Abstract
Background
Genomes of E. coli, including that of the human pathogen Escherichia coli O157:H7 (EHEC) EDL933, still harbor undetected protein-coding genes which, apparently, have escaped annotation due to their small size and non-essential function. To find such genes, global gene expression of EHEC EDL933 was examined, using strand-specific RNAseq (transcriptome), ribosomal footprinting (translatome) and mass spectrometry (proteome).
Results
Using the above methods, 72 short, non-annotated protein-coding genes were detected. All of these showed signals in the ribosomal footprinting assay indicating mRNA translation. Seven were verified by mass spectrometry. Fifty-seven genes are annotated in other enterobacteriaceae, mainly as hypothetical genes; the remaining 15 genes constitute novel discoveries. In addition, protein structure and function were predicted computationally and compared between EHEC-encoded proteins and 100-times randomly shuffled proteins. Based on this comparison, 61 of the 72 novel proteins exhibit predicted structural and functional features similar to those of annotated proteins. Many of the novel genes show differential transcription when grown under eleven diverse growth conditions suggesting environmental regulation. Three genes were found to confer a phenotype in previous studies, e.g., decreased cattle colonization.
Conclusions
These findings demonstrate that ribosomal footprinting can be used to detect novel protein coding genes, contributing to the growing body of evidence that hypothetical genes are not annotation artifacts and opening an additional way to study their functionality. All 72 genes are taxonomically restricted and, therefore, appear to have evolved relatively recently de novo.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2456-1) contains supplementary material, which is available to authorized users.
Keywords: Translatome, Riboseq, Proteome, Novel genes, Orphans, EHEC
Background
EHEC is an important human pathogen, which causes bloody diarrhea, hemolytic uremic syndrome and other sequelae [1]. Strain EDL933 has been isolated from ground beef in Michigan (USA) and has been linked to the first severe EHEC outbreak in 1982 in which this bacterium was shown to be the causative agent [2].
Next generation sequencing (NGS), especially if conducted strand-specifically on mRNA (RNAseq), has grown into a valuable tool to study bacterial transcriptomes [3]. However, transcripts represent mRNAs as well as non-coding RNAs (ncRNAs), which sometimes are hard to tell apart. Especially, transcripts of small proteins might be mistaken for short ncRNAs [4–6]. A much better tell-tale for protein-coding RNA is the translatome consisting of ribosomal footprints, which indicate mRNAs being in the process of translation [7–10]. To determine the translatome, ribosomes are stalled on the mRNA and RNA parts accessible to RNase are degraded. The parts of mRNA covered by ribosomes are protected and form “ribosomal footprints” which are sequenced [10]. In addition, a high-throughput method to interrogate the bacterial proteome is the mass-spectrometry-based analysis of proteolytically digested proteins, usually referred to as “bottom up” or “shotgun proteomics” [11]. This method is able to directly confirm the existence of proteins, albeit displaying a limited sensitivity for low abundance proteins [12].
It is surprising that a sizable fraction of genes in any bacterium is still of hypothetical status, which means that their function is unknown and it is even unclear whether these genes are expressed [13]. Some hypothetical proteins have no homology to any other predicted protein in any species [14], i.e., they are taxonomically restricted [15] and, therefore, belong to the orphan genes. Other hypothetical genes are found in genomes of many species and are termed “conserved hypotheticals” [13]. Some hypothetical genes are believed to be annotation artifacts, suggesting that genomes are “over-annotated” and several hypothetical genes may not be protein-coding genes at all. For instance, after genome sequencing, genes are often predicted by computational annotation tools like GLIMMER [16]. These automatic tools might proliferate annotation errors. Yu et al. [17] state: “Previous works show that a significant fraction of annotated short ORFs may be not true genes, which is one of the major causes that account for the over-annotation of microbial genomes.” This reality has influenced the annotation of genomes from the release of the first eukaryotic yeast genome that began with over 9000 ORFs and shrunk to about 6000 when the authors realized a high error rate removing many ORFs shorter than 100 residues [18]. However, the above statements are based on publications from an era in which short proteins were largely dismissed as not functional, hence, not existing [19, 20]. Today, we control the technology to dig deeper: we add 72 short protein coding genes to the E. coli EHEC gene repertoire by using a multi-omics approach, including translatomic data and transcription patterns specific for different growth conditions.
Methods
No ethics approval was required for any aspect of this study.
Strain and culture conditions
The derivative WS4202 (Weihenstephan Microbial Strain Collection, Lehrstuhl für Mikrobielle Ökologie, Technische Universität München, Germany = CIP106327, Collection de l’Institut Pasteur, Paris, France) of Escherichia coli O157:H7 EDL933 (EHEC) was used in this study. Bacteria were incubated in 10-fold diluted lysogeny-broth [21] at 37 °C with shaking (180 rpm). The optical density of the samples was monitored at 600 nm to determine the growth phase. Before harvesting, an aliquot was plated on CHROMagar O157 (CHROMagar, France) to confirm identity. Cells were harvested by centrifugation (10,000×g, 1 °C, 3 min) in the transition from late exponential to early stationary phase. The pellet was frozen in liquid nitrogen and stored at -80 °C for RNA extraction. For ribosomal footprints, corresponding transcriptomes, and mass spectrometry, the strain was also grown in 10-times diluted LB. For the condition LB-Nitrite, 200 mg/L sodium nitrite was added and the pH adjusted to 6 using HCl. Briefly, bacteria were grown in LB medium at pH4, pH9, or at 15 °C; in LB with nitrite or trimethoprim-sulfamethoxazole; in LB-agar surface, M9 minimal medium, spinach leaf juice, surface of living radish sprouts, or in cattle feces. Further details about the growth conditions are described in [9].
RNA-footprints and transcriptomes
RNA-footprints and transcriptomes (two biological replicates in each case) were analyzed as described by Landstorfer et al. [22] using 170 μg/mL chloramphenicol to stall the ribosomes, which is about 6-times above the concentration at which trans-translation occurs [23]. Isolated ribosomes were incubated with RNase I. Intact ribosomes were enriched twice by gradient centrifugation. Isolated footprints and isolated total RNA for transcriptome sequencing (rRNAs removed) were processed strand-specifically with the TruSeq Small RNA Sample Preparation Kit (Illumina) according to the manual and sequenced on an Illumina MiSeq.
Illumina FASTQ files were mapped to the genome using Bowtie [24] either in Galaxy [25, 26] or as standalone with default settings. Output SAM files were filtered for mappable reads using Samtools and further converted and indexed to BAM and BAM.BAI files [27]. Visualization of the data was carried out using our own NGS-Viewer [28] or BamView [29] implemented in Artemis 12.0 [30]. Transcription and translation levels were evaluated using RPKM values [31]. The RPKM value is defined as “reads per kilobase-gene length per million mapped reads”. In our calculations, we excluded rRNA reads since this value is compromised by the rRNA removal. All new open reading frames (ORFs) suspected of being translated (at least 10 RPKM translatome) were inspected manually [28] to exclude false positives, and the ribosomal coverage value (RCV) was calculated. The RCV is defined as the RPKM ratio of the translatome per transcriptome for each gene [31], both derived from the same biological experiment [22].
Protein isolation
Cells were harvested by centrifugation at 4 °C for 4 min at 10.000 × g and washed five times with cooled Ringer solution. The pellet was resuspended in cold lysis buffer (50 mM Tris/HCl pH 7.5, 5 % glycerol, 1.5 mM MgCl2, 150 mM NaCl, 1 mM Na3VO4, 25 mM NaF, 0,8 % NP40/Igepal, 1 mM DTT and 1 tablet / 25 mL buffer of EDTA-free tablets of the protease inhibitor cocktail; Roche Diagnostics). Cells were disrupted using a Fast-Prep (MP Biomedicals) with six runs for 20 s each at a shaking speed of 5 m/s and cooling on ice between each run. The suspension was then incubated on ice for 30 min and subsequently centrifuged for 10 min at 20.000 × g at 4 °C. The supernatant was transferred to an ultracentrifuge tube and centrifuged 1.5 h at 4 °C at 141,000 × g in a Beckmann L7 ultracentrifuge. The supernatant was transferred into fresh tubes and stored at −80 °C. Protein concentration was determined by Bradford assays (RotiQuant, Roth), measuring extinction on a Victor3 1420 multilabel counter (Perkin Elmer). The standard curve was generated using BSA (bovine serum albumin). The lysate was fractionated by SDS gel electrophoresis into 12 fractions. Each gel-fraction was washed and digested with trypsin for mass spectrometry (MS) analysis.
For the protein fraction “LB-small”, small proteins were fractionated by separating 300 μl of the above protein solution on two SDS gels. The proteins below 12 kDa were excised from the gel and transferred to a 0.5 ml microcentrifuge tube, which was pierced with a 20-gauge needle at the bottom. This tube was placed in a 1.5 ml microcentrifuge tube and centrifuged for 2 min at 13,000 × g at room temperature. The resultant gel debris was transferred to 500 μl elution buffer, rotated over night at room temperature in a microcentrifuge tube and then filtered with a 0.22 μm Spin-X spin filter (Corning, USA) for 2 min at 10,000 × g. Next, the proteins were precipitated using four-volumes of cold acetone (−20 °C), incubated at −20 °C for 60 min and then centrifuged at 15,000 × g for 10 min at −20 °C. The supernatant was decanted and remaining liquid evaporated at room temperature. Finally, the small protein fraction was treated with trypsin for MS analysis.
Mass spectrometry
The digested protein fractions were subjected to an Eksigent nanoLC-Ultra 1D+ (Eksigent, Dublin, CA) coupled to an Orbitrap Velos (Thermo Scientific, Bremen, Germany). Peptides were delivered to a trap column (100 μm inner diameter × 2 cm, packed with 5 μm C18 resin, ReproSil-Pur AQ (Dr. Maisch, Ammerbuch, Germany) at a flow rate of 5 μl per min in 100 % buffer A (0.1 % formic acid in HPLC-grade water). After 10 min of loading and washing, peptides were transferred to an analytical column (75 μm × 40 cm C18 column, ReproSil-Pur AQ, 3 μm, Dr. Maisch) and separated using a 110-min gradient from 2 % to 35 % of buffer B (0.1 % formic acid in acetonitrile) at a 300 nl per min flow rate. Full-scan mass spectrometric spectra were acquired in the Orbitrap at mass resolution of 30,000. The five most intense precursors were selected for HCD fragmentation (isolation width, 2.0 Th) with a normalized collision energy of 40 % at an AGC target setting of 50,000. HCD spectra were acquired in the Orbitrap at a mass resolution of 7,500. Dynamic exclusion was enabled for a 10-s repeat duration and a 10-s exclusion duration with a repeat count of one. The MS results were based on three biological experiments including LB-standard, LB-nitrite and LB-small.
Raw mass spectrometric data files were converted into Mascot generic format files (MGF) using Mascot Distiller (2.4.2.0, Matrix Science). The MGF files were searched against the Escherichia coli O157:H7 EDL933 non-redundant NCBI database (version 03.05.2011) and the six-frame translated genome (NC_002655) using the Mascot search engine (2.3.1, Matrix Science). Mascot parameters were: an enabled decoy search using a randomized database; monoisotopic peptide mass (considering up to two 13C isotopes); trypsin/P as protease; a maximum of two missed cleavages; peptide charges +2 and +3; peptide tolerance ± 5 ppm.; MS/MS tolerance ± 0.005 Da; instrument type ESI-Trap; fixed modification: carbamidomethyl (cysteine) and variable modification: oxidation (methionine).
The results from Mascot were further processed with the software Scaffold [32] for statistical validation and better visualization (parameters used were: peptide probability ≥ 80.0 %, protein probability ≥ 99.0 %, minimum two peptides resulting in zero hits for decoys in peptide spectra or protein. The identified proteins and peptides were visualized and investigated using Artemis [30].
Computational biology
The search engine PlatProm [33] was used to find potential promoters nearby the candidate genes. PlatProm scores were calculated for each nucleotide in the genome to estimate the probability for being the starting point of transcription. Scores exceeding the background level by four standard deviations (SD; score ≥ 7.44) were considered as statistically significant (p < 0.00004). While most bacterial promoters are located within the 250 bp region upstream of the initiation codon, about 10 % of the transcription start sites are within a more upstream region of 250–650 bp from the start codon [34]. Therefore, we searched promoters within 650 bp upstream of the start codons of the ORFs. The position with the highest potentiality to initiate transcription within this range was taken.
Homologues protein and gene sequences were searched using blastp and tblastn, respectively [35]. PredictProtein [36, 37] was used to generate predictions of protein functional and structural features. In particular, the following tools were applied: PROFphd (secondary structure and solvent accessibility, [38]), PROFtmb (transmembrane strands, [39]), TMSEG (transmembrane helices) and COILS (coiled-coil regions, [40]), ScanProsite (functional motifs, [41]), HMMER (PFAM domains, [42]), SomeNA (protein, DNA and RNA binding sites, [37]), PSI-BLAST [43] and HHblits (homology to known proteins, [44]), SEG (low- and high-complexity regions, [45]), ConSurf (evolutionary conservation of amino acids, [46]), DISULFIND (disulfide bonds, [47]). For disordered region predictions, PROFbval [48], UCON [49] and METADISORDER [50] were used. Further, PROFtmb (bacterial transmembrane β-barrels, [51]), Metastudent (Gene Ontology terms, [52]), and LocTree3 (subcellular localization, [53]) were applied. SignalP4.1 was used for the prediction of signal peptides [54]. In all cases, default settings were used.
In order to check whether the functional and structural features of the 72 novel proteins resemble those of known annotated proteins, we assembled a positive set of “real proteins” by randomly choosing four length-matched annotated EHEC-proteins for each of the 72 novel proteins. Of 288 proteins, one was dropped later since it was duplicated in the genome. The negative comparison set was generated by shuffling each of the 287 annotated comparison proteins 100-times, i.e. generating 100 new random sequences with the same amino acid distribution as in the original sequence, but destroying any positional signal. PredictProtein was applied to protein sequences of both sets (i.e. “real” and “shuffled”) and the result was then provided to the Support Vector Machine (SVM, [55]) implementation of WEKA [56] and the Radial Basis Function [57] to discriminate automatically between proteins of both sets. The SVM was trained on features predicted by PredictProtein for 287 annotated (positive data) and 2870 shuffled (negative data) protein sequences. A similar negative set of shuffled proteins for the 72 novel proteins was generated in the same way as for the 287 annotated proteins. The trained SVM (“real” versus “shuffled”) was applied to classify each of the 72 novel and corresponding 7200 shuffled novel proteins. The total sets of 100 shuffled proteins for the 72 novel and the 287 annotated proteins were used to calculate error bars for the predicted protein features.
Repeat sequences of X002 were detected using REPFIND [58] and its RNA was folded with mfold [59], both used with default parameters.
Results
Ribosomal footprinting reveals 72 novel short protein-coding genes
We performed ribosomal footprinting which detects only RNA covered by ribosomes, i.e., mRNA. All intergenic, non-annotated ORFs of at least 153 bp (≥50 aa) were extracted from the translated-mRNA data set if a minimal threshold of 10 RPKM for the translatome was reached. This value is about 10-fold above background [9]. Each ORF of this subset was visually screened for its translational signal in the translatome to exclude false positives (e.g., translation of preceding or subsequent genes). This procedure yielded 72 previously un-annotated ORFs with an RPKM translatome between 13 and 2974. The mean for this value was 327 comparing to 404 of all annotated genes [not shown; 22]. Similarly, the ribosomal coverage value (RCV) was between 0.02 and 3.6 (Table 1) for the novel genes with an average of 0.9. The average RCV of annotated genes in this experiment was about 1.1 [not shown; 22]. Thus, the novel proteins are produced in lower abundance compared to annotated genes. In Fig. 1, the ribosomal footprinting pattern of four examples is shown in detail. The mRNA of the 72 genes was under translation to various degrees (Table 1) and, therefore, the genes received tentative gene names starting from X001. This labeling indicates that they are of unknown function, although their differential expression under diverse conditions was determined and for some a phenotype was found (see below). Most of the 72 new genes were short (≤315 bp, mean 210 bp), but three were longer (384, 465, and 804 bp).
Table 1.
Novel genes detected in EHEC
| Gene description | Ribosomal footprintse | MSh | PlatProm predictioni | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Namea | Classificationb | Startc | Stopc | Length [bp] | Origind | RPKM | Gene coveragef | Ribosomal coverage valueg (RCV) | LB | LB-Nit | LB-small | Upstream of start codon [bp] | Score |
| X001 | real | 217270 | 217488 | 219 | 1690 | 0.99 | 2.35 | −460 | 9.00 | ||||
| X002 yahH | real | 391261 | 391725 | 465 | 56 | 0.61 | 0.64 | −211(yahF) | 8.93 | ||||
| X003 | real | 570516 | 570710 | 195 | 102 | 0.72 | 0.69 | -- | -- | ||||
| X004 | real | 667557 | 667805 | 249 | 18 | 0.59 | 0.51 | 2 | 2 | −287 | 7.90 | ||
| X005 | real | 713269 | 713421 | 150 | 190 | 0.92 | 0.89 | −54(cstA) | 7.61 | ||||
| X006 | real | 713433 | 713630 | 198 | 166 | 0.86 | 0.77 | −54(cstA) | 7.61 | ||||
| X007* | 790488 | 790682 | 195 | 79 | 0.65 | 0.80 | −563 | 9.15 | |||||
| X008 | real | 902889 | 903083 | 195 | phage | 678 | 0.71 | 0.82 | −14 | 7.63 | |||
| X009* | real | 978607 | 978747 | 141 | 17 | 0.52 | 0.50 | −129 | 7.63 | ||||
| X010a | real | 1112292 | 1112471 | 180 | 35 | 0.75 | 0.71 | −297 | 8.28 | ||||
| X010b | 1508079 | 1507899 | duplicate of X010a | ||||||||||
| X011* | real | 1146872 | 1147027 | 156 | 13 | 0.53 | 0.38 | -- | -- | ||||
| X012 | real | 1152583 | 1152795 | 213 | 57 | 0.51 | 0.42 | −2 | 7.66 | ||||
| X013 | real | 1256680 | 1256967 | 288 | phage | 230 | 0.89 | 0.92 | 2 | −590 | 7.73 | ||
| X014a | real | 1267635 | 1267820 | 186 | phage | 552 | 0.66 | 0.26 | −67 | 8.07 | |||
| X014b | 2314896 | 2314711 | duplicate of X014a | ||||||||||
| X015 | real | 1334776 | 1334931 | 156 | phage | 35 | 0.84 | 0.32 | −70(trxB) | 7.84 | |||
| X016a | real | 1346825 | 1347184 | 360 | phage | 58 | 0.65 | 0.69 | −20 | 9.29 | |||
| X016b | 3000443 | 3000802 | duplicate of X016a | ||||||||||
| X017 | real | 1353605 | 1353772 | 168 | phage | 23 | 0.52 | 0.21 | −91 | 9.27 | |||
| X018 | real | 1411438 | 1411557 | 120 | 49 | 0.8 | 0.37 | −30 | 10.92 | ||||
| X019 | real | 1680779 | 1680967 | 189 | phage | 242 | 0.77 | 3.51 | −269 | 9.01 | |||
| X020 | real | 1772962 | 1773144 | 183 | 53 | 0.6 | 1.04 | −24(dadA) | 10.52 | ||||
| X021 | real | 1843458 | 1843622 | 165 | 1029 | 0.65 | 2.56 | −625 | 7.74 | ||||
| X022* | real | 1866296 | 1866505 | 210 | phage | 2169 | 0.82 | 0.73 | −6 | 7.63 | |||
| X023* | 1866493 | 1866648 | 156 | phage | 280 | 0.88 | 1.30 | −203 | 7.63 | ||||
| X024 | real | 1881598 | 1881819 | 222 | phage | 21 | 0.37 | 0.60 | 2 | 2 | 2 | −76 | 8.93 |
| X025a | 1389500 | 1389288 | duplicate of X015b | ||||||||||
| X025b | real | 1888594 | 1888806 | 213 | phage | 524 | 0,95 | 0,08 | −112 | 8,27 | |||
| X026 | real | 1905731 | 1905850 | 120 | phage | 622 | 0.7 | 1.01 | −77(Z2121) | 12.18 | |||
| X027 | real | 2038161 | 2038382 | 222 | 75 | 0.51 | 1.46 | −313 | 7.47 | ||||
| X028* | 2101101 | 2101247 | 147 | 131 | 0.61 | 1.70 | 0 | 8.00 | |||||
| X029 | real | 2109655 | 2109921 | 267 | 629 | 0.97 | 0.85 | -- | -- | ||||
| X030 | real | 2138823 | 2139137 | 315 | phage | 1520 | 0.98 | 1.35 | −53 | 16.96 | |||
| X031 | real | 2168349 | 2168567 | 219 | 77 | 0.66 | 0.60 | −110 | 8.48 | ||||
| X032a | 1269797 | 1269913 | duplicate of X032c | ||||||||||
| X032b | 1868589 | 1868705 | duplicate of X032c | ||||||||||
| X032c | real | 2312618 | 2312734 | 117 | 650 | 0.81 | 0.74 | −447 | 7.83 | ||||
| X033* | 2379507 | 2379659 | 153 | 348 | 0.87 | 1.50 | −77 | 12.26 | |||||
| X034 | real | 2430386 | 2430598 | 213 | 47 | 0.53 | 0.22 | −9 | 11.59 | ||||
| X035* | 2480019 | 2480177 | 159 | 25 | 0.52 | 0.20 | −63 | 10.51 | |||||
| X036 | real | 2584677 | 2584847 | 171 | 52 | 0.66 | 0.17 | −162 | 12.18 | ||||
| X037 | real | 2663871 | 2664122 | 252 | 14 | 0.53 | 0.58 | −243 | 12.65 | ||||
| X038 | real | 2670869 | 2671075 | 207 | phage | 1209 | 0.8 | 0.69 | −28 | 11.39 | |||
| X039 | real | 2742703 | 2742918 | 216 | 90 | 0.58 | 0.61 | −103 | 7.60 | ||||
| X040 | real | 2777135 | 2777347 | 213 | phage | 37 | 0,57 | 0,02 | -- | -- | |||
| X041 | real | 2779284 | 2779508 | 225 | phage | 57 | 0.73 | 1.32 | -- | -- | |||
| X042 | real | 2844454 | 2844606 | 153 | 768 | 0.84 | 0.83 | −295(X043) | 8.26 | ||||
| X043 | real | 2844640 | 2844804 | 165 | 212 | 0.92 | 0.44 | −295 | 8.26 | ||||
| X044 | real | 2844865 | 2845074 | 210 | 36 | 0.53 | 0.17 | −210 | 11.00 | ||||
| X045 | real | 2845149 | 2845358 | 210 | 163 | 0.9 | 0.16 | −23 | 9.54 | ||||
| X046* | 2845408 | 2845602 | 195 | 145 | 0.69 | 0.35 | −33 | 9.54 | |||||
| X047 | real | 2966787 | 2966987 | 201 | phage | 34 | 0.71 | 0.17 | −21 | 8.08 | |||
| X048 | real | 3003688 | 3003945 | 258 | phage | 40 | 0.65 | 1.96 | −353 | 8.18 | |||
| X049 | real | 3004951 | 3005067 | 117 | phage | 241 | 0.75 | 1.39 | 3 | 2 | −93 | 9.71 | |
| X050 | real | 3013440 | 3013694 | 255 | phage | 28 | 0.64 | 0.47 | −71(Z3371) | 8.46 | |||
| X051 | real | 3261588 | 3261758 | 171 | 89 | 0.86 | 0.35 | -- | -- | ||||
| X052* | 3271689 | 3271820 | 132 | 34 | 0.79 | 0.32 | −95 | 9.93 | |||||
| X053 suhB |
real | 3453780 | 3454583 | 804 | 41 | 0.53 | 0.20 | 9 | 13 | 2 | −36 | 9.48 | |
| X054* | 3894853 | 3894993 | 141 | 98 | 0.86 | 0.56 | −220 | 8.25 | |||||
| X055 | real | 3918141 | 3918344 | 204 | 47 | 0.56 | 0.31 | -- | -- | ||||
| X056 | real | 4207372 | 4207641 | 270 | 725 | 0.92 | 0.66 | −52 | 10.58 | ||||
| X057 | real | 4240665 | 4240883 | 219 | 2974 | 0.88 | 2.01 | −24 | 13.80 | ||||
| X058* | 4441485 | 4441643 | 159 | 359 | 0.98 | 0.64 | −569 | 9.75 | |||||
| X059 | real | 4449723 | 4449821 | 99 | 19 | 0.6 | 0.08 | −96 | 7.97 | ||||
| X060 | real | 4468299 | 4468592 | 294 | 639 | 0.84 | 2.99 | −253 | 9.57 | ||||
| X061 | real | 4585965 | 4586174 | 210 | 202 | 0.92 | 1.98 | 2 | 2 | −67 | 9.03 | ||
| X062 | real | 4654347 | 4654490 | 144 | phage | 29 | 0.73 | 0.89 | −393 | 8.17 | |||
| X063* | 4730352 | 4730537 | 186 | 15 | 0.51 | 0.95 | −533 | 11.48 | |||||
| X064 | real | 4793504 | 4793737 | 234 | 20 | 0.53 | 0.28 | -- | -- | ||||
| X065 | real | 4870817 | 4870978 | 162 | 38 | 0.74 | 1.28 | −90(pldA) | 8.1 | ||||
| X066* | 4873916 | 4874122 | 207 | 117 | 0.84 | 2.58 | −104 | 7.92 | |||||
| X067 | real | 4916583 | 4916756 | 174 | 162 | 0.84 | 0.64 | −22(yihI) | 11.84 | ||||
| X068* | 5077694 | 5077831 | 138 | 2040 | 0.97 | 0.55 | −368(nfi) | 7.61 | |||||
| X069 | real | 5369765 | 5369998 | 234 | 141 | 0.94 | 0.33 | −159(pepA) | 11.47 | ||||
| X070 | real | 5456776 | 5457042 | 267 | 53 | 0.52 | 3.58 | −163(yjiM) | 8.02 | ||||
| X071 | real | 5494158 | 5494394 | 237 | 45 | 0.57 | 2.82 | −27 | 8.35 | ||||
| X072 | real | 5515374 | 5515541 | 168 | 38 | 0.69 | 0.80 | −39(serB) | 7.9 | ||||
a The asterisk indicates genes not annotated in any other organism (blastp against GenBank, threshold E-value ≤10−10)
b Machine learning classification based on the set of annotated proteins (“real”) and their shuffled counterparts as training set
c The positions are given in relation to GenBank accession no. NC_002655, the original genome sequence of strain EDL933. Only very recently, the genome has been updated (GenBank accession no. CP008957)
d Genes originating from prophages are indicated
e The RPKM footprint and coverage of the actual ORF with footprints is given as average of two replicate experiments for bacteria grown in LB medium
f Fraction of the ORF covered with one or more footprint reads
g Ratio of RPKM footprints to RPKM transcriptome
h Indicated is the number of individual peptide spectra gained by mass spectrometry
i Putative promoters have been predicted using PlatProm. The position of the assumed transcription start site upstream of the start codon and the quality of the prediction (score) are given
Fig 1.
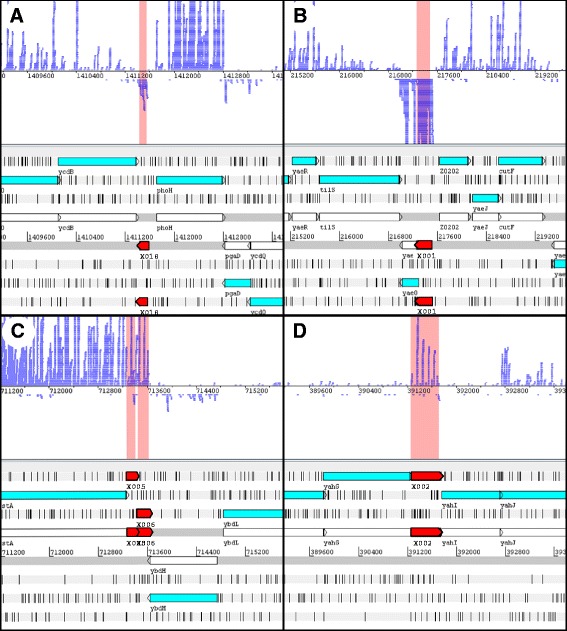
Four examples of new EHEC protein-coding ORFs (red arrows) discovered by ribosomal footprinting and visualized using Artemis [30]. Protein-coding ORFs are indicated by cyan arrows in the lower part of each panel. Blue lines in the upper part of each panel represent ribosomal footprint reads. a X018 is an example for a single (monocistronic) gene. b X001 is located in the upstream part of yaeO. These two genes might form a translationally coupled operon. c Two short genes, X005 and X006 are located downstream of cstA, maybe also translationally coupled. d X002 might be part of the operon yahDEFGIJ spanning from yahD to yahJ (only partly shown). The missing gene yahH had been annotated at first but was rejected later due to its structure (see Discussion and Fig 4)
Bioinformatics analysis of the proteins encoded in the novel genes
Suitable σ70-dependent promoters were predicted by PlatProm within the potential regulatory region of 50 candidate genes, while additional 14 genes are possibly transcribed as polycistronic units together with upstream genes (Table 1). Most novel genes, therefore, appeared to be driven by the housekeeping form of the RNA polymerase [34].
Protein sequences of the 72 new genes were submitted to PredictProtein [36], a powerful protein-analysis tool, which provides predictions of various aspects of protein structure and function (see Material & Methods and Additional file 1). The goal of this study was to compare the newly discovered proteins (“novel”) at a broader scale with gene products of annotated genes. Towards this end, a random choice of length-matching annotated proteins from EHEC was used as a control (“annotated”). To exclude excessive bias using randomly chosen annotated proteins, each novel protein of the 72 was length-matched with four annotated gene products.
The secondary structure prediction (helix, H; beta-sheet, E; loop, L; Fig. 2a) did not show any conspicuous difference between novel and annotated proteins. This was also true for the percentage of buried (b) versus exposed (e) residues (Fig. 2a). About 40 % of the proteins in both groups were predicted to contain transmembrane helices (mostly single-span membrane proteins, Fig. 2b). Furthermore, only one protein (≈1.4 %) from the set of 72 novel proteins had a predicted coiled-coil (of 14 residues), compared to 8 % of the annotated proteins (Fig. 2c, Additional file 1).
Fig 2.
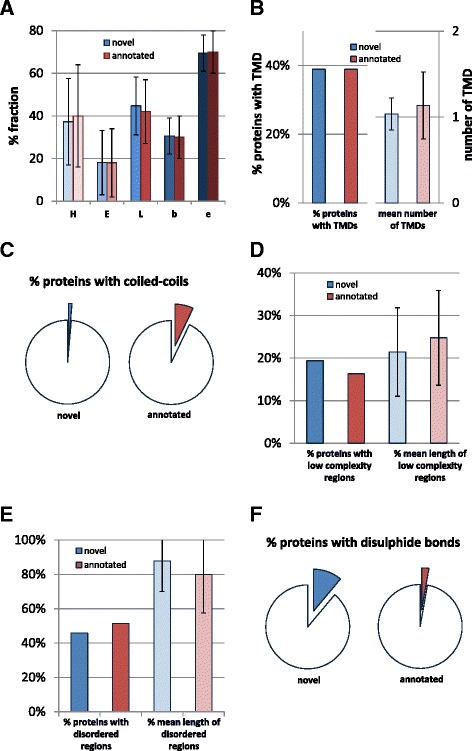
Graphical overview of PredictProtein values for the novel and length-matched annotated proteins. Error bars (if given) show the SD. a Shown is the predicted percentage of the protein length comprised of helices H, sheets E, and loops L. Furthermore, the percentage of buried and exposed amino acids is given (b and e). b On the left side, the fraction of proteins possessing at least one predicted transmembrane domain (TMD) is shown. On the right side, the mean number of TMDs per possessing proteins is shown. c The fraction of proteins having a coiled-coil prediction using a window of 14 amino acids is given. d The left bars show the fraction of proteins with a low-complexity region, the right bars give the mean length of this region compared to the overall length of the proteins for those possessing such a region. e The left bars show the fraction of proteins with a disordered region, the right bars give the mean length of this region compared to the overall length of the proteins for those possessing such a region. f The fraction of proteins having at least one Cys = Cys bond predicted
Low complexity segments circumscribe protein areas of “low information content” [45, 60]. Of the novel proteins, 19 % contained low complexity regions versus 16 % of the annotated. In the novel protein group, these regions tended to be marginally shorter (on average about 21 % of the protein length) compared to the annotated proteins (on average about 25 % of their length), but their distributions overlapped largely, thus, the difference was insignificant (Fig. 2d). Disordered regions were counted if MetaDisorder predicted intrinsically disordered stretches of 30 or more consecutive residues. About 45 % of the novel proteins contained such a disordered region, encompassing on average 88 % of the protein length. In the control set, 51 % of the proteins contained a disordered region, encompassing on average about 80 % of the protein length. Thus, slightly fewer of the novel proteins possessed a disordered region, but these fewer regions tended to be slightly longer than those in the annotated proteins (Fig. 2e). β-barrels are generally rare in proteins and none was detected in both groups (not shown).
Interestingly, about 11 % of the novel proteins were predicted to contain disulfide bond-forming cysteine residues compared to 3 % in the control set of annotated proteins (Fig. 2f). The higher number was not explained by an over-representation of cysteine (which was observed but only an over-representation by a factor of 1.5, i.e. maximally explaining 2.25 times more disulfide bridges, not 3.6 times more). Instead, the high number of disulfide bridges might suggest an abundance of secreted proteins. To test if the single-span membrane helices found above might be signal peptides, we examined those proteins which have one predicted transmembrane region using SignalP [54] to discriminate between true transmembrane domains and signal peptides. We found a lower percentage of the novel proteins to possess a signal peptide (22 %) compared to the annotated proteins (33 %; Additional file 2). However, using LocTree3 that combines homology-based inferences with de novo predictions of sub-cellular localization [53], we found that the novel proteins contain a larger fraction of proteins predicted to be secreted than the annotated set: over 75 % proteins in the novel set and only about 50 % in the annotated group were predicted as secreted (Fig. 3a). This fits to the above observation of an over-representation of disulfide bridges, typical for secreted proteins.
Fig 3.
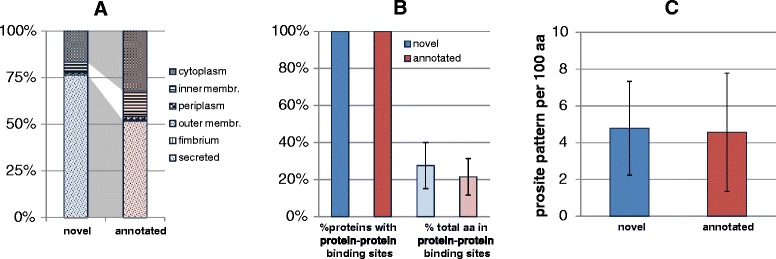
Graphical overview of the PredictProtein values for the novel and length-matched annotated proteins comparing localization, protein-protein binding sites, and PROSITE pattern. a Subcellular localization has been predicted using LocTree3 and is shown in per cent for the different compartments (membr., membrane). b The left bars show that all proteins have predicted protein-protein binding sites. The right bars show the percentage of the predicted number of amino acids involved in this type of interaction. c Given is the predicted number of PROSITE patterns per 100 aa
All proteins were predicted to contain protein-protein binding sites under consideration of proximal residues, varying between 1 to 54 residues per protein. The percentage of residues predicted to be involved in protein-protein binding was slightly larger for the novel proteins than for the annotated (28 % vs. 22 %; Fig. 3b). ScanProsite predicted functional domains and motifs [41]. Since they are of various length, we determined how many PROSITE patterns [61] exist per 100 residues. Interestingly, both the novel and annotated proteins have almost five of such patterns (Fig. 3c).
ConSurf estimates the evolutionary conservation of residues based on the phylogenetic relations between homologous sequences [62]. We counted all residues with a ConSurf value of 5 or higher. Interestingly, there was virtually no difference between both protein groups (not shown). For some of the novel proteins detected, Gene Ontology (GO) terms were predictable using Metastudent [52]. Predicted functions included stress response, protein binding, transcription regulation and metabolic processes for X001, X006, X030, X042, X043, X044, X047, X053, and X061 (Additional file 3).
Comparison of “real” versus “shuffled” protein sequences
To evaluate if real versus shuffled can be distinguished using computational predictions, we randomly shuffled each novel protein sequence as well as each annotated protein sequence from the comparison set 100-times. Structure and function features of the shuffled sequences were predicted with PredictProtein as before. For all features the mean ± 1 SD for all 100 shuffled sequences was compared to the value for the native protein sequences (if applicable; raw data in Additional file 4). Surprisingly, real proteins and their shuffled counterparts superficially showed similar distributions for most predictions (Additional file 5) and several of the real proteins had values within the mean ± 1 SD of their shuffled counterparts (Additional file 6). To gain further insight, we used machine learning to distinguish between 72 real and 7200 shuffled sequences. The machine was trained using the set of “real” proteins and their shuffled counterparts. Of 72 novel proteins, 61 (true positives; 85 %) were recognized as real, while of 7200 shuffled, only 8 (false positives; 1 %) were classified as such. Interestingly, all seven novel proteins with MS data (see below) were classified as “real” (Additional File 7).
Environmental regulation of transcription under eleven different growth conditions
To check for specific transcription of the newly discovered genes in comparison to the standard LB medium, we analyzed transcriptome data of the strain grown under a variety of different conditions [9]. Briefly, the bacteria were grown in LB medium at pH4, pH9, or at 15 °C; in LB with nitrite or trimethoprim-sulfamethoxazole; on the surface of LB-agar, in M9 minimal medium, in spinach leaf juice, on the surface of living radish sprouts, or in cattle feces. We found specific transcription profiles for each gene in comparison to standard LB, e.g., X071 was only induced in minimal medium, X059 only on radish sprouts, and X031 only in spinach leaf juice. X062 was up-regulated in minimal medium, LB with nitrite, and LB pH9; while X060 was induced in spinach leaf juice or at 15 °C. Cow dung, LB agar surface, and LB at pH4 did not show up regulation of any of the new genes, but rather a down regulation of several of them (Table 2, Additional file 8).
Table 2.
Transcriptome data of selected novel genes regulated under specific conditions given as fold-change compared to standard LBa. Data are taken from [9]
| Name b | Minimal medium | LB-Nit | pH9 | Radish sprouts | Spinach leaf juice | 15 °C | Amoeba | Antibiotics | Cow dung | Agar surface | pH4 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| X009* | u/c | n.r. | 9 | u/c | u/c | n.r. | 70 | u/c | u/c | n.r. | n.r. |
| X011* | 12 | u/c | 6 | 8 | 26 | 13 | 151 | n.r. | n.r. | 19 | 21 |
| X031 | u/c | u/c | u/c | u/c | 26 | u/c | u/c | u/c | u/c | u/c | −18 |
| X037 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | 213 | n.r. | n.r. | n.r. | n.r. |
| X048 | n.r. | - u/c | u/c | u/c | n.r. | u/c | n.r. | 48 | n.r. | n.r. | u/c |
| X052* | n.r. | −6 | −5 | n.r. | u/c | u/c | 12 | n.r. | n.r. | n.r. | −5 |
| X060 | u/c | 7 | u/c | 5 | 10 | 18 | u/c | −17 | u/c | u/c | u/c |
| X062 | 25 | 14 | 12 | n.r. | u/c | 7 | 23 | n.r. | n.r. | n.r. | n.r. |
| X070 | n.r. | n.r. | u/c | n.r. | n.r. | u/c | 25 | n.r. | n.r. | n.r. | n.r. |
| X071 | 122 | 14 | 9 | 5 | u/c | u/c | n.r. | n.r. | 5 | n.r. | n.r. |
apositive values, up regulated; negative values, down regulated; n.r., no reads under this condition; u/c, unchanged (threshold ≥5-fold regulation)
bThe asterisk indicates genes not annotated in any other organism (see Table 1)
We further performed a transcriptome analysis of EHEC grown in the presence of amoeba (Acanthamoeba castellanii; data not shown). This experiment yielded not enough sequencing reads for a proper global comparison to the other conditions, but still allowed to deduce specific up regulation of transcription: > 10-fold compared to LB for X009, X011, X037, X052, X062, and X070. These results show that gene expression of the novel genes changes in a diverse array of conditions which might indicate functionality.
MS data confirm expression of seven novel genes
When evaluating MS data of cells grown under the same conditions used for transcriptome and translatome analyses, as well as evaluating a six-frame translation of the EHEC-genome, we observed peptide signals belonging to seven of the new ORFs (Table 1).
An REP-element containing ORF, X002, is translated
While screening the genome for the novel genes, X002 piqued our specific interest (Fig. 1d), since it falls within a gap located between yahG and yahI. Presumably, this gene had been annotated as yahH but was removed later. X002 contains a sequence, which is a REP element, belonging to the group of bacterial interspersed mosaic elements (BIME). The amino acid sequence of X002 matched REP23 from E. coli K-12 in a blastp search [35] with an E-value of 3 × 10−174 [52, 63, 64]. The gene locus of X002 contains a long ORF (465 nt) and its transcript was well covered by ribosomal footprints (Fig. 1d). Using REPFIND [58], we discovered a block-like structure of five sequence repeats within the ORF. One of these repeated elements was predicted to fold in a relatively stable stem-loop structure according to mfold [59; Fig. 4]. The highly repetitive nature of this ORF was also visible in the footprint signal, which appeared to be very regular (Fig. 1d). The translation of such REP-elements is somewhat unexpected.
Fig 4.
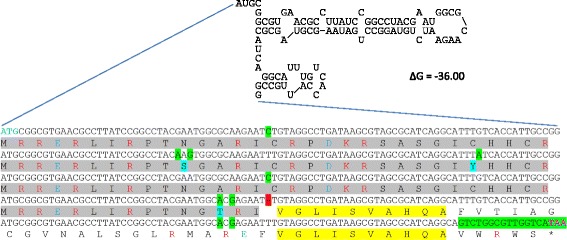
Repeat structure of the REP23 containing gene yahH (the same as X002) and its protein YahH [58]. The upper part shows one repeat block folded as mRNA [59]. The DNA sequence (lower part) basically consists of five of such repeated blocks, with only minor differences (when compared to each other – single nt differences are in green) and a short unique sequence at the 3’-end (green stretch). When comparing the fourth block to the other, a base appears to be missing (red marked gap) causing a change in the reading frame visible in the protein structure. Thus, the protein contains three large repeats and a fourth truncated one (grey blocks, few differences in aa indicated in blue). Downstream of the “frame shift” mutation, a different structure of two blocks is found (yellow). The protein contains many charged amino acids, either positive (RK, red print) or negative (DE, blue print)
Phenotypes of novel genes in cattle
Transposon (Tn) or signature-tagged mutagenesis (STM) studies of EHEC [65–74] were evaluated to find possible insertion mutants of the novel genes conferring a phenotype. Only few studies contained phenotypic data of intergenic insertions. We used a stringent threshold of 5-fold difference between the wild type and the transposon-mutated strain. Nevertheless, three novel genes could be linked to a phenotype in cattle: an existing EHEC-transposon mutant library [74] had been re-screened for mutants changed at least 5-fold in their ability to colonize the ruminant intestine [72]. These novel ORFs (i.e., X033, X036, and X045) were found to either have a direct transposon hit or a hit shortly upstream of its frame, hence, supposedly in their promoter region (Table 3). Transposon hits of genes X036 and X045 had already been detected in the original STM study looking only for a decrease in cattle colonization [74]. The finding that some of the novel genes display a phenotype in cattle is interesting, as it indicates our fragmentary knowledge about EHEC-host relationships and highlights the importance of short genes in general. However, future research must show if the changed colonization of the transposon-mutants is indeed caused by the novel genes hit or are only a byproduct of the transposon inserted in these positions.
Table 3.
Phenotype in calves of transposon hits in or nearby the novel genes. Threshold is defined as a 5-fold or higher regulation. Negative values indicate down-regulation. Data are taken from [72]
| Namea | Position of Tn insertion | Direct hit [H] or bases upstream [b] | Fold-change output versus input |
|---|---|---|---|
| X033* | 2379421 | 86 | −33 |
| X036 | 2584780 | H | −13 |
| X045 | 2845234 | H | −50 |
aThe asterisk indicates genes not annotated in any other organism (see Table 1)
Taxonomic distribution of the novel genes
For all 72 genes, homologous genome regions could be detected in E. coli O157:H7 Sakai using tblastn [35]. Twenty-eight of the 72 genes had been annotated in Sakai as protein-coding ORFs. Fifty-seven of the 72 genes were found to be annotated within the enterobacteriaceae using blastp (E-value threshold ≤10−10, E-value median for all top hits of the novel proteins was 5.5 × 10−34). Fifteen out of the 72 genes were unique which means that these had not been annotated as protein-coding genes before (marked by an asterisk in Table 1). Significantly, all 72 genes had no blastp (E-value ≤10-10) hit beyond the enterobacteriaceae in GenBank [75]. Thus, these genes appear to be taxonomically restricted to this single family of the order enterobacteriales, sometimes even to the species E. coli. Further, for 40 out of 72 proteins, the family members found using blastp in GenBank (40 genes) were exclusively labeled as “hypothetical” or “conserved hypothetical”. We substantiated this trend using various thresholds for defining families with PSI-BLAST [43] and HHblits [44]. For instance, at PSI-BLAST E-values ≤10−3 and HHblits E-values ≤10-10, about 40 % of the 72 novel genes were found to be novel (i.e., orphans), while the corresponding fraction for the control set was at least 20-times smaller (<2 %). Another 10–20 % had families smaller than the corresponding control set. However, the remaining 40–50 % of the 72 had families of similar sizes as the proteins in the control set (Additional file 9). Sixteen of 72 proteins had, at least to some extent, a functional annotation and 21 were of prophage origin (see Additional file 8).
Discussion
The short hypothetical genes are no annotation artifacts
Although E. coli is probably the bacterial species researched best, many environmental growth conditions have never been evaluated and many cannot be evaluated easily in the laboratory. This fact may partly explain why a sizable fraction of genes in any bacterium is still of hypothetical status. “Our lack of fundamental knowledge about the function of so many of the building blocks of cells“, as stated by Roberts [76], hampers downstream research and other –omics efforts [77], since only what is known will be examined. Unfortunately, the smaller the protein-coding ORFs, the more likely it is that such genes are either ignored based on the assumption that short ORFs are highly unlikely to be functional [19, 78], not predicted due to the bias towards longer ORFs [79] or evade detection due to technical difficulties [20, 80]. In addition, many of the novel proteins are supposedly secreted according to our LocTree3 data and, thus, may be missed by the proteomics approach.
Ribosomal footprinting provides a high-throughput method, which indicates that also short genes encode proteins, as a footprint fragment (i.e., a nucleic acid) can be detected much easier than a short protein [81] and independent of its final destination within or outside the cell. The mRNA of the novel genes described here was clearly covered by ribosomes, thus indicating translation [82, 83]. This hypothesis was corroborated by bioinformatics analysis: only few of the general protein structure and function traits were predicted to differ between the novel 72 proteins described here and a set of annotated proteins with similar length distributions. The exceptions were disulfide bonds and coiled-coils, but these two parameters mattered only for a small subset of the proteins. The largest difference was obtained in the predicted sub-cellular protein location using LocTree3. Interestingly, when examining the “dark proteome” – that is proteins never observed by experimental structure determination and, therefore, inaccessible to homology modeling, similar trends were observed [84]. For instance, these proteins were short, often secreted, and had a higher amount of disulfide bonds [84]. However, in most parameters investigated, differences appeared to be minor. This was corroborated by a machine learning approach, able to classify 61 of the 72 novel proteins to resemble known annotated proteins but not random sequences. These results suggest that the protein sequences encoded by the novel genes described here show the same structural features and, thus, functional traits as well-known annotated proteins [85], validating the idea that such sequences form the raw material for evolutionary optimization of novel proteins [86].
We found that several of the new genes are specifically induced only under one or a few growth conditions hardly ever tested in the lab. According to Hemm et al. [4] and Hobbs et al. [87], short proteins seem to be important for the stress response of E. coli. Indeed, the novel genes discovered in this study were found to be induced under specific and sometimes adverse culture conditions such as minimal medium, pH9, radish sprouts, spinach leaf juice, antibiotics, cow dung, and the presence of amoeba (Table 2, Additional file 8). Furthermore, transposon-mutants derived in a previous study conferred a phenotype for three novel genes detected. The mutants had a decreased ability to colonize the cattle intestine [72]. The fact that genes detected in our study were connected to a phenotype in a cattle study shows that not only well-known genes of “standard properties” play an important role in the bacterial life cycle, but also such short novel proteins, maybe as a toxin [88]. Indeed, we predicted an unusual abundance of secretion in the set of 72 novel proteins (ca. 75 % secreted proteins, Fig. 3a).
Seven out of 72 genes were validated by proteome analysis. However, the probability to detect a protein via MS decreases with the size of the protein. Peptide fragments between 7 and 13 residues have the highest probability of detection, whereas fragments below 5 or above 40 residues are missed [89]. Short proteins are less likely to be detected by MS due to possibly missing tryptic cleavage sites. The tryptic cleavage sites typically occur C-terminally of an arginine (R) or lysine (K). If none of these amino acids is present, no fragmentation occurs and the peptides are too long for successful detection [90]. No R or K are found in five of the novel proteins, but none of the annotated. Thus, not surprising, most MS spectra originated from the largest of the 72 proteins. In addition, the probability to detect a protein is strongly dependent on its abundance [91], but the novel proteins are less abundant proteins (lower RPKM and RCV values compared to the annotated).
Based on the multi-omics approach by combining data of transcriptomes, translatomes, mass spectrometry, bioinformatics analyses and phenotype searches, we suggest that these 72 short genes are an overlooked fraction of genes in the EHEC genome, which should be added to the genetic map of this bacterium. We showed that even densely covered genomes like those of bacteria (in which about 90 % of the genome is covered by annotated protein-coding genes) still provide room for new protein-coding genes. This finding also adds to the growing evidence that even short hypothetical genes of bacterial genomes are no artifacts [19]. However, there might be coding sequences for even shorter polypeptides [our unpublished data; 78]. Their detection is at the resolution limits of most experimental techniques and only targeted multi-omics approaches may resolve the problem in the future.
yahH – a gene locus with a potential triple function?
The novel gene X002 turned out to be a REP-element belonging to the bacterial interspaced mosaic elements. These elements play several roles based on their repetitive DNA sequence [64]. They are believed to be topological insulators for transcription-induced positive supercoiling and may bind proteins such as IHF, PolI and DNA gyrase to structure the DNA [63, 64]. Further, such elements can initiate a Rho-dependent transcription attenuation [92] and may stabilize RNA by inhibiting its degradation in vivo [93, 94]. Therefore, Gonnet et al. [95] suggested that it is highly unlikely that the REP-element yahH is translated, and, consequently, this gene was removed from the annotation [52] based on its unusual gene structure (Fig. 4). Interestingly, the Rho-dependent transcription attenuation of REP-elements is abolished, if the repeat element is translated [92]. Using our assay, we could show that X002, which is equivalent to yahH, is not only transcribed, but quite probably also translated. The resulting protein has a high number of charged amino acids and is of unknown function. If true, this gene locus would carry a triple function, i.e., as regulatory DNA element, as a regulatory RNA element, and in addition, as a protein.
The novel genes evolved recently
All novel genes described here are restricted to the enterobacteriaceae or even to taxa closer related, and, therefore, are taxonomically restricted genes (TRGs [96]). They appear only in higher phylostrata (i.e., closer relatives; [97]), which is evidence for their relatively recent origin [98]. This hypothesis is supported by several findings: the novel genes described here apparently use more often one of the rare start codons (e.g., GTG, TTG), which are translationally less optimal [99] and, therefore, may not yet be evolutionarily optimized. In addition, the novel genes are clearly shorter than the average E. coli genes and some are not classified as “real” by our bioinformatics approach. All of the above corroborates the findings of Tautz & Domazet-Lošo [15], who also observed that TRGs are generally shorter than conserved genes and confer weak phenotypes. Most of the newly discovered proteins are located directly up- or downstream of annotated, “established” genes (compare to Fig. 1b-d), perhaps contributing a (minor?) constituent to already known operons [19]. This particular arrangement may indicate a potential evolutionary mechanism to sample genetic regions, which may form a coding reserve, i.e., short ORFs are tested for their usefulness for the cell. A minor upstream promoter activity (or a weak terminator site) would permit the formation of polycistronic RNA carrying additional ORFs upstream (or downstream, respectively). Thus, ribosomes may bind “too early” or ribosomes which are already bound to the mRNA while translating an established upstream gene will initiate and translate a downstream short ORF with a higher probability, respectively. If the resulting protein provides a significant fitness gain, it may promote strain survival and subsequent improvements by classical Darwinian evolution.
Conclusion
It has been suggested by Carvunis et al. [86] that genes form de novo from non-coding DNA in yeast. Although prokaryotes possess much less non-coding intergenic DNA due to their dense gene content, such a mechanism as detailed above might be active in bacteria as well. However, the DNA sequence features which would allow for the formation of a protein sequence functional ab initio upon accidental expression of an intergenic, non-protein coding DNA sequence (i.e., being of supposedly random amino acid content) remain unknown [100].
Availability of supporting data
All additional files supporting the results of this article are available in the repository labarchives.com (http://www.labarchives.com/) using the link http://dx.doi.org/10.6070/H4610XB9. All the supporting data are included as Additional files.
Acknowledgment
This work was funded by the Deutsche Forschungsgemeinschaft DFG (SCHE316/3-1, KE740/13-1).
Abbreviations
- AA
amino acids
- AGC
automatic gain control
- b
buried or base
- BLAST
basic local alignment search tool
- BIME
bacterial interspersed mosaic elements
- BP
base pairs
- BSA
bovine serum albumin
- e
exposed
- E
beta-sheet
- EHEC
enterohemorrhagic E. coli
- GO
gene ontology
- H
helix
- HCD
Higher-energy collisional dissociation
- HPLC
high pressure liquid chromatography
- kDa
kilo Dalton
- L
loop
- LB
Luria-Bertani medium
- membr
membrane
- MGF
Mascot generic files
- MS
mass spectrometry
- ncRNA
non-coding RNA
- NGS
next generation sequencing
- n.r.
no reads
- ORF
open reading frame
- RCV
ribosomal coverage value
- REP
repetitive extragenic palindrome
- RNAseq
mRNA sequencing
- RPKM
reads per kilobase per million mapped reads
- RPM
revolutions per minute
- SD
standard deviation
- SDS
sodium dodecyl sulfate
- STM
signature-tagged mutagenesis
- SVM
support vector machine
- TMD
transmembrane domain
- TN
transposon
- TRG
taxonomically restricted genes
- u/c
unchanged
Additional files
Summarized values of PredictProtein for the novel proteins and length-matched annotated proteins. (XLSX 81 kb)
SignalP prediction of signal peptides for proteins with one transmembrane domain. (PDF 74 kb)
Gene Ontology terms predicted using Metastudent for the novel genes. (XLSX 12 kb)
Raw data of PredictProtein for all natural sequences (72 novel and 288 annotated) and their 100-times shuffled counterparts. (XLSX 16697 kb)
Prediction values for the novel and annotated proteins compared to their shuffled counterparts in dependence of the protein length. (PDF 443 kb)
Prediction values for the novel proteins compared to their shuffled counterparts for each protein individually. (PDF 144 kb)
Overview of the machine learning results. (XLSX 10 kb)
Expanded data set for all novel genes. (XLSX 56 kb)
Cumulative distributions of PSI-Blast and HHblits family sizes. (PDF 8541 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KN conceived and supervised the study, and wrote the manuscript with the help of all other authors. RL conducted transcriptome and translatome experiments, LF analyzed the data with help of SSi. Proteome data were conducted by HM and BK, promoter analysis by ONO. AS, TG, and BR provided the protein predictions. SSch helped to conceive the study which was critically attended by DK and SSch. All authors have read and approved the manuscript.
Contributor Information
Klaus Neuhaus, Email: neuhaus@wzw.tum.de.
Richard Landstorfer, Email: r.landstorfer@gmx.de.
Lea Fellner, Email: fellnerlea@hotmail.com.
Svenja Simon, Email: simon@dbvis.inf.uni-konstanz.de.
Andrea Schafferhans, Email: andrea.schafferhans@rostlab.org.
Tatyana Goldberg, Email: goldberg@rostlab.org.
Harald Marx, h4r4ld.marx@googlemail.com.
Olga N. Ozoline, Email: ozoline@rambler.ru
Burkhard Rost, Email: rost@rostlab.org.
Bernhard Kuster, Email: kuster@wzw.tum.de.
Daniel A. Keim, Email: keim@informatik.uni-konstanz.de
Siegfried Scherer, Email: siegfried.scherer@wzw.tum.de.
References
- 1.Mayer CL, Leibowitz CS, Kurosawa S, Stearns-Kurosawa DJ. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins. 2012;4(11):1261–87. doi: 10.3390/toxins4111261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409(6819):529–33. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 3.Mutz K-O, Heilkenbrinker A, Lönne M, Walter J-G, Stahl F. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol. 2013;24(1):22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Hemm MR, Paul BJ, Miranda-Rios J, Zhang A, Soltanzad N, Storz G. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J Bacteriol. 2010;192(1):46–58. doi: 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol. 2008;70(6):1487–501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomassen GO, Weel-Sneve R, Rowe AD, Booth JA, Lindvall JM, Lagesen K, et al. Tiling array analysis of UV treated Escherichia coli predicts novel differentially expressed small peptides. PLoS One. 2010;5(12):e15356. doi: 10.1371/journal.pone.0015356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33(9):981–93. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 2014;15(3):205–13. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 9.Landstorfer R, Simon S, Schober S, Keim D, Scherer S, Neuhaus K. Comparison of strand-specific transcriptomes of enterohemorrhagic Escherichia coli O157:H7 EDL933 (EHEC) under eleven different environmental conditions including radish sprouts and cattle feces. BMC Genomics. 2014;15:353. doi: 10.1186/1471-2164-15-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuersten S, Radek A, Vogel C, Penalva LO. Translation regulation gets its ‘omics’ moment. Wiley Interdiscip Rev: RNA. 2013;4(6):617–30. doi: 10.1002/wrna.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 12.Mallick P, Kuster B. Proteomics: a pragmatic perspective. Nat Biotechnol. 2010;28(7):695–709. doi: 10.1038/nbt.1658. [DOI] [PubMed] [Google Scholar]
- 13.Hu P, Janga SC, Babu M, Diaz-Mejia JJ, Butland G, Yang W, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7(4):e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias DA, Mukhopadhyay A, Joachimiak MP, Drury EC, Redding AM, Yen HC, et al. Expression profiling of hypothetical genes in Desulfovibrio vulgaris leads to improved functional annotation. Nucleic Acids Res. 2009;37(9):2926–39. doi: 10.1093/nar/gkp164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tautz D, Domazet-Loso T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12(10):692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 16.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu JF, Xiao K, Jiang DK, Guo J, Wang JH, Sun X. An integrative method for identifying the over-annotated protein-coding genes in microbial genomes. DNA Res. 2011;18(6):435–49. doi: 10.1093/dnares/dsr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goffeau A. Four years of post-genomic life with 6,000 yeast genes. FEBS Lett. 2000;480(1):37–41. doi: 10.1016/S0014-5793(00)01775-0. [DOI] [PubMed] [Google Scholar]
- 19.Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annu Rev Biochem. 2014;83:753–77. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramamurthi KS, Storz G. The small protein floodgates are opening; now the functional analysis begins. BMC Biol. 2014;12:96. doi: 10.1186/s12915-014-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landstorfer RB: Comparative transcriptomics and translatomics to identify novel overlapping genes, active hypothetical genes, and ncRNAs in Escherichia coli O157:H7 EDL933. München: Technische Universität München; 2014.
- 23.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J Biol Chem. 2004;279(15):15368–75. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 24.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010; Chapter 19: Unit 19 10 11–21. [DOI] [PMC free article] [PubMed]
- 26.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon S, Oelke D, Landstorfer R, Neuhaus K, Keim D. Visual Analysis of Next-Generation Sequencing Data to Detect Overlapping Genes. IEEE Symp Biol Data Vis. 2011;1:47–54. [Google Scholar]
- 29.Carver T, Bohme U, Otto TD, Parkhill J, Berriman M. BamView: viewing mapped read alignment data in the context of the reference sequence. Bioinformatics. 2010;26(5):676–7. doi: 10.1093/bioinformatics/btq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–5. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 31.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 32.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10(6):1265–9. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 33.Ozoline ON, Deev AA. Predicting antisense RNAs in the genomes of Escherichia coli and Salmonella typhimurium using promoter-search algorithm PlatProm. J Bioinform Comput Biol. 2006;4(2):443–54. doi: 10.1142/S0219720006001916. [DOI] [PubMed] [Google Scholar]
- 34.Shavkunov KS, Masulis IS, Tutukina MN, Deev AA, Ozoline ON. Gains and unexpected lessons from genome-scale promoter mapping. Nucleic Acids Res. 2009;37(15):4919–31. doi: 10.1093/nar/gkp490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Rost B, Yachdav G, Liu J. The predictprotein server. Nucleic Acids Res. 2004;32(suppl 2):W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, et al. PredictProtein--an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014;42(Web Server issue):W337–43. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins: Struct Funct Bioinformatics. 1994;19(1):55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 39.Bigelow HR, Petrey DS, Liu J, Przybylski D, Rost B. Predicting transmembrane beta-barrels in proteomes. Nucleic Acids Res. 2004;32(8):2566–77. doi: 10.1093/nar/gkh580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 41.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34(Web Server issue):W362–5. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43(W1):W30–8. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2012;9(2):173–5. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 45.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–71. doi: 10.1016/S0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 46.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19(1):163–4. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 47.Ceroni A, Passerini A, Vullo A, Frasconi P. DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 2006;34(suppl 2):W177–81. doi: 10.1093/nar/gkl266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlessinger A, Yachdav G, Rost B. PROFbval: predict flexible and rigid residues in proteins. Bioinformatics. 2006;22(7):891–3. doi: 10.1093/bioinformatics/btl032. [DOI] [PubMed] [Google Scholar]
- 49.Schlessinger A, Punta M, Rost B. Natively unstructured regions in proteins identified from contact predictions. Bioinformatics. 2007;23(18):2376–84. doi: 10.1093/bioinformatics/btm349. [DOI] [PubMed] [Google Scholar]
- 50.Schlessinger A, Punta M, Yachdav G, Kajan L, Rost B. Improved disorder prediction by combination of orthogonal approaches. PLoS One. 2009;4(2):e4433. doi: 10.1371/journal.pone.0004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bigelow H, Rost B. PROFtmb: a web server for predicting bacterial transmembrane beta barrel proteins. Nucleic Acids Res. 2006;34(suppl 2):W186–8. doi: 10.1093/nar/gkl262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2012;41(Database issue):D613–24. doi: 10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldberg T, Hecht M, Hamp T, Karl T, Yachdav G, Ahmed N, et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014;42(Web Server issue):W350–5. doi: 10.1093/nar/gku396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 55.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–97. [Google Scholar]
- 56.Frank E, Hall M, Trigg L, Holmes G, Witten IH. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20(15):2479–81. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 57.Buhmann MD. Radial basis functions: theory and implementations, vol. 12: Cambridge university press; 2003.
- 58.Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol. 2002;12(20):1756–61. doi: 10.1016/S0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 59.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coletta A, Pinney JW, Solis DY, Marsh J, Pettifer SR, Attwood TK. Low-complexity regions within protein sequences have position-dependent roles. BMC Syst Biol. 2010;4:43. doi: 10.1186/1752-0509-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41(Database issue):D344–7. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Web Server issue):W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee N, Chidambarathanu N, Michael D, Balakrishnan N, Sekar K. An algorithm to find all identical internal sequence repeats. Curr Sci. 2008;95(2):188–95. [Google Scholar]
- 64.Ton-Hoang B, Siguier P, Quentin Y, Onillon S, Marty B, Fichant G, et al. Structuring the bacterial genome: Y1-transposases associated with REP-BIME sequences. Nucleic Acids Res. 2012;40(8):3596–609. doi: 10.1093/nar/gkr1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bai J, McAteer SP, Paxton E, Mahajan A, Gally DL, Tree JJ. Screening of an E. coli O157:H7 bacterial artificial chromosome library by comparative genomic hybridization to identify genomic regions contributing to growth in bovine gastrointestinal mucus and epithelial cell colonization. Front Microbiol. 2011;2:168. doi: 10.3389/fmicb.2011.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A. 2004;101(10):3597–602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamner S, McInnerney K, Williamson K, Franklin MJ, Ford TE. Bile salts affect expression of Escherichia coli O157:H7 genes for virulence and iron acquisition, and promote growth under iron limiting conditions. PLoS One. 2013;8(9):e74647. doi: 10.1371/journal.pone.0074647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lio JC, Syu WJ. Identification of a negative regulator for the pathogenicity island of enterohemorrhagic Escherichia coli O157:H7. J Biomed Sci. 2004;11(6):855–63. doi: 10.1007/BF02254371. [DOI] [PubMed] [Google Scholar]
- 69.Puttamreddy S, Cornick NA, Minion FC. Genome-wide transposon mutagenesis reveals a role for pO157 genes in biofilm development in Escherichia coli O157:H7 EDL933. Infect Immun. 2010;78(6):2377–84. doi: 10.1128/IAI.00156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tatsuno I, Kimura H, Okutani A, Kanamaru K, Abe H, Nagai S, et al. Isolation and characterization of mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect Immun. 2000;68(10):5943–52. doi: 10.1128/IAI.68.10.5943-5952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torres AG, Kaper JB. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect Immun. 2003;71(9):4985–95. doi: 10.1128/IAI.71.9.4985-4995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eckert SE, Dziva F, Chaudhuri RR, Langridge GC, Turner DJ, Pickard DJ, et al. Retrospective application of transposon-directed insertion site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J Bacteriol. 2011;193(7):1771–6. doi: 10.1128/JB.01292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Diemen PM, Dziva F, Stevens MP, Wallis TS. Identification of enterohemorrhagic Escherichia coli O26:H- genes required for intestinal colonization in calves. Infect Immun. 2005;73(3):1735–43. doi: 10.1128/IAI.73.3.1735-1743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dziva F, van Diemen PM, Stevens MP, Smith AJ, Wallis TS. Identification of Escherichia coli O157: H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology. 2004;150(Pt 11):3631–45. [DOI] [PubMed]
- 75.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37(Database issue):26–31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roberts RJ. Identifying protein function--a call for community action. PLoS Biol. 2004;2(3):E42. doi: 10.1371/journal.pbio.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frishman D. Protein annotation at genomic scale: the current status. Chem Rev. 2007;107(8):3448–66. doi: 10.1021/cr068303k. [DOI] [PubMed] [Google Scholar]
- 78.Chu Q, Ma J, Saghatelian A. Identification and characterization of sORF-encoded polypeptides. Crit Rev Biochem Mol Biol. 2015;50(2):134–41. doi: 10.3109/10409238.2015.1016215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mathé C, Sagot MF, Schiex T, Rouzé P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 2002;30(19):4103–17. doi: 10.1093/nar/gkf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kemp G, Cymer F. Small membrane proteins–elucidating the function of the needle in the haystack. Biol Chem. 2014;395(12):1365–77. doi: 10.1515/hsz-2014-0213. [DOI] [PubMed] [Google Scholar]
- 81.Smith JE, Alvarez-Dominguez JR, Kline N, Huynh NJ, Geisler S, Hu W, et al. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 2014;7(6):1858–66. doi: 10.1016/j.celrep.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jackson R, Standart N. The awesome power of ribosome profiling. RNA. 2015;21(4):652–4. doi: 10.1261/rna.049908.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakahigashi K, Takai Y, Shiwa Y, Wada M, Honma M, Yoshikawa H, et al. Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC Genomics. 2014;15:1115. doi: 10.1186/1471-2164-15-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perdigão N, Heinrich J, Stolte C, Sabir KS, Buckley MJ, Tabor B, et al. Unexpected features of the dark proteome. Proc Natl Acad Sci. 2015;112(52):15898–903. doi: 10.1073/pnas.1508380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lobb B, Kurtz DA, Moreno-Hagelsieb G, Doxey AC. Remote homology and the functions of metagenomic dark matter. Front Genet. 2015;6:234. doi: 10.3389/fgene.2015.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, et al. Proto-genes and de novo gene birth. Nature. 2012;487(7407):370–4. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hobbs EC, Astarita JL, Storz G. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J Bacteriol. 2010;192(1):59–67. doi: 10.1128/JB.00873-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev. 2008;72(4):579–89. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J Proteome Res. 2010;9(3):1323–9. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olsen JV, Ong S-E, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3(6):608–14. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 91.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–73. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 92.Espeli O, Moulin L, Boccard F. Transcription attenuation associated with bacterial repetitive extragenic BIME elements. J Mol Biol. 2001;314(3):375–86. doi: 10.1006/jmbi.2001.5150. [DOI] [PubMed] [Google Scholar]
- 93.Bachellier S, Clement JM, Hofnung M. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res Microbiol. 1999;150(9–10):627–39. doi: 10.1016/S0923-2508(99)00128-X. [DOI] [PubMed] [Google Scholar]
- 94.Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol. 2004;51(3):777–90. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- 95.Gonnet P, Rudd KE, Lisacek F. Fine-tuning the prediction of sequences cleaved by signal peptidase II: a curated set of proven and predicted lipoproteins of Escherichia coli K-12. Proteomics. 2004;4(6):1597–613. doi: 10.1002/pmic.200300749. [DOI] [PubMed] [Google Scholar]
- 96.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25(9):404–13. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 97.Domazet-Lošo T, Brajković J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23(11):533–9. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 98.Tautz D. The discovery of de novo gene evolution. Perspect Biol Med. 2014;57(1):149–61. doi: 10.1353/pbm.2014.0006. [DOI] [PubMed] [Google Scholar]
- 99.Nie L, Wu G, Zhang W. Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics. 2006;174(4):2229–43. doi: 10.1534/genetics.106.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leisola M, Turunen O. Protein engineering: opportunities and challenges. Appl Microbiol Biotechnol. 2007;75(6):1225–32. doi: 10.1007/s00253-007-0964-2. [DOI] [PubMed] [Google Scholar]


