Abstract
The integration of membrane proteins into “lipid raft” membrane domains influences many biochemical processes. The intrinsic structural properties of membrane proteins are thought to mediate their partitioning between membrane domains. However, whether membrane topology influences the targeting of proteins to rafts remains unclear. To address this question, we examined the domain preference of three putative raft-associated membrane proteins with widely different topologies: human caveolin-3, C99 (the 99 residue C-terminal domain of the amyloid precursor protein), and peripheral myelin protein 22. We find that each of these proteins are excluded from the ordered domains of giant unilamellar vesicles containing coexisting liquid-ordered and liquid-disordered phases. Thus, the intrinsic structural properties of these three topologically distinct disease-linked proteins are insufficient to confer affinity for synthetic raft-like domains.
Many biochemical processes are believed to be mediated by the partitioning of integral membrane proteins into so-called “lipid raft” nanodomains enriched with saturated lipids, sphingolipids, cholesterol, and assorted proteins.1 These membrane domains are thought to persist in a state resembling the liquid-ordered (Lo) phase. Preferential interactions between certain lipids and proteins are believed to drive the condensation of lipid rafts.2 However, emerging evidence suggests that, unlike those in model membranes, Lo domains are small and transient in cellular membranes,1,3,4 which complicates their direct characterization.5 Nevertheless, it seems clear that biochemically defined lipid rafts contain a specific subset of proteins,6 which is indicative of an underlying physical mechanism for membrane protein integration. Experimental evidence has suggested that certain structural and biochemical properties of membrane proteins help modulate their partitioning between coexisting liquid-disordered (Lα) and Lo membrane domains.7−9 However, the structural basis for this selectivity remains poorly understood.
A wealth of insight has emerged from investigations of synthetic liposomes, the precise composition of which can be controlled and manipulated. Ternary liposomal membranes provide a stringent system for the exploration of membrane protein partitioning between chemically pure, coexisting Lo and Lα domains at equilibrium.10,11 Several previous investigations have revealed that membrane proteins are often excluded from synthetic Lo domains.11 However, these investigations have been restricted to a limited number of integral and peripheral membrane proteins amenable to purification and reconstitution,12−19 many of which are bacterial, viral, or synthetic peptides. Furthermore, several putative raft-associated proteins have distinctive structural features that have been proposed to facilitate their raft partitioning.7−9 Therefore, in the current study we characterized the phase partitioning of membrane proteins with unusual topological features in order to determine whether these motifs can function as specific raft targeting mechanisms.
We focused on three human integral membrane proteins with distinct topologies (Supplementary Figure 1), which are known to reside in lipid rafts under physiological conditions: caveolin-3 (Cav3),20 C99—the 99-residue C-terminal domain of the amyloid precursor protein,21 and peripheral myelin protein 22 (PMP22).22 Here, we characterize the partitioning of non-post-translationally modified forms of these proteins in phase-separated giant unilamellar vesicles (GUVs). In addition to the biophysical relevance of these studies, each of these proteins are directly related to human disease (Cav3—muscle disorders,23 C99—Alzheimer’s disease,24 PMP22—Charcot-Marie-Tooth disease25). Therefore, these results may also illuminate disease mechanisms and inform the development of novel therapeutics.
The partitioning of membrane proteins between coexisting membrane domains can be directly assessed in phase-separated GUVs. We first generated GUVs exhibiting robust separation of liquid phases of known composition. The ternary phase diagram of membranes containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), N-palmitoyl-d-erythro-sphingosylphosphorylcholine (PSM), and cholesterol has been characterized in great detail.26,27 Macroscopic phase separation was consistently observed in GUVs composed of 2:2:1 POPC:PSM:cholesterol. This composition lies at the center of a tie line within the portion of the phase diagram in which Lo and Lα phases coexist.27 Therefore, equal fractions of Lo and Lα phases should persist at equilibrium in these membranes. Indeed, confocal images of these membranes containing trace amounts of fluorescent lipid marker Rh-DOPE confirm the coexistence of two phases with roughly equal proportions under these conditions (Figure 1A). Rh-DOPE consistently exhibited a strong preference for one of the two phases—likely the Lα phase. To confirm the identity of this phase we incorporated 0.1 mol % ganglioside GM1, which exhibits a strong preference for the Lo phase, into the membrane in the presence of fluorescently labeled GM1-binding protein cholera enterotoxin subunit B (ctxB). Consistent with previous observations,28 CtxB binds GM1 in the phase opposite of that marked by Rh-DOPE (Figure 1B), confirming that Rh-DOPE specifically marks the Lα phase. Together, these findings illustrate the robust phase separation and probe partitioning in these GUVs.
Figure 1.
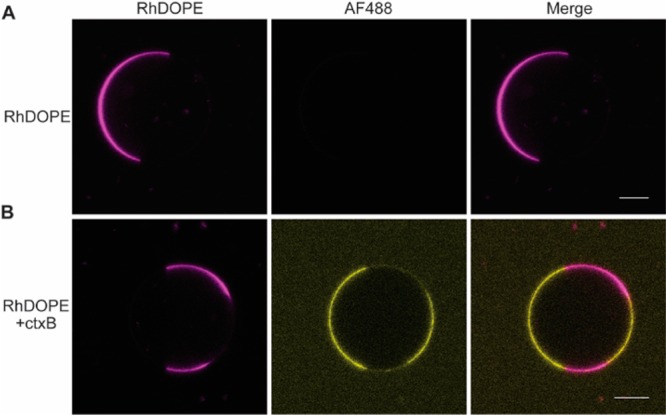
Phase-separated giant unilamellar vesicles. GUVs were formed from liposomes containing a 2:2:1 molar ratio of POPC:PSM:cholesterol. (A) To detect phase separation, 0.1 mol % of the fluorescent lipid tracer rhodamine DOPE (magenta) was incorporated to detect phase separation using confocal fluorescence microscopy. A representative image along with a 10 μm scale bar are shown for reference. (B) To identify the Lo phase, 0.1 mol % of the fluorescent lipid tracer rhodamine DOPE along with 0.1 mol % GM1 ganglioside were incorporated into the liposomes prior to GUV formation. GUVs were then mixed with 1 μg/ μL of AF488-labeled cholera enterotoxin subunit B (yellow) in order to mark the Lo phase. A representative image along with a 10 μm scale bar is shown for reference.
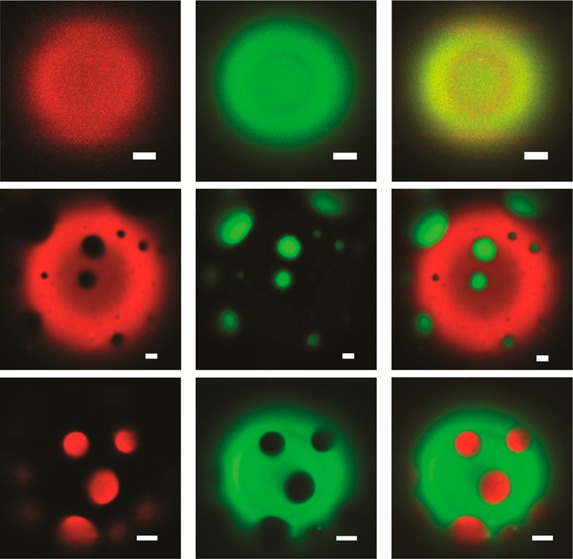
We first surveyed the partitioning of full-length human Cav3 in its non-lipidated form. Cav3 exhibits an uncommon topology featuring a single kinked helical hairpin that enters and exits the inner membrane leaflet (Supplementary Figure 1). Caveolins are critical components of caveolae,29 which are small raft-like membrane invaginations. Furthermore, Cav3 contains a CRAC motif30 and is highly enriched in biochemically extracted lipid raft preparations known as detergent resistant membranes (DRM).20 To determine whether Cav3 exhibits a thermodynamic preference for Lo domains, we first reconstituted fluorescently labeled Cav3 into proteoliposomes containing 2:2:1 POPC:PSM:cholesterol and the Lα phase marker Rh-DOPE at a molar lipid:protein ratio (LPR) of 400:1. Proteoliposomes were then electroswelled into GUVs and imaged using confocal fluorescence microscopy (see Supplementary Methods). We observed consistent colocalization of Cav3 and Rh-DOPE (3 trials, n = 38) (Figure 2A), which indicates the protein exhibits a strong preference for the Lα phase. Indeed, the partition coefficient of Cav3 could not be accurately determined due to the absence of detectable protein signal in the Lo domain of the vast majority of these GUVs. Similar results were obtained even when an alternative fluorophore, labeling position, and reconstitution protocol were employed (Supplementary Figure 2). We recognize that the oligomerization state of Cav3 is another variable that may be critical for regulating its phase preference. However, our studies were carried out at a bulk LPR of 400:1, which is almost certainly higher than the physiological concentration and potentially sufficient to promote oligomerization. Taken together, these results demonstrate that non-lipidated Cav3 is excluded from Lo domains.
Figure 2.
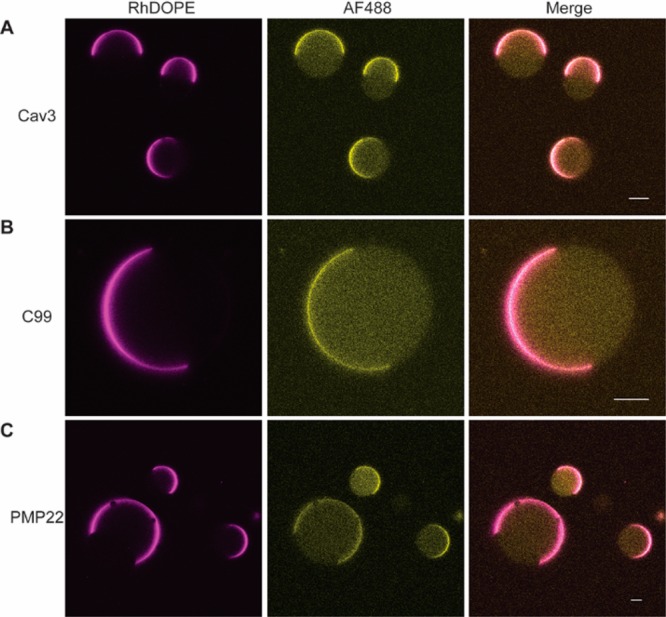
Partitioning of raftophillic integral membrane proteins in giant unilamellar vesicles. GUVs were formed from liposomes containing a 2:2:1 molar ratio of POPC:PSM:cholesterol along with 0.1 mol % of the fluorescent tracer lipid rhodamine-DOPE (magenta, Lα phase) and various membrane proteins at a bulk molar lipid: protein ratio of 400:1. (A) A representative image of GUVs containing AF488-labeled caveolin-3 (yellow) are shown along with a 10 μm scale bar for reference. (B) A representative image of GUVs containing AF488-labeled C99 (yellow) are shown along with a 10 μm scale bar for reference. (C) A representative image of GUVs containing AF488-labeled PMP22 (yellow) are shown along with a 10 μm scale bar for reference.
We next assessed the partitioning of C99, which consists of a single helical transmembrane domain flanked by short N- and C-terminal surface-associated helices (Supplementary Figure 1).31 C99 is found in DRMs.21 Additionally, the proteolytic processing of C99 is thought to depend on its association with lipid rafts.31−35 C99 is also known to bind cholesterol,31,36 which might promote partitioning into cholesterol-rich membrane domains. To determine whether C99 exhibits a thermodynamic preference for Lo domains, we reconstituted fluorescently labeled C99 into GUVs composed of 2:2:1 POPC:PSM:cholesterol and trace amounts of Rh-DOPE at an LPR of 400:1. Similar to our observations for Cav3, we also found C99 to consistently colocalize with the Lα domain marker (3 trials, n = 24) (Figure 2B). Again, the partition coefficient could not be accurately determined due to a lack of detectable C99 in the Lo phase in all cases. The partitioning of C99 was found to be identical even when an alternative fluorophore and lipid tracer were employed (Supplemental Figure 3). Together, these results demonstrate that C99 is excluded from Lo domains under these conditions.
Given its propensity to bind cholesterol, the exclusion of C99 from cholesterol-enriched Lo domains is somewhat surprising. However, such domains are only moderately enriched with cholesterol relative to the disordered phase. On the basis of the position of the tie line in the phase diagram,27 we expect cholesterol concentrations of 38 mol % in the Lo phase versus 10 mol % in the Lα phase under this condition. Furthermore, the difference in the concentration of free cholesterol in the two phases may be minimal due to favorable interactions between sphingomyelin and cholesterol in the Lo phase. Even if the activity coefficient for cholesterol approached unity in each phase, the difference in the free energy of binding would still only amount to roughly 0.6 kcal/mol given the experimentally determined equilibrium dissociation constant of 5 mol %.31 Our results suggest that this energetic contribution must be insignificant relative to the intrinsic transfer free energy of the protein. In this regard, it should also be noted that C99 likely persists as a mixture of monomers and dimers under this condition, the latter of which competes with cholesterol binding.36 Though we reconstituted C99 at a bulk LPR of 400:1, a condition under which C99 is predominantly monomeric,31,36 the effective LPR is likely to be closer to 200:1 because it is excluded from the lipids in the Lo phase. On the basis of our previous measurements,36 we expect C99 to be approximately 50% monomeric under this condition. Nevertheless, considering the absence of detectable signal in the Lo phase, it seems likely that both species exhibit a pronounced thermodynamic preference for the Lα phase.
Finally, we examined the partitioning of the multispan integral membrane protein PMP22. PMP22 forms a transmembrane four helix bundle that is highly abundant in the peripheral myelin sheath (Supplementary Figure 1).37,38 Its transmembrane helices are unusually long and hydrophobic. Like Cav3 and C99, PMP22 is found within cellular DRMs.22 Furthermore, its native membranes in the myelin sheath are highly enriched in both sphingolipids and cholesterol.39 To determine whether PMP22 partitions into Lo domains, we reconstituted fluorescently labeled PMP22 into GUVs containing 2:2:1 POPC:PSM:cholesterol and trace amounts of Rh-DOPE at a bulk LPR of 400:1. Again, PMP22 was only present at detectable levels in the Lα domains (three trials, n = 51) (Figure 2C). Similar results were obtained even when an alternative fluorophore and lipid tracer were employed (Supplemental Figure 3). These results demonstrate that PMP22 does not exhibit an intrinsic affinity for Lo domains under these conditions. This is particularly interesting in light of the fact that the “hydrophobic matching” of long transmembrane helices to thicker membranes has been found to mediate raft partitioning in some cases.9 Nevertheless, this protein is still apparently incompatible with the thicker Lo phase membranes within these GUVs.
The results of this study very clearly reveal that Cav3, C99, and PMP22 exhibit little tendency to partition into the Lo domain of phase-separated GUVs, despite their unusual structural features. This is contrary to an abundance of evidence suggesting that these proteins preferentially reside in the raft domains of cellular membranes: PMP22 in myelin membranes, Cav3 in membranes of caveolae, and C99 in raft-like membrane nanodomains.20−22 We suggest three nonmutually exclusive hypotheses to account for these GUV results.
First, the lack of appropriate post-translational modifications could potentially account for the difference between observations in GUVs relative to those in cellular membranes. It has been established that raft-association of several proteins is promoted by thiopalmitoylation.11,40 While C99 is not lipidated, both Cav3 and PMP22 are known to be natively thiopalmitoylated; Cav3 has three palmitoylation sites and PMP22 has one.41,42 The recombinant forms of Cav3 and PMP22 used in these studies were not lipidated. Thus, it is possible that the partitioning of the thiopalmitoylated forms of Cav3 and PMP22 may be distinct from that of the unmodified forms of these proteins. Indeed, the reversibility of palmitoylation makes it a potentially versatile biochemical mechanism for the dynamic remodeling of biological membranes. While the isolation and characterization of palmitoylated PMP22 and Cav3 are beyond the scope of this paper, this represents an important hypothesis for future investigations.
A second possible explanation involves differences in the physical and chemical properties of synthetic Lo domains relative to those of cellular lipid rafts. Because of their homogeneous lipid compositions, these liposomal Lo domains are more ordered than those found within natural membranes.10,11 It may be that the interactions between sphingomyelin and cholesterol are too strong in these synthetic membranes to accommodate the disruptive influence of a flexible polypeptide.43 In synthetic membranes, the characteristic features of Lo domains are also likely to diverge from those of native membranes in several ways including but not limited to their material properties,44 stoichiometric ratio of sphingomyelin and cholesterol,45 symmetric distribution of lipids in the inner and outer leaflets,4,46 and lack of a cytoskeletal scaffold.47
A third possibility is that integration of these proteins into cellular lipid rafts hinges upon specific binding interactions that are not present under these conditions. Cav3, C99, and PMP22 each form complexes with other proteins in the cell.48−51 Additionally, cellular lipid rafts contain a host of chemically distinct lipids that are not present in the ternary lipid mixture utilized herein. It cannot be excluded that raft association of Cav3, C99, and PMP22 is dependent on association with specific raft-resident lipids and/or proteins in the cell.
From our results we can conclude that the intrinsic structural properties of these topologically diverse proteins are insufficient to endow them with an intrinsic affinity for a simplified liquid-ordered membrane domain. Further studies on the targeting mechanisms of Cav3, C99, and PMP22 are needed to gain insight into the regulation of their spatiotemporal distribution in biological membranes. Such insights will also fortify our understanding of both the native functions and the disease mechanisms associated with each protein, insight that could potentially translate to new therapeutic strategies.
Acknowledgments
We thank Ajit Tiwari, Courtney Copeland, and Krishnan Raghunathan for technical assistance, as well as Patricia Bassereau (Institut Curie), Daniel Huster (Universität Leipzig), Sarah Veatch (University of Michigan), Ilya Levental (University of Texas Health Science Center), Andrew Beel (Stanford University), and Catherine Deatherage (Vanderbilt University) for helpful discussions.
Glossary
Abbreviations
- Cav3
caveolin 3
- C99
the 99-residue C-terminal fragment of the amyloid precursor protein
- APP
the amyloid precursor protein
- PMP22
peripheral myelin protein 22
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PSM
N-palmitoyl-d-erythro-sphingosylphosphorylcholine
- GUV
giant unilamellar vesicle
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications Web site. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b01154.
Supplemental Methods, Supplemental References, and Supplemental Figures 1–3 (PDF)
This work was supported by U.S. NIH Grants RO1 GM106672 and U54 GM094608. J.P.S. was supported by NIH F32 GM110929, while P.J.B. was supported by F31 NS077681.
The authors declare no competing financial interest.
Supplementary Material
References
- Lingwood D.; Simons K. (2010) Science 327, 46–50. 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Simons K.; Ikonen E. (1997) Nature 387, 569–572. 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Hancock J. F. (2006) Nat. Rev. Mol. Cell Biol. 7, 456–462. 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolfsson H. I.; Melo M. N.; van Eerden F. J.; Arnarez C.; Lopez C. A.; Wassenaar T. A.; Periole X.; de Vries A. H.; Tieleman D. P.; Marrink S. J. (2014) J. Am. Chem. Soc. 136, 14554–14559. 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- Kraft M. L. (2013) Mol. Biol. Cell 24, 2765–2768. 10.1091/mbc.E13-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A.; Chen D.; Boda A. R.; Foster L. J.; Davis M. J.; Hill M. M. (2015) Nucleic Acids Res. 43, D335–338. 10.1093/nar/gku1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P.; Roth M. G.; Simons K. (1997) EMBO J. 16, 5501–5508. 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benslimane N.; Hassan G. S.; Yacoub D.; Mourad W. (2012) PLoS One 7, e43070. 10.1371/journal.pone.0043070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q.; London E. (2013) J. Biol. Chem. 288, 1340–1352. 10.1074/jbc.M112.415596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser H. J.; Lingwood D.; Levental I.; Sampaio J. L.; Kalvodova L.; Rajendran L.; Simons K. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 16645–16650. 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent J. H.; Levental I. (2015) Chem. Phys. Lipids 192, 23–32. 10.1016/j.chemphyslip.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Nikolaus J.; Scolari S.; Bayraktarov E.; Jungnick N.; Engel S.; Plazzo A. P.; Stockl M.; Volkmer R.; Veit M.; Herrmann A. (2010) Biophys. J. 99, 489–498. 10.1016/j.bpj.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson L. D.; Chiantia S.; London E. (2010) Biophys. J. 99, 3255–3263. 10.1016/j.bpj.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacia K.; Schuette C. G.; Kahya N.; Jahn R.; Schwille P. (2004) J. Biol. Chem. 279, 37951–37955. 10.1074/jbc.M407020200. [DOI] [PubMed] [Google Scholar]
- Kahya N.; Brown D. A.; Schwille P. (2005) Biochemistry 44, 7479–7489. 10.1021/bi047429d. [DOI] [PubMed] [Google Scholar]
- Kalvodova L.; Kahya N.; Schwille P.; Ehehalt R.; Verkade P.; Drechsel D.; Simons K. (2005) J. Biol. Chem. 280, 36815–36823. 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- Shogomori H.; Hammond A. T.; Ostermeyer-Fay A. G.; Barr D. J.; Feigenson G. W.; London E.; Brown D. A. (2005) J. Biol. Chem. 280, 18931–18942. 10.1074/jbc.M500247200. [DOI] [PubMed] [Google Scholar]
- Tong J.; Briggs M. M.; Mlaver D.; Vidal A.; McIntosh T. J. (2009) Biophys. J. 97, 2493–2502. 10.1016/j.bpj.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal A.; McIntosh T. J. (2005) Biophys. J. 89, 1102–1108. 10.1529/biophysj.105.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F.; Engelman J. A.; Volonte D.; Zhang X. L.; Minetti C.; Li M.; Hou H. Jr.; Kneitz B.; Edelmann W.; Lisanti M. P. (2001) J. Biol. Chem. 276, 21425–21433. 10.1074/jbc.M100828200. [DOI] [PubMed] [Google Scholar]
- Urano Y.; Hayashi I.; Isoo N.; Reid P. C.; Shibasaki Y.; Noguchi N.; Tomita T.; Iwatsubo T.; Hamakubo T.; Kodama T. (2005) J. Lipid Res. 46, 904–912. [DOI] [PubMed] [Google Scholar]
- Erne B.; Sansano S.; Frank M.; Schaeren-Wiemers N. (2002) J. Neurochem. 82, 550–562. 10.1046/j.1471-4159.2002.00987.x. [DOI] [PubMed] [Google Scholar]
- Gazzerro E.; Sotgia F.; Bruno C.; Lisanti M. P.; Minetti C. (2010) Eur. J. Hum. Genet. 18, 137–145. 10.1038/ejhg.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M. S. (2012) Biol. Chem. 393, 899–905. 10.1515/hsz-2012-0140. [DOI] [PubMed] [Google Scholar]
- Li J.; Parker B.; Martyn C.; Natarajan C.; Guo J. (2013) Mol. Neurobiol. 47, 673–698. 10.1007/s12035-012-8370-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch S. L.; Keller S. L. (2005) Phys. Rev. Lett. 94, 148101. 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- Ionova I. V.; Livshits V. A.; Marsh D. (2012) Biophys. J. 102, 1856–1865. 10.1016/j.bpj.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonov A. V.; Mihalyov I.; Cohen F. S. (2001) Biophys. J. 81, 1486–1500. 10.1016/S0006-3495(01)75803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parat M. O. (2009) Int. Rev. Cell Mol. Biol. 273, 117–162. 10.1016/S1937-6448(08)01804-2. [DOI] [PubMed] [Google Scholar]
- Li H.; Papadopoulos V. (1998) Endocrinology 139, 4991–4997. 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- Barrett P. J.; Song Y.; Van Horn W. D.; Hustedt E. J.; Schafer J. M.; Hadziselimovic A.; Beel A. J.; Sanders C. R. (2012) Science 336, 1168–1171. 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.; Kenworthy A. K.; Sanders C. R. (2014) Protein Sci. 23, 1–22. 10.1002/pro.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel A. J.; Sakakura M.; Barrett P. J.; Sanders C. R. (2010) Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1801, 975–982. 10.1016/j.bbalip.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel A. J.; Mobley C. K.; Kim H. J.; Tian F.; Hadziselimovic A.; Jap B.; Prestegard J. H.; Sanders C. R. (2008) Biochemistry 47, 9428–9446. 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehehalt R.; Keller P.; Haass C.; Thiele C.; Simons K. (2003) J. Cell Biol. 160, 113–123. 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.; Hustedt E. J.; Brandon S.; Sanders C. R. (2013) Biochemistry 52, 5051–5064. 10.1021/bi400735x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittendorf K. F.; Kroncke B. M.; Meiler J.; Sanders C. R. (2014) Biochemistry 53, 6139–6141. 10.1021/bi500809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snipes G. J.; Suter U.; Welcher A. A.; Shooter E. M. (1992) J. Cell Biol. 117, 225–238. 10.1083/jcb.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. J.; Iwamori M.; Kishimoto Y.; Ostroff S. M.; Moser H. W.; Asbury A. K. (1979) Ann. Neurol. 5, 239–244. 10.1002/ana.410050305. [DOI] [PubMed] [Google Scholar]
- Levental I.; Lingwood D.; Grzybek M.; Coskun U.; Simons K. (2010) Proc. Natl. Acad. Sci. U. S. A. 107, 22050–22054. 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzen D. J.; Hastings W. R.; Lublin D. M. (1995) J. Biol. Chem. 270, 6838–6842. 10.1074/jbc.270.12.6838. [DOI] [PubMed] [Google Scholar]
- Zoltewicz S. J.; Lee S.; Chittoor V. G.; Freeland S. M.; Rangaraju S.; Zacharias D. A.; Notterpek L. (2012) ASN Neuro 4, 409–421. 10.1042/AN20120045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida P. F. (2009) Biochim. Biophys. Acta, Biomembr. 1788, 72–85. 10.1016/j.bbamem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- McIntosh T. J.; Simon S. A. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 177–198. 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- Frisz J. F.; Klitzing H. A.; Lou K.; Hutcheon I. D.; Weber P. K.; Zimmerberg J.; Kraft M. L. (2013) J. Biol. Chem. 288, 16855–16861. 10.1074/jbc.M113.473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain N. F.; Siegel A. P.; Ge Y.; Jordan R.; Naumann C. A. (2013) Biophys. J. 104, 2212–2221. 10.1016/j.bpj.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller V.; Ringemann C.; Honigmann A.; Schwarzmann G.; Medda R.; Leutenegger M.; Polyakova S.; Belov V. N.; Hell S. W.; Eggeling C. (2011) Biophys. J. 101, 1651–1660. 10.1016/j.bpj.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H. H.; Murray F.; Insel P. A. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 359–391. 10.1146/annurev.pharmtox.48.121506.124841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasse B.; Bosse F.; Hanenberg H.; Müller H. W. (2004) Mol. Cell. Neurosci. 27, 370–378. 10.1016/j.mcn.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Tamayev R.; Zhou D.; D’Adamio L. (2009) Mol. Neurodegener. 4, 1–14. 10.1186/1750-1326-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T.; Pietrzik C. U. (2012) Exp. Brain Res. 217, 377–387. 10.1007/s00221-011-2876-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


