Abstract
APOL1 variants have been associated with renal phenotypes in blacks. To refine clinical outcomes and discover mechanisms of APOL1-associated kidney injury, we analyzed clinical and genomic datasets derived from 90 black subjects in the Nephrotic Syndrome Study Network (NEPTUNE), stratified by APOL1 risk genotype. Ninety subjects with proteinuria ≥0.5 g/d were enrolled at first biopsy for primary nephrotic syndrome and followed. Clinical outcomes were determined, and renal histomorphometry and sequencing of Mendelian nephrotic syndrome genes were performed. APOL1 variants were genotyped, and glomerular and tubulointerstitial transcriptomes from protocol renal biopsy cores were analyzed for differential and correlative gene expression. Analyses were performed under the recessive model (high-risk genotype defined by two risk alleles). APOL1 high-risk genotype was significantly associated with a 17 ml/min per 1.73 m2 lower eGFR and a 69% reduction in the probability of complete remission at any time, independent of histologic diagnosis. Neither APOL1 risk group was enriched for Mendelian mutations. On renal biopsy, high-risk genotype was associated with increased fractional interstitial area, interstitial fibrosis, and tubular atrophy. Risk genotype was not associated with intrarenal APOL1 mRNA expression levels. Differential expression analysis demonstrated an increased steady-state level of five genes associated with the high-risk genotype (CXCL9, CXCL11, and UBD in glomerulus; SNOR14B and MUC13 in tubulointerstitium). APOL1 tubulointerstitial coexpression analysis showed coexpression of APOL1 mRNA levels with a group of intrarenal transcripts that together were associated with increased interstitial fibrosis and tubular atrophy. These data indicate the high-risk APOL1 genotype confers renal risk across histopathologic diagnoses.
Keywords: glomerular disease, nephrotic syndrome, genetic renal disease, epidemiology and outcomes, transcriptional profiling, APOL1
In 2010, two common exonic variants within apolipoprotein L1 (APOL1), termed G1 and G2 were associated with greatly increased risk of focal segmental glomerulosclerosis (FSGS) in blacks.1 Subsequent epidemiologic studies have shown that harm associated with these APOL1 risk variants in blacks extends to renal phenotypes beyond FSGS.2–7 This includes nondiabetic CKD, HIV-associated nephropathy, progression to ESRD for patients with lupus nephritis or CKD, and development of poor renal outcomes over time in population controls.
The absence of the endogenous APOL1 gene in laboratory animal species has led to an increased importance of analyzing human data to understand the biologic mechanism underlying APOL1-related renal disease. Earlier work in humans has localized APOL1 to the podocyte in the glomerulus, the proximal tubule epithelium, and the arteriolar endothelium.8 APOL1 renal expression is reduced in subjects with CKD compared with healthy kidneys but is independent of the known risk variants,8 and ApoL1 plasma levels do not associate with kidney disease phenotypes.9 Transplantation studies indicate that the increased risk of allograft loss follows the kidney from the APOL1 risk allele-positive donor.6,10
Studying APOL1-related kidney disease in the Nephrotic Syndrome Study Network (NEPTUNE)11 provides a unique opportunity to derive novel insights about the effect of these risk variants across proteinuric glomerular diseases. We genotyped 90 self-reported black subjects for the G1 and G2 risk alleles and classified these subjects with a low-risk (LR) or high-risk (HR) genotype. We report on the associations with clinical characteristics at enrollment and on clinical outcomes over time. Taking advantage of each subject’s paired molecular and histologic data, we also report on associations with genetic, gene expression, and histologic parameters.
Results
APOL1 Allele and Genotype Frequency in NEPTUNE Subjects
The minor allele frequencies for G1 and G2 were 0.32 and 0.23, respectively (Supplemental Table 1). The two risk allele (HR) genotype was considered any combination of G1 and G2 and was found in 43% of subjects. In a population-based study of blacks, the two risk allele genotype frequency was 13%.12 The APOL1 risk allele frequency in our subjects was significantly elevated in the FSGS and other glomerulopathy cohorts, but not in those with minimal change disease (MCD) or membranous nephropathy (MN) (Table 1).
Table 1.
Frequency of HR genotype in NEPTUNE versus Dallas Heart Study
| Cohort | Frequency of HR Genotype | P Value |
|---|---|---|
| Dallas Heart Study | 0.13 (236/1776) | ref |
| NEPTUNE all | 0.43 (39/90) | <0.01 |
| NEPTUNE FSGS | 0.60 (25/42) | <0.01 |
| NEPTUNE MCD | 0.25 (5/20) | 0.17 |
| NEPTUNE MN | 0.09 (1/11) | 1.00 |
| NEPTUNE other | 0.47 (8/17) | <0.01 |
Proportion of subjects with two risk alleles (HR genotype) in NEPTUNE by histologic diagnosis as compared with a population cohort of blacks (Dallas Heart Study). The P values were determined using Fisher’s exact test. other, other glomerulopathies; ref, reference.
Genetic Model for Association Studies
For all association studies, we used the recessive model of risk inheritance as the primary model: 0 or 1 risk alleles (LR) versus HR genotype.
Monogenic Forms of Steroid-Resistant Nephrotic Syndrome
Causative mutations in known monogenic steroid-resistant nephrotic syndrome genes were not found in any individuals (see Supplemental Table 2 for genes sequenced). The frequency of rare, heterozygous, protein-altering variants in known recessive steroid-resistant nephrotic syndrome genes did not differ by risk genotype.
Subject Genetic Ancestry
Genotype-based ancestry determination using principal component analysis methods showed that 80 of the 82 self-reported black subjects with genome-wide data clustered with the African continental ancestry group of 1000 Genomes Project Subjects (Supplemental Figure 1). The remaining two subjects deviated further from this cluster, indicative of greater admixture.
Baseline Characteristics
Table 2 summarizes selected characteristics of the 90 black subjects stratified by LR versus HR. Treating clinicians of these subjects were not aware of the APOL1 status. There was no significant difference between LR and HR subjects in demographic characteristics, duration of disease before onset, duration of follow-up, administration of immunosuppressive medication, or renin-angiotensin blockade.
Table 2.
Characteristics of 90 black NEPTUNE subjects
| Characteristic | LR (n=51) | HR (n=39) | P Value |
|---|---|---|---|
| Age (y) | 17.0 (12.0–46.5) | 17.0 (14.5–32.5) | 0.80 |
| Pediatric | 27 (53) | 21 (54) | 1.00 |
| Age of onset (y) | 15.5 (10.0–40.8) | 17.0 (14.0–28.2) | 0.60 |
| Duration disease (y) | 1.0 (0.0–3.0) | 0.0 (0.0–1.8) | 0.40 |
| Male | 32 (63) | 25 (64) | 1.00 |
| Histopath | <0.01 | ||
| FSGS | 17 (33) | 25 (64) | |
| MCD | 15 (29) | 5 (13) | |
| MN | 10 (20) | 1 (3) | |
| Other | 9 (18) | 8 (21) | |
| Clinical characteristics at baseline | |||
| RAAS blockade | 14 (27) | 11 (28) | 1.00 |
| eGFR | 83.7 (65.8–105.6) | 67.2 (42.9–84.5) | 0.02 |
| UPC | 2.9 (1.9–7.2) | 3.1 (1.3–6.9) | 0.94 |
Data presented as median (interquartile range) for quantitative variables and as count (%) for categorical characteristics. The P values were determined using Wilcoxon rank-sum test or Fisher’s exact test. Age, patient age at enrollment; Other, other glomerulopathies.
There was a significant difference in the distribution of the histopathologic diagnoses between the HR and LR groups. The largest differences were a higher prevalence of FSGS and a lower prevalence of MN in the HR group. The median eGFR at baseline was significantly lower in the HR group, at 67 versus 84 ml/min per 1.73 m2 (P=0.02).
Of the 90% of subjects providing birth history information, 17% (14/81) were preterm (Supplemental Table 3). Nine of 14 preterm subjects were in the HR group. FSGS was the histologic diagnosis in eight of the nine HR preterm subjects. There was an increased, but not statistically significant, odds of prematurity associated with the HR genotype (3 [95% confidence intervals, 0.8–12.7], P=0.08). There was no significant difference in birthweight between the HR and LR groups, at 2856 versus 3176 g, respectively (P=0.10).
Analysis of Primary Clinical Outcomes
Given its deleterious effects across renal phenotypes, our primary hypothesis was that the HR APOL1 genotype is associated with worse clinical outcomes in all NEPTUNE diagnoses. We therefore studied the full cohort rather than stratifying by histopathologic diagnosis (histopath). We did include histopath as a covariate in our multivariate models. Of note, an interaction term between risk-genotype status and histopath was evaluated in each model. It was not significant in any of the models tested. However, our power calculations suggest we are underpowered to detect interactions (Supplemental Appendix 1).
eGFR
Fitting a multivariable linear regression using generalized estimating equations (GEEs), the HR genotype was significantly associated with a 17 ml/min per 1.73 m2 lower eGFR (P<0.01) (Table 3). For those subjects whose first and last serum creatinine measurements were taken at least 8 months apart, we also analyzed the rate of change in eGFR over time. We computed the slope of the least-squares regression line of eGFR against time for each subject whose first and last serum creatinine levels were obtained at least 8 months apart (n=73). Across these subjects, the eGFR decreased modestly over time (approximately 8 ml/min per y, P<0.01). However, this was not associated with risk genotype in unadjusted or adjusted models (Supplemental Appendix 2, Supplemental Table 4).
Table 3.
Multivariate GEE analysis of eGFR and AP0L1 risk status adjusting for histopath, age, baseline UPC, and RAAS blockade
| Covariate | β | 95% CI | P Value |
|---|---|---|---|
| HR | −17.2 | –29.0 to 5.4 | <0.01 |
| Histopath | |||
| FSGS | ref | — | — |
| MCD | 22.0 | 6.7 to 37.2 | <0.01 |
| MN | 8.5 | –8.9 to 25.9 | 0.34 |
| Other | −1.4 | –15.8 to 13.1 | 0.85 |
| Age (y) | −1.1 | –1.4 to –0.7 | <0.01 |
| UPC baseline | −0.8 | –1.6 to 0.0 | 0.04 |
| RAAS | 2.6 | –3.7 to 8.9 | 0.42 |
Parameter estimates and P values determined using GEEs. Other, other glomerulopathies; Age, patient age at enrollment; ref, reference; 95% CI, 95% confidence interval.
Urine Protein to Creatinine Ratio
A multivariable linear regression using GEEs showed a 52% increase in the urine protein to creatinine ratio (UPC) in HR subjects (P=0.06) (Table 4).
Table 4.
Multivariate GEE analysis of repeated log2 UPC and AP0L1 risk status adjusting for histopath, baseline eGFR, and RAAS blockade
| Covariate | β | 2βa | 95% CI | P Value |
|---|---|---|---|---|
| HR | 0.6 | 1.5 | –0.0 to 1.2 | 0.06 |
| Histopath | ||||
| FSGS | ref | — | — | — |
| MCD | −0.7 | 0.6 | –1.8 to 0.4 | 0.22 |
| MN | 1.3 | 2.5 | 0.7 to 1.9 | <0.01 |
| Other | −0.1 | 0.9 | –0.8 to 0.6 | 0.72 |
| RAAS | −0.6 | 0.7 | –1.2 to –0.1 | 0.03 |
| eGFR baseline | −0.0 | 1.0 | –0.0 to –0.0 | 0.01 |
Parameter estimates and P values determined using GEEs. Other, other glomerulopathies; ref, reference; 95% CI, 95% confidence interval.
2β is the factor by which UPC increases in HR subjects compared with LR subjects. For example, if 2β is 1.5, the average UPC in HR subjects is 1.5 times that in LR subjects.
Complete Remission
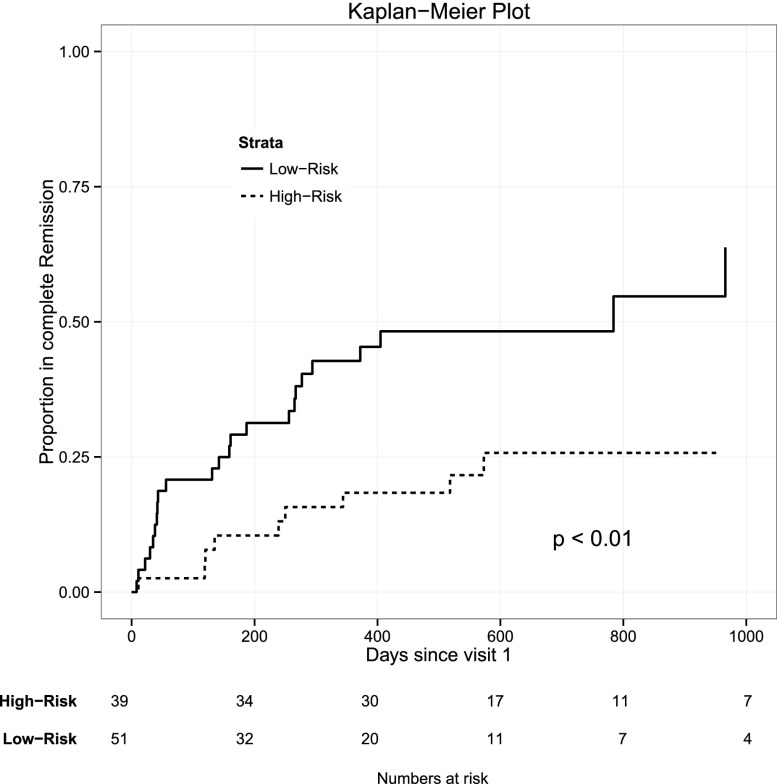
Of the subjects, 49% of LR versus 23% of HR subjects achieved complete remission (CR) of proteinuria (P=0.02) (Supplemental Table 5a). Additionally, risk genotype was a significant predictor of time to first CR in an unadjusted Cox proportional hazards model (Figure 1). This remained significant after adjusting for age, histopathologic diagnosis, and baseline eGFR and UPC (hazard ratio, 0.31; 95% confidence interval, 0.14 to 0.68; P<0.01) (Supplemental Table 5b).
Figure 1.
Unadjusted Kaplan–Meier plot of proportion of CR stratified by APOL1 risk status. Plot is truncated at day 1000. P value determined with an unadjusted Cox proportional hazards model of CR and APOL1 risk status.
A priori calculations also suggested that we were underpowered for stratified analyses (Supplemental Appendix 1). However, to explore clinical outcomes associated with the HR genotype for each histopath, we performed stratified analyses. These are presented in Supplemental Appendix 2, emphasizing that the results should be interpreted cautiously.
Histomorphometric Differences between LR and HR Status
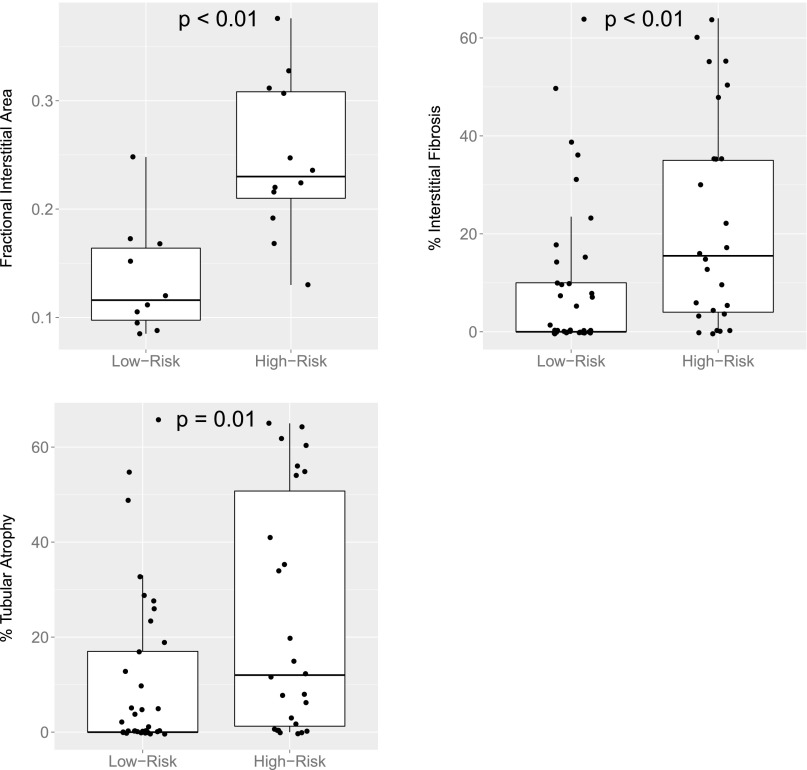
Average glomerular volume, fractional interstitial area, and cortical glomerular density were measured in the 22 subjects with available slides (LR: n=11, HR: n=12). Additionally, the percentages of interstitial fibrosis (IF) and tubular atrophy (TA) were measured in 63 subjects. Clinical characteristics of these subgroups are provided in Supplemental Table 6. Although average glomerular volume and cortical glomerular density did not show significant association with genotype, those with HR genotypes had a significantly increased percentage of fractional interstitial area (25% versus 13%, P<0.01) (Figure 2, Supplemental Table 7a). Percentages of IF and TA were elevated in HR subjects (Supplemental Table 7b). Average IF was 9.4% in LR subjects versus 22.4% in the HR group (P<0.01). Similarly, average TA was 10.5% versus 23.6% in the LR and HR groups, respectively (P=0.01).
Figure 2.
Fractional interstitial area, percentage IF, and percentage TA by APOL1 risk status. P values determined using Wilcoxon rank-sum test.
Glomerular and Tubulointerstitial Gene Expression
Intrarenal gene expression studies were performed using manually dissected glomeruli (GLOM) and tubulointerstitium (TI) from 44 and 55 subjects, respectively. These sample sizes reflect the biopsy samples that were available for expression analysis and which passed subsequent quality control measures at the time of study. Availability of samples was not biased toward any demographic or clinical metrics (Supplemental Table 6). As seen in the parent cohort, those in the HR subgroup had point estimates that were lower for eGFR and higher for percentage of patients that were FSGS. For both differential expression and correlation analyses, we first applied a variance filter to our gene set, reducing the pool of genes for testing to 3796 in GLOM and 3761 in TI (see the methods section for details).
APOL1 Differential Expression
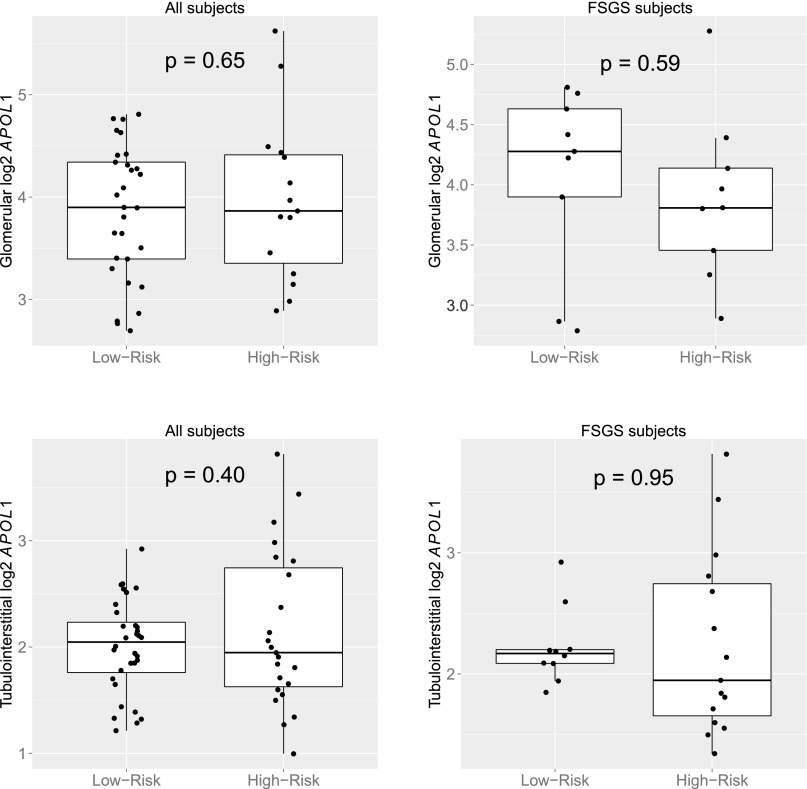
APOL1 HR status was not significantly associated with APOL1 expression in the GLOM (fold change, 1.0; P=0.65) or TI (fold change, 1.1; P=0.40) (Figure 3).
Figure 3.
log2 APOL1 expression by genotype in glomerulus and TI. P values determined with independent two-sample t test.
The increased IF and TA in the HR subjects raised the question of whether the association between TI APOL1 expression and risk genotype is confounded by differential degree of TI injury. IF and TA were available for 42 genotyped TI samples. There was no significant association between TI APOL1 expression and IF or TA (Supplemental Appendix 3).
There was no analogous measure of glomerular damage, however, as detailed in the methods section, only GLOM with open Bowman’s space are isolated across samples. Even when only studying subjects with FSGS, there were no significant differences in GLOM APOL1 expression between the HR and LR groups (Figure 3).
Genome-Wide Differential Expression
Three GLOM and two TI genes had significantly higher expression levels in HR subjects (FDR<0.05) (Tables 5 and 6). The most significantly differentially expressed gene was CXCL9 in GLOM, whose average expression in HR subjects was 4.1 times that of LR subjects. Expression was also higher in GLOM for another member of the CXCL family, CXCL11, and the proteosomal degradation pathway gene, ubiquitin D (UBD) also known as FAT10. Small nucleolar RNA 14B and mucin 13 (MUC13) transcript levels were significantly higher in the HR TI compartment. Quantile-quantile plots, volcano plots, and boxplots of expression by risk genotype for significant genes are provided in Supplemental Appendix 5.
Table 5.
Differentially expressed genes in glomerulus
| Gene | Fold Change | SAM Score | q Value |
|---|---|---|---|
| CXCL9 | 4.1 | 2.9 | 0 |
| CXCL11 | 2.7 | 2.4 | 0 |
| UBD | 3.0 | 2.4 | 0 |
SAM Score, T statistic value computed using Significance Analysis of Microarrays q value, lowest false discovery rate at which the gene is called significant.
Table 6.
Differentially expressed genes in TI
| Gene | Fold Change | SAM Score | q Value |
|---|---|---|---|
| MUG13 | 2.3 | 1.9 | 0 |
| SNORA14B | 1.9 | 1.9 | 0 |
SNORA14B, small nucleolar RNA 14B; SAM Score, T statistic value computed using Significance Analysis of Microarrays; q value, lowest false discovery rate at which the gene is called significant.
APOL1 Coregulation Analysis
Concordant changes in mRNA transcript levels can identify gene sets that underlay a unified regulatory mechanism and function. In fact, mRNA coexpression patterns can result from common upstream regulatory events that determine the transcriptional response in the functional context of disease.13–15 Although our data indicate that APOL1 risk genotypes do not influence the magnitude of its expression, we hypothesized that these exonic variants alter the function of the APOL1 protein and result in changes in the expression of other genes.
Identifying sets of genes whose expression is differentially correlated with HR versus LR APOL1 could help elucidate the biologic mechanism by which APOL1 risk variants contribute to disease. To model coexpression of genes with the HR and LR APOL1 transcripts, we used a Weighted Gene Correlation Network Analysis (WGCNA).16 In the WGCNA, a gene expression network is constructed using coexpression relationships in a weighted manner. Distinct coregulated gene sets (modules) are identified within this network. The genes contained in these modules can then be explored for biologic concepts. We used the WGCNA in TI to identify the coexpression network of the HR APOL1 transcript and compared it with that of the LR transcript. For this analysis, we adjusted for age, sex, and histologic diagnosis.
In the analysis using TI data from only HR subjects, the APOL1 transcript was part of a coexpression module containing 579 genes. This module’s expression was positively correlated with tissue injury (IF: r=0.43, P=0.04; TA: r=0.42, P=0.05). When analyzing LR subjects, TI APOL1 was part of a 681-gene module. Although the point estimate showed a positive correlation of lower magnitude with IF and TA, neither was statistically significant (IF: r=0.33, P=0.06; TA: r=0.3, P=0.09).
We next evaluated molecular pathways, concepts, and upstream regulators associated with the gene modules containing TI APOL1 from the HR and LR group analyses. Both HR and LR modules had overlapping canonical pathways and predicted upstream regulators and many innate immunity, adaptive immunity, and immune costimulation pathways. However, the two modules also had a substantial set of distinct pathways and regulators (Supplemental Figure 2, Supplemental Table 8). For instance, the HR module was specifically enriched for the pathways of role of Jak family kinases in IL-6-type cytokine signaling, ILK signaling, and JAK/Stat signaling.
Discussion
Through its longitudinal design and collection of detailed and diverse data and biosamples, the NEPTUNE provided a unique laboratory to discover molecular mechanisms underlying the pathogenicity of APOL1 in human subjects with proteinuric glomerular diseases. In addition, we used this unique cohort of deeply phenotyped individuals to more fully plumb the depths of the clinical correlates of this risk genotype.
This study was designed to test the hypothesis that, independent of classic histopath (e.g., MCD, FSGS, MN), the HR genotype resulted in worse phenotypic outcomes and a distinct intrarenal gene expression profile. In fact, we demonstrated that, in our combined cohort of 90 subjects, the HR genotype was associated with a lower eGFR and a lower likelihood of attaining CR. In multivariable analyses, these differences were not attributable to differences in age, sex, duration of disease, or duration of follow-up time.
Although not the primary question of this study, a distinct and interesting question is whether the HR genotype had a differential effect, depending on specific histopathologic diagnoses. Although underpowered, we performed this stratified analysis for eGFR, UPC, and CR. In regard to eGFR and CR, these exploratory analyses showed the same direction of effect associated with the HR genotype in FSGS, MCD, and other strata. For UPC, subjects with the HR genotype and MCD had a lower UPC than the other groups. In other cohorts, or as NEPTUNE expands its enrollment, future studies with adequate power can be specifically designed to study the HR genotype in distinct glomerular conditions.
The results of the primary analyses, and the suggestive support of stratified analyses in terms of direction of effect, support the concept that the HR genotype confers risk across histopathologic diagnoses. Even if HR genotype frequency is not enriched in MCD, MN, or other glomerulopathies, those who have it may represent a more vulnerable population. This seems to be an important area of further research because the field seeks to determine those populations and renal phenotypes in which there is clinical utility in ascertaining the APOL1 genotype.
Other studies have found that HR subjects have a more rapid decline in eGFR compared with those without it.3 In regression analysis of the rate of change in eGFR adjusted for its baseline levels, we did not observe this pattern. It may be that insufficient time had passed at time of analysis to discern differences in eGFR trajectory between groups. To sort this out, it will be necessary to continue to follow this over time in the NEPTUNE and in other clinical studies with similar intensity of follow-up.
HR subjects had significantly more IF and TA on biopsy at enrollment. Worse histologic metrics of renal tubulointerstitial disease with the HR genotype were also recently reported in a group of blacks with non-nephrotic CKD.17 Increased TI damage may, in part, explain the eGFR differences observed. The lack of difference in prior duration of disease implies that, from the outset, an HR kidney’s response to challenge or injury is accompanied by an increased inflammatory response. There were no quantitative differences in glomerular morphometry at the time of initial biopsy. It will be important to discern whether glomerular histomorphometric differences are present at baseline with effect sizes too small to detect at this sample size or if they only become apparent later in the disease course.
The HR genotype did not alter APOL1 steady-state mRNA levels in either compartment. A previous report showed that the HR genotype is not associated with circulating APOL1 protein levels either.9 Together, this provides additional support for the concept that the G1 and G2 genotypes of APOL1 result in damage by a mechanism other than altering their overall expression. Potential mechanisms could be through altering the protein’s overall conformation or the structure of critical epitopes.
CXCL9, a leukocyte secreted chemokine induced by IFN-γ, showed the highest risk genotype-specific expression difference in GLOM, with a fold change of 4.1 in HR versus LR subjects. Along with CXCL11 (also with higher expression in the HR subjects), this chemokine mediates leukocyte migration to areas of tissue damage and has been described in intrinsic renal cell lineages.18 UBD was significantly increased in GLOM of HR subjects. Upregulated by INF-γ and TNF-α, UBD targets proteins for proteasomal degradation by a ubiquitin-independent mechanism19,20 and can induce apoptosis.21 In addition to its role in innate immunity, the function of APOL1 as a proapoptotic and autophagic cell death gene has been described.22 It is possible that increased UBD-mediated proteasomal degradation is one mechanism that could link an aggravated immune response in HR APOL1 subjects with downstream renal damage. MUC13, a transmembrane mucin gene, was more highly expressed in TI. MUC13 has a role in protection of epithelial cells from pathogen infection and also functions in cell signaling.23
Correlated expression levels among genes in a disease- and compartment-specific manner have been used to identify functional relationships, including physical interactions and regulatory mechanisms.24 For the HR and LR genotypes separately and independent of histologic diagnosis, we found a single coexpression module of genes that included the expression of APOL1 in TI. The HR module was significantly correlated with increased tissue injury. This gene set’s functions were then analyzed in silico, revealing enrichment of specific immune canonical pathways. We also identified potential regulators of these HR correlated genes. These findings fit in well with emerging concepts of drivers of APOL1-mediated toxicity across renal phenotypes,25,26 while also providing new molecular targets for further investigation.
Through differential and intercorrelation analyses, we found sets of intrarenal transcripts associated with the HR genotype. Our studies do have several limitations. We showed that some of these transcripts, such as MUC13, are associated with the HR genotype, independent of tissue injury. However, not all coexpressed or differentially expressed genes will be mediators of the damage caused by the HR genotype. Alternatively, the increased tissue injury associated with the HR genotype may itself result in differential regulation or coexpression of sets of transcripts. Whether identified as potential mediators of the association between genotype and tissue damage or a response to the HR genotype-mediated tissue damage, each of these transcripts can serve to illuminate important molecular mechanisms involved in the renal damage associated with APOL1. In addition, the HR gene models identified by the WGCNA will require additional empirical confirmation. For example, protein-based assays and functional studies in model systems will be essential to validate the associations that we have observed. This vital step will also allow to more confidently delineate those causal pathways in these relationships.
Our study also did not have a measurement of tissue injury from each patient in whom expression analyses were performed, which prevented us from adjusting for tissue injury in each subject when relating HR genotype to gene expression. As the NEPTUNE cohort matures, each subject will have morphologic descriptors of tissue injury measured, which will allow us to account for this specific covariate in our models. Finally, we were underpowered to stratify by standard pathologic diagnosis. However, as the cohort expands, we will be empowered to perform expression analyses stratified by conventional diagnoses.
Using integrative genomics, we have shown that individuals with the HR APOL1 genotype enrolled in the NEPTUNE study present, from a clinical and histologic perspective, with more advanced disease and have less remission of proteinuria over time. This appears to be independent of histopath. If independently confirmed, these observations may imply that defining a patient’s APOL1 genotype could allow better stratification of subjects for trials which could eventually result in more precise and improved clinical management. Finally, transcriptomic data suggest that intrarenal APOL1 functions in the context of pathways of innate immunity and inflammation. It appears that genotype-associated expression differences reflect an aggravated inflammatory response to a challenge rather than activation of a de novo APOL1-specific program. Combining these emerging human tissue–based hypotheses with further longitudinal analyses and investigations of candidate molecules in experimental model systems should aid in defining the causal mechanism of APOL1.
Concise Methods
Study Participants
There were 92 subjects of self-reported black ancestry from the NEPTUNE. Two subjects did not give consent for genetic analyses and were not included in the genetic and transcriptomic analyses. The design of the NEPTUNE has been previously described.11 Subjects were recruited at the time of their first clinically indicated biopsy for suspicion of primary proteinuric glomerular disease. Study pathologists classified subjects into MCD, FSGS, MN, and other glomerulopathies histopathologic groups as previously defined.11
APOL1 Genotyping
The APOL1 SNPs corresponding to G1 (rs73885319 and rs60910145) and the G2 indel (rs71785313) were genotyped using Sanger sequencing of the last 250 bases of exon 7. Individuals were classified as having 0, 1, or 2 risk genotypes. The rare G+ allele,7 with rs73885319 being variant and rs60910145 being reference, was classified as a risk allele. There was no distinction made between G1 or G2 in subjects with 1 or 2 risk genotypes.
Mendelian FSGS Gene Sequencing
Using established protocols and gene targets, targeted multiplex PCR paired with next-generation sequencing27,28 was used to determine if blacks had rare, deleterious alleles in 20 genes known to cause Mendelian forms of FSGS. A deleterious allele was defined as meeting the following three criteria: (1) allele frequency <0.1% for dominant genes or <0.5% for recessive genes, (2) present in <5% of our cohort, and (3) a PolyPhen229 score of damaging. The R229Q polymorphism of podocin was also considered in compound heterozygosity with rare podocin mutations.30
Clinical Characteristics at Presentation
Data are summarized as mean±SD. Fisher’s exact test was used to compare count characteristics of individuals with the LR versus HR genotypes. For numerical variables, Wilcoxon rank-sum tests were used to test for differences in distributions between the two groups. Fisher’s exact test was used to assess the association between risk genotype and preterm status.
Longitudinal Analysis of eGFR and UPC
eGFR was calculated using the Schwartz formula31 for the pediatric population and the Chronic Kidney Disease Epidemiology Collaboration equation32 for adults. UPC was calculated from a random collection on a milligram/milligram ratio.
Two generalized estimating equation linear regressions were fit to assess the association of eGFR and UPC with risk genotype over the duration of subjects’ follow-up. Covariates to include in the models were chosen on the basis of a priori determination of clinically meaningful factors. The GEE analysis of eGFR was adjusted for histopath, age, baseline UPC, and use of renin-angiotensin aldosterone system (RAAS) blocker. The GEE analysis of UPC was adjusted for histopath, RAAS blocker, and baseline eGFR. In addition to fitting for marginal effect models, we also explicitly tested for interaction between histopath and risk genotype in both.
An analysis of the effect of risk genotypes on the rate of decline in eGFR over time was also performed. First, the slope of the least-squares regression line of eGFR versus time was calculated in all subjects whose first and last serum creatinine levels were measured at least 8 months apart. Then, two linear regressions of the eGFR slope by risk genotype were fit: one adjusting for baseline eGFR, age, baseline UPC, RAAS blocker, and histopath and another adjusting for baseline eGFR only.
Time to Event Analysis of CR
A Cox proportional hazards model was used to determine association between time to first CR of proteinuria and APOL1 risk genotype, adjusting for cohort and baseline age, UPC, and eGFR. Model selection was performed using the same approach as previously described.
Power and Stratified Analyses
Analogous analyses of eGFR, UPC, and CR were also performed stratified by histologic cohort. Because of the small sample sizes, stratified longitudinal models were considered exploratory in nature. Simplified a priori power calculations for both combined and stratified analyses were performed. Details of the methods used to estimate power are described in Supplemental Appendix 1, and the results are presented in Supplemental Appendix 2.
Histomorphometric Measurement
During annotation of biopsies, individual GLOM were identified throughout all sections in which they appeared. Measurements were made using the SlidePath platform (Leica Microsystems, Dublin) by a single individual (K.L.). Average glomerular tuft area was determined in patients with at least four GLOM represented. Tuft profiles were chosen randomly (uniform random generator) from all of the sections in which a particular glomerulus was sectioned. A built-in planimetry program in SlidePath was used for all area measurements. Cortical glomerular density was determined in a single section, usually one near the middle of the stack of slides, usually with periodic acid–Schiff staining. The number of patent tufts in the section was counted and was divided by the cortical area (as determined by planimetry) to give the density. Fractional interstitial area was measured by imposing a 5×5 grid in five different places over the cortex. The definition of the structure on which the 25 points of the grid fell (excluding the upper and right lines) was interstitium (including matrix and cells), blood vessels (capillaries, veins/venules, arteries, arterioles), intact tubules, atrophic tubules, patent GLOM, or globally sclerotic GLOM. This was done on one section each stained with periodic acid–Schiff and with Masson’s trichrome stain. Less than 125 points per stained section were rarely counted, if the amount of tissue was insufficient for five distinct cortical locations to be chosen.
Quantitation of IF and TA
Digital images of the biopsies were examined for scoring by pathologists, and for each biopsy whole-slide images of glass slides were scanned using Aperio technology at 40× for light microscopy.33 The degree of cortical IF and TA was scored as the percentage of cortex by each reader, and the average of the scores from at least two readers was used.
Gene Expression
RNA Preparation
Renal biopsy–derived gene expression data were collected and analyzed from subjects who had APOL1 genotype data available. There were 44 subjects with glomerular (LR: n=29; HR: n=15), and 55 subjects with tubulointerstitial transcriptomic data (LR: n=32; HR: n=23). The renal biopsy sample was manually dissected and isolated into tubulointerstitial and glomerular compartments per established protocol.34,35
In terms of glomerular injury, manual glomerular microdissection captures GLOM with an open Bowman’s space. Therefore, across samples, our GLOM expression studies are enriched for the transcriptomes of functioning GLOM, rather than for those that are globally sclerosed. RNA was purified according to the manufacturer’s instructions.
Microarray Expression
Renal biopsy specimens were processed and analyzed using Affymetrix 2.1 ST chip as previously described.34,35 Gene expression was normalized and quantitated on a probeset level as previously described.36
The 44 GLOM and 55 TI expression datasets generated are being uploaded to the Gene Expression Omnibus (reference series GSE68127). These data are stratified by APOL1 risk genotype status and are available at the time of publication. All analyses were performed using R software.
APOL1 Expression by Risk Genotype
Using a t test, we compared microarray-based gene-level expression of APOL1 between individuals with the LR versus HR genotypes. Expression levels were log2 transformed.
Genome-wide Expression by Risk Genotype
To identify differentially expressed genes in HR versus LR cohorts, we first calculated the variance of expression levels for all probesets on the array. This was done in a GLOM- and TI-specific manner. We subsequently restricted the set of transcripts analyzed to those corresponding to the 15% of probesets with the largest amount of variation in expression levels across samples. This resulted in 4012 transcripts. Differential expression analysis was then performed on these transcripts using Significance Analysis of Microarrays methods37 to properly account for multiple testing and correlation among genes. In this approach, permutations of the observed data are used to determine the expected distribution of test statistics under the assumption of no association between expression and APOL1 genotype. Then a significance threshold is empirically determined such that FDR is less than a desired value. In this framework, FDR is defined as the median number of genes called significant across the permutations divided by the number of genes called significant in the observed data. Extensive documentation of the statistical methods and computational tools used are provided in the Significance Analysis of Microarrays user guide (http://statweb.stanford.edu/~tibs/SAM/sam.pdf) in addition to the cited article.37 The analysis underwent 1000 permutations, and an FDR threshold of 0.05 was used to determine statistical significance.
Power calculations for this analysis were performed. For genes with a pooled SD≤0.9, we had an estimated 80% power to detect fold changes of 3.2 and 2.7 in glomerular and tubular expression, respectively (Supplemental Appendix 4).
APOL1-correlated Expression
Identifying genes coexpressed with APOL1 and performing functional annotation can aid in illuminating the mechanism of this molecule in the LR and HR genotypes.
After adjusting gene expression values for age, sex, and histopath, and again restricting our analyses to probesets with the most variation across samples, we used the published method weighted gene correlation analysis and its corresponding R package WGCNA.16
For the HR transcript analysis, the following parameters were used: (1) sample cut height=83, (2) soft thresholding parameter=12, and (3) merge module cut height=0. The LR transcript analysis parameters are as follows: (1) sample cut height=95, (2) soft thresholding parameter=7, and (3) merge module cut height=0. On visual inspection, none of the samples in the HR group for GLOM or TI appeared to be a substantial outlier. Therefore, the sample cut height was set to be just larger than the maximum height observed in the dendogram. Meanwhile, both dendograms for GLOM and TI expression in the LR group demonstrated one subject that was significantly separated from other samples. The selected cut height of 95 resulted in these samples being excluded from the analysis. The soft thresholding parameter for each analysis was chosen to achieve a scale free topology model fit (R2) ≥0.9 and such that a reasonable number of distinct gene modules were identified from the gene network (between 8 and 18). A merge module cut height of 0 was chosen to preserve the modules identified using the Dynamic Tree Cut (see WGCNA documentation for more details http://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA).
Using this method, a gene expression network was constructed in a manner designed to optimize biologic relevance, and then distinct gene sets were identified within the network. Association between the resulting gene sets and other traits were then tested using the eigengene of each gene set.
Functional Analysis
Gene sets emerging from correlation analysis using WGCNA were uploaded to Ingenuity Pathway Analysis (QIAGEN, Redwood City, CA) for functional annotation.
Statistical Analyses
All analyses were performed using R software. Additionally, two-sided tests of hypotheses were used, and P<0.05 was the criterion for statistical significance unless otherwise specified.
Disclosures
None
Supplementary Material
Acknowledgments
M.S. is a Carl Gottschalk Research Scholar of the American Society of Nephrology and is supported by the Charles Woodson Clinical Research Fund and by 1K08-DK100662-01 and M.K. is supported by U54-DK083912 Nephrotic Syndrome Study Network Consortium and P30-DK081943 George M. O’Brien Kidney Research Core Center at the University of Michigan. The Nephrotic Syndrome Study Network Consortium (NEPTUNE), U54-DK-083912, is a part of the National Center for Advancing Translational Sciences (NCATS) Rare Disease Clinical Research Network (RDCRN), supported through a collaboration between the Office of Rare Diseases Research (ORDR), NCATS, and the National Institute of Diabetes, Digestive, and Kidney Diseases. RDCRN is an initiative of ORDR, NCATS. Additional funding and/or programmatic support for this project has also been provided by the University of Michigan, NephCure Kidney International, and the Halpin Foundation.
Parts of this study were presented at the 2014 American Society of Nephrology Annual Meeting, November 13–16, in Philadelphia, PA.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014111131/-/DCSupplemental.
References
- 1.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito K, Bick AG, Flannick J, Friedman DJ, Genovese G, Parfenov MG, Depalma SR, Gupta N, Gabriel SB, Taylor HA, Jr, Fox ER, Newton-Cheh C, Kathiresan S, Hirschhorn JN, Altshuler DM, Pollak MR, Wilson JG, Seidman JG, Seidman C: Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ Res 114: 845–850, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, Fink JC, Anderson AH, Choi MJ, Wright JT, Jr, Lash JP, Freedman BI, Ojo A, Winkler CA, Raj DS, Kopp JB, He J, Jensvold NG, Tao K, Lipkowitz MS, Appel LJ, AASK Study Investigators. CRIC Study Investigators : APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 369: 2183–2196, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzur S, Rosset S, Skorecki K, Wasser WG: APOL1 allelic variants are associated with lower age of dialysis initiation and thereby increased dialysis vintage in African and Hispanic Americans with non-diabetic end-stage kidney disease. Nephrol Dial Transplant 27: 1498–1505, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reeves-Daniel AM, DePalma JA, Bleyer AJ, Rocco MV, Murea M, Adams PL, Langefeld CD, Bowden DW, Hicks PJ, Stratta RJ, Lin JJ, Kiger DF, Gautreaux MD, Divers J, Freedman BI: The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant 11: 1025–1030, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA: APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22: 2129–2137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madhavan SM, O’Toole JF, Konieczkowski M, Ganesan S, Bruggeman LA, Sedor JR: APOL1 localization in normal kidney and nondiabetic kidney disease. J Am Soc Nephrol 22: 2119–2128, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruggeman LA, O’Toole JF, Ross MD, Madhavan SM, Smurzynski M, Wu K, Bosch RJ, Gupta S, Pollak MR, Sedor JR, Kalayjian RC: Plasma apolipoprotein L1 levels do not correlate with CKD. J Am Soc Nephrol 25: 634–644, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee BT, Kumar V, Williams TA, Abdi R, Bernhardy A, Dyer C, Conte S, Genovese G, Ross MD, Friedman DJ, Gaston R, Milford E, Pollak MR, Chandraker A: The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant 12: 1924–1928, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadegbeku CA, Gipson DS, Holzman LB, Ojo AO, Song PX, Barisoni L, Sampson MG, Kopp JB, Lemley KV, Nelson PJ, Lienczewski CC, Adler SG, Appel GB, Cattran DC, Choi MJ, Contreras G, Dell KM, Fervenza FC, Gibson KL, Greenbaum LA, Hernandez JD, Hewitt SM, Hingorani SR, Hladunewich M, Hogan MC, Hogan SL, Kaskel FJ, Lieske JC, Meyers KE, Nachman PH, Nast CC, Neu AM, Reich HN, Sedor JR, Sethna CB, Trachtman H, Tuttle KR, Zhdanova O, Zilleruelo GE, Kretzler M: Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int 83: 749–756, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR: Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol 22: 2098–2105, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fessele S, Maier H, Zischek C, Nelson PJ, Werner T: Regulatory context is a crucial part of gene function. Trends Genet 18: 60–63, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Martini S, Nair V, Patel SR, Eichinger F, Nelson RG, Weil EJ, Pezzolesi MG, Krolewski AS, Randolph A, Keller BJ, Werner T, Kretzler M: From single nucleotide polymorphism to transcriptional mechanism: a model for FRMD3 in diabetic nephropathy. Diabetes 62: 2605–2612, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martini S, Nair V, Keller BJ, Eichinger F, Hawkins JJ, Randolph A, Böger CA, Gadegbeku CA, Fox CS, Cohen CD, Kretzler M, European Renal cDNA Bank. C-PROBE Cohort. CKDGen Consortium : Integrative biology identifies shared transcriptional networks in CKD. J Am Soc Nephrol 25: 2559–2572, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langfelder P, Horvath S: WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen CP, Beggs ML, Saeed M, Ambruzs JM, Cossey LN, Messias NC, Walker PD, Freedman BI: Histopathologic findings associated with APOL1 risk variants in chronic kidney disease. Mod Pathol 28: 95–102, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romagnani P, Lazzeri E, Lasagni L, Mavilia C, Beltrame C, Francalanci M, Rotondi M, Annunziato F, Maurenzig L, Cosmi L, Galli G, Salvadori M, Maggi E, Serio M: IP-10 and Mig production by glomerular cells in human proliferative glomerulonephritis and regulation by nitric oxide. J Am Soc Nephrol 13: 53–64, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G: FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol Cell Biol 25: 3483–3491, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raasi S, Schmidtke G, de Giuli R, Groettrup M: A ubiquitin-like protein which is synergistically inducible by interferon-gamma and tumor necrosis factor-alpha. Eur J Immunol 29: 4030–4036, 1999 [DOI] [PubMed] [Google Scholar]
- 21.Raasi S, Schmidtke G, Groettrup M: The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J Biol Chem 276: 35334–35343, 2001 [DOI] [PubMed] [Google Scholar]
- 22.Wan G, Zhaorigetu S, Liu Z, Kaini R, Jiang Z, Hu CA: Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J Biol Chem 283: 21540–21549, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maher DM, Gupta BK, Nagata S, Jaggi M, Chauhan SC: Mucin 13: Structure, function, and potential roles in cancer pathogenesis. Mol Cancer Res 9: 531–537, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95: 14863–14868, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith EE, Malik HS: The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res 19: 850–858, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols B, Jog P, Lee JH, Blackler D, Wilmot M, D’Agati V, Markowitz G, Kopp JB, Alper SL, Pollack MR, Friedman DJ: Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int 87: 332–342, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halbritter J, Diaz K, Chaki M, Porath JD, Tarrier B, Fu C, Innis JL, Allen SJ, Lyons RH, Stefanidis CJ, Omran H, Soliman NA, Otto EA: High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet 49: 756–767, 2012 [DOI] [PubMed] [Google Scholar]
- 28.Lovric S, Fang H, Vega-Warner V, Sadowski CE, Gee HY, Halbritter J, Ashraf S, Saisawat P, Soliman NA, Kari JA, Otto EA, Hildebrandt F, Nephrotic Syndrome Study Group : Rapid detection of monogenic causes of childhood-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 9: 1109–1116, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR: A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukaguchi H, Sudhakar A, Le TC, Nguyen T, Yao J, Schwimmer JA, Schachter AD, Poch E, Abreu PF, Appel GB, Pereira AB, Kalluri R, Pollak MR: NPHS2 mutations in late-onset focal segmental glomerulosclerosis: R229Q is a common disease-associated allele. J Clin Invest 110: 1659–1666, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz GJ, Muñoz A, Schneider MF, Mak RH, Kaskel F, Warady BA, Furth SL: New equations to estimate GFR in children with CKD. J Am Soc Nephrol 20: 629–637, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J, CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barisoni L, Nast CC, Jennette JC, Hodgin JB, Herzenberg AM, Lemley KV, Conway CM, Kopp JB, Kretzler M, Lienczewski C, Avila-Casado C, Bagnasco S, Sethi S, Tomaszewski J, Gasim AH, Hewitt SM: Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE). Clin J Am Soc Nephrol 8: 1449–1459, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen CD, Frach K, Schlöndorff D, Kretzler M: Quantitative gene expression analysis in renal biopsies: A novel protocol for a high-throughput multicenter application. Kidney Int 61: 133–140, 2002 [DOI] [PubMed] [Google Scholar]
- 35.Cohen CD, Klingenhoff A, Boucherot A, Nitsche A, Henger A, Brunner B, Schmid H, Merkle M, Saleem MA, Koller KP, Werner T, Gröne HJ, Nelson PJ, Kretzler M: Comparative promoter analysis allows de novo identification of specialized cell junction-associated proteins. Proc Natl Acad Sci U S A 103: 5682–5687, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lockstone HE: Exon array data analysis using Affymetrix power tools and R statistical software. Brief Bioinform 12: 634–644, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tusher VG, Tibshirani R, Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.