Abstract
The gut microbiota influences many aspects of host metabolism. We have previously shown that the presence of a gut microbiota remodels lipid composition. Here we investigated how interaction between gut microbiota and dietary lipids regulates lipid composition in the liver and plasma, and gene expression in the liver. Germ-free and conventionally raised mice were fed a lard or fish oil diet for 11 weeks. We performed lipidomics analysis of the liver and serum and microarray analysis of the liver. As expected, most of the variation in the lipidomics dataset was induced by the diet, and abundance of most lipid classes differed between mice fed lard and fish oil. However, the gut microbiota also affected lipid composition. The gut microbiota increased hepatic levels of cholesterol and cholesteryl esters in mice fed lard, but not in mice fed fish oil. Serum levels of cholesterol and cholesteryl esters were not affected by the gut microbiota. Genes encoding enzymes involved in cholesterol biosynthesis were downregulated by the gut microbiota in mice fed lard and were expressed at a low level in mice fed fish oil independent of microbial status. In summary, we show that gut microbiota-induced regulation of hepatic cholesterol metabolism is dependent on dietary lipid composition.
Keywords: cholesterol/biosynthesis, fish oil, gene expression, lipidomics, liver, microarrays, germ-free
The composition of dietary fat is known to affect metabolic health. Consumption of saturated lipids is associated with metabolic perturbations such as obesity, hyperglycemia, dyslipidemia, increased hepatic lipogenesis, and liver steatosis (1). In contrast, consumption of polyunsaturated fatty acids is associated with a lean and metabolically healthy phenotype (2–4). Emerging evidence suggests that some of the effects of dietary lipids on host metabolism may be mediated by modifications of the gut microbiota composition, which result in altered metabolic properties of the gut microbiota (5–8) and/or by alterations in gut integrity, which affect the extent of leakage of microbially derived metabolites into the circulation (6).
Metabolites produced by the gut microbiota may be utilized as a source of energy by the host or may act as signaling molecules that influence host metabolism (8). These metabolites can have a local effect in the intestine, but may also be transferred from the gut into the circulation, thereby affecting peripheral tissues (8). Indeed, germ-free (GF) mice have been shown to have improved glucose sensitivity and to be protected against obesity and dyslipidemia (9–12), exemplifying the systemic importance of the microbiota. Because of its proximity to the gastrointestinal tract, the liver is strongly affected by the microbiota, and bacterial status has been shown to affect both hepatic lipid biosynthesis (9) and lipid degradation (13). Several mechanisms linking bacterial metabolic processes with liver metabolism have been described (14); for example, microbial processing of dietary fibers, bile acids, and choline produces metabolites that regulate the activity of hepatic transcription factors and, hence, metabolic processes.
Here, we aimed to determine how the gut microbiota affects hepatic lipid metabolism during the metabolic challenge of a high-fat diet and how interaction between dietary lipids and the gut microbiota influences lipid composition and regulation of metabolic pathways.
MATERIALS AND METHODS
Mice
C57Bl/6 mice were maintained under standard specific-pathogen free or GF conditions, as described previously (10). All mice were males, age matched, and 12–14 weeks of age. Mice were fed irradiated isocaloric diets (45% kcal fat) of identical composition except for the source of fat, which was either menhaden fish oil (Research Diets, D05122102) or lard (Research Diets, D10011202) for 11 weeks (7). Fatty acid composition of the diets is displayed in supplementary Table 1. The mice were fasted for 4 h before they were euthanized. Blood samples and liver samples were harvested at the end of the experiment. All experiments were performed with protocols approved by the University of Gothenburg Animal Studies Committee.
Lipidomics analysis
Lipids were analyzed by ultra-performance LC coupled with TOFMS using a Waters Q-TOF Premier (Waters, Milford, MA) with a methodology described earlier (15). Briefly, the samples were extracted with a chloroform:methanol mixture after addition of internal standards containing lysophosphatidylcholine (17:0), phosphatidylcholine (17:0/17:0), phosphatidylethanolamines (17:0/17:0), ceramide (d18:1/17:0) (Avanti Polar Lipids, Alabaster, AL), and triglyceride (17:0/17:0/17:0) (Larodan Fine Chemicals, Malmö, Sweden). Finally, an additional external standard solution composed of three stable isotope-labeled reference compounds [lysophosphatidylcholine (16:1-D3), phosphatidylcholine (16:1/16:1-D6), and triglyceride (16:0/16:0/16:0-13C3] was added. The data were processed using MZmine 2 (16), which included alignment of peaks, peak integration, isotopic grouping, normalization, and peak identification. Lipids were identified using an in-house lipid species database and based on their MS/MS spectra. The data were normalized using the internal standards representative of each class of lipid present in the samples, as previously described (15). Sphingomyelins were normalized to the phosphatidylcholine standard.
The serum cholesterol concentrations were obtained by a metabolomics method based on 2D GC-MS. Briefly, each serum sample (30 μl) was spiked with 10 μl of internal standard solution (valine-d8, c = 37 mg/l; heptadecanoic acid, c = 187 mg/l; and succinic-d4 acid, c = 63 mg/l), extracted with 400 μl of methanol, centrifuged, and evaporated to dryness. Cholesterol was converted into its methoxime and TMS derivative(s) by two-step derivatization using methoxime and n-methyl-n-trimethylsilyl trifluoroacetamide reagents, and a retention index standard mixture (n-alkanes) in hexane was added. The analysis was performed as previously described (17) using a Leco Pegasus 4D GC×GC-TOFMS instrument (Leco Corp., St. Joseph, MI) equipped with a cryogenic modulator and with an Agilent 6890 gas chromatograph (Agilent Technologies, Palo Alto, CA) as the GC part of the instrument. The raw data processing was done using ChromaTOF version 4.22 (Leco Corp.) software. Finally, alignment and normalization of the data were done using an in-house developed program, Guineu (17), and concentrations of cholesterol were calculated using a calibration curve. Hepatic free cholesterol was extracted using the Folch method (18) and quantified using straight-phase HPLC coupled to evaporative light scattering detection (19).
Microarray expression analysis
RNA was isolated from the left hepatic lobe using RNeasy lipid tissue mini kit (Qiagen, Hilden, Germany). cDNA synthesis, labeling, hybridization to GeneChip® ST arrays (GeneChip® Mouse Gene 1.0 ST array), washing, and scanning were performed as previously described (7). The raw data were normalized using the robust multi-array average method (20). Differential expression of microarray data was evaluated by Student’s t-test followed by correction for false discovery rate (FDR). Analysis of interactions, using colonization status and diet as independent variables, was performed by two-way analysis of interaction followed by correction for FDR. Analysis of enrichment of regulated genes within functional categories [gene ontology (GO) categories] (21) was performed using the software DAVID (http://david.abcc.ncifcrf.gov/) (22). The results of the enrichment calculations were filtered for GO categories that were significantly enriched (P < 0.05) after Bonferroni correction. Redundancy within lists of GO terms was reduced by the Revigo software (http://revigo.irb.hr/) (23) with a similarity score set to 0.5. Principle component analysis (PCA) was performed in MultiExperiment Viewer. Microarray data have been uploaded to the Gene Expression Omnibus (GEO) database with accession number GSE73195.2
Immunoblot
Protein extracts (20 μg) were loaded onto 10% NuPAGE Bis-Tris gels (Invitrogen, Carlsbad, CA) and blotted to Hybond P polyvinylidene difluoride membranes (Invitrogen). After 1 h blocking in PBS Tween-20 (PBST) supplemented with 5% fat-free milk powder, the membranes were incubated overnight with primary antibodies [sterol regulatory element-binding protein 2 (SREBP2) (ab30682; Abcam, Cambridge, UK) or actin (1615; Santa Cruz Biotechnology, Santa Cruz, CA)] diluted 1:1,000 in blocking buffer. The membranes were subsequently washed in PBST, incubated for 1 h with secondary antibodies [donkey anti-rabbit lgG (NA934V; GE Healthcare, Little Chalfont, UK), or donkey anti-goat lgG (Sc2056; Santa Cruz Biotechnology)] diluted 1:2,000 in blocking buffer, and finally washed in PBST. Proteins were detected with LumiGLO blotting detection kit (Cell Signaling, Danvers, MA).
Statistical analysis
Data are represented as mean ± SEM. Statistical comparison of two groups was performed by Student’s t-test. Analysis of datasets with 2 × 2 factorial design (comparing bacterial status and diet) was performed by two-way ANOVA with Tukey’s multiple comparison test. Statistical analysis was performed in GraphPad Prism 6 unless otherwise stated.
RESULTS
Interaction between dietary lipids and gut microbiota regulates the levels of cholesteryl esters in the liver
We have previously shown that the gut microbiota affects liver and serum lipid composition in mice fed a chow diet (24) and that interaction between gut microbiota and dietary lipids influences adiposity, adipose tissue inflammation, and systemic glucose metabolism (7). Here we performed lipidomics analysis of liver and serum from conventionally raised (CONV-R) and GF mice fed a lard or fish oil diet for 11 weeks to study how interaction between the gut microbiota and dietary lipids affects hepatic and serum lipid composition.
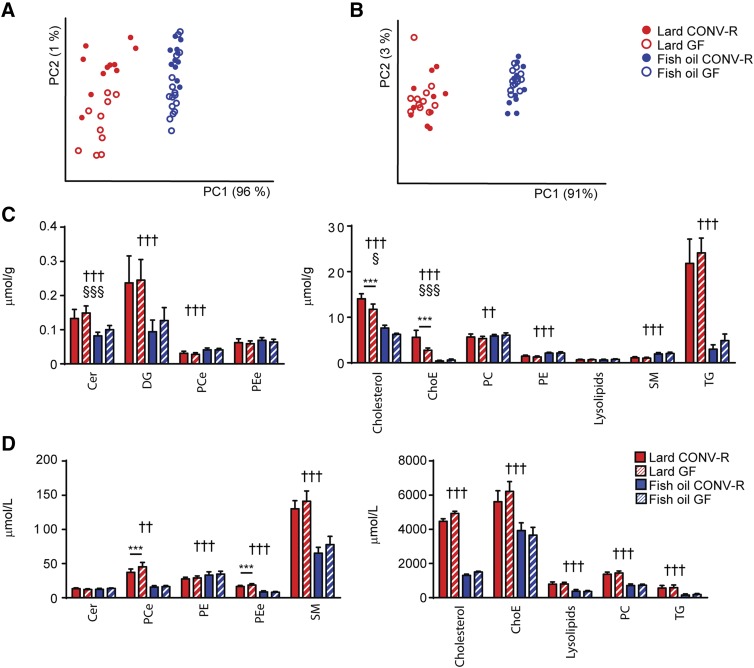
GF mice gained approximately 30% less weight than CONV-R mice (data not shown). To investigate the influence of gut microbiota on lipid metabolism independent of differences in body weight, we used GF mice that were slightly heavier than their CONV-R counterparts at the beginning of the feeding period. At the end of the experiment the CONV-R and GF mice had matching body weights (supplementary Fig. 1A). Diet consumption was similar between CONV-R and GF mice, while mice fed lard consumed more food than mice fed fish oil (supplementary Fig. 1B). PCA of lipidomics data showed that most of the variation in the dataset was induced by diet. Samples separated by diet in the first dimension, which accounted for 96% of the variation among liver samples (Fig. 1A) and for 91% of the variation among serum samples (Fig. 1B). Liver samples from mice fed lard separated on microbial status in the second dimension (Fig. 1A), which represented approximately 1% of the total variation in the dataset. Serum samples did not separate on microbial status in either of the dietary groups (Fig. 1B).
Fig. 1.
Lipidomics analysis of liver and serum from CONV-R and GF mice fed lard or fish oil for 11 weeks. PCA of lipidomics data from liver (A) and serum (B) samples from CONV-R and GF mice fed lard or fish oil. Liver samples: n = 11 (CONV-R lard); 10 (GF lard); 11 (CONVR fish oil); 18 (GF fish oil). Serum samples: n = 11 (CONV-R lard); 10 (GF lard); 10 (CONV-R fish oil); 15 (GF fish oil). Abundance of lipid classes in the liver (C) and in serum (D) of CONV-R and GF mice fed lard or fish oil. Liver cholesterol: n = 9 mice per group; other liver lipids: n = 11 (CONV-R lard); 10 (GF lard); 11 (CONVR fish oil); 18 (GF fish oil); all serum lipids: n = 11 (CONV-R lard); 10 (GF lard); 10 (CONVR fish oil); 15 (GF fish oil). Cer, ceramide; DG, diglyceride; PCe, ether phosphatidylcholines; PEe, ether phosphatidylethanolamine; ChoE, cholesteryl ester; PC, phosphatidylcholines; PE, phosphatidylethanolamine. Mean values ± SEM are plotted. Variation induced by the diet: ††P < 0.01, †††P < 0.001. Variation induced by the gut microbiota: §P < 0.05, §§§P < 0.001. Post hoc multiple comparison analysis: ***P < 0.001.
Dietary lipids affected the abundance of the majority of lipid classes both in liver and in serum (Fig. 1C, D). However, the gut microbiota only induced increased hepatic levels of cholesterol and cholesteryl esters (Fig. 1C) and decreased serum levels of ether phosphatidylcholines and ether phosphatidylethanolamines in mice fed lard, but not in mice fed fish oil (Fig. 1D).
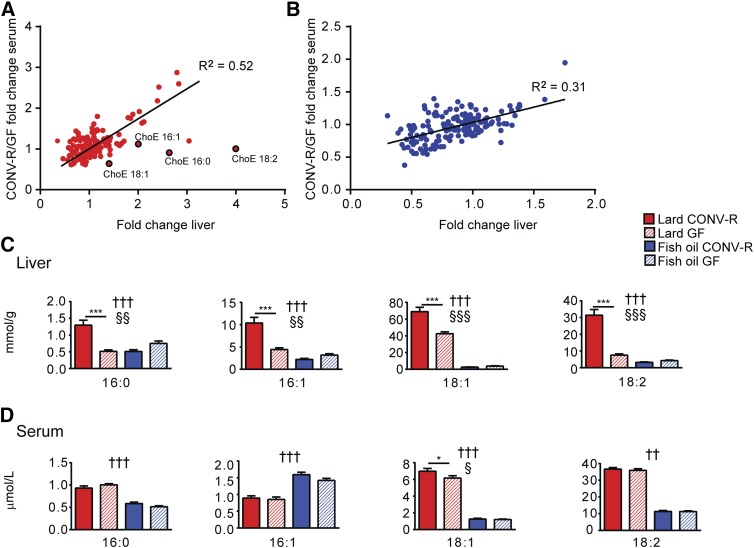
Despite the relatively small contribution by the microbiota to the overall variation in the lipidomics dataset, many individual lipid species in the liver and in serum were regulated by the gut microbiota (supplementary Tables 1, 2). To investigate whether the effect of the gut microbiota on individual lipid species in the liver and serum was similar, we plotted regulation induced by the gut microbiota in the liver against regulation induced by the gut microbiota in serum and observed a positive linear relationship in both dietary groups (Fig. 2A, B). An exception to this trend was observed for cholesteryl ester species, which were upregulated by the gut microbiota in the liver, but not in serum (except for a small increase in 18:1) of mice fed lard (Fig. 2A, C, D), but were not influenced by the gut microbiota in either the liver or serum of mice fed fish oil (Fig. 2B–D).
Fig. 2.
Cholesteryl esters in the liver are increased by the gut microbiota in mice fed lard for 11 weeks. Microbial regulation of liver and serum lipid species in mice fed lard (A) and fish oil (B). Fold changes for lipid species significantly regulated by the microbiota in serum and liver are plotted on the y axis and x axis, respectively. ChoE, cholesteryl ester. Abundance of cholesteryl ester species in the liver (C) and in serum (D) from CONV-R and GF mice fed lard or fish oil. Liver samples: n = 11 (CONV-R lard); 10 (GF lard); 11 (CONVR fish oil); 18 (GF fish oil). Serum samples: n = 11 (CONV-R lard); 10 (GF lard); 10 (CONVR fish oil); 15 (GF fish oil). Mean values ± SEM are plotted. Variation induced by the diet: ††P < 0.01, †††P < 0.001. Variation induced by the gut microbiota: §P < 0.05, §§P < 0.01, §§§P < 0.001. Post hoc multiple comparison analysis: *P < 0.05, ***P < 0.001.
Interaction between dietary lipids and gut microbiota regulates hepatic cholesterol biosynthesis
To study how the gut microbiota and dietary lipids regulate liver lipid metabolism, we performed microarray analysis of liver samples from CONV-R and GF mice fed lard or fish oil for 11 weeks.
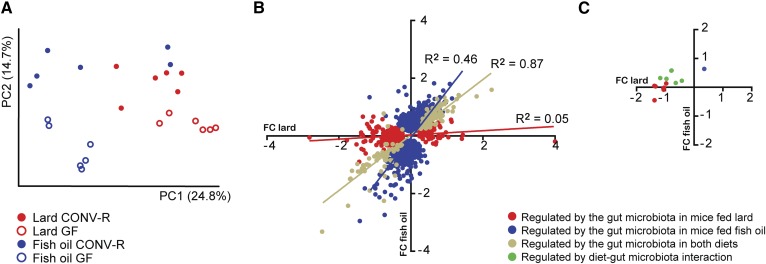
PCA of hepatic gene expression data revealed that mice separated on diet in the first dimension and on microbial status in the second dimension (Fig. 3A). To compare how the gut microbiota affects hepatic gene expression in mice fed lard or fish oil, we plotted differences in hepatic gene expression level that were significant between CONV-R and GF mice, with values for mice fed fish oil on the y axis and values for mice fed lard on the x axis (Fig. 3B). We found a linear relationship between microbiota-induced gene regulation in liver from mice fed lard or fish oil (Fig. 3B), indicating that many genes are regulated by the gut microbiota independently of dietary lipids.
Fig. 3.
Gene expression in the liver of CONV-R and GF mice fed lard or fish oil for 11 weeks. A: PCA of global hepatic gene expression in CONV-R and GF mice fed lard or fish oil (n = 6 mice per group). B: Microbial regulation of hepatic genes in mice fed lard (x axis) or fish oil (y axis). Fold changes (FC) for genes significantly regulated (P < 0.05, corrected for FDR) are displayed (n = 6 mice per group). C: Microbial regulation of hepatic genes within GO category GO:016126 (sterol biosynthesis) in mice fed lard (x axis) or fish oil (y axis). Fold changes for genes significantly regulated (P < 0.05, corrected for FDR) are displayed (n = 6 mice per group).
GO analysis showed that the microbiota induced hepatic expression of genes involved in the immune response in mice fed fish oil (Table 1). Most of these genes also showed a tendency toward being upregulated in mice fed lard, but did not reach significance in these mice (data not shown). Gene expressions of the macrophage markers Cd68, Emr1 (encoding F4/80), and Clec4f were upregulated by the gut microbiota on both diets (supplementary Fig. 2A–C) indicating that the microbiota induces infiltration of macrophages to the liver. However, the expression of genes encoding interleukins was not affected by either dietary lipids or microbial status (data not shown). Genes in functional categories associated with cholesterol biosynthesis were downregulated by the gut microbiota in mice fed lard (Table 1, Fig. 3C). Two-way ANOVA also showed that genes regulated by the interaction between dietary lipids and the gut microbiota were highly enriched in processes associated with cholesterol biosynthesis (Table 1, Fig. 3C).
TABLE 1.
Gene categories enriched in genes regulated by the gut microbiota during lard and fish oil diet
| GO | Term | Fold Enrichment | FDR (Adjusted P value) |
| Enriched in genes induced by the gut microbiota in mice fed lard | |||
| — | — | — | — |
| Enriched in genes reduced by the gut microbiota in mice fed lard | |||
| 16126 | Sterol biosynthetic process | 30.4 | 1.31E-09 |
| 6695 | Cholesterol biosynthetic process | 36.0 | 1.67E-09 |
| 8202 | Steroid metabolic process | 8.7 | 3.36E-08 |
| 16125 | Sterol metabolic process | 14.0 | 3.17E-08 |
| 8203 | Cholesterol metabolic process | 14.2 | 1.42E-07 |
| 6694 | Steroid biosynthetic process | 14.0 | 1.38E-07 |
| 8610 | Lipid biosynthetic process | 4.9 | 5.25E-05 |
| 7155 | Cell adhesion | 3.4 | 1.36E-04 |
| 22610 | Biological adhesion | 3.4 | 1.24E-04 |
| 8299 | Isoprenoid biosynthetic process | 18.8 | 1.40E-02 |
| 6720 | Isoprenoid metabolic process | 10.1 | 2.98E-02 |
| Enriched in genes induced by the gut microbiota in mice fed fish oil | |||
| 6955 | Immune response | 3.1 | 5.79E-03 |
| Enriched in genes reduced by the gut microbiota in mice fed fish oil | |||
| — | — | — | — |
| Enriched in genes regulated by interaction between dietary lipids and gut microbiota | |||
| 6694 | Steroid biosynthetic process | 15.0 | 1.10E-04 |
| 8202 | Steroid metabolic process | 7.3 | 3.45E-03 |
| 16126 | Sterol biosynthetic process | 19.7 | 2.99E-02 |
| 30198 | Extracellular matrix organization | 8.2 | 4.15E-02 |
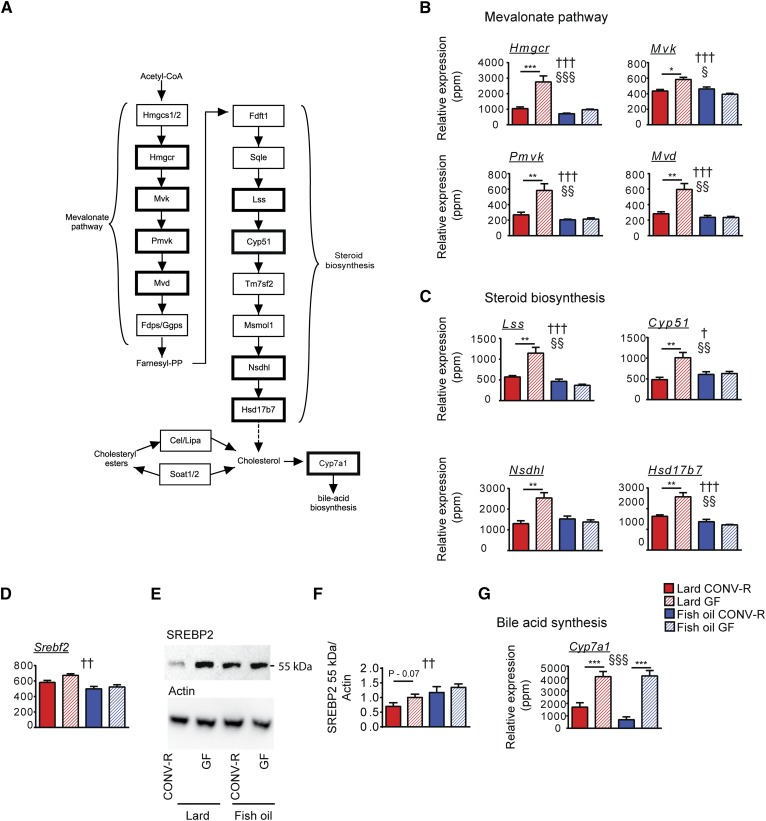
We found that the gut microbiota reduced the hepatic expression of many genes encoding enzymes in: 1) the mevalonate pathway, including the rate-limiting enzyme HMG-CoA reductase encoded by Hmgcr (Fig. 4A, B); and 2) the steroid biosynthesis pathway (Fig. 4A, C) in mice fed lard. The expression levels of these genes were low in mice fed fish oil independent of microbial status (Fig. 4B, C).
Fig. 4.
Expression of genes involved in cholesterol biosynthesis is decreased by the gut microbiota in mice fed lard. A: Schematic overview of de novo cholesterol biosynthesis pathways, including the mevalonate pathway and steroid biosynthesis, and synthesis of cholesteryl esters. Genes reported in (B) and (C) are indicated by bold borders. B: Genes in the mevalonate pathway significantly regulated by the gut microbiota in mice fed lard (n = 6 mice per group). C: Genes in the steroid biosynthesis pathway significantly regulated by the gut microbiota in mice fed lard (n = 6 mice per group). D: Expression of Srebf2 encoding the transcription factor SREBP2 (n = 6 mice per group). E: Representative immunoblot showing the 55 kDa fragment of SREBP2 and actin. F: Quantification of SREBP2 determined by the ratio between SREBP2 and actin (n = 9 mice per group). G: Expression of Cyp7a1 (n = 6 mice per group). Mean values ± SEM are plotted. Variation induced by the diet: †P < 0.05, ††P < 0.01, †††P < 0.001. Variation induced by the gut microbiota: §P < 0.05, §§P < 0.01, §§§P < 0.001. Post hoc multiple comparison analysis: *P < 0.05, **P < 0.01, ***P < 0.001.
To study the mechanisms underlying the regulation of cholesterol biosynthesis by gut microbiota and dietary lipids, we analyzed gene expression and protein cleavage of sterol regulatory element-binding protein 2 (SREBP2), a key regulator of cholesterol homeostasis. Gene expression of Srebf2 (encoding SREBP2) was increased in mice fed lard compared with mice fed fish oil (Fig. 4D). The bioactive cleaved form of SREBP2 was higher in mice fed fish oil than in mice fed lard, and there was a trend toward a reduction by the microbiota in mice fed lard (Fig. 4E, F).
The rate-limiting enzyme of bile acid synthesis, CYP7A1, which uses cholesterol as substrate, was downregulated by the gut microbiota both in mice fed lard and in mice fed fish oil (Fig. 4G).
Taken together, we found that genes involved with cholesterol biosynthesis were downregulated: 1) by the gut microbiota in mice fed lard; and 2) by fish oil independent of microbial status.
DISCUSSION
In the present study, we demonstrate that the interaction between gut microbiota and dietary lipids regulates cholesterol metabolism in the liver. The gut microbiota increased hepatic levels of cholesterol and cholesteryl esters and decreased the expression of genes involved in cholesterol biosynthesis in mice fed lard, but not in mice fed fish oil.
Fatty acids are absorbed from the intestine and utilized as building-blocks in the biosynthesis of lipids. In accordance with previous reports (25), we found that dietary lipids strongly affected the abundance and composition of lipids in the liver and in serum. We have previously shown that the presence of a gut microbiota in mice fed a chow diet regulates lipid composition in serum and liver and decreases abundance of hepatic cholesteryl esters (24). The present study demonstrates that the gut microbiota also influences lipid composition in mice on a high-fat diet. The gut microbiota regulated the abundance of many lipid species, but had a modest effect on the abundance of lipid classes with the exception of cholesterol and cholesteryl esters, which were upregulated by the gut microbiota in the liver of mice fed lard. The genes encoding sterol O-acyltransferase (Soat1/2), carboxyl ester lipase (Cel), and lipase A (Lipa), which convert cholesterol into cholesteryl esters, were not regulated by the gut microbiota (data not shown). The increased levels of cholesteryl esters in CONV-R mice fed lard may instead result from the increased availability of hepatic free cholesterol.
Depending on dietary composition, GF mice may be protected against obesity and associated conditions such as increased levels of triglycerides in the liver (9, 12). However, this is not always the case and Fleissner et al. (26) have shown that CONV-R and GF mice differ in adiposity and hepatic triglyceride levels on a Western diet, but not on a high-fat diet. Here we show that weight-matched GF and CONV-R mice on a high-fat diet have similar levels of lipids in the liver.
In accordance with Rabot et al. (12), we found that genes involved in cholesterol biosynthesis were downregulated by the gut microbiota in liver from mice fed lard. Interestingly, we also found that expression of genes involved in cholesterol biosynthesis was low in mice fed fish oil, independent of microbial status. Cholesterol biosynthesis is controlled by negative feedback through inhibition of SREBP2 processing. We found that the increased levels of hepatic cholesterol induced by the gut microbiota and by a lard diet were inversely related to the levels of activated SREBP2. Microbiota-induced inhibition of SREBP2 was paralleled by decreased expression of genes involved in cholesterol biosynthesis in mice on a lard diet. Surprisingly, however, mice fed fish oil had low expression of genes encoding cholesterol biosynthesis enzymes despite high levels of activated SREBP2. Saturated lipids have been shown to induce cholesterol biosynthesis (27), while lipids rich in polyunsaturated fatty acids have been shown to reduce cholesterol biosynthesis in the liver (28). Moreover, in vitro experiments have previously demonstrated that the polyunsaturated fatty acid, DHA, activates SREBP2 without increasing cholesterol biosynthesis (29, 30), suggesting that polyunsaturated fatty acids regulate cholesterol biosynthesis through alternative mechanisms.
Cholesterol is the precursor for bile acids, and hence bile acid production is a potential sink for the hepatic cholesterol pool. In accordance with previous reports on mice fed a chow diet (31), we found that expression of CYP7A1, the rate-limiting enzyme in the bile acid biosynthesis pathway, was downregulated by the gut microbiota in mice fed lard and in mice fed fish oil. The decreased levels of hepatic cholesterol in GF mice may therefore be related to increased bile acid production associated with suppression of the FXR-mediated negative feedback loop due to increased levels of the FXR antagonist, β-muricholic acid (31). Previous investigators have observed increased expression of genes encoding the membrane transporters ABCG5 and ABCG8 in the liver of GF mice (12) and suggested that these proteins may mediate increased cholesterol secretion and decreased hepatic cholesterol levels in GF mice. However, we did not find any differences in the expression of Abcg5 or Abcg8 between GF and CONV-R mice (data not shown). The dissimilarity in regulation of ABCG5 and ABCG8 may be related to differences in experimental design between the studies. In the present study, diets with 45% energy from fat were used and CONV-R and GF mice were matched for body weight, while in the previous study diet, 60% energy from fat was used and GF mice were leaner than their CONV-R counterparts (12).
In summary, we show that interaction between the gut microbiota and dietary lipids regulates hepatic cholesterol biosynthesis and that the influence of gut microbiota on hepatic cholesterol metabolism is greater in mice fed a lard diet compared with those on a fish oil diet.
Supplementary Material
Acknowledgments
The authors thank Rosie Perkins (Wallenberg Laboratory, University of Gothenburg) for editing the manuscript; Marcus Ståhlman for analysis of hepatic cholesterol; Anna Hallén and Carina Arvidsson for superb technical assistance; and Rozita Akrami for bioinformatics assistance.
Footnotes
Abbreviations:
- CONV-R
- conventionally raised
- FDR
- false discovery rate
- GF
- germ-free
- GO
- gene ontology
- PBST
- PBS Tween-20
- PCA
- principle component analysis
- SREBP2
- sterol regulatory element-binding protein 2
This work was supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, the Human Frontier of Science Program, the Swedish Heart Lung Foundation, the Torsten Söderbergs Foundation, the Ragnar Söderbergs Foundation, the Novo Nordisk Foundation, the Knut and Alice Wallenberg Foundation, the EU-funded ETHERPATHS project (FP7-KBBE-222639; http://www.etherpaths.org), and a LUA-ALF grant from Västra Götalandsregionen. F.B. is a recipient of a European Research Council (ERC) Consolidator grant (615362-METABASE).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Kennedy A., Martinez K., Chuang C-C., LaPoint K., and McIntosh M.. 2009. Saturated fatty acid-mediated inflammation and insulin resistance in adipose tissue: mechanisms of action and implications. J. Nutr. 139: 1–4. [DOI] [PubMed] [Google Scholar]
- 2.Oh D. Y., Talukdar S., Bae E. J., Imamura T., Morinaga H., Fan W., Li P., Lu W. J., Watkins S. M., and Olefsky J. M.. 2010. Gpr120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 142: 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calder P. C. 2006. N−3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am. J. Clin. Nutr. 83: 1505S–1519S. [DOI] [PubMed] [Google Scholar]
- 4.Buckley J. D., and Howe P. R.. 2009. Anti-obesity effects of long-chain omega-3 polyunsaturated fatty acids. Obes. Rev. 10: 648–659. [DOI] [PubMed] [Google Scholar]
- 5.Devkota S., Wang Y., Musch M. W., Leone V., Fehlner-Peach H., Nadimpalli A., Antonopoulos D. A., Jabri B., and Chang E. B.. 2012. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in il10-/- mice. Nature. 487: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam Y. Y., Ha C. W., Hoffmann J. M., Oscarsson J., Dinudom A., Mather T. J., Cook D. I., Hunt N. H., Caterson I. D., Holmes A. J., et al. . 2015. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity (Silver Spring). 23: 1429–1439. [DOI] [PubMed] [Google Scholar]
- 7.Caesar R., Tremaroli V., Kovatcheva-Datchary P., Cani P. D., and Bäckhed F.. 2015. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 22: 658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tremaroli V., and Backhed F.. 2012. Functional interactions between the gut microbiota and host metabolism. Nature. 489: 242–249. [DOI] [PubMed] [Google Scholar]
- 9.Bäckhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., Semenkovich C. F., and Gordon J. I.. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 101: 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caesar R., Reigstad C. S., Bäckhed H. K., Reinhardt C., Ketonen M., Östergren Lundén G., Cani P. D., and Bäckhed F.. 2012. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 61: 1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding S., Chi M. M., Scull B. P., Rigby R., Schwerbrock N. M. J., Magness S., Jobin C., and Lund P. K.. 2010. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS One. 5: e12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabot S., Membrez M., Bruneau A., Gerard P., Harach T., Moser M., Raymond F., Mansourian R., and Chou C. J.. 2010. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 24: 4948–4959. [DOI] [PubMed] [Google Scholar]
- 13.Bäckhed F., Manchester J. K., Semenkovich C. F., and Gordon J. I.. 2007. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 104: 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arslan N. 2014. Obesity, fatty liver disease and intestinal microbiota. World J. Gastroenterol. 20: 16452–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nygren H., Seppänen-Laakso T., Castillo S., Hyötyläinen T., and Orešič M.. 2011. Liquid chromatography-mass spectrometry (LC-MS)-based lipidomics for studies of body fluids and tissues. In Metabolic Profiling: Methods and Protocols. T. O. Metz, editor. Humana Press, New York. 247–257. [DOI] [PubMed] [Google Scholar]
- 16.Pluskal T., Castillo S., Villar-Briones A., and Oresic M.. 2010. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 11: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castillo S., Mattila I., Miettinen J., Orešič M., and Hyötyläinen T.. 2011. Data analysis tool for comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry. Anal. Chem. 83: 3058–3067. [DOI] [PubMed] [Google Scholar]
- 18.Folch J., Lees M., and Stanley G. H. S.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 19.Homan R., and Anderson M. K.. 1998. Rapid separation and quantitation of combined neutral and polar lipid classes by high-performance liquid chromatography and evaporative light-scattering mass detection. J. Chromatogr. B Biomed. Sci. Appl. 708: 21–26. [DOI] [PubMed] [Google Scholar]
- 20.Li C., and Wong W. H.. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 98: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., et al. . 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W., Sherman B. T., and Lempicki R. A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 23.Supek F., Bošnjak M., Škunca N., and Šmuc T.. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velagapudi V. R., Hezaveh R., Reigstad C. S., Gopalacharyulu P., Yetukuri L., Islam S., Felin J., Perkins R., Borén J., Orešič M., et al. . 2010. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 51: 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaqoob P., Sherrington E. J., Jeffery N. M., Sanderson P., Harvey D. J., Newsholme E. A., and Calder P. C.. 1995. Comparison of the effects of a range of dietary lipids upon serum and tissue lipid composition in the rat. Int. J. Biochem. Cell Biol. 27: 297–310. [DOI] [PubMed] [Google Scholar]
- 26.Fleissner C. K., Huebel N., Abd El-Bary M. M., Loh G., Klaus S., and Blaut M.. 2010. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br. J. Nutr. 104: 919–929. [DOI] [PubMed] [Google Scholar]
- 27.Lin C. C., and Yin M. C.. 2008. Effects of cysteine-containing compounds on biosynthesis of triacylglycerol and cholesterol and anti-oxidative protection in liver from mice consuming a high-fat diet. Br. J. Nutr. 99: 37–43. [DOI] [PubMed] [Google Scholar]
- 28.Rossmeisl M., Medrikova D., van Schothorst E. M., Pavlisova J., Kuda O., Hensler M., Bardova K., Flachs P., Stankova B., Vecka M., et al. . 2014. Omega-3 phospholipids from fish suppress hepatic steatosis by integrated inhibition of biosynthetic pathways in dietary obese mice. Biochim. Biophys. Acta. 1841: 267–278. [DOI] [PubMed] [Google Scholar]
- 29.Jakobsen C. H., Størvold G. L., Bremseth H., Follestad T., Sand K., Mack M., Olsen K. S., Lundemo A. G., Iversen J. G., Krokan H. E., et al. . 2008. DHA induces ER stress and growth arrest in human colon cancer cells: associations with cholesterol and calcium homeostasis. J. Lipid Res. 49: 2089–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Størvold G. L., Fleten K. G., Olsen C. G., Follestad T., Krokan H. E., and Schønberg S. A.. 2009. Docosahexaenoic acid activates some SREBP-2 targets independent of cholesterol and ER stress in SW620 colon cancer cells. Lipids. 44: 673–683. [DOI] [PubMed] [Google Scholar]
- 31.Sayin S. I., Wahlström A., Felin J., Jäntti S., Marschall H. U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., and Bäckhed F.. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.