Abstract
Nuclear reprogramming induced by somatic cell nuclear transfer is an inefficient process, and donor cell DNA methylation status is thought to be a major factor affecting cloning efficiency. Here, the role of donor cell DNA methylation status regulated by 5-aza-2'-deoxycytidine (5-aza-dC) or 5-methyl-2'-deoxycytidine-5'-triphosphate (5-methyl-dCTP) in the early development of porcine cloned embryos was investigated. Our results showed that 5-aza-dC or 5-methyl-dCTP significantly reduced or increased the global methylation levels and altered the methylation and expression levels of key genes in donor cells. However, the development of cloned embryos derived from these cells was reduced. Furthermore, disrupted pseudo-pronucleus formation and transcripts of early embryo development-related genes were observed in cloned embryos derived from these cells. In conclusion, our results demonstrated that alteration of the DNA methylation status of donor cells by 5-aza-dC or 5-methyl-dCTP disrupted nuclear reprogramming and impaired the developmental competence of porcine cloned embryos.
Keywords: DNA methylation, Donor cell, Nuclear reprogramming, Somatic cell nuclear transfer, Pig
Though somatic cell nuclear transfer (SCNT) has been achieved in many species, overall cloning efficiency is still low, and this limits the application of cloning technology in basic research, agriculture, medicine, etc [1,2,3].
The most important event during SCNT is nuclear reprogramming, and it is believed that the DNA methylation status of donor cells could influence nuclear reprogramming efficiency [4,5,6,7]. Generally, it is thought that DNA hypomethylation of donor cells could lead to the high developmental competence of cloned embryos; thus, various strategies, such as application of hypomethylating agents or knockdown of DNA methyltransferase 1, are employed to reduce the DNA methylation level of donor cells, and to some extent, the developmental competence of cloned embryos is improved by these methods [4, 8,9,10]. However, some studies reported that 5-aza-dC, a widely used hypomethylating agent, could not efficiently improve the developmental competence of cloned embryos [11,12,13]. Moreover, the similarity of the methylation profiles of donor cells and in vivo produced embryos is negatively correlated with the the blastocyst rate of cloned embryos [5]. Thus, the effect of the DNA methylation status of donor cells on the developmental competence of cloned embryos still needs further investigation.
In this study, the DNA methylation status of donor cells regulated by 5-aza-dC or 5-methyl-dCTP and its effect on the developmental competence of cloned embryos were evaluated in pigs. Our results demonstrated that alteration of the DNA methylation status of donor cells impaired the developmental competence of porcine cloned embryos due to disturbance of nuclear remodeling and the expression patterns of early embryo development-related genes. This work reveals that alteration of donor cell DNA methylation status induced by DNA methylation modification agents might not be an ideal choice for improving cloning efficiency.
Materials and Methods
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA), and disposable and sterile plasticware was obtained from Nunclon (Roskilde, Denmark), unless otherwise stated.
All experiments were approved by the Animal Care Commission of Northeast Agriculture University, according to animal welfare laws, guidelines and policies.
Porcine fetal fibroblasts (PFFs) culture and treatment
PFF culture has been described previously [14]. Briefly, PFFs were isolated from a 35-day-old fetus. After removal of the head, internal organs and limbs, the remaining tissues were finely minced into pieces, digested with 0.25% trypsin-0.04% ethylenediaminetetraacetic acid solution (GIBCO, CA, USA) and then dispersed in high glucose-enriched Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) containing 10% fetal bovine serum (FBS, GIBCO) and 1% penicillin-streptomycin (GIBCO). The dispersed cells were centrifuged, resuspended and cultured in DMEM. Until confluence, PFFs were digested, centrifuged, resuspended in FBS containing 10% dimethyl sulfoxide and stored in liquid nitrogen until use.
For 5-aza-dC treatment, PFFs in the 3rd passage were cultured in DMEM supplemented with 10 nM (the optimal concentration) 5-aza-dC without any antibiotics for 72 h. For 5-methyl-dCTP treatment, PFFs in the 3rd passage were resuspended in electroporation buffer containing 0 µM, 1 µM, 2 µM or 5 µM 5-methyl-dCTP (TriLink BioTechnologies, CA, USA), incubated on ice for 15 min and transferred to an electroporation cuvette with a 4 mm gap, which was subjected to a single 20 msec pulse of 250 V with an ECM 2001 Electro Cell Manipulator. Following electroporation, PFFs were resuspended and cultured in DMEM for 72 h.
Cellular genomic methylation level
Measurement of the cellular genomic methylation level with flow cytometry has been reported [15]. Briefly, PFFs treated with 5-aza-dC or 5-methyl-dCTP were trypsinized, resuspended in cold saline GM and fixed in 100% ethanol at 4 C for at least 12 h. Then, PFFs were incubated in anti-5-methylcytosine antibodies (Santa Cruz, TX, USA) at 4 C overnight, washed with DPBS plus 5% FBS and resuspended in FITC-conjugated secondary antibodies (Santa Cruz) at 37 C for 30 min. After PFFs were filtered through a 30 μm nylon mesh, a FACSCalibur flow cytometer was used to measure the fluorescence level (the fluorescence intensity of PFFs not treated with the secondary antibody was considered to be zero).
Bisulfite sequencing
Genomic DNA was extracted from PFFs treated with 5-aza-dC or 5-methyl-dCTP using a Universal Genomic DNA Extraction Kit (Takara, Tokyo, Japan), and this was followed by treatment with sodium bisulfite to convert all unmethylated cytosine to uracil using an EZ DNA Methylation-GoldTM Kit (Zymo Research, CA, USA) according to the manufacturer’s protocol and our previous report [16]. Briefly, a CT (cytosine to thymine) conversion reagent was added to purified genomic DNA at 98 C for 10 min and 64 C for 2.5 h. Then, the samples were desalted, purified and diluted with M-Elution Buffer. Subsequently, nested PCR was carried out to amplify the target regions using the primers described in Supplementary Table 1 (online only) and Hot Start TaqTM Polymerase (Takara) with a profile of 94 C for 5 min; 40 cycles of 94 C for 30 sec, 55 C for 30 sec and 72 C for 1 min; and then 72 C for 10 min. Products from the first amplification reaction of the studied genes, not including Centromeric repeat DNA (CenRep), were used in the second PCR reaction, and the optimal annealing temperatures of inner primers were 53 C for Thy1, 51 C for Oct4 and 50 C for DMR3 Igf2/H19. Then, the amplified products were verified by electrophoresis and purified using an Agarose Gel DNA Purification Kit (Takara), and the purified fragments were cloned into a pMD18-T Vector (Takara) and subjected to sequence analysis.
Oocyte collection and in vitro maturation
Oocyte maturation has been described previously [14]. Briefly, porcine ovaries were collected from a local slaughterhouse and transported to the laboratory. Follicles were aspirated, and follicular contents were washed with HEPES-buffered Tyrode's lactate. Cumulus-oocyte complexes (COCs) with at least three uniform layers of compact cumulus cells and a uniform cytoplasm were recovered, washed and cultured in maturation medium under mineral oil at 38.5 C in a 5% CO2 atmosphere and saturated humidity. After 42 h, COCs were vortexed in 1 mg/ml hyaluronidase to remove cumulus cells. Only oocytes with a visible polar body, regular morphology and a homogenous cytoplasm were used.
In vitro fertilization (IVF) and SCNT, embryo culture and collection
The procedures for porcine IVF and SCNT were described in one of our previous reports [17]. Briefly, for IVF, the semen was incubated, resuspended and washed in DPBS supplemented with 0.1% (w/v) BSA. The spermatozoa were diluted with modified Tris-buffered medium (mTBM) to the appropriate concentration. Matured oocytes were washed in mTBM, transferred into fertilization medium and co-incubated with spermatozoa. Then, the embryos were washed and cultured in porcine zygote medium-3 (PZM-3) for subsequent development. For SCNT, matured oocytes and PFFs treated with 5-aza-dC or 5-methyl-dCTP were placed in manipulation medium. After enucleation, donor cells were placed into the perivitelline space. Fusion and activation of the cell-cytoplast complexes were induced by electroporation, and the fusion rate was confirmed by microscopic examination. Then, reconstructed embryos were cultured in PZM-3 for subsequent development. The cleavage and blastocyst rates of IVF and SCNT embryos were evaluated at 48 h and 156 h, and for embryo collection, embryos at the 4-cell and blastocyst stages were pooled at 48 h and 156 h, respectively.
Nuclear staining
Embryos at 6 h and 156 h post activation were treated with acidic Tyrode's solution to remove the zona pellucida, fixed in 4% paraformaldehyde for 30 min and stained in DPBS containing 10 µg/ml Hoechst 33342 for 5 min in the dark. After staining, cloned embryos were washed and mounted on slides. Then, the nuclear status and blastocyst cell number were examined under ultraviolet light from a fluorescence microscope.
Quantitative real-time PCR
Measurement of gene expression with quantitative real-time PCR has been applied in our previous studies [14, 17]. Briefly, total RNA was extracted from 104 PFFs or 50 embryos at each stage using an RNeasy Mini Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instruction. Reverse transcription was performed using a PrimeScript® RT Reagent Kit (Takara) with the following parameters: 37 C for 15 min and 85 C for 5 sec, with the cDNA then stored at –20 C until use. For quantitative real-time PCR, reactions were performed in 96-well optical reaction plates using SYBR® Premix ExTaqTM II (Takara) and a 7500 Real-Time PCR System with the following conditions: 95 C for 30 sec, followed by 40 two-step cycles of 95 C for 5 sec and 60 C for 34 sec, and finally a dissociation stage consisting of 95 C for 15 sec, 60 C for 1 min and 95 C for 15 sec. For every sample, the cycle threshold (CT) values were obtained from three replicates. The primers used for amplification of target and internal reference genes are presented in Supplementary Table 2 (online only). The relative expression levels of target genes were analyzed using the 2−ΔΔCT method.
Statistical analysis
Differences in data (mean ± SEM) were analyzed with the SPSS statistical software. Statistical analysis of data regarding embryo development was performed using the general linear model (GLM). The data for DNA methylation, gene expression and nuclear morphology remodeling were analyzed with one-way analysis of variance (ANOVA). For all analyses, differences were considered to be statistically significant when P < 0.05.
Results
5-aza-dC or 5-methyl-dCTP altered the genomic methylation status of PFFs
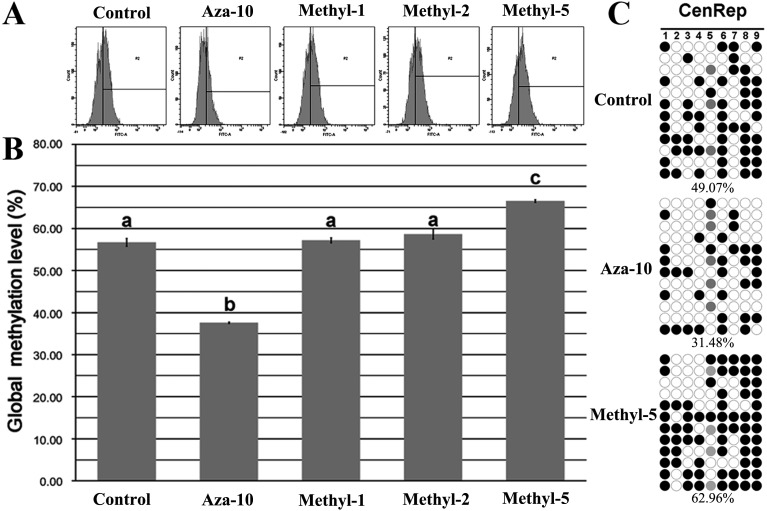
After PFFs were treated with 5-aza-dC or 5-methyl-dCTP, the genomic methylation status was investigated (Fig. 1). Compared with the control group (Fig. 1A and 1B), the genomic methylation level of PFFs treated with 10 nM 5-aza-dC (aza-10) was significantly decreased (37.60% vs. 56.73%, P < 0.05), and 5 µM 5-methyl-dCTP (methyl-5) significantly increased the global methylation levels of PFFs (66.57% vs. 56.73%, P < 0.05). To reconfirm the alteration of the global methylation status of PFFs treated with 5-aza-dC or 5-methyl-dCTP, the methylation levels of the CenRep region were detected (Fig. 1C), which revealed a lower or higher methylation level in the aza-10 or methyl-5 group in comparison with the control group (31.48% or 62.96% vs. 49.07%). Thus, our results revealed that 5-aza-dC or 5-methyl-dCTP could successfully alter the global methylation status of PFFs.
Fig. 1.
Global DNA methylation levels in PFFs. A, flow cytometry analysis of global DNA methylation in PFFs treated with 5-aza-dC or 5-methyl-dCTP; B, levels of global DNA methylation in PFFs treated with 5-aza-dC or 5-methyl-dCTP; C, methylation status of the CenRep region in PFFs treated with 5-aza-dC or 5-methyl-dCTP. The control, aza-10, methyl-1, methyl-2 and methyl-5 groups represent PFFs not treated or treated with 10 nM 5-aza-dC and 1 µM, 2 µM and 5 µM 5-methyl-dCTP for 72 h, respectively. Black and white circles represent methylated and unmethylated CpG sites, and gray circles represent mutated and/or single nucleotide polymorphism (SNP) variation at specific CpG sites. Percentages (% ± SEM) in columns with different superscripts differed significantly (P < 0.05).
5-aza-dC or 5-methyl-dCTP regulated gene methylation and transcription in PFFs
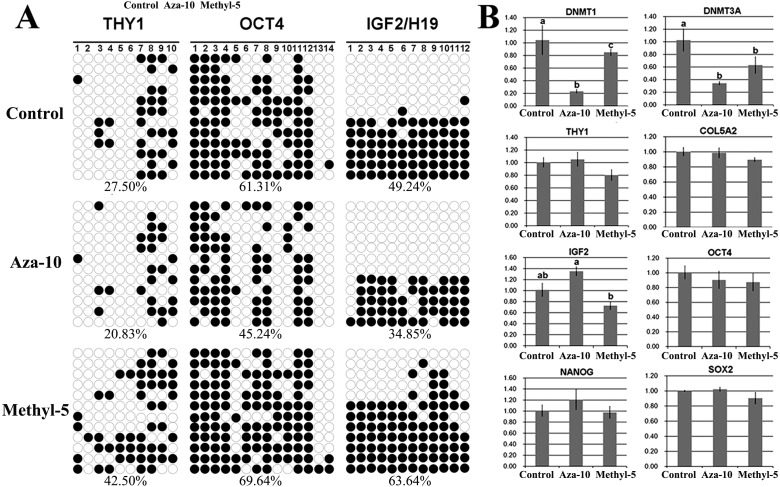
The DNA methylation and transcription levels of genes related to early development of cloned embryos from PFFs treated with 5-aza-dC or 5-methyl-dCTP were examined (Fig. 2). Compared with the control group, reduced methylation levels of Thy1, Oct4 and DMR3 Igf2/H19, a higher Igf2 transcription level and significantly (P < 0.05) lower expression of Dnmt1 and Dnmt3a were observed in the aza-10 group. In the methyl-5 group, the methylation levels of Thy1, Oct4 and DMR3 Igf2/H19 were increased, and a lower expression level of Igf2 and significantly (P < 0.05) reduced transcripts of Dnmt1 and Dnmt3a were detected. Thus, our results showed that 5-aza-dC or 5-methyl-dCTP could regulate DNA methylation and transcripts of genes related to early development of cloned embryos.
Fig. 2.
Specific gene methylation and transcription in PFFs. A, methylation status of Thy1, Oct4 and Igf2/H19 in PFFs after treatment with 10 nM 5-aza-dC (aza-10) or 5 µM 5-methyl-dCTP (methyl-5); B, relative transcription levels of specific genes in the aza-10 or methyl-5 group. The data are expressed as means ± SEM. a–c Values in columns for a given gene with different superscripts differ significantly (P < 0.05).
Alteration of the DNA methylation status of donor cells reduced the developmental competence of porcine cloned embryos
The developmental competence of cloned embryos generated with PFFs treated with 5-aza-dC or 5-methyl-dCTP as donor cells was examined (Table 1 and Supplementary Fig. 1: online only). Our results showed that the fusion and cleavage rates in the aza-10 and methyl-5 groups were significantly (P < 0.05) lower than those in the control group. Furthermore, the blastocyst rate in the methyl-5 group was also significantly (P < 0.05) reduced in comparison with the control group. Therefore, alteration of the DNA methylation status of donor cells reduced the developmental competence of porcine cloned embryos.
Table 1. Development of cloned embryos derived from donor cells treated with 5-aza-dC or 5-methyl-dCTP.
| Group | No. embryos (Rep.) | No. embryos fused (% ± SEM) |
No. embryos cleaved (% ± SEM) * |
No. blastocysts (% ± SEM) * |
Total cell numbers of blastocysts (mean ± SEM) & |
| IVF | 224 (5) | — | 175 (78.07 ± 2.22) abc | 52 (23.28 ± 0.88) a | 39 ± 3 (n = 52) |
| Control | 279 (5) | 222 (79.71 ± 1.99) a | 201 (90.31 ± 1.44) d | 48 (21.43 ± 1.20) ab | 37 ± 2 (n = 47) |
| Aza-10 | 278 (5) | 192 (69.46 ± 1.37) b | 153 (79.54 ± 1.35) ace | 36 (18.87 ± 0.72) bc | 35 ± 2 (n = 36) |
| Methyl-1 | 297 (5) | 216 (72.64 ± 3.47) b | 181 (83.50 ± 1.00) e | 40 (18.95 ± 1.12) bc | 35 ± 2 (n = 39) |
| Methyl-2 | 312 (5) | 216 (69.10 ± 1.87) b | 174 (80.79 ± 1.17) ae | 38 (17.63 ± 0.79) c | 36 ± 2 (n = 36) |
| Methyl-5 | 320 (5) | 193 (60.47 ± 2.30) c | 146 (75.77 ± 2.47) c | 28 (14.70 ± 1.17) d | 34 ± 3 (n = 28) |
* Cleavage and blastocyst rates of cloned embryos were adjusted for fusion rates. & Blastocyst cell numbers of less than 16 were not included. a–c Values in the same column with different superscripts differ significantly (P < 0.05).
Alteration of the DNA methylation status of donor cells impaired nuclear remodeling in porcine cloned embryos
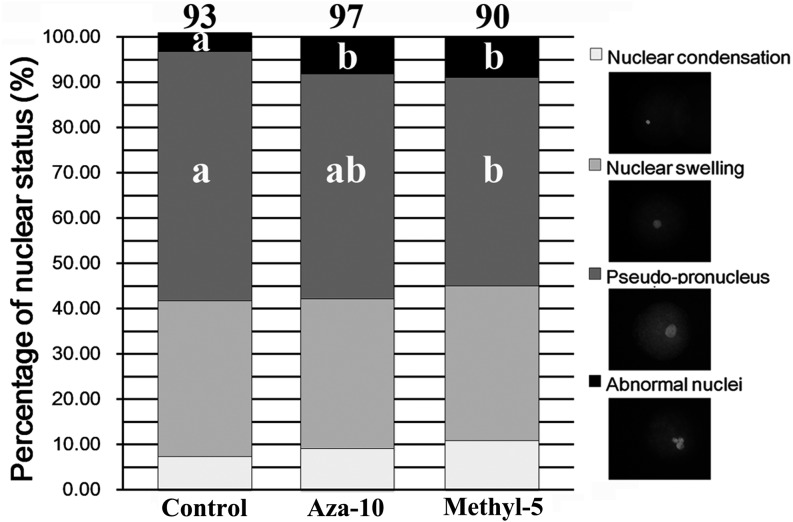
Morphological remodeling of the donor nucleus plays a key role in successful nuclear reprogramming [18]. Pseudo-pronucleus formation in cloned embryos was examined in the present study (Fig. 3). The results demonstrated significantly reduced pseudo-pronucleus proportions in the aza-10 and methyl-5 groups (49.67% and 46.06%, respectively, vs. 55.08%, P < 0.05), and the proportions of abnormal nuclei were significantly increased (8.19% and 8.93%, respectively, vs. 4.21%, P < 0.05) in comparison with the control group. These results revealed that abnormal nuclear remodeling in cloned embryos derived from donor cells treated with 5-aza-dC or 5-methyl-dCTP resulted in low developmental competence of porcine cloned embryos.
Fig. 3.
Pseudo-pronucleus formation. Pseudo-pronucleus formation of cloned embryos from PFFs treated with 5-aza-dC (aza-10) or 5-methyl-dCTP (methyl-5) at 6 h post activation. The images of nuclear morphology of cloned embryos were magnified (400 ×). a–b Percentages at a given nuclear status in columns with different superscripts differed significantly (P < 0.05).
Alteration of the DNA methylation status of donor cells disrupted the expression patterns of early embryo development- related gene in porcine cloned embryos
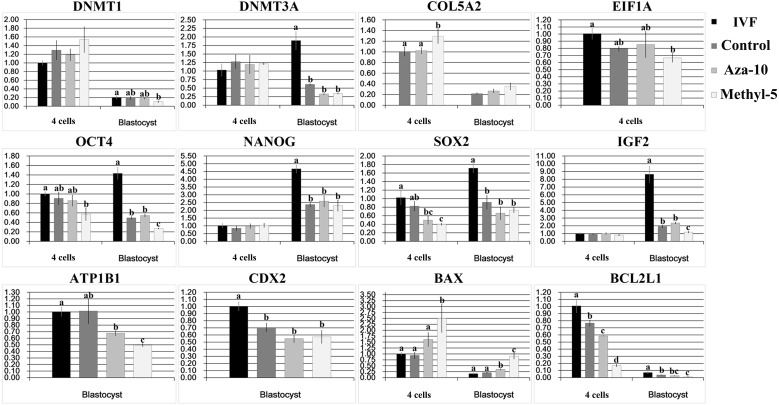
The expression pattern of early embryo development-related genes is correlated with embryonic developmental competence [19]. Here, we investigated transcripts of DNA methyltransferase (Dnmt1 and Dnmt3a), fibroblast marker (Col5a2), apoptosis (Bax and Bcl2 l1) and development-related (Eif1a, Oct4, Nanog, Sox2, Igf2, ATP1b1 and Cdx2) genes in early embryos (Fig. 4). In the aza-10 group, though the transcript levels of most of the studied genes were not significantly different from those in the control NT group, a significantly lower expression of Bcl2 l1 in 4-cell embryos and a significantly higher transcript of Bax in blastocysts were observed (P < 0.05), suggesting that the treatment of donor cells with 5-aza-dC induces apoptosis during early development of cloned embryos. In the methyl-5 group, significantly increased expression of Col5a2 at the 4-cell stage and significantly decreased transcripts of Sox2 at the 4-cell stage and of Oct4, Igf2 and ATP1b1 at the blastocyst stage were detected (P < 0.05) in comparison with the control NT group. Additionally, significantly reduced expression of Bcl2 l1 and significantly increased transcripts of Bax were also observed at the 4-cell and blastocyst stages in the methyl-5 group (P < 0.05). These results demonstrated that the transcripts of early embryo development-related genes in the aza-10 and methyl-5 groups were disturbed, resulting in low developmental competence of porcine cloned embryos.
Fig. 4.
Relative transcripts of embryo development-related genes in cloned embryos. Relative transcripts of embryo development-related genes in 4-cell embryos and blastocysts from PFFs treated with 5-aza-dC (aza-10) or 5-methyl-dCTP (methyl-5). The data are expressed as means ± SEM. a–d Values in columns for a given gene with different superscripts differed significantly (P < 0.05).
Discussion
Previous studies have shown that the DNA methylation status of donor nuclei could determine the developmental competence of cloned embryos [4, 5]. Here, we demonstrated that alteration of the DNA methylation status of donor cells impaired the developmental competence of porcine cloned embryos, and this could be due to the disturbance of nuclear remodeling and the expression patterns of early embryo development-related genes in cloned embryos.
To clarify the relationship between the DNA methylation status of donor cells and the developmental competence of cloned embryos, 5-aza-dC or 5-methyl-dCTP was employed to regulate the DNA methylation status of donor cells, as 5-aza-dC and 5-methyl-dCTP could be incorporated into the genome during DNA synthesis, resulting in DNA hypomethylation or hypermethylation, respectively [20,21,22]. In this study, the appropriate concentration of 5-aza-dC or 5-methyl-dCTP to treat donor cells was first determined with no obvious changes of cell proliferation and the cell cycle (Supplementary Fig. 2: online only), and then we found that 5-aza-dC successfully downregulated the DNA methylation levels of the genome and candidate genes in donor cells; conversely, the DNA methylation levels were upregulated by 5-methyl-dCTP. Moreover, both agents reduced the expression of DNA methyltransferases, seeming to be beneficial for nuclear reprogramming [9]. However, neither downregulation nor upregulation of the DNA methylation status of donor cells by 5-aza-dC and 5-methyl-dCTP, respectively, had a positive effect on the developmental competence of porcine cloned embryos. As we know, both 5-aza-dC and 5-methyl-dCTP have a fast metabolic rate [21], and only a trace amount of free 5-aza-dC or 5-methyl-dCTP possibly existed in donor cells, if any, when donor cells were fused with enucleated oocytes. Furthermore, this trace amount of free 5-aza-dC or 5-methyl-dCTP could be further diluted, as the volume of an oocyte can be approximately equal to that of one thousand donor cells. Thus, it is likely that free 5-aza-dC or 5-methyl-dCTP would have no detrimental effects on cloned embryos. These results indicated that regulation of the donor cell methylation status may be not an ideal choice for improving cloning efficiency.
Actually, several studies have reported that 5-aza-dC or 5-methyl-dCTP could cause some defects during nuclear reprogramming, leading to low developmental competence of early embryos [11, 20], and it has been reported that the detrimental effects could be reflected by the nuclear morphology remodeling and gene expression patterns [14, 18, 23, 24]. It is known that pseudo-pronucleus formation from nuclear morphology remodeling is fundamental to nuclear reprogramming in cloned embryos [18]; however, in this study, 5-aza-dC or 5-methyl-dCTP treatments did not facilitate pseudo-pronucleus formation but instead increased the proportion of abnormal nuclei in cloned embryos, suggesting that 5-aza-dC or 5-methyl-dCTP may lead to genome instability, further leading to the poor development of cloned embryos [14]. As we know, many molecules are involved in pseudo-pronucleus formation during nuclear morphology remodeling [25], and determination of the molecular mechanism underlying the disruption of nuclear remodeling induced by 5-aza-dC or 5-methyl-dCTP needs further study. In addition, the expression patterns of early embryo development-related genes are correlated with the developmental competence of cloned embryos [18, 19]. Though no significant changes of genomic methylation levels were observed (Supplementary Fig. 3: online only), somatic cell-specific genes were not effectively silenced, and the expression patterns of early embryo development-related genes were also disturbed in cloned embryos after treatment of donor cells with 5-aza-dC or 5-methyl-dCTP. More importantly, the transcripts of an antiapoptotic gene (Bax) and a proapoptotic gene (Bcl2l1) suggested that treatment of donor cells with 5-aza-dC or 5-methyl-dCTP may activate apoptosis-related signals in cloned embryos, and a TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay further proved that 5-aza-dC or 5-methyl-dCTP treatment significantly promoted embryo apoptosis (Supplementary Fig. 4: online only); thus, apoptosis induced by 5-aza-dC or 5-methyl-dCTP could inhibit nuclear reprogramming and challenge the survival of cloned embryos [20, 26,27,28]. Regarding how 5-aza-dC or 5-methyl-dCTP disrupts the expression patterns of genes related to early embryo development in cloned embryos, it could be that the alteration of DNA methylation induced by 5-aza-dC or 5-methyl-dCTP possibly causes genomic instability and induces DNA damage, further leading to the disrupted expression; however, the detailed mechanism still needs to be investigated. The results of the present study demonstrated that the low developmental competence of porcine cloned embryos induced by 5-aza-dC or 5-methyl-dCTP treatment of donor cells could result from the reduction of pseudo-pronucleus formation during nuclear reprogramming and the disturbance of the expression patterns of somatic cell-specific genes, embryo development-related genes and apoptosis-related genes during early embryo development.
In conclusion, our study demonstrated that alteration of the DNA methylation status of donor cells by 5-aza-dC or 5-methyl-dCTP impaired the developmental competence of porcine cloned embryos, and that low developmental competence could be due to the failure of nuclear remodeling and disturbance of the expression patterns of early embryo development-related genes in cloned embryos.
Supplementary Materials
Acknowledgments
This work was supported by grants from the National High Technology Research and Development Program of China, Program 863 (2012AA020601), National Natural Science Foundation of China, NSFC (31371457), and Shandong Provincial Natural Science Foundation, China (BS2015SW014). The authors declare that no conflicting financial interests exist.
References
- 1.Lee K, Prather RS. Advancements in somatic cell nuclear transfer and future perspectives. Anim Front 2013; 3: 56–61. [Google Scholar]
- 2.Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci 2013; 368: 20110329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizutani E, Wakayama S, Wakayama T. Treatment of donor cell/embryo with different approaches to improve development after nuclear transfer. Methods Mol Biol 2015; 1222: 101–111. [DOI] [PubMed] [Google Scholar]
- 4.Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells 2006; 24: 2007–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonk AJ, Cheong HT, Li R, Lai L, Hao Y, Liu Z, Samuel M, Fergason EA, Whitworth KM, Murphy CN, Antoniou E, Prather RS. Correlation of developmental differences of nuclear transfer embryos cells to the methylation profiles of nuclear transfer donor cells in Swine. Epigenetics 2007; 2: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peat JR, Reik W. Incomplete methylation reprogramming in SCNT embryos. Nat Genet 2012; 44: 965–966. [DOI] [PubMed] [Google Scholar]
- 7.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet 2013; 14: 204–220. [DOI] [PubMed] [Google Scholar]
- 8.Ding X, Wang Y, Zhang D, Wang Y, Guo Z, Zhang Y. Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology 2008; 70: 622–630. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka K, Sakatani M, Kubota K, Balboula AZ, Sawai K, Takahashi M. Effects of downregulating DNA methyltransferase 1 transcript by RNA interference on DNA methylation status of the satellite I region and in vitro development of bovine somatic cell nuclear transfer embryos. J Reprod Dev 2011; 57: 393–402. [DOI] [PubMed] [Google Scholar]
- 10.Huan YJ, Zhu J, Wang HM, Wu ZF, Zhang JG, Xie BT, Li JY, Kong QR, Liu ZH, He HB. Epigenetic modification agents improve genomic methylation reprogramming in porcine cloned embryos. J Reprod Dev 2014; 60: 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright BP, Sung LY, Chang CC, Yang X, Tian XC. Methylation and acetylation characteristics of cloned bovine embryos from donor cells treated with 5-aza-2′-deoxycytidine. Biol Reprod 2005; 72: 944–948. [DOI] [PubMed] [Google Scholar]
- 12.Jafarpour F, Hosseini SM, Hajian M, Forouzanfar M, Ostadhosseini S, Abedi P, Gholami S, Ghaedi K, Gourabi H, Shahverdi AH, Vosough AD, Nasr-Esfahani MH. Somatic cell-induced hyperacetylation, but not hypomethylation, positively and reversibly affects the efficiency of in vitro cloned blastocyst production in cattle. Cell Reprogram 2011; 13: 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ning SF, Li QY, Liang MM, Yang XG, Xu HY, Lu YQ, Lu SS, Lu KH. Methylation characteristics and developmental potential of Guangxi Bama minipig (Sus scrofa domestica) cloned embryos from donor cells treated with trichostatin A and 5-aza-2′-deoxycytidine. Zygote 2013; 21: 178–186. [DOI] [PubMed] [Google Scholar]
- 14.Huan YJ, Zhu J, Xie BT, Wang JY, Liu SC, Zhou Y, Kong QR, He HB, Liu ZH. Treating cloned embryos, but not donor cells, with 5-aza-2′-deoxycytidine enhances the developmental competence of porcine cloned embryos. J Reprod Dev 2013; 59: 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohana Kumar B, Jin HF, Kim JG, Song HJ, Hong Y, Balasubramanian S, Choe SY, Rho GJ. DNA methylation levels in porcine fetal fibroblasts induced by an inhibitor of methylation, 5-azacytidine. Cell Tissue Res 2006; 325: 445–454. [DOI] [PubMed] [Google Scholar]
- 16.Wei Y, Zhu J, Huan Y, Liu Z, Yang C, Zhang X, Mu Y, Xia P, Liu Z. Aberrant expression and methylation status of putatively imprinted genes in placenta of cloned piglets. Cell Reprogram 2010; 12: 213–222. [DOI] [PubMed] [Google Scholar]
- 17.Wei Y, Huan Y, Shi Y, Liu Z, Bou G, Luo Y, Zhang L, Yang C, Kong Q, Tian J, Xia P, Sun QY, Liu Z. Unfaithful maintenance of methylation imprints due to loss of maternal nuclear Dnmt1 during somatic cell nuclear transfer. PLoS ONE 2011; 6: e20154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Whyte J, Prather RS. Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res 2010; 341: 13–21. [DOI] [PubMed] [Google Scholar]
- 19.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat Struct Mol Biol 2013; 20: 282–289. [DOI] [PubMed] [Google Scholar]
- 20.Kaito C, Kai M, Higo T, Takayama E, Fukamachi H, Sekimizu K, Shiokawa K. Activation of the maternally preset program of apoptosis by microinjection of 5-aza-2′-deoxycytidine and 5-methyl-2′-deoxycytidine-5′-triphosphate in Xenopus laevis embryos. Dev Growth Differ 2001; 43: 383–390. [DOI] [PubMed] [Google Scholar]
- 21.Szyf M.DNA methylation and demethylation probed by small molecules. Biochim Biophys Acta 2010; 1799: 750–759. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y, Hu Z. Genome-wide demethylation by 5-aza-2′-deoxycytidine alters the cell fate of stem/progenitor cells. Stem Cell Rev 2015; 11: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long CR, Westhusin ME, Golding MC. Reshaping the transcriptional frontier: epigenetics and somatic cell nuclear transfer. Mol Reprod Dev 2014; 81: 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Y, Hai T, Wang Y, Guo R, Li W, Wang L, Zhou Q. Epigenetic reprogramming, gene expression and in vitro development of porcine SCNT embryos are significantly improved by a histone deacetylase inhibitor-m-carboxycinnamic acid bishydroxamide (CBHA). Protein Cell 2014; 14: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prather RS, Kühholzer B, Lai L, Park KW. Changes in the structure of nuclei after transfer to oocytes. Cloning 2000; 2: 117–122. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Maxson R, Jones PA. Direct induction of DNA hypermethylation in sea urchin embryos by microinjection of 5-methyl dCTP stimulates early histone gene expression and leads to developmental arrest. Dev Biol 1993; 155: 75–86. [DOI] [PubMed] [Google Scholar]
- 27.Nieto M, Samper E, Fraga MF, González de Buitrago G, Esteller M, Serrano M. The absence of p53 is critical for the induction of apoptosis by 5-aza-2′-deoxycytidine. Oncogene 2004; 23: 735–743. [DOI] [PubMed] [Google Scholar]
- 28.Oka M, Meacham AM, Hamazaki T, Rodić N, Chang LJ, Terada N. De novo DNA methyltransferases Dnmt3a and Dnmt3b primarily mediate the cytotoxic effect of 5-aza-2′-deoxycytidine. Oncogene 2005; 24: 3091–3099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.