Abstract
Infections caused by bacteria remain a leading cause of death worldwide. While antibiotics remain a key clinical therapy, their effectiveness has been severely compromised by the development of drug resistance in these pathogens. A common and powerful resistance mechanism, multi-drug efflux transporters are capable of extruding a number of structurally unrelated antimicrobials from the bacterial cell, including antibiotics and toxic heavy metal ions, facilitating their survival in noxious environments. Those transporters belonging to the resistance-nodulation-cell division (RND) superfamily typically assemble as tripartite efflux complexes, spanning the inner and outer membranes of the cell envelope. In Escherichia coli, the CusCFBA complex, which mediates resistance to copper(I) and silver(I) ions, is the only known RND transporter with a specificity for heavy metals. Herein, we describe the current knowledge of individual pump components of the Cus system, a paradigm for efflux machinery, and speculate on how RND pumps assemble to fight diverse antimicrobials.
Keywords: multi-drug resistance, heavy metal resistance, resistance-nodulation-cell division, CusCFBA efflux system
Multi-drug resistance in bacteria
Antibiotics and other antimicrobial therapies have been used to treat infections for over a century. It is widely accepted that the increased use of these drugs has resulted in bacteria that are adapted and more resistant to the typical treatments. Until recently, the major means of antimicrobial resistance in bacteria was believed to be similar to the breakdown of penicillin by β-lactamase, i.e., modification of the offending compound by a drug-specific enzyme (57, 101). However, as antibiotic engineering advances, so too do the methods of resistance. Currently, there are three major mechanisms bacteria use to avoid the toxic effects of biocides (84, 101, 106).
The first, enzymatic alteration, can be divided into two classes: 1) those enzymes that degrade the drug, like β-lactamase, and 2) those that chemically modify it. An example of the second class is the periplasmic copper oxidase CueO in Escherichia coli, which relieves the copper sensitivity of the cell by oxidizing toxic Cu(I) to less toxic Cu(II) (117, 118). As metals cannot be chemically degraded, bacteria must employ alternative methods of detoxification in the case of copper or silver poisoning (67, 82). Accordingly, the second method of bacterial drug resistance involves enzymatic alteration of the target of the drug, rather than the drug itself. Fluoroquinolone resistance in bacteria is most commonly attributed to this mechanism; specifically, modification of the enzyme DNA gyrase (43, 101). While the first two methods are limited due to their specificity for a single drug or drug-binding site, the third mechanism is more general. By expelling a toxin before it can act, or by preventing it from entering the cell altogether, a bacterium utilizes its most potent form of resistance.
The first line of cellular defense is either its surrounding peptidoglycan web (Gram-positive) or the outer lipid membrane (Gram-negative). However, the peptidoglycan shell is too coarse to limit the diffusion of very small molecules. Thus, Gram-negative bacteria are inherently less susceptible to toxins (85). Pseudomonas aeruginosa is one example of a pathogen with exceptionally high resistance to a wide range of antibiotics. It also possesses exceptionally narrow porins—outer-membrane proteins that allow passive diffusion of small molecules like sugars and amino acids. Thus, P. aeruginosa possesses an exceptionally-impermeable outer membrane (5, 84, 85). Despite this fact, neither simple nor porin-facilitated diffusion across the cell membrane is enough to explain the intrinsic antimicrobial resistance of certain bacteria. In two related studies, the minimum inhibitory concentration (MIC) of antibiotics tetracycline and cephalothin in E. coli increased by only factors of 1.5 and 2, respectively, when porin expression was interrupted (45, 58, 95). Furthermore, it is common for the most effective antibiotics to exhibit some hydrophobicity, which increases the permeability of the lipid bilayer to these drugs (81).
Not diffusion, but active transport is now recognized as the major player in antimicrobial resistance (101). To remove toxic molecules that enter via porins or porin-like transport systems, the cell utilizes a powerful pumping mechanism. Composed of one or more protein components, these multi-drug efflux pumps traverse the cell membrane, bind and actively pump out a broad range of noxious agents; in some cases, from the cytoplasm all the way to the outside of the cell. Metal-ion and antibiotic resistance systems of this type have been found encoded on plasmids of every eubacterial group tested, from E. coli to Streptomyces sp. (114). For example, E. coli’s pco (for plasmid-borne copper resistance determinant) and P. syringae’s cop operon (14). Many important resistance systems, including the subject of this review, are chromosomally represented, as well.
Heavy-metal resistance in bacteria
In addition to preventing the action of drugs, bacteria utilize efflux pumps to regulate the delicate cellular levels of metal ions, which are among the most common enzymatic cofactors. Around 40% of all enzymes contain transition metals such as Mg, Zn, Fe, Mn, Ca, Co and Cu, and even Cd has been found in the carbonic anhydrase of a certain algae (55, 82). Cytochrome c oxidase, the last step in the mitochondrial and bacterial electron transport chain; prokaryotic superoxide dismutase, which detoxifies dangerous superoxide radicals; lysyl oxidase, a collagen-elastin cross-linker; and tyrosinase, a melanin producer, are all enzymes that require bound copper ion to function (113). However, while transition metals such as copper are important and necessary components of the cellular environment, in excess they are extremely toxic. In recent experiments, E. coli cultures showed diminished to no growth at CuCl2 concentrations as low as 1 mM (9, 81), and in another study growth defects were observed at concentrations exceeding 8 μm (67). Non-essential metals can be toxic at even lower concentrations. Silver, for example, is such an efficient biocide that its effects can be observed on the nM scale (102).
The primary cause of this toxicity is the high reactivity of transition metals; e.g., cations in particular have a high affinity for thiol and thioether residues, imidazoles, sulfides and nucleic acids (25, 82). Furthermore, most transition metals have similar binding affinities for these groups. Copper, however, is exceptional for its ability to push nearly any other metal out of complex (81), and a mismatch of enzyme and metal ion cofactor will typically result in inactivation of the enzyme. Cytoplasmic copper interferes with the formation of iron-sulfur clusters, necessary for the activity of, for example, the citric acid cycle enzyme aconitase or the heme group of cytochrome c (81, 82). Finally, due to their high redox potential, copper and iron are able to participate in the following Fenton-type reaction:
| (1) |
which generates dangerous hydroxyl radicals from hydrogen peroxide and Cu+ or Fe2+ (36, 67, 82). These radicals are capable of reacting with basically anything and can cause damage to DNA, lipids and proteins.
While the mechanisms of heavy-metal toxicity have only been elucidated over the last century, the biocidal properties of copper and silver have been exploited for millennia (31, 99, 112). The use of silver is increasingly common, and both copper and silver are popular and effective sterilizers, viz., brass doorknobs, silverware, copper and silver water filters, silver-coated catheters, silver-containing bandages, mineral sanitizers in pools and silver nitrate and silver sulfadiazine, which are common antiseptics used to treat burns (31, 93, 115, 116). Many transition metal ions are essential cellular ingredients, however the efficacy of these metal-based antimicrobial treatments has forced bacteria to handle copper and silver with the same machinery as they do to antibiotics.
Classes of multi-drug efflux pumps
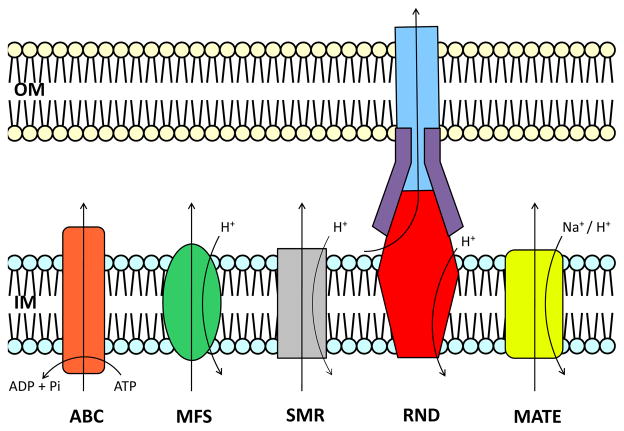
Based on sequence similarity, transport function, substrate specificity and energy coupling, ten families of efflux proteins have been classified as members of five superfamilies (104, 105): 1) the adenosine triphosphate (ATP)-binding cassette (ABC) superfamily; 2) the major facilitator superfamily (MFS); 3) the small multi-drug resistance (SMR) superfamily; 4) the resistance-nodulation-cell division (RND) superfamily and 5) the multi-drug and toxic compound extrusion (MATE) superfamily (Fig. 1) (15, 90, 103, 129). As an energy source, ABC transporters utilize ATP to drive toxins from the cell, while the other four superfamilies rely on an electrochemical gradient. Specifically, MFS, RND and SMR proteins employ the proton-motive force and the MATE superfamily is characterized by either Na+- or H+-substrate antiport (106). Gram-negative bacteria have been found to contain members of all five superfamilies (87), which contribute to their intrinsic resistance to diverse antimicrobials. RND transporters are capable of forming powerful cooperative multi-protein structures that bridge both the inner and outer membranes. They are essential to the multi-drug resistance observed in many pathogens (101).
Fig. 1. Functional diversity among efflux proteins.
Based on mode of transport, energy coupling mechanism and phylogeny (104, 105) these proteins are divided into five superfamilies. The ABC (left) superfamily proteins utilize ATP to transport diverse antimicrobials across the cellular inner-membrane. SMR, MFS and RND proteins function via a H+-substrate antiport mechanism. MATE proteins have been found to utilize both H+ and Na+ as an energy source (106). RND transporters, in particular, are capable of forming multi-protein structures which bridge the inner- and outer-membrane.
Unfortunately, structural information is available for few members of the RND superfamily. Whereas only two have resolved crystal structures, including AcrB (73) and CusA (64), there are a total of seven known RND proteins in E. coli. These seven transporters can be categorized into two distinct sub-families. AcrB (22, 73, 74, 88, 109, 120, 128, 134, 135), AcrD (1, 88, 100), AcrF (56, 66, 88), MdtB (11, 50, 76, 88), MdtC (11, 50, 76, 88) and YhiV (13, 52, 88) are multi-drug efflux pumps of the hydrophobic and amphiphilic efflux RND (HAE-RND) protein family (128) and CusA (64, 88, 124) belongs to the heavy-metal efflux RND (HME-RND) family (80, 128).
CusA, which specifically recognizes and confers resistance to Ag(I) and Cu(I) ions, is one of few known members of the HME-RND family, which includes CusA homolog SilA in Salmonella typhimurium (34) and CzcA of Ralstonia sp. CH34 (29, 77). CusA is unique not only as the only known E. coli HME-RND transporter, but also as the only HME-RND protein, among all organisms, with available crystal structures. Among all RND efflux systems, CusCFBA is one of just three, including MexAB-OprM (2, 3, 39, 110) and AcrAB-TolC (53, 71, 107, 128), with structures for each component. It is also the first efflux complex for which the adaptor and transporter were co-crystallized (123).
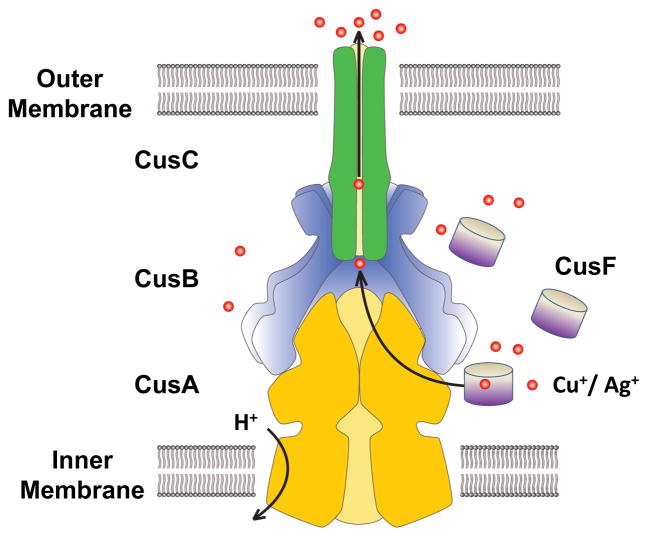
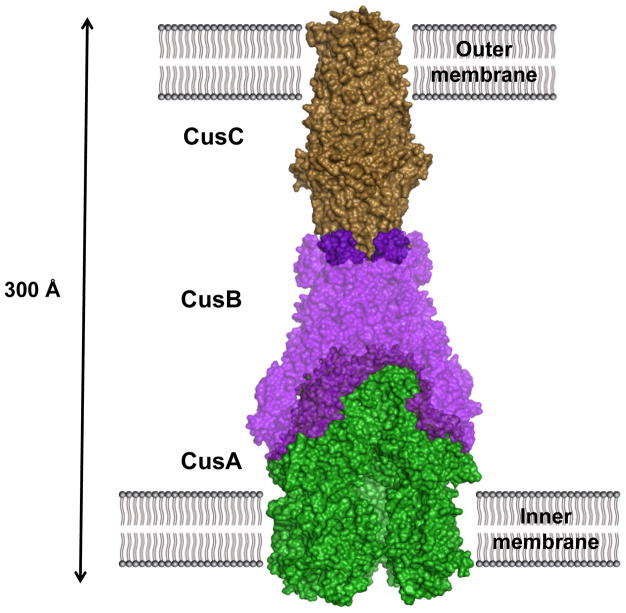
Four proteins are found to operate in conjunction with CusA, making it only one of two known tetrapartite efflux systems. The fully-assembled complex consists of the inner-membrane, substrate-binding transporter, or pump, CusA; the periplasmic membrane fusion protein (MFP), or adaptor, CusB; the outer membrane factor (OMF), or channel, CusC; as well as the small, periplasmic, metal-binding protein, or chaperone, CusF, a component that has no analog in HAE-RND complexes (Fig. 2). While HAE-RND proteins are typically of broad specificity, HME-RND proteins have been shown to be very specific, with the ability to distinguish between monovalent and divalent ions (9, 128).
Fig. 2. Model of the fully-assembled CusCFBA tetrapartite efflux system.
In E. coli, this HME-RND complex is responsible for extruding Cu(I) and Ag(I) ions directly from the cell with use of the proton-motive force. It consists of the inner-membrane transporter CusA (yellow), the membrane-fusion adaptor CusB (blue), the outer-membrane channel CusC (green) as well as the periplasmic metallochaperone CusF (grey). Cu(I) and Ag(I) ions have been found in complex with each of CusA, CusB and CusF.
Despite their differences, metal and antibiotic resistance systems are thought to have much in common (10). In fact, the line that distinguishes one from another is sometimes blurred. In addition to copper and silver, the cus determinant has been implicated in E. coli resistance to drugs dinitrobenzene, dinitrophenol and fosmomycin (20, 88). Thus, both HAE- and HME-RND proteins are believed to be key components of the acquired and intrinsic resistance of Gram-negative pathogens to antimicrobials. The acquisition of even one RND system by a bacterium can increase its tolerance to a broad spectrum of drugs. For example, the Acr system in E. coli has been linked with resistance to tetracycline, chloramphenicol, fluoroquinolones, novobiocin, erythromycin, fusidic acid, rifampin, ethidium bromide, acriflavin, crystal violet, sodium dodecyl sulfate (SDS) and deoxycholate—an unprecedented number of antibiotics, chemotherapeutics, detergents and dyes (85). Perhaps as a direct consequence of the use of these compounds, including metal-based antimicrobial therapies, RND resistance systems are being discovered with increasing frequency (9, 51, 115). Thus, these resistance systems pose a formidable clinical threat (90). As their prevalence in pathogens increases, so too does the need to understand them.
Although this review will focus on the details of the CusCFBA tetrapartite Cu(I)/Ag(I) transporter, many similarities are expected to be found between heavy-metal extrusion and antibiotic extrusion in general. Where appropriate throughout the article and as much as possible, parallels will be drawn between Cus and other known RND systems, with emphasis on the well-characterized MexAB-OprM and AcrAB-TolC efflux pumps, model systems for bacterial drug resistance.
Copper and silver resistance in E. coli
By the time the E. coli genome was completely sequenced (12), already two HME-RND pumps and their corresponding complexes had been found in other organisms: CzcCBA in Ralstonia sp. CH34 (29, 77, 78) and SilCBA in S. typhimurium (34, 115, 116). The earliest example SilA, with nearly 87% sequence identity to CusA, was purified from a hospital strain of S. typhimurium at Massachusetts General in 1975. S. typhimurium expressing this plasmid-encoded transporter were found to be resistant to a “disturbing” number of antibiotics and metal ions, including the popular silver nitrate (68). Ultimately, infection led to the deaths of three patients and the closing of the burn ward (115). Based in part on their sequence similarity to the deadly sil, czc and other known antimicrobial efflux pumps, 37 open reading frames in the E. coli genome were identified as putative drug transporters. In addition to CusA, this list includes six other RND proteins (88, 91).
The architecture of the cus locus consists of two back-to-back operons, cusCFBA and cusRS (formerly ylcBCD-ybdE and ylcA-ybcZ, respectively). The cusA gene is preceded by cusB, encoding the MFP, and cusC, encoding the OMF. Between cusB and cusC is the short ORF cusF, homologous to the corresponding gene in sil, for a small periplasmic chaperone protein (26). Transcribed in the opposite direction are cusR and cusS, encoding a response regulator and histidine kinase, respectively. Upstream of cusA and cusB is a promoter region with high sequence similarity to those of pcoAp, pcoEp, silCp, silEp, copAp and copHp—all copper and silver resistance genes of Gram-negative bacteria (26, 34, 77). Although the function of each cus gene was unknown at the time of discovery, all previously determined cusA homologs were found to be preceded by a cusB homolog and organized as an operon, including the acr and mex loci. In the closely-related heavy-metal resistance domains czc and cnr of R. metallidurans and the sil determinant of Salmonella, the OMF gene is also part of the operon. However, in the case of acr and mex, the outer membrane channel genes (tolC and oprM, respectively) were found elsewhere.
Several early experiments sought to characterize the new genes and their results led to the discovery that the cus locus encodes, primarily, a copper- and silver-transporting machine. This makes cus special in E. coli as one of only two chromosomally-encoded copper resistance systems and the only one implicated in silver resistance. The other copper locus, cue, encodes the membrane protein CopA and soluble protein CueO. Both would be found to share the substrate Cu(I), however CopA is a P-type ATPase, resting in the inner membrane, while CueO is a periplasmic multi-copper oxidase. CopA and CueO are both regulated by CueR, which is a transcriptional activator induced by copper (18). The role of cue and the proteins it encodes provide key insights to the cooperative nature of copper resistance in E. coli (30, 67, 79, 81, 89, 92, 97, 98, 99, 117, 118).
At first, the CusCFBA efflux system was not believed to be related to copper-resistance at all. Franke et al. were not able to distinguish the sensitivity of ΔcusA to CuCl2 from that of the wild-type, under aerobic conditions (26). Grass and Rensing also showed that the MICs of CuCl2 in ΔcusA and wild-type strains were indistinguishable (3.5 mM) and, additionally, that there was no observed difference between the growth of ΔcusA::cm and the double mutant ΔcusA::cmcopA::km in the copper medium (30). However, their susceptibility assays indicated a slight MIC difference of CuCl2 in copA::kmΔcueO::cm and copA::km (2.5 mM compared to 2.25 mM, respectively), a large difference in ΔcueO::cm and ΔcusA::cmΔcueO::cm (2.75 mM compared to 1.3 mM, respectively) and no difference at all between ΔcueOΔcusA::cm, copA::kmΔcueOcusA::cm and copA::kmΔcueOcusCFBA::cm.
It is clear from these results that CueO and particularly CopA each have a major role in conferring aerobic copper resistance in E. coli, whereas CusA alone had little to no role. Either the expression of CueO and CopA should be much higher at the copper concentrations tested or they should be much more efficient transporters. Indeed, in P. syringae, CopA was shown to bind as many as 11 atoms of copper (16). In addition, E. coli CueO was found to have at least five copper binding sites per molecule, with Km nearly 4-fold higher and kcat 8- and 30-fold higher, respectively, than for yeast Fet3 and human ceruloplasmin (117). Expression levels seem to fit this scheme, as well. Thieme et al. found that after addition of 1mM CuCl2 expression reached a maximum of 334, 180 and 8 transcripts per cell, of CopA, CueO and CusA, respectively (126).
Although CusA expression had no noticeable effect on aerobic copper resistance compared to the wild-type cell, a striking difference was observed between the aerobic and anaerobic growth of E. coli knockout strain ΔcusR (89). In the absence of oxygen, the ΔcusR strain was remarkably sensitive to Cu(I); more than the wild-type and ΔcueO, and nearly identical to ΔcopA. Under aerobic conditions, the half-maximal inductions of both cueO and copA were found to occur at 3 μM CuSO4 and cusC at 200 μM (89). In stark contrast to the aerobic case, the half-maximal anaerobic inductions of each protein were found to be 60, 70 and 3 μM for cueO, cusC and copA, respectively. While the expression level of CopA was unchanged, CueO expression fell and CusC expression increased dramatically in the absence of oxygen.
In an oxidizing environment, the dominant copper species should be Cu(II) and in the reducing environment it should be Cu(I). However, because Ag(I) is the dominant silver ion species with or without oxygen, there should be no difference in the aerobic or anaerobic silver-resistance conferred by CusA. The susceptibility experiments of Franke et al. were among the first to confirm the importance of CusA in Ag(I) resistance. They observed that while the wild-type E. coli strain grew in the presence of up to 25 μM Ag(I), the ΔcusA knockout strain did not (27). Additionally, ΔcusA, ΔcusAΔcopA and ΔcusCFBA knockout strains were very sensitive to AgNO3, compared to ΔcopA knockouts and the wild-type strain, agreeing with the experiments of Gupta et al. (26, 35).
Surprisingly, neither CopA nor CueO were implicated in silver resistance. While copper resistance in E. coli can be understood in terms of three synergistic components, regarding silver tolerance CusCFBA has no peers. In fact, the cus domain was even prematurely named agr before its role in copper-resistance was known (115). The presence of periplasmic Ag(I) actually inhibits the activity of the copper transporter CueO (118).
To determine the importance of each component of the tetrapartite efflux system in conferring copper resistance, various knockout strains of E. coli were prepared and grown in a medium containing CuCl2. The MIC was determined for each strain and the results were as follows: ΔcueO grew uninhibited in up to 3.25 mM CuCl2, ΔcueOΔcusA in 1.5, ΔcueOΔcusB in 1.5, ΔcueOΔcusC in 1.75, ΔcueOΔcusCΔtolC in 1.75 and, finally, ΔcueOΔcusF in 2.25 (27). Thus, all of the proteins in the cus operon were found to be necessary in conferring the maximal copper resistance in vivo. The small effect of CusC deletion was initially proposed to be due to TolC replacement, as in the Acr tripartite efflux system, however the ΔcueOΔcusCΔtolC knockout strain was no less resistant to copper. Therefore, CusC should be specific to the CusCFBA pump and not replaceable—at least not by TolC.
That copper resistance was strong no matter which subset of components were expressed, and that full copper resistance could not be conferred without expressing the entire pump, suggests that each piece has an active role in heavy-metal efflux. In the following sections, we will examine these roles, describing the structure and proposed function of each component, piece-by-piece.
CusA—inner-membrane transporter, “the pump”
Prior to the successful x-ray experiments of Long et al. in 2010, diffraction-quality crystals had been obtained for only two other RND pumps: E. coli AcrB (73) and P. aeruginosa MexB (110). Based in part on this experiment, CusA was revealed to be the main substrate-binding transporter of this efflux system.
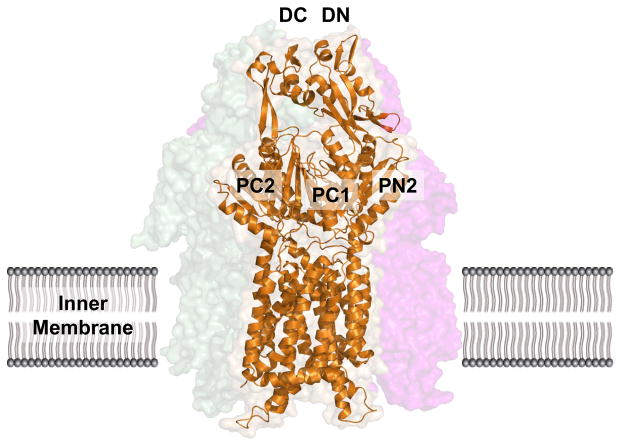
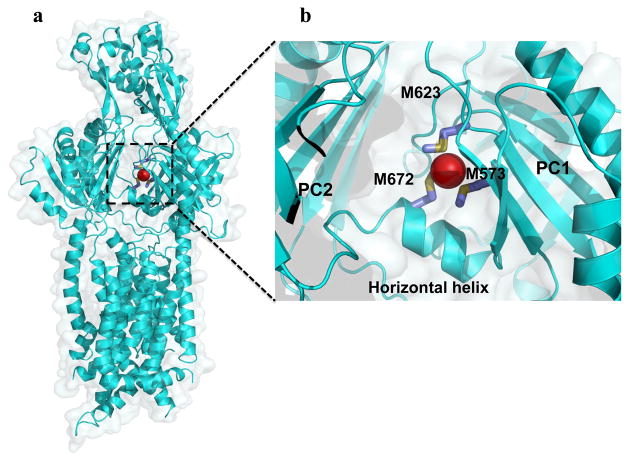
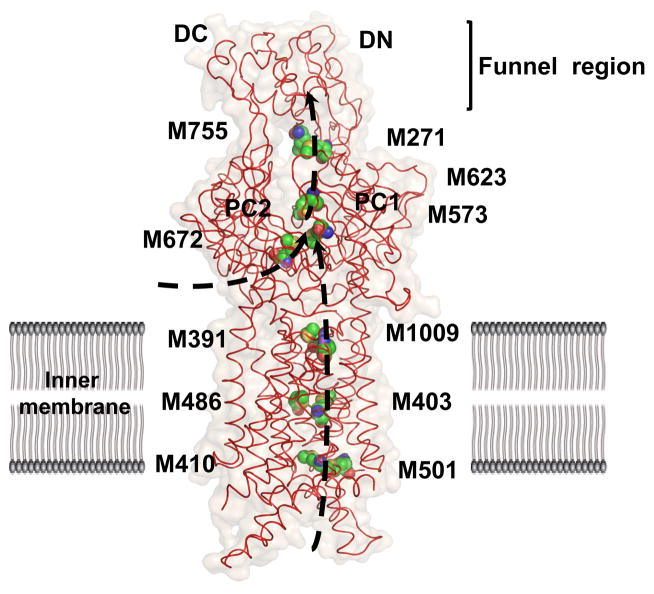
The apo-crystal structure of CusA was first determined to a resolution of 3.52 Å with 98% of the 1,047 residues (residues 5-504 and 516-1040) included in the final model (pdb: 3K07), and it suggests that CusA exists as a homotrimer (Fig. 3) (64). The tertiary structure contains 12-transmembrane α-helices (TMs), typical of membrane transport proteins (39). TMs 4, 5, 10 and 11 are confined to the cytoplasm and 2 and 8 to the periplasm, while the rest are embedded within the inner-membrane. Like AcrB and MexB before it, CusA can be divided into six sub-domains: PN1, PN2, PC1 and PC2 which comprise the pore domain and DN and DC which are presumed to form the CusB docking domain. Perhaps the most striking feature of CusA is one α-helix that extends horizontally into the bottom of a cleft created by PC1 and PC2 of the periplasmic domain, roughly dividing the transmembrane and periplasmic domains. Sitting atop this helical divider are three adjacent methionine residues (M573, M623 and M672). These same methionines were identified earlier as three of nine total conserved methionines among similar putative drug transporters, including SilA (98). Although the exact role of these methionines was not yet clear, they were found to be indispensable with respect to copper resistance. When mutated to isoleucines, the M573I, M623I and M672I mutants were entirely unable to relieve the copper sensitivity of the ΔcueO knockout strain in vivo (98). In the E. coli ΔacrBΔcueO knockout strain, Long et al. found that the MICs for CuSO4 in mutants M573I, M623I and M672I were drastically lower than in the wild-type (0.50 mM and 2.25 mM, respectively), while there was no noticeable difference in the MIC level of these mutants when compared to the complete knockout ΔcusA. For silver ions, the same effect was also observed. The MIC of AgNO3 in E. coli expressing CusA mutants M573I, M623I and M672I was 12.5 μM, compared to 30 μM in the wild-type and 10 μM in the ΔcusA knockout strain. In fact, the three-methionine motif is especially common among Cu(I)-resistance proteins (48, 99, 131), which suggests the importance of M573, M623 and M672 to the specificity of the CusA pump.
Fig. 3. Crystal structure of the apo-CusA efflux pump.
The CusA protomer found in the asymmetric unit of the crystal lattice is depicted by ribbon diagram (orange). The surface rendering corresponds to the CusA homotrimer, which is formed by symmetry within the crystal. Sub-domains PN1, PN2, PC1 and PC2 form the pore domain, while subdomains DN and DC comprise the docking domain, presumably interacting with the CusC channel.
An open question for the almost eight years between experiments was answered when the ion-bound structures of CusA were obtained, by soaking the apo-crystals in Cu(I) and Ag(I) solutions, individually (pdbs: 3KSS and 3KSO, respectively) (64). The root mean square deviation (RMSD) between the Cu(I)- and Ag(I)-bound structures was 1 Å, suggesting that these two structures are nearly identical. Accordingly, Cu(I) and Ag(I) were found to coordinate M573, M623 and M672 in each structure (Fig. 4). In addition to the Cu(I)- and Ag(I)-binding sites, they revealed that metal binding triggers a large conformational change in the pump. The RMSD between the ligand-bound and apo-CusA structures was 3.9 Å. This deviation is mainly attributed to a 30° opening between sub-domains PC1 and PC2, creating a kind of doorway for metal ions to enter the periplasmic domain of the pump (Fig. 4). The cleft was found closed in the apo-crystal and open in the ligand-bound crystal, suggesting that the binding site is revealed in the presence of Ag(I) or Cu(I) and hidden in their absence. As well as exposing it to the periplasm, the shift more closely coordinates the three methionines involved in binding. In comparison with the apo and ion-bound CusA structures, it is found that transmembrane helices TM1, TM2, TM3, TM6 and TM8 and pore domain PN1 also move noticeably toward the direction of the outer membrane upon metal binding.
Fig. 4. Crystal structure of the metal ion-bound CusA efflux pump.
a) The ribbon diagram and surface rendering correspond to the Cu(I)- or Ag(I)-CusA subunit (blue). The binding of Cu(I) or Ag(I) by CusA is correlated with a major conformational change in the pump. The separation between the PC1 and PC2 sub-domains of each Cu(I)- and Ag(I)-CusA subunit is exaggerated compared to that of apo-CusA. b) The metal ion binding site of CusA. The bound Ag(I) and Cu(I) ions are closely coordinated by methionine residues M623, M672 and M573.
Besides the main three-methionine binding site, four distinct methionine pairs were identified from each protomer of CusA (Fig. 5). Prior to this experiment, two-methionine or two-cysteine binding sites for copper were identified in at least four other copper tolerance proteins: CusF (62), CueR (18), Atx1 (6) and CopC (7). In CusA, three of these pairs, M410-M501, M403-M486 and M391-M1009, are found below the main binding site in the transmembrane domain and one, M271-M755, is positioned in the periplasm above the three-methionine site. All of these methionine coordination sites were found on the inside of the channel formed by each protomer of the CusA pump. Based on their locations, it was proposed that the methionine ladder created by these pairs could shuttle ions in a stepwise fashion for ion extrusion. Thus, the periplasmic methionines may serve as a transition of the transported metal ion from CusA to the putative outer-membrane channel CusC.
Fig. 5. Metal ion extrusion pathways of the CusA efflux pump.
Four methionine pairs (C, green; O, orange; N, blue; S, yellow) in each protomer of CusA form the path of Cu(I) or Ag(I) transport. Metal ions may enter from the cytoplasmic side of the channel, via M410-M501, or through the periplasmic cleft between the sub-domains PC1 and PC2, via the three-methionine binding site M573, M623 and M672. These pathways are illustrated in black.
Susceptibility assays were carried out to determine the utility of these ladder methionines (64). When one methionine in each pair was mutated to an isoleucine, the MIC of CuSO4 dropped to 1.25 mM for M391I and 1.75 mM for M410I, M486I and M755I, compared to 2.25 mM in the wild-type CusA. Silver resistance was also stunted, as the ladder mutants brought the MIC for AgNO3 to 10 μM, 17.5 μM, 17.5 μM and 12.5 μM for M391I, M410I, M486I and M755I, respectively.
In addition to the in vivo susceptibility assays, which demonstrated the importance of each residue in conferring copper and silver resistance, in vitro transport assays were employed to determine their roles in metal ion transport. CusA was reconstituted into liposomes containing Phen Green SK and the fluorescent signal was measured in the presence of extravesicular Ag+. In the wild-type CusA-loaded liposomes, quenching of the fluorescent signal was detected as soon as the silver ions were introduced into the medium. However, in the case of three-methionine binding site mutants M573I, M623I and M672I, and the methionine ladder mutants M391I, M486I and M755I, no quenching was detected, indicating that the extravesicular Ag+ could not traverse the membrane. Thus, these methionine residues were also implicated in CusA transport, and CusA was shown to have the ability to transport silver directly from the cytoplasm. It should be noted that a similar transport capability has also been found with CzcA (29) and AcrD (1).
Based on the crystal structure, the dynamics of the trimeric CusA pump was calculated using the elastic network model (8, 64). The result indeed suggests that CusA functions through three coupled motions in which the periplasmic cleft formed by subdomains PC1 and PC2 alternately open and close, similar to AcrB (74, 107, 109) and MexB (110).
CusB—membrane fusion protein, “the adaptor”
The structure of apo-CusB, the first of any HME-RND family MFP, was solved two years prior to that of CusA (64, 121). Before the experiments of Su et al. in 2009, both AcrA (71) and MexA (3, 41) had produced well-diffracting crystals. Although the CusCFBA complex has many similarities to AcrAB-TolC and MexAB-OprM, CusB itself shares only 13% identity and 52% similarity with MexA and 16% identity and 54% similarity with AcrA (121). Therefore, we should expect significant differences between these membrane fusion proteins.
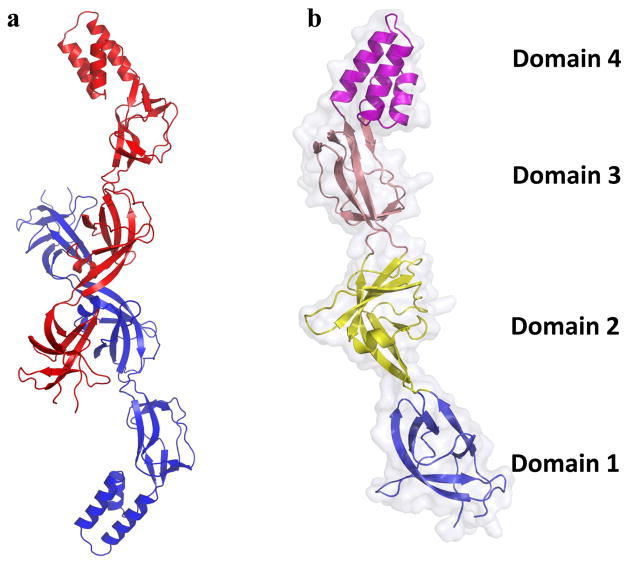
The crystal structure of CusB was resolved to 3.4 Å with 78.1% of the total residues (89-385) included (pdb: 3OOC) (Fig. 6) (117). Like AcrA, multiple conformations (here denoted A and B) of CusB were observed inside a single crystal, highlighting the flexibility of this MFP (71, 125). The overall fold, like that of MexA (3, 39, 125), can be divided into four domains: three β-domains and one α-helical domain. The first β-domain was identical in both structures, consisting of the N- and C-terminal ends of the protein arranged in a six β-stranded fashion. This domain was believed to interact directly with the CusA pump. Domain 2 in conformations A and B are quite dissimilar, consisting of two β-strands, four β-sheets and one short α-helix in A and six β-strands and two α-helices in B. Domains 3 and 4 were largely unchanged between each conformation, consisting of eight β-strands and an all α-helical three-helix bundle, respectively. When conformations A and B were superimposed, the overall RMSD calculated between alpha-carbons was 2.6 Å, indicating a significant structural difference between the two molecules. However, the superimposition of domains 1 and 2 of each conformation resulted in an RMSD of only 0.8 Å. Similarly, domains 3 and 4 of A and B could be superimposed to give an RMSD of only 0.8 Å. Therefore, the difference between conformations can be summarized as a shift of domains 1 and 2 with respect to 3 and 4. Best described as a rotation of ~20° about a hinge between the two groups, the difference between conformations results in a shift from what was called the “open” conformation of molecule A to the “closed” conformation of molecule B.
Fig. 6. Crystal structures of the CusB adaptor.
a) Molecule A (blue) and molecule B (red) of each subunit are depicted by ribbon diagram. The two distinct structures of the elongated CusB molecule suggest the flexible nature of this protein. b) Each protomer of CusB can be divided into four domains. An effective hinge between domains 2 and 3 is responsible for the conformational change between molecule A and molecule B of CusB.
Several years earlier, in the absence of structural information, Bagai et al. attempted to characterize the binding of Ag(I) and Cu(I) by CusB using isothermal titration calorimetry (ITC), x-ray absorption spectroscopy and in vitro susceptibility assays (9). Titration of Ag(I) into apo-CusB noticeably affected the measured binding enthalpy in a way that was not seen in the control, indicating that CusB does indeed bind Ag(I) in vitro. The equilibrium dissociation constant Kd for CusB-Ag(I) binding was measured to be 24.7 nM with a 1:1 Ag(I)-to-CusB molar ratio. Previously, this group had used x-ray absorption spectroscopy to characterize the binding environment of Cu(I) in the CusB-Cu(I) complex and found that the data was consistent with a three-sulfur coordination. Since there are no cysteine residues in CusB, the bound Cu(I) ion should be coordinated by three proximal methionines, as in CusA.
Of the nine total methionines in CusB, only four were found to be well-conserved and these residues are M21, M36, M38 and M283. Bagai et al. relied on ITC and site-directed mutagenesis to determine if these methionines are important for metal binding. Methionine mutants M21I, M36I, M38I and M283I were prepared, titrated with Ag(NH3)2+ and the binding enthalpy was measured. Compared to the wild-type, the mutation of M21 to isoleucine resulted in a 10-fold reduction in binding affinity of CusB for Ag(I). The M36I and M38I mutants resulted in no detectable affinity at all, while M283I had a similar affinity to the wild-type.
This experiment was supplemented with an in vivo susceptibility assay using the same CusB mutants, indicating that M21, M36 and M38 probably form a specific binding site for metal ions (9). Tragically, this three-methionine metal binding site identified by Bagai et al. is located in a region too disordered to obtain meaningful x-ray diffraction data. In combination with their results, the structure of the CusBA adaptor-transporter complex (123), which we will discuss in the next section, strongly supports the N-terminal residues M21, M36 and M38 as the specific Cu(I)/Ag(I) binding site of CusB.
CusBA—adaptor-transporter complex, “the co-crystal”
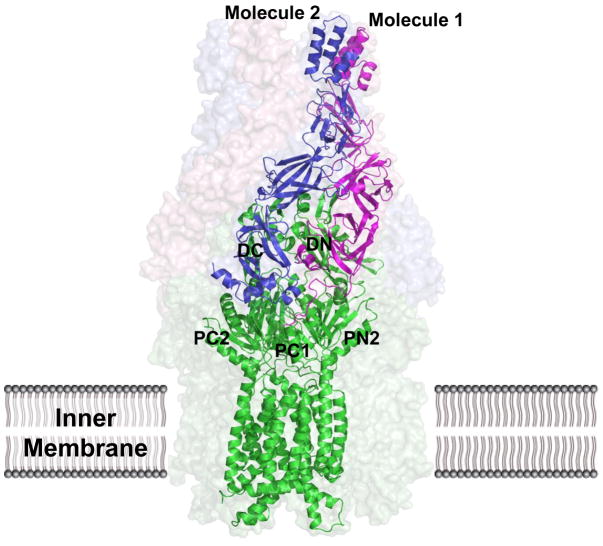
When it was published in 2011, the CusBA co-crystal structure was the first of its kind. This experiment was critical in elucidating specific interactions between the pump and adaptor molecules as well as developing detailed mechanisms of metal ion extrusion by the CusCFBA tetrapartite efflux complex. Its structure was first resolved to 2.90 Å (pdb: 3NE5), revealing that each CusA monomer interacts with each of two protomers of CusB (Fig. 7) (123). The structure included 1,686 amino acid residues of the total 1,861 (residues 4-1,043 of CusA, residues 79-400 of molecule 1 of CusB and residues 79-402 of molecule 2 of CusB). Based on the co-crystal structure, the trimeric CusA pump is found to contact six CusB adaptor molecules, which form a hexameric channel right above the periplasmic domain of CusA. The structure of the CusB hexamer mimics an inverted funnel. Domain 1 and the lower half of domain 2 of CusB create a cap-like structure, whereas domains 3, 4, and the upper half of domain 2 form the central channel of the funnel. The inner surface of the cap fits closely on top of the periplasmic domain of the CusA trimer. The funnel-like structure of hexameric CusB creates a channel extending contiguously from the top of the trimeric CusA pump. The narrowest section of the channel is at the hinge of CusB, between domains 2 and 3. The hexameric CusB channel formed above the cap of the adaptor is ~62 Å in length with an average internal diameter of ~37 Å. Thus, the interior of the channel gives rise to a large elongated cavity with a volume of ~65,000 Å3. The lower half of the channel is primarily created by β-barrels, whereas the upper half is an entirely α-helical tunnel. The inner surface of the channel is predominantly negatively charged, as indicated by the electrostatic surface distribution, which suggests that the interior surface of the channel may have the capacity to bind to positively charged metal ions. One striking feature of this possible extrusion pathway is that the inner surface of the hexameric CusB channel is predominantly composed of negatively charged residues, suggesting that the interior surface of the channel may have the capacity to bind to positively charged metal ions.
Fig. 7. Co-crystal structure of the CusBA adaptor-transporter complex.
The CusBA protomer found in the asymmetric unit of the crystal lattice is depicted by ribbon diagram and the surface rendering corresponds to the CusBA efflux complex. Each subunit of CusBA consists of one CusA molecule (green) and two CusB molecules (magenta and blue). The full structure includes the hexameric CusB adaptor as well as the trimeric CusA transporter.
It is worth noting that the 1:2 relationship of CusA to CusB has led to the conclusion that that the entire assembled structure should be CusA3-CusB6-CusC3. This assembly is indeed in good agreement with the predicted 3:6:3 polypeptide ratios of these three-part complexes (47, 96, 119).
ITC was used to determine the binding affinity of CusB to the CusA pump, indicating a tight interaction with a Kd of 5.1 μM. Molecule 1 of CusB interacts predominantly with CusA through charge–charge interactions, with residues K95, D386, E388 and R397 forming four salt bridges with D155, R771, R777 and E584 of CusA, respectively. Interestingly, the interaction between molecule 2 of CusB and CusA is mediated not by charge-charge but by charge–dipole and dipole–dipole interactions. Specifically, Q108, S109, S253 and N312 of molecule 2 of CusB form hydrogen bonds with Q785, Q194, D800 and Q198 of CusA, respectively (123).
Recently, artificial peptides, DARPins, have been introduced to bind and effectively inhibit the AcrB transporter (109). The inhibitor-binding site is found right above the cleft formed between PC1 and PC2 of AcrB. Interestingly, the location of this inhibitor-binding site corresponds to the binding site for molecule 2 of CusB in the CusBA complex. Thus, it is likely that the mechanism of action for these inhibitors may be the disruption of the adaptor-transporter interaction by competition.
By soaking the apo-crystals in Cu(I) solution, three distinct Cu(I)-bound structures were obtained, designated as the pre-extrusion 1, pre-extrusion 2 and extrusion states (pdbs: 3T56, 3T51 and 3T53, respectively) (124). The conformation of the pre-extrusion 1 structure was most similar to the binding state observed in CusA-Cu(I), where the periplasmic cleft formed by PC1 and PC2 was open. In the pre-extrusion 2 state, the conformation of the CusA molecule was more similar to the apo-CusA form (designated as the resting state), with a closed periplasmic cleft and disassembled three-methionine binding site. The extrusion form of CusBA-Cu(I) was nearly identical to the structure of apo-CusBA; similar to the pre-extrusion 2 state, if not for a subtle motion of a short C-terminal helix of molecule 1 of CusB (residues 391-400), which appears to push against another helix of CusA (582-589) and in turn a loop (residues 609-626) directly below the PC1 helix. Based on these interactions, it was proposed that CusB may be able to tune the width of the channel formed by the CusA pump, optimizing the process of metal ion extrusion.
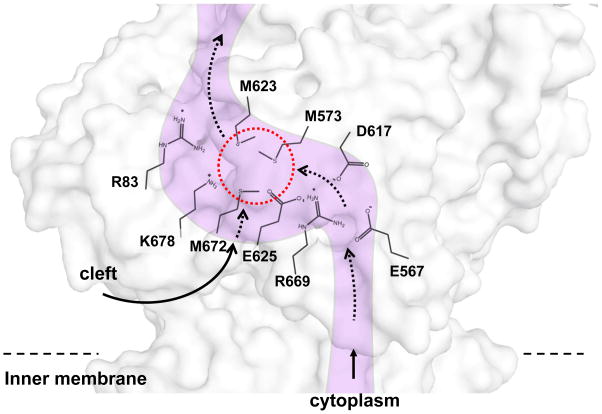
The conserved charged residues R83, E567, D617, E625, R664 and K678 in the periplasmic domain CusA can be seen in the CusBA structure to form a line along the inner wall of the methionine ladder (Fig. 8). These intriguingly positioned residues were mutated to alanines and expressed in the BL21(DE3) ΔcueOΔcusA knockout strain, to determine their importance in copper resistance. The MICs of CuSO4 in E. coli carrying these mutant CusA pumps were uniformly 0.5 mM; identical to the empty vector and much less than the 2.25 mM of the wild-type. An in vitro fluorescence assay was also used to determine these residues’ importance in silver transport. When liposomes containing the mutant proteins and loaded with fluorescent indicator Phen Green SK were exposed to extravesicular Ag+ ions no quenching of the signal was observed, suggesting that these mutant transporters were unable to transport Ag+ across the membrane.
Fig. 8. Cu(I) binding site and conserved charged residues.
This is a schematic representation of the CusA channel. The conserved residues R83, E567, D617, E625, R669 and K678, lining the channel at the periplasmic domain are indicated. The dotted red circle marks the location of the Cu(I) binding site formed by the methionine triad M573, M623 and M672. The paths for metal transport through the periplasmic cleft and transmembrane region are illustrated with black curves.
In the earlier work, it was shown that the methionine residues M21, M36 and M38 of CusB represent the most likely Cu(I)- and Ag(I)-binding site. In both crystal structures of the CusB protein and CusBA adaptor-transporter complex, these methionines were located in the disordered N-terminus and their locations could not be resolved. The CusBA co-crystal structure revealed that the disordered N-terminal regions of both CusB molecules are located directly outside the periplasmic cleft of CusA, created by helices PC1 and PC2 (123). Previously, that same cleft was shown to house CusA’s single three-methionine binding site (64). Therefore, it was suggested that CusB may transfer bound Cu(I) and Ag(I) at this location from the periplasm to the CusA pump for extrusion.
CusC—outer membrane factor, “the channel”
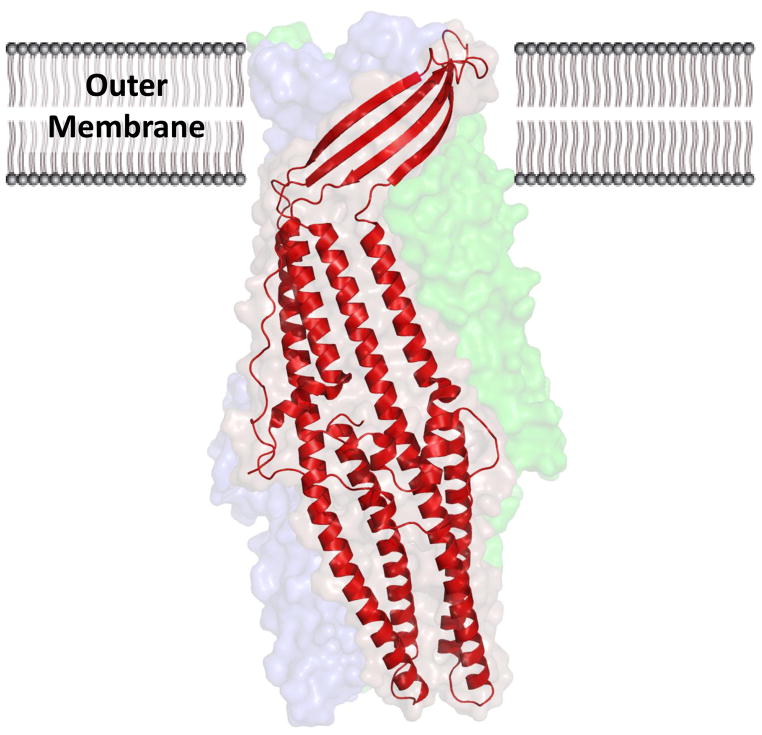
Kulathila et al. published the complete structure (with the exception of disordered residues 21-31) in 2011, to a resolution of 2.3 Å (pdb: 3PIK) (54). Based on the symmetry of the crystal and on gel-filtration measurements, CusC should assemble as a homotrimer, described as “cannon-shaped.” Each subunit of CusC in the trimer forms a ~130 Å long α/β barrel, containing four β-strands (contributing to the 12-stranded outer membrane β-barrel) and nine α-helices (forming the elongated periplasmic α-barrel) (Fig. 9). The trimeric CusC channel creates a large cylindrical internal cavity of ~28,000 Å3. Interestingly, the structure of CusC suggests that the N-terminal cysteine residue of each protomer is covalently linked to the outer membrane via a thioester bond. This N-terminal cysteine residue may play an important role in protein-membrane interactions and could be critical for the insertion of this channel protein into the outer membrane.
Fig. 9. Crystal structure of the CusC outer-membrane transporter.
The CusC monomer (red) within the asymmetric unit of the crystal is depicted by ribbon diagram. The surface rendering corresponds to the biological CusC trimer, which is created by crystal symmetry. Each subunit of CusC consists of a four β-strands atop nine α-helices, which are arranged as a barrel in the trimeric structure.
The cannon shape is remarkably similar to that of both TolC (53) and OprM (2), considering that TolC could not replace CusC in the mature pump (27). Many outer-membrane channels exhibit the character of interchangeability. For example, AcrAB shares TolC with AcrEF and a hemolysin secretion system in E. coli (56), and in R. metallidurans cnrC deletion could be functionally restored by czcC or nccC expression (81). However, CusC is believed to only work within the Cus efflux system. This irreplaceability may be partly due to the unique secondary structure of CusB among members of the MFP family. Specifically, CusB forms a three α-helical domain, which is supposed to directly interact with the CusC channel.
The RMSD calculated over alpha-carbons between CusC and TolC was measured at only 1.6 Å, and between CusC and OprM it was only 1.3 Å. Much of this difference is due to the exceptionally wide extracellular opening of CusC (~30 Å in diameter) compared to those of OprM or TolC (~12 Å). While the extracellular end of each channel is quite different in size, the periplasmic sides are all nearly closed in their respective crystal structures. Therefore, the CusC channel may require a substantial conformational change before allowing extrusion of substrates. Thus far, almost all available crystal structures of outer membrane channels are closed at one or both sides. It would be interesting to visualize the open conformational state of these channel proteins by crystallography.
Although CusC was found to be highly specific to the CusCFBA pump as its outer membrane factor, there was no evidence of copper or silver specificity in the channel. The effects of soaking apo-CusC crystals in high concentrations of Cu(I) or Ag(I), even exceeding 10 mM, could not be observed by x-ray diffraction. Both CusA and CusB were found to coordinate their bound Cu(I) and Ag(I) with methionine residues, however the five methionines in CusC are far removed from one another as seen in the apo-structure. It is unlikely that CusC possesses any binding sites or specificity for Ag(I) and Cu(I) ions, however, as seen in the electrostatic charge distribution, the interior surface of the channel is “strikingly” electronegative (41). Although not an indication of specificity for metal ions, a similar but less electronegative feature was also observed in the hexameric CusB channel of the crystal structure of the CusBA complex. If the hexameric CusB and trimeric CusC channels form the extrusion pathway of the pump, the electrostatic gradient created by the two interior surfaces may provide a possible pathway for ion extrusion.
To understand how the CusBA complex interacts with CusC, a CusCBA fitting model was constructed based on the crystal structures of CusBA complex and CusC protein. The final CusC3-CusB6-CusA3 structural model represents an assembled tripartite efflux complex spanning both the inner and outer membranes of E. coli to extrude metal ions (Fig. 10).
Fig. 10. Docking of CusC to CusBA.
The α-helical end of CusC interacts with the α-helices (Domain 4) of CusB in the CusBA complex. The surface rendering of the CusC3-CusB6-CusA3 complex is colored as follows: brown, CusC trimer; purple, CusB hexamer; green, CusA trimer.
CusF—small periplasmic metal-binding protein, “the chaperone”
So far, each component (CusA, CusB and CusC) of our tetrapartite complex has a counterpart in the protypical multi-drug resistance systems AcrAB-TolC and MexAB-OprM. Then, there is CusF. When the cusF gene was first sequenced, nothing like it had been found in any known RND systems or any drug transporters (115). However, by the time its crystal structure was published, more than 35 CusF homologues had been identified among Gram-negative bacteria, including two putative heavy-metal resistance determinants: czc of R. metallidurans and sil of S. typhimurium (61). The function of these proteins was largely unknown; nonetheless, three conserved residues M47, M49 and H36 were identified among their sequences, suggesting that these residues may represent a potential binding site.
The crystal structure of apo-CusF was published by Loftin et al. in 2005 (pdb: 1ZEQ) (61), and two years later the CusF-Ag(I) complex structure was solved by the same group (pdb: 2QCP) (62). The apo-structure was resolved to 1.5 Å, including ~93% of the total residues (residues 6-88). Based on the diffraction results, the structure of CusF can be simply described as a small five-stranded β-barrel, composed of two anti-parallel three-stranded β-sheets which were packed orthogonally. The CusF-Ag(I) complex was resolved to 1.0 Å, including residues 10-88 (62). An RMSD of only 0.5 Å was calculated between the apo and ion-bound structures, indicating that these two structures are essentially the same. The bound Ag(I) in CusF was found to be indeed coordinated by H36, M47 and M49.
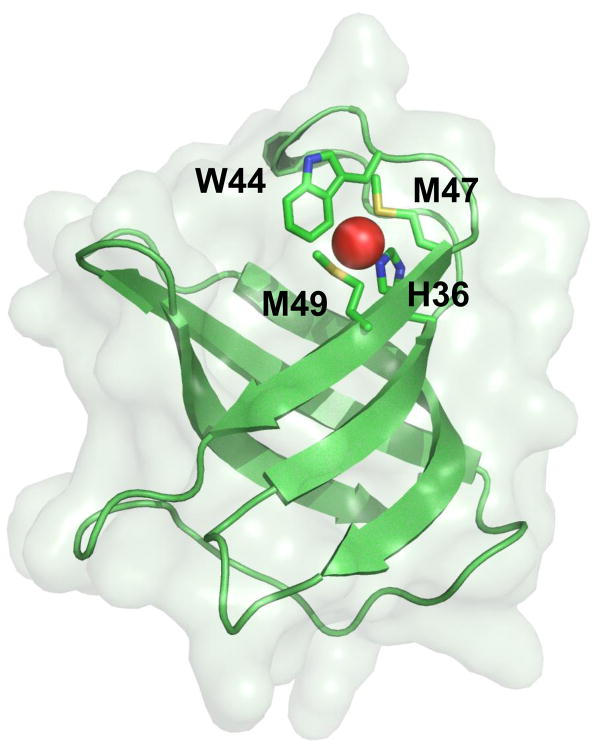
The crystal structure of the CusF-Cu(I) complex (pdb: 2VB2) (Fig. 11) was published by Xue et al., one year later, with a resolution of 1.7 Å and including residues 13-87 (131). Their structure was nearly identical to both the previous CusF-Ag(I) complex and apo-CusF structures. However, there was a slight conformational change observed upon Cu(I) binding, which includes a twist of residues M47 and M49 to accommodate the bound metal ion. Upon ion-binding, these side chains swiveled to more closely coordinate the bound Cu(I), with a fourth residue, W44, implicated in binding. The proximity of this tryptophan residue to Cu(I) (~2.7 Å as determined by x-ray absorption spectroscopy) was “unprecedented” among previous metal-binding sites, and introduces a novel motif for Cu(I)-binding.
Fig. 11. Crystal structure of the metal ion-bound CusF metallochaperone.
The binding of Cu(I) and Ag(I) is coordinated by M47, M49, W44 and H36, which form a novel metal ion-binding site in CusF.
Although it was found to bind both Cu(I) and Ag(I), it was yet unclear what role CusF might have in the efflux process. Recent experiments strongly suggest the possibility of a hand-off of bound Cu(I) or Ag(I) from CusF to CusB (17). Presumably, the specific interaction between CusF and CusB is critical for a direct transfer of Cu(I) and Ag(I) ions before extrusion via the CusCFBA efflux complex. Thus, the interaction between these two components may mark the initial step for metal ion extrusion by the CusCFBA tetrapartite efflux complex.
Concluding remarks
It has been well established that overexpression of RND multi-drug efflux pumps led to a resistant phenotype in pathogenic organisms. This problem is exacerbated by the ease with which many of these resistance genes can pass from one microorganism to another through plasmid transfer (21). Because of the fact that these multi-drug efflux pumps are able to respond to a wide spectrum of substrates, pathogenic bacteria that overexpress these pumps can be selected for by many different agents. For example, the organic compound nonoxynol-9, which is widely used as an active ingredient in most spermicide creams and jellies, can select Neisseria gonorrhoeae mutants that overexpress the MtrCDE RND-type multi-drug efflux pump (130). Thus, it is very important to understand the molecular mechanism as well as detailed structural information of these efflux pumps to combat infectious diseases.
The availability of the three-dimensional structures of these efflux transporters potentially allows us to rationally design agents that block their function. However, there is still quite a mountain to climb in the development toward achieving this goal. For instance, a crystallographic model of these RND tripartite efflux complexes has not yet been reported. One direct approach to obtain the complete picture of these efflux complexes is to elucidate the structures of individual components as well as the assembled complexes of these efflux systems. This invaluable structural information should be able to provide a platform for producing new drugs and inhibitors to heighten bacterial sensitivity to these antimicrobials. In addition, the outer membrane channel proteins of these RND efflux complexes that span the outer membrane and expose to the surface of bacterial cells make them ideal targets for vaccines. Thus, the knowledge of these outer membrane channels is essential for the development of vaccines against bacterial diseases.
To date, no efflux pump inhibitor has been licensed for use in the clinical treatment of bacterial infections. Bacterial efflux pump inhibitors may probably simultaneously inhibit human detoxification systems and bacterial multi-drug efflux pumps. This will require a thorough research to ensure that these inhibitors are specific solely for the bacterial efflux pumps, minimizing their side effects to the host. As RND pumps assemble as tripartite complexes, one method to inhibit the drug-resistant phenotype is to prevent the assembly of complex formation by blocking the different components from forming a functional complex. The possibility of this approach has been demonstrated by the crystal structure of AcrB in complex with the designed ankyrin repeat protein inhibitor (DARPin) (109). Based on the structural information of the CusBA adaptor-transporter complex (123), the inhibitor in principle is bound in such a way that the AcrB transporter is not able to form a functional complex with the AcrA adaptor. Therefore, the structural information of multi-drug efflux pumps, particularly with their assembled complex models, is of importance. In the case of the Cus efflux system, the availability of the co-crystal structures of the adaptor-transporter CusBA, channel-adaptor CusCB, and complete tetrapartite CusCFBA efflux complexes are essential in order to achieve this task although co-crystallization of different components of these RND efflux systems have been proven to be extremely difficult.
Another potential approach is to target the transcriptional regulators that modulate the expression of these RND pumps. For example, the global regulator CmeR controls the expression of several genes, including cmeABC which produces the CmeABC RND efflux complex (31, 57, 58), and the transcriptional repressor MtrR which regulates the expression level of the MtrCDE tripartite multi-drug efflux pump (37, 42, 111, 130). By inhibiting the interactions between the regulators and their corresponding substrates, expression of these transporter genes could potentially be blocked. Recently, more examples of RND transporters regulated by two-component systems have been identified, including E. coli CusRS, which controls the expression of the CusCFBA efflux complex (33, 72). The structures of different components of these regulatory systems may potentially allow us to rationally design inhibitors that reduce the level of efflux pump expression.
References
- 1.Aires JR, Nikaido H. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J Bacteriol. 2005;187(6):1923–29. doi: 10.1128/JB.187.6.1923-1929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, et al. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J Biol Chem. 2004;279(51):52816–9. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 3.Akama H, Matsuura T, Kashiwagi S, Yoneyama H, Narita S, et al. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J Biol Chem. 2004;279(25):25939–42. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez-Ortega C, Olivares J, Martínez JL. RND multidrug efflux pumps: what are they good for? Front Microbiol. 2013;4:7. doi: 10.3389/fmicb.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angus BL, Carey AM, Caron DA, Kropinski AM, Hancock RE. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother. 1982;21(2):299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnesano F, Banci L, Bertini I, Huffman DL, O’Halloran TV. Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1. Biochemistry. 2001;40(6):1528–39. doi: 10.1021/bi0014711. [DOI] [PubMed] [Google Scholar]
- 7.Arnesano F, Banci L, Bertini I, Mangani S, Thompsett AR. A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc Natl Acad Sci USA. 2003;100(7):3814–19. doi: 10.1073/pnas.0636904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, et al. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80(1):505–15. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem. 2007;282(49):35695–702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 10.Bagai I, Rensing C, Blackburn NJ, McEvoy MM. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry. 2008;47(44):11408–14. doi: 10.1021/bi801638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baranova N, Nikaido H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol. 2002;184(15):4168–76. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–62. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 13.Bohnert JA, Schuster S, Fähnrich E, Trittler R, Kern WV. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF) J Antimicrob Chemother. 2007;59(6):1216–22. doi: 10.1093/jac/dkl426. [DOI] [PubMed] [Google Scholar]
- 14.Brown NL, Rouch DA, Lee BTO. Copper resistance determinants in bacteria. Plasmid. 1992;27:41–51. doi: 10.1016/0147-619x(92)90005-u. [DOI] [PubMed] [Google Scholar]
- 15.Brown MH, Paulsen IT, Skurray RA. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31:394–95. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- 16.Cha J-S, Cooksey DA. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci USA. 1991;88:8915–19. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravorty DK, Wang B, Ucisik MN, Merz KM., Jr Insight into the cation-p interaction at the metal binding site of the copper metallochaperone. J Am Chem Soc. 2011;133(48):199330–3. doi: 10.1021/ja208662z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Changela A, Chen K, Xue Y, Holschen J, Outten CE, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301(5638):1383–87. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 19.Chopra I. The increasing use of silver-based products as antimicrobial agents: a useful development or a cause for concern? J Antimicrob Chemother. 2007;59:587–90. doi: 10.1093/jac/dkm006. [DOI] [PubMed] [Google Scholar]
- 20.Conroy O, Kim E-H, McEvoy MM, Rensing C. Differing ability to transport non-metal substrates by two RND-type metal exporters. FEMS Microbiol Lett. 2010;308(2):115–22. doi: 10.1111/j.1574-6968.2010.02006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courvalin P. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 1994;38:1447–51. doi: 10.1128/aac.38.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das D, Xu QS, Lee JY, Ankoudinova I, Huang C, et al. Crystal structure of the multidrug efflux transporter AcrB at 3.1 Å resolution reveals the N-terminal region with conserved amino acids. J Struct Bio. 2007;158(3):494–502. doi: 10.1016/j.jsb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delmar JA, Su C-C, Yu EW. Structural mechanisms of heavy-metal extrusion by the Cus efflux system. Biometals. 2013;26(4):593–607. doi: 10.1007/s10534-013-9628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eicher T, Cha HJ, Seeger MA, Brandstätter L, El-Delik J, et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci USA. 2012;109(15):5687–92. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Festa RA, Thiele DJ. Copper: an essential metal in biology. Curr Biol. 2011;21(21):R877–83. doi: 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology. 2001;147:965–72. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 27.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185(13):3804–12. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaetke LM, Chow CK. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology. 2003;189:147–63. doi: 10.1016/s0300-483x(03)00159-8. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg M, Pribyl T, Juhnke S, Nies DH. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J Biol Chem. 1999;273(37):26065–70. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 30.Grass G, Rensing C. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol. 2001;183(6):2145–47. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grass G, Rensing C, Solioz M. Metallic copper as an antimicrobial surface. Appl Environ Microb. 2011;77(5):1541–47. doi: 10.1128/AEM.02766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu R, Su C-C, Shi F, Li M, McDermott G, et al. Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni. J Mol Biol. 2007;372(3):583–93. doi: 10.1016/j.jmb.2007.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gudipaty SA, Larsen AS, Rensing C, McEvoy MM. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol Lett. 2012;330:30–7. doi: 10.1111/j.1574-6968.2012.02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta A, Matsui K, Lo J-F, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5(2):183–88. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 35.Gupta A, Phung LT, Taylor DE, Silver S. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology. 2001;147:3393–402. doi: 10.1099/00221287-147-12-3393. [DOI] [PubMed] [Google Scholar]
- 36.Haber F, Weiss J. On the catalysis of hydroperoxides. Naturwissenschaften. 1932;20(51):948–50. [Google Scholar]
- 37.Hagman KE, Shafer WM. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol. 1995;177(14):4162–5. doi: 10.1128/jb.177.14.4162-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henderson PJ. The 12-transmembrane helix transporters. Curr Opin Cell Biol. 1993;5(4):708–21. doi: 10.1016/0955-0674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 39.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci. 2004;101(27):9995–99. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. 2007;446:749–57. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 41.Hinchliffe P, Symmons MF, Hughes C, Koronakis V. Structure and operation of bacterial tripartite pumps. Annu Rev Microbiol. 2013;67:221–42. doi: 10.1146/annurev-micro-092412-155718. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann KM, Williams D, Shafer WM, Brennan RG. Characterization of the multiple transferable resistance repressor MtrR, from Neisseria gonorrhoea. J Bacteriol. 187(14):5008–12. doi: 10.1128/JB.187.14.5008-5012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooper DC. Mechanisms of fluoroquinolone resistance. Drug Resist Update. 1999;2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 44.Irving H, Williams RJP. Order of stability of metal complexes. Nature. 1948;162(4123):746–47. [Google Scholar]
- 45.Jaffé A, Chabbert YA, Derlot E. Selection and characterization of beta-lactam-resistant Escherichia coli K-12 mutants. Antimicrob Agents Chemother. 1983;23(4):622–25. doi: 10.1128/aac.23.4.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janganan TK, Zhang L, Bavro VN, Matak-Vinkovic D, Barrera NP, et al. Opening of the outer membrane protein channel in tripartite efflux pumps is induced by interaction with the membrane fusion partner. J Biol Chem. 2011;286(7):5484–93. doi: 10.1074/jbc.M110.187658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janganan TK, Bavro VN, Zhang L, Matak-Vinkovic D, Barrera NP, et al. Evidence for the assembly of a bacterial tripartite multidrug pump with a stoichiometry of 3:6:3. J Biol Chem. 2011;286(30):26900–12. doi: 10.1074/jbc.M111.246595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang J, Nadas IA, Kim MA, Franz KJ. A Mets motif peptide found in copper transport proteins selectively binds Cu(I) with methionine-only coordination. Inorg Chem. 2005;44(26):9787–94. doi: 10.1021/ic051180m. [DOI] [PubMed] [Google Scholar]
- 49.Kim E-H, Nies DH, McEvoy MM, Rensing C. Switch or Funnel: How RND-Type Transport Systems Control Periplasmic Metal Homeostasis. J Bacteriol. 2011;193(10):2381–87. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HS, Nikaido H. Different function of MdtB and MdtC subunits in the heterotrimeric efflux transporter MdtB2C complex of Escherichia coli. Biochemistry. 2012;51(20):4188–97. doi: 10.1021/bi300379y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kittleson JT, Loftin IR, Hausrath AC, Engelhardt KP, Rensing C, McEvoy MM. Periplasmic metal-resistance protein CusF exhibits high affinity and specificity for both Cu(I) and Ag(I) Biochemistry. 2006;45(37):11096–102. doi: 10.1021/bi0612622. [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183(19):5639–44. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–19. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 54.Kulathila R, Kulathila R, Indic M, van den Berg B. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PLoS One. 2011;6(1):e15610. doi: 10.1371/journal.pone.0015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lane TW, Saito MA, George GN, Pickering IJ, Prince RC, et al. Biochemistry: a cadmium enzyme from a marine diatom. Nature. 2005;435:42. doi: 10.1038/435042a. [DOI] [PubMed] [Google Scholar]
- 56.Lau SY, Zgurskaya HI. Cell division defects in Escherichia coli deficient in the multidrug efflux transporter AcrEF-TolC. J Bacteriol. 2005;187(22):7815–25. doi: 10.1128/JB.187.22.7815-7825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levy SB. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Ch. 1992;36(4):695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64(2):159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 59.Lin J, Michel LO, Zhang Q. CmeABC Functions as a Multidrug Efflux System in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46(7):2124–31. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin J, Akiba M, Sahin O, Zhang Q. CmeR Functions as a Transcriptional Repressor for the Multidrug Efflux Pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother. 2005;49(3):1067–75. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loftin IR, Franke S, Roberts SA, Weichsel A, Héroux A, Montfort WR, Rensing C, McEvoy MM. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry. 2005;44(31):10533–40. doi: 10.1021/bi050827b. [DOI] [PubMed] [Google Scholar]
- 62.Loftin IR, Franke S, Blackburn NJ, McEvoy MM. Unusual Cu(I)/Ag(I) coordination of Escherichia coli CusF as revealed by atomic resolution crystallography and X-ray absorption spectroscopy. Prot Sci. 2007;16:2287–93. doi: 10.1110/ps.073021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loftin IR, McEvoy MM. Tryptophan Cu(I)-π interaction fine-tunes the metal binding properties of the bacterial metallochaperone CusF. J Biol Inorg Chem. 2009;14(6):905–12. doi: 10.1007/s00775-009-0503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long F, Su C-C, Zimmermann MT, Boyken SE, Rajashankar KR, et al. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467(7314):484–88. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Long F, Su C-C, Lei H-T, Bolla JR, Do SV, Yu EW. Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Phil Trans R Soc B. 2012;367:1047–58. doi: 10.1098/rstb.2011.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, et al. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. 1993;175(19):6299–313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci. 2009;106(20):8344–49. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McHugh GL, Moellering RC, Hopkins CC, Swartz MN. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet. 1975;1(7901):235–40. doi: 10.1016/s0140-6736(75)91138-1. [DOI] [PubMed] [Google Scholar]
- 69.Mealman TD, Zhou M, Affandi T, Chacón KN, Aranguren ME, et al. N-terminal region of CusB is sufficient for metal binding and metal transfer with the metallochaperone CusF. Biochemistry. 2012;51:6767–75. doi: 10.1021/bi300596a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mealman TD, Blackburn NJ, McEvoy MM. Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. Curr Top Membr. 2012;69:163–96. doi: 10.1016/B978-0-12-394390-3.00007-0. [DOI] [PubMed] [Google Scholar]
- 71.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14(3):577–87. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munson GP, Lam DL, Outten FW, O’Halloran TV. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol. 2000;182(20):5864–71. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–93. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 74.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–79. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 75.Murakami S. Multidrug efflux transporter, AcrB—the pumping mechanism. Curr Opin Struc Biol. 2008;18:459–65. doi: 10.1016/j.sbi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 76.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol. 2002;184(15):4161–67. doi: 10.1128/JB.184.15.4161-4167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nies DH, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–55. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nies DH. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177(10):2707–12. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nies DH. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51:730–50. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 80.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. 2003. FEMS Microbiol Rev. 2003;27(2):313–39. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 81.Nies DH. Bacterial transition metal homeostasis. In: Nies DH, Silver S, editors. Molecular Microbiology of Heavy Metals. Vol. 6. Berlin Heidelberg: Springer; 2007. pp. 117–42. [Google Scholar]
- 82.Nies DH. RND efflux pumps for metal cations. In: Yu EW, Zhang Q, Brown MH, editors. Microbial Efflux Pumps: Current Research. Vol. 1. Caister Academic Press; 2013. pp. 79–121. [Google Scholar]
- 83.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Ch. 1989;33(11):1831–36. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–88. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 85.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178(20):5853–59. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 2009;1794:769–81. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol. 2011;77:1–60. doi: 10.1002/9780470920541.ch1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183(20):5803–12. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Outten FW, Huffman DL, Hale JA, O’Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. 2001;276(33):30670–77. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 90.Paulsen IT, Brown MH, Skurray RA. Proton-dependent multi-drug efflux systems. Microbiol Rev. 1996;60(4):575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paulsen IT, Nguyen L, Sliwinski MK, Rabus R, Saier MH., Jr Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J Mol Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 92.Peña MM, Lee J, Thiele DJ. A delicate balance: homeostatic control of copper uptake and distribution. J Nutr. 1999;129(7):1251–60. doi: 10.1093/jn/129.7.1251. [DOI] [PubMed] [Google Scholar]
- 93.Percival SL, Bowler PG, Russell D. Bacterial resistance to silver in wound care. J Hosp Inf. 2005;60:1–7. doi: 10.1016/j.jhin.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 94.Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794(5):782–93. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 95.Pugsley AP, Schnaitman CA. Identification of three genes controlling production of new outer membrane pore proteins in Escherichia coli K-12. J Bacteriol. 1978;135(3):1118–29. doi: 10.1128/jb.135.3.1118-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rensing C, Pribyl T, Nies DH. New functions for the three subunits of the CzcCBA cation-proton antiporter. J Bacteriol. 1997;179:6871–79. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci. 2000;97(2):652–56. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 99.Roberts SA, Weichsel A, Grass G, Thakali K, Hazzard JT, et al. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc Natl Acad Sci. 2002;99(5):2766–71. doi: 10.1073/pnas.052710499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rosenberg EY, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol. 2000;182(6):1754–56. doi: 10.1128/jb.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Routh MD, Zalucki Y, Su C-C, Long F, Zhang Q, et al. Efflux pumps of the resistance-nodulation-division family: a perspective of their structure, function, and regulation in gram-negative bacteria. In: Toone EJ, editor. Advances in Enzymology and Related Areas of Molecular Biology. Vol. 77. John Wiley & Sons; 2011. pp. 109–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Russell AD, Hugo WB. Antimicrobial activity and action of silver. Prog Med Chem. 1994;31:351–70. doi: 10.1016/s0079-6468(08)70024-9. [DOI] [PubMed] [Google Scholar]
- 103.Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58(1):71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saier MH., Jr Molecular phylogeny as a basis for the classification of transport proteins from bacteria, archaea and eukarya. Adv Microb Physiol. 1998;40(81):81–136. doi: 10.1016/s0065-2911(08)60130-7. [DOI] [PubMed] [Google Scholar]
- 105.Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1(2):257–79. [PubMed] [Google Scholar]
- 106.Saier MH, Jr, Paulsen IT. Phylogeny of multidrug transporters. Semin Cell Dev Biol. 2001;12:205–13. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 107.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313(5791):1295–98. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 108.Seeger MA, Diederichs K, Eicher T, Brandstätter L, Schiefner A, et al. The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance. Curr Drug Targets. 2008;9(9):729–49. doi: 10.2174/138945008785747789. [DOI] [PubMed] [Google Scholar]
- 109.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS One. 2007;5(1):e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sennhauser G, Bukowksa MA, Briand C, Grütter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol. 2009;389:134–45. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 111.Shafer WM, Qu X-D, Waring AJ, Lehrer RI. Modulation of the Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA. 1998;95(4):1829–33. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Silver S, Misra TK. Plasmid-mediated heavy metal resistances. Ann Rev Microbiol. 1988;42:717–43. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- 113.Silver S. Bacterial heavy metal resistance: new surprises. Ann Rev Microbiol. 1996;50:753–89. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 114.Silver S. Bacterial resistances to toxic metal ions—a review. Gene. 1996;179:9–19. doi: 10.1016/s0378-1119(96)00323-x. [DOI] [PubMed] [Google Scholar]
- 115.Silver S. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol Rev. 2003;27:341–53. doi: 10.1016/S0168-6445(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 116.Silver S, Phung LT, Silver G. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J Ind Microbiol Biotechnol. 2006;33:627–34. doi: 10.1007/s10295-006-0139-7. [DOI] [PubMed] [Google Scholar]
- 117.Singh SK, Grass G, Rensing C, Montfort WR. Cuprous oxidase activity of CueO from Escherichia coli. J Bacteriol. 2004;186(22):7815–17. doi: 10.1128/JB.186.22.7815-7817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Singh SK, Roberts SA, McDevitt SF, Weichsel A, Wildner GF, et al. Crystal structures of multicopper oxidase CueO bound to Copper(I) and Silver(I) J Biol Chem. 2011;286(43):37849–57. doi: 10.1074/jbc.M111.293589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stegmeier JF, Polleichtner G, Brandes N, Hotz C, Andersen C. Importance of the adaptor (membrane fusion) protein hairpin domain for the functionality of multidrug efflux pumps. Biochemistry. 2006;45:10303–12. doi: 10.1021/bi060320g. [DOI] [PubMed] [Google Scholar]
- 120.Su C-C, Li M, Gu R, Takatsuka Y, McDermott G, et al. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J Bacteriol. 2006;188(20):7290–96. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Su C-C, Yang F, Long F, Reyon D, Routh MD, et al. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J Mol Biol. 2009;393(2):342–55. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Su C-C, Long F, Yu EW. The Cus efflux system removes toxic ions via a methionine shuttle. Prot Sci. 2011;20(6):6–18. doi: 10.1002/pro.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Su C-C, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, et al. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011;470:558–63. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Su C-C, Long F, Lei H-T, Bolla JR, Do SV, et al. Charged amino acids (R83, E567, D617, E625, R669, and K678) of CusA are required for metal ion transport in the Cus efflux system. J Mol Biol. 2012;422:429–41. doi: 10.1016/j.jmb.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci USA. 2009;106(17):7173–78. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thieme D, Neubauer P, Nies DH, Grass G. Sandwich hybridization assay for sensitive detection of dynamic changes in mRNA transcript levels in crude Escherichia coli cell extracts in response to copper ions. Appl Environ Microb. 2008;74(24):7463–70. doi: 10.1128/AEM.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Toes A-CM, Daleke MH, Kuenen JG, Muyzer G. Expression of copA and cusA in Shewanella during copper stress. Microbiol. 2008;154:2709–18. doi: 10.1099/mic.0.2008/016857-0. [DOI] [PubMed] [Google Scholar]
- 128.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1(1):107–25. [PubMed] [Google Scholar]
- 129.Van Bambeke F, Balzi E, Tulkens PM. Antibiotic efflux pumps. Biochem Pharmacol. 2000;60:457–70. doi: 10.1016/s0006-2952(00)00291-4. [DOI] [PubMed] [Google Scholar]
- 130.Veal WL, Nicholas RA, Shafer WM. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J Bacteriol. 2002;184(20):5619–24. doi: 10.1128/JB.184.20.5619-5624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wells TNC, Scully P, Paravicini G, Proudfoot AEI, Payton MA. Mechanism of irreversible inactivation of phosphomannose isomerases by silver ions and flamazine. Biochemistry. 1995;34:7896–903. doi: 10.1021/bi00024a014. [DOI] [PubMed] [Google Scholar]
- 132.Xue Y, Davis AV, Balakrishnan G, Stasser JP, Staehlin BM, et al. Cu(I) recognition via cation-π and methionine interactions in CusF. Nature Chem Biol. 2008;4(2):107–109. doi: 10.1038/nchembio.2007.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external copper. Mol Microbiol. 2005;56(1):215–27. doi: 10.1111/j.1365-2958.2005.04532.x. [DOI] [PubMed] [Google Scholar]
- 134.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE., Jr Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–80. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 135.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J Bacteriol. 2005;187(19):6804–15. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zgurskaya HI, Nikaido H. Multidrug resistance mechanisms: drug efflux across two membranes. Mol Microbiol. 2000;37(2):219–25. doi: 10.1046/j.1365-2958.2000.01926.x. [DOI] [PubMed] [Google Scholar]
- 137.Zhang L, Koay M, Maher MJ, Xiao Z, Wedd AG. Intermolecular transfer of copper ions from the CopC protein of Pseudomonas syringaeCrystal structures of fully loaded CuICuII forms. J Am Chem Soc. 2006;128:5834–50. doi: 10.1021/ja058528x. [DOI] [PubMed] [Google Scholar]