Highlight
CHIL is essential for the accumulation of proanthocyanidins and flavonols in seeds of Arabidopsis and it functions as a unique enhancer in the flavonoid pathway.
Key words: Arabidopsis, chalcone isomerase-like, flavonoids, flavonols, proanthocyanidins, seed coat.
Abstract
Flavonoids are important natural products for plant defence and human health. Although almost all the flavonoid pathway genes have been well-documented by biochemical and/or genetic approaches, the role of the Arabidopsis chalcone isomerase-like (CHIL) gene remains unclear. Two chil mutants with a seed colour similar to that of wild-type Arabidopsis have been identified here, but in sharp contrast to the characteristic transparent testa seed phenotype associated with other known flavonoid pathway genes. CHIL loss-of-function mutations led to a strong reduction in the proanthocyanidin and flavonol levels in seeds, but not in the anthocyanin levels in leaves. CHIL over-expression could partially recover the mutant phenotype of the chil mutant and increased both proanthocyanidin and flavonol accumulation in wild-type Arabidopsis. However, the CHIL gene could not rescue the mutant phenotype of TT5 that encodes the intrinsic chalcone isomerase in Arabidopsis. Parallel phenotypical and metabolic analyses of the chil, tt5, chs, and f3h mutants revealed that, genetically, CHIL functions at the same step as TT5. Moreover, it is demonstrated that CHIL co-expresses, co-localizes, and interacts with TT5 in Arabidopsis for flavonoid production. Based on these genetic and metabolic studies, it is concluded that CHIL functions with TT5 to promote flavonoid production, which is a unique enhancer in the flavonoid pathway.
Introduction
Flavonoids are a large group of natural products that are widely present in the leaves, flowers, fruits, and seeds of various plants. Based on their basic chemical structures, flavonoids can be further divided into different subgroups, including the three major subgroups of flavonols, anthocyanins, and proanthocyanidins (PAs) (Winkel-Shirley, 2001; Lepiniec et al., 2006). Flavonoids play important roles in many aspects of plant development and response to stress, including UV light protection, auxin transport, and resistance to pathogens (Lattanzio et al., 2006; Ferreyra et al., 2012; Hassan and Mathesius, 2012). Meanwhile, flavonoids are also known to have beneficial effects on human health, such as anti-viral, antioxidant, and anticancer activities and they are also known to lower the incidence of cardiovascular disease, obesity, and diabetes (Yao et al., 2004; Lee et al., 2007; Kozlowska and Szostak-Wegierek, 2014). Given their importance in both plant physiology and human nutrition, it is not surprising that there has been a great deal of research efforts focused on the flavonoid biosynthetic pathway in a range of plant species over the last several decades.
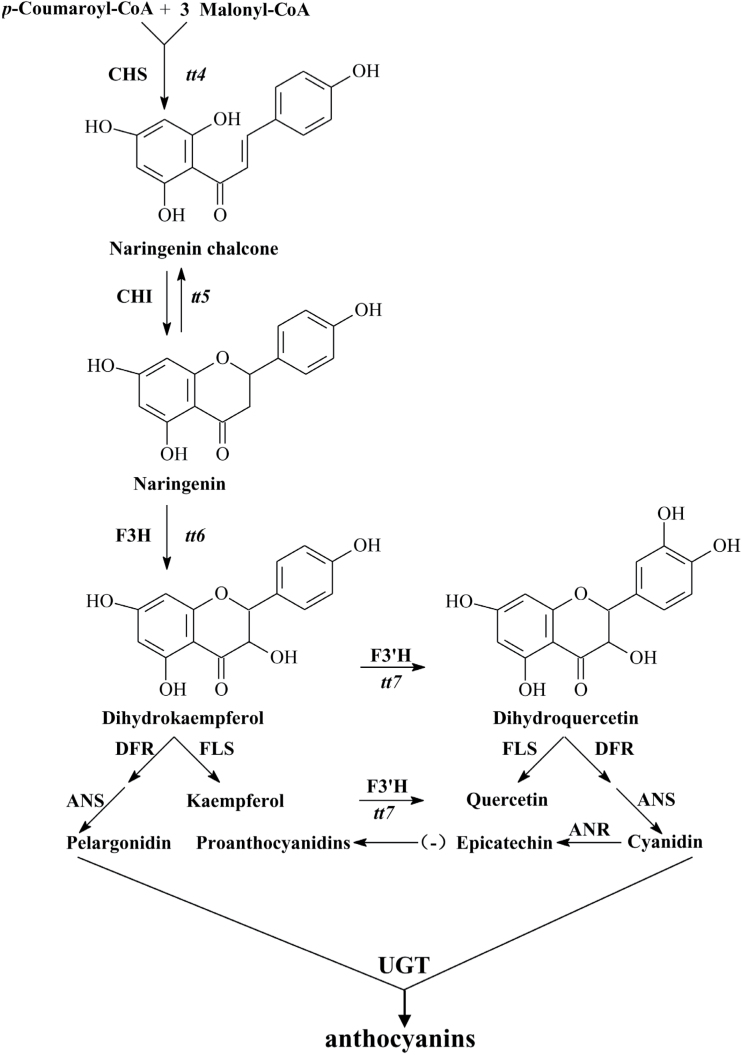
In the core flavonoid pathway, chalcone synthase (CHS, encoded by the TT4 locus) catalyses the first committed step to form 4,2′,4′,6′-tetrahydroxychalcone (naringenin chalcone), by the stepwise condensation of three molecules of malonyl-CoA with one molecule of p-coumaroyl-CoA. Chalcone isomerase then catalyses the isomerization of naringenin chalcone into 5,7,4′-trihydroxyflavanone (naringenin), which is considered to be the earliest intermediate with the flavonoid core. Naringenin can be hydroxylated by flavanone 3-hydroxylase (F3H) to form dihydrokaempferol, and dihydrokaempferol can be further hydroxylated by flavanone 3′-hydroxylase (F3′H) to form dihydroquercetin. These intermediates are further converted to flavonols, anthocyanins, and PAs through distinct branches of the flavonoid pathway (Fig. 1). Flavonols, anthocyanins, and PAs are three major subgroups of flavonoid molecules known to occur in Arabidopsis thaliana, where they accumulate in an organ-specific manner (Lepiniec et al., 2006). Among them, flavonols are accumulated in almost all organs, including leaves and seeds, whereas anthocyanins are present in all organs with the exception of the immature seeds. Unlike flavonols and anthocyanins, PAs and their precursor (-)-epicatechin are specifically accumulated in the seed coat (Lepiniec et al., 2006; Routaboul et al., 2006; Saito et al., 2013). PAs can react with p-dimethylaminocinnamaldehyde (DMACA) to form a blue to purple stain (Li et al., 1996). Typically, mutations in the genes encoding the enzymes of the core flavonoid pathway usually lead to transparent testa seed phenotypes in Arabidopsis due to the lack of oxidized flavonoid compounds (mainly PAs).
Fig. 1.
Flavonoid biosynthetic pathway in A. thaliana. Abbreviations: ANR, anthocyanidin reductase; ANS, anthocyanidin synthase; CHI, chalcone isomerase; CHS, chalcone synthase; DFR, dihydroflavonol 4-reductase; F3H, flavanone 3-hydroxylase; F3′H, flavanone 3′-hydroxylase; FLS, flavonol synthase; tt, the corresponding transparent testa mutants; UGT, UDP-glucosyltransferase.
The chalcone isomerase-like protein (CHIL, At5g05270) is probably the least well-documented among the potential functional proteins in the flavonoid pathway of Arabidopsis; only a few characteristics of CHIL have been reported, often in parallel with studies of TT5 (At3g55120). Both CHIL and TT5 belong to the CHI super-family that comprises four subclasses in Arabidopsis. Type I CHI proteins are ubiquitous in vascular plants and are responsible for the formation of 5,7,4′-trihydroxyflavanone. Type II CHIs are mostly found in leguminous plants and catalyse the formation of 5-deoxy(iso)flavonoids (Shimada et al., 2003; Ralston et al., 2005; He et al., 2011). Both Type I and II CHI proteins are bona fide enzymes with catalytic activity. Type III CHIs, of which there are three in Arabidopsis, were recently shown to be fatty acid-binding proteins (FAPs) that are present widely in land plants and green algae (Ngaki et al., 2012). There is only one member of Type IV CHI proteins in Arabidopsis known as CHIL, and it shares less than 30% identity with TT5 at the amino acid level. Type IV CHI proteins are only found in land plants (Ralston et al., 2005; Ngaki et al., 2012).
In a previous study that used comprehensive flavonoid profiling and transcriptome co-expression analyses, the CHIL gene was found to be co-expressed with 9 out of the 19 fully characterized flavonoid pathway genes (Yonekura-Sakakibara et al., 2008), including Anthocyanidin Reductase (ANR), a PA-specific pathway gene (Xie et al., 2003). Similarly, in several other studies, CHIL was also found to be co-expressed with one or more flavonoid pathway genes, including Flavonol Synthase (FLS) and/or TT5 (Ma, 2002; Oravecz et al., 2006; Pan et al., 2009). These data indicated that CHIL is an expressed gene in Arabidopsis and that it is potentially associated with flavonoid biosynthesis. To date, many of the known mutants of flavonoid pathway genes, with the exception of FLSs (Wisman et al., 1998) and glycosyltransferase genes required for the formation of flavonol glycosides and anthocyanins, exhibited a transparent testa seed phenotypes with a yellow or pale brown seed coat colour (Saito et al., 2013). However, no transparent testa mutation has been shown to be associated with a loss-of-function of CHIL and, importantly, no in vitro catalytic activity has been reported for CHIL (Jez et al., 2000; Ngaki et al., 2012). Therefore, at present, neither the genetic nor the biochemical role(s) of CHIL is clear.
In the present study, it was found that the expression level of the CHIL gene was down-regulated in a tt2 mutant that failed to produce PAs, implying that CHIL is involved in PA biosynthesis. Mutants for CHIL in Arabidopsis were further identified using a reverse genetics approach; interestingly, the seeds exhibited a brown seed colour phenotype similar to the wild-type. CHIL was not able to rescue the tt5 phenotype, but it did enhance the accumulation of flavonol and PA compounds in the presence of TT5. Moreover, the CHIL protein co-expresses and co-localizes with TT5, which implies that CHIL may enhance flavonoid production by co-operation with TT5 in Arabidopsis. Our results indicate a functional role for CHIL gene with a distinct characteristic different from other known genes in the flavonoid pathway.
Materials and methods
Plant materials and growth conditions
The Arabidopsis mutants chil-1 (SALK_096551C, Alonso et al., 2003), chil-2 (CS418063, Rosso et al., 2003), chi (tt5, SALK_034145, Alonso et al., 2003), chs (tt4, CS66119, Zhang et al., 2010), and tt2 (SALK_005260 and CS83, Koornneef, 1981; Alonso et al., 2003) were obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus, OH, USA). The tt8 mutant (SALK_063334, Alonso et al., 2003) was a gift from Professor Lixi Jiang (Zhejiang University, Hangzhou, China). The ttg1-9 (Pang et al., 2009) and tt2 (CS83) mutants are in the Landsberg erecta background, other mutants and transgenic Arabidopsis plants are in the Col-0 background. Arabidopsis seeds were surface-sterilized with 10% bleach and stratified on plates with 1/2 strength Murashige and Skoog (1/2 MS) medium containing 5% (w/v) sucrose. For the observation of anthocyanins in 4-d-old seedlings, the seeds were kept in the dark at 4 °C for 3 d and then transferred to a tissue culture room at 22 °C under constant light (150 μmol photons m–2 s–1) for 4 d. For the flavonoid analysis and DMACA staining, plants were grown at 22 °C with 16/8h light/dark cycles. The seeds were harvested when the plants were completely mature and were dried at 37 °C for 7 d after harvest. For the analysis of anthocyanin content, the seeds were stratified on plates with 1/2 MS medium containing 1% (w/v) sucrose in the dark at 4 °C for 3 d, and then transferred to a tissue culture room at 22 °C under constant light (40 μmol photons m–2 s–1) for 14 d. These seedlings were transferred to plates with 1/2 MS medium containing 12% (w/v) sucrose and grown under constant light (80 μmol photons m–2 s–1) for 3 d (Yonekura-Sakakibara et al., 2012). For Norflurazon treatment, seeds were stratified on plates with 1/2 MS medium containing 100 μM Norflurazon in the dark at 4 °C for 3 d, and grown at 22 °C with 16/8h light/dark cycles for 4 d. For the detection of flavonol content, sterile seeds were grown on MS medium lacking sucrose. The plates were kept for 5 d in the dark at 4 °C and the seedlings were grown in a tissue-culture room at 22 °C under constant light (100 μmol photons m–2 s–1) for 21 d.
Microarray hybridization and data analysis
The seedlings of wild-type (Ler) and tt2 (CS83) Arabidopsis were grown under the conditions described above. The flowers were tagged with different coloured tape to indicate the days after pollination. The siliques (6 d after pollination) were conducted for Ler and CS83 in the microarray experiment with three independent biological replicates. Probe labelling and chip hybridization followed the procedures suggested by the manufacturer (http://www.affymetrix.com/support/technical/manual/expr-ession_manual.affx). Statistical analysis of microarray data and gene selection followed the procedures detailed in Pang et al. (2008). All microarray data have been deposited in ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under the accession number E-MTAB-3659.
Identification of the chil T-DNA insertion mutants
Identification of the Arabidopsis T-DNA insertion mutants was performed as previously described (Jiang and Yu, 2009). Homozygous plants of the chil mutant were identified by two round PCRs. In the first round PCR, a pair of CHIL-specific primers U1 (forward, 5′-TGAGTGGTTGTGGACTGAGTG-3′) and U2 (reverse, 5′-AAGTGATGATCTGTGGAGGA-3′) were designed to anneal to a location outside the T-DNA insertion in order to ensure the production of a band with an expected size in the non-homozygosity. In the subsequent PCR, the T-DNA border primer (5′-ATTTTGCCGATTTCGGAAC-3′) and primer U1 were used. Homozygous chil-2 T-DNA insertion lines were obtained by PCR screening using primer U2 (forward, 5′-AGTTCACTGCGATCGGAGTT-3′) and L2 (reverse, 5′-TCCTCAGCCAAACGATCTCT-3′). To confirm the nature and location of the T-DNA insertion, the PCR products were sequenced. To remove additional T-DNA loci or mutations from the chil mutants, backcrosses to wild-type plants were performed, and plants homozygous for the T-DNA insertion were identified again.
Gene expression analysis
Total RNA preparation, first-strand cDNA synthesis, and qRT-PCR were performed as previously described (Jiang et al., 2013), with minor modifications. The DNase I-treated total RNA (2 μg) was denatured and subjected to reverse transcription using Moloney Murine Leukemia Virus Reverse Transcriptase (Promega). Semi-quantitative RT-PCR was carried out on Applied Biosystems Veriti Thermal Cycler. The PCR conditions were 94 °C for 3min; 28 cycles of 94 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s; followed by a final extension of 72 °C for 10min. Quantitative RT-PCR analyses were carried out on Mx3000P (Stratagene) by using the SYBR Green reagent (Kapa) according to the manufacturer’s instructions. The ACTIN2 gene was used as a control in the qRT-PCR in Fig. 2. The UBQ10 gene was used as a control in the other qRT-PCRs. The primer sequences used for qRT-PCR were all listed in Supplementary Table S2 at JXB online. Data were calculated from three biological replicates, and each biological replicate was examined in triplicate.
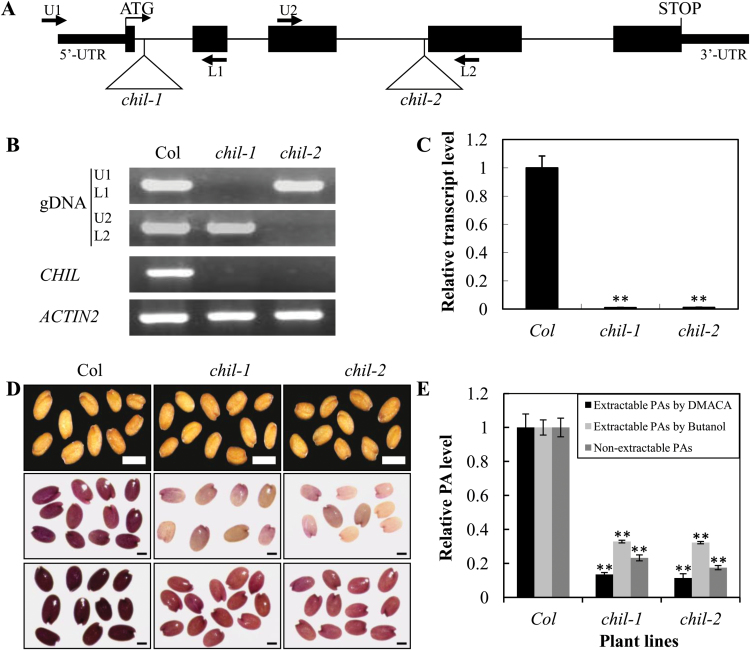
Fig. 2.
Characterization of the two chil mutants. (A) Diagram of CHIL gene structure [thick lines, untranslated regions (UTRs); thin lines, introns; black boxes, exons] with the relative position of the two T-DNA insertions corresponding to the chil-1 and chil-2 mutants. The positions of the U1/L1 and U2/L2 primer pairs used in the screening of homozygous lines are shown, respectively. (B) Molecular characterization of the two chil mutant lines at the genomic (confirmation by using U1/L1 and U2/L2 primer pairs for the chil-1 and chil-2 lines, respectively) and transcript levels (by using semi-quantitative RT-PCR). (C) Relative transcript levels of the CHIL gene in 4-d-old siliques detected by qRT-PCR analysis. Data are presented as mean ±SD, Student’s t test (n=3, *P <0.05, **P <0.01). (D) Seed phenotype of the chil mutants, compared with the wild-type Arabidopsis ecotype Col. Seed phenotype before staining with DMACA (upper). Bars=0.5mm. Seed phenotype after staining with DMACA for 2 d (middle) and 20 d (lower), respectively. Bars=0.2mm. (E) Relative proanthocyanidin levels in seeds of Col and chil mutants detected by different methods. The extractable PAs measured by DMACA (2.64mg epicatechin equivalence g–1) and by butanol (1.08mg B2 equivalence g–1); non-extractable PAs (2.50mg B2 equivalence g–1) in Col were set as a value of 1.0, respectively. Data are presented as mean ±SD, Student’s t test (n=3, *P <0.05, **P <0.01). (This figure is available in colour at JXB online.)
Generation of transgenic plants
To produce constitutively CHIL-expressing Arabidopsis plants, the 630-bp CDS (coding sequence) fragment was amplified by PCR with primers CHILCF and CHILR and then cloned into the Gateway Entry vector pENTR/D/TOPO (Invitrogen) and confirmed by sequencing. For stable transformation by Agrobacterium tumefaciens, the CDS of CHIL was first transferred into the Gateway plant transformation destination vector pB2GW7 (Karimi et al., 2002) by performing an LR recombination reaction with the resulting entry vector pENTR-CHIL according to the manufacturer’s instructions (Invitrogen). The resulting pB2GW7-CHIL vector was then transformed into A. tumefaciens strain GV3101 for Arabidopsis transformation. In the same way, the 1742-bp promoter fragment of CHIL was cloned into entry vector with primers CHILPF and CHILPR and transferred to vector pBGWFS7 for GUS staining. The resulting vector pBGWFS7-CHIL was transformed into A. tumefaciens strain GV3101 for Arabidopsis transformation. Transformation of Arabidopsis was performed by the floral dip method (Clough and Bent, 1998). T2 generation seeds were germinated on plates with MS containing 2.5mg ml–1 phosphothricin, and the resistant seedlings were transferred to soil to obtain homozygous T3 generation seeds. For more detailed phenotypic analysis, three independent T3 generation homozygous lines were used for further analysis.
Purification and detection of recombinant proteins
The recombinant CHIL and TT5 proteins were obtained by cloning its ORF into the expression vector pQE-30 (Qiagen), followed by transformation into E. coli strain M15. The primers used were CHILBF and CHILSR, and TT5BF and TT5BR (see Supplementary Table S2 at JXB online). The soluble histidine (His)-tagged fusion protein was purified using Ni-NTA affinity resin (Qiagen) according to the manufacturer’s instruction. The reaction were conducted at 25 °C for 10min in a 100 µl-volume reaction with the following components: 50mM TRIS-HCl buffer (pH 7.6) containing 1% ethanol, 100 µM naringenin chalcone, and 10 µg recombinant protein. The reaction with the same above-mentioned components but 10 µg boiled recombinant protein was used as the control.
Enzymatic reactions were stopped with 100 µl methanol, followed by centrifugation at 12 000rpm for 30min. One-hundred microlitre samples were then run on an HPLC 1260 (Agilent) system with an Eclipse XDB-C18 reverse phase column (4.6×150mm, particle size 5 µm). Compounds were separated with a linear eluting gradient (5–70% solvent B over 30min) with solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) at flow rate of 1ml min–1. A photodiode array detector (Agilent) was used for the detection of UV-visible absorption from 190–600nm.
Extraction and quantification of anthocyanins, flavonols, and proanthocyanidins
Anthocyanins were extracted from leaf tissue with methanol containing 0.1% HCl, followed by the addition of the same amount of water and chloroform to remove chlorophyll, and then measured spectrophotometrically at 530nm. The anthocyanin level was calculated and compared with the wild-type control Col in biological triplicates (1.57mg g–1 dry weight in the wild-type control Col was set as a value of 1.0 in Fig. 3B and in Supplementary Fig. S4 at JXB online).
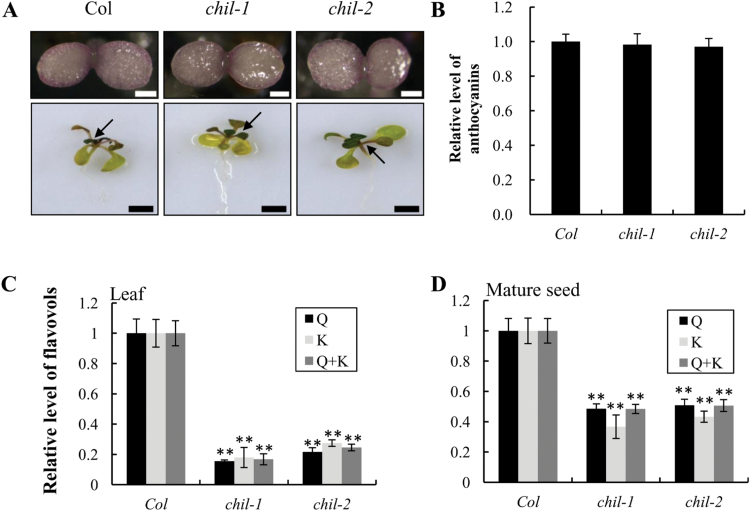
Fig. 3.
Flavonoid accumulation in the chil mutants. (A) Anthocyanin accumulation in the 4-d-old Arabidopsis seedlings (upper, bars=0.5mm) and 17-d-old seedlings (bottom, bars=5mm). (B) Relative level of anthocyanins in the 17-d-old seedlings of Col and the chil-1 and chil-2 mutants. The level of anthocyanins in the wild-type control Col (1.57mg g–1 dry weight) was set as a value of 1.0. (C, D) Relative level of the major flavonols [quercetin (Q) and kaempferol (K) aglycones] in 21-d-old seedlings (C) and mature seeds (D) of Col and the chil-1 and chil-2. Levels of quercetin (4.79 μg g–1 dry weight and 584.27 μg g–1 dry weight in seedlings and seeds, respectively) and kaempferol (4.90 μg g–1 dry weight and 11.38 μg g–1 dry weight in seedlings and seeds, respectively) from wild-type control Col were set as a value of 1.0. Data are presented as mean ±SD, Student’s t test (n=3, *P <0.05, **P <0.01). (This figure is available in colour at JXB online.)
Flavonols were extracted with 80% methanol overnight, the extract was hydrolysed with the same amount of 6 N HCl at 70 °C for 40min, followed by the addition of the same amount of methanol. Twenty to forty microlitres of the extract was run on the HPLC with the same conditions as mentioned above. Flavonols standards were purchased from the Shanghai Tongtian Biotechnology Company. Levels of quercetin (4.79 µg g–1 dry weight and 584.27 µg g–1 dry weight in seedlings and seeds, respectively) and kaempferol (4.90 µg g–1 dry weight and 11.38 µg g–1 dry weight in seedlings and seeds, respectively) from the wild-type control Col were set as a value of 1.0 in Figs 3C, D, 4D, E, and 6D, E).
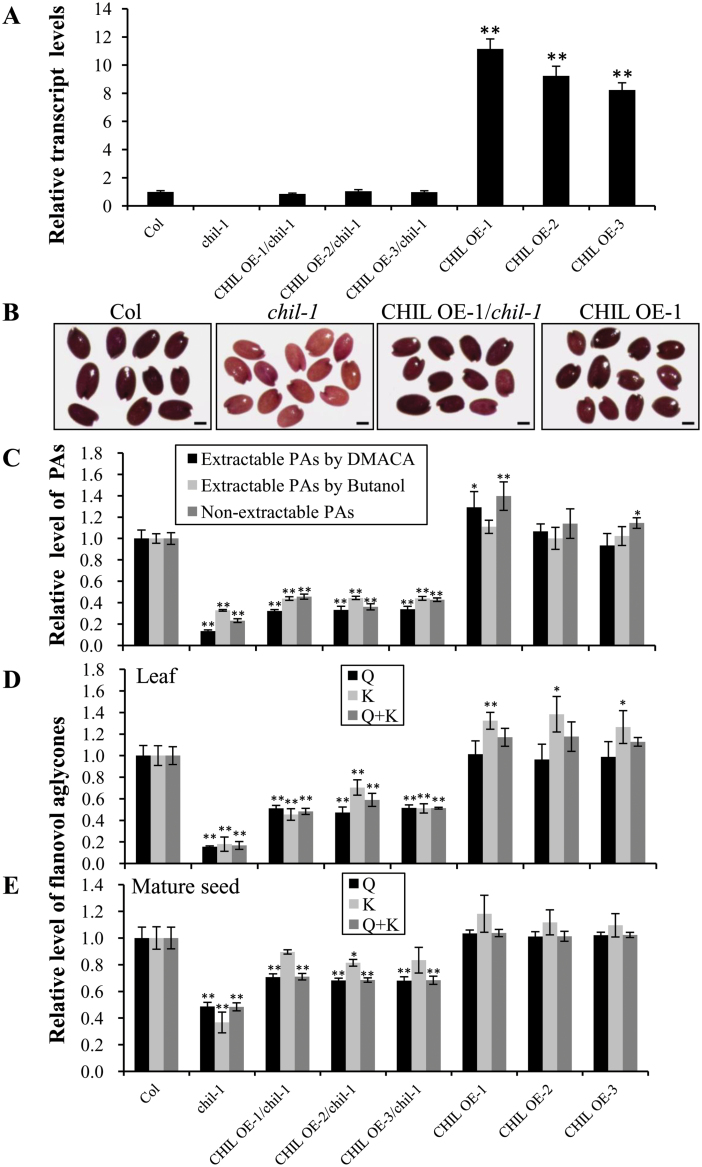
Fig. 4.
Over-expression of the CHIL gene in the chil and Col background. (A) Relative transcript levels of the CHIL gene in the Col wild-type, chil-1, three CHIL OE/chil-1 lines, and three CHIL OE lines detected by qRT-PCR analysis. (B) Seed phenotypes of Col, chil-1, CHIL OE-1/chil-1, and CHIL OE-1 after staining with DMACA for 20 d. Bars=0.2mm. (C) Relative level of proanthocyanidins in mature seeds of Col, chil-1, three CHIL OE/chil-1 lines, and three CHIL OE lines. The extractable PAs measured by DMACA (2.64mg epicatechin equivalence g–1) and by butanol (1.08mg B2 equivalence g–1); non-extractable PAs (2.50mg B2 equivalence g–1) in Col were set as a value of 1.0, respectively. (D, E) Relative levels of flavonols [quercetin (Q) and kaempferol (K) aglycones] in 21-d-old seedlings (D) and mature seeds (E) of Col, chil-1, three CHIL OE/chil-1 lines, and three CHIL OE lines. Levels of quercetin (4.79 µg g–1 dry weight and 584.27 µg g–1 dry weight in seedlings and seeds, respectively) and kaempferol (4.90 µg g dry weight and 11.38 µg g–1 dry weight in seedlings and seeds, respectively) from wild-type control Col were set as a value of 1.0. (A, C, D, E) Data are presented as mean ±SD, Student’s t test (n=3, *P <0.05, **P <0.01). (This figure is available in colour at JXB online.)
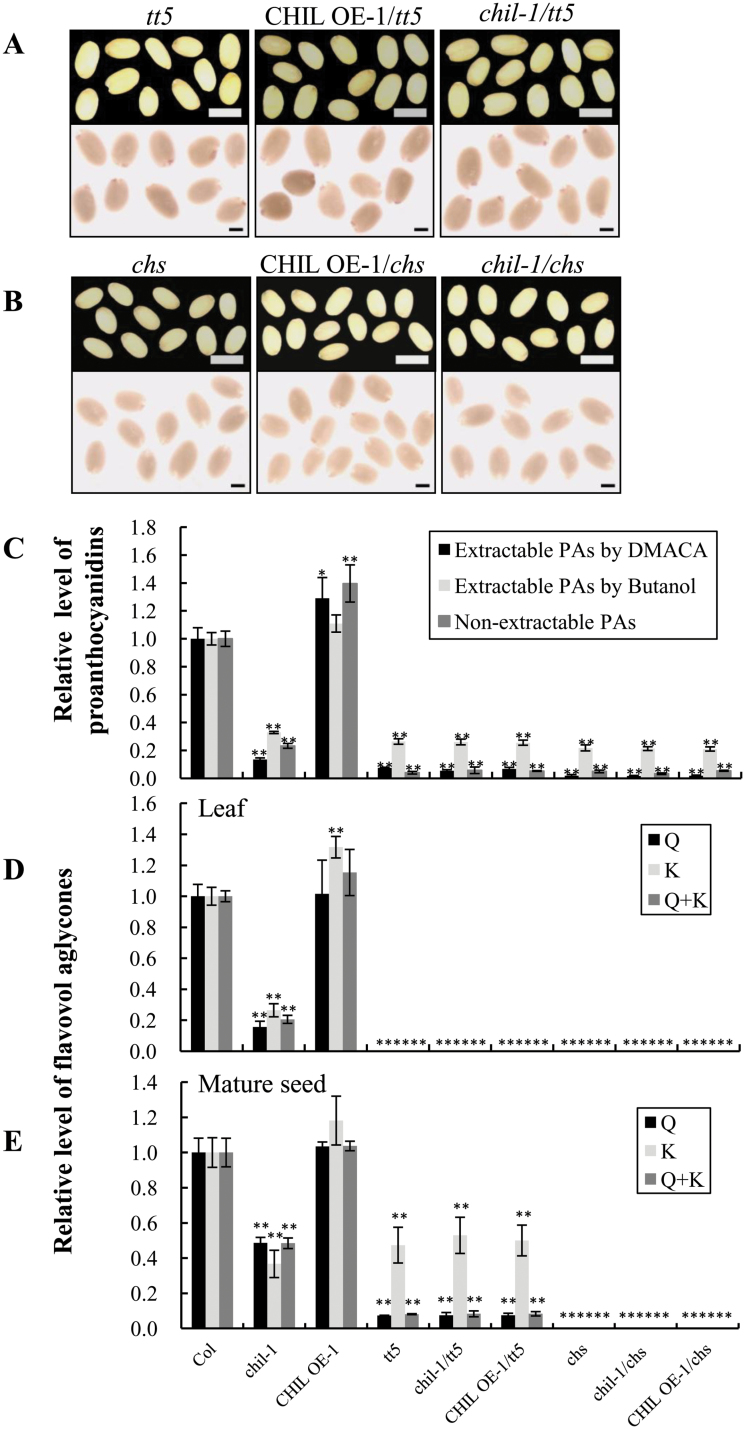
Fig. 6.
Characteristics of CHIL knockout and over-expression in the tt5 and chs background. (A) Seed phenotype of the lines tt5, chil-1/tt5, CHIL OE-1/tt5 (upper), stained with DMACA (lower). Bars=0.5mm. (B) Seed phenotype of chs, chil-1/chs, CHIL OE-1/chs (upper), stained with DMACA (lower). Bars=0.5mm. (C) Relative levels of proanthocyanidins in mature seeds of the above-mentioned plants. The extractable PAs measured by DMACA (2.64mg epicatechin equivalence g–1) and by butanol (1.08mg B2 equivalence g–1); non-extractable PAs (2.50mg B2 equivalence g–1) in Col were set as a value of 1.0, respectively. (D, E) Relative levels of the major flavonols [quercetin (Q) and kaempferol (K) aglycones] in 21-d-old seedlings (D) and mature seeds (E) of the above-mentioned plants. Levels of quercetin (4.79 µg g–1 dry weight and 584.27 µg g–1 dry weight in seedlings and seeds, respectively) and kaempferol (4.90 µg g–1 dry weight and 11.38 µg g–1 dry weight in seedlings and seeds, respectively) from wild-type control Col were set as a value of 1.0. (C, D, E) Data are presented as mean ±SD, Student’s t test (n=3, *P <0.05, **P <0.01). (This figure is available in colour at JXB online.)
For proanthocyanidin staining, mutants and wild-type seeds were stained with 0.1% DMACA reagent in methanol:6 N HCl (1:1 v/v) for different time points. Extractable proanthocyanidins were extracted from the powder of 100mg of dry seeds three times with 70% acetone containing 0.5% acetic acid, three times with chloroform, and twice with hexanes; the aqueous phase was then freeze-dried and resuspended in 70% acetone containing 0.5% acetic acid. The proanthocyanidin levels were determined by reaction with 0.2% DMACA (methanol:6 N HCl=1:1) at 640nm. The extractable PA level in seeds of the wild-type control Col (2.64mg epicatechin equivalence g–1) was set as a value of 1.0. The extractable PA levels were also measured by heating with butanol/HCl (95:5 v/v) at 550nm (Pang et al., 2007), and the PA level (1.08mg B2 equivalence g–1) in the wild-type control Col was set as a value of 1.0. Non-extractable PAs were measured by heating with butanol/HCl and determining the absorption at 550nm. The non-extractable PA level in the wild-type control Col (2.50mg B2 equivalence g–1) was set as a value of 1.0. All the assays were conducted in triplicate, and the relative levels were compared with that of the Col control as a value of 1.0 in Figs 2E, 4C, and 6C).
Promoter analysis with GUS staining
Arabidopsis tissue for GUS staining was immersed in 90% ice-cold acetone for 20min and then incubated in GUS staining solution (100mM sodium phosphate, pH 7.0; 10mM EDTA; 0.5mM potassium ferricyanide; 0.5mM potassium ferrocyanide; 1mM 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid; 0.1% (v/v) Triton X-100) at 37 °C. Tissues were cleared in 70% ethanol before observation.
Transient expression of CHIL-GFP and TT5-GFP fusion proteins in Arabidopsis leaf mesophyll protoplasts
The coding sequences of CHIL and TT5 genes were amplified with primer pairs CHIL-1302-F/CHIL-1302-R for CHIL and TT5-1302-F/TT5-1302-R for TT5, respectively. The restriction sites NcoI and SpeI were introduced into the PCR products of both CHIL and TT5. The PCR products were digested with NcoI and SpeI, and cloned into the pCAMBIA1302 plasmid digested with the same enzymes. After confirmation by sequencing, the recombinant constructs were used in protoplast transformation, and pCAMBIA1302 was used as the positive control.
Isolation and PEG-mediated transformation of Arabidopsis protoplasts by the constructs mentioned above were as described by Sheen (2002). After incubation for 16-24h, GFP fluorescence in Arabidopsis protoplast cells was detected by laser scanning confocal microscopy using Leica TCS SP5. The emission from 500nm to 560nm was collected for GFP and from 605nm to 700nm for chlorophyll.
Arabidopsis gene IDs studied
Gene IDs in the Arabidopsis Genome Initiative database are as follows: ACTIN2 (At3G18780), UBQ10 (At4G05320), CHIL (At5G05270), CHS (At5G13930), TT5 (At3G55120), F3H (At3G51240), F3′H (At5G07990), DFR (At5G42800), ANS (At4G22880), ANR (At1G61720), FLS (At5G08640), TT2 (At5G35550), TT8 (At4G09820), and TTG1 (At5G24520), UGT79B1 (At5G54060), UGT84A2 (At3G21560), and UGT78D2 (At5G17050).
Results
Identification and characterization of the chil mutants in Arabidopsis
TT2, an R2-R3 MYB type transcription factor, is a master regulator of PA biosynthesis in the seed coat of Arabidopsis (Nesi et al., 2001). In order to screen for genes involved in PA biosynthesis, a tt2 mutant line (CS83) with a pale yellow seed colour phenotype was used for global gene expression analysis using the Arabidopsis Affymetrix microarray. The result revealed that 255 probe sets were down-regulated by 2.0-97.4-fold in the tt2 mutant (see Supplementary Table S1 at JXB online). As expected, several PA-specific pathway genes, including ANR (At1g61720) and AHA10 (At1g17260) were down-regulated (see Supplementary Table S1 at JXB online). Interestingly, CHIL expression was reduced by 2.3-fold in the mutant, indicating that CHIL may be involved in PA biosynthesis.
To investigate whether the uncharacterized CHIL gene was indeed involved in PA biosynthesis, two independent T-DNA insertion mutant alleles, chil-1 and chil-2, were obtained from ABRC. It was further confirmed by PCR screening that the T-DNAs were inserted into the first and the third introns of the CHIL gene in the chil-1 and the chil-2 mutants, respectively (Fig. 2A), disrupting transcription of the CHIL gene. This was also confirmed with PCR using gene-specific primer pairs (Fig. 2B). No CHIL transcripts were detected by reverse transcription PCR or quantitative Real Time PCR (qRT-PCR) in homozygous genotypes of either mutant line (Fig. 2B, C).
The seed colour of the two mutant lines was essentially the same as that of the wild-type Col (Fig. 2D, upper panel), a finding in sharp contrast to the pale yellow seed colour phenotypes of the mutants of many other flavonoid pathway genes. The fact that the seed coats were the same colour suggests insignificant changes in levels of the major seed coat pigments. However, when stained with DMACA, a dye that is specific for PAs, seeds of the chil-1 and chil-2 mutants exhibited a lighter blue-purple colour than the wild-type seed (Fig. 2D, middle and lower panels). This strongly suggests that the chil mutants accumulate lower levels of PA compounds than does the wild-type; this result was further confirmed by a determination of the extractable PA content by either the DMACA or acid butanol hydrolysis methods (Fig. 2E). In addition, the non-extractable PA levels were greatly reduced in the mutant, to about 20% of the level of the wild-type control (Fig. 2E).
To evaluate whether flavonoid compounds other than PAs are affected by a loss-of-function of CHIL, seedlings of the chil-1 and chil-2 mutants were observed for anthocyanin accumulation compared with the wild-type control. Anthocyanins normally accumulated in the hypocotyls of 4-d-old seedlings of both the wild-type and the mutants under normal growth conditions (Fig. 3A, upper panels). The accumulation of anthocyanins is known to be stress inducible. To visualize the accumulation of anthocyanins in Arabidopsis better, 17-d-old seedlings were therefore grown under stress conditions (increased sucrose concentration in the growth medium and higher light intensity) as previously described by Yonekura-Sakakibara et al. (2012). These experiments revealed that the two chil mutants accumulated similar levels of anthocyanins in the true leaves as did the wild-type (Fig. 3A, lower panels). Spectrometric quantification further confirmed that the anthocyanin levels in the two chil mutants did not differ significantly from the level of the wild-type (Fig. 3B). These results indicated that CHIL does not affect the accumulation level of anthocyanins in Arabidopsis leaves.
Flavonols are among the major flavonoid compounds accumulating in leaves and seeds of Arabidopsis. To evaluate whether CHIL loss-of-function mutants affected flavonol accumulation, flavonoids extracted from leaves and seeds of mutant and wild-type plants were hydrolysed and analysed by HPLC. Levels of the two major flavonol aglycones, quercetin and kaempferol, were decreased greatly (down to 17-25%) in the chil mutants in both leaves and mature seeds compared with the wild-type (Fig. 3C, D; see Supplementary Fig. S1A–C at JXB online), indicating that CHIL affects flavonol accumulation in both seeds and leaves of Arabidopsis.
CHIL is a functional gene that enhances flavonoid production in Arabidopsis
To verify whether CHIL is functional in vivo, and to confirm that the phenotype of the chil-1 mutant was indeed the consequence of loss-of-function of the CHIL gene, CHIL was over-expressed in the chil-1 mutant background. PCR-positive transgenic lines were further evaluated by qRT-PCR to determine the transcript level of the CHIL gene in the transgenic lines. The transcript levels of CHIL in the three lines over-expressing the CHIL gene (in the chil-1 background) were all increased to a level that is comparable with the wild-type control (Fig. 4A). The CHIL over-expressing lines in the chil-1 background produced brown coloured seeds, which was the same as the wild-type (see Supplementary Fig. S2A at JXB online). Further analysis showed that the seed appeared to accumulate a similar level of PAs as visualized by DMACA staining (Fig. 4B; see Supplementary Fig. S2B at JXB online). The extractable and non-extractable PA levels in the mature seeds of the over-expression lines were all increased (Fig. 4C), but were not restored to the level found in the wild-type, which may be because the promoter used (the cauliflower mosaic virus 35S promoter) for the construct may be less strong than the endogenous promoter. Similarly, the levels of flavonol aglycones in the CHIL over-expression lines were increased, but not to levels comparable with those of the wild-type in both leaves and mature seeds (Fig. 4D, E). The flavonol profiles in mature seeds of these lines were qualitatively the same as that of the wild-type (see Supplementary Fig. S3 at JXB online). In addition, the accumulation of anthocyanins in leaves of over-expression lines did not appear to be affected (see Supplementary Fig. S4 at JXB online). In conclusion, over-expression of the CHIL gene in the chil-1 mutant background could partially rescue the chil mutant phenotype.
To investigate whether higher over-expression of the CHIL gene could further impact flavonoid accumulation in wild-type Arabidopsis, transgenic lines over-expressing the CHIL gene in the Col background were generated and their flavonoid levels were analysed. The CHIL over-expression lines exhibited the same intense dark blue-purple DMACA staining compared with that of the wild-type (Fig. 4B), and the extractable and non-extractable PA contents were increased compared with Col (Fig. 4C, 30% and 40% increase in line 1). Kaempferol levels increased in the leaves and seeds of the over-expression lines compared with Col control plants (Fig. 4D, E). However, there was no change in the anthocyanin levels although the CHIL gene was highly expressed (see Supplementary Fig. S4 at JXB online). Taken together, these results indicate that over-expression of CHIL enhances the accumulation of both flavonol (kaempferol) and PAs in Arabidopsis.
Expression levels of other flavonoid pathway genes in the chil-1 mutant
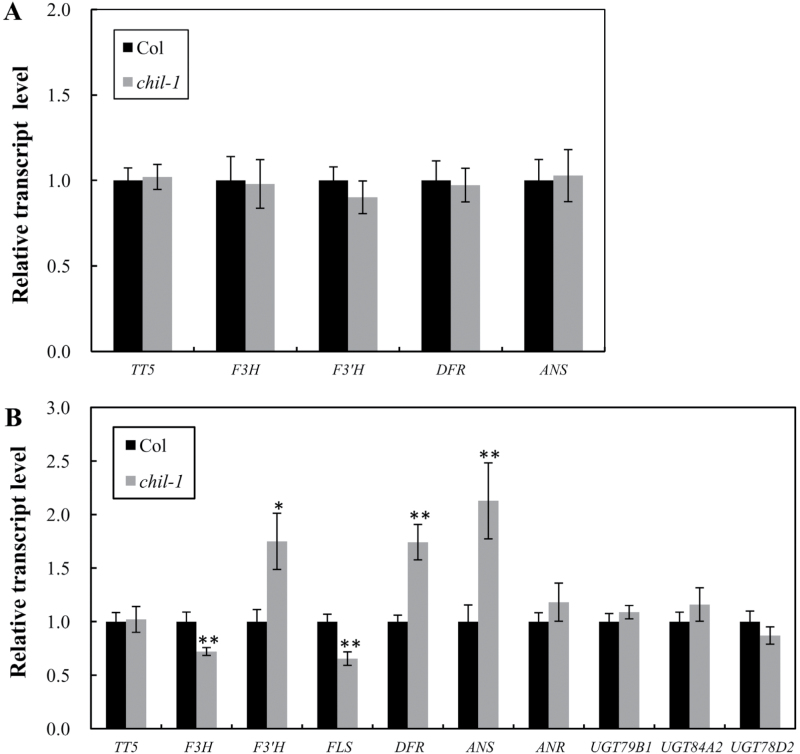
To investigate whether the transcript levels of other pathway genes were affected by the loss-of-function of CHIL in Arabidopsis, qRT-PCR was performed to determine the transcript levels of key flavonoid pathway genes in both seedlings and developing seed tissues. The transcript levels of the TT5, F3H, F3′H, DFR, and ANS genes were almost unchanged in 17-d-old seedlings of the chil-1 mutant, compared with the wild-type (Fig. 5A), consistent with the unaffected anthocyanin levels. In the developing seeds (4 d after flowering), none of the above genes changed by more than 2-fold in the chil-1 mutant, although there were slight changes in the transcript levels of F3′H, DFR, and ANS (Fig. 5B). Taken together, the data suggest that CHIL does not function as a transcription factor in the flavonoid pathway.
Fig. 5.
Relative transcript levels of flavonoid pathway genes in the chil-1 mutant compared with the wild-type Col control. (A) Relative transcript levels of TT5, F3H, F3′H, DFR, and ANS in the 17-d-old seedlings of Col and chil-1 mutant detected by qRT-PCR analysis. (B) Relative transcript levels of TT5, F3H, F3′H, FLS, DFR, ANS, ANR, and three UGTs in 4-d-old developing seed detected by qRT-PCR analysis. Data are presented as mean ±SD, Student’s t test (n=3, *P <0.05, **P <0.01).
Neither loss- nor gain-of-function of CHIL affects the tt5 and chs mutant phenotypes
In previous biochemical studies, CHIL could not be shown to have catalytic activity in vitro (Ngaki et al., 2012). To confirm this further, the recombinant CHIL protein in E. coli was purified to high purity (see Supplementary Fig. S5A at JXB online) and its in vitro catalytic activity was tested with naringenin chalcone as the substrate. As reported previously, no activity was detected in this assay. Interestingly, when run on a non-denaturing PAGE gel, the recombinant CHIL protein displayed several bands with multiple integer sizes, indicating that CHIL can form different multimers (see Supplementary Fig. S5B at JXB online). By contrast, recombinant TT5 protein only appeared to form a tetramer (see Supplementary Fig. S5C at JXB online).
To determine if CHIL has isomerase activity under in vivo conditions, an over-expressing line (CHIL OE-1) was crossed with the tt5 mutant to investigate whether CHIL could rescue the tt5 mutant phenotype. The CHIL OE-1/tt5 line produced seeds with a transparent testa phenotype (as in the tt5 mutant), and the seeds were not stained blue by DMACA, indicating that no PAs were produced in the CHIL OE-1/tt5 line (Fig. 6A, upper panel). The flavonol profile of the tt5 mutant on HPLC is similar to that of the chil-1 mutants, except for the absence of quercetin and kaempferol in the tt5 mutant (see Supplementary Figs S1 and S6 at JXB online). The PA and flavonol contents, and flavonol profiles, in CHIL OE-1/tt5 were the same as those of the tt5 mutant (Fig. 6C–E; see Supplementary Fig. S6 at JXB online). In addition, the anthocyanin levels did not differ between the CHIL OE-1/tt5 and tt5 seedlings (see Supplementary Fig. S7 at JXB online). Taken together, these results strongly indicated that CHIL cannot rescue the tt5 phenotype and that CHIL does not encode an enzyme with CHI activity in vivo.
To investigate the genetic position of CHIL in the flavonoid pathway further, the chil-1 mutant was next crossed with the tt5 mutant. The chil/tt5 double mutant produced progeny with a tt seed phenotype (Fig. 6, upper panel), and the levels of PA, flavonol, and anthocyanin, as well as the flavonol profiles, were not changed compared with the tt5 mutant (Fig. 6C–E; see Supplementary Figs 5 and 6 at JXB online), indicating that CHIL does not function in the same way as TT5 in the flavonoid pathway.
Similarly, the chil-1 mutant was also crossed with a chs mutant to produce the chil/chs double mutant, and the CHIL gene was also over-expressed in the chs mutant background by crossing the chs mutant with the CHIL OE-1 line. Both chil/chs double mutants and CHIL OE-1/chs lines produced seeds with transparent testa phenotypes, as in the chs mutant. Moreover, over-expression of the CHIL gene in the chs background could not rescue the tt phenotype, as measured by DMACA staining (Fig. 6B). In addition, both the double mutant and the CHIL over-expressing lines showed no difference in the accumulation levels of PAs, flavonols, and anthocyanins, as well as the flavonol profiles, compared with the chs mutant (Fig. 6B-D; see Supplementary Figs 7 and 8 at JXB online). All of these results indicate that CHIL does not function as a CHS in the flavonoid pathway.
Temporal and spatial pattern of CHIL expression
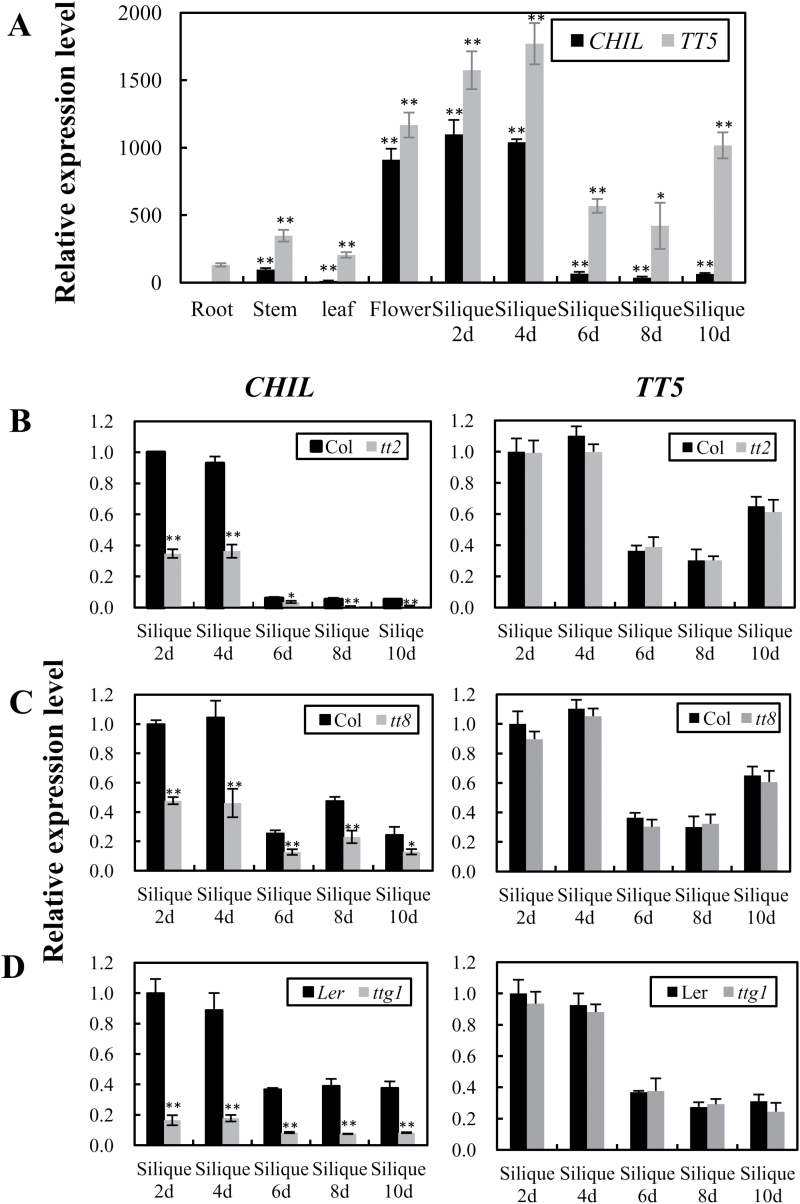
To evaluate the expression pattern of the CHIL gene in Arabidopsis, its transcript levels were analysed in various organs and seed developmental stages using qRT-PCR. The CHIL gene was highly expressed in inflorescences and expressed at relatively lower levels in stems and rosette leaves. CHIL transcripts were hardly detectable in roots (Fig. 7A). Since CHIL affects the accumulation of flavonoids in seeds, CHIL transcript levels were also evaluated during different seed developmental stages (as represented by siliques). CHIL transcripts were at their highest level during the early stages of seed development (days 2 and 4), followed by a dramatic reduction at the later stages (days 6 and 10, Fig. 7A). The expression of the TT5 gene showed a similar expression pattern in various organs and seed developmental stages (Fig. 7A). Nevertheless, the transcript pattern of CHIL is similar to the accumulation pattern of extractable PAs and a quercetin-rhamnoside-hexoside during seed development as previously reported by Routaboul et al. (2006), suggesting that CHIL is primarily involved in the biosynthesis of PAs and flavonols.
Fig. 7.
Expression pattern of CHIL and TT5 genes in various tissues and mutants. (A) Relative transcript levels of CHIL and TT5 in root, stem, rosette leaf, inflorescence, 2-, 4-, 6-, 8-, and 10-d-old siliques of wild-type Arabidopsis Col. (B–D) Relative transcript levels of the CHIL gene in developing seeds (represented by siliques) of tt2 (B), tt8 (C), and ttg1 (D) mutants compared with the corresponding wild-type control. Data are presented as mean ±SD, Student’s t test (n=3, *P <0.05, **P <0.01).
As it is known that the TT2/TT8/TTG1 transcription factor complex regulates the biosynthesis of PAs in the developing seeds of Arabidopsis (Baudry et al., 2004), it was also evaluated whether CHIL was regulated by TT2, TT8 or TTG1 in the corresponding mutants. The CHIL transcript level was greatly reduced in the tt2, tt8, and ttg1 mutants at all of the seed developmental stages compared with the wild-type (Fig. 7B-D). The reduction was particularly pronounced at the early stages of seed development (days 2 and 4), indicating that CHIL expression is regulated by TT2, TT8, and TTG1 in the developing Arabidopsis seed.
Next, the 1742bp intergenic region of At5g05260 and At5g05270 (CHIL) was cloned and analysed using the Database of Plant Cis-acting Regulatory DNA Elements (PLACE, http://www.dna.affrc.go.jp/PLACE/) (Higo et al., 1999) (see Supplementary Fig. S9 at JXB online). The S000144 element (CANNTG) is recognized by a transcription factor that controls the light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes (Hartmann et al., 2005), and this element is present in several positions in the promoters of both CHIL and the Arabidopsis ANR gene (Debeaujon et al., 2003). Another element, S000176 (CNGTTR), identified in the Petunia hybrida CHSJ gene is recognized by regulators of flavonoid biosynthesis (Solano et al., 1995) and this element is also present in the promoters of CHIL and ANR (Debeaujon et al., 2003). Analysis of the promoter region of the TT5 gene revealed that it also has these two elements (ten for S000144 and three for S000176) that are shared by CHIL and ANR (see Supplementary Fig. S10 at JXB online), implying these genes may be co-ordinately regulated.
To examine the location of CHIL gene expression further, its promoter was fused to the gusA gene. This construct was introduced into Arabidopsis and various tissues were examined histochemically (see Supplementary Fig. S11 at JXB online). Staining of the CHIL promoter:GUS transgenic Arabidopsis plants with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid revealed CHIL expression in young seedlings, the leaves of mature plants, the inflorescences, the peduncle of mature siliques, and immature seeds (before 6 d; see Supplementary Fig. S11 at JXB online). The expression pattern of CHIL detected by GUS staining was consistent with its transcript level pattern measured by qRT-PCR during seed development.
CHIL is co-localized in the endoplasmic reticulum and physically interacts with TT5
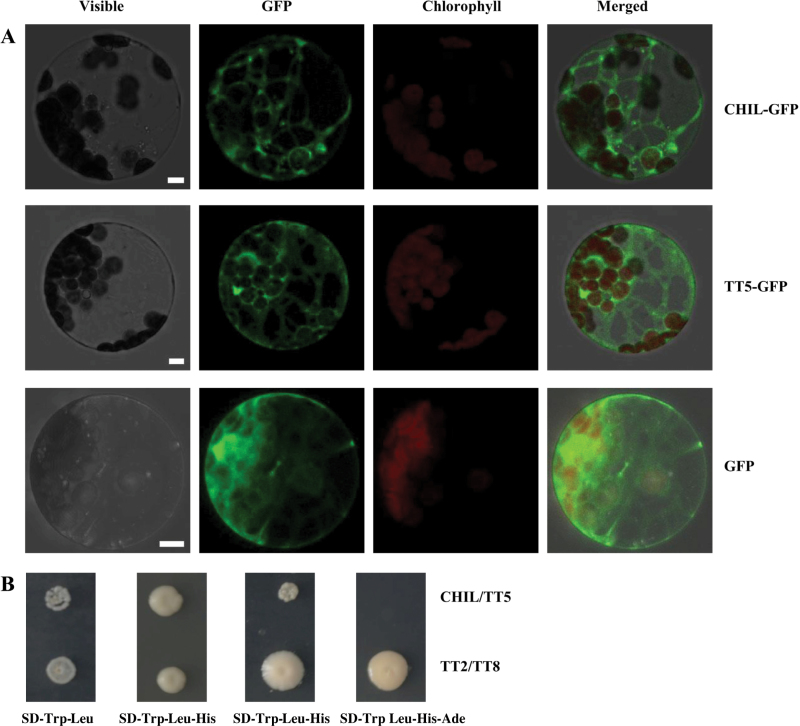
TT5 was reported to be localized to the endoplasmic reticulum and to act as an ER-membrane anchor for a metabolon involved in flavonoid biosynthesis (Saslowsky and Winkel-Shirley, 2001). To determine whether CHIL is localized to the endoplasmic reticulum, CHIL together with TT5 were fused to green fluorescent protein (GFP) for transient expression experiments using Arabidopsis leaf protoplasts (Fig. 8). Clear fluorescence signals for CHIL:GFP were detected in the endoplasmic reticulum (Fig. 8A); the same signal pattern was observed for TT5:GFP (Fig. 8B). The fluorescence signal patterns of CHIL:GFP and TT5:GFP were clearly distinct from that of control GFP (Fig. 8C). These results indicate that CHIL is localized to the endoplasmic reticulum as well as TT5 for flavonoid production.
Fig. 8.
Co-localization and interaction of CHIL with TT5. (A) Subcellular localization of CHIL and TT5 by GFP assays in Arabidopsis protoplasts. Fluorescence signals were visualized using confocal laser scanning microscopy. From left to right: bright field, the green fluorescence, autofluorescence of the chloroplast, and merged images of CHIL-GFP fusion protein (upper panel), TT5-GFP fusion protein (middle panel), and GFP (lower panel). Bars=5 µm. (B) Interaction between CHIL and TT5 in yeast two hybrid assays. The yeast cells grew on the medium SD-Trp-Leu, SD-Trp-Leu-His, SD-Trp-Leu-His, and SD-Trp Leu-His-Ade. The interaction between TT2 and TT8 served as a positive control. (This figure is available in colour at JXB online.)
The co-expression and co-localization of CHIL and TT5 suggest that the CHIL protein possibly interacts with TT5. In order to verify this hypothesis, a yeast two hybrid approach was taken to assess the CHIL and TT5 interaction, and TT2 and TT8 were used as positive controls. Yeast co-transformed with BD-CHIL and AD-TT5, BD-TT2 and AD-TT8, respectively, were both able to grow in the absence of Trp, Leu, and His, indicating that CHIL could indeed physically interact with TT5. However, CHIL/TT5 was not able to grow in the absence of Ade in contrast to TT2/TT8, indicating that the interaction of CHIL with TT5 was not as strong as that of TT2 with TT8 (Fig. 8D).
Discussion
CHIL is a functional gene involved in flavonoid biosynthesis in Arabidopsis
The biosynthesis of flavonoids has been well established in Arabidopsis. This was mainly achieved through the screening and characterization of a number of transparent testa mutants. In comparison with the dark brown seed colour in the wild-type, these tt mutants exhibit a pale yellow seed colour due to deficiencies in oxidized flavonoid compounds (mainly PAs), (Shirley et al., 1995; Routaboul et al., 2006; Appelhagen et al., 2014). Nearly all of the tt mutants have been characterized via biochemical and/or genetic approaches and they have been shown to have mutations in structural, transporter or regulatory genes related to flavonoid production (Shirley et al., 1992; Dixon et al., 2005; Appelhagen et al., 2014).
Research on PA biosynthesis is currently one of the hot spots in flavonoid metabolism research. Such research has advanced our understanding of the modification and transport of PA precursors in the pathway (Pang et al., 2008; Zhao and Dixon, 2009; Zhao et al., 2010), but there are still many unanswered questions regarding their biosynthetic mechanism. To explore further the candidate genes responsible for PA biosynthesis in Arabidopsis, a global gene expression analysis was performed in a tt2 mutant that is deficient in PA biosynthesis in the seed coat. It was found that the CHIL gene was down-regulated in the tt2 mutant, suggesting a potential role for CHIL in PA biosynthesis. The two null mutant alleles (chil-1 and chil-2) produced dark brown seeds, as in the wild- type (Fig. 2A), a finding in sharp contrast to many other tt mutants with pale coloured seed phenotypes. Obviously, from the point view of the seed phenotype, the CHIL gene is different from all of the other known flavonoid pathway genes.
Several tt mutants that affect the loci specific for seed coat pigmentation are deficient in anthocyanins in the leaves, including tt4, tt5, tt6, and tt7, but the anthocyanin accumulation in the chil mutants was not affected in the leaves, another finding that was obviously distinct from these tt mutants (Appelhagen et al., 2014). The accumulation levels of anthocyanins in the two chil mutants were similar to those of the wild-type, so CHIL is not involved in anthocyanin production. Unlike other flavonoid pathway genes, the CHIL gene may act in flavonoid biosynthesis via an as yet uncharacterized mechanism.
Further analysis showed that the seeds of the two chil mutant lines accumulated markedly reduced levels of PAs and flavonols when compared with the wild-type (Figs 1, 2), but the phenotype of the chil mutant could be recovered via complementation (Fig. 4). This observation unambiguously shows that CHIL is involved in flavonoid biosynthesis and plays a major role in PA biosynthesis in Arabidopsis.
Both CHIL and TT5 belong to the CHI superfamily of Arabidopsis (Ngaki et al., 2012), and a strong association of CHIL expression levels with the expression levels of TT5 was observed in this study and elsewhere (Ma, 2002; Oravecz et al., 2006; Yonekura-Sakakibara et al., 2008; Pan et al., 2009). However, the CHIL gene could not rescue the deficiency of flavonoid accumulation and failed to recover the seed colour phenotypes of the tt5 mutant (Fig. 6). Therefore, the CHIL gene is not a redundant TT5 gene, but rather a new functional gene in the flavonoid biosynthetic pathway, particularly in the PA branch.
CHIL functions with TT5 for enhancing flavonoid accumulation in Arabidopsis
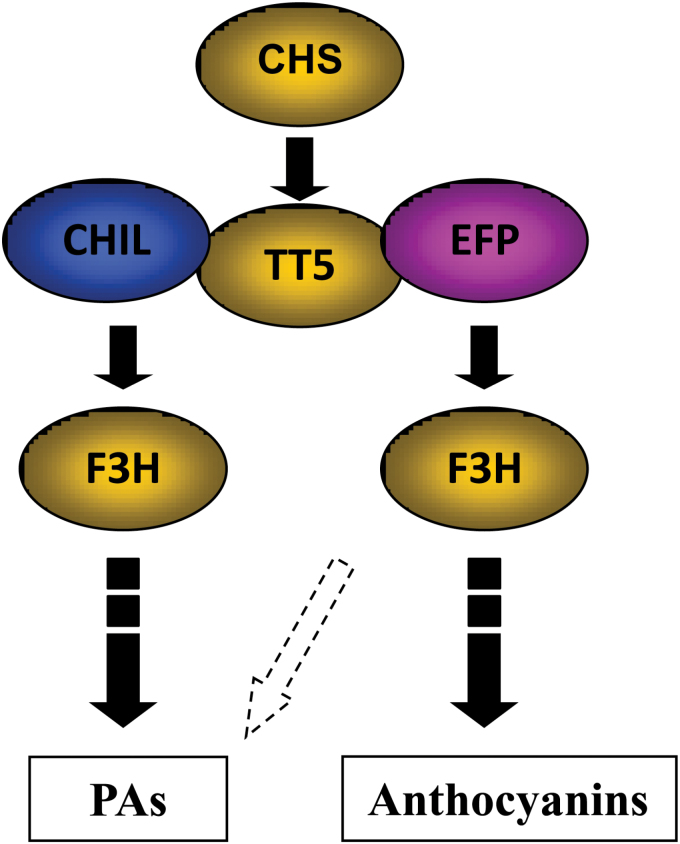
Generally, loss-of-function mutations in genes at different steps in the same pathway produce different metabolite profiles (Yonekura-Sakakibara et al., 2008). In the present study, it was found that chil mutants displayed a similar flavonol profile to tt5 (see Supplementary Figs S1 and S6 at JXB online), the profile of which is largely distinct from those of the chs and f3h mutants (see Supplementary Figs S8 and S12 at JXB online). These results suggest that CHIL most probably acts at the same step as TT5 and that it is located immediately downstream of CHS and upstream of F3H (Fig. 9).
Fig. 9.
Hypothetical model illustrating the role of CHIL and EFP proteins in flavonoid biosynthesis. CHIL and EFP proteins are downstream of CHS, and interact with TT5 in the pathway for proanthocyanidin and anthocyanin biosynthesis, respectively. They may function as anchor, enhancer or chaperone for specific flavonoid accumulation. (This figure is available in colour at JXB online.)
Apart from a strong association of CHIL expression with that of TT5, CHIL and TT5 are also co-expressed in various organs and seed developmental stages, which is consistent with the presence of two cis-acting elements shared by CHIL and TT5 in their promoter regions (Fig. 7; see Supplementary Fig. S11 at JXB online). In addition, both CHIL and TT5 are localized to the endoplasmic reticulum, potentially associated with a metabolon for flavonoid synthesis (Winkel-Shirley et al., 2001).
Most importantly, CHIL was observed to interact physically with TT5 in yeast two-hybrid assays. X-ray crystallography analyses of the CHI-related proteins revealed that the structure of CHIL is much more similar to TT5 than to those of the other CHI-related proteins (Jez et al., 2000; Ngaki et al., 2012). CHIL may thus be a more suitable partner for heterodimerization with TT5 than any of the other CHI-related proteins. Taken together, these lines of evidence suggest that CHIL may conceivably act as a partner for TT5 and may thus enhance TT5 catalytic activity.
A unique role of the CHIL proteins in flavonoid biosynthesis
Unfortunately, no catalytic activity has been detected for recombinant CHIL protein in this or previous studies (Jez et al., 2000; Ngaki et al., 2012). As with the CHIL protein, no catalytic activity with naringenin chalcone was observed for recombinant EFP (Enhance Flavonoid Production) protein, a homologue of CHIL in Ipomoea nil (Japanese morning glory) (Morita et al., 2014). Therefore, the CHIL proteins (CHIL and EFP) do not seem to have catalytic activity with naringenin chalcone, although they have enzyme-like structures (Morita et al., 2014).
In addition, expression analysis showed that the transcript levels of several investigated structural genes of the flavonoid pathway were not affected (namely TT5, ANR and three UGTs), or did not change by more than 2.1-fold (namely F3H, F3′H, FLS, DFR, and ANS) in the chil-1 mutant. Similar results were observed for EFP, as deficiency of the EFP gene did not affect the transcript levels of the other pathway genes (CHS, DFR, and 3GT) in the efp mutant (Morita et al., 2014). Thus, CHIL does not appear to function as a transcription factor in flavonoid biosynthesis.
It is possible that the contrast between the pale-coloured flowers of the efp mutant and the colourless flowers of the chi mutant in I. nil plant, was due to differential accumulation levels of anthocyanins (Iida et al., 2004; Morita et al., 2014), a situation similar to the differential accumulation levels of PAs and flavonols in the Arabidopsis chil and tt5 mutants. The transcript levels of both CHIL and EFP genes were completely absent in the corresponding mutants (Morita et al., 2014), indicating that they are not weak alleles of CHIL genes. On the other hand, loss-of-function of CHIL in Arabidopsis is different from loss-of-function of EFP of I. nil in that it affected the accumulation of PAs and flavonols rather than anthocyanins. It is possible that there may be another EFP orthologue in Arabidopsis that is involved in anthocyanin accumulation (Fig. 9).
As hypothesized by Morita et al. (2014), the CHIL proteins may function in guiding the proper folding of polyketide intermediates into homo- or hetero-multimeric proteins with TT5. Alternatively, CHIL may function as an anchor, chaperone or enhancer to promote the accumulation of flavonoids by stabilizing or enhancing the activity of the corresponding CHI proteins (Fig. 9). Although the detailed molecular mechanism remains to be elucidated of how CHIL and EFP promote flavonoid production, they appear to be unique positive regulator proteins that lack catalytic activities. It remains to be seen whether these hypothesized regulation mechanisms underlying CHIL and EFP are universal in plants.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Representative HPLC chromatograms of the chil-1 mutants and Col.
Supplementary Fig. S2. Over-expression of the CHIL gene in the chil-1 mutant and Col background.
Supplementary Fig. S3. Representative HPLC chromatograms of the over-expression lines in the chil-1 mutant and Col backgrounds.
Supplementary Fig. S4. Anthocyanin accumulation in the CHIL over-expression lines in the chil and Col backgrounds.
Supplementary Fig. S5. Purified recombinant CHIL and TT5 protein on denaturing and native PAGE.
Supplementary Fig. S6. Representative HPLC chromatograms of CHIL loss-of-function and over- expression lines in the tt5 background.
Supplementary Fig. S7. Anthocyanin accumulation in the CHIL loss-of-function and over-expression lines in the tt5 and chs mutant backgrounds.
Supplementary Fig. S8. Representative HPLC chromatograms of CHIL loss-of-function and over-expression lines in the chs background.
Supplementary Fig. S9. Cis-acting elements upstream the ORF of the CHIL gene as predicted by PLACE.
Supplementary Fig. S10. Cis-acting elements upstream the ORF of the TT5 gene as predicted by PLACE.
Supplementary Fig. S11. Tissue-specific promoter activity of the CHIL promoter fused with GUS.
Supplementary Fig. S12. Representative HPLC chromatograms of F3H loss-of-function compared with the chil-1 mutant in the Col background.
Supplementary Table S1. The probe sets with expression more than 2 fold down-regulated in seeds of the tt2 mutant of Arabidopsis.
Supplementary Table S2. The primer sequences used in the present study.
Acknowledgements
Naringenin chalcone was kindly provided by Dr Peng Nan from Fudan University and Professor Liping Gao from Anhui Agriculture University, China. We thank Professor Lixi Jiang from Zhejiang University, China for providing the tt8 mutant seeds and Dr Yuhong Tang from the SR Noble Foundation for analysing the microarray data. This work was supported by the Major State Basic Research Development Program (2013CB127002), the National Natural Science Foundation of China (31370335), and the ‘Hundred Talents Program’ of the Chinese Academy of Sciences (39391503-7).
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Appelhagen I, Thiedig K, Nordholt N, Schmidt N, Huep G, Sagasser M, Weisshaar B. 2014. Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta 240, 955–970. [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. 2004. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . The Plant Journal 39, 366–380. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Joural 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. 2003. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. The Plant Cell 15, 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Xie DY, Sharma SB. 2005. Proanthocyanidins: a final frontier in flavonoid research? New Phytologist 165, 9–28. [DOI] [PubMed] [Google Scholar]
- Ferreyra FML, Rius SP, Casati P. 2012. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Frontiers in Plant Science 3, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. 2005. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Molecular Biology 57, 155–171. [DOI] [PubMed] [Google Scholar]
- Hassan S, Mathesius U. 2012. The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions. Journal of Experimental Botany 63, 3429–3444. [DOI] [PubMed] [Google Scholar]
- He X, Blount JW, Ge S, Tang Y, Dixon RA. 2011. A genomic approach to isoflavone biosynthesis in kudzu (Pueraria lobata). Planta 233, 843–855. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S, Morita Y, Choi JD, Park KI, Hoshino A. 2004. Genetics and epigenetics in flower pigmentation associated with transposable elements in morning glories. Advances in Biophysics 38, 141–159. [PubMed] [Google Scholar]
- Jez JM, Bowman ME, Dixon RA, Noel JP. 2000. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nature Structural and Molecular Biology 7, 786–791. [DOI] [PubMed] [Google Scholar]
- Jiang WB, Huang HY, Hu YW, Zhu SW, Wang ZY, Lin WH. 2013. Brassinosteroid regulates seed size and shape in Arabidopsis . Plant Physiology 162, 1965–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WB, Yu DQ. 2009. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Koornneef M. 1981. The complex syndrome of ttg mutants. Arabidopsis Information Service 18, 45–51. [Google Scholar]
- Kozlowska A, Szostak-Wegierek D. 2014. Flavonoids: food sources and health benefits. Roczniki Państwowego Zakładu Higieny 65, 79–85. [PubMed] [Google Scholar]
- Lattanzio V, Lattanzio VMT, Cardinali A. 2006. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. Phytochemistry: Advances in Research 23–67. [Google Scholar]
- Lee ER, Kang GH, Cho SG. 2007. Effect of flavonoids on human health: old subjects but new challenges. Recent Patents in Biotechnology 1, 139–150. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. 2006. Genetics and biochemistry of seed flavonoids. Annual Review of Plant Biology 57, 405–430. [DOI] [PubMed] [Google Scholar]
- Li Y, Tanner G, Larkin P. 1996. The DMACA-HC protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. Journal of the Science of Food and Agriculture 70, 89–101. [Google Scholar]
- Ma L. 2002. Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis . The Plant Cell 14, 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Takagi K, Fukuchi-Mizutani M, et al. 2014. A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. The Plant Journal 78, 294–304. [DOI] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. 2001. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. The Plant Cell 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngaki MN, Louie GV, Philippe RN, Manning G, Pojer F, Bowman ME, Li L, Larsen E, Wurtele ES, Noel JP. 2012. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 485, 530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Mate Z, Brzezinska A, Molinier J, Oakeley EJ, Adam E, Schafer E, Nagy F, Ulm R. 2006. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis . The Plant Cell 18, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Michael TP, Hudson ME, Kay SA, Chory J, Schuler MA. 2009. Cytochrome P450 monooxygenases as reporters for circadian-regulated pathways. Plant Physiology 150, 858–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA. 2008. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula . Proceedings of the National Academy of Sciences, USA 105, 14210–14215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Wright E, Wang Z, Dixon RA. 2007. Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula . Plant Physiology 145, 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Wenger JP, Saathoff K, et al. 2009. A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiology 151, 1114–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston L, Subramanian S, Matsuno M, Yu O. 2005. Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiology 137, 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L. 2006. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana . Planta 224, 96–107. [DOI] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR. 2013. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiology and Biochemistry 72, 21–34. [DOI] [PubMed] [Google Scholar]
- Saslowsky D, Winkel-Shirley B. 2001. Localization of flavonoid enzymes in Arabidopsis roots. The Plant Journal 27, 37–48. [DOI] [PubMed] [Google Scholar]
- Sheen J. 2002. A transient expression assay using Arabidopsis mesophyll protoplasts http://genetics.mgh.harvard.edu/sheenweb/.
- Shimada N, Aoki T, Sato S, Nakamura Y, Tabata S, Ayabe S. 2003. A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus . Plant Physiology 131, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM. 1992. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. The Plant Cell 4, 333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. 1995. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. The Plant Journal 8, 659–671. [DOI] [PubMed] [Google Scholar]
- Solano R, Nieto C, Avila J, Canas L, Diaz I, Paz-Ares J. 1995. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida . The EMBO Journal 14, 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. 1998. Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proceedings of the National Academy of Sciences, USA 95, 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. 2003. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299, 396–399. [DOI] [PubMed] [Google Scholar]
- Yao LH, Jiang YM, Shi J, Tomas-Barberan FA, Datta N, Singanusong R, Chen SS. 2004. Flavonoids in food and their health benefits. Plant Foods for Human Nutrition 59, 113–122. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, et al. 2012. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana . The Plant Journal 69, 154–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. 2008. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis . The Plant Cell 20, 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, et al. 2010. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proceedings of the National Academy of Sciences, USA 107, 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. 2009. MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis . The Plant Cell 21, 2323–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Pang Y, Dixon RA. 2010. The mysteries of proanthocyanidin transport and polymerization. Plant Physiology 153, 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.