Abstract
Background
DNA methylation at a gene promoter region has the potential to regulate gene transcription. Patterns of methylation over multiple CpG sites in a region are often complex and cell type specific, with the region showing multiple allelic patterns in a sample. This complexity is commonly obscured when DNA methylation data is summarised as an average percentage value for each CpG site (or aggregated across CpG sites). True representation of methylation patterns can only be fully characterised by clonal analysis. Deep sequencing provides the ability to investigate clonal DNA methylation patterns in unprecedented detail and scale, enabling the proper characterisation of the heterogeneity of methylation patterns. However, the sheer amount and complexity of sequencing data requires new synoptic approaches to visualise the distribution of allelic patterns.
Results
We have developed a new analysis and visualisation software tool “Methpat”, that extracts and displays clonal DNA methylation patterns from massively parallel sequencing data aligned using Bismark. Methpat was used to analyse multiplex bisulfite amplicon sequencing on a range of CpG island targets across a panel of human cell lines and primary tissues. Methpat was able to represent the clonal diversity of epialleles analysed at specific gene promoter regions. We also used Methpat to describe epiallelic DNA methylation within the mitochondrial genome.
Conclusions
Methpat can summarise and visualise epiallelic DNA methylation results from targeted amplicon, massively parallel sequencing of bisulfite converted DNA in a compact and interpretable format. Unlike currently available tools, Methpat can visualise the diversity of epiallelic DNA methylation patterns in a sample.
Electronic supplementary material
The online version of this article (doi:10.1186/s12859-016-0950-8) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, software, visualization, bisulfite, targeted amplicon, epigenetics, epiallele
Background
In mammals, the predominant and most widely studied DNA methylation mark occurs at CpG dinucleotide (CpG) palindromic sequences [1]. The vast majority of methods that investigate DNA methylation utilise bisulfite treatment of genomic DNA followed by PCR amplification to distinguish methylated from unmethylated CpG sites [2–5]. Bisulfite treatment discriminates methylated from unmethylated cytosines by selectively reacting with unmethylated cytosines to generate uracil. During the subsequent first step of PCR amplification, the uracils are read as thymine. Conversely, methylated cytosines do not react with the bisulfite reagent and remain as cytosines after PCR amplification [6]. DNA methylation readouts at single sites employing bisulfite conversion become analogous to genotyping assays by detecting either a cytosine or thymidine at the C position of a CpG site and are interpreted as methylated or unmethylated cytosines respectively.
An epiallele refers to a distinct pattern of methylation, typically over a short genomic region [7, 8]. In addition to the methylation state given for each CpG site, the pattern of DNA methylation of all CpG sites across the epiallelic or clonal template can also be characterised [7]. Indeed, in terms of biological function, CpG methylation should be often considered in an allelic fashion over multiple adjacent CpG sites [9, 10].
However, currently most studies summarise data into average percentage values at each CpG site thus losing the positional pattern information of DNA methylation across each clonal template [9]. Analysis platforms such as the Illumina Infinium BeadArray [11], bisulfite pyrosequencing [12] and SEQUENOM™ EpiTYPER™ [13] use bisulfite mediated chemistry to discriminate the methylation state of CpG sites but summarise measurements into percentage values across each CpG site or region of interest. Percentage methylation described in most DNA methylation studies hides important pattern and positional information of DNA methylation with potential functional and regulatory relevance [7]. It is only with clonal sequencing approaches [1, 14, 15], whole genome bisulfite sequencing [16] or reduced representation bisulfite sequencing [17], that the methylation state of individual CpG sites within a genomic DNA template can be readily measured in a digital sense, as methylated or not, allele by allele.
Imprinted regions of the genome such as IGF2/H19 and MEST typically display two epialleles, where one is completely methylated and the other is unmethylated. The loss of imprinting at such loci leads to syndromic complications [18, 19]. Average DNA methylation across these loci are typically presented as 50 % methylation but the pattern of DNA methylation at each epiallele is lost [7].
Heterogeneous DNA methylation describes the phenomenon where different contiguous CpG sites have different levels of methylation. DNA methylation heterogeneity can arise in a variety of ways including but not limited to: (i) more than a single population of cells is analysed that differ in DNA methylation at the locus of interest, (ii) the locus of interest is imprinted i.e. two different epialleles are present in each cell or, (iii) the locus is inherently heterogeneous in its DNA methylation composition. It is only using clonal sequencing approaches with allelic outputs, high resolution melting (HRM) [7, 20], or a novel ligation mediated approach [10] that heterogeneous DNA methylation can be detected. It is also inferred by varying methylation at CpG sites e.g. from Pyrosequencing. Importantly, the number of methylated alleles can be substantially underestimated unless clonal approaches are used [20]. Clonal sequencing is currently the best method to investigate heterogeneous DNA methylation and the extent of epiallelic methylation patterns that exist within a single sample [15].
Until recently, it has been cost prohibitive to assess the complexity of methylation patterns, as large number of clones need to be individually sequenced to determine the extent of heterogeneous DNA methylation. As one clone represents a single epiallele, many tens to hundreds of clones need to be sequenced to gain a true representation of different epialleles in a sample. The introduction of massively parallel sequencing enables the sequencing of many thousands of DNA templates from multiple regions simultaneously providing a true representation of the diversity and extent of heterogeneous DNA methylation patterns derived from a given sample. However, as the number of clones sequenced increases, the ability to analyse and present this type of data then becomes a significant challenge, and at this time, there are very few software tools available to manage such data from massively parallel sequencing experiments [21, 22]. Some visualisation and analysis tools are available for Bisulfite Sanger Sequencing including BiQ Analyzer [23], MethVisual [24], QUMA [25], BISMA [26]. However, these tools do not scale up with massively parallel sequencing having been designed for Sanger sequencing. BiQ Analyser HiMod is a tool that enables visualisation of high throughput sequencing of 5-methylcytosine and other methyl-variant modifications [27] however, results are expressed in percentage methylation values masking allelic methylation patterns.
In this study, we have developed Methpat, a software tool which processes bisulfite sequencing data following Bismark alignment [28] and summarises DNA methylation according to epiallelic methylation patterns. This software has been used to analyse multiplex bisulfite amplicon PCR coupled to massively parallel deep sequencing on a range of primary haematopoietic tissue samples and model cancer cell lines to observe the extent of heterogeneous DNA methylation. Methpat is also able to create publication-ready, compact visualisations of the summarised data showing heterogeneous DNA methylation patterns in a space efficient and comprehensible manner.
Materials, methods and implementation
Samples, library preparation, sequencing and sequence alignment. Details of sample preparation, library generation, sequencing and sequence alignment protocol employed are summarised in the Additional file 1. Human samples used in this study were approved for research by The Royal Children’s Hospital Human Research Ethics Committee (RCH HREC#27138E).
Methpat—a tool to summarise epiallelic DNA methylation patterns
We have developed the software tool, Methpat to summarise and visualise the resultant epiallelic DNA methylation patterns from multiplex bisulfite amplicon experiments. Source code is available on GitHub (http://bjpop.github.io/methpat/). Methpat takes the output from bismark_methylation_extractor and summarises the methylation state of each CpG site within each amplicon template sequenced. DNA methylation patterns are then counted and their abundance is summarised into a tab delimited text file amenable for further downstream statistical analyses. Methpat also outputs a standalone HTML file that provides a visualisation of the DNA methylation pattern of each amplicon of interest and a visual summary of their abundance in each sample. A range of visualisation settings are customisable so that the end-user can change the settings to facilitate interpretation of the data and generate publication-ready figures. These options include presenting pattern counts as a percentage of the total, as absolute count or log-scaled counts (Additional file 2: Figure S1). Patterns can be arranged in order either by count abundance or by DNA methylation state. Colours within the visualisation can also be modified (Additional file 3: Figure S2), and the image saved as a PNG file for presentation or publication.
Results
Bismark alignment of sequencing data and statistics
After evaluating a range of bisulfite-aware massively parallel alignment software [29], we decided to use Bismark [28] with the highest mapping efficiency and highest proportion of concordantly mapped reads across the aligners compared to unique alignments in our previous study [29]. In addition, Bismark produces an output string that enables the processing of epiallelic DNA methylation patterns when parsed. , We developed Methpat to read this output and summarise the data in a compact and interpretable manner.
Using the stringent criterion of no mismatches within the initial 28 nt seed sequence during alignment and discarding non-unique alignments, the range of unique read alignments among the samples analysed ranged from 3,691 to 275,040 reads in total, corresponding to a mapping efficiency ranging from 7.9 to 55.3 % (Table 1). The total number of cytosine residues analysed within each sample ranged from 151,722 to 11,313,285 and includes CpG dinucleotide and non-CpG cytosine residues (Table 1). An indirect measure of bisulfite conversion efficiency was calculated by determining the percentage methylation at CHG and CHH residues in each sample. This was possible as the amplicons used in this study do not target loci where such non-CpG methylation is known to occur in humans [16] nor had human stem cells been used that are known to contain non-CpG DNA methylation [30]. CHG and CHH methylation was observed at a frequency of 0.1 to 1.0 % and 0.2 to 1.3 %, respectively, which corresponds to 98.7 to 99.9 % bisulfite conversion efficiency. This finding provides high confidence in our dataset for scoring DNA methylation states.
Table 1.
Mapping statistics of bisulfite amplicon libraries
| Sample | Mapping Efficiency | Unique Hits | Methylated CpG | Methylated CHG | Methylated CHH | Total C’s analysed |
|---|---|---|---|---|---|---|
| 293 | 52.2 % | 7539 | 64.9 % | 0.2 % | 0.3 % | 316211 |
| 40424 | 55.3 % | 9414 | 37.5 % | 0.2 % | 0.2 % | 351086 |
| 910046 | 42.0 % | 7060 | 32.6 % | 0.2 % | 0.3 % | 299795 |
| 12a-cd19 | 14.9 % | 48648 | 47.9 % | 0.4 % | 0.5 % | 1933767 |
| 12a-cd34 | 30.3 % | 85049 | 36.5 % | 0.1 % | 0.2 % | 3703147 |
| 12a-cd45 | 32.4 % | 109173 | 32.6 % | 0.1 % | 0.2 % | 4714744 |
| 12acd33 | 36.2 % | 161885 | 32.8 % | 0.2 % | 0.2 % | 6997070 |
| 6-mda453 | 54.6 % | 201660 | 84.4 % | 0.8 % | 1.3 % | 9179816 |
| 6c-cd19 | 7.9 % | 22258 | 77.8 % | 0.2 % | 0.3 % | 777739 |
| 6c-cd33 | 27.9 % | 20071 | 35.2 % | 0.2 % | 0.2 % | 851116 |
| 6c-cd34 | 19.5 % | 36928 | 49.7 % | 0.2 % | 0.2 % | 1628107 |
| 6ccd45 | 33.0 % | 31087 | 39.5 % | 0.1 % | 0.2 % | 1314281 |
| 9a-cd19 | 21.2 % | 39352 | 48.7 % | 0.2 % | 0.3 % | 1638757 |
| 9a-cd33 | 31.9 % | 125884 | 35.8 % | 0.2 % | 0.2 % | 5459419 |
| 9a-cd34 | 26.2 % | 77870 | 43.4 % | 0.2 % | 0.2 % | 3321993 |
| 9a-cd45 | 46.6 % | 28085 | 29.8 % | 0.2 % | 0.2 % | 1211803 |
| 9awholeblood | 31.5 % | 97532 | 30.8 % | 0.2 % | 0.2 % | 4081834 |
| brl | 49.3 % | 9107 | 32.7 % | 0.2 % | 0.4 % | 398977 |
| caco | 19.6 % | 129536 | 78.1 % | 0.2 % | 0.2 % | 4512574 |
| dg75 | 51.7 % | 10827 | 57.2 % | 0.3 % | 0.3 % | 489096 |
| ekvx | 23.0 % | 115915 | 63.1 % | 0.2 % | 0.2 % | 4494359 |
| hela | 43.1 % | 41650 | 55.9 % | 0.2 % | 0.2 % | 1731811 |
| hepg2 | 39.2 % | 24667 | 63.4 % | 0.3 % | 0.3 % | 971693 |
| ht1080 | 40.7 % | 4586 | 67.0 % | 0.2 % | 0.4 % | 176188 |
| htb22-col | 30.9 % | 45576 | 79.9 % | 0.2 % | 0.2 % | 1863098 |
| jwl | 31.3 % | 18814 | 42.7 % | 0.2 % | 0.2 % | 771188 |
| k562 | 49.7 % | 144791 | 55.9 % | 0.3 % | 0.3 % | 6230391 |
| ls174t | 41.2 % | 3691 | 57.2 % | 0.2 % | 0.3 % | 151722 |
| mcf7 | 30.0 % | 87404 | 71.6 % | 0.8 % | 0.8 % | 3786412 |
| mda-mb231-bag | 29.0 % | 94811 | 77.3 % | 1.0 % | 1.1 % | 4171147 |
| nalm6 | 43.6 % | 37669 | 85.8 % | 0.2 % | 0.2 % | 1569041 |
| nccit | 44.0 % | 31656 | 45.7 % | 0.4 % | 0.3 % | 1406165 |
| ovcar8 | 32.3 % | 46864 | 63.4 % | 0.3 % | 0.3 % | 1917527 |
| sknas | 21.6 % | 275040 | 27.7 % | 0.1 % | 0.2 % | 11313285 |
| u231 | 14.0 % | 123302 | 74.8 % | 0.4 % | 0.2 % | 4389352 |
Furthermore, two amplicons targeting unique regions within the human genome that contain no CpG sites were used to determine the bisulfite conversion efficiency in an orthogonal manner. Of the reads that passed alignment criteria for a subset of samples, we found that all non-CpG cytosines were converted in our experiment (Additional file 4: Figure S3). Mapping efficiency is one of many metrics used to determine the quality of the data and would suggest data from 6c-cd19 was not nominal. However, across all samples analysed, the bisulfite conversion efficiency was very high and was therefore included for visualisation using Methpat.
For the target regions analysed, an overall DNA methylation level ranging from 27.7 to 85.8 % was observed. In the lower ranges, the samples were mainly primary human tissue and non-cancerous cell lines while many model cancer cell lines demonstrated higher overall DNA methylation levels. This observation was expected, given that the amplicons selected for analysis were predominantly from promoter regions of genes known to be hypermethylated in cancer (Additional file 5: Table S2).
Methpat analysis of DNA methylation demonstrates a wide diversity of DNA methylation patterns
DNA methylation of FOXP3 in primary haematopoietic cells
The promoter region of FOXP3 was analysed for DNA methylation to validate the amplicon next generation sequencing, bioinformatics analysis and Methpat visualisation pipeline. Amplicons obtained from whole blood and subpopulations of cells from bone marrow were analysed from a single individual, from which, a diverse range of DNA methylation states and their abundance was observed. Analysis of whole blood showed that although the majority of epialleles were either completely methylated or completely unmethylated at CpG sites (Fig. 1), there were a diverse array of methylation patterns present (62 in total). This could reflect the cellular composition of whole blood, such that a number of cell types exist with a variable DNA methylation state at FOXP3. In contrast, DNA extracted from CD34, CD19 and CD33 positive subpopulations were found to be largely methylated at FOXP3. The CD45 positive compartment was unmethylated (Fig. 1). This was in line with previous investigations on similar sample types [31].
Fig. 1.
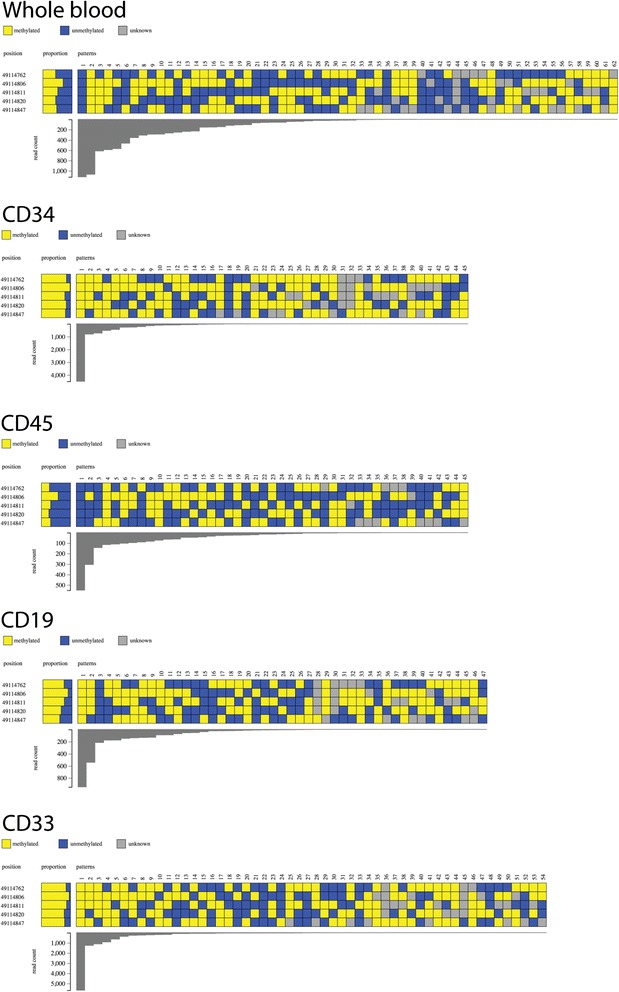
Methpat visualisation of DNA methylation at the FOXP3 gene promoter region. Samples from one individual (blood) fluorescence activated cell sorted (FACS) into various haematopoetic compartments were assessed for DNA methylation and analysed by Methpat. DNA methylation across this locus varies according to cell type. Furthermore, the diversity of epialleles within each cell type analysed also varies with one or two patterns dominating the read counts
Methpat can visualise imprinted loci
The extent of DNA methylation at a known imprinted locus, MEST, was investigated. This locus also served as a PCR amplification bias control as the DNA methylation state was expected to be 50 %, as this locus is comprised of two populations of epialleles where one is completely methylated while the other is completely unmethylated. Both epialleles were clearly identified in whole blood, CD34, CD33, CD19 and CD45 positive samples (Fig. 2) with the unmethylated epiallele more abundant than the methylated epiallele. Additional epialleles of varying DNA methylation patterns were also identified but at a significantly lower abundance (Fig. 2). The same imprinted state was also observed in the lymphoblastoid cell line, BRL (Fig. 2). The imprinting of MEST is known to be disrupted in model cancer cell lines [32]; HeLa and MDA-MB-231-BAG cell lines were observed to have predominantly hypermethylated epialleles at this locus (Fig. 2) and is in keeping with publically available datasets with these cell lines found on ENCODE [33].
Fig. 2.
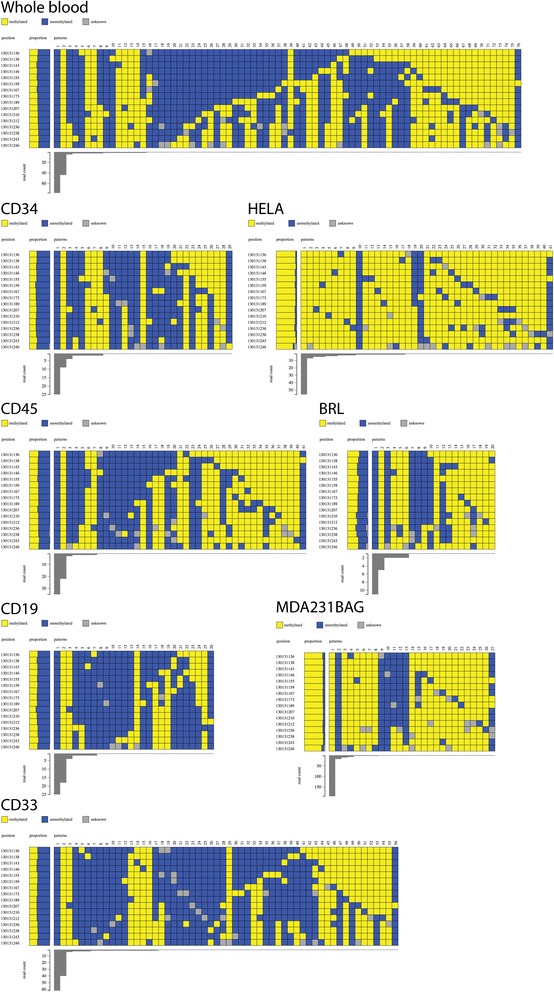
Methpat visualisation of DNA methylation at the MEST imprinted region on a range of primary cells (CD34, CD45, CD19 and CD33) and tissue (Whole blood), model cancer cell lines (HeLA and MDA-MB-231-BAG) and a normal lymphoblast cell line (BRL). The methylation status of MEST, expected to be ~50 % was observed in all normal sample types. The cancer cell lines demonstrate methylated MEST. In addition, Methpat visualizes the epiallelic diversity of MEST in all these samples
Methpat visualisation of gene promoters associated with cancer
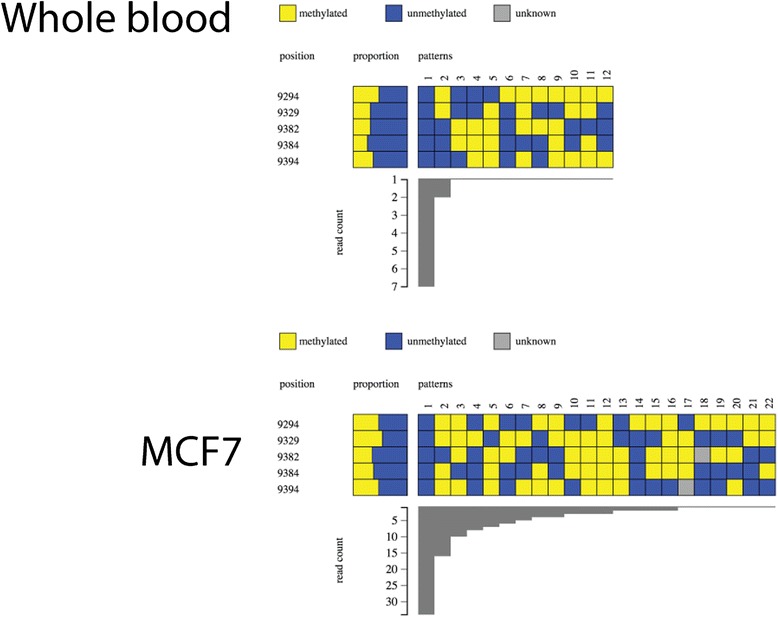
The methylation state of the RASSF1A gene promoter, which is known to be methylated in cancer [34, 35], was determined. In wild-type whole blood and the lymphoblast cell line JWL, unmethylated epialleles were primarily observed with a significant number of other much lower abundance epiallele states with varying patterns of DNA methylation (Fig. 3). HeLa was also unmethylated at RASSF1A while other cancer cell lines, HEPG2, NALM6, Caco (Fig. 3), MCF7 and NCCIT (Additional file 6: Figure S4) were predominantly hypermethylated. Of note, the diversity and range of the DNA methylation state of epialleles are much greater than might be expected of cell lines.
Fig. 3.
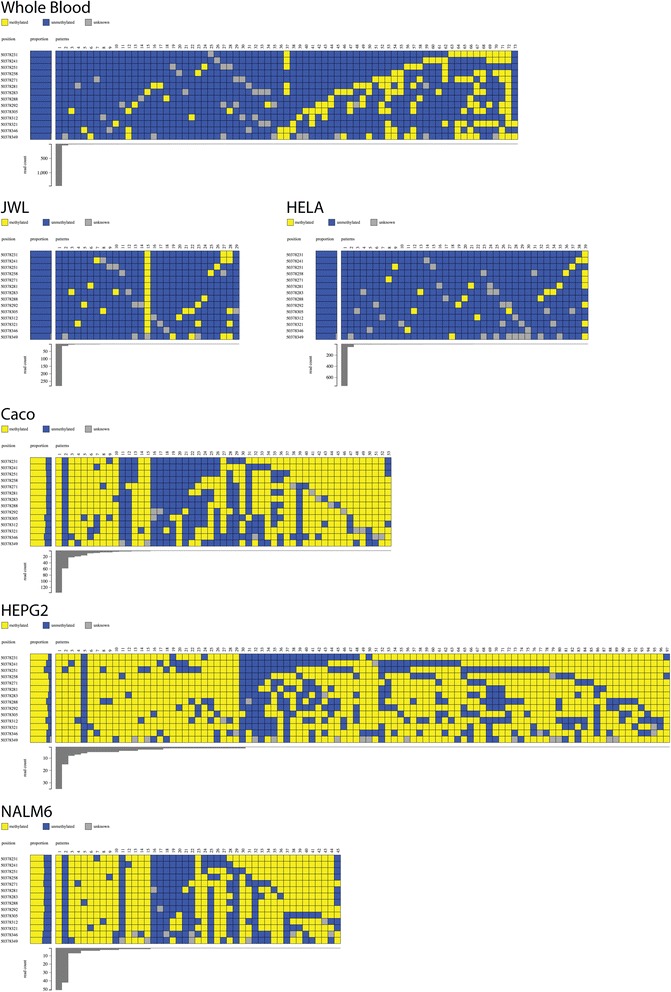
Methpat visualisation of DNA methylation at the RASSF1A gene promoter region. Methylation of RASSF1A is present in cancer cell lines (Caco, HEPG2 and NALM6) with the exception of HeLa. Examples of RASSF1A methylation in whole blood and a normal lymphoblast cell line (JWL) are also shown
We also investigated DNA methylation of the gene promoter of CDKN2A, at which DNA methylation is also seen in many cancers [36] (Fig. 4). We found that the unmethylated epiallele was most abundant in normal whole blood, HeLa, HEPG2, JWL, MCF7 and NCCIT. In contrast, Caco was hypermethylated at this locus. Interestingly, in wildtype whole blood and the cell lines HEPG2, JWL, and NCCIT, the completely methylated epiallele could be observed but was at very low abundance compared to the unmethylated epiallele (Fig. 4). We confirmed that these alleles did not arise from incomplete bisulfite conversion artefacts as all non-CpG cytosines were converted to thymidine.
Fig. 4.
Methpat visualisation of DNA methylation at the CDKN2A gene promoter region
Methpat visualisation of mitochondrial genome DNA methylation
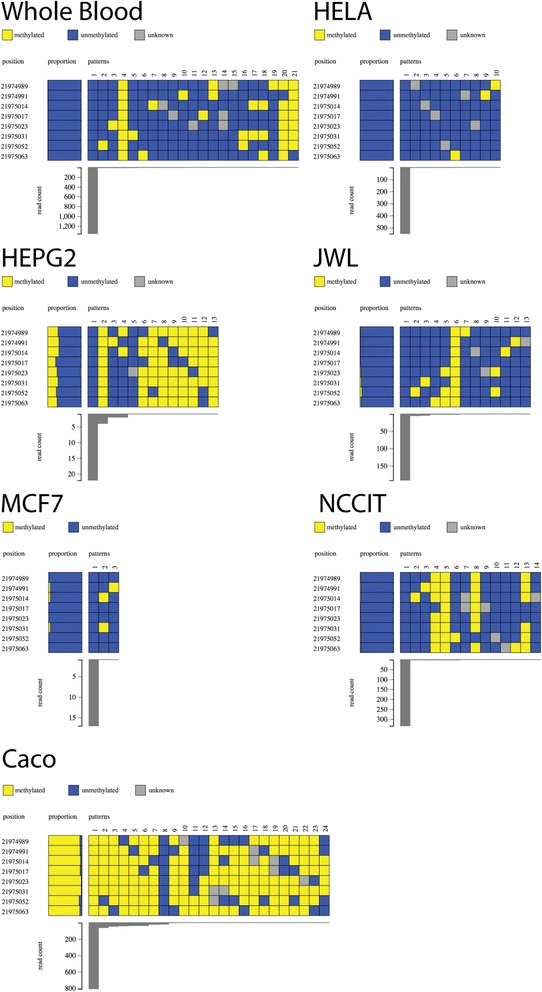
Bisulfite amplicon primers to the mitochondrial DNA D-loop regulatory sequence were included in the analysis to determine the DNA methylation state of the mitochondrial genome. The predominant epiallele was found to be unmethylated across most samples analysed; however, there was a significant range in the abundance of epialleles with variable DNA methylation state across all samples (Fig. 5, Additional file 7: Figure S5), suggesting that DNA methylation of the mitochondrial genome was present [37] but appeared to be independent of the disease status of the sample. This is in keeping with recent observations of mitochondrial genomic DNA methylation in human cells [38, 39]. We again confirmed that these alleles did not arise from incomplete bisulfite conversion artefacts as all non-CpG cytosines were converted to thymidine.
Fig. 5.
Methpat visualisation of DNA methylation within the D-Loop regulatory region of the mitochondrial genome
Discussion
Most studies investigating DNA methylation using conventional sequencing approaches represent DNA methylation into percentage values at each CpG site and in turn, do not show important positional information encoded within the epiallelic DNA methylation patterns. A comparison of features between methylation visualisation tools is summarised in Table 2. We have developed a new software tool called Methpat that processes output files from Bismark to visualise DNA methylation sequencing data by epialleles. Methpat facilitates visualisation of high throughput sequencing data after Bismark analysis and does not attempt to determine the success of a particular experiment. This is left to the investigator to interpret the metrics from Bismark prior to Methpat visualisation. We demonstrate the utility of Methpat by examining the DNA methylation pattern abundance and epiallelic DNA methylation states that are lost when DNA methylation is summarised as percentage DNA methylation.
Table 2.
Alternative DNA methylation Analysis and Visualisation Tools
| Software | Program Language and Implementation | Analysis Process | Visual Output | Input file | Output file | Epiallelic Counts | Experiment Quality Check |
|---|---|---|---|---|---|---|---|
| Methpat | Python, pip install, URL available to install files locally | Summarises Bismark output | Interactive HTML and summary text file of epiallele counts. Scalable PNG file | Bismark methylation extractor output, user-defined BED format file | HTML and tab delimited text file | Yes | No, leverages Bismark |
| Bismark | command line,Python, requires bwa | Performs alignment to bisulfite reference genome | None, generates BAM files for visualisation with SeqMonk or IGV | fastq file | BAM and tab deliminted text files | No | Yes calculates C to T conversion |
| BSPAT | Java/JSP web interface | Visualisation and summarisation of Bismark output | PNG file and UCSC Genome Browser file | Bismark output, fastq files | Text file summary, PNG and UCSC Genome Browser BED file | Yes | No |
| MPFE | R library, Bioconductor | Calculates probabilities that epialleles are true | R image outputs | Table of read counts from bisulfite sequencing data | Derived statistics and plots | Yes | Yes |
| Methylation plotter | R library, shiny interactive web application | Visualises beta DNA methylation values | Interactive webpage with setting options to adjust a static image of DNA methylation values for each sample. PNG and PDF output. | Text file containing matrix of sample vs beta value at each CpG of interest | PDF and PNG image file | No | No |
| RnBeads | R library, Bioconductor | Processes summary data from other software for visualisation | Interactive HTML and UCSC Genome browser track hub files. PNG files | BED file | HTML summary | No | Yes |
| coMET | R library, Webserver for analysis | For EWAS studies. Analyses derived matrix files | Image files of plots with genomic locations. | Text matrix files | Image files | No | No |
EWAS epigenome-wide association studies using Illumina Infinium HM450 BeadArrays
Methpat operates on Bismark output files and further summarizes this data into an interactive visualization that can be quickly interpreted within a web-browser. It can be executed locally to generate an HTML file which can be hosted remotely through the Internet or visualized locally on the most common web browsers (Chrome, Safari, Firefox, Internet Explorer). This feature which is unique to Methpat, is a major advantage. At this stage, Methpat does not have capability as a “genome-browser” to look at DNA methylation patterns at a genome-scale because it was designed for targeted deep sequencing of amplicons, however, we have made the source code available for further development by the research community to further improve Methpat (http://bjpop.github.io/methpat/).
We demonstrated the importance of calculating epiallelic abundance on the imprinted locus MEST, where we showed two predominant populations of epiallelic DNA methylation patterns, one completely methylated and the other completely unmethylated. Such patterns cannot be interpreted with percentage values at each CpG site as heterogeneous DNA methylation or, a sample containing a heterogeneous population of cells with variable DNA methylation states could give rise to the same percentage value [7]. Using Methpat to visualise the diversity of epialleles enables the inference at least of the existence of heterogeneous DNA methylation, or, the detection of heterogeneous populations of cells as demonstrated by investigating FOXP3 in whole blood and subpopulations of the haematopoietic compartment.
Of interest, in some model cancer cell lines, we observed a wide and diverse range of methylated epialleles. Having ruled out to the best of our ability any bisulfite conversion or PCR amplification artefacts, our results suggest that even within apparently homogeneous cell lines, the methylation state at a subset of gene promoters analysed is heterogeneous. This could be due to the nature of cell culture where the phenomenon of increasing DNA methylation is observed with increasing passage [40, 41], plasticity, or the setting of epigenetic memory of a sub-population of cells in the culture [42]. The detection of completely methylated epialleles of the CDKN2A gene promoter in whole blood and in other samples interrogated supports the validity of our approach, and indicates that Methpat provides a new tool to enable the detection of low level DNA methylation [43, 44]. The functional and biological implications of our current findings remain unclear, however, further investigation with appropriate specimens using Methpat is warranted.
We investigated mitochondrial DNA methylation and believe our analysis is one of the first accounts of characterising epiallelic DNA methylation within the D-loop regulatory region of the mitochondrial genome. Our study confirms observations of DNA methylation within the mitochondria [37–39]. Given there can be many thousands of copies of the mitochondrial genome per cell, it is not possible at this stage to determine the providence of the methylation states we have identified. The issue of heteroplasmy for mutations in the mitochondrial genome [45] apply for DNA methylation and techniques to address heteroplasmy could be applied to investigate DNA methylation within the mitochondrial genome further [46]. By visualising DNA methylation patterns within the mitochondrial genome, Methpat can facilitate insight towards new biomarkers of disease [47].
While our current strategy and experimental results are unable to resolve PCR amplification artefacts (over-representation of particular sequence reads because of amplification), incorporation of unique molecular identifiers [48] could resolve this in future studies.
Conclusions
In summary, we demonstrate the feasibility of multiplex bisulfite amplicon deep sequencing to identify the extent of DNA methylation epialleles in a range of human samples. We have developed a software tool, called Methpat, which enables the summarisation and visualisation of DNA methylation sequencing data in the context of epiallelic information.
Availability of data and materials
The raw amplicon sequencing data, Bismark alignments and Methpat output files associated with this manuscript have been published with the DOI 10.1186/s13742-015-0098-x.
Methpat software can be obtained from this URL. (http://bjpop.github.io/methpat/)
Acknowledgements
Illumina Australia Pty Ltd for a MiSeq Pilot Sequencing Grant for next generation sequencing reagents.
Funding
This work was supported, in part, by National Breast Cancer Foundation of Australia (NCBF) grants to AD, DK and MT (CG-08-07, CG-10-04 and CG-12-07), the Cancer Council of Victoria to AD, and by grants from the Victorian Cancer Agency to NW and AD. SW was supported by the Melbourne Melanoma Project funded by the Victorian Cancer Agency Translational Research program and established through support of the Victor Smorgon Charitable Fund. Computation time was granted by the Life Sciences Computation Centre (LSCC) at the Victorian Life Sciences Computational Initiative (VLSCI) under grant VR0002. The Murdoch Childrens Research Institute and the Olivia Newton-John Cancer Research Institute are supported by the Victorian Government Operational and Infrastructure Support Grant.
Additional files
Sample preparation, library preparation and sequencing methods. (DOCX 132 kb)
Example of a screenshot of Methpat visualisation. A. Epiallele representation of the patterns of DNA methylation for respective amplicon in respective sample. B. Count histogram, the abundance of each epiallele represented in A. C. Genomic co-ordinate and position of CpG of interest. D. Proportion of DNA methylation at each CpG position. E. Save button, export visualisation as a PNG file. F. Amplicon of interest. G. Legend depicting DNA methylation status. (PNG 202 kb)
Example of a screenshot of the settings page for each Methpat visualisation. A number of parameters can be changed and the visualisation replotted for ease of interpretation. (PNG 92 kb)
IGV screenshot of two amplicon regions used in this study that target DNA sequences with no CpG sites within the RANBP17 locus. Therefore it is expected that all cytosines within this region of interest are completely converted by bisulfite treatment. This is shown here for MCF7 and MDA-MB-231-BAG. (PNG 135 kb)
Bisulfite PCR primers used in this study. (XLS 15 kb)
Diverse and wide ranging epiallelic DNA methylation patterns of RASSF1A in MCF7 and NCCIT model cancer cell lines. (PNG 434 kb)
Epiallelic DNA methylation patterns of the D-loop regulatory region of the mitochondrial genome. (PNG 163 kb)
Human Samples used in this study. (XLS 8 kb)
Amplicon details required for Methpat input (hg19 coordinates). (XLS 9 kb)
Description of Methpat options. (DOCX 118 kb)
Footnotes
Competing interests
XZ is a salaried employee of BioInfoRx Inc. MP is a salaried employee of BioResearch Software Consultants. NW is currently a salaried employee of Pacific Edge Biotechnology Limited however, performed this work prior to joining Pacific Edge. Next generation sequencing reagents used in this study were kindly supplied by Illumina Australia Pty Ltd as a part of their MiSeq Pilot Sequencing Grant Program.
Authors’ contributions
NCW designed the study, performed the experiments, analysed the data and wrote the paper, BJP developed the software and wrote the paper, ILC designed the study, performed initial pilot experiments and wrote the paper, DK designed the study, analysed the data and wrote the paper, MT designed the study and wrote the paper, SQW designed the study, performed initial pilot experiments and wrote the paper, THM designed the study, performed initial pilot experiments and wrote the paper, XZ analysed the data and created the pilot visualisation software and wrote the paper, MP analysed the data and created the pilot visualisation software and wrote the paper, SE performed the experiments, analysed the data and wrote the paper, SRD performed the experiments, analysed the data and wrote the paper, AD conceptualised the study, designed the study, analysed the data and wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Nicholas C. Wong, Email: nwon@unimelb.edu.au
Bernard J. Pope, Email: bjpope@unimelb.edu.au
Ida L. Candiloro, Email: ic85@hotmail.com
Darren Korbie, Email: d.korbie@uq.edu.au.
Matt Trau, Email: m.trau@uq.edu.au.
Stephen Q. Wong, Email: Stephen.Wong@petermac.org
Thomas Mikeska, Email: Thomas.Mikeska@onjcri.org.au.
Xinmin Zhang, Email: xinmin@bioinforx.com.
Mark Pitman, Email: mark@bioresearchsoftware.com.
Stefanie Eggers, Email: steffi.eggers@gmail.com.
Stephen R. Doyle, Email: s.doyle@latrobe.edu.au
Alexander Dobrovic, Phone: +61 3 9496 9689, Email: alex.dobrovic@onjcri.org.au.
References
- 1.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 2.Hayatsu H. Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis--a personal account. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:321–330. doi: 10.2183/pjab.84.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobrovic A, Kristensen LS. DNA methylation, epimutations and cancer predisposition. Int J Biochem Cell Biol. 2009;41:34–39. doi: 10.1016/j.biocel.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. Biotechniques. 2002;33:632–634. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- 5.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikeska T, Candiloro IL, Dobrovic A. The implications of heterogeneous DNA methylation for the accurate quantification of methylation. Epigenomics. 2010;2:561–573. doi: 10.2217/epi.10.32. [DOI] [PubMed] [Google Scholar]
- 8.Finer S, Holland ML, Nanty L, Rakyan VK. The hunt for the epiallele. Environ Mol Mutagen. 2011;52:1–11. doi: 10.1002/em.20590. [DOI] [PubMed] [Google Scholar]
- 9.Mikeska T, Bock C, Do H, Dobrovic A. DNA methylation biomarkers in cancer: progress towards clinical implementation. Expert Rev Mol Diagn. 2012;12:473–487. doi: 10.1586/erm.12.45. [DOI] [PubMed] [Google Scholar]
- 10.Wee EJH, Rauf S, Shiddiky MJA, Dobrovic A, Trau M. DNA Ligase-Based Strategy for Quantifying Heterogeneous DNA Methylation without Sequencing. Clin Chem. 2014;61:163–171. doi: 10.1373/clinchem.2014.227546. [DOI] [PubMed] [Google Scholar]
- 11.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan J-B, Shen R: High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288-95 [DOI] [PubMed]
- 12.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 13.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat Protoc. 2006;1:2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 15.Stirzaker C, Millar DS, Paul CL, Warnecke PM, Harrison J, Vincent PC, Frommer M, Clark SJ. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229–2237. [PubMed] [Google Scholar]
- 16.Lister R, Pelizzola M, Dowen R, Hawkins R, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315-22. [DOI] [PMC free article] [PubMed]
- 17.Meissner, Mikkelsen T, Gu H, Wernig, Hanna J, Sivachenko A, Zhang X, Bernstein B, Nusbaum, Jaffe D, Gnirke A, Jaenisch R, Lander E: Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008. [DOI] [PMC free article] [PubMed]
- 18.Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, Rogers J, Pask AJ, Shaw G, VandeBerg JL, McCarrey JR, SAVOIR Consortium. Renfree MB, Reik W, Dunham I. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- 19.Lambertini L, Diplas A, Lee M, Sperling R, Chen J, Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Cancer Biol Ther. 2008;3:261-9. [DOI] [PMC free article] [PubMed]
- 20.Candiloro I, Mikeska T, Hokland P: Rapid analysis of heterogeneously methylated DNA using digital methylation-sensitive high resolution melting: application to the CDKN2B (p15) gene. Epigenetics & … 2008. [DOI] [PMC free article] [PubMed]
- 21.Candiloro ILM, Mikeska T, Dobrovic A. Assessing combined methylation-sensitive high resolution melting and pyrosequencing for the analysis of heterogeneous DNA methylation. Epigenetics. 2011;6:500–507. doi: 10.4161/epi.6.4.14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutsik P, Feuerbach L, Arand J, Lengauer T, Walter J, Bock C. BiQ Analyzer HT: locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing. Nucleic Acids Res. 2011;39(Web Server issue):W551–W556. doi: 10.1093/nar/gkr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 24.Zackay A, Steinhoff C. MethVisual - visualization and exploratory statistical analysis of DNA methylation profiles from bisulfite sequencing. BMC Res. Notes. 2010;3:337. doi: 10.1186/1756-0500-3-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36(Web Server):W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA - Fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230–12. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker D, Lutsik P, Ebert P, Bock C, Lengauer T, Walter J. BiQ Analyzer HiMod: an interactive software tool for high-throughput locus-specific analysis of 5-methylcytosine and its oxidized derivatives. Nucleic Acids Res. 2014;42:W501–W507. doi: 10.1093/nar/gku457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong NC, Ng J, Hall NE, Lunke S, Salmanidis M, Brumatti G, Ekert PG, Craig JM, Saffery R. Exploring the utility of human DNA methylation arrays for profiling mouse genomic DNA. Genomics. 2013;102:38–46. doi: 10.1016/j.ygeno.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 30.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:1–16. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakanishi H, Suda T, Katoh M, Watanabe A, Igishi T, Kodani M, Matsumoto S, Nakamoto M, Shigeoka Y, Okabe T, Oshimura M, Shimizu E. Loss of imprinting of PEG1/MEST in lung cancer cell lines. Oncol Rep. 2004;12:1273–1278. [PubMed] [Google Scholar]
- 33.The ENCODE Project Consortium A User's Guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hesson LB, Cooper WN, Latif F. The role of RASSF1A methylation in cancer. Dis Markers. 2007;23:73–87. doi: 10.1155/2007/291538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saelee P, Wongkham S, Chariyalertsak S, Petmitr S, Chuensumran U. RASSF1A promoter hypermethylation as a prognostic marker for hepatocellular carcinoma. Asian Pac J Cancer Prev. 2010;11:1677–1681. [PubMed] [Google Scholar]
- 36.Candiloro ILM, Mikeska T, Hokland P, Dobrovic A. Rapid analysis of heterogeneously methylated DNA using digital methylation-sensitive high resolution melting: application to the CDKN2B (p15) gene. Epigenetics Chromatin. 2008;1:7. doi: 10.1186/1756-8935-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace DC, Fan W. Mitochondrion. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellizzi D, D'Aquila P, Scafone T, Giordano M, Riso V, Riccio A, Passarino G. The Control Region of Mitochondrial DNA Shows an Unusual CpG and Non-CpG Methylation Pattern. DNA Res. 2013;20:537–547. doi: 10.1093/dnares/dst029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, Absher DM, Wold BJ, Myers RM. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bork S, Pfister S, Witt H, Horn P, Korn B, Ho AD, Wagner W. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell. 2010;9:54–63. doi: 10.1111/j.1474-9726.2009.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 43.Snell C, Krypuy M, Wong EM, Loughrey MB, Dobrovic A. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res. 2008;10:R12. doi: 10.1186/bcr1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong EM, Southey MC, Fox SB, Brown MA, Dowty JG, Jenkins MA, Giles GG, Hopper JL, Dobrovic A. Constitutional Methylation of the BRCA1 Promoter Is Specifically Associated with BRCA1 Mutation-Associated Pathology in Early-Onset Breast Cancer. Cancer Prev. Res. 2011;4:23–33. doi: 10.1158/1940-6207.CAPR-10-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiner JE, Kishore RB, Levin BC, Albanetti T, Boire N, Knipe A, Helmerson K, Deckman KH. Detection of Heteroplasmic Mitochondrial DNA in Single Mitochondria. PLoS One. 2010;5:e14359. doi: 10.1371/journal.pone.0014359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iacobazzi V, Castegna A, Infantino V, Andria G. Molecular Genetics and Metabolism. Mol Genet Metab. 2013;110:25–34. doi: 10.1016/j.ymgme.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw amplicon sequencing data, Bismark alignments and Methpat output files associated with this manuscript have been published with the DOI 10.1186/s13742-015-0098-x.
Methpat software can be obtained from this URL. (http://bjpop.github.io/methpat/)


