Abstract
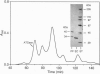
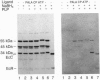
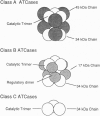
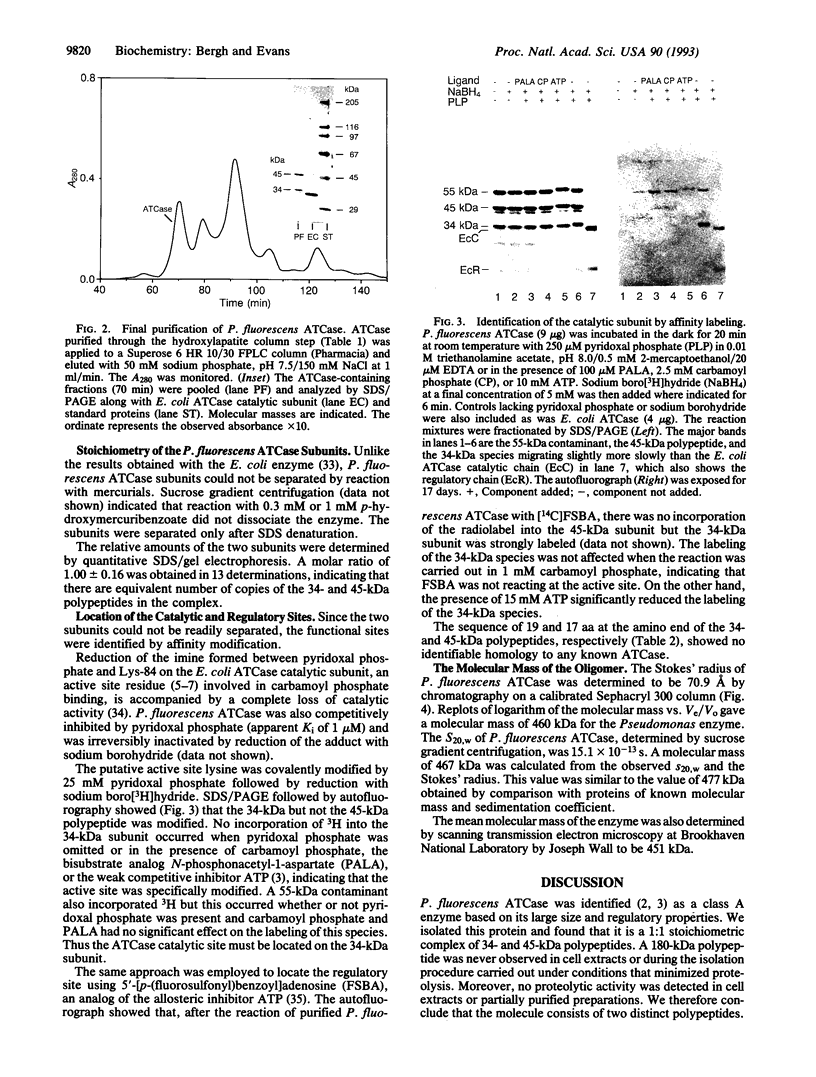
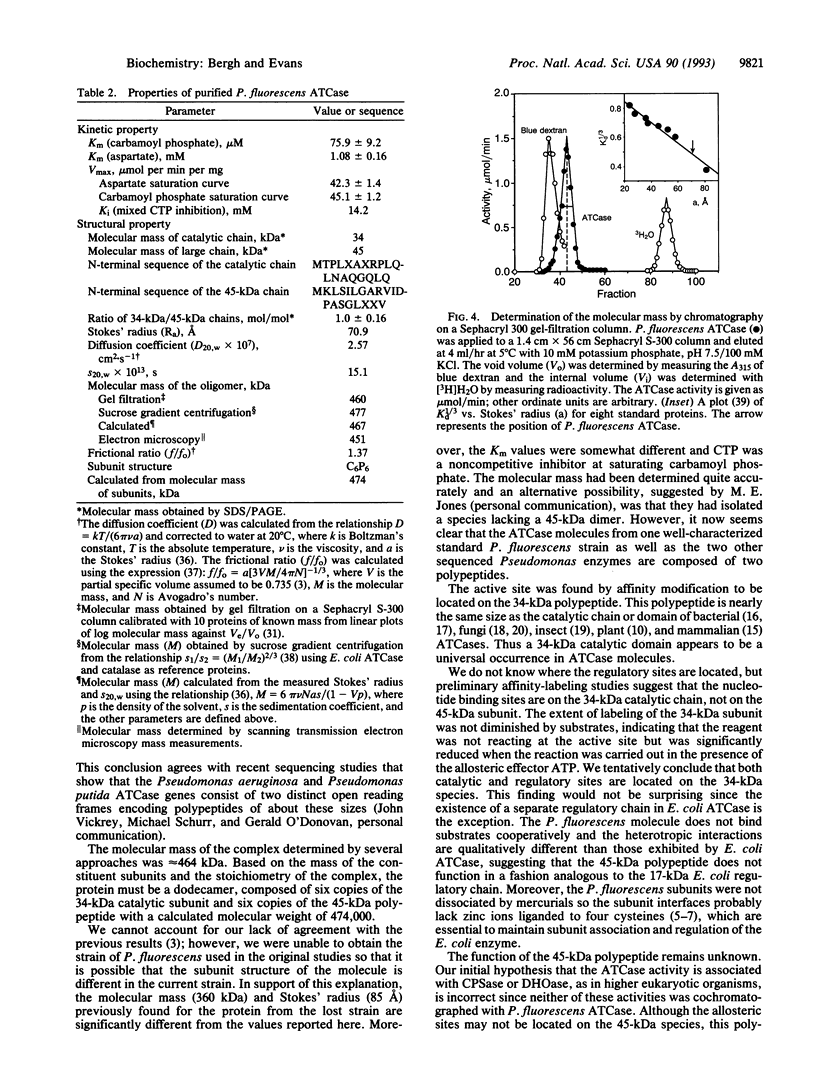
The class A aspartate transcarbamoylase (ATCase, EC 2.1.3.2) from Pseudomonas fluorescens was purified to homogeneity with retention of full catalytic and regulatory functions. Careful determinations under conditions that minimized proteolysis showed that the molecule is a 1:1 stoichiometric complex of two polypeptide chains of 34 and 45 kDa. Pyridoxal phosphate is a competitive inhibitor of the enzyme (Ki = 1 microM). Reduction of the pyridoxal phosphate enzyme adduct with sodium boro[3H]hydride showed that the active site is located on the 34-kDa polypeptide. Affinity labeling with 5'-[p-(fluorosulfonyl)benzoyl]adenosine, an ATP analog, suggested that the regulatory site is also located on the 34-kDa species. While the function of the 45-kDa subunit is unknown, neither carbamoyl phosphate synthetase nor dihydroorotase activities are associated with the ATCase. The molecular mass of the enzyme was determined by gel filtration, sedimentation velocity, and electron microscopy to be 464 kDa. Thus the enzyme is composed of six copies of the 34-kDa polypeptide and six copies of the 45-kDa polypeptide. The molecule has a Stokes' ratio of 70.9 A and a frictional ratio of 1.37, suggesting a compact globular shape. We propose that the P. fluorescens ATCase is composed of two trimers of 34-kDa catalytic chains and is likely to be a D3 dodecamer with an arrangement of subunits analogous to that of the class B ATCase molecules.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adair L. B., Jones M. E. Purification and characteristics of aspartate transcarbamylase from Pseudomonas fluorescens. J Biol Chem. 1972 Apr 25;247(8):2308–2315. [PubMed] [Google Scholar]
- Bethell M. R., Jones M. E. Molecular size and feedback-regulation characteristics of bacterial asartate transcarbamulases. Arch Biochem Biophys. 1969 Nov;134(2):352–365. doi: 10.1016/0003-9861(69)90294-x. [DOI] [PubMed] [Google Scholar]
- Brabson J. S., Switzer R. L. Purification and properties of Bacillus subtilis aspartate transcarbamylase. J Biol Chem. 1975 Nov 25;250(22):8664–8669. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Coleman P. F., Suttle D. P., Stark G. R. Purification from hamster cells of the multifunctional protein that initiates de novo synthesis of pyrimidine nucleotides. J Biol Chem. 1977 Sep 25;252(18):6379–6385. [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Davidson J. N., Rumsby P. C., Tamaren J. Organization of a multifunctional protein in pyrimidine biosynthesis. Analyses of active, tryptic fragments. J Biol Chem. 1981 May 25;256(10):5220–5225. [PubMed] [Google Scholar]
- Faure M., Camonis J. H., Jacquet M. Molecular characterization of a Dictyostelium discoideum gene encoding a multifunctional enzyme of the pyrimidine pathway. Eur J Biochem. 1989 Feb 1;179(2):345–358. doi: 10.1111/j.1432-1033.1989.tb14560.x. [DOI] [PubMed] [Google Scholar]
- Freund J. N., Jarry B. P. The rudimentary gene of Drosophila melanogaster encodes four enzymic functions. J Mol Biol. 1987 Jan 5;193(1):1–13. doi: 10.1016/0022-2836(87)90621-8. [DOI] [PubMed] [Google Scholar]
- Gerhart J. C., Schachman H. K. Allosteric interactions in aspartate transcarbamylase. II. Evidence for different conformational states of the protein in the presence and absence of specific ligands. Biochemistry. 1968 Feb;7(2):538–552. doi: 10.1021/bi00842a600. [DOI] [PubMed] [Google Scholar]
- Grayson D. R., Evans D. R. The isolation and characterization of the aspartate transcarbamylase domain of the multifunctional protein, CAD. J Biol Chem. 1983 Apr 10;258(7):4123–4129. [PubMed] [Google Scholar]
- Greenwell P., Jewett S. L., Stark G. R. Aspartate transcarbamylase from Escherichia coli. The use of pyridoxal 5'-phosphate as a probe in the active site. J Biol Chem. 1973 Sep 10;248(17):5994–6001. [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Hoover T. A., Roof W. D., Foltermann K. F., O'Donovan G. A., Bencini D. A., Wild J. R. Nucleotide sequence of the structural gene (pyrB) that encodes the catalytic polypeptide of aspartate transcarbamoylase of Escherichia coli. Proc Natl Acad Sci U S A. 1983 May;80(9):2462–2466. doi: 10.1073/pnas.80.9.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. E. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu Rev Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- Kantrowitz E. R., Lipscomb W. N. Escherichia coli aspartate transcarbamylase: the relation between structure and function. Science. 1988 Aug 5;241(4866):669–674. doi: 10.1126/science.3041592. [DOI] [PubMed] [Google Scholar]
- Ke H. M., Honzatko R. B., Lipscomb W. N. Structure of unligated aspartate carbamoyltransferase of Escherichia coli at 2.6-A resolution. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4037–4040. doi: 10.1073/pnas.81.13.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K. L., Volz K. W., Lipscomb W. N. 2.5 A structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-L-aspartate. J Mol Biol. 1987 Feb 5;193(3):527–553. doi: 10.1016/0022-2836(87)90265-8. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lerner C. G., Switzer R. L. Cloning and structure of the Bacillus subtilis aspartate transcarbamylase gene (pyrB). J Biol Chem. 1986 Aug 25;261(24):11156–11165. [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Mally M. I., Grayson D. R., Evans D. R. Controlled proteolysis of the multifunctional protein that initiates pyrimidine biosynthesis in mammalian cells: evidence for discrete structural domains. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6647–6651. doi: 10.1073/pnas.78.11.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Mori M., Ishida H., Tatibana M. Aggregation states and catalytic properties of the multienzyme complex catalyzing the initial steps of pyrimidine biosynthesis in rat liver. Biochemistry. 1975 Jun 17;14(12):2622–2630. doi: 10.1021/bi00683a010. [DOI] [PubMed] [Google Scholar]
- Nagy M., Le Gouar M., Potier S., Souciet J. L., Hervé G. The primary structure of the aspartate transcarbamylase region of the URA2 gene product in Saccharomyces cerevisiae. Features involved in activity and nuclear localization. J Biol Chem. 1989 May 15;264(14):8366–8374. [PubMed] [Google Scholar]
- Nowlan S. F., Kantrowitz E. R. Superproduction and rapid purification of Escherichia coli aspartate transcarbamylase and its catalytic subunit under extreme derepression of the pyrimidine pathway. J Biol Chem. 1985 Nov 25;260(27):14712–14716. [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Penefsky H. S. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1977 May 10;252(9):2891–2899. [PubMed] [Google Scholar]
- Prescott L. M., Jones M. E. Modified methods for the determination of carbamyl aspartate. Anal Biochem. 1969 Dec;32(3):408–419. doi: 10.1016/s0003-2697(69)80008-4. [DOI] [PubMed] [Google Scholar]
- Robey E. A., Schachman H. K. Regeneration of active enzyme by formation of hybrids from inactive derivatives: implications for active sites shared between polypeptide chains of aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 1985 Jan;82(2):361–365. doi: 10.1073/pnas.82.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully J. L., Evans D. R. Comparative modeling of mammalian aspartate transcarbamylase. Proteins. 1991;9(3):191–206. doi: 10.1002/prot.340090305. [DOI] [PubMed] [Google Scholar]
- Shoaf W. T., Jones M. E. Uridylic acid synthesis in Ehrlich ascites carcinoma. Properties, subcellular distribution, and nature of enzyme complexes of the six biosynthetic enzymes. Biochemistry. 1973 Oct 9;12(21):4039–4051. doi: 10.1021/bi00745a004. [DOI] [PubMed] [Google Scholar]
- Siegel L. M., Monty K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966 Feb 7;112(2):346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Simmer J. P., Kelly R. E., Scully J. L., Grayson D. R., Rinker A. G., Jr, Bergh S. T., Evans D. R. Mammalian aspartate transcarbamylase (ATCase): sequence of the ATCase domain and interdomain linker in the CAD multifunctional polypeptide and properties of the isolated domain. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4382–4386. doi: 10.1073/pnas.86.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R. C., Reinisch K. M., Lipscomb W. N. Molecular structure of Bacillus subtilis aspartate transcarbamoylase at 3.0 A resolution. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6087–6091. doi: 10.1073/pnas.88.14.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yon R. J., Grayson J. E., Chawda A., Butterworth P. J. The quaternary structure of wheat-germ aspartate transcarbamoylase. Biochem J. 1982 May 1;203(2):413–417. doi: 10.1042/bj2030413. [DOI] [PMC free article] [PubMed] [Google Scholar]