Abstract
An imaging-coupled 3D printing methodology for the design, optimization, and fabrication of a customized nerve repair technology for complex injuries is presented. The custom scaffolds are deterministically fabricated via a microextrusion printing principle which enables the simultaneous incorporation of anatomical geometries, biomimetic physical cues, and spatially controlled biochemical gradients in a one-pot 3D manufacturing approach.
Keywords: 3D printing, 3D scanning, tissue engineering, neural engineering, nerve regeneration
Graphical Abstract
Personalized tissue engineering and regeneration strategies offer a potential remedy for currently untreatable injuries and diseases via the development of customized screening approaches, tissue scaffolds, and advanced biomedical devices.[1–4] Such strategies are a fundamental requirement for regenerating tissues which contain complexity in geometry and heterogeneity in composition. Particularly useful would be novel biomanufacturing approaches capable of generating geometrically programmable architectures augmented with biomimetic physical, biochemical, and cellular components.[5,6] Indeed, integrating such diverse functionalities into a unified, customizable manufacturing scheme remains a critical challenge due to the constraints of conventional approaches. Here we introduce a 3D printing methodology for the design, optimization, and fabrication of a custom nerve repair technology for the regeneration of complex peripheral nerve injuries containing bifurcating sensory and motor nerve pathways. The custom scaffolds are deterministically fabricated via a microextrusion printing principle using 3D models which were reverse engineered from patient anatomies by 3D scanning. The bifurcating pathways are augmented with 3D printed biomimetic physical cues (microgrooves) and path-specific biochemical cues (spatially controlled multicomponent gradients). In vitro studies reveal that 3D printed physical and biochemical cues provide axonal guidance and chemotractant/chemokinetic functionality. In vivo studies examining the regeneration of bifurcated injuries across a 10 mm complex nerve gap in rats showed that the 3D printed scaffolds achieve successful regeneration of complex nerve injuries, resulting in enhanced functional return of the regenerated nerve. This approach suggests the potential of 3D printing toward advancing tissue regeneration in terms of: 1) the customization of scaffold geometries to match inherent tissue anatomies; 2) the integration of biomanufacturing approaches with computational modelling for design, analysis, and optimization; and 3) the enhancement of device properties with spatially controlled physical and biochemical functionalities, all enabled by the same 3D printing process.
1. Introduction
Efforts to personalize regenerative approaches, therapeutics, and biomedical devices are catalyzing major advances in the treatment of serious injuries and chronic diseases.[7–9] State of the art efforts in personalized medicine, already common within certain areas in the biomedical space (e.g., dentistry), are being extended toward the development of patient-specific tissues and organs,[1,5] drug screening approaches,[2] and advanced biomedical devices[10–14] (e.g. advanced prosthetics and biointerfaces). Customization of medical treatments could convey significant advantages by targeting treatment directly to a specific injury or disease profile of a patient, which is critical due to inherent variance in patient anatomies, injury profiles, and genetic and proteomic structures.[2,15] Recent advances in genome and proteome mapping are enabling advances in personalized treatment approaches at the molecular level. Yet, it remains a critical challenge to provide customized treatments at the tissue level which address patient-to-patient variances in disease and injury profiles, particularly in neuroregeneration.
Indeed, nerve regeneration is a complex biological phenomenon which often requires a balance of molecular- and tissue-level repair strategies, depending on the nature of the particular injury or neurological disorder.[16] Peripheral nerve regeneration is a particularly important field, as damage to peripheral nerves occurs via various mechanisms. These include disease and traumatic injuries (e.g., from car accidents and battlefield wounds) which result in greater than 200,000 annual nerve repair procedures performed in the United States alone.[17] Conventional nerve repair techniques center on grafting approaches, such as autografts and de-cellularized allografts, which have the major advantages of closely mimicking natural nerve characteristics.[18] However, grafting approaches also present various drawbacks and limitations, including the need for an additional harvesting surgery, chronic pain and morbidity at the donor site, limitations on graft size and geometry, and potential immune response.[17] This has motivated alternative nerve repair strategies, such as the use of nerve guidance channels constructed from synthetic and biological polymers, which provide conduits for the regeneration of damaged nerves.[17] Nerve guidance channels possess various advantages, including flexibility in material choice, avoidance of additional surgeries, and the ability to control physical, mechanical, cellular, and biochemical properties of the guide, each of which affect cell and tissue fate.[19–26] Existing nerve guide technologies have facilitated the regeneration of linear nerve injuries;[16,17] however, it is challenging to apply conventional fabrication approaches to the regeneration of large, geometrically complex injuries.
2. Concept of Anatomical 3D Printed Nerve Regeneration
3D printing is a computer-driven, robotics based biomanufacturing approach which is becoming a valuable tool in the development of customized biomedical devices. For example, 3D printing has catalyzed novel efforts in the manufacturing of artificial tissues and organs,[1,5,27] electronics,[28,29] nanotechnology,[27,30] and biomedical devices,[12–14] with particular utility in customized manufacturing and biomedical applications.[31,32] This is further enabled by the coupling of 3D printing approaches with 3D imaging technologies.[15] 3D printing provides compatibility with a multivariate material set, including metals,[33,34] synthetic polymers,[35,36] biomaterials,[37,38] and nanomaterials,[39,40] which could make it a particularly exciting and expansive tool in next-generation fabrication initiatives. 3D printing thus offers compatibility with computer modelling, choice of input material, and control over material integration. Further, the mechanical properties of 3D printed devices may be optimized, which is critical in many applications including neuroregeneration.[26] For example, a 3D printing approach to the design and manufacturing of nerve guidance pathways could provide new opportunities for advanced nerve repair via the production of anatomically-accurate, complex scaffold geometries, as well as programmable incorporation of biomimetic physical and biochemical functionalities.
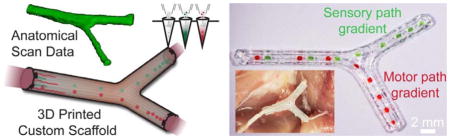
Here we demonstrate the combination of 3D imaging and 3D printing for the design, optimization, and biomanufacturing of anatomically-true biomimetic custom nerve regeneration pathways. Specifically, we demonstrate the creation of custom pathways generated from non-linear anatomical geometries, and the incorporation of physical cues in the form of microgrooves and path-specific multicomponent biochemical cues in the form of spatiotemporal growth factor gradients. We examine the application of this nerve guide technology to the regeneration of bifurcating mixed nerve pathways using the original tissue structure as a template for the pathway geometry. Important features offered by the new manufacturing approach include biomimicry of natural anatomical structure, introduction of guiding physical cues, and the implementation of path-specific biomimetic multicomponent gradients within the scaffold architecture. This approach ultimately could provide a general strategy for the regeneration of complex injury types.
3. Reverse Engineering of Nerve Pathways via 3D Structured Light Scanning
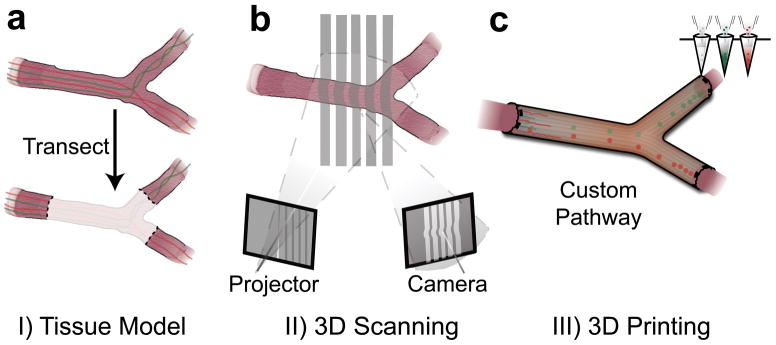
Conventional nerve guidance channels are typically fabricated around cylindrical substrates, so the resultant devices are inherently restricted to linear structures. Alternatively, it would be highly desirable to develop strategies which allow for fabrication of pathways with complex anatomical structures and internal biofunctionalization. Our strategy to accomplish this in a one-pot 3D printing process is outlined in Figure 1. The approach encompasses three critical steps: 1) accessing the nerve pathway for imaging, and transecting the bifurcating nerve, which consists of bifurcating motor and sensory nerve branches from a mixed nerve source (Figure 1a), 2) imaging the original nerve structure either in situ or ex situ, to generate a corresponding 3D computer model (Figure 1b), and 3) 3D printing a geometrically matching network of pathways which are functionalized with physical cues and path-specific biochemical gradients (Figure 1c). This approach provides a mechanism for regenerating damaged nerve plexuses, which is difficult to accomplish using conventional nerve guidance channels.
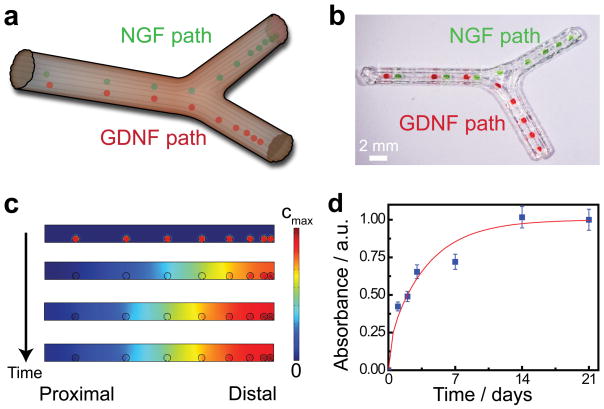
Figure 1.
Personalized nerve regeneration pathways enabled by 3D scanning and printing. a) A tissue model of the nerve pathway to be constructed is prepared for subsequent imaging by either incision (in situ approach) or transection (ex situ approach). b) The intact or transected tissue is imaged using structured light scanning (SLS), which results in a high precision 3D model of the nerve pathway. c) The reverse engineered nerve pathway is 3D printed, to realize a device which mimics the original nerve in terms of geometry, physical cues, and path-specific biochemical cues in the form of gradient distributions.
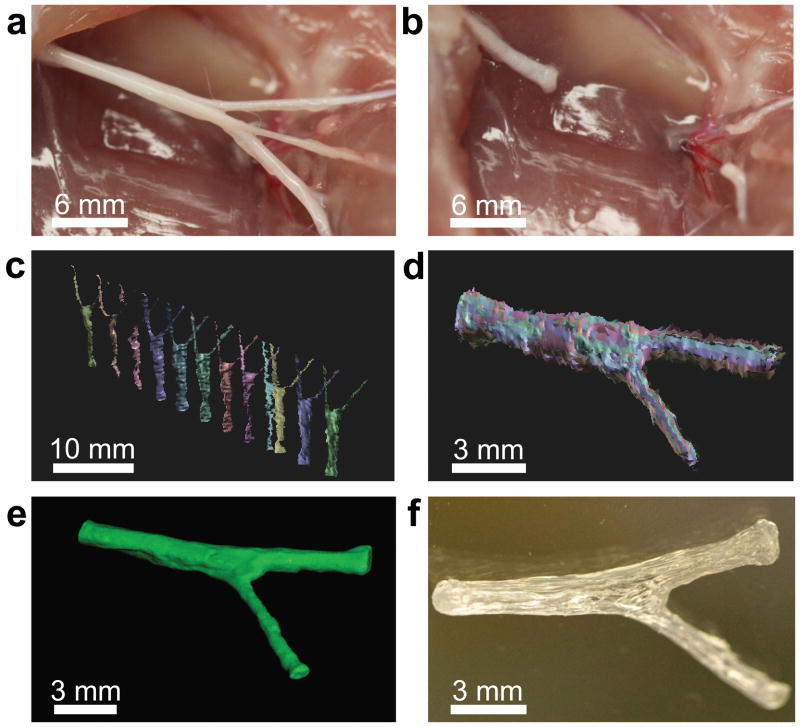
We selected the sciatic nerve bifurcation as the model for regeneration, as it contains a mixed nerve system which bifurcates into sensory (source sural nerve) and motor (source tibial-peroneal nerve) branches (Figure 2a). Following exposure of the bifurcating nerve via an incision in the superior muscle tissue, we transected the sciatic nerve above and below the bifurcation point (Figure 2b). Having selected the bifurcating mixed nerve pathway as a model and acquired the representative tissue sample, we then prepared a cast of the nerve which was subsequently imaged using a 3D structured light scanning (SLS) technique. SLS is a valuable imaging technique for reverse engineering of geometrically complex free-form objects, including body parts and teeth.[41]
Figure 2.
3D printed complex nerve pathways from 3D scanned bifurcating nerves. a) The sciatic nerve provides a bifurcating mixed nerve model which contains branching sensory (derived from the sural nerve; top) and motor nerves (derived from the peroneal and tibial nerves; bottom). b) The complex nerve pathway is transected, providing a tissue template for ex situ scanning measurements. c) Scans are conducted from various perspectives to assemble a 3D model which describes the geometry of the nerve pathway (sural and tibial nerve motor branches). d) The individual scans are aligned to replicate the 3D geometry of the nerve tissue. e) The aligned scans are assembled into a water-tight 3D model, leading to a full reconstruction of the nerve pathway geometry, which provides a template for 3D printing. f) The 3D model is printed into a hollow silicone pathway which is customized to fit the exact geometry of the original tissue.
As shown in Figure 2c, the SLS technique couples well with tissue cast imaging, ultimately generating multiple data sets which collectively describe the 3D geometry of the imaged nerve. Following scanning of the cast from multiple perspectives, the individual scans were then aligned into a full 3D reconstruction of the original nerve pathway (Figure 2d), indicating that SLS can be used to accurately reverse engineer the structure of internal tissues. As shown in Figure 2e, we then demonstrated that the reconstructed 3D model could be used to directly manufacture an anatomically accurate nerve pathway for regeneration of the complex nerve gap by using the scanned model as a template for generating 3D printer path information (Figure 2f). We verified that various neurocompatible materials could be used, including polycaprolactone, alginate, poly(lactic-co-glycolic acid), and silicone. For the present study, silicone was selected as the guide material due to its previous use in conduit-based peripheral nerve regeneration applications.[42,43] As SLS is a highly sensitive imaging technology, we also conducted proof-of-concept studies to determine whether in situ imaging could be performed without the use of a cadaver model, which would potentially enable point-of-care applications of our general strategy. As shown in Supporting Figure S1, we found that SLS can also be used to acquire 3D information from nerves in situ. This process is also compatible with conventional imaging technologies, such as computed tomography and magnetic resonance imaging.[44,45]
4. Biomechanical Analysis
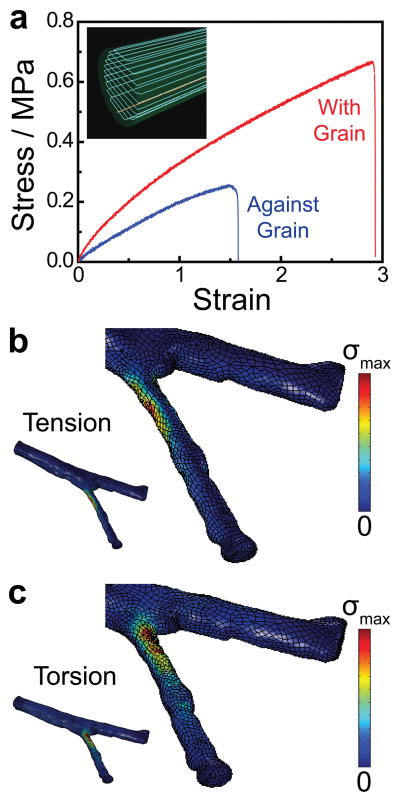
Previous work has shown that the cellular components of peripheral nerve tissue respond to the mechanical properties of materials and devices[26,46] as well as to biomechanical stresses.[47] Therefore, we analyzed the mechanical properties of the devices via computational techniques in order to characterize and optimize the mechanical responses of the 3D printed nerve pathways. We first examined the mechanical properties of the 3D printed silicone to obtain fundamental parameters and determine the effect of the printing orientation on the mechanical response. As shown in Figure 3a, the 3D computer template of the nerve pathway was first sliced into discrete layers to generate path information for the printer, which creates an inherent grain in the sliced structure due to printing artifacts. For instance, the inset of Figure 3a shows the grain orientation resulting from slicing in the radial dimension (in contrast to slicing in the axial dimension; data not shown). The effect of this grain orientation on the mechanical response of the material was tested using tensile strength measurements, in which the tensile load was applied either with or against the grain. As shown in Figure 3a, the samples exhibited linear strain-strain responses up to 8% strain. Although the Young’s modulus of silicone rubbers can vary depending on type and processing method, the Young’s modulus here was calculated to be 0.44 MPa, which compares well with previously published values of bulk silicone,[48–50] and with the modulus of the peripheral nerve (0.45 MPa).[51] This is important for maintaining elastic continuity between the nerve guide and the repaired tissue.[26]
Figure 3.
Mechanical characterization and computational analysis of the pathways. a) Tensile strength measurements on 3D printed materials reveal the influence of the printing orientation (physical cue direction) on the ultimate tensile strength. b) Von Mises stress (σ) distribution in the nerve pathway under a tensile load applied to the distal ends of the nerve (σmax = 0.41 MPa). c) Von Mises stress (σ) distribution in the nerve pathway under a torsional load applied to the distal ends of the nerve (σmax = 0.61 MPa).
Since material defects are often pronounced at high strains, where non-linear material behavior occurs, we also examined the effect of the 3D printed grain orientation on the ultimate tensile strength (UTS). As shown in Figure 3a, the 3D printed material contains anisotropy with respect to its high strain response and ultimate properties. The UTS was significantly higher when the load was applied with the grain than when the load was applied against the grain (0.57 MPa compared with 0.25 MPa, respectively, p < 0.05). The UTS observed when the load was applied with the grain compared favorably with previously published values of bulk silicone.[48] We also observed a difference in the failure mechanism of the samples, which was characterized by a tear in the case of the load applied against the grain, in contrast to a rapid snap in the case of the load applied with the grain (Supporting Figure S2). These results suggest that the 3D printed device will perform similarly to a bulk silicone device if loads are applied with the grain, but would be relatively weaker with loads applied against the grain. Since axial loads on the implanted nerve pathways from the surgical process are expected to dominate radial biomechanical loads which arise during regeneration, the guides were printed based on path information generated from slicing the model in the radial dimension, for primary loading with the grain. A representative video of the printing process is provided in Supporting Movie 1. Likewise, the material properties obtained from measurements in which the load was applied with the grain were used in subsequent finite element analysis (FEA) of nerve pathway mechanics.
We next conducted FEA on the nerve pathway under both tensile and torsional loading conditions (Figures 3b and c, respectively), which occur during both surgery and the subsequent regeneration phase. Applied boundary conditions included regions of zero displacement and regions of applied load (see Experimental Section and Supporting Figure S3). Surgical and biomechanical loads are challenging to estimate due to the complex geometry of the nerve guide, so the mechanical response of the device to a representative basis load was examined to characterize the global strain distributions and regions of concentrated strain. As shown in Figures 3b and c, both loading conditions generated concentrated stress regions near the bifurcation point. This information is useful from two perspectives: 1) it enables one to determine if complex loading conditions will cause the nerve pathway to fail, which would compromise the resultant regeneration process, and 2) it enables one to identify regions in the device which undergo significant mechanical deformations. Although the former is unlikely in the present study given that silicone is a compliant material, the latter is especially useful for gaining a complete understanding of the outcomes of in vivo studies. For future 3D printed nerve pathways fabricated from relatively less compliant materials, such as polycaprolactone or poly(lactic-co-glycolic acid), the former information may be especially useful.
5. In Vitro Studies
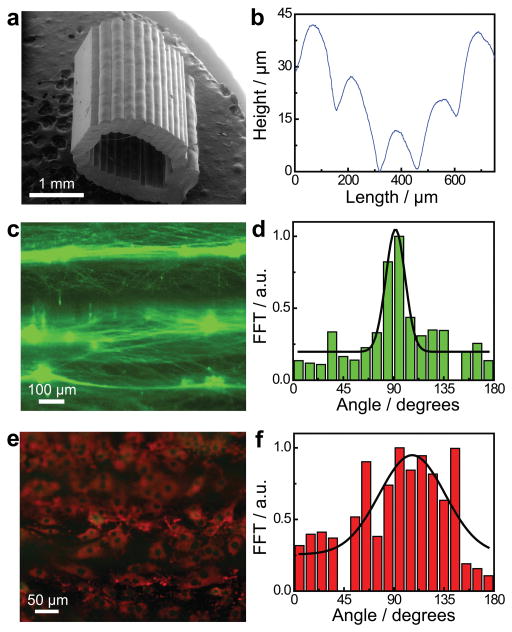
In addition to providing a material grain which can be rationally selected to optimize the mechanical performance of the nerve pathway, the layer-by-layer fused deposition approach also generates a luminal physical cue which is axially-oriented due to model slicing in the radial dimension (Figure 4a). As shown in Figure 4b, a measurement of the luminal surface profile perpendicular to the physical cue orientation reveals that the 3D printed nerve pathways contain an axial physical cue with microgroove architecture (see also Supporting Figure S4). The mean spacing between grooves was 156 ± 27 μm and the mean depth of the grooves on the luminal surface was 17 ± 8 μm. It is interesting to note that this physical cue qualitatively resembles naturally occurring physical cues present in degraded nerve pathways, known as the bands of Büngner, which guide regenerating axons in vivo. We also verified that the physical cue extends along the entire length of the nerve guide, and that the lumen remains open for regenerating nerve along the entire length of the device (Figure S10).
Figure 4.
Characterization and influence of the 3D printed physical cue. a) Scanning electron micrograph of a 3D printed hollow nerve pathway displaying an axially-oriented physical cue on the luminal surface. b) Profilometry measurement performed on the luminal surface of the 3D printed nerve pathway shows a distinct microgroove structure. c) Cultured primary embryonic neurons on the 3D printed horizontally-oriented physical cue (90° reference angle) stained for tau (green). d) Corresponding orientation analysis showing a coincidence of the neurite network alignment with the physical cue. e) Cultured Schwann cells on the horizontally-oriented physical cue (90° reference angle) stained for GFAP (green) and laminin (red). f) Corresponding orientation analysis showing a coincidence of the cytoskeleton and extracellular matrix alignment with the physical cue.
It has previously been shown that physical cues in the form of microfibers and microgrooves affect the orientations of the two main components of regenerating peripheral nerve, axons and Schwann cells.[22,52] Here, we examined whether these 3D printed microgrooves affected the structures of regenerating axons and Schwann cells in vitro. Importantly, Figure 4c shows that the neurite network established by dissociated primary superior cervical ganglion (SCG) neurons on the 3D printed physical cue was highly aligned, with an orientation which coincided with the printed physical cue (oriented at a 90° basis angle). This orientation was quantified via a fast Fourier transform (FFT) analysis, as shown in Figure 4d. Control studies done in the absence of the 3D printed physical cue showed no such alignment, resulting in a randomly distributed neurite network (Supporting Figure S5). Additional experiments were also done with dorsal root ganglia (DRG) instead of dissociated SCG neurons, which similarly exhibited alignment of the neurite network with the physical cue (Supporting Figure S6). Schwann cells cultivated on the 3D printed physical cue also aligned with the physical cue (Figures 4e and f), especially in the vicinity of the microgroove.
End-organ connections have been shown to act as guides for selective reinnervation of sensory and motor paths.[53] We sought to further optimize the potential for reinnervation of each path by using 3D printing to introduce supporting biochemical gradients within each path according to the strategy illustrated in Figure 5a. We selected nerve growth factor (NGF) as the supporting sensory path cue and glial cell line-derived neurotrophic factor (GDNF) as the motor path cue, given their differential expression profiles in sensory and motor nerves.[54] Gelatin methacrylate hydrogel was chosen as the encapsulation medium, given its previously demonstrated use for controlled drug release and high degree of neurocompatibility.[55,56] As representatively shown in Figure 5b, the hydrogel was printed in a spatial gradient distribution concentrated towards the distal end of the each path, to provide a continuously enriching attractant. The hydrogel in the sensory path was loaded with NGF, while the hydrogel in the motor path was loaded with GDNF.
Figure 5.
Functionalization of nerve pathways with path-specific biochemical gradients. a) Schematic of the path-specific incorporation of gradient distributions of supporting biochemical cues – nerve growth factor, NGF, and glial cell line-derived neurotrophic factor, GDNF – in the sensory and motor paths, respectively. b) Representative photograph of the 3D printed gradient pattern achieved using a protein-loaded hydrogel. Green and red dyes were added to the hydrogel to enhance the image contrast. c) Results from finite element analysis (FEA) of transient drug release showing the establishment of an axially-oriented concentration gradient which results from the 3D printed luminal hydrogel pattern over time. d) Experimental drug release studies showing the protein release kinetics from the gelatin methacrylate hydrogel system.
We next confirmed that the 3D printed biochemical cues would establish an axial spatiotemporal gradient within each nerve pathway. The release of protein from the hydrogel occurs via a diffusive release mechanism. As shown in Figure 5c, FEA of a representative 3D printed hydrogel configuration indeed results in an axial gradient inside the nerve pathway, which is concentrated at the distal end and stretches across the thickness of the guide. The typical regeneration period of a peripheral nerve injury is at least three to four weeks, so we also examined the kinetics of protein release from the hydrogel system to confirm that the time scale would be suitable. As shown in Figure 5d, the controlled protein release was maintained over a three week period. A standard drug release model previously used for the analysis of thin film hydrogel systems was also fit to the data:[57,58]
| (1) |
where θ(t) is the fraction released, t is the time, L is the film thickness, and D is the diffusivity. Using the diffusivity as a single fitting parameter, we were able to estimate the effective protein diffusivity in the hydrogel system for use in FEA studies, which agreed with previously published values under similar conditions.[59]
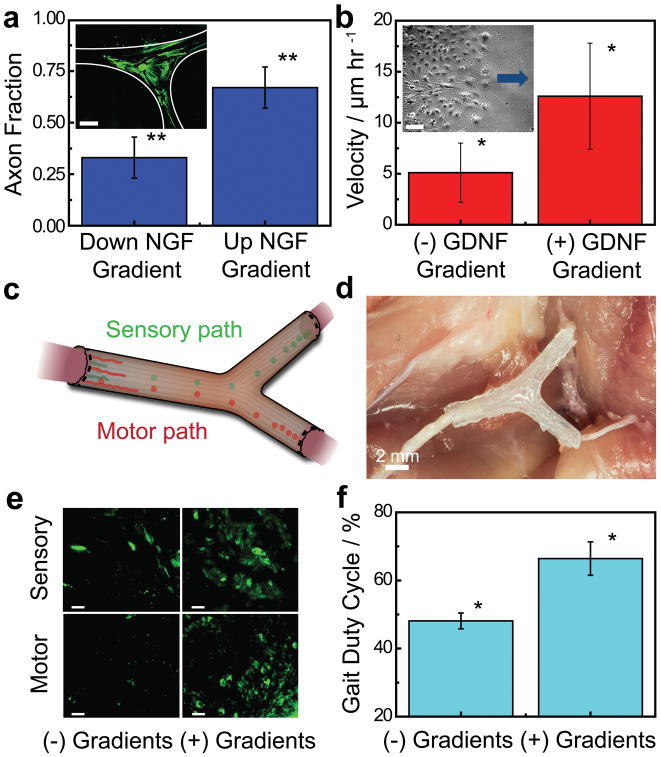
Although it is well established that NGF and GDNF are critical supporting factors of sensory and motor neuron health, respectively, it was of interest to examine their effects on axon and Schwann cell responses when implemented in diffusive gradients. Longitudinal imaging is a useful tool for examining nerve regeneration over large length scales both in vitro and in vivo.[60,61] We first examined the effect of a diffusive NGF gradient on the formation of sensory neurite networks in vitro via longitudinal imaging of axon growth. As shown in Figure 6a, we found that 67% of axons were attracted towards the source, in comparison with 49% observed in control studies lacking the gradient, indicating that diffusive NGF gradients act as a chemotractant for sensory axons (p < 0.01). This result is consistent with previous literature which examined the effects of immobilized and diffusive NGF gradients using both primary and model sensory neurons.[21,62,63]
Figure 6.
In vitro and in vivo characterization of regeneration with 3D printed nerve pathways. a) Effect of the diffusive NGF gradient on the guidance of the sensory neurite network growth (inset scale bar = 1,000 μm; a full size image is provided in Supporting Figure S7). b) Effect of the diffusive GDNF gradient on the migration velocity of Schwann cells (inset scale bar = 100 μm; arrow indicates direction of source and migration direction). c) Schematic of implanted nerve guide showing bifurcation into sensory and motor nerve paths. d) Photograph of an implanted 3D printed nerve guide prior to suturing. e) Histology of regenerated nerve showing cross-sections of regenerated nerves stained for tubulin (green) (scale bar = 50 μm). f) Comparison of the functional return in regenerated rat hind limbs (* indicates p-value < 0.05, ** indicates p-value < 0.01).
Having established that the NGF gradient attracts sensory axons, we next examined the potential for the GDNF gradient to support regeneration via its effect on Schwann cells. It is known that in some organisms there is a higher abundance of Schwann cells in sensory nerve pathways, which act as a potential mechanism by which motor axons improperly enter sensory pathways during regeneration.[53] Thus, it was important to determine whether the GDNF gradient affected the Schwann cell migration velocity, as the use of gradients within bifurcating conduits may offer a means of controlling the repopulation level of each pathway in vivo. As shown in Figure 6b, the diffusive GDNF gradient acted as a chemokinetic cue which caused an increase in the Schwann cell migration velocity from 5.1 to 12.6 μm hr−1 (p < 0.05). This result is consistent with previous literature, which found that GDNF acts as a chemokinetic agent for glial cells.[64,65]
6. In Vivo Studies
Having demonstrated various advantages offered by the 3D printing approach in terms of geometric, physical, and biochemical customization in vitro, we next carried out a small animal study to investigate the use of this technology in vivo. Specifically, we examined the regeneration of a 10 mm complex nerve gap injury to the sciatic nerve bifurcation over a 3 month period. Figure 6c shows a schematic while Figure 6d shows a representative image of implanted nerve guide. The nerve regeneration was characterized by the histology of the regenerated nerve in the bifurcated motor and sensory distal paths, at cross-sections located 2 mm proximal to the distal suture site in both paths. As shown in Figure 6e, histological analysis of the regenerated nerve revealed the incorporation of 3D printed gradients led to a greater degree of nerve regeneration in both paths. We note that histological analysis based on S100 staining also verified the presence of Schwann cells in the regenerated nerve (Supporting Figure S8). It was of interest to determine whether the gradients provided any significant improvement in the functional return of the regenerated nerve. As shown in Figure 6f, detailed gait analysis of the rats revealed significant improvement in the functional return of the limbs treated with the 3D printed nerve scaffolds which were augmented with biochemical gradients. Significant improvement was observed with respect to the gait duty cycle, which showed an improvement by a factor of 1.4 (p < 0.05) relative to performance in guides lacking the 3D printed gradient. While our in vitro studies demonstrated that the GDNF gradient increased the Schwann cell migration velocity (Figure 6b), we did not observe significant differences in the numbers of Schwann cells that infiltrated both the motor and sensory pathways of our conduit as indicated by S100 staining (Figure S8). This suggests that the enhanced functional regeneration may be via a different mechanism to differential Schwann cell migration. Importantly, our technique is, for the first time, capable of providing custom bifurcating, and even multi-branched, designs for future research in customized neuroregeneration. Importantly, the results of this in vivo study suggest two major conclusions: 1) 3D printing can be applied to successfully regenerate complex bifurcating mixed nerve injuries in vivo, and 2) functionalizing the nerve pathways with 3D printed path-specific biochemical gradients enhanced the functional return of regenerated nerve.
7. Conclusions
In summary, we have developed a novel 3D printing approach for manufacturing a custom nerve repair technology which is personalized to anatomical geometries, and augmented with physical and biochemical cues to promote the regeneration of multiple nerve pathways. This demonstration represents a proof-of-concept illustration that imaging-coupled 3D printing approaches can facilitate customized neuroregeneration in previously inaccessible ways. 3D scanning allowed us to adapt the final pathway geometry to the original tissue structure. Mechanical and computational tools allowed us to design, analyze, and optimize the integrity of the pathways. A one-pot 3D printing process provided the ability to introduce advantageous physical and biochemical cues in the form of microgrooves and multicomponent diffusive biomolecular gradients. This combination of complex geometries and sophisticated supporting cues offers a proof-of-concept as a biomimetic approach for the regeneration of complex mixed nerve injuries. Future studies will focus on several additional goals, including: 1) replacement of silicone with biodegradable alternatives; 2) optimization of the diffusive gradient structure; and 3) examination of the underlying mechanisms of biochemical gradient-enhanced functional return. Overall, we expect that this platform will ultimately have significant impact in both the fundamental understanding of neuroregeneration and clinical treatment of complex nerve injuries.
Experimental Section
In situ 3D scanning
Imaging of the sciatic nerve bifurcation via structured light scanning (SLS) was done using Sprague-Dawley rats (Hilltop Labs Inc., Pittsburgh, PA). For each study, the animal was euthanized and handled in strict accordance with good animal practice as defined by the relevant national and local animal welfare bodies, and approved by the Princeton University Institutional Animal Care and Use Committee (IACUC). The sciatic nerve bifurcation was exposed for imaging by making an incision at the base of the gastrocnemius and carefully cutting along the outside edge of the muscle and along the biceps femoris. The muscle was then pulled back to expose the underlying sciatic nerve. A thin film of a scanning contrast agent (Magnaflux) was then applied to the nerve while masking the surrounding tissue to provide enhanced contrast for the nerve during scanning. The contrast agent was later removed with a saline solution wash following scanning. Subsequently, a clean low-contrast wound dressing was re-applied around and underneath the exposed nerve which further enhanced the contrast for the nerve and reduced the signal from the surrounding muscle tissue. The animal was then placed on a motorized stage (CR1/M-Z7E, ThorLabs), which allowed the tissue to be imaged from various vantage points over a full rotational angle. The single camera-projector SLS system (SLS-1, David Vision) was then calibrated according to a vendor-provided protocol. Subsequently, the nerve was imaged by performing multiple scans over a full rotational angle. Scan data was collected without the use of scanning software-associated data smoothing or alignment. The above protocol was repeated multiple times over the course of two months using randomly selected animals (n = 4) in order to simulate application in the point-of-care which presents inherent patient-to-patient variance.
Ex situ 3D scanning
Similar to the in situ SLS protocol, the sciatic nerve bifurcation was exposed in euthanized Sprague-Dawley rats (n = 3) by first making an incision at the base of the gastrocnemius and carefully cutting along the outside edge of the muscle and along the biceps femoris. The muscle was then pulled back to expose the sciatic nerve. Subsequently, the nerve was transected 2 cm proximal and distal to the bifurcation site in strict accordance with good animal practice as defined by the relevant national and local animal welfare bodies, and approved by the Princeton University IACUC. The tissue was subsequently trimmed and fixed by immersion in a room temperature 4% paraformaldehyde (PFA, Affymetrix)-PBS solution for 20 minutes, and blotted dry to begin the molding-casting process. A resin cast of the fixed nerve was then made from a silicone mold to provide a rigid and anatomically-consistent model for SLS. Prior to scanning, the cast was coated with a thin film of a scanning contrast agent, and the single camera-projector SLS system was calibrated according to a vendor-provided protocol. Subsequently, the nerve cast was mounted vertically on the motorized stage and was imaged by performing multiple scans over a full rotational angle. Scan data was collected without the use of scanning software-associated data smoothing or alignment. For high resolution imaging, scanning was also performed using a single camera-light emitting diode (LED) SLS system (COMET L3D, Steinbichler Optotechnik). The single camera-LED SLS system was calibrated according to a vendor-provided protocol. Subsequently, the nerve cast was mounted on a custom goniometer and imaged by performing multiple scans over a full hemispherical angle. Scan data was collected without the use of scanning software-associated data smoothing or alignment.
Reverse engineering of 3D nerve geometry from scan data
The individual scans obtained using the SLS-1 system were aligned and assembled using 3D mesh processing software (MeshLab) and 3D printing software (Netfabb, FIT GmbH), which resulted in a water-tight 3D model of the imaged nerve. The individual scans obtained using the COMETL3D system were aligned and assembled using reverse engineering software (Geomagic Design X, 3DSystems) and additive manufacturing software (Magics, Materialise) using software-provided alignment and assembly algorithms, which resulted in a water-tight 3D model of the imaged nerve.
Finite element analysis of nerve guide solid mechanics
All studies were performed using commercially available finite element analysis (FEA) software (COMSOL Multiphysics, Version 4.4). Stationary studies were conducted in 3D using the Structural Mechanics – Solid Mechanics module. The domain was created by importing the SLS-generated 3D models to the COMSOL modelling environment. All simulations were done assuming a linear elastic material property relationship. The material properties were taken from silicone vendor-provided data (density = 1,010 kg m−3), previously published literature values of bulk silicone (Poisson ratio = 0.45),[66] and experimental mechanical testing studies conducted on the printed materials (Young’s modulus = 0.44 MPa). Applied boundary conditions included regions of zero displacement and regions of applied load (Supporting Figure S3). The regions of zero displacement condition served to model constrained regions arising from surgical implantation, while the regions of applied load served to model potential loading conditions which may arise during implantation or post-implantation as a result of mechanical limb motion. Two different loading conditions were examined, which represented tensile or torsional loads. In both cases, a load of 1% of the experimentally measured ultimate tensile strength (UTS) was applied as a basis to demonstrate the value of the approach. The value of UTS was taken from mechanical testing studies conducted on the printed materials, for experiments done with loading applied with the grain, since this printing direction was used for animal studies. Initial values corresponded to zero displacement and velocity fields across the entire domain. The model was discretized using a physics-controlled mesh (normal element size), which consisted of 34,374 domain, 9,982 boundary, and 8,474 edge elements. The von Mises stress profile under both loading conditions was then calculated using a stationary solver. Proper density of the mesh was checked by examining convergence of the maximum von Mises stress by iterating from an extremely coarse to a normal mesh element size, which led to convergence within 4–10% of the previous mesh iterate.
Methacrylated gelatin hydrogel synthesis
Gelatin methacrylate was synthesized according to an earlier protocol.[56] Briefly, a 10% w/v gelatin (porcine skin, Sigma) solution in phosphate buffered saline (PBS) was prepared and heated to 60 °C with constant mixing. After the gelatin was dissolved completely, the temperature was reduced to 50 °C and allowed to reach steady state. After the solution temperature reached 50 °C, methacrylic anhydride (Sigma) was slowly added to the solution to achieve a 5:1 volumetric ratio of gelatin solution : methacrylic anhydride solution. The typical basis reaction volume consisted of 50 ml gelatin solution. Subsequently, the mixture was allowed to react for one hour at 50 °C with continual mixing. After one hour, warmed PBS (40 °C) heated in a secondary beaker was added at a 4:1 volume ratio to the gelatin-methacrylic anhydride solution to deactivate the reaction. Subsequently, the resulting mixture was added to 10 kDa dialysis tubing and the tubing was placed in reverse osmosis water and allowed to dialyze for one week. To ensure effective separation, the dialysis solution was replaced with fresh solution daily. Following the dialysis procedure, the gelatin mixture was lyophilized until dry.
Gelatin methacrylate hydrogel protein release studies
Controlled drug release studies using 50 mg ml−1 gelatin methacrylate hydrogels were done using glial cell line-derived neurotrophic factor (GDNF, Sigma) to characterize the kinetics of the protein release from the gelatin methacrylate hydrogel matrix. The gelatin methacrylate hydrogel was prepared on a 1 ml basis containing 400 ng GDNF ml−1, 5 mg ml−1 photoinitiator (Irgacure 2959, BASF), 0.1% bovine serum albumin (BSA) w/w (UltraPure non-acylated, Life Technologies), 1 mg ml−1 heparin (Sigma), and 0.05% w/w sodium azide (Sigma) in Dulbecco’s PBS (DPBS). For drug release measurements, 150 μl of gelatin methacrylate hydrogel was added to the bottom of a clean containment vessel, cross-linked using a hand-held UV lamp (UVL-56, UVP) and 600 μl DPBS was added to the vessel. The containment vessel was then sealed and maintained at 37 °C with gentle shaking over the course of a three week period. Multiple identical vessels were prepared which enabled collection of samples at various time points over the course of the three week release study. Samples were collected by removing the GDNF containing-DPBS which were stored at -4 °C until the end of the study. After three weeks, the level of released protein in the samples was characterized using enzyme-linked immunosorbent assay (ELISA, GDNF mouse ELISA kit, Abcam) according to vendor-provided protocols.
Finite element analysis of nerve guide diffusive biochemical gradient
All studies were performed using commercially available FEA software (COMSOL Multiphysics, Version 4.4). Transient studies were conducted in 2D using the Chemical Species Transport – Transport of Diluted Species module. The domain was created using the software-provided geometry toolbox based on the printed nerve guide and gelatin methacrylate gradient pattern dimensions. All simulations were done assuming a Fickian transport property relationship. The isotropic protein diffusivity in the gelatin methacrylate hydrogel system was taken from experimental drug release studies (3 × 10−13 m2 s−1), and the isotropic protein diffusivity in the aqueous phase was assumed to be 1 × 10−10 m2 s−1. Applied boundary conditions included regions of no flux and regions of flux continuity. The regions with no flux boundary condition served to model the outer edges of the nerve guide, while the regions of flux continuity served to model the interfaces of the gelatin methacrylate droplets and the surrounding aqueous domain. Applied initial conditions included regions of zero concentration and regions of defined concentration (10 μg ml−1), which represented the initially un-loaded aqueous and protein-loaded hydrogel domains, respectively. The model was discretized using a physics-controlled mesh (normal element size), which consisted of 3,644 domain and 467 boundary elements. The concentration profile was then calculated using a time-dependent solver. Proper density of the mesh was checked by examining convergence of the steady state concentration by iterating from an extremely coarse to a normal mesh element size, which led to convergence within 0.2% of the previous mesh iterate.
3D printing of bifurcated nerve pathways
Water-tight 3D models of the sciatic nerve bifurcation were imported to 3D printing software (Netfabb, FIT GmbH) and residual errors were repaired. The repaired models were subsequently exported to commercially available 3D CAD software (SolidWorks) for final optimization. Resultant models were then validated using a commercially available plastic 3D printer (Dimension Elite, Alleghaney Ed. Systems). Following validation, the 3D models were then converted to printer path information using model slicer software (KISSlicer). Devices were then printed using a custom microextrusion-based 3D printing system, which has been described previously.[28] Briefly, the system consisted of a three axis dispensing robot (Fisnar), pneumatic dispensing system (Nordson EFD), vision system, and personal computer (see Supporting Figure S9). Printing speeds ranged from ca. 0.1 – 1 mm s−1. Alginate (alginic acid sodium salt from brown algae, medium viscosity, Sigma), calcium chloride (Sigma), poly(lactic-co-glycolic acid) (75:25, Mw ~ 76,000–115,000; Sigma), polycaprolactone (average Mn ~ 10,000; average Mn ~ 80,000; Sigma), silicone (Superflex Clear RTV, Loctite), and gelatin hydrogel served as the printed materials. For fabrication of devices with luminal path-specific gradients of nerve growth factor (NGF, mouse, Life Technologies) and glial cell line-derived neurotrophic factor (GDNF, mouse, Sigma), methacrylated gelatin methacrylate hydrogel containing either NGF or GDNF served as the printed luminal supplement. Hydrogel preparation was consistent with drug release studies, except for the azide component, which was removed, and the protein concentration, which was increased to 10 μg ml−1. The heparin component was also incorporated into both hydrogels, which acted to both stabilize the protein and inhibit binding of the growth factor to the channel walls.[67] For printing the luminal gradient functionalized devices, the gradient pattern corresponded to hydrogel droplets spaced at sequentially increasing gap sizes of n × 100 μm, where n represents the droplet number starting at the distal end of the pathway. This sequentially increasing spacing was truncated at 700 μm, after which constant 1 mm spacing was employed. NGF-loaded hydrogel was printed along the bottom luminal wall of the sensory pathway, and GDNF-loaded hydrogel was printed along the bottom luminal wall of the motor pathway. The result of the printing process was a hollow 3D printed silicone conduit that contained gradient patterns of hydrogel droplets printed along the inner silicone wall, which is adjacent to regenerating nerve on the inside of the conduit. Subsequent to the hydrogel printing, the hydrogel was partially cross-linked by a UV irradiation period (UVL-56, UVP). Following completion of the silicone printing process, the printed devices were irradiated with UV and sprayed with ethanol to complete crosslinking and sterilization.
Mechanical testing of the 3D printed material anisotropy
In order to examine the effect of the physical cue orientation on the nerve guide mechanical properties, mechanical testing was done on 3D printed silicone samples which contained different orientations of the physical cue. Flat rectangular silicone samples were printed with identical layer spacing as used for the surgically implanted 3D printed nerve guides (150 μm), allowed to completely cure at room temperature, and subsequently diced into consistently sized samples. Tensile tests were performed using a commercially available mechanical testing unit (Instron 5865 with Instron 3111 temperature-controlled chamber, 1 kN load cell) under a constant strain rate of 2 in min−1. Prior to all tests, the load was reset and the load cell was calibrated. Multiple 3D printed samples were characterized allowing the stress-strain characteristics to be measured with the tensile load applied both with (n = 8) and against the grain (n = 4) of the 3D printed material. The Young’s modulus was taken as the slope of the linear region of the stress-strain curve. The ultimate tensile strength (UTS) was taken as the maximum stress reached before failure. Analysis of the statistical significance in the difference of the UTS between the two groups was done using a two-tailed student t-test.
Characterization of the 3D printed physical cue morphology
Printed nerve pathways were fabricated and subsequently diced into thirds along either the circumference or the length for profilometry and environmental scanning electron microscopy (ESEM) measurements, respectively. The axially-diced samples (n = 8) were anchored to SEM specimen mounts using double-sided carbon tape and coated with a thin gold layer for imaging. All measurements were done using a commercially available ESEM platform (FEI Quanta 200). The circumferentially-diced samples (n = 4) were mounted on a glass slide using double-sided carbon tape, with the luminal surface facing up. All measurements were done using a commercially available surface profiler (KLA-Tencor P-15).
Cell culture
Primary embryonic sensory neurons were obtained from Sprague-Dawley rats (embryonic day 15.5 – 16.5). Superior cervical ganglia (SCG) and dorsal root ganglia (DRG) were first harvested. Ganglia were then either maintained in whole form or dissociated. A detailed procedure can be found elsewhere.[68,69] Briefly, whole ganglia were suspended in 1 ml trypsin (Life Technologies) in a 15 ml conical centrifuge tube and incubated at 37 °C for 15 minutes. The solution was then centrifuged at 1 krcf for 1 min to pellet the ganglia, the supernatant was removed, and the ganglia were resuspended in soybean trypsin inhibitor (2 mg ml−1 in neurobasal media; Sigma) for 10 min. The solution was then centrifuged, the supernatant was removed, and the ganglia were resuspended in warm growth medium which consisted of neurobasal medium (Life Technologies) containing B-27 medium supplement (Life Technologies), 1% Pen Strep Glutamine (Life Technologies), and nerve growth factor 2.5S (NGF, 100 ng ml−1, mouse, Life Technologies). The medium was filtered using a 0.22 μm filter prior to addition of NGF (Stericup filter unit, Millipore). The ganglia were dissociated by repeated aspiration using a flame-polished pipette. Culture surfaces were pre-coated with poly-L-ornithine (500 μg ml−1, Sigma) overnight at 37 °C, rinsed 3 times with DPBS, coated in laminin (10 μg ml−1, Sigma) overnight at 37 °C, and rinsed 3 times with DPBS. Whole DRG or dissociated SCG were seeded with an inoculum of 1–3 DRG or SCG per 35 mm dish and cultured at 37 °C and 5% CO2 in complete growth medium. Antimitotic agent cytosine arabinoside (AraC, Sigma) was added to the medium 2 days after plating to inhibit fibroblast outgrowth. Two-thirds of medium was exchanged every 7 – 10 days.
Schwann cells derived from rat sciatic nerves were purchased from a culture collection organization (S16, CRL-2941, ATCC). Culture surfaces were pre-coated with poly-L-lysine (0.1 ml cm−2 surface area with a 15 μg ml−1 solution, Sigma) overnight at 37 °C, rinsed 3 times with DPBS, and allowed to dry for a minimum of 30 minutes at room temperature. Schwann cells were then seeded with an average inoculum of 4 × 103 to 6 × 104 cells cm−2 and cultivated over 5 – 7 days at 37 °C and 5% CO2 in Dulbecco’s modified Eagles medium (DMEM, Life Technologies) which contained 10% fetal bovine serum (FBS, Life Technologies) and 1% Pen Strep (Life Technologies). The medium was filtered using a 0.22 μm filter. Two-thirds of medium was exchanged every 2 – 3 days.
Immunofluorescence
Following cultivation, cells were fixed in a PFA solution (4% in PBS, Electron Microscopy Sciences) for 20 minutes at room temperature, rinsed with DPBS, permeabilized in a Triton X-100 solution (0.1% in DPBS, Sigma) for 30 min at room temperature, rinsed with DPBS, and blocked with a BSA solution (3% in DPBS, Sigma) overnight at 4 °C. Subsequently, the BSA solution was removed and the cells were stained using primary antibodies. Axons were stained for tau marker using anti-tau (1:1000 in diluted blocking solution, monoclonal, mouse, Life Technologies) overnight at 4 °C. Schwann cells were stained for glial fibrillary associated protein (GFAP) and laminin markers using anti-GFAP (1:1000 in diluted blocking solution, monoclonal, mouse, Sigma) and anti-laminin (1:1000 in diluted blocking solution, polyclonal, rabbit, Sigma) overnight at 4 °C, respectively. Subsequently, the samples were rinsed 3 times in DPBS and exposed to labeled secondary antibody against mouse (1:1000 in diluted blocking solution, Alexa Flour 488 anti-mouse, Life Technologies) and rabbit (1:1000 in diluted blocking solution, Alexa Flour 568 anti-rabbit, Life Technologies) overnight at 4 °C. Following secondary antibody labeling, the samples were rinsed 3 times in DPBS and coated with a thin layer of mountant (ProLong Gold Antifade with DAPI, Life Technologies). Imaging was then carried out using a fluorescence microscope (Nikon Eclipse 50i, X-Cite 120 Fluorescence Illumination Source [EXFO]).
Characterization of the 3D printed physical cue on neurite network orientation
Circumferentially-diced 3D printed nerve pathways were anchored at the well bottom in uncoated 35 mm dishes using a silicone adhesive layer with the luminal surface facing up and subsequently allowed to cure overnight. The exposed luminal surface was then pre-coated and whole DRG or dissociated SCG were then seeded, cultivated for 3 weeks, and the cultures were then imaged. Similar experiments were done in the 3D printed bifurcating nerve pathways, in which the printing was interrupted halfway through the program, resulting in an open half-shell geometry. The devices were then pre-coated, whole DRG were seeded near the center of the bifurcation point, cultivated over a 3 week period, and the cultures were then imaged. A minimum amount of medium was added such that the liquid height just reached the top of the silicone pathway (2 ml). The images were then processed so that the printed physical cue was oriented horizontally across the image providing a basis for the printed physical cue orientation at 90°. The orientation of neurite network was then examined using ImageJ software. Images were processed using a fast Fourier transform (FFT) and resultant power spectrums were then generated by plotting the FFT signal intensity along a circumferential profile as done in previous studies.[70]
Characterization of the 3D printed physical cue effect on Schwann cell orientation
Circumferentially-diced 3D printed nerve pathways were anchored at the well bottom in uncoated 6 well plates using a silicone adhesive layer with the luminal surface facing up and subsequently allowed to cure overnight. The exposed luminal surface was then pre-coated, Schwann cells were seeded, cultivated over 5 – 7 days, and the cultures were then imaged. The images were then processed so that the printed physical cue was oriented horizontally across the image providing a basis for the printed physical cue orientation at 90°. The orientation of Schwann cell structure was then examined using ImageJ software. Images were processed using a FFT and resultant power spectrums were then generated by plotting the FFT signal intensity along a circumferential profile as done in previous studies.[70]
Characterization of the GDNF gradient effect on Schwann cell migration
Individual wells of 6-well plates were first pre-coated. Following the pre-coating steps, one sterile 3D printed silicone compartmented neural chamber was placed in each well to form a seal along the bottom of the well. Schwann cells were then seeded in the middle compartment and allowed to settle overnight under growth conditions. The medium was then aspirated and the silicone compartment was peeled off, resulting in seeded cells concentrated in a rectangular configuration at the center of the well. Subsequently, 50 μl of GDNF-containing gelatin methacrylate hydrogel (1 μg ml−1) was added along the edge of the well, the seeded cells were masked, and the hydrogel was cross-linked with UV irradiation for 10 minutes. The mask was then removed, 1 ml of fresh warm medium was added to the well, and the culture was returned to growth conditions for 2 days to establish the diffusive gradient. Live imaging was then done the following day using an inverted phase contrast microscope (Nikon Eclipse TS100) and a live cell perfusion chamber (Quorum Technologies) at 37 °C and 5% CO2. Time-lapse videos captured by recording sequential phase-contrast micrographs at the cell-free surface interface at five minute intervals over a 2 hr period facilitated calculation of the migration velocity, which was defined as the horizontal distance travelled divided by the time interval. The population average migration velocity was calculated by averaging the migration velocity of the leading cells (n = 8 – 12) along the imaged interface. Control studies were done by carrying out repeated experiments, which lacked the diffusive GDNF gradient. Analysis of the statistical significance in difference of the migration velocity between the two groups was done using a two-tailed student t-test.
Characterization of the NGF gradient effect on neurite outgrowth
35 mm dishes were first pre-coated. Subsequently, 10 μl of NGF-containing gelatin methacrylate hydrogel (1 μg ml−1) was added along the edge of the dish, and the hydrogel was cross-linked with UV irradiation for 10 min. Whole DRG were then seeded near the center of the dish, cultivated over a 1 week period, and the cultures were then imaged. Similar experiments were done in the 3D printed bifurcating nerve pathways in which the printing was interrupted halfway through the program resulting in open half-shell geometry. The devices were first pre-coated. Subsequently, 10 μl of hydrogel was added to the distal end of the left path, and UV cross-linked for 10 minutes. Whole DRG were then seeded near the center of the bifurcation point, cultivated over a 3 week period, and the cultures were then imaged. A minimum amount of medium was added such that the liquid height just reached the top of the silicone pathway (2 ml). For experiments done in 35 mm dishes which lacked the silicone pathway, the hydrogel source was oriented to the right of the captured image for subsequent analysis. The orientation of the neurite network was then characterized using ImageJ software by counting the number of axon bundles growing towards the source (referred to as up the gradient) and away from the source (referred to as down the gradient). The fraction of axon outgrowth was calculated as the number of directionally growing axon bundles divided by the total number of bundles. Control studies were done by carrying out repeated experiments which lacked the diffusive NGF gradient. Analysis of the statistical significance in difference of the axon outgrowth between the two groups was done using a two-tailed student t-test.
Sciatic nerve transection and repair model
The experimental protocol was reviewed and approved by the IACUC of the University of Maryland School of Medicine. Male Wistar rats (300 – 350g, Charles River) were used in this study. All animals (n = 3) were maintained in the Animal Facility of the University of Maryland School of Medicine. Both limbs underwent sciatic nerve transection. The right limb was randomly selected to be repaired with the bifurcated nerve guides, which contained no added biochemical functionality, and the left limb was repaired with bifurcated nerve guides which contained path-specific gradients of NGF and GDNF in the respective sensory and motor channels. Sciatic nerve transection and repair were done as we previously described.[71] Prior to implantation, the nerve guides were sterilized via ethylene oxide treatment. Briefly, 1 cm of the sciatic nerve was transected 0.5 cm both proximal and distal to the bifurcation point of the sciatic nerve. The epineurium of all nerve stumps was sutured to the nerve scaffold four times on each side using 9-0 sutures under a microscope. The proximal channel was sutured to the sciatic nerve, the distal sensory channel was sutured to the sural nerve, and the distal motor channel was sutured to the tibial nerve motor braches to the gastrocnemius muscle. The muscular and skin incisions were then closed in layers. The animals had free access to food and water before and after the experiments, and were subjected to a 12 hr day/night cycle in a quiet environment.
Immunohistochemistry
After three months, rats were perfused transcardially with 4% PFA and the nerve graft was harvested. Nerve segments at 2 mm proximal to the distal suture site in both branches (sensory and motor channels) were sectioned at 25 μm on a cryostat. The nerves were washed with PBS three times then permeabilized in 0.2% Triton-X-PBS for 1 hr. After blocking for 1 hr in 5% normal goat serum in PBS, the sections were incubated overnight at 4 °C in mouse monoclonal antibodies to tubulin (Abcam) and rabbit polyclonal antibodies to S-100β (S100, Abcam) diluted in the blocking solution for neurofilament and S100 labeling, respectively. After incubation with the primary antibodies, nerve slices were rinsed in PBS and incubated with secondary antibodies (Alexa 488-conjugated goat anti-mouse antibody; Jackson Immunologicals and Alexa 555-conjugated goat anti-rabbit; Invitrogen) in the blocking solution for 1 hr at room temperature. After incubation with the secondary antibodies, the nerves were rinsed in PBS, mounted in Prolong Gold with DAPI, and subsequently imaged.
Gait analysis
Automated gait analysis was performed 12 weeks after surgery using a CatWalk system (Noldus). All experiments were performed during the same period of the day (1 – 4 p.m.) and analyzed as we previously described.[72] Briefly, we trained rats to cross the CatWalk walkway daily for 7 days before the surgery. During the test, each rat was placed individually in the CatWalk walkway, which consists of a glass plate (100 cm × 15 cm × 0.6 cm), plus two Plexiglas walls. The rat was allowed to walk freely and traverse from one side to the other of the walkway glass plate. Two infrared light beams spaced 90 cm apart were used to detect the arrival of the rat and control the start and end of data acquisition. The recordings were carried out when the room was completely dark, with the exception of the light from the computer screen. An LED light from an encased fluorescent lamp was emitted inside the glass plate and completely internally reflected. When the rat paws made contact with the glass plate, light was reflected down and the illuminated contact area was recorded with a high-speed color video camera positioned underneath the glass plate connected to a computer running Catwalk software v10.5 (Noldus). Comparison was made between the ipsilateral (left) and the contralateral (right) hind paw in each run of each animal at each time point. The duty cycle of the gait is defined as the ratio of stand time divided by the sum of stand and swing time (= stand /[stand + swing]). Stand time is the duration of time that the paw is in contact with the glass, and swing time is the duration of time that the paw is not in contact with the glass. The comparison of catwalk data between two groups was performed using a paired t-test.
Supplementary Material
Acknowledgments
The authors thank Ahmet Höke, Hai-Quan Mao, Winston Soboyejo, Howard Stone, Adam Burns, Richard Register, Kaiyan Qiu, ShuangZhuang Guo, Yifan Zhang, Leanne Young, Junfang Wu and Ruifa Mi for conversations and technical support. We acknowledge the use of the Princeton Institute for the Science and Technology of Materials Imaging and Analysis Center, which is supported by the National Science Foundation (NSF) Materials Research Science and Engineering Centers (MRSEC) Program via the Princeton Center for Complex Materials (Grant No. DMR-0819860). L.E. acknowledges the support of this work by the National Institutes of Health (NIH Grant No. R01NS033506 and Grant No. R01NS060699). X.J. acknowledges the support of this work by the United States National Institutes of Health (Grant No. R01HL118084) and Maryland Stem Cell Research Fund (Grant No. 2013-MSCRFE-146-00). M.C.M. acknowledges support of this work by the Defense Advanced Research Project Agency (DARPA; Award No. D12AP00245) and the Grand Challenges Program at Princeton University.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Prof. Blake N. Johnson, Department of Mechanical and Aerospace Engineering, Princeton University, Princeton, New Jersey 08544, United States, Department of Industrial and Systems Engineering, Virginia Tech, Blacksburg, Virginia 24061, United States
Dr. Karen Z. Lancaster, Department of Molecular Biology and Princeton Neuroscience Institute, Princeton University, Princeton, New Jersey 08544, United States
Dr. Gehua Zhen, Department of Orthopedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, United States
Dr. Junyun He, Department of Neurosurgery, University of Maryland School of Medicine, Baltimore, Maryland 21201, United States
Dr. Maneesh K. Gupta, Department of Mechanical and Aerospace Engineering, Princeton University, Princeton, New Jersey 08544, United States
Yong Lin Kong, Department of Mechanical and Aerospace Engineering, Princeton University, Princeton, New Jersey 08544, United States.
Dr. Esteban A. Engel, Department of Molecular Biology and Princeton Neuroscience Institute, Princeton University, Princeton, New Jersey 08544, United States
Kellin D. Krick, Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD 21205, United States
Alex Ju, Department of Mechanical and Aerospace Engineering, Princeton University, Princeton, New Jersey 08544, United States.
Dr. Fanben Meng, Department of Mechanical and Aerospace Engineering, Princeton University, Princeton, New Jersey 08544, United States
Prof. Lynn W. Enquist, Department of Molecular Biology and Princeton Neuroscience Institute, Princeton University, Princeton, New Jersey 08544, United States
Prof. Xiaofeng Jia, Email: xjia@smail.umaryland.edu, Department of Neurosurgery, Orthopedics, University of Maryland School of Medicine, Baltimore, Maryland 21201, United States, Department of Biomedical Engineering, Anesthesiology and Critical Care Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, United States
Prof. Michael C. McAlpine, Email: mcalpine@umn.edu, Department of Mechanical and Aerospace Engineering, Princeton University, Princeton, New Jersey 08544, United States, Department of Mechanical Engineering, University of Minnesota, Minneapolis, Minnesota 55455, United States
References
- 1.Murphy SV, Atala A. Nat Biotechnol. 2014;32:773. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 2.Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost J-P, Le Net J-L, Baker D, Walley RJ, Everett JR, Nicholson JK. Nature. 2006;440:1073. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 3.Borton D, Micera S, Millán JdR, Courtine G. Sci Transl Med. 2013;5:1. doi: 10.1126/scitranslmed.3005968. [DOI] [PubMed] [Google Scholar]
- 4.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. Adv Mater. 2014;26:3124. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 5.Derby B. Science. 2012;338:921. doi: 10.1126/science.1226340. [DOI] [PubMed] [Google Scholar]
- 6.Uygun BE, Yarmush ML, Uygun K. Nat Rev Gastroenterol Hepatol. 2012;9:738. doi: 10.1038/nrgastro.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volzke H, Schmidt CO, Baumeister SE, Ittermann T, Fung G, Krafczyk-Korth J, Hoffmann W, Schwab M, Meyer zu Schwabedissen HE, Dorr M, Felix SB, Lieb W, Kroemer HK. Nat Rev Cardiol. 2013;10:308. doi: 10.1038/nrcardio.2013.35. [DOI] [PubMed] [Google Scholar]
- 8.Milne CP, Garafalo S, Bryan C, McKiernan M. Nat Rev Drug Discov. 2014;13:324. doi: 10.1038/nrd4325. [DOI] [PubMed] [Google Scholar]
- 9.Hamburg MA, Collins FS. N Engl J Med. 2010;363:301. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 10.Ware T, Simon D, Rennaker RL, Voit W. Polym Rev. 2013;53:108. [Google Scholar]
- 11.Williams JC, Rennaker RL, Kipke DR. Neurocomputing. 1999;26–27:1069. [Google Scholar]
- 12.Kim DH, Ghaffari R, Lu N, Rogers JA. Annu Rev Biomed Eng. 2012;14:113. doi: 10.1146/annurev-bioeng-071811-150018. [DOI] [PubMed] [Google Scholar]
- 13.Xu L, Gutbrod SR, Bonifas AP, Su Y, Sulkin MS, Lu N, Chung HJ, Jang KI, Liu Z, Ying M, Lu C, Webb RC, Kim JS, Laughner JI, Cheng H, Liu Y, Ameen A, Jeong JW, Kim GT, Huang Y, Efimov IR, Rogers JA. Nat Commun. 2014;5:3329. doi: 10.1038/ncomms4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengsbach S, Lantada AD. Biomed Microdevices. 2014;16:617. doi: 10.1007/s10544-014-9864-2. [DOI] [PubMed] [Google Scholar]
- 15.Rengier F, Mehndiratta A, von Tengg-Kobligk H, Zechmann CM, Unterhinninghofen R, Kauczor HU, Giesel FL. Int J Comput Assist Radiol Surg. 2010;5:335. doi: 10.1007/s11548-010-0476-x. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt CE, Leach JB. Annu Rev Biomed Eng. 2003;5:293. doi: 10.1146/annurev.bioeng.5.011303.120731. [DOI] [PubMed] [Google Scholar]
- 17.Kehoe S, Zhang XF, Boyd D. Injury. 2012;43:553. doi: 10.1016/j.injury.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 18.Ray WZ, Mackinnon SE. Exp Neurol. 2010;223:77. doi: 10.1016/j.expneurol.2009.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y-t, Romero-Ortega MI. MRS Bull. 2012;37:573. [Google Scholar]
- 20.Madduri S, Gander B. J Control Release. 2012;161:274. doi: 10.1016/j.jconrel.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 21.Kapur TA, Shoichet MS. J Biomed Mater Res, Part A. 2004;68A:235. doi: 10.1002/jbm.a.10168. [DOI] [PubMed] [Google Scholar]
- 22.Miller C, Jeftinija S, Mallapragada S. Tissue Eng. 2004;8:367. doi: 10.1089/107632702760184646. [DOI] [PubMed] [Google Scholar]
- 23.Hadlock T, Sundback C, Hunter D, Cheney M, Vacanti JP. Tissue Eng. 2004;6:119. doi: 10.1089/107632700320748. [DOI] [PubMed] [Google Scholar]
- 24.Corey JM, Lin DY, Mycek KB, Chen Q, Samuel S, Feldman EL, Martin DC. J Biomed Mater Res, Part A. 2007;83A:636. doi: 10.1002/jbm.a.31285. [DOI] [PubMed] [Google Scholar]
- 25.Pateman CJ, Harding AJ, Glen A, Taylor CS, Christmas CR, Robinson PP, Rimmer S, Boissonade FM, Claeyssens F, Haycock JW. Biomaterials. 2015;49:77. doi: 10.1016/j.biomaterials.2015.01.055. [DOI] [PubMed] [Google Scholar]
- 26.Nectow AR, Marra KG, Kaplan DL. Tissue Eng Pt B. 2012;18:40. doi: 10.1089/ten.teb.2011.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mannoor MS, Jiang Z, James T, Kong YL, Malatesta KA, Soboyejo WO, Verma N, Gracias DH, McAlpine MC. Nano Lett. 2013;13:2634. doi: 10.1021/nl4007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong YL, Tamargo IA, Kim H, Johnson BN, Gupta MK, Koh TW, Chin HA, Steingart DA, Rand BP, McAlpine MC. Nano Lett. 2014;14:7017. doi: 10.1021/nl5033292. [DOI] [PubMed] [Google Scholar]
- 29.Adams JJ, Duoss EB, Malkowski TF, Motala MJ, Ahn BY, Nuzzo RG, Bernhard JT, Lewis JA. Adv Mater. 2011;23:1335. doi: 10.1002/adma.201003734. [DOI] [PubMed] [Google Scholar]
- 30.Gupta MK, Meng F, Johnson BN, Kong YL, Tian L, Yeh YW, Masters N, Singamaneni S, McAlpine MC. Nano Lett. 2015 doi: 10.1021/acs.nanolett.5b01688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bak D. Assembly Autom. 2003;23:340. [Google Scholar]
- 32.Hu SJ. Procedia CIRP. 2013;7:3. [Google Scholar]
- 33.Ladd C, So JH, Muth J, Dickey MD. Adv Mater. 2013;25:5081. doi: 10.1002/adma.201301400. [DOI] [PubMed] [Google Scholar]
- 34.Tabatabai A, Fassler A, Usiak C, Majidi C. Langmuir. 2013;29:6194. doi: 10.1021/la401245d. [DOI] [PubMed] [Google Scholar]
- 35.Cao T, Ho KH, Teoh SH. Tissue Eng. 2003;9:103. doi: 10.1089/10763270360697012. [DOI] [PubMed] [Google Scholar]
- 36.Hollister SJ. Nat Mater. 2005;4:518. doi: 10.1038/nmat1421. [DOI] [PubMed] [Google Scholar]
- 37.Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR. Trends Biotechnol. 2003;21:157. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 38.Fedorovich NE, Alblas J, Wijn JRd, Hennink WE, Verbout AJ, Dhert WJA. Tissue Eng Pt B. 2007;13:1905. doi: 10.1089/ten.2006.0175. [DOI] [PubMed] [Google Scholar]
- 39.Fan H, Lu Y, Stump A, Reed ST, Baer T, Schunk R, Perez-Luna V, Lopez GP, Brinker CJ. Nature. 2000;405:56. doi: 10.1038/35011026. [DOI] [PubMed] [Google Scholar]
- 40.Ahn JH, Kim HS, Lee KJ, Jeon S, Kang SJ, Sun Y, Nuzzo RG, Rogers JA. Science. 2006;314:1754. doi: 10.1126/science.1132394. [DOI] [PubMed] [Google Scholar]
- 41.Rusinkiewicz S, Hall-Holt O, Levoy M. ACM Trans Graphics. 2002;21:438. [Google Scholar]
- 42.Williams LR, Longo FM, Powell HC, Lundborg G, Varon S. J Comp Neurol. 1983;218:460. doi: 10.1002/cne.902180409. [DOI] [PubMed] [Google Scholar]
- 43.Labrador RO, Buti M, Navarro X. Exp Neurol. 1998;149:243. doi: 10.1006/exnr.1997.6650. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Cai H, Cui X, Cao P, Zhang J, Li G, Zhang J. Eur J Cardiothorac Surg. 2014;46:e67. doi: 10.1093/ejcts/ezu296. [DOI] [PubMed] [Google Scholar]
- 45.Pfister BJ, Gordon T, Loverde JR, Kochar AS, Mackinnon SE, Cullen DK. Crit Rev Biomed Eng. 2011;39:81. doi: 10.1615/critrevbiomedeng.v39.i2.20. [DOI] [PubMed] [Google Scholar]
- 46.Heidemann SR, Buxbaum RE. Cell Motil Cytoskelet. 1990;17:6. doi: 10.1002/cm.970170103. [DOI] [PubMed] [Google Scholar]
- 47.Topp KS, Boyd BS. Phys Ther. 2006;86:92. doi: 10.1093/ptj/86.1.92. [DOI] [PubMed] [Google Scholar]
- 48.Schneider F, Fellner T, Wilde J, Wallrabe U. J Micromech Microeng. 2008;18:065008. [Google Scholar]
- 49.Licari JJ, Swanson DW. Materials and Processes for Electronic Applications. Elsevier; 2011. p. 126. [Google Scholar]
- 50.Harrison C, Stafford CM, Zhang W, Karim A. Appl Phys Lett. 2004;85:4016. [Google Scholar]
- 51.Sundback CA, Shyu JY, Wang Y, Faquin WC, Langer RS, Vacanti JP, Hadlock TA. Biomaterials. 2005;26:5454. doi: 10.1016/j.biomaterials.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Miller C, Shanks H, Witt A, Rutkowski G, Mallapragada S. Biomaterials. 2001;22:1263. doi: 10.1016/s0142-9612(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 53.Madison RD, Sofroniew MV, Robinson GA. Neuroscience. 2009;163:213. doi: 10.1016/j.neuroscience.2009.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Höke A, Redett R, Hameed H, Jari R, Zhou C, Li ZB, Griffin JW, Brushart TM. J Neurosci. 2006;26:9646. doi: 10.1523/JNEUROSCI.1620-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wallace DG, Rosenblatt J. Adv Drug Delivery Rev. 2003;55:1631. doi: 10.1016/j.addr.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Nichol JW, Koshy S, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zarzycki R, Modrzejewska Z, Nawrotek K. Ecol Chem Eng S. 2010;17:117. [Google Scholar]
- 58.Lin CC, Metters AT. Adv Drug Delivery Rev. 2006;58:1379. doi: 10.1016/j.addr.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 59.Sutter M, Siepmann J, Hennink WE, Jiskoot W. J Control Release. 2007;119:301. doi: 10.1016/j.jconrel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 60.Vyas A, Li Z, Aspalter M, Feiner J, Hoke A, Zhou C, O’Daly A, Abdullah M, Rohde C, Brushart TM. Exp Neurol. 2010;223:112. doi: 10.1016/j.expneurol.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saijilafu, Hur EM, Liu CM, Jiao Z, Xu WL, Zhou FQ. Nat Commun. 2013;4:2690. doi: 10.1038/ncomms3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao X, Shoichet MS. Neuroscience. 2001;103:831. doi: 10.1016/s0306-4522(01)00029-x. [DOI] [PubMed] [Google Scholar]
- 63.Gundersen RW, Barrett JN. J Cell Biol. 1980;87:546. doi: 10.1083/jcb.87.3.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cornejo M, Nambi D, Walheim C, Somerville M, Walker J, Kim L, Ollison L, Diamante G, Vyawahare S, de Bellard M. Neurochem Res. 2010;35:1643. doi: 10.1007/s11064-010-0225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin YC, Ramadan M, Hronik-Tupaj M, Kaplan DL, Philips BJ, Sivak W, Rubin JP, Marra KG. Ann Plast Surg. 2011;67:147. doi: 10.1097/SAP.0b013e3182240346. [DOI] [PubMed] [Google Scholar]
- 66.Biddis EA, Bogoch ER, Meguid SA. Int J Mech Mater Des. 2005;1:317. [Google Scholar]
- 67.Zhu G, Mallery SR, Schwendeman SP. Nat Biotechnol. 2000;18:52. doi: 10.1038/71916. [DOI] [PubMed] [Google Scholar]
- 68.Curanović D, Ch’ng TH, Szpara M, Enquist L. Current Protocols in Cell Biology. Vol. 43. John Wiley & Sons, Inc; 2009. p. 26.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ch’ng TH, Enquist LW. J Virol. 2005;79:10875. doi: 10.1128/JVI.79.17.10875-10889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ayres C, Bowlin GL, Henderson SC, Taylor L, Shultz J, Alexander J, Telemeco TA, Simpson DG. Biomaterials. 2006;27:5524. doi: 10.1016/j.biomaterials.2006.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewitus D, Vogelstein RJ, Zhen G, Choi YS, Kohn J, Harshbarger S, Jia X. IEEE Trans Neural Syst Rehabil Eng. 2011;19:204. doi: 10.1109/TNSRE.2010.2098047. [DOI] [PubMed] [Google Scholar]
- 72.Zhen G, Wen C, Jia X, Li Y, Crane JL, Mears SC, Askin FB, Frassica FJ, Chang W, Yao J, Carrino JA, Cosgarea A, Artemov D, Chen Q, Zhao Z, Zhou X, Riley L, Sponseller P, Wan M, Lu WW, Cao X. Nat Med. 2013;19:704. doi: 10.1038/nm.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.