SUMMARY
Targeting of Endoplasmic Reticulum (ER)-synthesized membrane proteins to the Inner Nuclear Membrane (INM) has long been explained by the ‘diffusion-retention model’. However, several INM proteins contain non-classical Nuclear Localization Signal (NLS) sequences that, in a few instances, have been shown to promote importin α/β– and Ran–dependent translocation to the INM. Here, using structural and biochemical methods, we show that yeast INM proteins Heh2 and Src1/Heh1 contain bipartite import sequences that associate intimately with the minor NLS-binding pocket of yeast importin α and unlike classical NLSs efficiently displace the IBB-domain in the absence of importin β. In vivo, the intimate interactions at the minor NLS-binding pocket make the h2NLS highly efficient at recruiting importin α at the ER and drive INM localization of endogenous Heh2. Thus, h1/h2NLSs delineate a novel class of super-potent, IBB-like membrane protein NLSs, distinct from classical NLSs found in soluble cargos, and of general interest in biology.
Keywords: nuclear transport, Inner Nuclear Membrane, Src1/Heh1, Heh2, membrane protein NLS, IBB-autoinhibition
Graphical Abstract
INTRODUCTION
Transport of soluble cargos through the Nuclear Pore Complex (NPC) is typically an active, signal-mediated and highly regulated process that requires soluble transport factors of the importin βsuperfamily (also known as β-karyopherins) and the small GTPase Ran (Bednenko et al., 2003; Cook et al., 2007; Nardozzi et al., 2010; Stewart, 2007). Transport factor-cargo complexes move through the NPC interior by interaction with phenylalanine-glycine-rich repeats present on disordered NPC proteins, the FG-Nups. Import complexes usually assemble in the cytoplasm upon recognition of a cargo NLS by β-karyopherins (Cingolani et al., 1999). This interaction can be direct (Cingolani et al., 2002), or mediated by transport adaptors such as importin α and snurportin (Lott et al., 2010). Importin α is made up of 10 stacked Armadillo (Arm) repeats, each formed by three α-helices (Goldfarb et al., 2004; Pumroy and Cingolani, 2015) and binds classical NLS (cNLS) substrates, exemplified by the SV40 T-large antigen monopartite NLS and the nucleoplasmin bipartite NLS. The basic side chains of an NLS occupy a shallow groove within the Arm repeats 2-4 of importin α known as the ‘major’ binding site, as well as a minor binding site between Arm repeats 7-8. At each site, as many as five points of contact between NLS and importin α have been identified (referred to as P1-P5 and P1’-P5’ at major and minor binding site, respectively) (Chang et al., 2012; Chang et al., 2013; Chen et al., 2005; Conti and Kuriyan, 2000; Conti et al., 1998; Fontes et al., 2003; Fontes et al., 2000; Giesecke and Stewart, 2011; Lott et al., 2011; Marfori et al., 2012; Roman et al., 2013).
Unlike soluble cargos, significantly less is known about trafficking of membrane-embedded cargos to the nuclear envelope (NE) (Antonin et al., 2011; Burns and Wente, 2012; Laba et al., 2014; Zuleger et al., 2012). Proteomic approaches have identified close to 100 nuclear envelope transmembrane proteins (NETs) (Schirmer et al., 2003), many linked to genetic diseases known as laminopathies (Capell and Collins, 2006), but to date only for a few proteins specific localization at the INM has been proven. Morphologically, the nuclear envelope is composed of an outer and an inner membrane that have distinct protein composition. The outer nuclear membrane (ONM) is contiguous with the ER so that membrane proteins destined for the INM and synthesized in the ER can diffuse laterally through the ER membrane system and the ONM until they encounter NPCs. At the NPCs, the INM and ONM are continuous to the pore membrane, and so the transmembrane (TM)-domain of a membrane protein can pass from the ONM to the pore membrane and hence to the INM (Powell and Burke, 1990), where it is finally retained upon binding to other NE-components (also known as the ‘diffusion-retention model’ (Ellenberg et al., 1997; Smith and Blobel, 1993; Soullam and Worman, 1993)).
Over the past decade, several lines of evidence have suggested that, in addition to diffusion-retention, other mechanisms must exist whereby the NPC plays an active role in trafficking membrane proteins to the INM (Ohba et al., 2004). In higher eukaryotes, several important INM-localized membrane proteins such as POM121, UNC-84 and Sun2 were shown to use importin α-dependent NLSs (Funakoshi et al., 2011; Tapley et al., 2011; Turgay et al., 2010; Yavuz et al., 2010). In Saccharomyces cerevisiae (S. cerevisiae), INM-proteins Src1/Heh1 and Heh2 (orthologs of mammalian MAN1 and LEM2) have NLSs that, like soluble proteins, bind importin-α/β (named Kap60/Kap95 in yeast) to promote nuclear translocation; deletion of such NLSs or lack of functional Kap60, Kap95 or Ran hydrolysis results in mislocalization (King et al., 2006). More recently, it was found (Meinema et al., 2011) that the NLS together with a ~180-230 aminoacids long intrinsically disordered (ID) linker in the extra-luminal surface of Heh1 and Heh2 is essential and sufficient for INM targeting. The long ID linkers are proposed to facilitate recruitments of importin-α/β and project the highly basic NLSs inside the NPC, allowing for importin α/β-mediated nuclear import. As for cNLS-bearing cargos, nuclear targeting depends on importin β interaction with FG-repeats inside the NPC and RanGTP hydrolysis (Meinema et al., 2011).Truncated isoforms of Kap60 lacking the Importin-β binding (IBB) domain have been implicated in nuclear import of Heh2 (Liu et al., 2010), although it is unclear how these isoforms can promote passage through the NPC in the absence of Kap95, since Kap95 is absolutely essential for Heh2 localization to the INM (King et al., 2006; Meinema et al., 2011; Meinema et al., 2013). Thus, there is mounting evidence in the literature for the existence of a dedicated import pathway for INM proteins that requires importin α/β binding to a ‘special’ NLS exposed on the extra-luminal domain of INM proteins. To obtain a quantitative description of the structure, recognition and potency of a membrane protein NLS, in this paper, we have carried out a structural biochemical analysis of Heh2 and Heh1 NLS sequences (abbreviated as h1NLS and h2NLS) complemented by an in vivo study of h2NLS karyophilic properties.
RESULTS
Crystallization of Heh1 and Heh2 NLS sequences with Kap60
S. cerevisiae INM proteins Heh1 and Heh2 contain long NLSs characterized by highly basic NLS-boxes and a variable intra-NLS sequence, possibly longer than 8-12 residues commonly found in classical bipartite NLSs (Jans et al., 2000) (Figure 1A), but falling well within a more recent description (Lange et al., 2010). In vitro, peptides encoding h1NLS and h2NLS are prone to aggregation and highly susceptible to proteolysis, which hampers structural analysis. To study the interaction with Kap60, we co-expressed plasmids encoding Kap60 lacking the IBB (ΔIBB-Kap60) and GST-tagged h1NLS (res. 171-221) or h2NLS (res. 100-137), followed by one-step affinity purification of homogeneous ΔIBB-Kap60:NLS complexes. Co-expression was effective in preventing proteolytic degradation of the highly basic NLSs, essential to obtain well-ordered crystals. The structures of ΔIBB-Kap60 bound to h2NLS and h1NLS were solved by molecular replacement and refined to an Rwork/free of 18.9/22.7% at 2.50 Å resolution and 19.6/21.5% at 2.25 Å, respectively (Table 1). Both crystal structures revealed strong S-shaped electron density running along Kap60 concave surface, mainly localized at the major and minor NLS-binding boxes and weak density between these two boxes. We will first describe the structure of h2NLS that has continuous density between the two boxes, and then that of h1NLS.
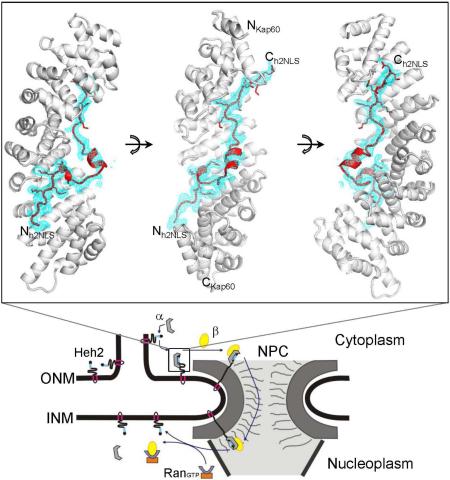
FIGURE 1. Crystal structure of h2NLS bound to ΔIBB-Kap60.
(A) Aminoacid sequence of Heh2 and Heh1 peptides co-crystallized with ΔIBB-Kap60. In black and gray are residues visible and invisible in the crystal structures, respectively; bolded are basic residues occupying minor (left) and major (right) NLS-binding boxes; underlined are residues at position P2’ and P2. (B) Crystal structure of ΔIBB-Kap60 (gray surface) in complex with h2NLS (red ribbon). (C) Schematic diagram of the interactions between h2NLS (in red) and Kap60 residues (in gray) in a distance range of 2.5-4.5 Å. See also Figure S1.
Table 1.
Data collection and refinement statistics.
| ΔIBB-Kap60:h2NLS | ΔIBB-Kap60:h1NLS | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 49.5, 105.3, 224.9 | 129.7, 58.3, 95.4 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 129.3, 90.0 |
| Resolution (Å) | 50-2.50 (2.59-2.50) | 30-2.25 (2.33-2.25) |
| R sym | 8.8 (52.2) | 7.1 (55.6) |
| I / σI | 22.5 (3.5) | 31.4 (3.3) |
| Completeness (%) | 97.0 (97.1) | 98.8 (98.1) |
| Redundancy | 4.4 (4.2) | 3.8 (3.7) |
| Refinement | ||
| Resolution (Å) | 30-2.20 | 30-2.25 |
| No. reflections | 40,407 | 25,900 |
| Rwork / Rfree | 18.9/22.7 | 19.6/21.5 |
| No. atoms | ||
| Protein | 6,556 | 3,285 |
| Ligand (h1/h2NLS) | 449 | 148 |
| Water | 277 | 113 |
| B-factors (Å2) | ||
| Protein | 45.7 | 62.6 |
| Ligand/ion | 72.2 | 85.9 |
| Water | 40.0 | 54.0 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.003 |
| Bond angles (°) | 1.00 | 0.8 |
Values in parentheses are for highest-resolution shell.
The Rfree was calculated using 5% of randomly selected reflections.
h2NLS binds the Arm-core of Kap60 like an IBB-domain
The structure of h2NLS bound to Kap60 can be divided in three regions that make over 50 close contacts with Kap60 Arm-core, burying 3,510 Å2 of solvent-accessible surface area (Figure 1B, 1C, S1). The first region include h2NLS residues 100-105 that bind within (and downstream of) Kap60 minor NLS-binding site (ARM 7-8). This region has the lowest refined B-factor (~36.2 A2) in the h2NLS model. It is superimposable to the smaller NLS-box of NP-NLS (Conti and Kuriyan, 2000) and to other non-classical NLSs that bind exclusively (or preferentially) to importin α minor NLS-site (Chang et al., 2012; Chang et al., 2013; Giesecke and Stewart, 2011; Lott et al., 2011) (Table S1). Unlike cNLSs that usually have only two basic residues at the minor NLS-binding site, four basic amino acids in h2NLS (102-KRKR-105) insert their side chains deeply inside Kap60 groove, making ~15 close contacts (Figure 1C), of which R103 occupies position P2′. The second region starts after R105, where the NLS backbone makes a 90° turn to form a 3/10 helix, H1 (105-REQ-107) that connects via a short linker (108-ISTDNE-113) to a second helix, H2 (114-AKMQI-118), followed by a short stretch (119-IEEKS-123) (Figure 1B, 1C). Both helices and linker have weak electron density (Figure S1) and high B-factor in our final model (~108.9 Å2). This region of h2NLS makes minor contacts with Kap60 surface and is highly variable in other putative membrane protein NLSs (Lusk et al., 2007). The h2NLS's third structural region contains seven consecutive basic residues (124-PKKKRKKRS-132) that span within (and upstream of) the major NLS-binding site of Kap60 (ARMs 1-4) (Figure 1B, 1C). The average refined B-factor of this region is ~68 Å2, higher than at the minor NLS-box: only residues 125-KKKR-128 at position P1-P4 (Table S1) have clear side-chain density (Figure S1), while only main-chain atoms are visible for the residues 129-KKR-131. Thus, h2NLS binds Kap60 like a classical bipartite NLS but makes more extensive contacts at the minor NLS-box than seen in the structure of Kap60 bound to NPNLS (Conti and Kuriyan, 2000).
Heh1 NLS makes strong contacts at the minor NLS-binding box
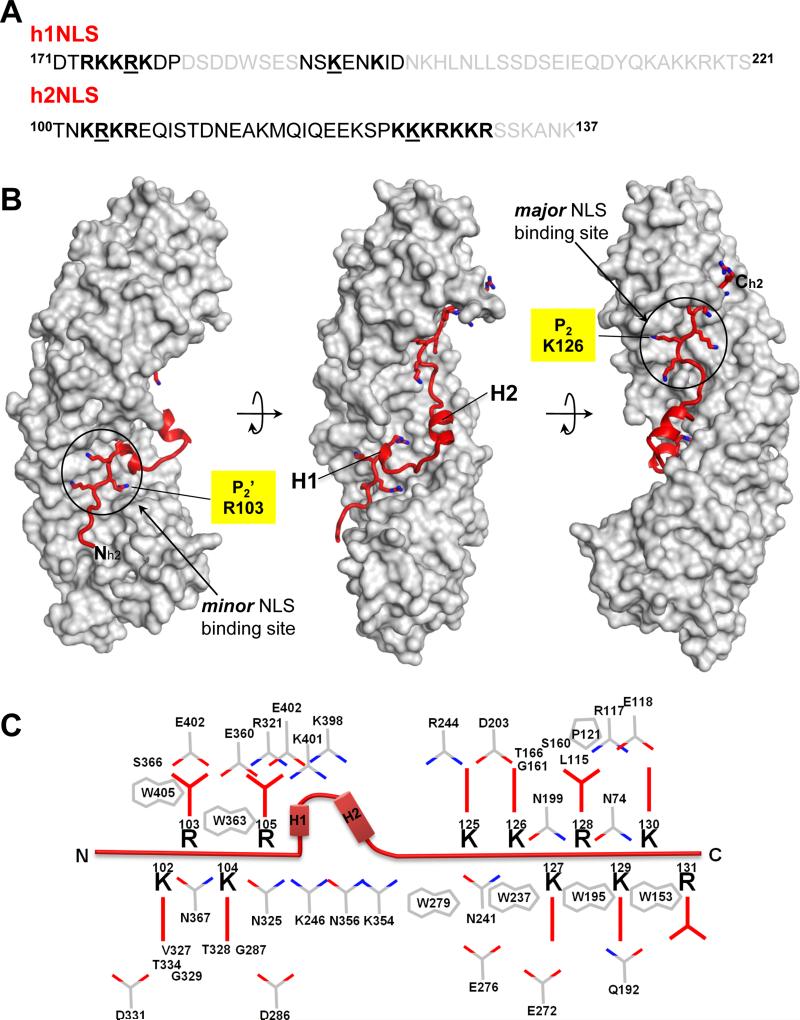
The exact boundaries of the Heh1 NLS were unknown before this study, though it was shown that a region between residues 173-220, encoding several basic patches similar to a cNLS, and a ~200 residues unfolded linker were required and sufficient for nuclear import (Meinema et al., 2011). The crystal structure of ΔIBB-Kap60 crystallized in complex with a 50-mer spanning Heh1 resides 171-221 (Figure 1A) has density only for the first half of the Heh1 construct (residues 173-195) (Figure 2A), while no discernable electron density was observed for residues 196-221. h1NLS resembles closely to h2NLS and occupies both minor and major NLS-binding sites. h1NLS’ first basic box (173-RKKRK-177) binds intimately the minor NLS-pocket, while, unexpectedly, a minimally basic stretch of residues (189-SKENKID-195) occupies the major NLS-box. The canonical intra-NLS spacer of 11 residues has poor density and some of its residues (180-DSDDWSES-187) were not modeled in the final structure. Noticeably, the five basic amino acids in h1NLS that bind the minor NLS-pocket engage in nearly 20 close contacts with Kap60 Arm 6-8 (Figure 2B), of which R176 occupies position P2′ (Table S1). Instead, at the major NLS-binding pocket, only P2 and P5 are occupied by lysines (Figure 2B), whereas non-basic side chains interact at P1, P3 and P4, as previously seen for PLSCR1 NLS (Chen et al., 2005) (Table S1). Overall, Kap60 recognizes the NLS of Heh1 and Heh2 using a combination of electrostatic and hydrophobic contacts with main- and side-chains atoms. Arrays of Asn (Conti et al., 1998) projecting from Kap60 Arm-core stabilize the NLS backbone, while conserved Trps (Figure 1C, 2B) engage in hydrophobic and cation-π interactions (Koerner et al., 2003) with the critical side chains of R103/176 and K126/190 that occupy P2’ and P2 positions at minor and major NLS-binding boxes, respectively.
FIGURE 2. Mapping crystallographically h1NLS in complex with the Arm-core of Kap60.
(A) Crystal structure of ΔIBB-Kap60 (gray surface) in complex with h1NLS (red ribbon). The dotted line indicated residues in the intra-NLS linker that is poorly visible in the electron density and that was not included in the final model. (B) Schematic diagram of the interactions between h1NLS (in red) and Kap60 residues (in gray) in a distance range of 2.5-4.5 Å.
h1NLS and h2NLS bind ΔIBB-Kap60 with nanomolar affinity
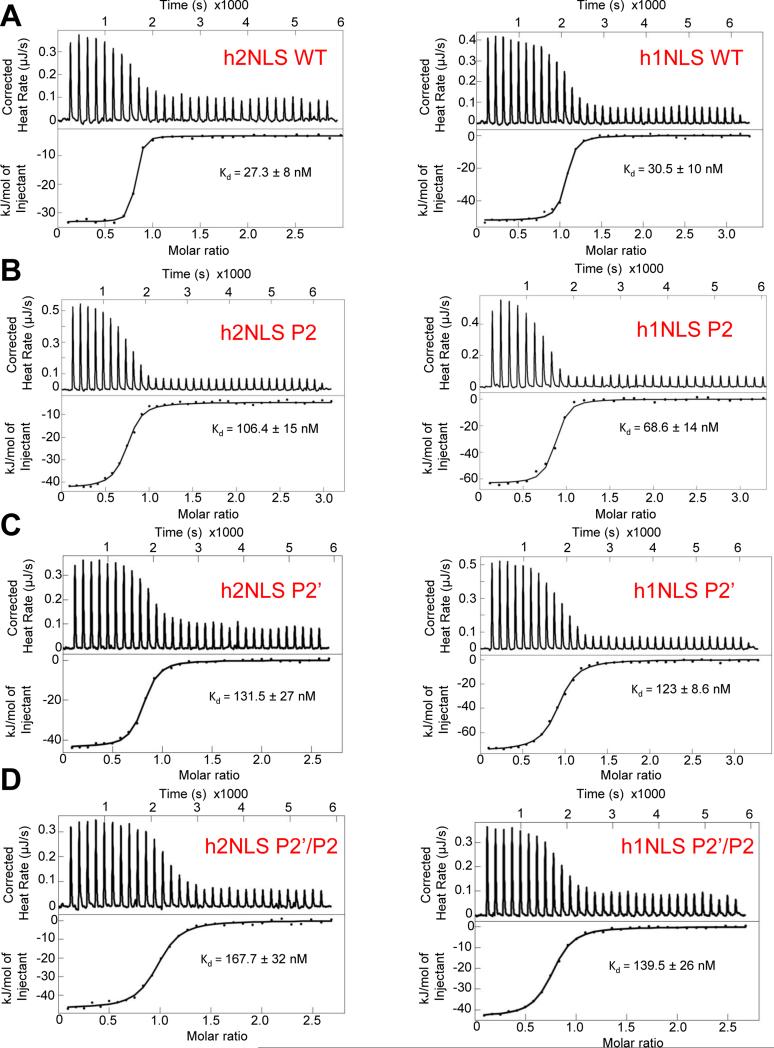
The intimate association of h1 and h2NLSs with Kap60 observed crystallographically prompted us to measure their binding affinity for Kap60. Using nano Isothermal Titration Calorimetry (nano-ITC), we measured the heat released upon titration of increasing concentrations of MBP-tagged h1NLS (MBP-h1NLS) or h2NLS (MBP-h2NLS) into a cell containing ΔIBB-Kap60 (Figure 3A). This analysis yielded an equilibrium dissociation constant (Kd) of 27.3 ± 8 nM for h2NLS and 30.5 ± 10 nM for h1NLS, slightly lower than the Kd of a control NP-NLS for ΔIBB-Kap60 measured under identical experimental conditions (Kd = 46.0 ± 14 nM) (Figure S2). The observation that the two membrane protein NLSs bind Kap60 with similar affinity, though h1NLS has only 2 basic residues at the major NLS-binding box (Figure 2B) versus seven in h2NLS (Figure 1C), suggests a minimal contribution of this moiety in the overall binding affinity for Kap60. This is clearly not the case for cNLSs, which are disrupted by a single point mutation at P2 in the major NLS-binding box (Colledge et al., 1986; Kalderon et al., 1984). To test this idea, we introduced Ala-mutations at position P2’ and P2 of h1/h2NLSs and measured their effect on the overall equilibrium binding affinity for ΔIBB-Kap60. A mutation at position P2 reduced moderately (~2-fold) h1NLS affinity for ΔIBB-Kap60 (Kd = 68.6 ± 14 nM), consistent with the small number of contacts made at the major NLS-binding site (Figure 3B), whereas a 4-fold drop in affinity was caused by an Ala-substitution at P2’ (Kd = 123.0 ± 8.6 nM) (Figure 3C). A similar effect was seen in h2NLS, where a mutation at P2 yielded a 4-fold drop in binding affinity for ΔIBB-Kap60 (Kd = 106.4 ± 15 nM) (Figure 3B) while a 5-fold destabilization was caused by an Ala-substitution at P2’ (Kd = 131.5 ± 27 nM) (Figure 3C). Combining mutations at P2’ and P2 did not significantly aggravate loss of binding affinity for ΔIBB-Kap60 (Kd = 139.5 ± 26 nM and 167.7 ± 32 nM for h1NLS and h2NLS, respectively) (Figure 3D) as compared to single point mutants at P2’, confirming that the overall affinity of membrane protein NLSs for Kap60 depends primarily on structural determinants at P2’, in the minor NLS-binding box.
FIGURE 3. Calorimetric analysis of the interaction of h1/h2NLSs with ΔIBB-Kap60.
ITC analysis of the interaction of ΔIBB-Kap60 (in cell) with in syringe (A) WT h2NLS and h1NLS; (B) h2NLS(P2’) and h1NLS(P2’); (C) h2NLS(P2) and h1NLS(P2); (D) h2NLS(P2’/P2) and h1NLS(P2’/P2). Raw data are in the top panel, and the integrated enthalpy plotted as a function of the NLS:ΔIBB-Kap60 molar ratio is shown in the bottom panel. See also Figure S2.
h1NLS and h2NLS compete off the IBB-domain in the absence of importin β
Superimposition of ΔIBB-Kap60 bound to Heh2 or Heh1 NLSs with FL-Kap60 previously solved as part of an export complex (Matsuura and Stewart, 2004) revealed a striking structural resemblance between the membrane protein NLSs and the IBB-domain (rmsd 1.1 Å) (Figure 4A). The h2NLS, which has a continuous trace between NLS-boxes, the h1NLS and the IBB adopt a nearly identical conformation at the minor and major NLS-binding pockets of Kap60 with a striking conserved lysine at position P2 (IBB-54/h2NLS-126/h1NLS-190) and an arginine at P2’ (IBB-34/h2NLS-103/h1NLS-176). In contrast, the intra-NLS regions are partially helical in h2NLS (res. 106-120), not visible in the structure with h1NLS (res. 182-187) and random coiled in IBB (res. 37-49), suggesting this region makes non-essential contacts with Kap60. However, despite the structural similarity to an IBB, h1/h2NLSs do not associate directly with Kap95 (data not shown) suggesting these NLSs mimic only the importin α-bound conformation of IBB, which is mainly unstructured, but cannot adopt the helical conformation of IBB induced upon binding to importin β (Cingolani et al., 2000; Mitrousis et al., 2008).
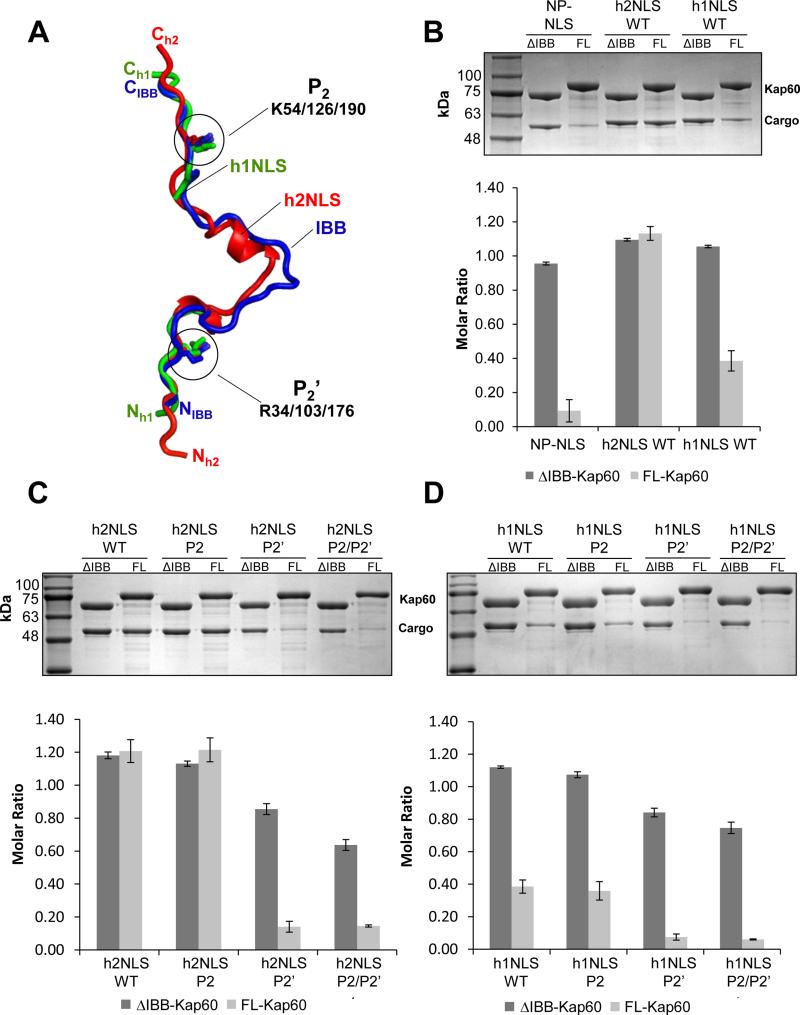
FIGURE 4. h2NLS and h1NLS compete off the IBB-domain of Kap60.
(A) Superimposition of ΔIBB-Kap60 bound to h2NLS or h1NLS with FL-Kap60 (pdb id 1WA5) (Matsuura and Stewart, 2004). h1NLS and h2NLS are colored in green and red, respectively, while the IBB-domain is blue. For clarity, Kap60 has been omitted. Only the Arg at P2’ and Lys at P2 are modelled as sticks. (B) Pull-down analysis and quantification of the interaction of GST-tagged Kap60 lacking the IBB (ΔIBB-) or full length (FL-) immobilized on glutathione beads and incubated with NP-NLS, h2NLS and h1NLS. (C) Pull-down analysis and quantification of the interaction of GST-ΔIBBKap60 or FL-Kap60 with WT h2NLS and mutants at P2’, P2 and P2/P2’(D) Pull-down analysis and quantification of the interaction of GST-ΔIBB-Kap60 or FL-Kap60 with wt-h1NLS and mutants at P2’, P2 and P2/P2’. Pull-downs are shown as mean ± standard deviation for three experiments. See also Figure S3.
As it was previously shown that the h2NLS can bind FL-Kap60 in the absence of Kap95 (King et al., 2006), we hypothesized that Ala-mutation at P2’ affects the way h2NLS competes off the IBB-domain of Kap60. To test this hypothesis, we tried to measure association of h1/h2NLSs with FL-Kap60 using ITC but obtained uninterpretable binding data, likely due to the concomitant presence of two binding events, namely the intramolecular dissociation of IBB from Kap60 Arm-core and the intermolecular association of h1/h2NLS with Kap60. To overcome this problem, we turned to an ‘on-bead’ binding assay (Pumroy et al., 2015) where GST-tagged FL-Kap60 (GST-FLKap60) and GST-ΔIBB-Kap60 were immobilized on glutathione beads and incubated with a 2-fold molar excess of h1/h2NLSs or control NP-NLS. In the absence of importin β, the IBB-domain binds the Arm-core preventing association of cNLS-cargos (Kobe, 1999) (Figure 4B). Instead, h2NLS bound stoichiometrically both to FL-Kap60 and Kap60 Arm-core, confirming this NLS can efficiently bypass IBB-autoinhibition. The h1NLS was also able to overcome autoinhibition, yet to a lesser extent compared to h2NLS and as much as 40% of MBP-h1NLS was recovered bound to beads after 15 minutes incubation (Figure 4B). Mutation at P2′, but not P2 (Figure 4C) completely disrupted the interaction of h2NLS with FL-Kap60, rendering h2NLS indistinguishable from NPNLS. Similar results were obtained for h1NLS that, though less effective at displacing the IBB, was disrupted by a single point mutation at P2’ but not P2 (Figure 4D). Thus, the membrane protein NLSs of Heh1 and Heh2 adopt an IBB-like structure that combines binding determinants seen in the recognition of cNLSs, as well as a deeper interaction at the minor NLS-binding site and particularly the P2’ position, which make these NLSs able to bypass IBB autoinhibition.
In vivo potency of h2NLS depends on P2’ position
To complement our in vitro studies we sought to confirm the importance of the interaction with the minor NLS-binding site, particularly at the P2’ position, for transport of membrane proteins in vivo. We focused on h2NLS, which was previously characterized in detail in live cells, both in the context of the full length protein and in reporter proteins (King et al., 2006; Meinema et al., 2011; Meinema et al., 2013). The advantage of using Heh2-derived reporter proteins is that they are mobile within the network of NE and ER (Meinema et al., 2011; Meinema et al., 2013) because they lack domains that contribute to nuclear retention, such as the LEM domains found in Heh1 and Heh2 (Heh2 domain composition is schematically illustrated in Figure 5A). Being mobile, their distribution in the network of NE and ER reflects their nuclear import rates: for a Heh2-reporter with h2NLS and ID linker, we find a higher fluorescence at the NE than at the peripheral ER, while a reporter lacking the NLS or ID linker shows similar levels of fluorescence in the entire NE-ER network (Meinema et al., 2011). The nuclear location is completely dependent on Kap95, as demonstrated by conditionally tethering Kap95-FRB to Pma1-FKBP at the plasma membrane, which results in gradual decrease in the NE/ER ratio (Figure 5B) (Meinema et al., 2011; Meinema et al., 2013), arguing against a recent report that Kap60 isoforms lacking the IBB, and therefore unable to heterodimerize with Kap95, are responsible for Heh2 translocation to the INM (Liu et al., 2010).
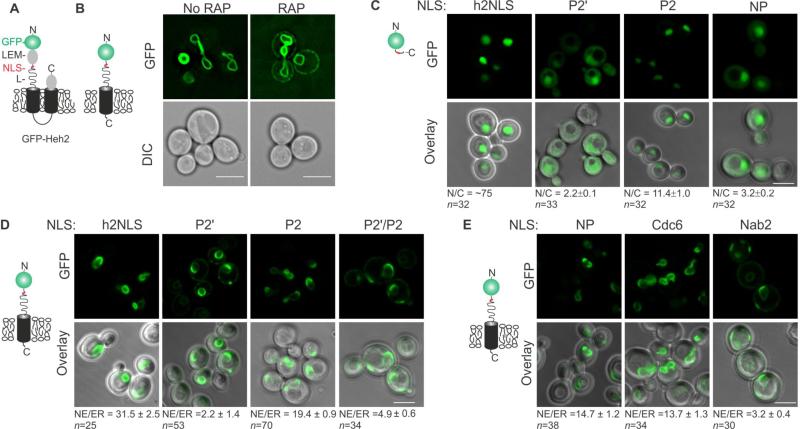
FIGURE 5. Quantitative analysis of h2NLS karyophilic properties.
(A) Cartoon showing the domain composition of Heh2 where GFP is in green, NLS in red, Heh2's ID linker represents a curved line and the TM domain is in black. (B) Deconvolved wide-field images of the Heh2-based transmembrane reporter protein expressed in the Kap95AA strain (Haruki et al., 2008) (No RAP) and when Kap95-FRB is conditionally trapped at Pma1-FKBP at the plasma membrane upon addition of rapamycin (RAP). (C) Confocal fluorescent images of yeast expressing GFP fused to indicated NLSs: WT h2NLS, h2NLS mutants at position P2′ and P2 and NP-NLS and quantification of average N/C-ratios over n cells. (D) Confocal fluorescent images of yeast expressing GFP-h2NLS-L-TM with mutations at position P2', P2, P2'/P2 and quantification of average NE/ER-ratios over n cells. (E) Same as (D) but with different indicated NLSs. Scale bar is 5 μm and SEM is indicated.
To test the importance of position P2’ in vivo, we introduced Ala-substitutions at position P2’, P2, and P2’/P2 in a soluble GFP-h2NLS fusions (Figure 5C) and in a membrane-embedded Heh2-based reporter (Figure 5D) and imaged their subcellular localization. As shown previously (Meinema et al., 2011), the karyophilic properties of the h2NLS are so strong that many cells showed no cytosolic h2NLS-GFP and the N/C ratio is very high (N/C~75 is likely an underestimation). The reduction in nuclear accumulation of soluble (GFP-NLS, Figure 5C) and transmembrane (G-h2NLS-L-TM, Figure 5D) reporter proteins was most severely affected by substitution at position P2’ in the minor NLS-box. In the case of the transmembrane reporter protein, the NE/ER ratio was moderately reduced when introducing the P2 mutation (NE/ER-ratio 21.6 ± 2.7 and 31.5 ± 2.5 for P2 and wild type), but mutation at P2’ resulted in complete loss of nuclear accumulation (NE/ER-ratio 2.8 ± 0.6), comparable to a ΔNLS mutant (NE/ER-ratio 2.3 ± 0.2 in (Meinema et al., 2011). Likewise, the double mutant (P2’/P2) localized similar as the P2’ mutant, confirming the dominant negative role of the P2’ mutation.
Knowing now that the interactions at the minor binding site of Kap60 are critical, we compared the karyophilic properties of the h2NLS with known NLSs. In the context of the transmembrane reporter proteins, the h2NLS lead to NE/ER-ratio's that were approximately 3-fold higher compared to a single partite variant of the h2NLS (lacking 102-KRKR105) or 8-fold higher compared to a cNLS (Meinema et al., 2011). Complementing these studies we also replaced the h2NLS with the NP NLS and observe GFP-NP-L-TM accumulates approximately 2.5 fold lower than with h2NLS (NE/ER-ratio 14.7 ± 1.2) (Figure 5E) consistent with the NE targeting of full length Heh2 with and NP NLS (King et al., 2006). The high affinity NLS of Cdc6 (Hahn et al., 2008) behaved similar to the NP-NLS with NE/ER-ratio 13.7 ± 1.3 (Figure 5E). Reinforcing the specific role for Kap60 and Kap95 in nuclear import of Heh2 (King et al., 2006), the high affinity Kap104-dependent NLS of Nab2 had a similar NE/ER-ratio (NE/ER-ratio 3.2 ± 0.4) (Figure 5D) as observed without an NLS (NE/ER-ratio 2.3 ± 0.2) (Meinema et al., 2011). Similarly, NP-NLS fused to a soluble import cargo (GFP) was much less efficient than h2NLS in promoting nuclear translocation (N/C-ratio 3.2 ± 0.2) (Figure 5C). Altogether we conclude that, in vivo, h2NLS is an exceptionally potent import signal.
R103 at position P2′ is critical for Heh2 function and translocation to the INM
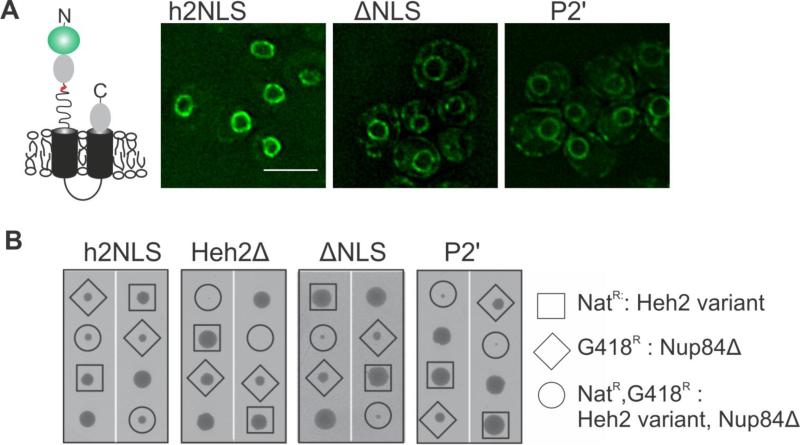
To test the importance of the interaction at the minor binding site P2’ position, we sought to determine the localization of full length N-terminally GFP-tagged Heh2 expressed from the chromosome from its endogenous promotor (Figure S4). This is more physiological than the Heh2-reporter, although this protein can engage in protein:protein interactions at the INM that retain it in the nucleus. Indeed, the sole mutation R103A at the P2’ resulted in a complete loss of the NE specific localization, indistinguishable from that of a ΔNLS mutant (Figure 6A).
FIGURE 6. Mutation of R103 at position P2′ abolishes NE accumulation of Heh2 and the double mutant with nup84Δ is synthetic sick.
(A) Deconvolved wide-field images of yeast expressing native levels of GFP-Heh2 with WT NLS (h2NLS), without the NLS (ΔNLS), and Heh2 with the P2’ mutation. Scale bar: 5 μm. (B) Synthetic sick/lethal interaction using tetrad dissection of nup84Δ expressing WT (h2NLS) and mutant variants of Heh2 (ΔNLS, P2’) or no Heh2 (Heh2Δ). Each tetrad is oriented vertically and represents the meiotic progeny of a heterozygous diploid between GFP-HEH2-NAT/NUP84 and HEH2/nup84::KANMX. Two representative tetrads for each double mutant are shown. The genetic background of each spore is identified by the presence of the NAT and KAN marker, respectively. The double mutant spore colonies are enclosed in circles whereas single mutants are enclosed in squares or diamonds, and WT strains are not enclosed. See also Figure S4.
To gain further insight into the in vivo relevance of the P2’mutation, the mutation was introduced in a strain lacking NUP84. Previously it was shown that the double mutant nup84Δheh2Δ (Yewdell et al., 2011), in contrast to the single mutants, fails to grow, and the double mutant nup84Δheh2Δh2NLS is synthetic sick compared to the single mutants. Consistent with the complete loss of accumulation of the P2’ mutant, and the in vivo relevance of this accumulation, the double mutant nup84Δheh2P2’ is also synthetic sick, indistinguishable from nup84Δheh2ΔNLS (Figure 6B). Thus mutation of the P2’position in h2NLS correlates with loss of function in vivo both on the level of cellular localization and cell fitness.
Position P2’ is critical to retain NLS-bound Kap60 at the ER
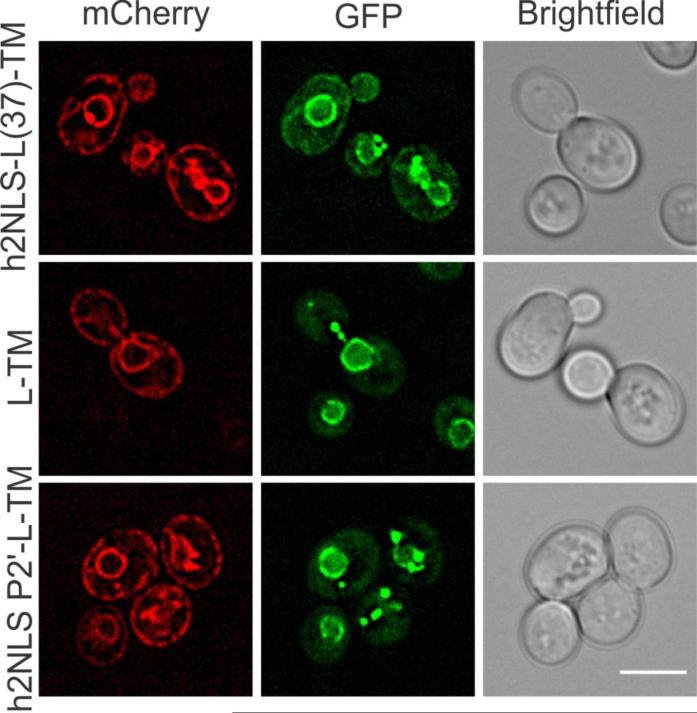
Next we aimed to confirm our in vitro data by showing that the binding to Kap60 depends on the interaction at the P2’ position. We thus assessed the binding of Kap60 (and Kap95) to the h2NLS and the P2’ mutant in vivo using an assay in which we monitor co-enrichment of Kap60-GFP with the membrane reporters (Meinema et al., 2013). While Kap60-GFP normally does not enrich at the peripheral ER, it did so in 45% of the cells (n=86) expressing an ER localized h2NLS-containing reporter protein (mCherry-h2NLS-L(37)-TM) (Figure 7). This protein lacks a functional linker domain so that it remains ER-localized (and does not accumulate at the INM) (Meinema et al., 2013). Co-enrichment of the reporter protein and Kap60-GFP reflects binding of Kap60 to the h2NLS because cells expressing a reporter that lacks an NLS (mCherry-L-TM) did not show Kap60-GFP at the peripheral ER (n=47) (Figure 7). Interestingly, cells that expressed the reporter protein with the mutant NLS (mCherry-h2NLSP2’-L-TM) also did not show Kap60 enriched at the peripheral (n=54) (Figure 7) consistent with the dramatic reduction in import efficiency to the INM. Thus, h2NLS recruits Kap60 by making a crucial contact with the minor NLS-binding box that is critically dependent on R103 at position P2’.
FIGURE 7. In vivo analysis of h2NLS interaction with Kap60.
Deconvolved wide-field images of cells co-expressing Kap60-GFP with mCherry-tagged reporter proteins mCh-h2NLS-L(37)-TM, mCh-h2NLS P2’-L-TM or mCh-L-TM. Scale bar is 5 μm and SEM is indicated. See also Figure S5 and Supplemental Experimental Procedures.
Nup2 and h2NLS compete at the minor NLS-binding site
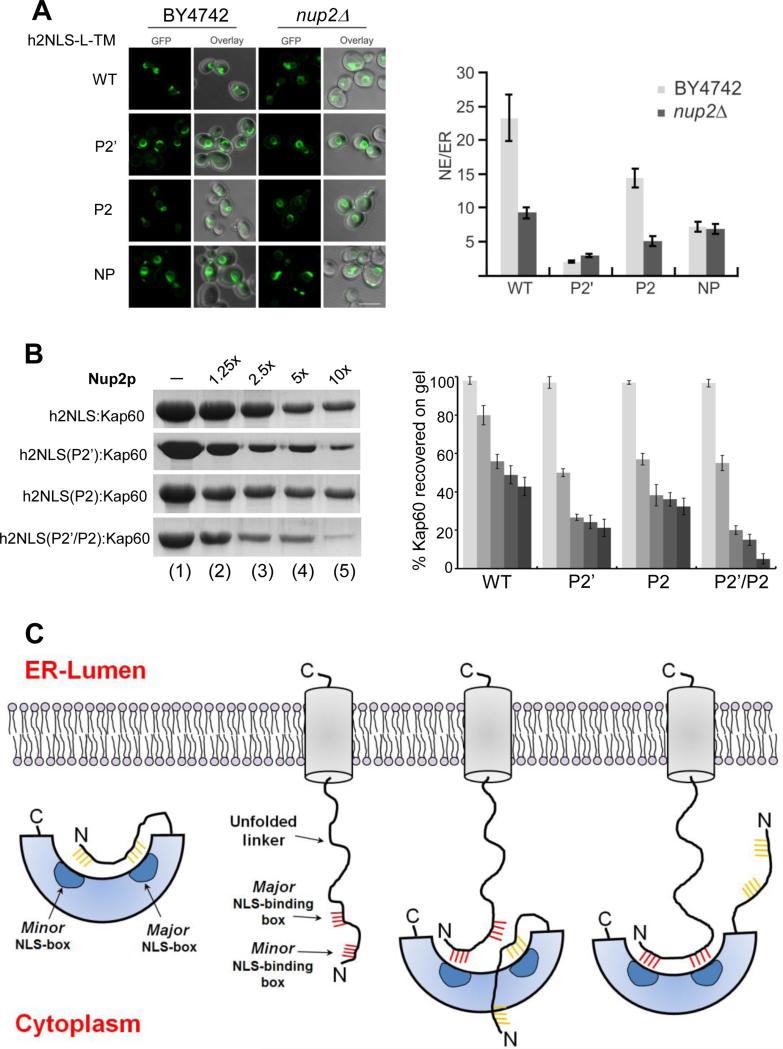
Next, we asked whether Nup2, a mobile nucleoporin that also binds the minor NLS-binding site of Kap60 (Matsuura et al., 2003; Pumroy et al., 2012) plays a role in disassembly of h2NLS from Kap60, as proposed for cNLS-cargos (Dilworth et al., 2001; Hood et al., 2000; Solsbacher et al., 2000). Accumulation of GFP-h2NLS-L-TM in a nup2Δ strain was approximately 2.5-fold decreased compared to a wild type (WT) strain, consistent with measurements on full length Heh2 (King et al., 2006) (Figure 8A). Since Nup2 also functions in the recycling of Kap60 back to the cytoplasm (Solsbacher et al., 2000), and the knockout suffers from other cellular effects like an mRNA export defect (Casolari et al., 2004; Dilworth et al., 2005) the results were not readily interpreted. However, the localization of a similar reporter protein containing NP-NLS (GFP-NP-L-TM) was not dramatically affected, pointing to a specific role for Nup2 in import of the h2NLS cargo (Figure 8A). To determine if the N-terminal 51 residues of Nup2 were sufficient to dissociate h2NLS from ΔIBB-Kap60, we immobilized a stoichiometric complex of ΔIBB-Kap60:h2NLS on glutathione beads and challenged it with increasing molar excess (from 1.25-10x) of purified Nup2 (res. 1-51), followed by SDS-PAGE and quantification (Figure 8B). Notably, a 10-fold excess of Nup2 dissociated as much as 60% of the otherwise very stable ΔIBB-Kap60:h2NLS complex. Ala-substitution at position P2’ enhanced Nup2-mediated displacement of ΔIBB-Kap60 from h2NLS more markedly than the mutant at P2 (Figure 8B). Thus, Heh2 association to Kap60 is affected by Nup2, consistent with an intimate interaction of both proteins with the minor NLS-binding site of Kap60.
FIGURE 8. Role of Nup2 in displacement of Heh2 from Kap60.
(A) Confocal fluorescence images of a wild type yeast strain (BY4742) and a nup2Δ knockout strain expressing GFP-h2NLS-L-TM and the reporter with mutations at position P2′, P2, and NP-NLS, as well as quantification of average NE/ER-ratios (average of ~30 cells). SEM, and 5 μm scale bar are indicated. (B) Nup2-mediated displacement of ΔIBB-Kap60 from GST-h2NLS (and its mutants at P2’, P2 and P2’/P2) coupled to glutathione beads. The complex was challenged with 1.25-10 fold molar excess of MBPNup2 (res. 1-51) and ΔIBB-Kap60 left on beads is quantified on the right panel (error bars from averaging three independent experiments). (C) Model for recognition and association of a membrane protein NLS to auto-inhibited FL-Kap60. From left to right are schematic illustrations of auto-inhibited FL-Kap60, an ER-synthesized membrane protein (like Heh2) projecting an h2NLS-like import sequence in the cytoplasm and two putative snapshots of FL-Kap60 partially and fully bound to the membrane protein NLS.
DISCUSSION
A long standing question in cell biology is how integral membrane proteins translocate from the ER to the INM. The current model is that INM proteins move from the ER to the INM by diffusion of the membrane spanning trans-membrane domains through the pore membrane. The extra-luminal soluble domains pass either along the membrane through lateral channels (Ellenberg et al., 1997; Smith and Blobel, 1993; Soullam and Worman, 1993, 1995), or, as proposed for Heh1 and Heh2 (Meinema et al., 2011), bind import factors and travel through the NPC making contact with the FG-Nups while long ID linkers project the NLS away from the membrane. Non-classical NLSs similar to h2NLS are not unique to yeast, but are also found in a variety of vertebrate INM proteins (Lusk et al., 2007). The exact role of these NLSs in nuclear translocation of ER-synthesized membrane proteins destined to the INM remains poorly understood.
Distinctive features of membrane protein NLSs
In this paper, we have characterized the NLS of yeast INM protein Heh2 and Heh1 and defined a set of molecular properties that we propose are distinctive of these membrane protein NLSs. First, h2NLS resembles the IBB-domain of importin α in the auto-inhibited conformation, as opposed to a bipartite NLS. Analogous to known IBBs (Lott and Cingolani, 2011), h2NLS accommodates intra-NLS residues as partially folded helices that make minimal contacts with Kap60. Second, both h1 and h2NLSs bind ΔIBB-Kap60 with low nanomolar affinity, comparable to NP-NLS but their assembly to Kap60 is different from cNLS. The dominant negative mutation in h2NLS that disrupts nuclear localization is at position P2′ of the minor NLS-binding site. This is distinct from a cNLS (Colledge et al., 1986; Kalderon et al., 1984), where mutation at P2’ marginally disrupts nuclear localization (Robbins et al., 1991), reinforcing the idea that h2NLS is not a simple variation of a classical bipartite NLS. Third, h2NLS and to a lesser extent h1NLS compete off the IBB-domain in the absence of importin β, which predicts a reduced auto-inhibitory role of IBB on membrane protein cargos trafficking from the ER to the INM. This is similar to the influenza polymerase subunit PB2 (Pumroy et al., 2015), which also overcomes IBB-autoinhibition by making strong contacts at the minor-NLS box. Fourth, nucleoporin Nup2 plays a critical role in displacement of h2NLS from Kap60 by directly competing for binding to the minor NLS-binding site, which provides an anchoring point to both h2NLS and Nup2's N-terminal NLS-like moiety (Matsuura et al., 2003).
A potential multi-step mechanism describing the recruitment of Heh2 membrane protein NLS by Kap60 can be hypothesized (Figure 8C). Recognition of h2NLS begins at the minor NLS-binding pocket, where h2NLS's basic box 102-KRKR-105 competes off the equivalent region of IBB (33-RRRR-36) (Table S1). Whereas all four basic residues in h2NLS insert at Kap60 helical interface between ARM 7-8, only three Args in IBB (at position P1’, P2’ and P4’) make contacts with the minor NLS-binding site, projecting the guanidinium group of R35 (at position P3’) at the surface of ARM 7 (Figure 3A). This initial interaction cements h2NLS to Kap60 minor NLS-binding pocket, increasing its local concentration and allows zippering to the major NLS-site, where the IBB's major-NLS box 54-KRR-56 is readily competed off, overcoming IBB-auto-inhibition and displacing the IBB in the absence of Kap95. We speculate that early recruitment of importin α could occur while a membrane protein is being synthesized and/or inserted at the ER-membrane.
Physiological significance of membrane protein NLSs
What is the advantage of bearing a membrane protein NLS instead of a classical bipartite NLS? Though a conclusive answer to this question will require further in-depth analysis of additional membrane protein NLSs, especially from higher eukaryotes (Lusk et al., 2007), and a complete understanding of inner membrane proteins full length 3D-structure (in addition to minimal NLS fragments), a few hypotheses can be formulated on the basis of the data presented in this paper. The karyophilic potency of an h2NLS-like import signal is likely to aid in all steps of membrane protein translocation to the INM, thereby providing a selective biological advantage over cNLSs. At first in the cytoplasm, during import complex assembly, we propose membrane protein NLSs facilitate recruitment of karyopherins and formation of a productive membrane-bound import complex. Unlike soluble NLS-cargos moving fast by 3D-diffusion, membrane proteins move much slower in the 2D-plane of the membrane (Meinema et al., 2013). They thus could have a reduced probability to encounter karyopherins, which are soluble factors. However, as shown for h1/h2NLSs in this paper, the ability of recruiting importin α in the absence of importin β possibly compensates for the restricted 2D-diffusion of membrane-embedded cargos providing a kinetic advantage over classical cargos that assemble into productive import complexes only when importin α and β are simultaneously present (Pumroy et al., 2015). During translocation through the NPC, though the actual mechanisms of passage are controversial and it is unclear if an import complex undergoes cycles of dissociation and re-association while moving inside the NPC (Bednenko et al., 2003), the advantage of a membrane protein NLS would be its ability to remain bound to importin α even when importin β has been displaced, possibly expediting re-formation of an import complex. Finally, membrane protein NLSs may provide a selective advantage to release cargos at the INM. After importin β– and Ran–dependent passage through the NPC, competition with Nup2 for binding to importin α minor NLS-binding pocket is likely to promote release of membrane-embedded cargos at the INM, where NETs can be retained by binding interactions with other NE-components.
In summary, the present work expands the definition of NLS and provides a framework to identify the molecular mechanisms by which ER-synthesized membrane proteins translocate to INM to play a critical role in nuclear signaling.
EXPERIMENTAL PROCEDURES
Biochemical techniques
ΔIBB-Kap60 was co-expressed with GST-h1/h2NLS in E. coli strain BL21-CodonPlus® (DE3)-RIL (Stratagene) for 6 h at 30° C. ΔIBB-Kap60 bound to GST-h1NLS or GST-h2NLS was purified on glutathione-resin (GenScript) and after cleaving off the GST with PreScission Protease, the complex was purified over a Superdex 200 column (GE Healthcare) equilibrated in G.F. buffer (20 mM Tris pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol, and 0.2 mM PMSF). All GST-tagged constructs used in this paper were purified as described above. GST-Nup2 was expressed as described for human Nup50 (Pumroy et al., 2012). All His-MBP-tagged constructs were purified over His-resin (GenScript) followed by gel filtration chromatography. For pull-down assays and ITC analysis, see Supplemental Experimental Procedures.
Crystallographic studies
Crystals of ΔIBB-Kap60 bound to h1NLS or h2NLS were obtained by mixing equal volume of gel filtration-purified complex at 12.5 mg/ml with 100 mM ammonium acetate, 20% PEG 8000, 100 mM BisTris (pH 6.0) and equilibrating the droplet against 600 μl of the same precipitant. 25% glycerol was added as cryo-protectant before flash-freezing at −170° C. Crystals were diffracted at beamlines X6A and X29 at the National Synchrotron Light Source (NSLS) on a Quantum Q270 and a Quantum-315r CCD detector, respectively. Data were processed using HKL2000 (Otwinowski and Minor, 1997) and initial phases calculated using Phaser (McCoy et al., 2007). Atomic models were built using Coot (Emsley and Cowtan, 2004) and refined with phenix.refine (Adams et al., 2002). Data collection and refinement statistics are summarized in Table 1 and additional methods are in Supplemental Experimental Procedures.
Yeast Cultivation and Microscopy
Yeast strains used in this study are listed in Table S2 and are isogenic to S288C except the Kap95-AA strain (Haruki et al., 2008) which is W303-based (Figure 5). Cells were grown at 30°C and kept at mid-log growth phase for 24 hours before imaging. Reporters were induced at mid-log phase with 0.1% galactose for 1.5 hours (GFP-reporters, Figure 5,8A) or 5 hours (mCherry-reporters, Figure 7). Imaging for Figures 5C,D,E was performed on a commercial LSM 710 confocal microscope (Carl Zeiss MicroImaging, Jena, Germany), using an objective C-Apochromat 40×/1.2NA, a solid state laser (488 nm) for excitation and a pixel dwell times of 101-177 μs. Imaging for Figures 5B,6,7,8A was on a wide-field deconvolution microscope (DeltaVision; Applied Precision/GE Healthcare), taking 60 × 0.2 μm sections, equipped with a 100×, 1.40 NA objective lens and solid state illumination; deconvolution was performed using Softworx, 10 iterations and medium noise filtering. The images were acquired using a charge-coupled device (CCD) camera (CoolSNAP HQ2; Photometrics). Data analysis is described in (Meinema et al., 2013) and in Supplemental Experimental Procedures.
Supplementary Material
Research Highlights.
Yeast membrane proteins Heh1 and Heh2 possess bipartite, IBB-like NLSs
Heh1 and Heh2 NLSs compete off the IBB of importin α in the absence of importin β
In vivo, importin α recruitment is critically dependent on P2’ in the minor-NLS box
Heh2 NLS is a super-potent membrane protein NLS
ACKNOWLEDGEMENTS
We thank the staff at NSLS beamlines X6A and X29, Bert Poolman, Michael Chang and the members of Veenhoff lab for valuable suggestions. This work was supported by NIH grant GM074846-01A1 (to G.C.), an NWO-VIDI grant from the Netherlands organization for scientific research (to L.M.V.) and a PhD fellowship from the Zernike Institute for Advanced Materials (to R.A.H.). R.A.P. is supported by NIH grant T32 GM100836. Research in this publication includes work carried out at the Sidney Kimmel Cancer Center X-ray Crystallography and Molecular Interaction Facility, which is supported in part by NCI grant P30 CA56036.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
R.K.L. and G.C. performed all biophysical/biochemical studies. A.B. and R.A.P helped with SPR and pull-downs, respectively. R.A.H., M.vR., A.S. and L.M.V. performed all in vivo studies. L.M.V. and G.C. wrote the manuscript with feedback from other authors.
Accession codes
The atomic coordinates and structure factors for Kap60 bound to h2NLS and h1NLS have been deposited in the protein Data Bank with accession codes 4PVZ and 4XZR.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Antonin W, Ungricht R, Kutay U. Traversing the NPC along the pore membrane: targeting of membrane proteins to the INM. Nucleus. 2011;2:87–91. doi: 10.4161/nucl.2.2.14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednenko J, Cingolani G, Gerace L. Nucleocytoplasmic transport: navigating the channel. Traffic. 2003;4:127–135. doi: 10.1034/j.1600-0854.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- Burns LT, Wente SR. Trafficking to uncharted territory of the nuclear envelope. Curr Opin Cell Biol. 2012;24:341–349. doi: 10.1016/j.ceb.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Chang CW, Counago RL, Williams SJ, Boden M, Kobe B. Crystal structure of rice importin-alpha and structural basis of its interaction with plant-specific nuclear localization signals. Plant Cell. 2012;24:5074–5088. doi: 10.1105/tpc.112.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Counago RM, Williams SJ, Boden M, Kobe B. Distinctive conformation of minor site-specific nuclear localization signals bound to importin-alpha. Traffic. 2013;14:1144–1154. doi: 10.1111/tra.12098. [DOI] [PubMed] [Google Scholar]
- Chen MH, Ben-Efraim I, Mitrousis G, Walker-Kopp N, Sims PJ, Cingolani G. Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin alpha. J Biol Chem. 2005;280:10599–10606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol Cell. 2002;10:1345–1353. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Lashuel HA, Gerace L, Muller CW. Nuclear import factors importin alpha and importin beta undergo mutually induced conformational changes upon association. FEBS Lett. 2000;484:291–298. doi: 10.1016/s0014-5793(00)02154-2. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Colledge WH, Richardson WD, Edge MD, Smith AE. Extensive mutagenesis of the nuclear location signal of simian virus 40 large-T antigen. Mol Cell Biol. 1986;6:4136–4139. doi: 10.1128/mcb.6.11.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure Fold Des. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–1478. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–965. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott- Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J Biol Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell. 2011;22:1058–1069. doi: 10.1091/mbc.E10-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesecke A, Stewart M. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J Biol Chem. 2011;285:17628–17635. doi: 10.1074/jbc.M110.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Hahn S, Maurer P, Caesar S, Schlenstedt G. Classical NLS proteins from Saccharomyces cerevisiae. J Mol Biol. 2008;379:678–694. doi: 10.1016/j.jmb.2008.04.038. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Hood JK, Casolari JM, Silver PA. Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-alpha export. J Cell Sci. 2000;113(Pt 8):1471–1480. doi: 10.1242/jcs.113.8.1471. [DOI] [PubMed] [Google Scholar]
- Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- Koerner C, Guan T, Gerace L, Cingolani G. Synergy of silent and hot spot mutations in importin beta reveals a dynamic mechanism for recognition of a nuclear localization signal. J Biol Chem. 2003;278:16216–16221. doi: 10.1074/jbc.M301137200. [DOI] [PubMed] [Google Scholar]
- Laba JK, Steen A, Veenhoff LM. Traffic to the inner membrane of the nuclear envelope. Curr Opin Cell Biol. 2014;28:36–45. doi: 10.1016/j.ceb.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Lange A, McLane LM, Mills RE, Devine SE, Corbett AH. Expanding the definition of the classical bipartite nuclear localization signal. Traffic. 2010;11:311–323. doi: 10.1111/j.1600-0854.2009.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Wu X, Summers MD, Lee A, Ryan KJ, Braunagel SC. Truncated isoforms of Kap60 facilitate trafficking of Heh2 to the nuclear envelope. Traffic. 2010;11:1506–1518. doi: 10.1111/j.1600-0854.2010.01119.x. [DOI] [PubMed] [Google Scholar]
- Lott K, Bhardwaj A, Mitrousis G, Pante N, Cingolani G. The importin beta binding domain modulates the avidity of importin beta for the nuclear pore complex. J Biol Chem. 2010;285:13769–13780. doi: 10.1074/jbc.M109.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Bhardwaj A, Sims PJ, Cingolani G. A Minimal Nuclear Localization Signal (NLS) in Human Phospholipid Scramblase 4 That Binds Only the Minor NLS-binding Site of Importin {alpha}1. J Biol Chem. 2011;286:28160–28169. doi: 10.1074/jbc.M111.228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Cingolani G. The importin beta binding domain as a master regulator of nucleocytoplasmic transport. Biochim Biophys Acta. 2011;1813:1578–1592. doi: 10.1016/j.bbamcr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk CP, Blobel G, King MC. Highway to the inner nuclear membrane: rules for the road. Nat Rev Mol Cell Biol. 2007;8:414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- Marfori M, Lonhienne TG, Forwood JK, Kobe B. Structural basis of high-affinity nuclear localization signal interactions with importin-alpha. Traffic. 2012;13:532–548. doi: 10.1111/j.1600-0854.2012.01329.x. [DOI] [PubMed] [Google Scholar]
- Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. Embo J. 2003;22:5358–5369. doi: 10.1093/emboj/cdg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk CP, Poolman B, Veenhoff LM. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333:90–93. doi: 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- Meinema AC, Poolman B, Veenhoff LM. Quantitative analysis of membrane protein transport across the nuclear pore complex. Traffic. 2013;14:487–501. doi: 10.1111/tra.12048. [DOI] [PubMed] [Google Scholar]
- Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J Biol Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- Nardozzi JD, Lott K, Cingolani G. Phosphorylation meets nuclear import: a review. Cell Commun Signal. 2010;8:32. doi: 10.1186/1478-811X-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Schirmer EC, Nishimoto T, Gerace L. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276: Macromolecular Crystallography. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Powell L, Burke B. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J Cell Biol. 1990;111:2225–2234. doi: 10.1083/jcb.111.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy RA, Cingolani G. Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem J. 2015;466:13–28. doi: 10.1042/BJ20141186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy RA, Ke S, Hart DJ, Zachariae U, Cingolani G. Molecular Determinants for Nuclear Import of Influenza A PB2 by Importin alpha Isoforms 3 and 7. Structure. 2015;23:374–384. doi: 10.1016/j.str.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy RA, Nardozzi JD, Hart DJ, Root MJ, Cingolani G. Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin alpha5. J Biol Chem. 2012;287:2022–2031. doi: 10.1074/jbc.M111.315838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Roman N, Christie M, Swarbrick CM, Kobe B, Forwood JK. Structural Characterisation of the Nuclear Import Receptor Importin Alpha in Complex with the Bipartite NLS of Prp20. PLoS One. 2013;8:e82038. doi: 10.1371/journal.pone.0082038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Smith S, Blobel G. The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J Cell Biol. 1993;120:631–637. doi: 10.1083/jcb.120.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J, Maurer P, Vogel F, Schlenstedt G. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol Cell Biol. 2000;20:8468–8479. doi: 10.1128/mcb.20.22.8468-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B, Worman HJ. The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J Cell Biol. 1993;120:1093–1100. doi: 10.1083/jcb.120.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B, Worman HJ. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Tapley EC, Ly N, Starr DA. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol Biol Cell. 2011;22:1739–1752. doi: 10.1091/mbc.E10-08-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29:2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuz S, Santarella-Mellwig R, Koch B, Jaedicke A, Mattaj IW, Antonin W. NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett. 2010;584:3292–3298. doi: 10.1016/j.febslet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Yewdell WT, Colombi P, Makhnevych T, Lusk CP. Lumenal interactions in nuclear pore complex assembly and stability. Mol Biol Cell. 2011;22:1375–1388. doi: 10.1091/mbc.E10-06-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleger N, Kerr AR, Schirmer EC. Many mechanisms, one entrance: membrane protein translocation into the nucleus. Cell Mol Life Sci. 2012;69:2205–2216. doi: 10.1007/s00018-012-0929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.