt protease inhibitors cocktail (Pierce Biotechnology, Rockford, IL). Nuclear extraction was prepared using NXTRACT CelLytic Nuclear Extraction Kit (Sigma, St. Louis, MO), following the manufacturer's instructions. Protein concentrations were determined using the BCA assay kit (Pierce). Samples with equal amounts of proteins were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with 5% non-fat milk in TBS for 1 hour at room temperature and then probed with primary antibodies against GR, Egr-1 (Santa Cruz Biotechnology, Santa Cruz, CA), Sp1 (Active Motif, Carlsbad, CA), CREB and phospho-CREB (Cell Signaling, Danvers, MA). After a brief wash, membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies. Protein signal was visualized with enhanced chemiluminescence reagents (Pierce) and blots were exposed to Hyperfilm. The results were quantified with the Kodak electrophoresis documentation and analysis system and Kodak ID image analysis software. The target protein abundance was normalized to the abundance of β-actin.
2.3. Real-time RT-PCR
Total RNA was isolated with TRIzol reagent (Life Technologies) and subjected to reverse transcription using Superscript III First-Strand Synthesis System (Life Technologies). The abundance of GR mRNA and the alternate exon 1 variants was determined with real-time PCR using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), as described previously [26]. Primers used are listed in Online Table I. Real-time PCR was performed in a final volume of 25 μl and the following protocol was used: 95°C for 5 minutes, followed by 40 cycles of 95°C for 20 seconds, annealing for 20 seconds at appropriate temperature depending on the primer sequence, 72°C for 20 seconds. Serial dilutions of the positive control were done on each plate to create a standard curve for the quantification. PCR was done in triplicate and threshold cycle numbers were averaged for each sample.
2.4. Reporter gene assay
Plasmids that fused the rat GR gene to a luciferase reporter gene were constructed into the pGL3-Basic vector (Promega, Madison, WI). Plasmids containing the following rat GR gene fragments were gifts from Dr. Karen Chapman [17]: P2, fragment encoding rat GR gene from −4572 to −9 (the ATG translation start is designated +1); P2(Rev), the same fragment of P2 in the reverse orientation with respect to luciferase; P16, fragment encoding −4572 to −3336; P17, fragment encoding −4572 to −2931; P110, fragment encoding −4572 to −2318; and P111, fragment encoding -4572 to -1767. In addition, rat genomic DNA was isolated from fetal heart tissue and the GR gene fragments P14 (−4571 to −4208) and P15 (−4571 to −3575) were cloned in MluI-XhoI orientation into pGL3-basic vector to drive the transcription of the luciferase reporter gene. Site-specific deletion mutations were constructed at the corresponding putative transcription factor binding sites with the QuikChange II Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). All promoter construct sequences were confirmed with DNA sequencing analyses. Embryonic rat ventricular H9c2 cells were obtained from ATCC (ATCC, Rockville, MD) and cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in 95% air/5% CO2. Cells were grown and sub-cultured in 24-well plates with experiments performed at 90% confluence. Cells were transfected using the Lipofectamine2000 Transfection Reagent (Life Technologies) following the manufacturer's instructions. 0.1 μg of internal control pRL-SV40 vectors were co-transfected with 1 μg of pGL3-Basic vector or an equal-molar amount of GR promoter-luciferase plasmids. After 48 hours, firefly and Renilla reniformis luciferase activities in cell extracts were measured in a luminometer using a dual-luciferase reporter assay system (Promega), as described previously [27]. The activities of the wild-type or site specific deletion constructs were then calculated by normalizing the firefly luciferase activities to R. reniformis luciferase activity, and expressed as relative to wild-type reporter activity (% WT).
2.5. Methylated DNA immunopreciptation (MeDIP)
MeDIP assays were performed with the MeDIP kit (Active Motif), following the manufacturer's instructions. Briefly, genomic DNA was extracted from heart tissues and sonicated to yield fragments ranging in size from 200 to 1000 base pairs. An additional quantity of fragmented DNA equivalent to 10% of the DNA being used in the IP reaction was saved as input DNA. The double strand DNA fragments were denatured at 95°C to produce single strand DNA, and a 5-methylcytosine (5-mC) antibody was then used to precipitate DNA containing 5-mC. Input DNA was heat-denatured and cooled following the same conditions. Both the 5-mC enriched DNA and input DNA were then purified with phenol/chloroform extraction and subjected to quantitative real-time PCR analysis. Sequence of primers flanking the appropriate GR promoters are listed in Online Table I.
2.6. Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from hearts using NXTRACT CelLytic Nuclear Extraction Kit (Sigma). The oligonucleotide probes with unmethylated CpG and methylated CmG of corresponding transcription factor binding sites at GR promoter regions (Online Table I) were labeled with a Biotin 3′ end labeling kit and subjected to gel shift assays using the LightShift Chemiluminescent EMSA Kit (Pierce), as described previously [27, 28]. Briefly, single strand oligos were incubated with Terminal Deoxynucleotidyl Transferase (TdT) and Biotin-11-dUTP in binding mixture for 30 minutes at 37°C. TdT adds a biotin-labeled dUTP to the 3’-end of the oligonucleotides. The oligos were extracted using chloroform and isoamyl alcohol to remove the enzyme and unincorporated biotin-11-dUTP. Dot blots were performed to ensure the oligos were labeled equally. Equal molar of sense and antisense oligos were mixed and annealed to make double strand oligos. The labeled oligonucleotides were then incubated with or without nuclear extracts in the binding buffer. Binding reactions were performed in 20 μl containing 50 fmol oligonucleotieds probes, 1× binding buffer, 1 μg of poly (dI-dC), and 5 μg of nuclear extracts. For cold competition reactions, 100-fold concentrations of non-labeled oligonucleotides were added to binding reactions. For super-shift assays, 2 μl of affinity purified primary antibodies were added to the binding reaction. The samples were separated by a native 5% polyacrylamide gel and transferred to a positively-charged nylon membrane (Pierce). The samples were crosslinked to the membrane with a UV crosslinker (125 mJoules/cm2), and membranes were then blocked and visualized using the reagents provided in the LightShift kit.
2.7. Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using the Chip-IT Express Kit (Active Motif), as described previously [11]. Briefly, tissues were minced and fixed with 1.5% formaldehyde to crosslink and maintain the DNA/protein interactions. After the reactions were stopped with glycine, tissues were washed with PBS. Chromatin extracts were sonicated to produce DNA fragments between 200 and 1000 base pairs. Antibodies against CREB (Cell Signaling), and Sp1 (Active Motif) were incubated with the chromatin extracts to precipitate the transcription factor/DNA complexes. Crosslinking was then reversed using a salt solution and proteins were digested with proteinase K. The antibody-pulled chromatin extracts were then subjected to real-time quantitative PCR analysis using two primers that flank the predicted transcription factor binding sites at GR promoters, as described above in MeDIP (Online Table I).
2.8. Site-directed CpG methylation mutagenesis
The effect of site-directed CpG methylation on GR promoter 16 activity was determined as described previously [27]. Briefly, two unique cutting sites were engineered at the Sp1 site, with EcoRI at 5’ upstream and PmeI at 3’ downstream. A customized 25 bp EcoRI/PmeI oligonucleotide fragment with methylation at the CpGs was synthesized by IDT, and ligated to the GR pGL3/P16 promoter-reporter plasmids digested with EcoRI and PmeI. An identical fragment with unmethylated CpGs at the Sp1 site was used as a control and ligated into GR pGL3/P16 promoter-reporter plasmids. Amount of formed ligation products was quantified by real-time PCR (forward primer: GGTGGGGGTTGAACTTGG; reverse primer: CTTTATGTTTTTGGCGTCTTCCA) and equal amounts of plasmids were used to transfect H9c2 cells. Activities of each promoter-reporter constructs were determined as described above. The sequence of Sp1 methylated oligos used for in vitro methylation assays is: AATTmCGGGGmCGmCGGGGGAGGGTGGGTTT.
2.9. 5-mC DNA ELISA for global methylation
DNA methylation in hearts was determined by measuring 5-methylcytosine (5-mC) using a 5-mC DNA ELISA kit (Zymo Research) following the manufacturer's instructions. Briefly, 100 ng of genomic DNA from hearts and standard controls provided by the kit were denatured and used to coat the plate wells. After incubation at 37°C for 1 hour, the wells were washed with 5-mC ELISA buffer and an antibody mix consisting of anti-5-mC and a secondary antibody was added to each well. The plate was covered with foil and incubated at 37°C for 1 hour. Wells were then washed with 5-mC ELISA buffer and an HRP developer was added to each well and incubated at room temperature for 1 hour. The absorbance at 405 nm was measured using an ELISA plate reader. The percent 5-mC was calculated using the second-order regression equation of the standard curve that was constructed with negative and positive controls in the same experiment.
2.10. Measurement of cardiac function and ischemia and reperfusion injury
Rats were anaesthetized with isoflurane (5% for induction, 2% for maintenance) in oxygen (2 L/min for induction, 1 L/min for maintenance) and hearts were removed. The adequacy of anaesthesia was determined by the loss of a pedal withdrawal reflex and any other reaction from the animal in response to pinching the toe, tail, or ear of the animal. Isolated hearts were retrogradely perfused via the aorta in a modified Langendorff apparatus, as described previously [11]. Left ventricle end-diastolic pressure (LVEDP) was set at 5 mmHg. After baseline recording for 60 minutes, hearts were subjected to 45 minutes of global ischemia followed by 30 minutes of reperfusion. Left ventricle developed pressure (LVDP), heart rate, dP/dtmax, dP/dtmin, and LVEDP were continuously recorded. At the end of reperfusion, left ventricles were collected and myocardial infarct size was measured with 1% triphenyltetrazolium chloride, as described previously [11]. Lactate dehydrogenase (LDH) activity was measured in the coronary effluent collected during reperfusion, using the TOX 7 assay kit (Sigma).
2.11. Statistical analysis
Data are expressed as mean ± SEM. Experimental number represents fetuses or offspring from different dams. Statistical significance (P < 0.05) was determined by analysis of variance (ANOVA) followed by Neuman-Keuls post hoc testing or Student's t test, where appropriate.
3. Results
3.1 Maternal hypoxia down-regulated GR expression in the developing heart
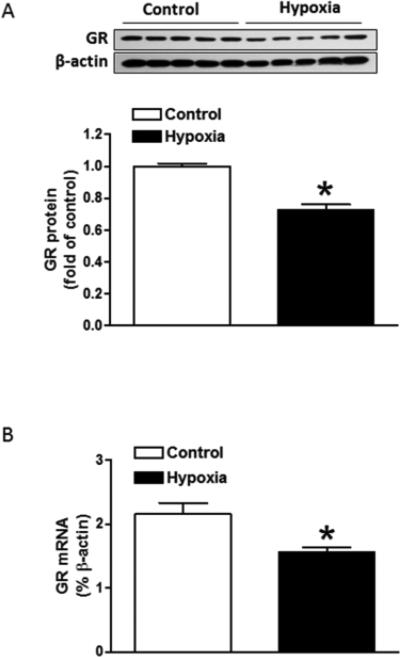
The effect of maternal hypoxia on GR expression was determined in E21 rat fetal hearts. Cardiac GR protein abundance was significantly decreased by maternal hypoxic treatment (Fig. 1a). In real-time quantitative PCR, a set of primers aligned to a fragment located at GR exon 8 and 9 were used to amplify the total GR mRNA transcripts. In accordance with decreased GR protein, maternal hypoxia resulted in a significant decrease in total GR mRNA in the fetal heart (Fig. 1b), indicating the regulation of GR at transcriptional level.
Fig. 1. Maternal hypoxia decreased expression of GR protein and mRNA in the rat fetal heart.
Hearts were isolated from E21 fetuses from pregnant rats treated with normoxia control or hypoxia at 10.5% O2 from day 15 to day 21 of gestation. (A) GR protein abundance was determined using Western blot analysis. (B) GR mRNA abundance was determined by quantitative real-time RT-PCR. Data are mean ± SEM, n = 5. * P < 0.05, hypoxia vs. control.
3.2. Relative transcriptional activity of GR exon 1 promoters
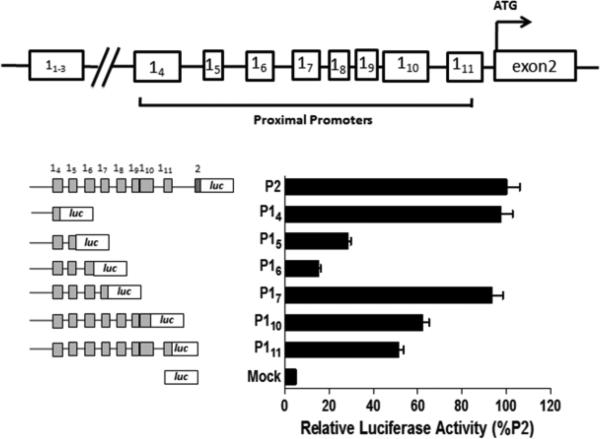
The rat GR gene has multiple alternative first exons that play important roles in regulating the tissue-specific expression of GR [17]. Among the multiple exon 1s, exon 11 to 13 are located in the distal region about −30Kb upstream of exon 2, and exon 14 to 111 in the proximal region about −5 Kb upstream of exon 2. Transcription of each first exon is believed to be controlled by its own promoter. To investigate whether the individual promoter activity is associated with GR exon 1 expression in fetal hearts, plasmids containing regions of the rat GR gene within each of the alternate exon 1s joined to a luciferase reporter gene were used to transfect H9c2 cells and promoter activities were measured. Detailed information of the plasmids can be found in a previous publication [17]. In brief, plasmid P2 (promoter 2) was constructed by joining the GR gene to the luciferase promoter within exon 2 (from −4572 to −9). Since each exon 1 contains a splicing donor site at the 3’-end that can be spliced onto the acceptor site at 5’-end of exon 2, the luciferase activity of P2 represents the promoter activity of the whole GR proximal promoter region. In accordance, luciferase activities of other promoter 1 plasmids represent promoter activities that promote the transcription of exon 14, 15 through 111. As shown in Fig. 2, P2 had the highest promoter activity, followed by promoter 14 (P14) and 17 (P17).
Fig. 2. Relative promoter activity associated with GR alternate first exons.
Top panel: diagrammatic representation of GR gene structure. Lower left panel: diagrammatic representation of GR promoter constructs. Fragments containing regions of the rat GR gene were fused, within specific exon 1s, to the luciferase reporter gene in pGL3-basic vector. Lower right panel: relative promoter activity of the GR promoter constructs in H9c2 cells, expressed as %P2. Data are mean ± SEM, n = 5.
3.3. Maternal hypoxia selectively decreased GR alternative exon 1 mRNA variants
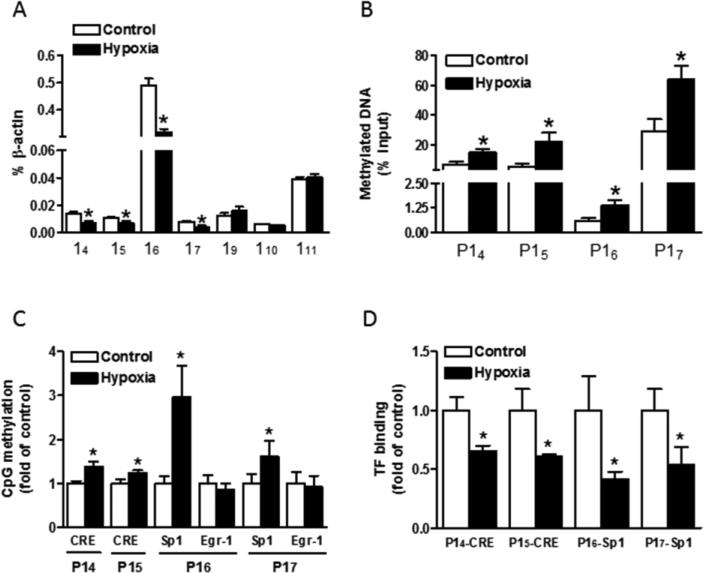
Considering that total GR mRNA consists of transcripts containing multiple exon 1 variants, we further investigated the role of hypoxia on the transcription of each GR exon 1 alternate. Since GR exons 11-3 were not expressed in the heart [17], our subsequent experiments were focused on the proximal exons 14-11. Quantitative RT-PCR assays were carried out using primers designed to amplify specific mRNA transcripts containing GR exon 14 to exon 111. For each set of primers, forward primers were located in exon 1, while reverse primers were located in the common exon 2 region. As shown in Fig. 3a, the abundance of exon 14, 15, 16 and 17 containing transcripts was significantly decreased by hypoxic treatment, while exon 19, 110 and 111 mRNA variants were not changed. Expression of exon 18 transcript was below the limit of detection by RT-PCR assay. Among all these exon 1 transcripts, the expression of 16 containing mRNA was predominant in the fetal heart, followed by expression of exon 111, while other exon1s were expressed at relatively low levels.
Fig. 3. Maternal hypoxia increased GR promoter methylation and decreased expression of exon 1 variants in the fetal heart.
Hearts were isolated from E21 fetuses from pregnant rats treated with normoxia control or hypoxia at 10.5% O2 from day 15 to day 21 of gestation. (A) mRNA abundance of GR exon 1 variants was determined by quantitative real-time RT-PCR. (B) Methylation of GR promoter 14, 15, 16 and 17, determined by MeDIP. (C) CpG methylation of specific transcription factor binding sites at GR promoters determined by methylation specific PCR. (D) Binding of CREB and Sp1 to GR promoters was determined by ChIP assays. TF, transcription factor. Data are mean ± SEM, n = 5. * P < 0.05, hypoxia vs. control.
3.4. Maternal hypoxia selectively increased methylation levels of GR exon 14, 15, 16 and 17 promoters
Increasing evidence indicates that prenatal stress may affect GR gene expression through epigenetic modification of promoter methylation. Therefore, we determined the role of hypoxia on methylation of GR exon 14 to 17 promoters by precipitation of methylated DNA and subsequent PCR analysis. As shown in Fig. 3b, the methylation level of promoter 14, 15, 16 and 17 was significantly increased by hypoxia. In addition, we found that compared to methylation of promoter 16 at less than 1%, methylation levels of promoter 14, 15 and 17 were relatively higher at about 7%, 5.5% and 29%, respectively.
The regulating region of GR gene is embedded in a CpG island with many CpGs that could potentially be regulated by methylation. Therefore, we further investigated the methylation levels of specific CpGs located in possible transcription factor binding sites. The sequences of GR promoter 14 to 17 were analyzed by MatInspector software for putative transcription factor binding sites, and multiple putative transcription factor binding sites with CpGs were found at the GR promoter, including CREB (cAMP response element-binding proteins) response elements (CRE), as well as Sp1 and Egr-1 binding sites. By using methylation-specific PCR, we found that CpG methylation levels at the putative CRE−4408 located in promoter 14, CRE−3896 located in promoter 15, Sp1−3425 site located in promoter 16 and Sp1−3034 site located in promoter 17 were significantly increased by hypoxia, while the methylation levels of CpGs at putative Egr-1 binding sites (at −3361 and −2996) were not significantly affected (Fig. 3c).
3.5. CpG methylation blocked CREB and Sp1 binding to the GR promoter
Given that maternal hypoxia selectively altered the methylation status at the CRE and Sp1 binding sites, our further investigation was focused on the CREs at promoter 14 and 15 and Sp1 binding sites at promoter 16 and 17. To determine whether transcription factors can bind these elements, EMSA and super-shift assays were performed with oligonucleotide probes encompassing the putative CRE and Sp1 binding sites, respectively. As shown in Online Fig. I, incubation of nuclear extracts from fetal hearts with double-stranded oligonucleotide probes encompassing the putative CREs at GR promoter 14 and 15 (Online Fig. Ia, lanes 2 and 6), as well as Sp1 binding sites at GR promoter 16 (Online Fig. Ib, lane 2) and 17 (Online Fig. Ic, lane 2) resulted in the appearance of DNA-protein complexes. The formation of these complexes was blocked in the presence of 100-fold excess of unlabeled probes (lanes 4 and 8 in Online Fig. Ia; lane 4 in Online Fig. Ib and Ic), demonstrating a specific binding of nuclear proteins to the oligonucleotide probes. Of importance, methylation of CpGs at the core binding sequences prevented the formation of DNA/protein complexes (lanes 5 and 9 in Online Fig. Ia; lane 5 in Online Fig. Ib and Ic), indicating that CpG methylation inhibits the binding of transcription factors to the DNA. Super-shift analyses showed that anti-CREB antibodies caused a super-shift of the CRE-DNA-protein complexes (Online Fig. Ia, lanes 3 and 7) and anti-Sp1 antibodies caused a super-shift of the Sp1-DNA-protein complexes (Online Fig. Ib and Ic, lane 3), demonstrating the binding of CREB and Sp1 to the corresponding binding sites. To further investigate the effect of hypoxia-induced hypermethylation on transcription factor binding in vivo in the context of intact chromatin, the binding of CREB to GR promoter 14 and 15 and binding of Sp1 to promoter 16 and 17 were determined in the fetal heart by ChIP assays. As shown in Fig. 3d, maternal hypoxia significantly decreased the binding of CREB to GR promoter 14 and 15, as well as the binding of Sp1 topromoter 16 and 17 in the fetal heart.
3.6. Deletion and CpG methylation of the CRE and Sp1 binding sites inhibited GR promoter activity
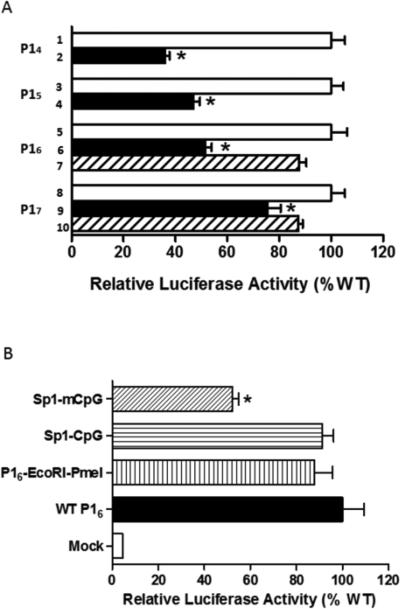
The transcriptional activity of CRE and Sp1 binding sites in the regulation of GR promoter activity was determined with a reporter gene assay in the rat embryonic ventricular myocyte cell line H9c2. GR exon 1 promoter constructs containing site-directed deletion at the CRE, Sp1 and Egr-1 sites (Online Fig. IIa) were transfected in H9c2 cells, and promoter activities were measured. As shown in Fig. 4a, deletion of CREs at promoter 14 and 15, respectively, resulted in a significant decrease in the promoter activity. For promoter 16 and 17, deletion of Sp1 sites, but not Egr-1 sites, significantly decreased the promoter activity. These results revealed the strong stimulatory role of CREs and Sp1 sites in driving the transcription of respective GR exon 1s.
Fig. 4. Deletion or methylation of CREs and Sp1 binding sites decreased GR promoter transcriptional activity.
Diagrammatic representation of GR promoter constructs of site-directed deletion of CRE−4408, CRE−3896, Sp1−3425, Egr-1−3361, Sp1−3034 and Egr-1−2996 in GR promoter 14, 15, 16 and 17, and site-specific CpG methylation at Sp1 (Sp1-mCpG) binding site in GR promoter 16 were shown in Online Figure II. Panel A shows that the deletion of transcription factor binding sites decreased transcriptional activity of the GR promoters. 1, 3, 5, 8: wild type GR promoters; 2: CRE−4408 deletion at promoter 14 (P14); 4: CRE−3896 deletion at promoter 15 (P15); 6: Sp1−3425 deletion at promoter 16 (P16); 7: Egr-1−3361 deletion at promoter 16 (P16); 9: Sp1−3034 deletion at promoter 17 (P17); 10: Egr-1−2996 deletion at promoter 17 (P17). Panel B shows that site-specific CpG methylation at Sp1 (Sp1-mCpG) decreased transcriptional activity of the GR promoter 16. Data are mean ± SEM, n = 4-6. * P < 0.05, vs. wild type.
Accumulating evidence indicates that methylation at core sequence of transcription factor binding sites results in loss of transcription activity of the corresponding site [27, 29]. Our results also showed an association between hypoxia-induced promoter hypermethylation and repression of GR transcription in the fetal heart. To demonstrate the effect of CpG methylation on the alteration of GR promoter activity, we generated a site-specific CpG methylation mutation at the Sp1−3425 site of GR promoter 16 and measured the promoter activity. To create a constitutive site-directed CmG at the Sp1 site, two restriction enzyme sites, with EcoRI site upstream at the 5’ end and PmeI site downstream at the 3’ end, were cloned into luciferase reporter gene constructs of GR promoter 16. Using these two sites, double strand oligonucleotides containing the Sp1 site with or without CmGs were engineered into GR promoter 16-luciferase reporter constructs (Online Fig. IIb). As shown in Fig. 4b, the mutation of CmG significantly decreased the transcriptional activity of GR promoter 16, while insertion of EcoRI and PmeI sites had no significant effect.
3.7. 5-Aza-2’-deoxycytidine reversed hypoxia-induced changes in GR expression in the offspring heart
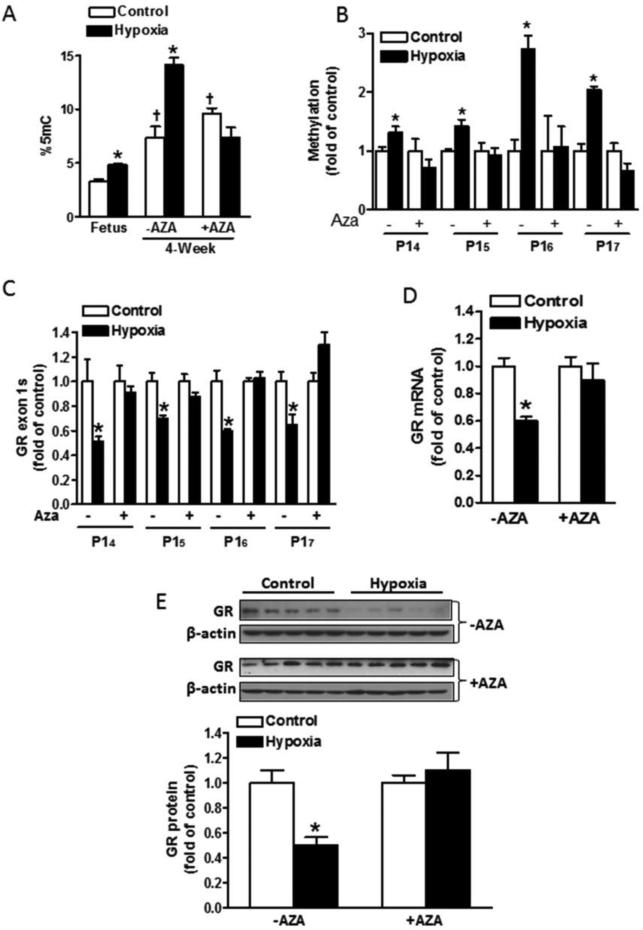
A development-dependent increase of global DNA methylation was demonstrated in the heart of 4 week old offspring as compared with the fetal heart (Fig. 5a). Maternal hypoxia caused a significant increase of global methylation in the fetal heart, which was sustained in 4 week old offspring (Fig. 5a). In addition to global methylation, the hypoxia-induced hypermethylation of GR promoter 14 through 17 was maintained in 4 week old rat offspring (Fig. 5b). Administration of two doses of 5-aza-2’-deoxycytidine at postnatal day 1 and day 3 completely reversed the hypoxia-induced DNA hypermethylation, both at the global level (Fig. 5a) and at GR promoter 14 to 17 (Fig. 5b) in the heart of 4 week old offspring. In accordance, the hypoxia-induced down-regulation of GR exon 14 through 17 containing mRNA transcripts (Fig. 5c), total GR mRNA (Fig. 5d), and protein (Fig. 5e) abundance were completely restored in 4 week old offspring hearts by the 5-aza-2’-deoxycytidine treatment.
Fig. 5. 5-Aza-2’-deoxycytidine reversed antenatal hypoxia-induced GR promoter hypermethylation and gene repression in offspring hearts.
Pregnant rats were treated with normoxia control or hypoxia at 10.5% O2 from day 15 to day 21 of gestation. Newborn rats were injected with saline (−AZA) or 5-aza-2’-deoxycytidine (+AZA) (1 μg/g/day, i.p.) at postnatal day 1 and day 3. At 4 week old, rats were sacrificed and hearts were isolated. Global DNA methylation levels were measured using a 5-mC DNA ELISA kit (A). DNA methylation at GR promoter 14, 15, 16 and 17 was determined by MeDIP (B). mRNA abundance of GR exon 14, 15, 16, 17 (C) and total GR mRNA (D) was measured using real-time RT-PCR. GR protein abundance was measured with Western blot (E). Data are mean ± SEM, n = 4-6. * P < 0.05, hypoxia vs. control, † P < 0.05, control 4 week offspring vs. control fetus.
3.8. 5-Aza-2’-deoxycytidine abrogated hypoxia-induced increase in heart susceptibility to ischemic injury
To determine the role of GR in cardiac function in vivo, the GR specific agonist dexamethasone was administered to 4 week old rats 24 hours before the hearts were isolated and subjected to ischemia and reperfusion injury in a Langendorff preparation. Dexamethasone treatment significantly improved the recovery of myocardial function by increasing LVDP, dP/dtmax and dP/dtmin after 45 minutes of global ischemia followed by 30 minutes of reperfusion (Online Fig. IIIa, IIIb, IIIc). Consistent with these findings, dexamethasone reduced ischemia and reperfusion-induced myocardial injury by decreasing LVEDP (Online Fig. IIId) and myocardial infarct size (Online Fig. IIIe).
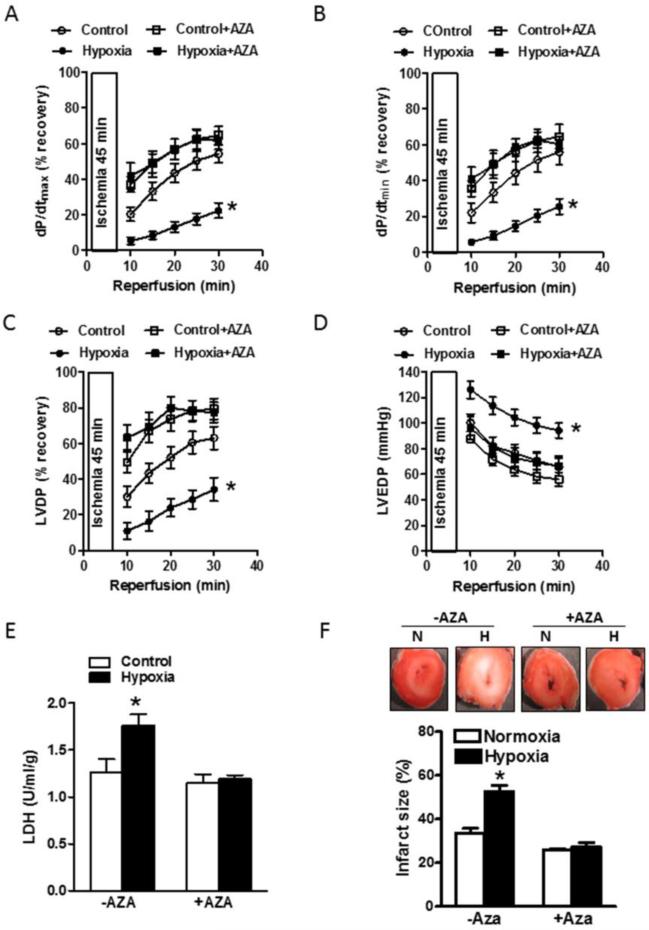
We then determined whether 5-aza-2’-deoxycytidine-mediated rescue of GR expression in the heart reversed antenatal hypoxia-induced increase in heart susceptibility to ischemic injury. There were no significant differences in baseline cardiac parameters among four experimental groups (Online Table II). As shown in Fig. 6, in 4 week old rat offspring antenatal hypoxia resulted in a significant decrease in post-ischemic recovery of the heart, manifesting as decreased dP/dtmax (Fig. 6a), dP/dtmin (Fig. 6b) and LVDP (Fig. 6c), and increased LVEDP (Fig. 6d), LDH release (Fig. 6e) and myocardial infarct size (Fig. 6f). Of importance, 5-aza-2’-deoxycytidine administration at postnatal day 1 and day 3 completely abrogated the hypoxia-mediated increase in heart susceptibility to ischemia and reperfusion injury in the offspring (Fig. 6).
Fig. 6. 5-Aza-2’-deoxycytidine abrogated antenatal hypoxia-induced increase in heart susceptibility to ischemic injury in offspring hearts.
Pregnant rats were treated with normoxia control (N) or hypoxia (H) at 10.5% O2 from day 15 to day 21 of gestation. Newborn rats were injected with saline (−AZA) or 5-aza-2’-deoxycytidine (+AZA) (1 μg/g/day, i.p.) at postnatal day 1 and day 3. At 4 week old, rats were sacrificed and hearts were subjected to 45 minutes of ischemia and 30 minutes of reperfusion in a Langendorff preparation. LVDP, left ventricle developed pressure; LVEDP, left ventricle end-diastolic pressure; LDH, lactate dehydrogenase. Data are mean ± SEM, n = 8-9. * P < 0.05, hypoxia vs. control, in the absence of 5-aza-2’-deoxycytidine.
4. Discussion
The present study provides novel evidence of DNA methylation as an important transcriptional regulation mechanism in maintaining not only the optimal development of the heart but also heart function after birth. Alteration of CpG methylation at the promoter region of the GR gene play a critical role in antenatal hypoxia-induced reprogramming of cardiac vulnerability to health challenges later in life. Maternal hypoxia results in increased methylation at GR promoter 14, 15, 16 and 17 and subsequent down-regulation of GR exon 14, 15, 16 and 17, but not exon 19 to 111 containing mRNA transcripts, suggesting selective transcriptional regulation of GR first exons as an important mechanism of GR expression in response to fetal stress. In addition, several functional transcription factor binding sites, including CREs at promoter 14 and 15 and Sp1 binding sites at promoter 16 and 17, have been identified at the GR promoter, and hypoxia-induced CpG hypermethylation at these sites is responsible for the decreased binding of transcription factors to proximal GR promoters 4-7 and repression of corresponding GR exons 14-7 mRNA variants, as well as total GR mRNA and protein. Inhibition of DNA methylation by 5-aza-2’-deoxycytidine completely reversed the hypoxia-induced GR gene repression and rescued the hypoxia-induced increase in heart vulnerability to ischemic injury.
The structure of the GR gene has been investigated in several animal species and the proximal CpG islands, ~5Kb upstream of the GR exon 2 is found to be highly structured and conserved [17, 30, 31]. This CpG-rich region encodes multiple alternative first exons that show remarkable similarity between the human and rat. In the human, exon 1s encoded by this region includes exon 1D, 1J, 1E, 1B, 1F, 1C and 1H. In the rat, exons 14 to 111 are encoded by this proximal region, with 14 being homologous to human 1D, 16 to human 1B, 17 to human 1F, 19/10 to human 1C, and 111 to human 1H [30]. McCormick and colleagues demonstrated that rat GR exon 1s were differentially expressed at variant levels in adult rat tissues. Among the relatively abundant GR exon 1s, exon 110 was found to be broadly and predominantly expressed, accounting for at least half of total GR transcripts [17, 30]. Although GR exon 16 is also broadly expressed in the rat, it only contributes about 10-20% of total GR mRNA in the adult heart [17]. Similar tissue-specific expression of GR exon 1s has been observed in the human [32]. Although exon 1B and 1C were shown to be broadly distributed in various adult tissues, expression of only exon 1C and a minor form 1G, but not exon 1B, was detected in human heart muscle [30]. In the present study, by using real-time PCR that is both sensitive and quantitative, we detected the expression of all proximal GR exon 1s, except for exon 18, in the near term fetal rat heart. Contrary to the previous findings in adult rat heart, GR exon 16 was found to be most abundant in fetal heart tissue, contributing to more than 80% of total GR mRNA, followed by exon 111 which contributes about 6%. The content of exon 110 represented only a minor portion of GR mRNA. This discrepancy of GR exon 1 expression levels in adult and fetal tissues strongly suggests a developmentally related fine-tuning of GR transcription regulation before and after birth. Indeed, while a remarkable elevation of GR protein levels was observed in neonatal and adult heart tissue compared to fetal hearts [8], the relative contributions of individual exon 1 isoforms remain elusive and further study is needed to demonstrate the role of selective usage of the alternate GR promoters in this process.
It is widely recognized that differential promoter usage can regulate the expression levels of GR exon 1 mRNA variants. In the present study, we establish that methylation levels of critical CpGs in the GR promoters are strongly associated with the transcriptional activity of corresponding exon 1s, influencing promoter usage in vivo. The proximal GR regulating region is located in a CpG island with multiple CpG sites that can be potentially methylated. By using an anti-5mC antibody that specifically recognizes methylated cytosine, the general methylation level of each promoter 1 was determined. Among these promoter 1s, we found that methylation levels of promoter 16 (less than 1%) is much lower than that of promoter 14, 15 and 17. This may in part explain the discrepancy between luciferace activities of individual promoters and expression levels of corresponding exon 1s. As we expected, constructs encoding the whole CpG island of the GR gene, which represents a sum of all the promoter 1s, exhibited substantial promoter activity in H9c2 cells; however, in the case of each individual exon 1, transcriptional activity of the regulating region failed to correlate with the expression level of the corresponding exon 1. For instance, exon 16 represents the most highly-expressed transcript, but the promoter activity was the lowest in the luciferase activity assay. In contrast, for the promoter of the lowly-expressed exon 1s such as 14 and 17, the intrinsic transcriptional activity was relatively high. Since hypermethylation of the GR promoter is related to suppression of GR expression, the remarkable lower methylation level of promoter 16 may indicate an active transcription status of exon 16 in the fetal rat heart. In contrast, the higher level of methylation at promoter 14, 15 and 17 can be, therefore, associated with their minor abundance in total GR mRNA transcripts. To date, there is little direct evidence of the regulation of minor GR exon 1 transcripts by perinatal stress, and most studies are focused on exon 17 in the rat or 1F in the human brain [17, 21, 33, 34]. To the best of our knowledge, the present study, for the first time, discloses the selective influence of maternal hypoxia on the expression of GR exon 14, 15, 16, and 17 in the developing heart. The contribution of these minor isoforms to the down-regulation of total GR may still be significant, especially since recent studies have highlighted the possible role of the variable 5’end in GR transcripts in the regulation of mRNA stability/half-life, protein translation and membrane GR production [18].
The role of epigenetic mechanisms, especially DNA methylation, in the regulation of GR expression by early life experiences has been extensively studied and has been shown to be highly site selective and tissue specific. For instance, low levels of maternal care are associated with hypermethylation of an NGFI-A binding site at the GR promoter 17, decreasing binding of NGFI-A, and resulting in subsequent down-regulation of GR in the hippocampus [21, 35]. Maternal hypoxia leads to decreased GR expression via increased methylation levels of GR promoter 17 and 111 in the developing brain, which enhances the susceptibility of neonatal rats to hypoxic-ischemic-induced brain injury [22]. The methylation level of GR gene promoter exon 110 is 33% lower and GR expression is 84% higher in the liver of rat offspring exposed to maternal protein restriction at postnatal day 34 [23]. Similarly, our findings showed that in the developing heart, exposure to maternal hypoxia in pregnancy induced selective down-regulation of GR exon 14, 15, 16 and 17. In accordance, a significant increase of DNA methylation levels at promoter 14, 15, 16 and 17 resulted from gestational hypoxia, further confirming individual GR promoter 1s as targets of epigenetic modification in the regulation of total GR expression.
The GR promoters lack typical TATA boxes and expression of GR is tightly associated with transcription factors that bind to the promoter region. Multiple transcription factors have been demonstrated or predicted to act at the GR promoter to regulate GR expression, including Sp1, Sp3, NGFI-A (Egr-1), NF-1 and Yin Yang1 (YY1) [19, 21, 22, 36, 37]. The role of the NGFI-A binding site at rat GR promoter 17 or human 1F in the regulation of brain GR expression has been extensively studied in response to different types of stress [21, 22, 34]. In the present study, we found that methylation of this site was not affected by maternal hypoxia in the fetal heart, suggesting this Egr-1 site may be used in brain-specific regulation of GR exon 17. On the contrary, the Sp1 binding site at promoter 17 is significantly methylated by hypoxia in the fetal heart, accompanied by decreased expression of GR exon 17, although it was not affected in the brain by the common stress maternal hypoxia [22]. In accordance, deletion of the Egr-1 binding sites failed to change the transcriptional activity of GR promoter in a cardiomyocyte cell line, whereas deletion or methylation mutation of Sp1 site significantly inhibited its transcriptional activity. Similar findings were observed for the YY1 binding sites at the human GR promoter, which functions distinctly in NIH3T3 and Hela cells [36]. The differential responses at DNA regulation elements despite a common stimulus may account for the tissue-specific regulation of GR expression in different organs. Sp1 is a ubiquitous transcription factor that plays an active role in hypoxia-driven gene expression [38-40]. Sp1 binding sites have been identified at the GR promoter and have been demonstrated to regulate GR expression [22, 36, 41]. Although the molecular mechanisms are not clear, maternal hypoxia has been shown to increase DNA methylation of Sp1 binding sites at different genes, and this epigenetic modification may be related to the corresponding sustained suppression of these genes from fetal to adult age [11, 22, 27]. In addition to the Sp1 sites at promoter 16 and 17, two CREs were identified at the GR promoter 14 and 15. Although the CREs (14:TGACGCCA; 15:TGACGTTT) show slight variations in sequence compared to the consensus CRE (TGACGTCA), both sites are functional and are capable of binding the regulating protein CREB. CREB signaling and transcriptional activities play essential roles in vascular smooth muscle cell and cardiomyocyte hypertrophy induced by angiotensin-II or acute hypoxia/reoxygenation [42, 43]. Acute hypoxia followed by re-oxygenation changes the DNA binding activity of CREB. However, the effect of prolonged hypoxia on CREB transcriptional regulation of the GR was heretofore unknown. Our results indicate that methylation of CREs at the GR promoter is significantly increased. Correspondingly, the binding of CREB to the GR promoter 14 and 15 is decreased. Our gel shift assay showed that CpG methylation at CRE abolished the binding of CREB and this is in accordance to the previous findings showing that CpG methylation blocks transcription factor binding and transcription activity of CRE [29, 44]. CpG methylation at different GR promoter 1s is highly variable between individuals [45, 46]. However, using different detection methods, we demonstrated that the increase of CpG methylation can be induced by maternal hypoxia both globally, as well as site-specifically at GR promoters.
Of importance, application of 5-aza-2’-deoxycytidine during the early postnatal period completely reversed the hypoxia-induced hypermethylation to levels comparable to the control, both globally and gene-specifically at the GR promoter. The functional significance of 5-aza-2-deoxycytidine-mediated demethylation in rescuing gene expression was demonstrated in the heart, in which 5-aza-2′-deoxycytidine reversed hypoxia-induced GR gene repression and recovered GR mRNA and protein expression in the offspring heart. Whether this transcriptional regulation of GR expression reflects GR in its active, dimerized state remains to be determined. The finding that inhibition of promoter methylation recovered GR expression is in agreement with previous findings showing that 5-aza-2′-deoxycytidine inhibited promoter hypermethylation and rescued PKCε gene expression in fetal hearts [11, 47, 48]. Given that 5-aza-2′-deoxycytidine induces a global change of DNA methylation and may affect other genes, potential involvement of other mechanisms in the 5-aza-2′-deoxycytidine-mediated effect may not be ruled out in the present study. Rat hearts continue to develop within the first two weeks after birth, and both proliferation and differentiation of cardiomyocytes take place in this time frame. Although it was not determined whether the effects can be reversed if 5-aza-2’-deoxycytidine was applied later than day 3 in the present study, we and others demonstrated previously that 5-aza-2′-deoxycytidine, given intraperitoneally (i.p. 1 mg/kg/day) for 3-7 days, caused demethylation of PKCε gene promoter in the heart and 11β-hydroxysteroid dehydrogenase type 2 (11βHSD2) gene promoter in the kidney, lung, and liver, resulting in increased expression of PKCε gene and 11βHSD2 gene in these organs [24, 25]. In addition to the regulation of GR expression, maternal hypoxia increased plasma glucocorticoid levels in the fetus in both sheep and rodents [49-51]. Both maternal hypoxia and antenatal glucocorticoid treatment result in down-regulation of GR and may thus account for similar effects in regulating the epigenome and gene expression, supporting a notion that glucocorticoids/GR may play a center role in programming models of fetal stress.
The antenatal hypoxia-induced down-regulation of GR gene in the heart is of functional significance. The pathophysiological significance of decreased GR expression levels in the heart is highlighted by the findings that demonstrated the cardioprotective effects of glucocorticoids in the acute setting of myocardial ischemia and reperfusion injury both in humans and in animals [52-63]. It has been demonstrated that glucocorticoid-GR-mediated cardioprotective effects are local and heart-specific, mainly through direct effects on cardiomyocytes rather than vasculature [53]. Consistent with these findings, the present study demonstrated that activation of GR by dexamethasone induced protection of the heart from ischemia and reperfusion injury in 4 week old offspring. Of importance, we demonstrated that consistent with the rescue of GR expression in the heart, inhibition of DNA methylation in early postnatal life by 5-aza-2′-deoxycytidine reversed the antenatal hypoxia-induced increase in heart susceptibility to ischemic injury in offspring. In rodents, cardiomyocyte proliferation and terminal differentiation continue throughout the first two weeks after birth [64], providing a possible window for intervening and rescuing the adverse effects caused by prenatal insults in the heart given that DNA methyltransferase activities are closely related with DNA synthesis in proliferative cells. Similar findings were obtained in adult rats, in which treatment of rats with 5-aza-2′-deoxycytidine (i.p. 1 mg/kg/day) resulted in a reversal of norepinephrine-induced hypermethylation in the heart and rescued the phenotype of heart hypertrophy and reduced contractility [25]. Thus, the present findings that 5-aza-2′-deoxycytidine abrogated GR promoter hypermethylation, rescued GR gene expression, and reversed antenatal hypoxia-induced increase in heart susceptibility to ischemic injury provide novel evidence of a causative mechanism of DNA hypermethylation and GR gene repression in programming of ischemic-sensitive phenotype in the heart. It is interesting that 5-aza-2′-deoxycytidine also improved cardiac performance following ischemia in the control group, given that it did not alter methylation patterns in this group. This is in accordance with previous findings, albeit the mechanisms remain largely elusive. It has been demonstrated that 5-azacytidine treatment improves cardiac performance following cardiac ischemia, and various mechanisms have been proposed including modulation of macrophage phenotype and inhibition of fibrosis [65]. It is possible that these mechanisms may also contribute to 5-aza-2′-deoxycytidine-mediated improvement of cardiac function in the hypoxic offspring. The present finding of no significant differences in cardiac function following ischemia between control and hypoxic offspring in 5-aza-2′-deoxycytidine-treated animals indicates that the specific effects of hypoxia were inhibited by 5-aza-2′-deoxycytidine.
In addition to a functional phenotype demonstrated in the present study, accumulating evidence has shown that gestational hypoxia induces morphological changes, including myocardial thinning, ventricle dilation and cardiomyocyte hypertrophy in the developing heart [7, 66, 67]. Our previous studies in the same animal model demonstrated that maternal hypoxia significantly decreased ventricular wall thickness in both fetal and neonatal rats [68]. In addition, we demonstrated that hypoxia and glucocorticoid treatments of newborn rats significantly decreased proliferation and increased binucleation of cardiomyocytes in the developing heart, resulting in reduced myocyte endowment in the heart, and these effects were reversed by 5-aza-2’-deoxycytidine [69-71].
The present study provides novel mechanistic evidence that antenatal hypoxia represses GR expression by increasing GR promoter methylation in the developing heart. In addition, this work highlights novel mechanism of controlling gene expression patterns during the developmental programming of the heart by adverse intrauterine environments through methylation of specific transcription factor binding sites. Multiple fetal stressors, including hypoxia, unbalanced nutrition, and antenatal exposure to synthetic glucocorticoids, are known to cause programming of cardiac dysfunction later in life. It is important to note that these stressors are not mutually exclusive and the regulation of the GR is likely to be a common pathway in mediating these programming effects. Therefore, our findings demonstrating the epigenetic regulation of GR gene expression by fetal stress are of critical importance in understanding the molecular mechanisms underlying developmental programming of health and disease in the heart.
Supplementary Material
Highlights.
Fetal hypoxia differentially regulates transcription of alternate GR first exons in the heart.
Hypoxia increases promoter methylation and represses GR expression.
5-Aza reverses hypoxia-induced GR gene repression and rescues heart susceptibility to ischemia.
Acknowledgments
The authors thank Dr. Karen E. Chapman for her generous sharing of luciferase-GR-promoter constructs with us.
Sources of Funding
This study was supported by the National Institutes of Health grant HL118861 (L.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None.
References
- 1.Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1:1077–81. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- 2.Bateson P, Barker D, Clutton-Brock T, Deb D, D'Udine B, Foley RA, et al. Developmental plasticity and human health. Nature. 2004;430:419–21. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- 3.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. The New England journal of medicine. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 5.Giussani DA, Davidge ST. Developmental programming of cardiovascular disease by prenatal hypoxia. J Dev Orig Health Dis. 2013;4:328–37. doi: 10.1017/S204017441300010X. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert RD, Pearce WJ, Longo LD. Fetal cardiac and cerebrovascular acclimatization responses to high altitude, long-term hypoxia. High Alt Med Biol. 2003;4:203–13. doi: 10.1089/152702903322022802. [DOI] [PubMed] [Google Scholar]
- 7.Ream M, Ray AM, Chandra R, Chikaraishi DM. Early fetal hypoxia leads to growth restriction and myocardial thinning. Am J Physiol Regul Integr Comp Physiol. 2008;295:R583–95. doi: 10.1152/ajpregu.00771.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue Q, Dasgupta C, Chen M, Zhang L. Foetal hypoxia increases cardiac AT(2)R expression and subsequent vulnerability to adult ischaemic injury. Cardiovasc Res. 2011;89:300–8. doi: 10.1093/cvr/cvq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae S, Xiao Y, Li G, Casiano CA, Zhang L. Effect of maternal chronic hypoxic exposure during gestation on apoptosis in fetal rat heart. Am J Physiol Heart Circ Physiol. 2003;285:H983–90. doi: 10.1152/ajpheart.00005.2003. [DOI] [PubMed] [Google Scholar]
- 10.Li G, Bae S, Zhang L. Effect of prenatal hypoxia on heat stress-mediated cardioprotection in adult rat heart. Am J Physiol Heart Circ Physiol. 2004;286:H1712–9. doi: 10.1152/ajpheart.00898.2003. [DOI] [PubMed] [Google Scholar]
- 11.Patterson AJ, Chen M, Xue Q, Xiao D, Zhang L. Chronic prenatal hypoxia induces epigenetic programming of PKC{epsilon} gene repression in rat hearts. Circulation research. 2010;107:365–73. doi: 10.1161/CIRCRESAHA.110.221259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowden AL, Li J, Forhead AJ. Glucocorticoids and the preparation for life after birth: are there long-term consequences of the life insurance? The Proceedings of the Nutrition Society. 1998;57:113–22. doi: 10.1079/pns19980017. [DOI] [PubMed] [Google Scholar]
- 13.Ren R, Oakley RH, Cruz-Topete D, Cidlowski JA. Dual role for glucocorticoids in cardiomyocyte hypertrophy and apoptosis. Endocrinology. 2012;153:5346–60. doi: 10.1210/en.2012-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakley RH, Ren R, Cruz-Topete D, Bird GS, Myers PH, Boyle MC, et al. Essential role of stress hormone signaling in cardiomyocytes for the prevention of heart disease. Proc Natl Acad Sci U S A. 2013;110:17035–40. doi: 10.1073/pnas.1302546110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertram C, Trowern AR, Copin N, Jackson AA, Whorwood CB. The maternal diet during pregnancy programs altered expression of the glucocorticoid receptor and type 2 11beta-hydroxysteroid dehydrogenase: potential molecular mechanisms underlying the programming of hypertension in utero. Endocrinology. 2001;142:2841–53. doi: 10.1210/endo.142.7.8238. [DOI] [PubMed] [Google Scholar]
- 16.Giraud GD, Louey S, Jonker S, Schultz J, Thornburg KL. Cortisol stimulates cell cycle activity in the cardiomyocyte of the sheep fetus. Endocrinology. 2006;147:3643–9. doi: 10.1210/en.2006-0061. [DOI] [PubMed] [Google Scholar]
- 17.McCormick JA, Lyons V, Jacobson MD, Noble J, Diorio J, Nyirenda M, et al. 5′-heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: differential regulation of variant transcripts by early-life events. Mol Endocrinol. 2000;14:506–17. doi: 10.1210/mend.14.4.0438. [DOI] [PubMed] [Google Scholar]
- 18.Turner JD, Vernocchi S, Schmitz S, Muller CP. Role of the 5′-untranslated regions in post-transcriptional regulation of the human glucocorticoid receptor. Biochim Biophys Acta. 2014;1839:1051–61. doi: 10.1016/j.bbagrm.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Suehiro T, Kaneda T, Ikeda Y, Arii K, Kumon Y, Hashimoto K. Regulation of human glucocorticoid receptor gene transcription by Sp1 and p53. Mol Cell Endocrinol. 2004;222:33–40. doi: 10.1016/j.mce.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Turner JD, Alt SR, Cao L, Vernocchi S, Trifonova S, Battello N, et al. Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochem Pharmacol. 2010;80:1860–8. doi: 10.1016/j.bcp.2010.06.037. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, et al. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–62. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Rodriguez PJ, Xiong F, Li Y, Zhou J, Zhang L. Fetal hypoxia increases vulnerability of hypoxic-ischemic brain injury in neonatal rats: role of glucocorticoid receptors. Neurobiology of disease. 2014;65:172–9. doi: 10.1016/j.nbd.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr. 2007;97:1064–73. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alikhani-Koopaei R, Fouladkou F, Frey FJ, Frey BM. Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J Clin Invest. 2004;114:1146–57. doi: 10.1172/JCI21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao D, Dasgupta C, Chen M, Zhang K, Buchholz J, Xu Z, et al. Inhibition of DNA methylation reverses norepinephrine-induced cardiac hypertrophy in rats. Cardiovasc Res. 2014;101:373–82. doi: 10.1093/cvr/cvt264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer K, Zhang H, Zhang L. Direct effect of cocaine on epigenetic regulation of PKCepsilon gene repression in the fetal rat heart. J Mol Cell Cardiol. 2009;47:504–11. doi: 10.1016/j.yjmcc.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dasgupta C, Chen M, Zhang H, Yang S, Zhang L. Chronic hypoxia during gestation causes epigenetic repression of the estrogen receptor-alpha gene in ovine uterine arteries via heightened promoter methylation. Hypertension. 2012;60:697–704. doi: 10.1161/HYPERTENSIONAHA.112.198242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Xiao D, Yang S, Zhang L. Promoter methylation represses AT2R gene and increases brain hypoxic-ischemic injury in neonatal rats. Neurobiology of disease. 2013;60:32–8. doi: 10.1016/j.nbd.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–9. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 30.Turner JD, Muller CP. Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: identification, and tissue distribution of multiple new human exon 1. J Mol Endocrinol. 2005;35:283–92. doi: 10.1677/jme.1.01822. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Z, Qian L, Zou H, Jia Y, Ni Y, Yang X, et al. Porcine glucocorticoid receptor (NR3C1) gene: tissue-specificity of transcriptional strength and glucocorticoid responsiveness of alternative promoters. J Steroid Biochem Mol Biol. 2014;141:87–93. doi: 10.1016/j.jsbmb.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Mata-Greenwood E, Jackson PN, Pearce WJ, Zhang L. Endothelial glucocorticoid receptor promoter methylation according to dexamethasone sensitivity. J Mol Endocrinol. 2015;55:133–46. doi: 10.1530/JME-15-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alt SR, Turner JD, Klok MD, Meijer OC, Lakke EA, Derijk RH, et al. Differential expression of glucocorticoid receptor transcripts in major depressive disorder is not epigenetically programmed. Psychoneuroendocrinology. 2010;35:544–56. doi: 10.1016/j.psyneuen.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 34.McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weaver IC, Diorio J, Seckl JR, Szyf M, Meaney MJ. Early environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic target sites. Ann N Y Acad Sci. 2004;1024:182–212. doi: 10.1196/annals.1321.099. [DOI] [PubMed] [Google Scholar]
- 36.Breslin MB, Vedeckis WV. The human glucocorticoid receptor promoter upstream sequences contain binding sites for the ubiquitous transcription factor, Yin Yang 1. J Steroid Biochem Mol Biol. 1998;67:369–81. doi: 10.1016/s0960-0760(98)00138-1. [DOI] [PubMed] [Google Scholar]
- 37.Nunez BS, Vedeckis WV. Characterization of promoter 1B in the human glucocorticoid receptor gene. Mol Cell Endocrinol. 2002;189:191–9. doi: 10.1016/s0303-7207(01)00676-1. [DOI] [PubMed] [Google Scholar]
- 38.Deacon K, Onion D, Kumari R, Watson SA, Knox AJ. Elevated SP-1 transcription factor expression and activity drives basal and hypoxia-induced vascular endothelial growth factor (VEGF) expression in non-small cell lung cancer. J Biol Chem. 2012;287:39967–81. doi: 10.1074/jbc.M112.397042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson AJ, Xiao D, Xiong F, Dixon B, Zhang L. Hypoxia-derived oxidative stress mediates epigenetic repression of PKCepsilon gene in foetal rat hearts. Cardiovasc Res. 2012;93:302–10. doi: 10.1093/cvr/cvr322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie L, Collins JF. Transcription factors Sp1 and Hif2alpha mediate induction of the copper-transporting ATPase (Atp7a) gene in intestinal epithelial cells during hypoxia. J Biol Chem. 2013;288:23943–52. doi: 10.1074/jbc.M113.489500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolla V, Litwack G. Upregulation of mineralocorticoid- and glucocorticoid-receptor gene expression by Sp-I. Mol Cell Biol Res Commun. 1999;1:44–7. doi: 10.1006/mcbr.1999.0110. [DOI] [PubMed] [Google Scholar]
- 42.Funakoshi Y, Ichiki T, Takeda K, Tokuno T, Iino N, Takeshita A. Critical role of cAMP-response element-binding protein for angiotensin II-induced hypertrophy of vascular smooth muscle cells. J Biol Chem. 2002;277:18710–7. doi: 10.1074/jbc.M110430200. [DOI] [PubMed] [Google Scholar]
- 43.El Jamali A, Freund C, Rechner C, Scheidereit C, Dietz R, Bergmann MW. Reoxygenation after severe hypoxia induces cardiomyocyte hypertrophy in vitro: activation of CREB downstream of GSK3beta. FASEB J. 2004;18:1096–8. doi: 10.1096/fj.03-1054fje. [DOI] [PubMed] [Google Scholar]
- 44.DiNardo DN, Butcher DT, Robinson DP, Archer TK, Rodenhiser DI. Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene. 2001;20:5331–40. doi: 10.1038/sj.onc.1204697. [DOI] [PubMed] [Google Scholar]
- 45.Turner JD, Pelascini LP, Macedo JA, Muller CP. Highly individual methylation patterns of alternative glucocorticoid receptor promoters suggest individualized epigenetic regulatory mechanisms. Nucleic Acids Res. 2008;36:7207–18. doi: 10.1093/nar/gkn897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao-Lei L, Suwansirikul S, Jutavijittum P, Meriaux SB, Turner JD, Muller CP. Glucocorticoid receptor gene expression and promoter CpG modifications throughout the human brain. J Psychiatr Res. 2013;47:1597–607. doi: 10.1016/j.jpsychires.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 47.Lawrence J, Chen M, Xiong F, Xiao D, Zhang H, Buchholz JN, et al. Foetal nicotine exposure causes PKCepsilon gene repression by promoter methylation in rat hearts. Cardiovasc Res. 2011;89:89–97. doi: 10.1093/cvr/cvq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong F, Xiao D, Zhang L. Norepinephrine causes epigenetic repression of PKCepsilon gene in rodent hearts by activating Nox1-dependent reactive oxygen species production. FASEB J. 2012;26:2753–63. doi: 10.1096/fj.11-199422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cuffe JS, Walton SL, Singh RR, Spiers JG, Bielefeldt-Ohmann H, Wilkinson L, et al. Mid- to late term hypoxia in the mouse alters placental morphology, glucocorticoid regulatory pathways and nutrient transporters in a sex-specific manner. J Physiol. 2014;592:3127–41. doi: 10.1113/jphysiol.2014.272856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giussani DA, Fletcher AJ, Gardner DS. Sex differences in the ovine fetal cortisol response to stress. Pediatr Res. 2011;69:118–22. doi: 10.1203/PDR.0b013e3182042a20. [DOI] [PubMed] [Google Scholar]
- 51.Alfaidy N, Gupta S, DeMarco C, Caniggia I, Challis JR. Oxygen regulation of placental 11 beta-hydroxysteroid dehydrogenase 2: physiological and pathological implications. The Journal of clinical endocrinology and metabolism. 2002;87:4797–805. doi: 10.1210/jc.2002-020310. [DOI] [PubMed] [Google Scholar]
- 52.Xue Q, Patterson AJ, Xiao D, Zhang L. Glucocorticoid modulates angiotensin II receptor expression patterns and protects the heart from ischemia and reperfusion injury. PLoS One. 2014;9:e106827. doi: 10.1371/journal.pone.0106827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tokudome S, Sano M, Shinmura K, Matsuhashi T, Morizane S, Moriyama H, et al. Glucocorticoid protects rodent hearts from ischemia/reperfusion injury by activating lipocalin-type prostaglandin D synthase-derived PGD2 biosynthesis. J Clin Invest. 2009;119:1477–88. doi: 10.1172/JCI37413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Libby P, Maroko PR, Bloor CM, Sobel BE, Braunwald E. Reduction of experimental myocardial infarct size by corticosteroid administration. J Clin Invest. 1973;52:599–607. doi: 10.1172/JCI107221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valen G, Kawakami T, Tahepold P, Dumitrescu A, Lowbeer C, Vaage J. Glucocorticoid pretreatment protects cardiac function and induces cardiac heat shock protein 72. Am J Physiol Heart Circ Physiol. 2000;279:H836–43. doi: 10.1152/ajpheart.2000.279.2.H836. [DOI] [PubMed] [Google Scholar]
- 56.Varga E, Nagy N, Lazar J, Czifra G, Bak I, Biro T, et al. Inhibition of ischemia/reperfusion-induced damage by dexamethasone in isolated working rat hearts: the role of cytochrome c release. Life Sci. 2004;75:2411–23. doi: 10.1016/j.lfs.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 57.Skyschally A, Haude M, Dorge H, Thielmann M, Duschin A, van de Sand A, et al. Glucocorticoid treatment prevents progressive myocardial dysfunction resulting from experimental coronary microembolization. Circulation. 2004;109:2337–42. doi: 10.1161/01.CIR.0000127961.66744.F4. [DOI] [PubMed] [Google Scholar]
- 58.Giugliano GR, Giugliano RP, Gibson CM, Kuntz RE. Meta-analysis of corticosteroid treatment in acute myocardial infarction. Am J Cardiol. 2003;91:1055–9. doi: 10.1016/s0002-9149(03)00148-6. [DOI] [PubMed] [Google Scholar]
- 59.Qin C, Buxton KD, Pepe S, Cao AH, Venardos K, Love JE, et al. Reperfusion-induced myocardial dysfunction is prevented by endogenous annexin-A1 and its N-terminal-derived peptide Ac-ANX-A1(2-26). Br J Pharmacol. 2013;168:238–52. doi: 10.1111/j.1476-5381.2012.02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spath JA, Jr., Lane DL, Lefer AM. Protective action of methylprednisolone on the myocardium during experimental myocardial ischemia in the cat. Circulation research. 1974;35:44–51. doi: 10.1161/01.res.35.1.44. [DOI] [PubMed] [Google Scholar]
- 61.Hammerman H, Kloner RA, Hale S, Schoen FJ, Braunwald E. Dose-dependent effects of short-term methylprednisolone on myocardial infarct extent, scar formation, and ventricular function. Circulation. 1983;68:446–52. doi: 10.1161/01.cir.68.2.446. [DOI] [PubMed] [Google Scholar]
- 62.Wynsen JC, Preuss KC, Gross GJ, Brooks HL, Warltier DC. Steroid-induced enhancement of functional recovery of postischemic, reperfused myocardium in conscious dogs. Am Heart J. 1988;116:915–25. doi: 10.1016/0002-8703(88)90141-x. [DOI] [PubMed] [Google Scholar]
- 63.Barzilai D, Plavnick J, Hazani A, Einath R, Kleinhaus N, Kanter Y. Use of hydrocortisone in the treatment of acute myocardial infarction. Summary of a clinical trial in 446 patients. Chest. 1972;61:488–91. doi: 10.1378/chest.61.5.488. [DOI] [PubMed] [Google Scholar]
- 64.Paradis AN, Gay MS, Zhang L. Binucleation of cardiomyocytes: the transition from a proliferative to a terminally differentiated state. Drug Discov Today. 2014;19:602–9. doi: 10.1016/j.drudis.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim YS, Kang WS, Kwon JS, Hong MH, Jeong HY, Jeong HC, et al. Protective role of 5-azacytidine on myocardial infarction is associated with modulation of macrophage phenotype and inhibition of fibrosis. J Cell Mol Med. 2014;18:1018–27. doi: 10.1111/jcmm.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma SK, Lucitti JL, Nordman C, Tinney JP, Tobita K, Keller BB. Impact of hypoxia on early chick embryo growth and cardiovascular function. Pediatr Res. 2006;59:116–20. doi: 10.1203/01.pdr.0000191579.63339.90. [DOI] [PubMed] [Google Scholar]
- 67.Martin C, Yu AY, Jiang BH, Davis L, Kimberly D, Hohimer AR, et al. Cardiac hypertrophy in chronically anemic fetal sheep: Increased vascularization is associated with increased myocardial expression of vascular endothelial growth factor and hypoxia-inducible factor 1. Am J Obstet Gynecol. 1998;178:527–34. doi: 10.1016/s0002-9378(98)70433-8. [DOI] [PubMed] [Google Scholar]
- 68.Tong W, Xue Q, Li Y, Zhang L. Maternal hypoxia alters matrix metalloproteinase expression patterns and causes cardiac remodeling in fetal and neonatal rats. Am J Physiol Heart Circ Physiol. 2011;301:H2113–21. doi: 10.1152/ajpheart.00356.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gay MS, Li Y, Xiong F, Lin T, Zhang L. Dexamethasone Treatment of Newborn Rats Decreases Cardiomyocyte Endowment in the Developing Heart through Epigenetic Modifications. PloS one. 2015;10:e0125033. doi: 10.1371/journal.pone.0125033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paradis A, Xiao D, Zhou J, Zhang L. Endothelin-1 promotes cardiomyocyte terminal differentiation in the developing heart via heightened DNA methylation. International journal of medical sciences. 2014;11:373–80. doi: 10.7150/ijms.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paradis AN, Gay MS, Wilson CG, Zhang L. Newborn hypoxia/anoxia inhibits cardiomyocyte proliferation and decreases cardiomyocyte endowment in the developing heart: role of endothelin-1. PloS one. 2015;10:e0116600. doi: 10.1371/journal.pone.0116600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.