Abstract
The Fc region of Immunoglobulin G (IgG) initiates inflammatory responses such as antibody-dependent cell-mediated cytotoxicity (ADCC) through binding to activating Fc receptors (FcγRI, FcγRIIa, FcγRIIIa). These receptors are expressed on the surface of immune cells including macrophages, dendritic cells, and natural killer cells. An inhibitory receptor, FcγRIIb, is expressed on macrophages and other myeloid leukocytes simultaneously with the activating receptor FcγRIIa, thereby setting a threshold for cell activation. The affinity of IgG Fc for binding activating Fc receptors depends on IgG subclass and the composition of N-linked glycans attached to a conserved asparagine in the Fc CH2 domain. For example, Fc regions with afucosylated glycans bind more tightly to FcγRIIIa than fucosylated Fc, and afucosylated Fcs exhibit enhanced ADCC activity in vivo and in vitro. Enhanced pro-inflammatory responses have also been seen for Fc regions with amino acid substitutions. GASDALIE Fc is an Fc mutant (G236A/S239D/A330L/I332E) that exhibits a higher affinity for FcγRIIIa and increased effector functions in vivo compared to wild-type Fc. To explore its altered functions, we compared the affinities of GASDALIE and wild-type Fc for activating and inhibitory FcγRs. We also determined the crystal structure of GASDALIE Fc alone and bound to FcγRIIIa. The overall structure of GASDALIE Fc alone was similar to wild-type Fc structures, however, increased electrostatic interactions in the GASDALIE Fc:FcγRIIIa interface compared with other Fc:FcγR structures suggest a mechanism for the increased affinity of GASDALIE Fc for FcγRIIIa.
Keywords: Affinity, Fc-FcR structure, Fc receptors, GASDALIE Fc, IgG Fc, SDALIE Fc
Introduction
Immunoglobulin G (IgG) plays critical roles in antibody-mediated immune responses through specific engagement of both antigens and immune effector cells. The fragment crystallizable (Fc) region specifically engages Fc gamma receptors (FcγRs), the initial step in effector functions such as antibody-dependent cell-mediated cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP) (Arnold et al., 2007). In humans, activating FcγR FcγRIIIa (CD16) is expressed on the surface of natural killer cells and monocytes while FcγRIIa (CD32) is found on a wider range of innate immune cells including macrophages, dendritic cells, and neutrophils (DiLillo and Ravetch, 2015b; Richards et al., 2008). Both are low-affinity receptors, therefore activation of cells results from crosslinking of these surface receptors upon engagement of clustered Fc regions in antibody-antigen immune complexes (ICs) (Nimmerjahn and Ravetch, 2008). Activation is mediated by immunoreceptor tyrosine-based activating motifs (ITAMs) found on the receptor-associated γ-chain. Upon engagement of ICs with FcγRs, these motifs become phosphorylated leading to a cascade of events that activate the cell to destroy an invading pathogen or infected cell (Nimmerjahn and Ravetch, 2008). An inhibitory receptor, FcγRIIb, is found on cells including B cells, macrophages, neutrophils, and mast cells (Hulett and Hogarth, 1994; Ravetch and Bolland, 2001; Ravetch and Kinet, 1991). Immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in the cytoplasmic tail of FcγRIIb mediate suppression of FcγR activation by activating phosphatases that reverse the effects of kinases in the ITAM signaling pathway (Nimmerjahn and Ravetch, 2008). Co-expression of activating and inhibitory receptors sets a threshold for activation of specific effector cells.
IgG Fc regions are independently-folded, stable homodimers comprising two N-terminal hinges (each connected to an antigen-binding Fab arm) followed by two Ig constant domains, CH2 and CH3, on each chain of the homodimer. FcγRs bind asymmetrically to Fc homodimers with 1:1 receptor:Fc stoichiometry to a site involving the hinge region and N-terminal portions of the CH2 domains (Ferrara et al., 2011). The Fc region includes a heterogeneous N-linked glycan attached to a conserved residue, Asn297, in each Fc CH2 domain. The Asn297-linked glycan is a complex carbohydrate composed of a mannose (Man) and N-acetylglucosamine (GlcNAc) core that is usually fucosylated. The glycan can be additionally modified with a terminal galactose or sialic acid. Typically, wild-type (wt) IgG is agalactosylated (G0; 35%), mono-galactosylated (G1; 35%), or digalactosylated (G2; 16%) and asialylated (S0; 85%), although mono- and disialylated IgG represent 11% and 4%, respectively, of IgG in human serum (Butler et al., 2003). Modification of the N-linked glycan of IgG Fc can alter its function. For example, terminally sialylating the glycan switches the Fc from a pro-inflammatory to anti-inflammatory molecule (Anthony et al., 2011; Anthony et al., 2008a; Anthony and Ravetch, 2010; Anthony et al., 2008b; Kaneko et al., 2006; Nimmerjahn and Ravetch, 2008; Samuelsson et al., 2001; Sondermann et al., 2013), which may correlate with increased conformational flexibility compared with wtFc regions (Ahmed et al., 2014). Afucosylation of the normally core-fucosylated glycan leads to enhanced binding to the activating receptor FcγRIIIa, resulting in enhanced ADCC (Ferrara et al., 2011; Matsumiya et al., 2007; Mizushima et al., 2011; Okazaki et al., 2004) due to increased protein-protein and protein-carbohydrate interactions between the Fc and receptor (Ferrara et al., 2011; Matsumiya et al., 2007; Okazaki et al., 2004). Removing the N-linked glycan leads to a closed Fc conformation that exhibits no binding to any FcγR (Shields et al., 2001); however, aglycosylated Fc can bind the FcRn protection receptor (Shields et al., 2001), which recognizes the interface between the CH2 and CH3 domains of each Fc chain (Burmeister et al., 1994), a site distant from the Asn297-linked N-linked glycan.
Previous efforts were made to alter the IgG Fc region to either enhance or suppress binding to FcγRs (Bournazos et al., 2014a; Duncan et al., 1988; Ferrara et al., 2011; Lazar et al., 2006; Lin et al., 2015; Matsumiya et al., 2007; Nimmerjahn and Ravetch, 2008; Oganesyan et al., 2008a; Oganesyan et al., 2008b; Richards et al., 2008; Sazinsky et al., 2008; Shields et al., 2001). In a 2006 study, IgG Fc with mutations S239D/I332E (SDIE) or S239D/A330L/I332E (SDALIE) showed enhanced ADCC and increased affinity for binding to FcγRIIIaF158 (30- and 60-fold for SDIE Fc and 60- and 170-fold for SDALIE Fc) (Lazar et al., 2006). In a separate study, SDIE Fc showed a 30-fold increased affinity for FcγRIIIAF158 (Smith et al., 2012). SDIE and SDALIE Fc also showed increased binding to the inhibitory receptor, FcγRIIb, by 70-fold and 40-fold, respectively (Lazar et al., 2006; Oganesyan et al., 2008a; Smith et al., 2012). A later study reported only a 14-fold increased affinity for SDIE binding to FcγRIIb (Smith et al., 2012). The crystal structure of unbound SDALIE Fc revealed a more open conformation compared with wtFc structures, but no major changes in the FcγR recognition interface (Oganesyan et al., 2008a). Related Fc mutants also showed increased activity and affinity for binding to FcγRs. A single G236A substitution in Fc showed 7-fold enhanced binding to FcγRIIa and enhanced phagocytosis in vitro, while an Fc with mutations G236A/S239D/I332E (GASDIE) exhibited a 70-fold phagocytosis enhancement (Richards et al., 2008). The G236A substitution alone did not cause enhanced binding to FcγRIIb, whereas the GASDIE mutant showed 14-fold enhanced affinity for this inhibitory receptor (Richards et al., 2008). GASDALIE Fc, a variant of SDALIE Fc that combined the G236A substitution with the SDALIE substitutions (G236A/S239D/A330L/I332E), showed increased affinity for activating receptor FcγRIIIaF158 by 20–30-fold (Bournazos et al., 2014b; DiLillo and Ravetch, 2015a; Smith et al., 2012). Unlike SDALIE Fc, GASDALIE Fc did not exhibit a large increase in affinity for the inhibitory receptor FcγRIIb (only a 2–3–fold increase for GASDALIE, as compared with a 40-fold increase for SDALIE), (Bournazos et al., 2014b; DiLillo and Ravetch, 2015a; Smith et al., 2012).
IgGs with the GASDALIE mutations have recently been shown to exhibit increased protection in animal models of cancer and infectious disease (Bournazos et al., 2014b; DiLillo and Ravetch, 2015a; Smith et al., 2012). Although affinities of GASDALIE and SDALIE Fcs have been measured for some of the FcγRs (Bournazos et al., 2014a; Bournazos et al., 2014b; DiLillo and Ravetch, 2015a; Oganesyan et al., 2008a; Richards et al., 2008; Sazinsky et al., 2008; Smith et al., 2012), they have not been directly compared in a single experiment. In addition, there are discrepancies in reported affinities of Fc mutants for activating and inhibitory FcγRs (Bournazos et al., 2014a; Bournazos et al., 2014b; DiLillo and Ravetch, 2015a; Richards et al., 2008; Sazinsky et al., 2008; Smith et al., 2012). Here we directly compared binding of GASDALIE and SDALIE Fc to FcγRs, finding that both Fcs exhibited large affinity increases for FcγRIIIa compared to wtFc, that GASDALIE exhibited a moderate affinity increase for FcγRIIa compared with wtFc, and that both Fcs showed only ~5-fold enhanced affinity for the inhibitory FcγRIIb receptor compared to wtFc. In order to determine the mechanism by which the GASDALIE substitutions enhanced binding to activating FcγRs, we solved crystal structures of GASDALIE Fc alone and in complex with FcγRIIIa, comparing the bound and free GASDALIE Fc structures to the previously-solved unbound SDALIE Fc structure (Oganesyan et al., 2008a; Smith et al., 2012) and other Fc structures. We find that the GASDALIE substitutions did not dramatically alter the overall Fc conformation; however, the GASDALIE Fc:FcγRIIIa structure revealed increased electrostatic interactions at the binding interface that may account for its increased affinity for FcγRIIIa when compared to wtFc structures. Our GASDALIE Fc:FcγRIIIa crystal structure, homology models of GASDALIE Fc bound to FcγRIIa, and affinity measurements suggested that the G236A substitution that distinguishes GASDALIE Fc from SDALIE Fc is localized in the FcγRIIa binding site, thereby enhancing binding of FcγRIIa to GASDALIE compared with binding to wtFc or SDALIE Fc.
Materials and Methods
Protein Expression and Purification
The S239D/A330L/I332E (SDALIE) and G236A/S239D/A330L/I332E (GASDALIE) mutations were introduced by site-directed mutagenesis in a pcDNA3.1 vector containing the IgG1 Fc gene (Agilent Technologies). A plasmid containing the GASDALIE mutations (G236A/S239D/A330L/I332E) introduced into Fc region of the gene encoding the heavy chain of an anti-HIV-1 antibody, 3BNC117, was the gift of Stylianos Bournazos and Jeffrey Ravetch (Rockefeller University) (Bournazos et al., 2014b; Scheid et al., 2011). IgG and Fc proteins were expressed in transiently-transfected HEK 293-6E cells and purified from harvested supernatants using protein A chromatography (GE Healthcare) followed by size exclusion chromatography in 25 mM Tris-Cl pH 7.5, 100 mM NaCl on a Superdex 200 10/300 gel-filtration column (GE Healthcare) as described (Diskin et al., 2010; Sprague et al., 2004). The Fc region from 3BNC117 IgG was isolated after papain digestion of 3BNC117 GASDALIE IgG as described (Ahmed et al., 2014) and used for crystallization of GASDALIE Fc alone; other experiments used GASDALIE Fc expressed as an Fc fragment.
Expression plasmids encoding the F158 variant of FcγRIIIa and the H131 variant of FcγRIIa (FcγRIIIaF158 and FcγRIIaH131) were obtained from Stylianos Bournazos and Jeffrey Ravetch (Rockefeller University) (Bournazos et al., 2014b). Site directed mutagenesis was used to remove three of five potential N-linked glycosylation sites on FcγRIIIa (Asn38, Asn74, Asn169) by changing asparagines to glutamines to make N38Q, N74Q, N169Q substitutions, as described for previous structural studies of FcγRIIIa (Ferrara et al., 2011). Potential N-linked glycosylations sites at sites 45 and 162 were retained due to their effects on protein expression and binding affinity, respectively (Ferrara et al., 2011; Ferrara et al., 2006). C-terminally 6x-His tagged FcγR ectodomains FcγRIIaH131 (residues 1–211) and FcγRIIIaF158 (residues 1–218) were expressed in transiently-transfected HEK293 cells as described for Fc proteins and isolated from supernatants using a HisTrap HP affinity column (GE Healthcare) followed by size exclusion chromatography in 25 mM Tris-Cl pH 7.5, 100 mM NaCl on a Superdex 200 16/ 60 gel-filtration column (GE Healthcare). To reduce carbohydrate heterogeneity that might impede crystallization, 2.5 μM kifunensine was added immediately before transfection of cells to inhibit processing of high-mannose N-linked glycans to their complex forms (Edberg and Kimberly, 1997).
Conventional SPR experiments
Biacore assays were performed on a Biacore T200 instrument. Proteins were immobilized on a CM5 sensor chip using primary amine coupling chemistry (amine-coupling wizard routine in the Biacore T200 software). Briefly, 1 μM FcγRIIIaF158, FcγRIIaH131, or FcγRIIb in 10 mM sodium acetate buffer at pH 5.0 or pH 4.5 were coupled at a flow rate of 1 μL/minute for 420 seconds followed by ethanolamine coupling to block excess reactive carboxyl groups on the sensor chip surface. The coupling density was equivalent to 3162 RU for FcγRIIIaF158, 1840 RU for FcγRIIaH131, and 1516 RU for FcγRIIb. Flow channel 1 served as the blank reference channel for bulk refractive index subtraction. Serial dilutions starting at a concentration of 10,000 nM of wtFc, SDALIE Fc, or GASDALIE Fc in HBSEP+ buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% (v/v) surfactant P20, pH 7.4) were injected at a flow rate of 30 μL/minute at 25 °C followed by a 200 second dissociation phase during which only HBSEP+ buffer was injected. 10 mM glycine pH 3 at 30 μL/minute for 30 seconds was used for regeneration.
Sensorgrams were globally fitted to a 1:1 binding model using nonlinear regression in the Biaevaluation software. The fits were evaluated by plotting residuals between modeled and experimental curves. For comparisons with previously reported affinity measurements (Bournazos et al., 2014a; Bournazos et al., 2014b; Bruhns et al., 2009; DiLillo and Ravetch, 2015a; Janeway et al., 2005; Lazar et al., 2006; Smith et al., 2012), KD values derived from these experiments are reported in Table 1 as conventional measurements and are calculated from the ratio of the kinetic constants as KD = kd/ka.
Table 1.
Comparison of Fc:FcγR affinities
| IgG Variant | Detection Method | FcγRIIIa KD (nM) | Variant | Fold improved affinity compared with wtFc | FcγRIIa KD (nM) | Variant | Fold improved affinity compared with wtFc | FcγRIIb KD (nM) | Fold improved affinity compared with wtFc | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| wtFc (IgG1) | Conventional SPR (FcR surface) | 2950 | F158 | 1380 | H131 | 2520 | Smith et al., 2012 | |||
| Conventional SPR (FcR surface) | 432 | V158 | 1550 | R131 | Smith et al., 2012 | |||||
| Conventional SPR (FcR surface) | 2220 | F158 | 1330 | R131 | 2700 | Bournazos et al., 2014a | ||||
| Conventional SPR (FcR surface) | 2200 | F158 | 1400 | H131 | 2400 | Bournazos et al., 2014b | ||||
| Conventional SPR (FcR surface) | 410 | V158 | 1500 | R131 | Bournazos et al., 2014b | |||||
| Conventional SPR (FcR surface) | 2950 | F158 | 1550 | R131 | 2520 | DiLillo et al., 2015 | ||||
| ? | 2000 | ? | 500 | ? | 500 | Janeway et al., 2005 | ||||
| Conventional SPR (FcR surface) | 854 | F158 | 192 | H131 | 8333 | Bruhns et al., 2009 | ||||
| Conventional SPR (FcR surface) | 500 | V158 | 286 | R131 | Bruhns et al., 2009 | |||||
| Competition SPR (FcR surface) | 252 | V158 | Lazar et al., 2006 | |||||||
| Conventional SPR (FcR surface) | 4454 | F158 | 4486 | H131 | 9275 | current publication | ||||
| SDALIE Fc | Conventional SPR (FcR surface) | 201 | F158 | 22 | 1487 | H131 | 3 | 2193 | 4 | current publication |
| Competition SPR (FcR surface) | – | F158 | 318 | – | H131 | 2 | – | 5 | current publication | |
| GASDALIE Fc | Conventional SPR (FcR surface) | 100 | F158 | 30 | 82 | H131 | 17 | 736 | 3 | Smith et al., 2012 |
| 124 | V158 | 3 | 63 | R131 | 25 | Smith et al., 2012 | ||||
| Conventional SPR (FcR surface) | 144 | F158 | 15 | 104 | R131 | 13 | 1670 | 2 | Bournazos et al., 2014a | |
| Conventional SPR (FcR surface) | 110 | F158 | 20 | 82 | H131 | 17 | 1500 | 2 | Bournazos et al., 2014b | |
| Conventional SPR (FcR surface) | 100 | V158 | 4 | 120 | R131 | 13 | Bournazos et al., 2014b | |||
| Conventional SPR (FcR surface) | 100 | F158 | 30 | 63 | R131 | 25 | 736 | 3 | DiLillo et al., 2015 | |
| Conventional SPR (FcR surface) | 213 | F158 | 21 | 1585 | H131 | 3 | 3882 | 2 | current publication | |
| Competition SPR (FcR surface) | – | F158 | 431 | – | H131 | 17 | – | 6 | current publication |
Competition SPR experiments
Competition SPR measurements were performed using methods similar to previously-reported competition Biacore studies (Nieba et al., 1996). Flow cell 1 (ethanolamine blank) and flow cell 2 (FcγRIIIaF158) from the CM5 sensor chip used in the conventional SPR experiments were used for competition Biacore experiments. All measurements were carried out at 25 °C and a flow rate of 10 μL/minute and an injection time period of 120 seconds using HBSEP+ buffer. The chip was regenerated by injecting 10 mM glycine pH 3 at 30 μL/minute for 30 seconds. Each experimental cycle evaluating the binding of an Fc protein to a receptor consisted of (i) Establishing a calibration curve to correlate the SPR response for each Fc protein binding to immobilized FcγRIIIaF158 as a function of concentration. For the calibration measurements, 2-fold dilution series of the Fc proteins including a no-protein standard (HBSEP+ buffer only) were used starting at the following concentrations: 803 nM (wtFc), 8 nM (SDALIE Fc), 5 nM (GASDALIE Fc). (ii) A measurement series evaluating the binding of an Fc protein to immobilized FcγRIIIaF158 in the presence of varying concentrations of competing FcγR in solution. For these measurements, a given Fc protein at a fixed concentration was first incubated for three hours with a 2-fold dilution series of a competitor receptor in solution. For wtFc injections, the concentration of wtFc was 401.5 nM and the competitor dilution series starting concentrations were 1396 nM (FcγRIIIaF158), 1692 nM (FcγRIIaH131), and 1756 nM (FcγRIIb). For SDALIE Fc injections, the concentration of SDALIE Fc was 4 nM and the competitor dilution series starting concentrations were 87 nM (FcγRIIIaF158), 1692 nM (FcγRIIaH131), and 3512 nM (FcγRIIb). For GASDALIE Fc injections, the concentration of GASDALIE Fc was 2.5 nM and the competitor dilution series starting concentrations were 44 nM (FcγRIIIaF158), 846 nM (FcγRIIaH131), and 3512 nM (FcγRIIb).
Data were analyzed using the Biacore T200 Evaluation software version 3.0. For the calibration curve, a 4-parameter fitting function was used. For calculation of the KD, the software uses a non-linear regression fit to the following function:
where variable Atot refers to the total concentration of the receptor in the Fc:FcγR mixture, Bfree refers to the concentration of the unbound Fc, and Btot refers to the total concentration of Fc in the mixture.
All curves were generated using the bulk refractive index subtracted response after an optimal injection time of 35 seconds (wtFc) or 144 seconds (SDALIE and GASDALIE Fc).
Crystallization
Crystals of GASDALIE Fc (space group P212121; a = 49.32 Å, b = 79.13 Å, c = 137.62 Å; one Fc dimer per asymmetric unit) were grown in sitting drop vapor diffusion by mixing equal volumes of GASDALIE Fc (7.36 mg/ml) with a solution containing 0.2 M ammonium formate and 20% (w/v) PEG 3350 at 20°C. Crystals were cryopreserved in well solution supplemented with 30% glycerol. Crystals of the GASDALIE Fc:FcγRIIIaF158 complex (space group P212121; a = 74.38 Å, b = 94.50 Å, c = 108.67 Å; one complex per asymmetric unit) were grown in sitting drop vapor diffusion by mixing equal volumes of protein (10 mg/ml) with a solution containing 0.04 M potassium dihydrogen phosphate and 16% (w/v) PEG 8000 at 20°C. The complex crystals were cryopreserved in well solution.
Data Processing and Structure Determination
Data were collected to 2.4 Å resolution (GASDALIE Fc alone) and 3.1 Å resolution (Fc:FcγRIIIaF158 complex) at beamline 8.2.1 of the Advanced Light Source (ALS) at Lawrence Berkeley National Laboratory. Diffraction data were processed, indexed, integrated and scaled using iMosflm (Leslie and Powell, 2007) POINTLESS and SCALA, respectively (Evans, 2006; Evans, 2011). To determine the high-resolution cutoff for datasets, we used I/σI ratios and completeness of the highest resolution shell in addition to the criterion that the CC1/2 statistic (correlation coefficient between two random halves of a data set) should be greater than 10% (Karplus and Diederichs, 2012). We used Phenix (Adams et al., 2010) to compute CC1/2 values. Structures were solved by molecular replacement using PHASER (McCoy et al., 2007) and published Fc and Fc:FcγRIIIa structures as search models (PDB codes 3DO3 and 3SGK). Modeling was done using COOT (Emsley et al., 2010). Crystallographic refinement was done using the Phenix crystallography package (Adams et al., 2010) by refining individual B factors for GASDALIE Fc and group B factors for the lower-resolution GASDALIE Fc:FcγRIIIaF158 structure. We used PyMol (Schrödinger, 2011) for superposition calculations and molecular representations. Protein interfaces, surfaces and assemblies’ service PISA at the European Bioinformatics Institute (Krissinel and Henrick, 2007) was used to determine hydrogen bonding and electrostatic interactions at the GASDALIE Fc:FcγRIIIaF158 interface.
The GASDALIE Fc model (Rfree = 25.6%; Rwork = 23.5%) included 416 protein residues in the GASDALIE Fc dimer (Leu235 – Ser444 on chain A and Gly237 – Ser444 on chain B), 18 glycan residues (GlcNAc1-Gal6, Fuc12 on chain A and GlcNAc1-GlcNAc8, Fuc12 on Chain B), and 105 water molecules.
The GASDALIE Fc: FcγRIIIaF158 complex (Rfree = 29.6%; Rwork = 27.1%) included 417 protein residues in the GASDALIE Fc dimer (Ala236-Ser444 on chain A and Ala236-Leu443 on chain B), 17 Fc glycan residues (GlcNAc1-Gal6, Fuc12 on chain A and GlcNAc1-Gal6, Fuc12 on Chain B), 169 protein residues in the FcγRIIIaF158 ectodomain, and FcγRIIIaF158 glycans GlcNAc1 attached to Asn162 and GlcNAc1 attached to Asn45. Side chains were disordered for residues Lys246, Lys248, Glu258, Glu269, Asp270, Glu272, Glu294, Tyr300, Lys326, and Arg355 on chain A, Lys246, Gln419 and Ser442 on chain B in the GASDALIE Fc dimer. No electron density was observed for side chains Phe11, Leu48, Ile49, Glu68, Gln72, Lys101, Glu103, Thr116, Lys128, Lys 143, Arg155, Gln174 and residues Tyr33-Ser39 and Ser75-Leu78 in the FcγRIIIaF158 ectodomain.
Homology Modeling
We used Pymol (Schrödinger, 2011) to superimpose the D2 domains of FcγRIIa (3RY4) or FcγRIIb (2FCB) to the D2 domain of FcγRIIIa in the GASDALIE Fc:FcγRIIIaF158 complex and the wtFc:FcγRIIIaV158 complex (3SGJ) in order to generate homology models of GASDALIE Fc:FcγRIIa, GASDALIE Fc:FcγRIIb, wtFc:FcγRIIa, and wtFc:FcγRIIb. The D2 domain of FcγRIIIa in the wtFc:FcγRIIIaV158 structure was onto V158 superimposed the D2 domain in the GASDALIE Fc:FcγRIIIaF158 complex to generate a GASDALIE Fc:FcγRIIIaV158 homology model. PISA (Protein interfaces, surfaces and assemblies) service at the European Bioinformatics Institute (Krissinel and Henrick, 2007) was used to evaluate hydrogen bonding and electrostatic interactions at the GASDALIE Fc:FcγRIIIaV158, GASDALIE Fc:FcγRIIa and wtFc:FcγRIIa interfaces.
Results
Comparison of Affinities of Fc Variants for Activating and Inhibitory FcγRs
Activating FcγRs exist in variant forms in the human population depending on the amino acid identity at positions 158 (Phe158 or Val158 for FcγRIIIa) or 131 (His131 or Arg131 for FcγRIIa) (Ackerman and Nimmerjahn, 2014). The polymorphism at residue 158 of FcγRIIIa was reported to affect binding to Fcs: e.g., FcγRIIIaF158 exhibited an ~5-fold lower affinity for wtFc compared to FcγRIIIaV158 (Bournazos et al., 2014b); however, there was no interaction between residue 158 of FcγRIIIa and wtFc in the wtFc:FcγRIIIaV158 complex structure (PDB 3SGJ). The histidine to arginine polymorphism at residue 131 of FcγRIIa did not affect binding to wtFc of human IgG1 (Bournazos et al., 2014b), although Arg131 of FcγRIIaR131 interacts with Fc in a wtFc:FcγRIIaR131 complex structure (PDB 3RY6). For our studies, we expressed the Phe158 and His131 forms of FcγRIIIa and FcγRIIa (FcγRIIIaF158 and FcγRIIaH131) for comparisons with previous in vitro and in vivo studies (Bournazos et al., 2014a; Bournazos et al., 2014b; DiLillo and Ravetch, 2015a; Oganesyan et al., 2008a; Richards et al., 2008; Sazinsky et al., 2008; Smith et al., 2012).
FcγR ectodomains, wtFc, and Fc variants were purified from supernatants of transiently-transfected mammalian cells as previously described (Diskin et al., 2010; Sprague et al., 2004). To attempt to resolve inconsistencies in the reported affinities of Fc mutants to FcγRs (Bournazos et al., 2014a; Bournazos et al., 2014b; Richards et al., 2008; Sazinsky et al., 2008; Smith et al., 2012) (Table 1), we first directly compared the affinities of wtFc, SDALIE Fc and GASDALIE Fc for binding to immobilized FcγRIIaH131, FcγRIIb, or FcγRIIIaF158 using a surface plasmon resonance (SPR)-based binding assay in which equilibrium dissociation constants (KD values) were calculated by determining the ratio of kinetic constants based on global fits of the association and dissociation phases of sensorgrams. However, we found that the binding data could not be accurately modeled by a 1:1 binding interaction (Fig. 1a). When we calculated KD values using the poorly-fit models, we obtained results similar to some of the previous reports that obtained affinities using a 1:1 binding interaction to model Fc:FcγR sensorgrams involving Fcs injected over immobilized FcγRs (Bournazos et al., 2014a; Bournazos et al., 2014b; Bruhns et al., 2009; DiLillo and Ravetch, 2015a; Janeway et al., 2005; Smith et al., 2012) (Table 1). However, higher affinities were reported for methods that did not involve measurements of affinities for immobilized FcγRs: for example, competition Biacore measurements (Lazar et al., 2006) (Table 1) or conventional SPR experiments in which the affinity of an injected FcγR for immobilized Fc was derived (Oganesyan et al., 2008a; Richards et al., 2008). These results suggested the possibility of artifacts in experiments that derive affinities for direct interactions with immobilized FcγRs.
Figure 1.
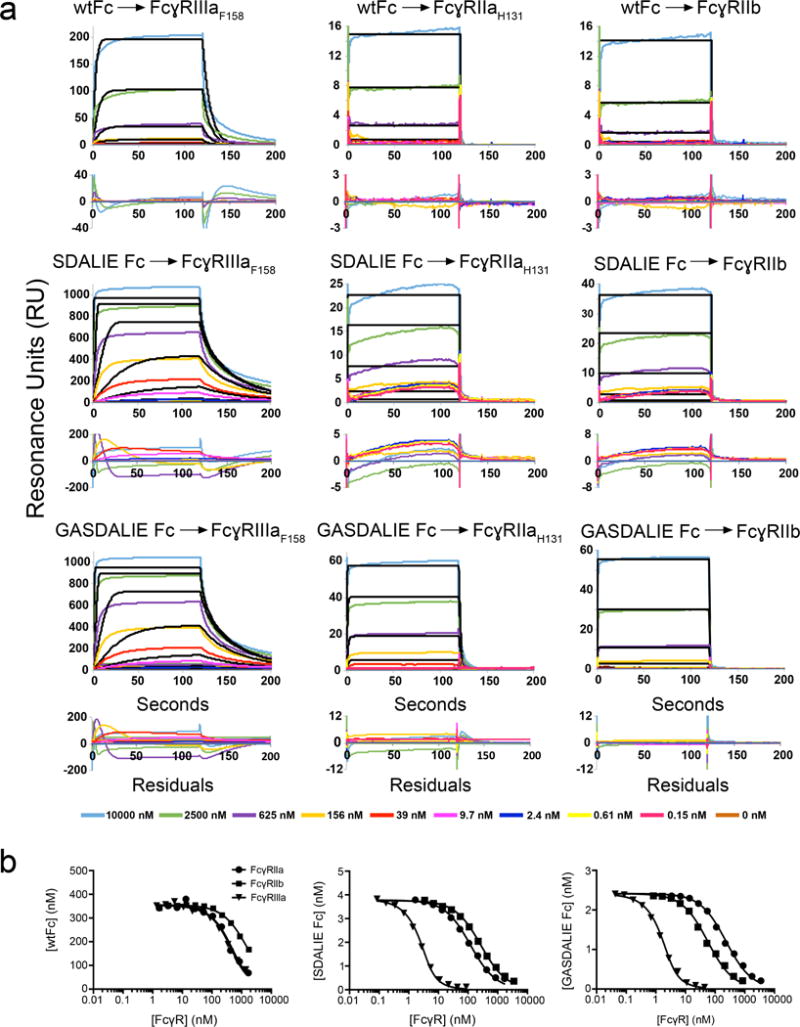
SPR binding assays of Fc:FcγR interactions. (a) Sensorgrams from conventional SPR experiments in which Fc proteins were injected over immobilized FcγRs. Experimental data (colored lines) were fit to a 1:1 binding model (black lines). Residual plots are shown below each set of sensorgrams. For most interactions, the association and/or dissociation phases of the sensorgrams do not fit a 1:1 binding model. In cases in which the association and dissociation rates are very fast, the 1:1 binding model appears to fit the sensorgrams, but the kinetic rate constants are outside of the detectable range of the instrument and the equilibrium RU values do not converge as the concentration of analyte is raised. (b) Competition SPR results. Equilibrium binding curves for interactions of the Fc variants (wtFc, SDALIE Fc, and GASDALIE Fc) with Fcγ receptors (FcγRIIIa, FcγRIIa, and FcγRIIb). Each curve represents an equilibrium binding experiment in which the free Fc concentration (y-axis) is plotted versus the competitor (FcγR) concentration (logarithmic x-axis). Data points (triangles, circles, or squares) were fit by non-linear regression to the second order root function to an equilibrium binding model (solid black line) as described in the Methods. The more a curve is shifted to the left, the stronger the Fc:FcγR binding (i.e., the higher the affinity).
To obtain consistent and more accurate comparisons of affinities, we used a measurement method similar to competition Biacore (Nieba et al., 1996) in which affinities are derived for Fc:FcγR interactions in solution. In this method, an FcγR (FcγRIIIaF158 in our experiments) is immobilized on a sensor chip, and a constant concentration of Fc incubated with varying amounts of a soluble FcγR (either FcγRIIIaF158, FcγRIIaH131, or FcγRIIb) is injected over the sensor chip (Fig. 1b). Fc proteins that are bound to the FcγR in solution are unable to bind the immobilized receptor. By measuring the amount of Fc that binds to immobilized receptor, we can calculate the concentration of Fc bound to the soluble FcγR. As the concentration of soluble receptor is decreased, fewer FcγR interactions with Fc occur in solution, and more of the unbound Fc is available to bind to the immobilized receptor. This allows for correlation of the SPR signal generated from unbound Fc in solution with a signal derived from an independent calibration experiment recording SPR signals from a series of known Fc concentrations. The concentration values derived this way are used together with known concentrations of total Fc and soluble FcγR to calculate the KD for a given Fc:FcγR interaction. Because the KD values obtained using this method were higher than those derived in our own (Table 1) and in others’ conventional SPR experiments (Bournazos et al., 2014a; Bournazos et al., 2014b; Bruhns et al., 2009; DiLillo and Ravetch, 2015a; Smith et al., 2012), we present the results as the fold improvement in the affinity of the SDALIE or GASDALIE Fc for a given FcγR in Table 1 (i.e., KD of Fc mutant/KD of wtFc). Given the poor fits of models to the conventional SPR measurements (Fig. 1a) compared with the excellent fits for the competition-based experiments (Fig. 1b), we based our comparisons of wtFc:FcγR and mutant Fc:FcγR affinities on the competition-based experiments.
We found that both GASDALIE and SDALIE Fc bound to FcγRIIIaF158 with approximately the same increased affinity compared with wtFc (431-fold and 318-fold increases for GASDALIE and SDALIE, respectively) (Table 1). GASDALIE Fc bound to FcγRIIaH131 7-fold more tightly than did SDALIE Fc, with GASDALIE Fc showing 17-fold increased affinity for FcγRIIaH131 compared to wtFc, but SDALIE Fc and wtFc showing approximately the same affinity for FcγRIIaH131 (within ~2-fold). Both GASDALIE Fc and SDALIE Fc had a similar increase in binding to the inhibitory receptor FcγRIIb when compared to wtFc (5–6-fold). Although GASDALIE Fc and SDALIE Fc showed similar binding to FcγRIIIaF158 and FcγRIIb, the additional G236A mutation (GA in GASDALIE) further enhanced the affinity of GASDALIE Fc for FcγRIIaH131 when compared to SDALIE Fc (Table 1).
Crystal Structure of GASDALIE Fc Bound to FcγRIIIa
To facilitate crystallization of the GASDALIE Fc:FcγRIIIaF158 complex, three of five potential N-linked glycosylation sites on FcγRIIIaF158 were removed by site directed mutagenesis (N38Q, N74Q, N169Q). N-linked glycans at asparagines 45 and 162 were retained due to their effects on protein expression and Fc binding affinity, respectively (Ferrara et al., 2011; Ferrara et al., 2006). The complex structure was solved by molecular replacement using an afucosylated Fc:FcγRIIIa complex structure (PDB 3SGK) as a search model and refined to 3.1 Å (Rcryst = 0.27; Rfree = 0.29) (Supplementary Table 1). N-linked glycans were built into electron density up to a terminal galactose on the 6-arm and up to a terminal GlcNAc on the 3-arm of Fc chains A and B (Supplementary Fig. 1a). On FcγRIIIaF158, ordered electron density was observed for a GlcNAc attached to Asn45 and for a GlcNAc attached to N162 (Supplementary Fig. 1b). The sites of the GASDALIE substitutions (positions 236, 239, 330, and 332) were visible in ordered electron density on Fc chains A and B (Supplementary Fig. 1c).
Similar to previous structures of Fc:FcγR complexes (Ferrara et al., 2011; Matsumiya et al., 2007; Mizushima et al., 2011; Radaev et al., 2001), the GASDALIE Fc:FcγRIIIaF158 structure revealed an asymmetric 1:1 interaction in which the region connecting FcγRIIIaF158 domains 1 and 2 (D1 and D2) together with D2 loops BC, C’E and FG interacted with the lower hinge region of Fc chains A and B and the CH2 domain BC, C’E and FG loops (Fig. 2a). Because the asymmetric binding of FcγRs to two-fold symmetric Fc regions results in different effects of the Fc substitutions on the two chains of the Fc homodimer, we will describe comparisons of the GASDALIE Fc:FcγRIIIaF158 and wtFc:FcγRIIIaV158 (PDB 3SGJ) structures separately for both Fc chains. Starting with the interactions of Fc chain A with FcγRIIIaF158, we found electrostatic interactions resulting from one of the GASDALIE substitutions, S239D, in which a positively-charged residue, Lys120, in the BC loop of FcγRIIIaF158 D2 was positioned in a pocket between Fc chain A residues Asp239 (SD in GASDALIE) and Asp265 (Fig. 2b). By comparison, in the wtFc:FcγRIIIaV158 complex, Fc residue Ser239 formed a hydrogen bond with FcγRIIIa Lys120. The introduced residues Leu330 and Glu332, the AL and IE substitutions of GASDALIE, did not play a role in the interface of FcγRIIIaF158 with chain A of GASDALIE Fc. Ala236 (the GA substitution in GASDALIE Fc) also was not at the binding interface; however, Gly236 in the wtFc:FcγRIIIaV158 complex formed a hydrogen bond with His135 of FcγRIIIaF158 (Fig. 2b). The salt bridge formed by Asp239 on Fc chain A (SD substitution in GASDALIE Fc and SDALIE Fc) and Lys120 of FcγRIIIa, which is not seen in wtFc:FcγRIIIaV158 complex structures (Ferrara et al., 2011; Mimoto et al., 2013; Mizushima et al., 2011; Radaev et al., 2001; Sondermann et al., 2000), may contribute to the increased affinity of GASDALIE Fc for FcγRIIIa, whereas the additional mutations G236A, A330L, and I332E do not participate in the interaction between GASDALIE Fc chain A and FcγRIIIaF158.
Figure 2.
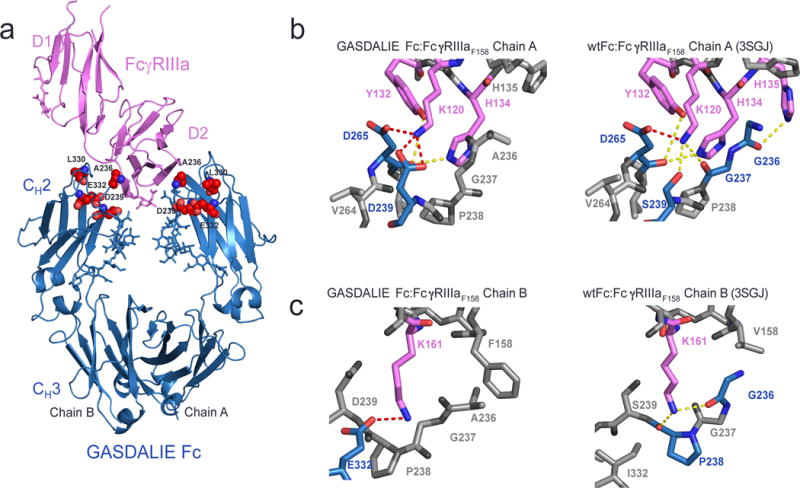
Structure of GASDALIE Fc:FcγRIIIaF158 complex. (a) Overview of the GASDALIE Fc:FcγRIIIaF158 complex. FcγRIIIaF158 is in pink ribbon representation and GASDALIE Fc is in blue ribbon representation with the G236/S239D/A330L/I332E mutations highlighted as red spheres and N-linked glycans shown as blue sticks. (b–c) Comparison of interfaces in the GASDALIE Fc:FcγRIIIaF158 and wtFc:FcγRIIIa structures on Fc chain A (panel b) and Fc chain B (panel c). Residues involved in interactions are shown in magenta (FcγRIIIa) and blue (Fc) with oxygen atoms in red and nitrogens in blue. Dotted lines between atoms represent lectrostatic interactions (red) or hydrogen bonds (yellow). Residues that do not participate in hydrogen bonds or electrostatic interactions are shown in gray.
On chain B of GASDALIE Fc, we found new interactions with the introduction of the GASDALIE substitutions that may also contribute to the enhanced affinity for FcγRIIIaF158 of GASDALIE compared to wtFc (Bournazos et al., 2014b; Smith et al., 2012) (Table 1). Lys161 in the FG loop of FcγRIIIaF158 formed an electrostatic interaction with Fc chain B residue Glu332, the IE substitution in GASDALIE Fc, an interaction not seen in wtFc:FcγRIIIaV158 complex structures since Ile332 of wtFc did not interact with FcγRIIIaF158 Lys161 (Fig. 2c). Ala236, the GA substitution in GASDALIE Fc, makes a hydrophobic contact with FcγRIIIa residue Phe158; however, Fc residue Gly236 in the wtFc:FcγRIIIaV158 complex formed a hydrogen bond with Lys161 of the receptor and did not contact receptor residue 158. Asp239 and Leu330 (the SD and AL substitutions in GASDALIE) on chain B were not at the binding interface with the receptor. The salt bridge formed by Glu332 on Fc chain B (IE substitution in GASDALIE Fc and SDALIE Fc) and Lys161 of FcγRIIIaF158, and the hydrophobic interaction between Fc residue Ala236 and FcγRIIIa residue Phe158, which is not seen in wtFc:FcγRIIIaV158 complex structures (Fig. 2c), may have contributed to the increased affinity of GASDALIE Fc to FcγRIIIa, whereas the additional mutations on chain B (S239D and A330L) did not participate in the interface.
Modeling of the interactions of GASDALIE Fc with FcγRIIa and FcγRIIb
To gain understanding of the enhanced affinity of GASDALIE Fc for FcγRIIa compared to SDALIE Fc and to wtFc (GASDALIE Fc binds FcγRIIa with 17-fold increased affinity compared with wtFc, whereas SDALIE Fc binds FcγRIIa with only 2-fold increased affinity compared with wtFc; Table 1 competition SPR results), we used homology modeling (Schrödinger, 2011) to create models of GASDALIE Fc:FcγRIIa, and wtFc:FcγRIIa complexes based on high-resolution structures of FcγRIIa (PDB 3RY4), the wtFc:FcγRIIIaV158 complex (PDB 3SGJ) and the GASDALIE Fc:FcγRIIIaF158 complex described in this study. We also created homology models of GASDALIE Fc:FcγRIIb and wtFc:FcγRIIb complexes using the FcγRIIb structure (PDB 2FCB) to look for a structural rationalization for the relatively small effect of the GASDALIE substitutions on Fc binding to the inhibitory FcγRIIb receptor (GASDALIE Fc and SDALIE Fc bind FcγRIIb with only 5–6–fold increased affinities compared with wtFc; Table 1 competition SPR results). The homology modeling was feasible because the Fc-binding D2 domains of FcγRIIa, FcγRIIb, and FcγRIIIa share sequence and structural homology (root mean square deviations, rmsds, ranging from 0.5 – 0.7 Å for pairwise combinations) (Fig. 3a,b) and bind to Fcs with similar orientations, as demonstrated by crystal structures (Ferrara et al., 2011; Maxwell et al., 1999; Mimoto et al., 2013; Mizushima et al., 2011; Radaev et al., 2001; Ramsland et al., 2011; Sondermann et al., 1999; Sondermann et al., 2000; Sondermann et al., 2001) and site-directed mutagenesis studies (Maxwell et al., 1999; Mimoto et al., 2013; Ramsland et al., 2011; Shields et al., 2001; Sondermann et al., 1999; Sondermann et al., 2001).
Figure 3.
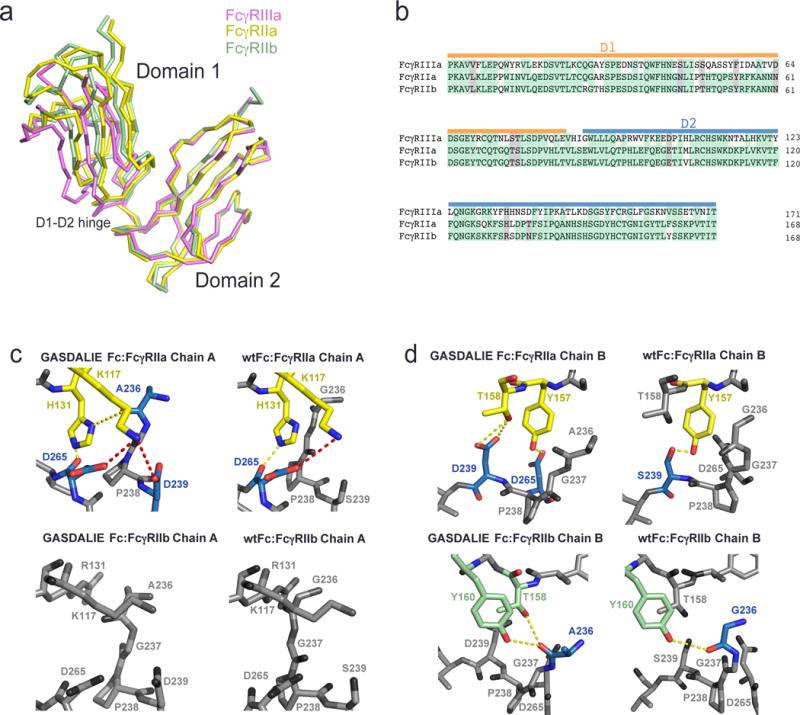
Comparison of the interfaces of FcγRIIa and FcγRIIb with GASDALIE Fc and wtFc using homology modeling. (a) Alignment of the D2 domains of FcγRIIa (PDB 3RY4, yellow), FcγRIIb (PDB 2FCB, green), and FcγRIIIa (pink; from the GASDALIE Fc:FcγRIIIa complex structure). (b) Sequence alignment of the D1 and D2 domains of FcγRIIa, FcγRIIb and FcγRIIIa. Identical residues are highlighted in green and similar residues are highlighted in gray. (c–d) Homology modeled interfaces of GASDALIE Fc and wtFc with FcγRIIa and FcγRIIb. Predicted electrostatic interactions (red) and hydrogen bonds (yellow) are shown as dashes. Residues that are not predicted to participate in the Fc:FcgR interfaces are shown in gray. (c) Interactions of chain A of GASDALIE Fc (left) and wtFc (right) with FcγRIIa (top) and FcγRIIb (bottom). (d) Interactions of chain B of GASDALIE Fc (left) and wtFc (right) with FcγRIIa (top) and FcγRIIb (bottom).
To validate the modeling methods, we first constructed a homology model of the GASDALIE Fc:FcγRIIIaF158 structure using the wtFc:FcγRIIIaV158 complex structure (PDB 3SGJ) as a starting reference. The GASDALIE Fc:FcγRIIIaF158 model exhibited a 1.2 Å rmsd compared with the GASDALIE Fc:FcγRIIIaF158 crystal structure (calculated for all Cα atoms) and reproduced the interactions of the GASDALIE substitutions with FcγRIIIaF158. The relatively large rmsd for the superposition is explained by the flexibility and partial disorder of the D1 domain of FcγRIIIa, which does not contact Fc in Fc:FcγRIIIa complex structures (Ferrara et al., 2011; Mizushima et al., 2011; Radaev et al., 2001; Sondermann et al., 2000) (Fig. 2a). By comparison, the D2 domain in the modeled GASDALIE Fc:FcγRIIIaF158 complex exhibited a lower rmsd (0.4 Å; calculated for 72 Cα atoms) compared with the GASDALIE Fc:FcγRIIIaF158 crystal structure. Having validated the homology modeling procedure, we generated homology models for GASDALIE Fc:FcγRIIa, GASDALIE Fc:FcγRIIb, wtFc:FcγRIIa, and wtFc:FcγRIIb complex structures by aligning the D2 domain of FcγRIIa or FcγRIIb with D2 of FcγRIIIa in the appropriate Fc:FcγRIIIa complex structure.
Several interactions predicted by the GASDALIE Fc:FcγRIIa and wtFc:FcγRIIa homology models gave insight into the 17-fold increased affinity of GASDALIE Fc versus wtFc for FcγRIIa. On Fc chain A, Asp239 (SD in GASDALIE) and Asp265 were predicted to form electrostatic interactions with FcγRIIa Lys117 (the counterpart of FcγRIIIa Lys120) (Fig. 3c, top left). Fc residue Asp265 may also hydrogen bond with FcγRIIa residue His131. By comparison, in the wtFc:FcγRIIa structure, Fc residue 239, a serine, did not contact FcγRIIa, although Fc residue Asp265 was predicted to form a salt bridge with FcγRIIa Lys117 and a hydrogen bond with FcγRIIa His131 (Fig. 3c, top right). Fc substitutions Leu330 and Glu332 (AL and IE in GASDALIE Fc) were not predicted to play a role in binding at the interface of FcγRIIa with GASDALIE Fc chain A. The effects of the GA substitution in GASDALIE Fc may be explained by a predicted hydrogen bond between GASDALIE Fc residue Ala236 (the site of the GA substitution) and FcγRIIa residue His131, whereas Fc residue Gly236 in wtFc is not predicted to interact with the FcγRIIa (Fig. 3c). Consistent with the similar binding affinities of GASDALIE Fc and wtFc for FcγRIIb (Table 1), the GASDALIE substitutions Ala236, Asp239, Leu330, and Glu332 in GASDALIE Fc chain A were not predicted to play a role at the binding interface with FcγRIIb in the GASDALIE Fc:FcγRIIb homology model (Fig. 3c, bottom left). Although FcγRIIa residue Lys117 was predicted to participate in electrostatic interactions and hydrogen bonding at the GASDALIE Fc:FcγRIIa interface, Lys117 of FcγRIIb was not predicted to interact with any of the Fc residues on chain A of either GASDALIE Fc or wtFc (Fig. 3c, bottom).
On Fc chain B, residues Asp239 (the SD substitution of GASDALIE) and Asp265 are predicted to hydrogen bond with FcγRIIa residue Thr158 (Fig. 3d, top left). By comparison, if there is a serine instead of an aspartate at Fc position 239, as in wtFc, a hydrogen bond is predicted to form between the serine and FcγRIIa residue Tyr157 (Fig. 3d, top right). The remaining GASDALIE substitutions (GA, AL, and IE substitutions) are not predicted to play a role at the interface in this region. In the GASDALIE Fc:FcγRIIb model, Fc residue Ala236 (GA substitution) is predicted to hydrogen bond with FcγRIIb residues Tyr160 and possible also Thr158 (Fig. 3d, bottom left). If there is a glycine at Fc position 236 instead of an alanine, as in the wtFc, a hydrogen bond is predicted to form with FcγRIIb residue Tyr160 (Figure 3d, bottom right). Asp239, Leu330 and Glu332 (GASDALIE SD, AL, and IE substitutions) are not predicted to play a role at the GASDALIE chain B Fc interface with FcγRIIb. Thus the GASDALIE Fc substitutions in Fc chain B do not exert major effects on interactions with FcγRIIb. The addition of the G236A mutation is predicted to result in an increase of hydrogen bonding at the GASDALIE Fc:FcγRIIa interface but not at the GASDALIE Fc:FcγRIIb interface, which may explain the 7-fold increase in affinity for FcγRIIa of GASDALIE Fc when compared to SDALIE Fc, which lacks the GA substitution in GASDALIE Fc.
Conformations of free and receptor-bound GASDALIE Fc
In order to determine if unbound GASDALIE Fc exhibits conformational changes associated with its increased affinity for FcγRIIIaF158 compared with wtFc, we solved a 2.4 Å resolution crystal structure of GASDALIE Fc alone by molecular replacement (Rcryst = 0.24; Rfree = 0.26). The structure revealed a typical Fc homodimer with N-glycans attached to CH2 domain residue Asn297. Ordered electron density was observed up to a terminal galactose on the 6-arm and up to a terminal N-acetyl glucosamine on the 3-arm of the N-linked glycan, as found in wtFc structures and the SDALIE Fc structure (Oganesyan et al., 2008a). The SDALIE substitutions were located in ordered electron density; however, the region containing Ala236 (the GA substitution in GASDALIE) was disordered on both chains A and B (Supplementary Fig. 1d).
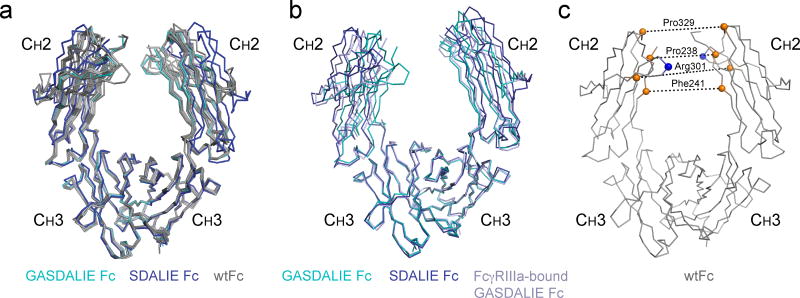
To assess potential differences between GASDALIE Fc and wtFc structures, the CH3 domains of each Fc were aligned and the positions of the CH2 domains were compared (Figure 4a,b). We monitored CH2 domain separations in the various Fc structures by measuring distances between the Cα atoms at CH2 positions 238, 241, 301, and 329 as described (Ahmed et al., 2014; Teplyakov et al., 2013) (Figure 4c). As previously noted, wtFc structures exhibit relatively small structural differences, showing similar CH2 domain separations despite differences in crystal packing (Ahmed et al., 2014). The separation between the CH2 domains of GASDALIE Fc structure fell within range of CH2 domain separations in wtFc (PDB 3DO3, 2DTS, 3AVE, 1HZH and 4Q7D); therefore the GASDALIE mutations did not greatly influence the overall CH2-CH2 domain arrangement in the unbound structure (Fig. 4a; Table 2). We also aligned the CH3 domains of the unbound GASDALIE Fc structure with the CH3 domains of the 2.5 Å SDALIE Fc structure (PDB 2QL1) (Oganesyan et al., 2008a). SDALIE Fc was in a more open conformation than GASDALIE with an rmsd of 5.8 Å calculated for the 201 CH2 domain Cα atoms (Fig. 4a).
Figure 4.
Structure of unbound GASDALIE Fc. (a) Comparison of structures of unbound GASDALIE Fc (cyan), SDALIE Fc (blue; PDB 2QL1), and wtFc (gray; PDB IDs: 3DO3, 2DTS, 3AVE, 4Q7D, 1HZH) after alignment of CH3 domains. (b) Comparison of unbound GASDALIE Fc (cyan), FcγRIIIa-bound GASDALIE Fc (blue-gray), and SDALIE Fc (blue) after alignment of CH3 domains. (c) wtFc structure (PDB: 3AVE) showing location of Cα atoms for Pro238, Phe241, Arg301, and Pro329 (orange spheres) used for CH2 domain separation distance measurements. CH2 domain separations in individual Fc structures in Table 2 were evaluated by measuring distances (dotted lines) between the corresponding orange spheres on each chain. Cα atoms for Asn297 (site of N-glycan attachment) indicated as blue spheres.
Table 2.
Comparison of CH2 domain separation in Fc structures
| PDB | Protein | Resolution (Å) | Space group | Unit cell a, b, c (Å) α, β, γ (°) |
Pro238 (Å) | Phe241 (Å) | Arg301 (Å) | Pro329 (Å) |
|---|---|---|---|---|---|---|---|---|
| 5D4Q | GASDALIE Fc | 2.4 | P212121 | 49, 79, 138 90, 90, 90 |
19 | 22 | 33 | 24 |
| 2QL1 | SDALIE Fc | 2.5 | C2221 | 50, 149, 76 90, 90, 90 |
28 | 29 | 37 | 39 |
| 5D6D | GASDALIE Fc: FcγRIIIa | 3.1 | P212121 | 74, 95, 108 90, 90, 90 |
25 | 25 | 38 | 31 |
| 3SGJ | afucosylated Fc: FcγRIIIa | 2.2 | P212121 | 67, 89, 140 90, 90, 90 |
26 | 26 | 37 | 32 |
| 3SGK | fucosylated Fc: FcγRIIIa | 2.4 | P212121 | 67, 88, 141 90, 90, 90 |
26 | 27 | 37 | 33 |
| 3AY4 | afucosylated Fc: FcγRIIIa | 2.2 | P41212 | 77,77,350 90, 90, 90 |
25 | 26 | 37 | 31 |
| 1E4K | fucosylated Fc: FcγRIIIb | 3.2 | P6522 | 115, 115, 299 90, 90, 120 |
25 | 26 | 37 | 30 |
| 1T89 | fucosylated Fc: FcγRIIIb | 3.5 | P6522 | 115, 115, 301 90, 90, 120 |
25 | 26 | 37 | 31 |
| 1T83 | fucosylated Fc: FcγRIIIb | 3.0 | P212121 | 73, 102,123 90, 90, 90 |
25 | 26 | 37 | 31 |
We next compared the free GASDALIE Fc structure to the structure of GASDALIE solved in complex with FcγRIIIaF158. No notable differences were seen in side chain conformations at the sites of the GASDALIE mutations (Supplementary Fig. 1c.d). The CH2 domains of the bound GASDALIE Fc were in a more open conformation compared with the unbound form (Fig. 4b, Table 2) (rmsd of 2.3 Å calculated for the 202 Cα atoms of the CH2 domains of one Fc after alignment of the CH3 domains in each structure) and the FcγRIIIaF158-bound GASDALIE Fc fell within the range of CH2 domain separations for receptor-bound Fc structures (Fig. 4b; Table 2). This result was expected since previously solved complexes of afucosylated and fucosylated Fc bound to FcγRIIIa did not show major changes in overall Fc conformation (Ferrara et al., 2011; Sondermann et al., 2000).
Discussion
The effector functions of IgG can be modulated with glycan and protein modifications to its Fc region (Ackerman and Nimmerjahn, 2014). Here we examined the effects of introducing amino acid substitutions in the Fc CH2 domain, either G236A/S239D/A330L/I332E (GASDALIE) or S239D/A330L/I332E (SDALIE), which increase binding to activating FcγR receptors but exhibit relatively small affinity increases for the FcγRIIb inhibitory receptor (Bournazos et al., 2014a; Bournazos et al., 2014b) (Table 1). These effects lead to enhanced Fc-mediated effector functions, of importance for the efficacies of therapeutic antibodies against cancer and infectious diseases (Ferrara et al., 2011; Lazar et al., 2006; Matsumiya et al., 2007; Mizushima et al., 2011; Richards et al., 2008).
Previous SPR-based measurements to derive affinities of wtFc and mutant Fcs for FcγRs used a 1:1 (Langmuir) binding model to derive kinetic constants for the interactions of Fc proteins with immobilized receptors (Bournazos et al., 2014b; Lazar et al., 2006; Richards et al., 2008; Smith et al., 2012). Here we showed that sensorgrams of Fc binding to immobilized FcγRs do not fit a 1:1 binding model, and are therefore unlikely to result in reliable binding affinity constants (Fig. 1a). In order to avoid inaccuracies resulting from poorly fit models of Fc binding to immobilized receptors, we chose a measurement method based on equilibrium binding in solution to derive affinities for FcγRs binding to the GASDALIE and SDALIE Fcs (Fig. 1b). The method is based on concentration measurements of unbound ligands after equilibrium binding to their binding partner (Nieba et al., 1996); in this case, Fc binding to an FcγR. Concentrations of free ligand were determined using a calibration measurement series carried out in the same experiment. Using this method, we found that GASDALIE Fc and SDALIE Fc showed over two orders of magnitude of enhanced binding compared with wtFc to the activating Fc receptor FcγRIIIaF158, but only a 5–6-fold increase in binding to the inhibitory receptor FcγRIIb. GASDALIE Fc also exhibited increased binding (17-fold higher affinity) to FcγRIIa, whereas the affinities of SDALIE Fc and wtFc for FcγRIIa were similar (Table 1). Although our structural studies showed that the G236A substitution that distinguishes GASDALIE and SDALIE formed a hydrophobic contact with residue Phe158 of FcγRIIIaF158 in the GASDALIE Fc:FcγRIIIaF158 complex structure, it did not cause a difference in increased binding affinity between SDALIE Fc and GASDALIE Fc to FcγRIIIaF158 when compared to wtFc (Fig. 2; Table 1); however, homology modeling of a GASDALIE Fc:FcγRIIa complex suggested that GASDALIE Fc residue Ala236 could enhance binding of GASDALIE Fc to FcγRIIa through increased H-bond interactions as compared with binding of SDALIE and wtFc (Fig. 3).
We used the GASDALIE Fc:FcγRIIIaF158 complex crystal structure to rationalize the increased affinities for activating FcγRs resulting from the GASDALIE substitutions. The structure revealed electrostatic interactions resulting from the interactions of negative charges introduced by the SD and IE GASDALIE substitutions with lysine residues in the receptor; e.g., the side chain of FcγRIIIa Lys120 extended into a pocket on Fc containing chain A residues Asp265 and Asp239 (SD in GASDALIE). Although not one of the GASDALIE substitutions, Fc residue Asp265 is important for interactions with FcγRs to mediate ADCC: when this residue was substituted for alanine, binding to both activating and inhibitory FcγRs was abrogated (DiLillo et al., 2014). The introduction of Asp239 (SD in GASDALIE and SDALIE) may further stabilize the Asp265Fc/Lys120FcγR interaction contributing to the higher than wtFc affinities of GASDALIE and SDALIE Fcs for FcγRIIIa. FcγRIIa also includes a lysine at position 120 predicted to form similar electrostatic interactions with Fc chain A Asp239 leading to an enhanced binding to activating FcγRs. Another receptor lysine, Lys161FcγRIIIa, formed an electrostatic interaction with GASDALIE Fc residue Glu332 (the IE portion of the GASDALIE substitutions), which cannot occur in wtFc containing an isoleucine at position 332. The greater increase in electrostatic interactions at the interface of GASDALIE Fc with FcγRIIIaF158 (GASDALIE Asp239 interactions with FcγRIIIaF158 Lys120 and GASDALIE Glu332 interactions with FcγRIIIaF158 Lys161) and FcγRIIa (GASDALIE Asp239 interactions with FcγRIIa Lys120) suggests that electrostatic interactions with receptor lysines are the main contributors in enhancement of GASDALIE Fc binding to activating receptors, with FcγRIIIa benefitting more than FcγRIIa from the SD and IE substitutions of GASDALIE Fc.
We also present the structures of GASDALIE Fc in its unbound state. Fc structures exhibit varying degrees of openness, as assessed by the separation of their CH2 domains (Ahmed et al., 2014; Ferrara et al., 2011; Mizushima et al., 2011; Oganesyan et al., 2008a; Radaev et al., 2001; Sondermann et al., 2001), although wtFc structures are relatively similar to each other (Ahmed et al., 2014). The GASDALIE Fc structure was similar to wtFc structures in that the separation of its CH2 domains fell within the range of CH2 domain separations in wtFc structures (Fig. 4A; Table 2). By contrast, the SDALIE Fc structure was in a more open conformation than wtFc structures (Oganesyan et al., 2008a). However, notable changes in the side chain conformations of the SDALIE substitutions were not observed when compared to GASDALIE Fc.
Conclusions
Here we present binding and structural data relevant to understanding the effects of the GASDALIE substitutions in Fc for its interactions with FcγRs. We found that both GASDALIE and SDALIE Fcs exhibited large increases in binding to activating FcγRs, most notably to FcγRIIIa. This property, when combined with their relatively small effects on binding to the inhibitory FcγRIIb receptor, is optimal for eliciting activation of FcγRIIIa- and FcγRIIa-expressing cells leading to ADCC. Further co-crystallization experiments with both FcγRIIa and FcγRIIb and binding experiments with other Fc variants will be essential for further understanding how these mutations allow selection for activating Fc receptors.
Supplementary Material
Acknowledgments
We thank Stylianos Bournazos, John Desjarlais, and Jeffrey Ravetch for helpful discussions, the Caltech Protein Expression Center for protein production, members of the Bjorkman lab for critical reading of the manuscript, Marta Murphy for help making figures, and Jens Kaiser and members of the staff at the Advanced Light Source (ALS) for help with data collection and processing. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This research was supported by the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health Grant HIVRAD P01 AI100148 (P.J.B.); (the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health) and the Molecular Observatory at Caltech supported by the Gordon and Betty Moore Foundation.
Abbreviations
- ADCC
antibody-dependent cell-mediated cytotoxicity
- ADCP
antibody-dependent cellular phagocytosis
- Fc
fragment crystallizable
- FcγR
Fc gamma receptor
- GlcNAc
N-acetylglucosamine
- KD
equilibrium dissociation constant
- IgG
immunoglobulin G
- IC
immune complex
- ITAM
immunoreceptor tyrosine-based activating motif
- ITIM
immunoreceptor tyrosine-based inhibitory motif
- SPR
surface plasmon resonance
- wt
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Atomic coordinates and structure factors have been deposited in the PDB with accession codes 5D4Q (GASDALIE Fc) and 5D6D (GASDALIE Fc:FcγRIIIa)
References
- Ackerman ME, Nimmerjahn F. Antibody Fc linking adaptive and innate immunity. Amsterdam; Burlington: Elsevier Science; 2014. p. 1. online resource (xii, 363 pages) [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AA, Giddens J, Pincetic A, Lomino JV, Ravetch JV, Wang LX, Bjorkman PJ. Structural characterization of anti-inflammatory immunoglobulin G Fc proteins. Journal of molecular biology. 2014;426:3166–3179. doi: 10.1016/j.jmb.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RM, Kobayashi T, Wermeling F, Ravetch JV. Intravenous gammaglobulin suppresses inflammation through a novel T(H)2 pathway. Nature. 2011;475:110–113. doi: 10.1038/nature10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008a;320:373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RM, Ravetch JV. A novel role for the IgG Fc glycan: the anti-inflammatory activity of sialylated IgG Fcs. J Clin Immunol. 2010;30(Suppl 1):S9–14. doi: 10.1007/s10875-010-9405-6. [DOI] [PubMed] [Google Scholar]
- Anthony RM, Wermeling F, Karlsson MC, Ravetch JV. Identification of a receptor required for the anti-inflammatory activity of IVIG. Proc Natl Acad Sci U S A. 2008b;105:19571–19578. doi: 10.1073/pnas.0810163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annual review of immunology. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- Bournazos S, Chow SK, Abboud N, Casadevall A, Ravetch JV. Human IgG Fc domain engineering enhances antitoxin neutralizing antibody activity. The Journal of clinical investigation. 2014a;124:725–729. doi: 10.1172/JCI72676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014b;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daeron M. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- Butler M, Quelhas D, Critchley AJ, Carchon H, Hebestreit HF, Hibbert RG, Vilarinho L, Teles E, Matthijs G, Schollen E, et al. Detailed glycan analysis of serum glycoproteins of patients with congenital disorders of glycosylation indicates the specific defective glycan processing step and provides an insight into pathogenesis. Glycobiology. 2003;13:601–622. doi: 10.1093/glycob/cwg079. [DOI] [PubMed] [Google Scholar]
- DiLillo DJ, Ravetch JV. Differential Fc-Receptor Engagement Drives an Anti-tumor Vaccinal Effect. Cell. 2015a;161:1035–1045. doi: 10.1016/j.cell.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, Ravetch JV. Fc-Receptor Interactions Regulate Both Cytotoxic and Immunomodulatory Therapeutic Antibody Effector Functions. Cancer Immunol Res. 2015b;3:704–713. doi: 10.1158/2326-6066.CIR-15-0120. [DOI] [PubMed] [Google Scholar]
- DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Marcovecchio PM, Bjorkman PJ. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol. 2010;17:608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AR, Woof JM, Partridge LJ, Burton DR, Winter G. Localization of the binding site for the human high-affinity Fc receptor on IgG. Nature. 1988;332:563–564. doi: 10.1038/332563a0. [DOI] [PubMed] [Google Scholar]
- Edberg JC, Kimberly RP. Cell type-specific glycoforms of Fc gamma RIIIa (CD16): differential ligand binding. Journal of immunology. 1997;159:3849–3857. [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta crystallographica Section D, Biological crystallography. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara C, Stuart F, Sondermann P, Brunker P, Umana P. The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms. The Journal of biological chemistry. 2006;281:5032–5036. doi: 10.1074/jbc.M510171200. [DOI] [PubMed] [Google Scholar]
- Hulett MD, Hogarth PM. Molecular basis of Fc receptor function. Advances in immunology. 1994;57:1–127. doi: 10.1016/s0065-2776(08)60671-9. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Travers P, Walport M, Schlomchik MJ. Immunobiology. 6th. New York, NY: Garland Science Publishing; 2005. [Google Scholar]
- Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lazar GA, Dang W, Karki S, Vafa O, Peng JS, Hyun L, Chan C, Chung HS, Eivazi A, Yoder SC, et al. Engineered antibody Fc variants with enhanced effector function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW, Powell HR. Processing DIffraction Data with Mosfilm. Evolving Methods for Macromolecular Crystallography. 2007:41–51. [Google Scholar]
- Lin CW, Tsai MH, Li ST, Tsai TI, Chu KC, Liu YC, Lai MY, Wu CY, Tseng YC, Shivatare SS, et al. A common glycan structure on immunoglobulin G for enhancement of effector functions. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1513456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, Kato K. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J Mol Biol. 2007;368:767–779. doi: 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Maxwell KF, Powell MS, Hulett MD, Barton PA, McKenzie IF, Garrett TP, Hogarth PM. Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa. Nature structural biology. 1999;6:437–442. doi: 10.1038/8241. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimoto F, Katada H, Kadono S, Igawa T, Kuramochi T, Muraoka M, Wada Y, Haraya K, Miyazaki T, Hattori K. Engineered antibody Fc variant with selectively enhanced FcgammaRIIb binding over both FcgammaRIIa(R131) and FcgammaRIIa(H131) Protein engineering, design & selection: PEDS. 2013;26:589–598. doi: 10.1093/protein/gzt022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima T, Yagi H, Takemoto E, Shibata-Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, Kato K. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes to cells: devoted to molecular & cellular mechanisms. 2011;16:1071–1080. doi: 10.1111/j.1365-2443.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieba L, Krebber A, Pluckthun A. Competition BIAcore for measuring true affinities: large differences from values determined from binding kinetics. Anal Biochem. 1996;234:155–165. doi: 10.1006/abio.1996.0067. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Anti-inflammatory actions of intravenous immunoglobulin. Annu Rev Immunol. 2008;26:513–533. doi: 10.1146/annurev.immunol.26.021607.090232. [DOI] [PubMed] [Google Scholar]
- Oganesyan V, Damschroder MM, Leach W, Wu H, Dall’Acqua WF. Structural characterization of a mutated, ADCC-enhanced human Fc fragment. Molecular immunology. 2008a;45:1872–1882. doi: 10.1016/j.molimm.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Oganesyan V, Gao C, Shirinian L, Wu H, Dall’Acqua WF. Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta Crystallogr D Biol Crystallogr. 2008b;64:700–704. doi: 10.1107/S0907444908007877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, Shitara K. Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcgammaRIIIa. J Mol Biol. 2004;336:1239–1249. doi: 10.1016/j.jmb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The structure of a human type III Fcgamma receptor in complex with Fc. The Journal of biological chemistry. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- Ramsland PA, Farrugia W, Bradford TM, Sardjono CT, Esparon S, Trist HM, Powell MS, Tan PS, Cendron AC, Wines BD, et al. Structural basis for Fc gammaRIIa recognition of human IgG and formation of inflammatory signaling complexes. Journal of immunology. 2011;187:3208–3217. doi: 10.4049/jimmunol.1101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch JV, Bolland S. IgG Fc receptors. Annual review of immunology. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- Ravetch JV, Kinet JP. Fc receptors. Annual review of immunology. 1991;9:457–492. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- Richards JO, Karki S, Lazar GA, Chen H, Dang W, Desjarlais JR. Optimization of antibody binding to FcgammaRIIa enhances macrophage phagocytosis of tumor cells. Molecular cancer therapeutics. 2008;7:2517–2527. doi: 10.1158/1535-7163.MCT-08-0201. [DOI] [PubMed] [Google Scholar]
- Samuelsson A, Towers TL, Ravetch JV. Anti-inflammatory activity of IVIG mediated through the inhibitory Fc receptor. Science. 2001;291:484–486. doi: 10.1126/science.291.5503.484. [DOI] [PubMed] [Google Scholar]
- Sazinsky SL, Ott RG, Silver NW, Tidor B, Ravetch JV, Wittrup KD. Aglycosylated immunoglobulin G1 variants productively engage activating Fc receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20167–20172. doi: 10.1073/pnas.0809257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L. The PyMOL Molecular Graphics System (The PyMOL Molecular Graphics System) 2011 [Google Scholar]
- Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. The Journal of biological chemistry. 2001;276:6591–6604. doi: 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- Smith P, DiLillo DJ, Bournazos S, Li F, Ravetch JV. Mouse model recapitulating human Fcgamma receptor structural and functional diversity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6181–6186. doi: 10.1073/pnas.1203954109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann P, Huber R, Jacob U. Crystal structure of the soluble form of the human fcgamma-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 A resolution. The EMBO journal. 1999;18:1095–1103. doi: 10.1093/emboj/18.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- Sondermann P, Kaiser J, Jacob U. Molecular basis for immune complex recognition: a comparison of Fc-receptor structures. Journal of molecular biology. 2001;309:737–749. doi: 10.1006/jmbi.2001.4670. [DOI] [PubMed] [Google Scholar]
- Sondermann P, Pincetic A, Maamary J, Lammens K, Ravetch JV. General mechanism for modulating immunoglobulin effector function. Proc Natl Acad Sci U S A. 2013;110:9868–9872. doi: 10.1073/pnas.1307864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague ER, Martin WL, Bjorkman PJ. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE–gI. J Biol Chem. 2004;279:14184–14193. doi: 10.1074/jbc.M313281200. [DOI] [PubMed] [Google Scholar]
- Teplyakov A, Zhao Y, Malia TJ, Obmolova G, Gilliland GL. IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface. Molecular immunology. 2013;56:131–139. doi: 10.1016/j.molimm.2013.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.