Abstract
Native proteins perform an amazing variety of biochemical functions, including enzymatic catalysis, and can engage in protein-protein and protein-DNA interactions that are essential for life. A key question is how special are these functional properties of proteins. Are they extremely rare, or are they an intrinsic feature? Comparison to the properties of compact conformations of artificially generated compact protein structures selected for thermodynamic stability but not any type of function, the artificial (ART) protein library, demonstrates that a remarkable number of the properties of native-like proteins are recapitulated. These include the complete set of small molecule ligand-binding pockets and most protein-protein interfaces. ART structures are predicted to be capable of weakly binding metabolites and cover a significant fraction of metabolic pathways, with the most enriched pathways including ancient ones such as glycolysis. Native-like active sites are also found in ART proteins. A small fraction of ART proteins are predicted to have strong protein-protein and protein-DNA interactions. Overall, it appears that biochemical function is an intrinsic feature of proteins which nature has significantly optimized during evolution. These studies raise questions as to the relative roles of specificity and promiscuity in the biochemical function and control of cells that need investigation.
Keywords: Protein structure, protein interactions, protein conformation, biochemical function, native proteins, Enzymatic active sites
Introduction
Often proteins adopt a unique, thermodynamically stable native conformation that can perform an amazing variety of biochemical functions ranging from enzyme catalysis and signal transduction to force generation 1. When one looks at the diversity of protein functions, one cannot but wonder how they came about. At first glance, the natural tendency is to assume that their remarkable properties mainly arise from evolutionary selection, with the inherent background features that reflect the physical properties of proteins playing a minor role. If so, proteins should exhibit little intrinsic background function, and those that do should be very rare 2– 8. The fundamental problem with this viewpoint is that for selection to occur, there must be some background function on which to select; in practice, low-level function emerges remarkably quickly in function design studies 9– 11. The key issue is how to estimate this random background probability for function. Here, computer experiments can provide important insights 12– 16. For function to occur, often there must be an interaction between molecules. Thus, in what follows, we examine the inherent ability of proteins to engage in small molecule protein-protein and protein-DNA interactions. Surprisingly many biochemical properties of native proteins are found in a library of stable artificial structures generated without any selection for biochemical function. Remarkably, this includes enzymatic active sites and, at much lower frequency, pockets that loosely resemble the enzymatic binding pocket. This suggests that functional selection by evolution is most likely involved in fine-tuning rather than in generation of intrinsic function. If so, marginally stable proteins are inherently ready to engage at low level in the biochemical functions necessary for life.
Generation of an artificial protein library to examine their intrinsic functional features
To separate out the intrinsic properties of proteins from those due to evolution, one could design proteins without selection for function, solve their structures, assay their function, and explore their similarity to native proteins 17– 19. To cover all representative protein functions would be a long, expensive process that is, at present, impractical. Rather, we chose to perform a series of computer experiments 12– 16, where a library of compact homopolypeptides from 40–250 residues in length were generated using the TASSER structure prediction algorithm 20. Then, sequences with protein-like composition were selected by optimizing their thermodynamic stability in the putative fold of interest 13. These artificial proteins are termed the “ART” protein library.
Small molecule ligand-binding pockets
Having the ART library in hand, we compared the small molecule ligand-binding pockets to those in native proteins. Remarkably, all ligand-binding pockets in native proteins have a statistically significant match to the pockets in the ART library. This suggests that the library of all ligand-binding pockets, the “pocketome” 21, is likely complete and arises from defects in packing of compact secondary structures, as proteins without secondary structure have tiny pockets that cannot bind biologically relevant molecules 22. In practice, for single-domain globular proteins, the space of protein pockets is covered by a remarkably small number (about 500) of representative pockets. These results are consistent with a large-scale study on a non-redundant set of ~20,000 known ligand-binding pockets that finds their structural space is crowded, likely complete, and represented by a similar number of pockets 23. Similar protein pockets occur in proteins that have globally unrelated folds. On the other hand, closely related proteins need not have similar pockets. The presence of similar pockets capable of binding similar, if not identical, ligands in multiple protein families rationalizes at least part of the reason why drugs have unintended side effects.
Ability of ART proteins to bind small molecule metabolites
A representative set of 1400 Kyoto Encyclopedia of Genes and Genomes (KEGG) molecules (clustered using Tanimoto coefficient TC=0.7 from a total 12,271 molecules 24) were screened against a representative set of ART proteins using the FINDSITE comb virtual ligand screening algorithm 25. FINDSITE comb has an average success rate of 21% at identifying micromolar or better binders when 50 or fewer small molecules are screened 26. Enrichment factors of the top 1% of ranked ligands relative to a set of 69,271 background molecules (the ZINC 8 library 27) culled with a TC 28 of 0.7 were 2.57, with 98.6% of ligands having an enrichment factor >1 (the random background result). We found that the median number of binding targets per KEGG molecule is 35, quite close to the number (38) of proteins predicted to bind to drugs in the human exome 29. Of these 1400 molecules, 1186 or 84.7% molecules have at least one binding target, and the median number of small molecules that bind per protein is 36 (as compared to 57 drugs per protein, but this discrepancy may be due to the small number of metabolites considered).
We next explored the enrichment factor of metabolites predicted to bind to proteins in a given metabolic pathway. We define the enrichment factor of a pathway as
The average enrichment factor of 238 KEEG pathways is 14.6 with 84.0% of pathways having an E p >1. Thus, there is a significant tendency for metabolites in existing pathways to bind to ART proteins even without any functional selection. As shown in Table 1, the top 18 most enriched pathways by FINDSITE comb include ancient pathways associated with glycolysis 30, the metabolism of ancient amino acids alanine, aspartate, and glutamate 31, 32, and glycerolipid metabolism 33. Thus, a subset of the top 18 pathways is believed to be ancient. However, the ability to bind a molecule is a necessary but insufficient condition for enzymatic activity, an issue we turn to next.
Table 1. Top 18 most enriched pathways by FINDSITE comb.
| Pathway | Enrichment factor | Number of proteins
in the pathway |
|---|---|---|
| Insulin signaling pathway | 77.5 | 67 |
| Alcoholism | 68.0 | 5 |
| Amphetamine addiction | 68.0 | 46 |
| Cocaine addiction | 68.0 | 42 |
| Huntington's disease | 68.0 | 36 |
| Amyotrophic lateral sclerosis (ALS) | 68.0 | 29 |
| GABAergic synapse | 68.0 | 28 |
| Taurine and hypotaurine metabolism | 68.0 | 27 |
| Proximal tubule bicarbonate reclamation | 67.3 | 14 |
| HMG-CoA reductase inhibitors | 59.8 | 4 |
| Galactose metabolism | 56.8 | 20 |
| Phosphotransferase system (PTS) | 54.1 | n.a. |
| Glycerolipid metabolism | 48.7 | 20 |
| Butirosin and neomycin biosynthesis | 45.9 | 3 |
| Glyoxylate and dicarboxylate metabolism | 45.9 | 18 |
| Alanine, aspartate, and glutamate metabolism | 45.6 | 27 |
| Glycolysis/gluconeogenesis | 39.7 | 30 |
| Retrograde endocannabinoid signaling | 39.6 | 23 |
Enzymatic active sites
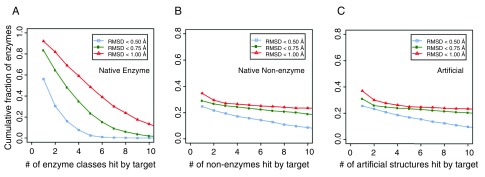
We next explored how special the active sites in enzymes are. To address this question, we undertook a large-scale search for amino acids with similar geometry and same residue identity as in enzyme active sites found in a manually curated set from the Catalytic Site Atlas (CSA) database 34. There, each entry corresponds to a protein chain with an experimentally determined structure in the Protein Data Bank (PDB) 35. In total, we studied 1373 protein chains that are annotated as being enzymes. For each target enzyme, we first detected pockets using a geometry-based method 36. We then scanned these pockets against known active sites of the template library of enzymes 37. If the target had an amino acid arrangement with a similar geometry as the active sites of a template enzyme whose root-mean-square-deviation (RMSD) from that of the known enzyme’s active site <1 Å RMSD and had 100% sequence identity, we considered it a hit. About 94% of the enzymes hit at least one template enzyme that had different first two-digit Enzyme Commission (EC) numbers, i.e. they are from very different enzyme classes. We further counted hits according to their enzyme classes at the four-digit EC level using various RMSD cutoffs ( Figure 1); 75% of target enzymes hit three or more enzyme classes below an RMSD of 1 Å, 54% below a RMSD of 0.75 Å, and 21% below a RMSD of 0.5 Å. Thus, in native proteins, the active sites of enzymes are not as rare nor as geometrically and chemically unique as previously thought; no more than 5000 or so ART structures were searched here.
Figure 1.
Cumulative fraction of enzymes whose active sites match pocket residues in ( A) other classes of enzymes in native structures with different first two digit Enzyme Commission (EC) numbers, ( B) in non-enzymes, and ( C) in ART structures. For each target enzyme, we count the number of alternative enzyme classes that contain at least a hit by the target enzyme at various root-mean-square-deviation (RMSD) cut-offs.
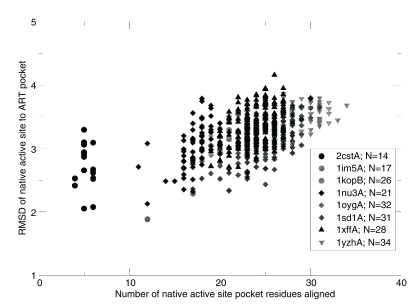
Next, we performed a search of enzyme-like active sites in native structures of non-enzymes ( Figure 1B) and in the ART library ( Figure 1C). From a set of 4609 non-enzymes 23 and a set of the same number of randomly selected artificial structures, we first identified the largest pocket in these structures, then searched in these pockets for residues that resemble active sites in native enzymes. We only considered hits that had a different global structure with a template modeling (TM)-score <0.4 38 (a threshold for structural significance) from a target native enzyme. Using the same criteria, at an RMSD <1 Å and 100% coverage and sequence identity, we found at least a hit for 35% of enzyme active sites in non-enzymes and a comparable value (37%) in artificial structures. For an RMSD <0.75 Å, 29% and 31% of native active sites were matched, respectively. Finally, at an RMSD <0.50 Å, 25% and 26% of native active sites were found for non-enzymes and artificial structures, respectively. Small-size active sites were mostly easy to find a hit: about 88% of three-residue active sites, 35% of four-residue active sites, and 0.3% of five-residue active sites were found in artificial structures. About 25% of enzymes had more than four hits in artificial structures. However, it should be pointed out that the global pockets in these matches usually did not have a significant similarity score to the native active site pocket, despite the high structural similarity of their active site residues. Whether these native non-enzymes could weakly catalyze a similar reaction in a different substrate is unknown, as there are other factors that could dictate enzymatic activity 39. To further investigate this issue, we froze the catalytic residues in the artificial structure of interest and generated sets of stable sequences for the given fold. We then examined whether artificial pockets globally similar to the active pocket in that native enzyme are generated. As shown in Table 2, depending on the particular ART structure, the success rates (p-values of the pockets <0.05) ranged from 0% to 1.5% of the sequences generated. Given a fixed orientation of the active site residues, there are certain backbone geometries that cannot accommodate the native pocket geometry in certain global folds. Consider, for example, a long narrow pocket. Given the location of the active site residues, it might have to penetrate the backbone for the pocket to be completely recapitulated; clearly, in such a situation, that native enzymatic pocket cannot occur. For successful cases, all of which have a globally unrelated fold to the native structure as assessed by their TM-score 40, one need only sample on the order of ~10 4–10 5 random sequences to generate a pocket that is at least weakly related to the native pocket. For these, the RMSDs of the aligned residues versus the number of aligned pocket residues for eight pairs of native enzymes-ART proteins are shown in Figure 2. The range of RMSD values is 2–4 Å and spans 4-35 residues. These pockets have p-values <0.05 associated with the pocket similarity (PS)-score 37. At this range of PS-scores 23, about 13% of ligands share significant chemical similarity as assessed by their TC 28.
Figure 2. Root-mean-square-deviation (RMSD) of native enzymatic to ART pockets versus the number of pocket residues aligned.
The number of residues in the native active site pocket, N, is shown in the figure legend.
Table 2. For a subset of ART proteins containing active site residues and geometrics, % of pockets that have a significant p-value to the native enzymatic pocket.
| Native enzyme | Enzyme Commission
(EC) number |
% (number) of ART
structures whose active site pocket has a p-value <0.05 |
Number of
residues in the native pocket |
Template modeling
(TM)-score of ART template to native 39 |
|---|---|---|---|---|
| 2a8yB-ART1 a | 2.4.2.28 | 0% (0/11,680) | 25 | 0.25 |
| 2cstA-ART1 | 2.6.1.1 | 0.33% (39/11,680) | 14 | 0.23 |
| 2fniA-ART1 a | 2.6.1.92 | 0 (0/11,680) | 45 | 0.25 |
| 1im5A-ART1 | 3.5.1.19 | 0.027% (3/11,520) | 17 | 0.27 |
| 1kopB-ART1 | 4.2.1.1 | 0.21% (24/11,680) | 26 | 0.28 |
| 1nu3A-ART1 | 3.3.2.8 | 0.38% (82/211,440) | 21 | 0.27 |
| 1oygA-ART1 a | 2.4.1.10 | 0.0% (4/11,680) | 32 | 0.23 |
| 1oygA-ART2 | 2.4.1.10 | 0.043% (5/11,680) | 32 | 0.17 |
| 1sd1A-ART1 | 2.4.2.28 | 1.4% (167/11,680) | 31 | 0.23 |
| 1sd1A-ART2 a | 2.4.2.28 | 0 (0/11,520) | 31 | 0.27 |
| 1w23B-ART1 b | 2.6.1.52 | 0% (0/221,280) | 10 | 0.19 |
| 1xffA-ART1 | 2.6.1.16 | 1.8% (211/11,680) | 28 | 0.34 |
| 1yxhA-ART1 | 3.1.1.4 | 0.45% (52/11,520) | 34 | 0.35 |
| 2z2xA-ART1 | 3.4.21.62 | 0% (0/11,680) | 27 | 0.26 |
aNo pockets matched even without the active site residue matching restraint imposed.
b8/221,280 pockets match without the active site residue matching restraint imposed.
ART protein-protein and protein-DNA complexes
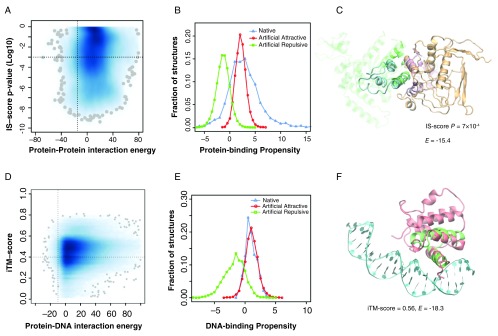
Not only do the ART structures resemble native proteins in terms of fold similarity and ligand-binding pocket but docked ART structures match native protein-protein interfaces, suggesting that the space of protein-protein interfaces is complete and covered by roughly 1000 distinct types of interfaces 15. Interestingly, they also possess the ability to form native-like protein-protein and protein-DNA complexes. To demonstrate this, we randomly selected 30,000 pairs of ART structures in representative native-like folds; each fold had 80 protein-like sequences predicted to be stable for that fold. This gives 192 million pairs of ART monomers. To find possible native-like complexes, a simple yet efficient strategy was adopted. First, we compared the backbone structural similarity of ART monomeric structures with monomeric structures found in a library of 1690 non-redundant native dimeric complexes 41, 42. Using structural alignments, we built putative complexes by superimposing individual ART structures onto their corresponding aligned monomers from the native templates 38. We only considered those putative complexes that had significant global structural similarity and were aligned to more than 50% of the native interface. This yielded 135,942 putative ART complexes, and each had a corresponding native protein complex as its template. As shown in Figure 3A, the vast majority were either energetically unfavorable or did not share significant structural similarity to their corresponding template. However, about 2584 ART monomer pairs, or 1.3×10 -5 of the total, had strongly favorable interactions and shared significant structural similarity with their templates. These ART complexes may be considered native-like. In general, attractive ART interfaces are enriched in hydrophobic residues. The protein-binding propensity scores of attractive ART complexes overlap with the scores of native complexes ( Figure 3B). An example is illustrated in Figure 3C. This ART complex has a favorable interaction energy of -15.4 43 and shares significant interface similarity (IS) at an IS-score p-value of 7×10 -4 with respect to the closest native protein complex 42, 44. Thus, putative native-like protein-protein complexes are found without any selection whatsoever for protein-protein interactions.
Figure 3. Artificial protein-protein, protein-DNA complexes.
( A) Statistics of putative artificial protein-protein complexes. Joint probability density of interaction energy E PP 43 and the p-value of the interface similarity (IS)-score 42, 44 between an artificial complex and its corresponding native template. Darker blue indicates higher density, with the 100 lowest density spots represented by grey spheres. A vertical/horizontal dashed line is placed at E PP = -15 (a cut-off for high likelihood of interaction) and P = 1×10 -3. ( B) Protein-binding propensity scores (>0 implies favorable binding) of native protein-protein interfaces versus putatively attractive ( E PP <-15) and repulsive ( E PP >10) artificial protein-protein interfaces. ( C) Example of an ART protein-protein complex. The complex was built by superimposing two artificial structures (cyan and orange) onto a native dimeric template (Protein Data Bank [PDB] code 2f4m, chain A and B, colored in green and purple). Interface alignment according to iAlign 42. Both structures are shown in line representations, with the non-interfacial regions of the native template shown in transparent mode for clarity. ( D) Statistics of artificial DNA-protein complexes. Joint probability density of DNA-protein interaction energy, E DP 46, and the interfacial template modeling (TM)-score 22 between an ART protein and its corresponding native template. A vertical/horizontal dashed line is placed at E DP = -10 and iTM-score = 0.4. ( E) DNA-binding propensity scores (>0 implies favorable binding) of native DNA-protein interfaces versus putatively attractive ( E DP <-10) and repulsive ( E DP >10) artificial DNA-protein interfaces. ( F) Example of an artificial DNA-protein complex. The complex was built by superimposing the ART structure (red) onto a native template (PDB code 1akh, the native protein and DNA are colored in green and cyan, respectively).
Similarly, we searched for ART structures with a strong native-like DNA-binding propensity. A set of 32,279 ART folds, each with 80 sequences selected for stability, was scanned. As above, we first performed all-against-all structural comparison between individual ART structures and native protein structures found in 1350 experimentally determined protein/DNA complexes 45. The vast majority had either energetically unfavorable DNA-protein interfaces or did not share significant structural similarity with their corresponding native protein templates ( Figure 3D). However, 2515 ART proteins, or 9.7×10 -4 of the total, had strongly favorable interactions and significant structural similarity to DNA-binding templates. These ART proteins may be considered to have native-like DNA-binding function. Analysis of their DNA-binding interface suggests that they have a large number of positively charged Arg and Lys residues, especially Arg, which is enriched at the DNA-binding interface. This is reasonable, as DNA molecules are negatively charged 46. By comparison, DNA-repulsive ART interfaces have a similar sequence composition as native non-DNA-binding surface residues. The DNA-binding propensity scores of DNA-attractive ART structures overlap with the scores of native DNA-binding proteins ( Figure 3E); an example is displayed in Figure 3F. Thus, intermolecular interactions between proteins or involving DNA and proteins could emerge without any selection.
Conclusion
Comparison of the properties of native proteins with those of ART structures selected for stability, but not function, shows that many of the properties seen in native proteins emerge as intrinsic features resulting from the packing of secondary structures. The space of small molecule ligand-binding sites found in native and artificial protein structures is shown to be complete, with about 500 representative pockets. Similarly, pockets can occur in proteins with different global folds, while dissimilar pockets are found in proteins that are closely related by evolution with similar structures. Thus, the geometry and amino acid composition of protein pockets are only weakly coupled to the global fold of a protein. The likelihood that a given small molecule differentially interacts with multiple proteins in different families is high. How nature gets around this promiscuity to generate and control cells is a key unanswered question. If cells operated on the basis of one small molecule-one protein target, it is easy to understand how the organized biochemical processes of life occur, but this is apparently not the case 29. In practice, the situation is possibly more complex.
Remarkably, ART proteins are predicted to bind weakly to a sufficient number of native metabolites that metabolic pathways are enriched relative to what would be expected at random. Moreover, the ART library has significant matches to the active sites and their associated pockets of enzymes in native proteins (which also are found in putative non-enzyme native proteins). Thus, active site geometry is not special, and it appears that a significant fraction of the biochemistry of life, at least at very low level, is encoded in the physical properties of proteins. If this view is true, and these observations need to be experimentally validated, this has significant implications for the origin of life.
Turning to the likelihood of protein-protein and protein-DNA interactions occurring at random, the strong implication is that a tiny fraction of proteins can engage in at least intermolecular interactions without functional selection. Once again, intermolecular interactions emerge as an inherent feature of proteins due to the packing of secondary structures 22. Again, there is the implication of weak omnipresent promiscuous interactions in a cell. How cells sort out the myriad of weak interactions relative to the small fraction of specific ones needs to be better clarified. Part of the answer may lie in subcellular localization.
Overall, these studies suggest that the “special” functional properties of proteins are not as special as commonly viewed. Pockets, enzymatic active sites, and native-like protein-protein and protein-DNA interactions are found in artificial protein structures that are selected for stability and nothing more. The packing of secondary structure is found to provide the geometric context for pockets and intermolecular interfaces. The requirements that a protein be compact and water soluble and adopt a thermodynamically unique conformation give rise to protein sequences that recapitulate the necessary functional features (at least at low level) of real native proteins. Overall, it appears that biochemical function is merely an intrinsic feature of proteins that nature has then significantly optimized.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Ron Elber, Department of Chemistry and Institute for Computational Engineering and Sciences, University of Texas at Austin, Austin, TX, USA
Vajda Sandor, Department of Biomedical Engineering, Boston University, Boston, MA, USA
Funding Statement
This research was supported in part by grant no. GM-48835 of the Division of General Medical Sciences of the National Institutes of Health.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Alberts B, Wilson JH, Hunt T: Molecular biology of the cell. 5th ed. New York, N.Y., Abingdon: Garland Science;2008. Reference Source [Google Scholar]
- 2. Khersonsky O, Malitsky S, Rogachev I, et al. : Role of chemistry versus substrate binding in recruiting promiscuous enzyme functions. Biochemistry. 2011;50(13):2683–90. 10.1021/bi101763c [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 3. Tawfik DS: Messy biology and the origins of evolutionary innovations. Nat Chem Biol. 2010;6(10):692–6. 10.1038/nchembio.441 [DOI] [PubMed] [Google Scholar]
- 4. Khersonsky O, Tawfik DS: Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. 10.1146/annurev-biochem-030409-143718 [DOI] [PubMed] [Google Scholar]
- 5. Khersonsky O, Roodveldt C, Tawfik DS: Enzyme promiscuity: evolutionary and mechanistic aspects. Curr Opin Chem Biol. 2006;10(5):498–508. 10.1016/j.cbpa.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 6. Khersonsky O, Kiss G, Röthlisberger D, et al. : Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc Natl Acad Sci U S A. 2012;109(26):10358–63. 10.1073/pnas.1121063109 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Ben-David M, Elias M, Filippi JJ, et al. : Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1. J Mol Biol. 2012;418(3–4):181–96. 10.1016/j.jmb.2012.02.042 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Bar-Even A, Noor E, Savir Y, et al. : The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50(21):4402–10. 10.1021/bi2002289 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Jürgens C, Strom A, Wegener D, et al. : Directed evolution of a (beta alpha) 8-barrel enzyme to catalyze related reactions in two different metabolic pathways. Proc Natl Acad Sci U S A. 2000;97(18):9925–30. 10.1073/pnas.160255397 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Song G, Lazar GA, Kortemme T, et al. : Rational design of intercellular adhesion molecule-1 (ICAM-1) variants for antagonizing integrin lymphocyte function-associated antigen-1-dependent adhesion. J Biol Chem. 2006;281(8):5042–9. 10.1074/jbc.M510454200 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Pande J, Szewczyk MM, Grover AK: Phage display: concept, innovations, applications and future. Biotechnol Adv. 2010;28(6):849–58. 10.1016/j.biotechadv.2010.07.004 [DOI] [PubMed] [Google Scholar]
- 12. Skolnick J, Gao M, Zhou H: On the role of physics and evolution in dictating protein structure and function. Isr J Chem. 2014;54(8–9):1176–88. 10.1002/ijch.201400013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skolnick J, Gao M: Interplay of physics and evolution in the likely origin of protein biochemical function. Proc Natl Acad Sci U S A. 2013;110(23):9344–9. 10.1073/pnas.1300011110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Gao M, Skolnick J: The distribution of ligand-binding pockets around protein-protein interfaces suggests a general mechanism for pocket formation. Proc Natl Acad Sci U S A. 2012;109(10):3784–9. 10.1073/pnas.1117768109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao M, Skolnick J: Structural space of protein-protein interfaces is degenerate, close to complete, and highly connected. Proc Natl Acad Sci U S A. 2010;107(52):22517–22. 10.1073/pnas.1012820107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skolnick J, Arakaki AK, Lee SY, et al. : The continuity of protein structure space is an intrinsic property of proteins. Proc Natl Acad Sci U S A. 2009;106(37):15690–5. 10.1073/pnas.0907683106 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Korendovych IV, Kim YH, Ryan AH, et al. : Computational design of a self-assembling β-peptide oligomer. Org Lett. 2010;12(22):5142–5. 10.1021/ol102092r [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Sheffler W, Baker D: RosettaHoles: rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 2009;18(1):229–39. 10.1002/pro.8 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Shu JY, Tan C, DeGrado WF, et al. : New design of helix bundle peptide-polymer conjugates. Biomacromolecules. 2008;9(8):2111–7. 10.1021/bm800113g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang C, Liu S, Zhou Y: Accurate and efficient loop selections by the DFIRE-based all-atom statistical potential. Protein Sci. 2004;13(2):391–9. 10.1110/ps.03411904 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Abagyan R, Kufareva I: The flexible pocketome engine for structural chemogenomics. Methods Mol Biol. 2009;575:249–79. 10.1007/978-1-60761-274-2_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brylinski M, Gao M, Skolnick J: Why not consider a spherical protein? Implications of backbone hydrogen bonding for protein structure and function. Phys Chem Chem Phys. 2011;13(38):17044–55. 10.1039/c1cp21140d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao M, Skolnick J: A comprehensive survey of small-molecule binding pockets in proteins. PLoS Comput Biol. 2013;9(10):e1003302. 10.1371/journal.pcbi.1003302 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Kanehisa M, Goto S: KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou H, Skolnick J: FINDSITE comb: a threading/structure-based, proteomic-scale virtual ligand screening approach. J Chem Inf Model. 2013;53(1):230–40. 10.1021/ci300510n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Srinivasan B, Zhou H, Kubanek J, et al. : Experimental validation of FINDSITE comb virtual ligand screening results for eight proteins yields novel nanomolar and micromolar binders. J Cheminform. 2014;6:16. 10.1186/1758-2946-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Irwin JJ, Shoichet BK: ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177–82. 10.1021/ci049714 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Tanimoto TT: An elementary mathematical theory of classification and prediction.1958. Reference Source [Google Scholar]
- 29. Zhou H, Gao M, Skolnick J: Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Sci Rep. 2015;5:11090. 10.1038/srep11090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romano AH, Conway T: Evolution of carbohydrate metabolic pathways. Res Microbiol. 1996;147(6–7):448–55. 10.1016/0923-2508(96)83998-2 [DOI] [PubMed] [Google Scholar]
- 31. Ouzounis C, Kyrpides N: The emergence of major cellular processes in evolution. FEBS Lett. 1996;390(2):119–23. 10.1016/0014-5793(96)00631-X [DOI] [PubMed] [Google Scholar]
- 32. Engel M, Randall P: Amino acids as probes for ancient life in the solar system. Proceedings of spie - the international society for optical engineering. 2006;6309:630907–6309075. 10.1117/12.678793 [DOI] [Google Scholar]
- 33. Caetano-Anollés G, Kim HS, Mittenthal JE: The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Natl Acad Sci U S A. 2007;104(22):9358–63. 10.1073/pnas.0701214104 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Furnham N, Holliday GL, de Beer TA, et al. : The Catalytic Site Atlas 2.0: cataloging catalytic sites and residues identified in enzymes. Nucleic Acids Res. 2014;42(Database issue):D485–9. 10.1093/nar/gkt1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rose PW, Beran B, Bi C, et al. : The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39(Database issue):D392–401. 10.1093/nar/gkq1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang B, Schroeder M: LIGSITE csc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. 10.1186/1472-6807-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Gao M, Skolnick J: APoc: large-scale identification of similar protein pockets. Bioinformatics. 2013;29(5):597–604. 10.1093/bioinformatics/btt024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, Skolnick J: TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–9. 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tian W, Arakaki AK, Skolnick J: EFICAz: a comprehensive approach for accurate genome-scale enzyme function inference. Nucleic Acids Res. 2004;32(21):6226–39. 10.1093/nar/gkh956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y, Skolnick J: The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci U S A. 2005;102(4):1029–34. 10.1073/pnas.0407152101 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Chen H, Skolnick J: M-TASSER: an algorithm for protein quaternary structure prediction. Biophys J. 2008;94(3):918–28. 10.1529/biophysj.107.114280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao M, Skolnick J: iAlign: a method for the structural comparison of protein-protein interfaces. Bioinformatics. 2010;26(18):2259–65. 10.1093/bioinformatics/btq404 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Lu H, Lu L, Skolnick J: Development of unified statistical potentials describing protein-protein interactions. Biophys J. 2003;84(3):1895–901. 10.1016/S0006-3495(03)74997-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao M, Skolnick J: New benchmark metrics for protein-protein docking methods. Proteins. 2011;79(5):1623–34. 10.1002/prot.22987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao M, Skolnick J: A threading-based method for the prediction of DNA-binding proteins with application to the human genome. PLoS Comput Biol. 2009;5(11):e1000567. 10.1371/journal.pcbi.1000567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao M, Skolnick J: DBD-Hunter: a knowledge-based method for the prediction of DNA-protein interactions. Nucleic Acids Res. 2008;36(12):3978–92. 10.1093/nar/gkn332 [DOI] [PMC free article] [PubMed] [Google Scholar]