Abstract
There is a growing interest in the photosynthetic carbon fixation by microalgae for the production of valuable products from carbon dioxide (CO2). Microalgae are capable of transporting bicarbonate (HCO3−), the most abundant form of inorganic carbon species in the water, as a source of CO2 for photosynthesis. Despite the importance of HCO3− as the carbon source, little is known about the chemotactic response of microalgae to HCO3−. Here, we showed the chemotaxis of a model alga, Chlamydomonas reinhardtii, towards HCO3− using an agarose gel-based microfluidic device with a flow-free and stable chemical gradient during the entire assay period. The device was validated by analyzing the chemotactic responses of C. reinhardtii to the previously known chemoattractants (NH4Cl and CoCl2) and chemotactically neutral molecule (NaCl). We found that C. reinhardtii exhibited the strongest chemotactic response to bicarbonate at the concentration of 26 mM in a microfluidic device. The chemotactic response to bicarbonate showed a circadian rhythm with a peak during the dark period and a valley during the light period. We also observed the changes in the chemotaxis to bicarbonate by an inhibitor of bicarbonate transporters and a mutation in CIA5, a transcriptional regulator of carbon concentrating mechanism, indicating the relationship between chemotaxis to bicarbonate and inorganic carbon metabolism in C. reinhardtii. To the best of our knowledge, this is the first report of the chemotaxis of C. reinhardtii towards HCO3−, which contributes to the understanding of the physiological role of the chemotaxis to bicarbonate and its relevance to inorganic carbon utilization.
I. INTRODUCTION
Microalgae, the aquatic photosynthetic microorganisms, have recently garnered a lot of interest for their potential to produce valuable products, such as feedstocks of biofuels1,2 and chemicals3 from CO2. They are recognized as a sustainable solution to the global warming and energy security caused by the consumption of fossil fuels. Despite the potential, it is still required to improve photosynthetic productivity for the economic viability of microalgal production systems.4 A deeper understanding of algal physiology is crucial for the realization of biotechnological application of microalgae.5,6 There have been a number of studies on the physiology related to photosynthesis,7 circadian rhythm,8 and tactic responses9–12 using a unicellular motile green alga, Chlamydomonas reinhardtii, as a model organism of microalgae.
One of the attractive properties of microalgae is the ability to take up inorganic carbon (Ci, CO2, and HCO3−) for the photosynthetic conversion into organic carbon,13 which accounts for a large portion of global biomass production.14 Due to the variability of dissolved Ci level and 10 000-fold lower diffusion rate of CO2 in aquatic conditions compared to atmospheric conditions, most microalgae developed the carbon concentrating mechanism (CCM) for acclimation to CO2-limiting conditions with active Ci uptake systems to accumulate an intracellular Ci pool.13–16 Under neutral to alkaline pH conditions, HCO3− is the predominant form of Ci species in the water.15 Because HCO3− is 1000-fold less permeable to lipid membranes relative to CO2, internal Ci is accumulated in the form of HCO3− to prevent unintended efflux of Ci with the assistance of various carbonic anhydrases (CA) and HCO3− transporters.14,16
Chemotaxis is the directional migration of organisms in response to the gradient of chemical stimuli including nutrient sources17 and toxic compounds.18 The organisms move towards a favorable chemical (positive chemotaxis) or away from an unfavorable chemical (negative chemotaxis). As a consequence, the organisms are able to find optimal conditions for their survival. In this context, chemotaxis may provide a significant fitness advantage.19 Since modern studies of bacterial chemotaxis,20,21 the chemotactic responses of bacteria to a wide range of chemical substances, such as sugars,17 amino acids,22 and signaling molecules,23 have been demonstrated. Chemotaxis of C. reinhardtii has been studied during the past few decades as well, but relatively less information is available for the chemotactic response of C. reinhardtii compared with other organisms (e.g., Escherichia coli). Till date, the chemotactic responses in this microalga have been investigated using a limited number of chemicals, including ammonium (NH4+) ion.8,10,12 Furthermore, the chemotaxis of C. reinhardtii to several chemicals (e.g., l-arginine and CoCl2) is disputable.10–12 Although HCO3− is an important inorganic carbon source in C. reinhardtii, it is not known that HCO3− triggers a chemotactic response of C. reinhardtii.24
In the chemotaxis assay, the traditional methods, including swarm plate assay20 and capillary assay,21 have been developed and extensively exploited in conventional biology laboratories. However, these approaches are unsuitable for an accurate analysis of chemotaxis, because it is difficult to obtain quantitative information on the extent of chemotactic migration and stable concentration gradient of chemicals.25–27 To overcome the shortcomings of the classical methods, various microfluidic systems have been recently developed.26,27 There are a number of advantages in the microfluidic assays over the conventional methods. Since microfluidics offers unprecedented level of controllability and stability, a precise manipulation of the fluidic properties, including the flow rate and the concentration, is possible.23,25,26 Furthermore, the microscale approach enables the behavior analysis of microorganisms at the single-cell level.27 Therefore, the microfluidic systems are considered to be highly suitable for the quantitative analysis of chemotactic response. In general, the microfluidic gradient generator can be classified into two groups, flow- and diffusion-based methods.25,26 The flow-based systems generate a gradient by mixing two or three parallel laminar flow of different concentrations.28,29 Even though the flow-based systems have several advantages, such as the capability of generating controllable and switchable gradients,30 the flowing nature limits the period for assay and may hinder the chemotaxis-induced migration of cells.23,26 Furthermore, the additional apparatus including syringe pumps and tubes are required to operate these systems, which cause inconveniences in system operation.26 Compared to the flow-based devices, the diffusion-based devices generate the gradient between several fluids which are separated by physical barriers (i.e., semi-permeable membrane).26,31–34 Through these systems, undisturbed long-term monitoring of the chemotaxis is possible without the additional auxiliary equipments.23,26 Consequently, the flow-free systems seem to be more suitable than the flow-based systems for an accurate analysis of chemotaxis.
In this study, we describe a microfluidic analysis of the chemotaxis of C. reinhardtii to HCO3−, an important carbon source for microalgal photosynthesis. For the quantitative analysis of a chemotactic response, we developed a modified diffusion-based microfluidic platform, which is a single-layer polydimethylsiloxane (PDMS) microfluidic device with microchannels filled with small agarose gel plugs as a semi-permeable membrane for diffusion. Agarose gel is a commonly used biocompatible material in general biology labs, and it is easy to cast in microchannels because of its thermal gelation property.32,33 In addition, agarose gel allows the free diffusion of small molecules but prevents the fluid flow.26 Although the system in this study has been developed based on the previous agarose gel-based microfluidic platform,26 it has unique structural advantages which make it easier to use in general biology labs: (1) a separated observation region useful for the quantitative analysis of chemotaxis at the single-cell level, (2) the system consisted of single-layer PDMS microchannels with a uniform height, which excludes the necessity of a complicated two-layer lithography process.26 Using the device, we demonstrated that HCO3− is a strong chemoattractant of C. reinhardtii and there exists the concentration at which C. reinhardtii shows the strongest chemotaxis. To the best of our knowledge, this is the first report on the chemotaxis of a model microalga, C. reinhardtii, to HCO3− and relation of Ci metabolism to this chemotactic behavior.
II. MATERIALS AND METHODS
A. Strains, culture media, and conditions
The wild-type (WT) strain of C. reinhardtii, CC-125 (mt + agg1 + nit1 nit2), and its mutant strain with a point mutation in CIA5, CC-2702 (cia5),14,35 were obtained from Chlamydomonas Resource Center (University of Minnesota). The wild-type microalgal cells were cultured under a low CO2 condition (ambient CO2 condition) and a high CO2 condition (air enriched with 5% (v/v) CO2). The cia5 mutant, which cannot acclimate to the low CO2 condition due to the lack of CCM, was grown in the presence of 5% CO2. All cultivations were conducted under a photoautotrophic condition in 100 ml Erlenmeyer flask, with 50 ml of Tris-phosphate medium (TP medium, Tris-Acetate-Phosphate (TAP) medium36 without acetate; 20 mM Tris base, 7 mM NH4Cl, 0.83 mM MgSO4·7H2O, 0.45 mM CaCl2·2H2O, 1.65 mM K2HPO4, 1.05 mM KH2PO4, 0.134 mM Na2EDTA·2H2O, 0.136 mM ZnSO4·7H2O, 0.184 mM H3BO3, 40 μM MnCl2·4H2O, 32.9 μM FeSO4·7H2O, 12.3 μM CoCl2·6H2O, 10 μM CuSO4·5H2O, 4.44 μM (NH4)6MoO3, pH 7.3), at 23 °C in a shaking CO2 incubator at an agitation rate of 120 rpm under continuous light condition (50 μmol photons m−2 s−1).
B. Design of the microfluidic device
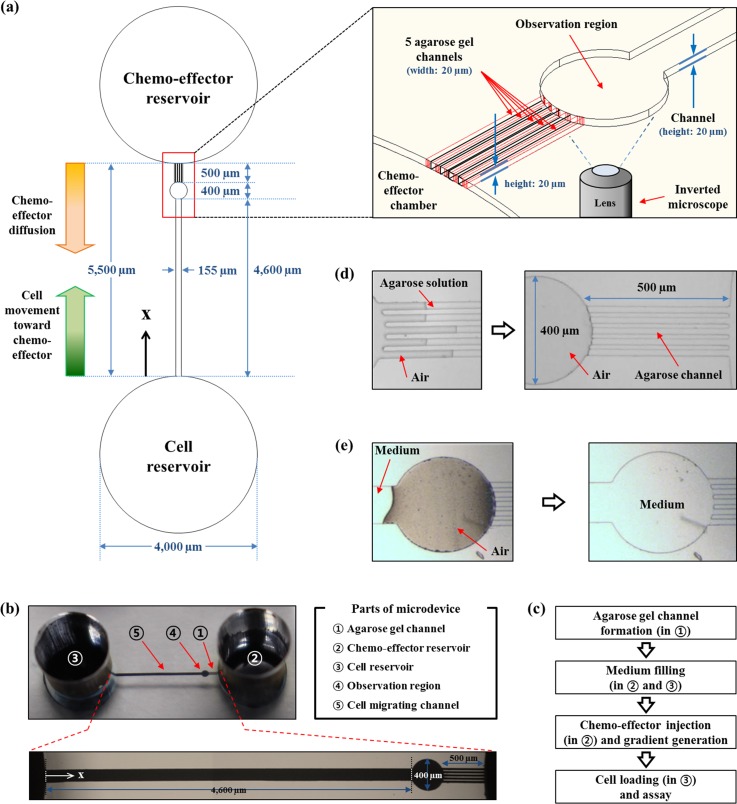
The schematic diagram and the image of the single-layer PDMS microfluidic device with small agarose gel plugs in the microchannels as a semi-permeable membrane for diffusion are presented in Fig. 1. As shown, there are two chambers which served as a cell reservoir and a chemo-effector reservoir on the both ends of the device. The reservoirs are connected with a concentration gradient generating channel with a length of 5500 μm and a uniform height of 20 μm. The height of the channel was determined to prevent congestion and overlap of the cells in the channel which disturb accurate analysis of the cell behavior (C. reinhardtii is about 10 μm in diameter). In detail, the chemo-effector reservoir is connected to 5 microchannels (20 μm wide and 500 μm long) filled with agarose gel (named as agarose gel channel). The channels function as a membrane for chemical diffusion and a barrier that simultaneously prevents the intrusion of cells and fluid flow. The opposite end of the agarose channels is connected to a circular observation region with a diameter of 400 μm, which provides a venue for cell accumulation in response to chemo-effectors. The observation region and the cell reservoir are connected to the long channel (named as cell migration channel; 155 μm wide and 4600 μm long) for the cell migration by generating a stable concentration gradient of chemo-effectors.
FIG. 1.
Schematic diagram of the device for chemotaxis assay. (a) Overall design of the device is presented with the direction of diffusion and cell migration (upper right box: a perspective view of the device mainly focused on the agarose channels and observation region). (b) Image and description of the microdevice with magnified view of cell migration channel, observation region, and agarose gel channels. (c) Flow chart of experimental process. (d) A filling process of agarose gel into microchannels which functions as a semi-permeable membrane allowing free diffusion of small molecules but preventing fluid flow in the microsystem. (e) A medium filling process in the microfluidic device. Due to their hydrophilic and gas-permeable properties of oxygen-treated PDMS microchannel, the channel was filled with a medium while pushing out the trapped air in the channel.
C. Microchip fabrication
The design of the device was generated using AutoCAD software (Autodesk) and printed on a transparent film as a photomask film. The master for PDMS cast was fabricated using SU-8 50 negative photoresist (MicroChem) on a silicon wafer by standard photolithography.37 The device was fabricated by pouring 10:1 w/w PDMS prepolymer mixture of 184 Sylgard base and curing agent (Dow Corning) onto the generated master and curing at 80 °C in an oven after degassing process. The two reservoirs were bored using a 4 mm size biopsy punch. The PDMS layer containing microchannels and chambers was bonded to a slide glass via oxygen plasma treatment. In order to make the microchannel of the device hydrophobic, the device was stored at 80 °C in an oven overnight. Subsequently, the device was treated with oxygen plasma for 10 s to convert the surface of the microchannel into partially hydrophilic state. This process is necessary for the control of the hydrophilicity of the surface of PDMS microchannels. After the plasma treatment, the microdevice was stored in atmosphere for several minutes to obtain thermal equilibrium with room temperature (about 25 °C).
TP medium-based 3% w/v agarose gel solution was then prepared, and 2.5 μl of the solution at 65 °C was injected into the agarose gel channel. Fig. 1(d) shows agarose gel filling process to make diffusion membrane channels. Due to the hydrophilicity of surface after the treatment of oxygen plasma for 10 s, the agarose solution was driven into the agarose channels by the surface tension.26 After flowing through the channels, the agarose solution suddenly stopped and solidified at the end of the agarose gel channels because of large cross sectional area difference between an agarose channel and the observation region as reported previously.26 Since the cross sectional area of the observation region is much larger than that of an agarose gel channel, the flow speed of agarose solution was suddenly retarded when the agarose solution flows into the observation channel. This concept is supported by the continuity equation
| (1) |
where ρ is the density of the fluid, A is the cross sectional area of the channel, and v is flow velocity. Due to the large temperature difference between the agarose solution and the surface of the channels of the PDMS chip, the solution turned into gel immediately after the flow slowed down. Therefore, the agarose channels were completely formed within 5 s. A small amount of agarose gel that is remained in the chemo-effector reservoir does not affect the diffusion of molecules because the diffusivity of molecules in low concentration of agarose gel is nearly the same as in the water as previously reported.34 After the semi-permeable channels were constructed, 35 μl of TP medium was loaded into both the cell and the chemo-effector reservoir. As the surface of the microchannel is hydrophilic and PDMS is a gas permeable material, the medium flowed into cell migration channel and the observation region from the cell reservoir while pushing out the trapped air in the channel (Fig. 1(e)).
D. Fluorescent dye diffusion analysis
Concentration gradient generation through the channel of the microfluidic device was analyzed using fluorescein sodium salt (Sigma-Aldrich) solution, since the diffusion behavior of small molecules is analogous to that of fluorescein.26 The time-lapse fluorescence image of the microchannel was obtained every 1 h after 35 μl of TP medium based 0.1 mM fluorescein solution was supplied into the chemo-effector channel. The images were taken with an exposure time of 3 s using Carl-Zeiss Axio Observer D1 inverted microscope equipped with a 5× objective lens, AxioCam CCD camera (Carl-Zeiss), and AxioVision image software (Carl-Zeiss). The fluorescent intensity was quantified using ImageJ software (National Institutes of Health).
E. Biological validation of microfluidic device for analysis of chemotaxis
Prior to the experiment, the wild-type cells were subcultured to OD800 of 0.1 every 4 days to maintain cells in exponential growth phase under the low CO2 condition. Cells were harvested during exponential growth phase by centrifugation at 3000 rpm for 5 min. Subsequently, the cell pellet was washed once and resuspended in the TP medium at an OD800 of 1.0 (∼1.65 × 106 cells/ml). To test the functionality of the microfluidic device, the TP medium in the chemo-effector reservoirs was replaced by chemical solutions in TP medium, including CoCl2 and NaCl with different concentrations (10, 50, and 100 mM, respectively). Due to the concentration difference between the TP medium in the cell reservoir and the chemical solutions in the chemo-effector reservoir, chemical concentration gradient generations were initiated. For each experiment, the diffusion process was done for 4 h to obtain the stable and the linear concentration gradients along the channel. While the diffusion process was in progress, the device was stored in a 100% humidity chamber to prevent evaporation. After the linear gradient in the channel was achieved, the TP medium in the cell reservoir was carefully removed, and 35 μl of the cell suspension was injected. In case of chemotaxis assay using NH4Cl, the harvested cells were resuspended in nitrogen-deprived TP medium (TP−N medium, TP medium without NH4Cl) at an OD800 of 1.0. The TP−N medium was also used as the basis of NH4Cl solution with different concentrations (10, 50, and 100 mM) for the chemotaxis analysis. To prevent unintended phototaxis-involved cell movement, the microfluidic device was placed in dark during the assay period. The number of cells in the observation region was determined through the manual counting and the use of software (ImageJ) to minimize errors in counting number of cells in the observation chamber. All reagents, including CoCl2, NaCl, and NH4Cl, were purchased from Sigma-Aldrich.
F. Analysis of chemotactic response to HCO3−
The wild-type cells were grown to exponential phase under the low CO2 and the high CO2 conditions. Chemotaxis to HCO3− was analyzed with various concentrations of NaHCO3 (from 10 mM to 100 mM at an interval of 10 mM). To analyze the effect of the inhibition of inorganic carbon utilization on chemotaxis to HCO3−, cells were treated with an inhibitor of external carbonic anhydrase, 5-acetamido-1,3,4-thiadiazole-2-sulfonamide (AZ),38,39 and a putative inhibitor of HCO3−/Cl− anion exchange, 4,4′-dithiocyanatostilbene-2,2′-disulfonate (DIDS).40,41 Prior to the analysis, the wild-type cells grown under the low CO2 condition were treated with 50 μM of AZ and 500 μM of DIDS solution for 3 h, respectively. Subsequently, the cells were harvested, washed, and resuspended in each inhibitor solution at an OD800 of 1.0. 35 μl of the cell solutions from each condition were loaded into the cell reservoir after HCO3− concentration gradient was generated for 4 h. The chemotaxis assays were performed under darkness. The cia5 mutant grown under the high CO2 condition was also used for chemotaxis analysis to investigate the relationship between CCM and chemotaxis to HCO3−.
G. Analysis of the effect of circadian rhythm on chemotaxis to HCO3−
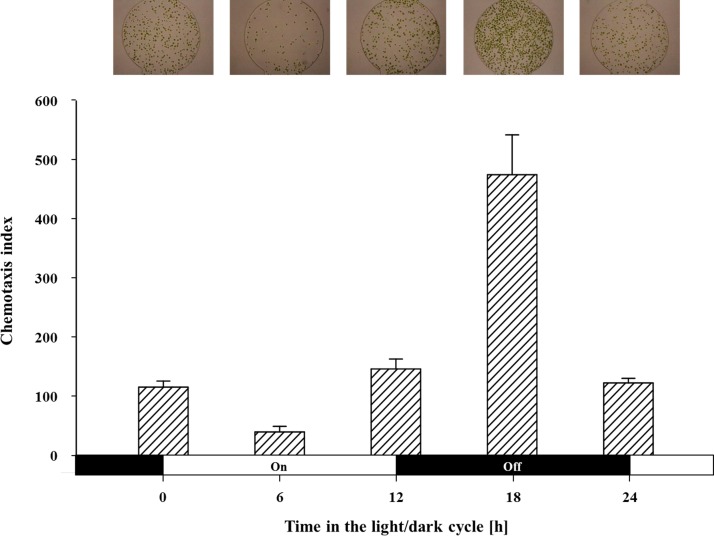
To synchronize circadian clock, the wild-type cells were placed under the cycle of 12 h-light and 12 h-dark in the low CO2 condition. After synchronization, the cells were harvested, washed, and resuspended as mentioned above. Chemotaxis was analyzed every 6 h for 24 h (i.e., 0 h, 6 h, 12 h, 18 h, and 24 h) using 30 mM of NaHCO3 solution, which triggered the strongest chemotactic response of the wild-type cells as the chemo-effector. To minimize the effect of time on the chemotactic response by circadian rhythm, the assays were conducted for a short period of time (1 h) under the dark condition.
H. Data analysis
We used the chemotaxis index, CI,42,43 to quantify the response of the cells to the chemo-effectors. In this study, CI is defined as
| (2) |
where Nchemo–effector is the number of cells accumulated in the observation region in response to a chemo-effector, and NControl is the number of spontaneously migrated cells to the observation region in the absence of a chemical gradient (i.e., only fresh TP medium was supplied in the chemo-effector reservoir; indicated by control experiments in all following studies). The negative control experiments were performed for each assay condition. The number of the cells was counted from the image of the observation region, which was captured every 1 h. For further analysis, the cell migration channel was observed in the HCO3− chemotaxis assay after the cell migration reached the steady-state. To determine the location of the cell band, the image of the overall channel was analyzed by densitometer using ImageJ software. The center position of the band was obtained from the point where the density was the highest. The chemotactic responses of the different assay conditions were determined and compared by the skewness, an index of asymmetry of distribution, which was calculated using the skew function in the Excel software (Microsoft). For all chemotactic response assays, the images were taken by an Olympus CKX 41 inverted microscope (Olympus) equipped with a Cannon EOS 700D CMOS camera (Cannon).
III. RESULTS AND DISCUSSION
A. Gradient generation and characterization
The diffusion of a chemical without fluid flow can be described with the Fick's second law
| (3) |
where c is the concentration, t is the diffusion time, and D is the diffusion coefficient of a molecule. If the diffusion only occurs along the x-axis, the solution of the equation is given by the following equation:
| (4) |
where c0 is the concentration at the chemo-effector reservoir, and L is the entire length of the channel (L0 = 5000 μm). Theoretically, the concentration at the end of the channel (x = 0) can always be regarded as zero since the diffused chemical immediately spreads into the cell reservoir whose volume is relatively large compared with that of the channel enough to offset the diffusion effect. As a result, the initial condition is c(x,0) = 0, and the boundary conditions of the system are c(0,t) = 0 and c(L0,t) = c0.34 When completely diffused along the long channel (i.e., the diffusion reached the steady-state), the gradient of the chemical becomes linear as follows:
| (5) |
We used fluorescein solution to characterize the diffusive property in the device since the fluorescein has been widely used to monitor and confirm the diffusion phenomena of small molecules in the microfluidic systems.26,27,34 The diffusivity of fluorescein is about 4.9 × 10−6 cm2 s−1 that was calculated from the Wilke-Chang correlation.44 The diffusivity of HCO3 at 25 °C, 1.19 × 10−5 cm2 s−1, is about 2.43-fold higher than fluorescein, which is estimated by the Nernst-Einstein equation45
| (6) |
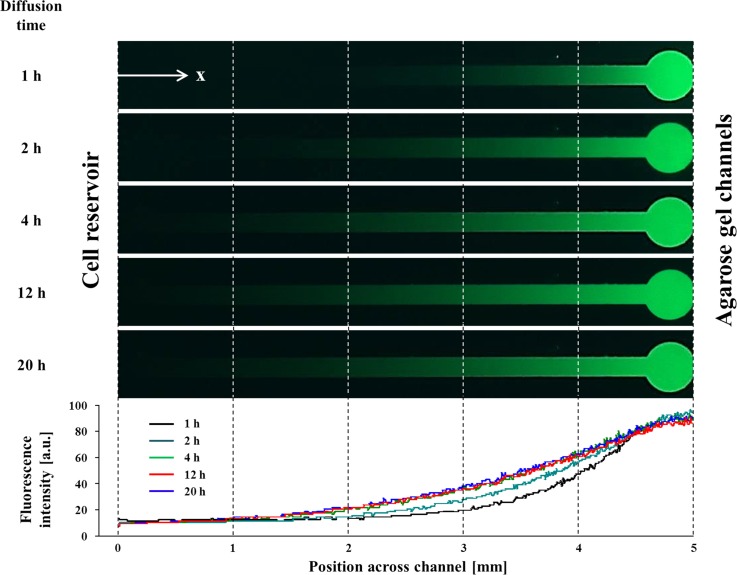
where D is the self-diffusion coefficient, R is the gas constant (8.3145 J K−1 mol−1), T is temperature in Kelvin, λ is the limiting conductivity (per mole), z is charge, and F is the Faraday constant (9.6485 × 104 C mol−1). The diffusion time of molecules can be estimated from t ∼ L2/D as described previously.26 Thus, the diffusion velocity of HCO3− is about two times faster than fluorescein in the microfluidic device. Therefore, the stable gradient of small molecules with higher diffusivity compared to fluorescein can be generated more quickly. For the comparison of the theoretical approach with our experimental results, we analyzed the diffusion profile of fluorescein using ImageJ software. The generation of the gradient was performed by providing a 0.1 mM fluorescein solution into the chemo-effector reservoir. We observed that the fluorescence signal consistently diffused from the chemo-effector reservoir to the cell reservoir through the agarose channels due to the concentration difference. The generated gradient along the channel of the device was measured at an interval of an hour. As shown in Fig. 2, we found that 4 h are enough to generate steady-state diffusion profile and the gradient was stably maintained over 20 h (longer than the entire assay period). In accordance with the theoretical analysis, the gradient profile was gradually linearized as the time passed (all of the r2 of the profiles after 4 h were >0.916).
FIG. 2.
Fluorescence microscopy images (the top panel) and their concentration gradient profiles (the bottom panel) across the channel at different times. 0.1 mM of fluorescein solution was used for the diffusion tracking. The steady-state diffusion was achieved after 4 h and maintained for the entire assay period (>20 h).
For the quantitative analysis of chemotaxis, we needed to estimate the steady-state concentration at the observation region. The relationship between the concentration of chemo-effector reservoir (CS) and the maximum concentration of the observation region (CM; indicating the concentration at x = 5000 μm) was calibrated in a previous study26
| (7) |
where Aa and A0 indicate the cross sectional area of an agarose gel channel and the following channel part (the observation region + the cell migration channel), respectively; La and L0 denote the length of an agarose gel channel and the following channel part, respectively; and N is the number of the agarose gel channels. For simple modeling, A0 and L0 represent the cross sectional area of the cell migration channel and length of the following part of the agarose gel channels (i.e., the observation region + the cell migration channel), respectively. Considering the parameters in this study, we could obtain CM = 0.866CS. Hence, the concentration of the chemicals at the particular position of the channel can be estimated from the relationship and its linearity.
B. Biological validation of microfluidic device for analysis of chemotaxis
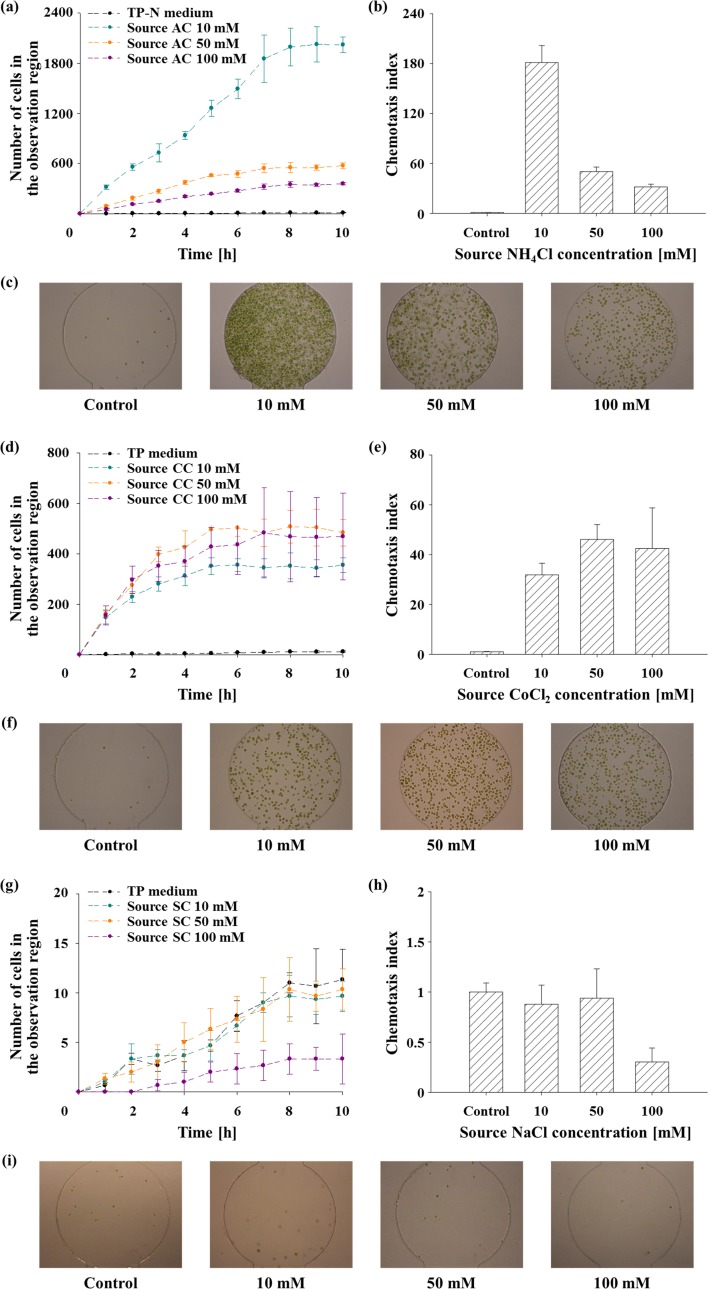
We validated the microfluidic device by analyzing chemotactic responses of C. reinhardtii to previously known chemoattractants (NH4Cl and CoCl2) and the chemotactically neutral molecule (NaCl).8,10–12 It was reported that ammonium ion (NH4+) is a strong chemoattractant for C. reinhardtii,8,10,12 but NaCl does not elicit any response (chemotactically neutral)10 of the alga. The chemotaxis of C. reinhardtii to cobalt ion (Co2+ ion from CoCl2) was also studied in several studies,10,11 but its effect is still disputable. We employed the CI given by Eq. (2) for the quantitative analysis of chemotaxis. We only considered the cells that arrived at the observation region to determine the chemotaxis index at the specific concentration in the observation region corresponding to different source concentrations. Due to the concentration gradient from the observation region to the cell reservoir, the concentration along the long cell migration channel changes according to the distance from the chemo-effector reservoir. Therefore, it is unsuitable to include cells remaining in the migration channel for the determination of chemotactic response to the specific concentration of chemicals in the observation region. As above mentioned, the index is defined as the number of attracted cells by chemotaxis in the observation region with chemo-effector divided by the number of randomly migrated cells into the observation region without chemo-effector. Therefore, the CI represents the intensity of the chemotactic response to various chemo-effectors quantitatively. After 4 h of diffusion with each chemical solution for establishing stable concentration gradient through the channel, 35 μl of the wild-type cells grown under atmospheric CO2 condition were loaded into the cell reservoir. All the assays were performed under darkness to prevent the unintended phototactic movement.
In response to the gradient of NH4+, the algal cells were attracted as shown in Figs. 3(a)–3(c), indicating NH4+ acts as a strong chemoattractant which are consistent with the results of previous works.8,10,12 Fig. 3(a) shows that the number of accumulated cells in the observation region increases with time and approached its maximum asymptotically. The cell response to TP−N medium was investigated as a control (i.e., in the absence of NH4+ gradient), and no chemotactic response was observed. After net cell migration reached steady-state after 8 h of observation, the CI was estimated to quantify the chemotactic response intensity. As a result, we found that 10 mM of NH4Cl solution induced the strongest chemotactic response. At the higher concentrations of the source NH4Cl than 10 mM, the number of accumulated cells and CI decreased (Figs. 3(b) and 3(c)). Considering diffusion characteristics of the device, the NH4+ concentration at the observation region was about 8.66 mM when 10 mM of NH4Cl solution was provided as a chemo-effector source. This result implies that the microalga prefers 8.66 mM of NH4+ rather than higher concentrations (i.e., 43.3 mM and 86.6 mM for the source concentration of 50 mM and 100 mM, respectively) because excessive ion (i.e., NH4+ and/or Cl−) supply might cause a negative influence on the cells,34 resulting in the decrease of the chemotactic response intensity.
FIG. 3.
The validation of agarose gel-based microfluidic device for chemotaxis assay of C. reinhardtii using NH4Cl (ammonium chloride; AC), CoCl2 (cobalt chloride; CC), and NaCl (sodium chloride; SC). All data and error bars are the means and standard deviations of the three biological replicates (n = 3). (a) The number of accumulated C. reinhardtii cells in the observation region in response to NH4Cl gradient according to time. (b) CI measured in the presence of NH4Cl gradient after 8 h of observation. (c) A representative microscopy image showing the cell response towards NH4+ after 8 h at different concentrations. (d) The number of the accumulated cells in the observation region towards CoCl2 according to time. (e) CI measured in the presence of CoCl2 gradient after 8 h. (f) A representative microscopy image showing the cell response towards Co2+ at 8 h at different concentrations. (g) The number of the accumulated cells in the observation region in response to NaCl gradient according to time. (h) CI measured in the presence of NaCl gradient after 8 h. (i) A representative microscopy image showing the cell response towards NaCl at 8 h at different concentrations.
Fig. 3(d) shows the number of accumulated cells in the observation region in response to CoCl2 over time. The cells showed no response to TP medium used as a negative control. The TP medium intrinsically contains 6.73 × 10−3 mM of Co2+ ion, but it is negligible compared to the chemotaxis assay conditions (i.e., 10 mM, 50 mM, and 100 mM of CoCl2), since the concentration is below the threshold concentration of Co2+ to elicit the chemotactic response.11 As a result of the experiment, C. reinhardtii showed a positive chemotaxis to CoCl2 as previously reported.11 However, CIs (Fig. 3(e)) and the number of accumulated cells (Fig. 3(f)) did not display a distinctive peak at a particular concentration between 10 mM and 100 mM. The Co2+-induced CI is 3.93-fold lower than that of NH4+, indicating NH4+ is a stronger chemoattractant than Co2+. Although it was reported that cobalt is toxic to C. reinhardtii at the concentration of 19 μM,46 it was also known as strong chemoattractant at 0.1 mM.11 Therefore, a further investigation related to cytotoxicity and chemotaxis of cobalt in C. reinhardtii is required. The chemotactic responses of the cells to various concentrations of NaCl solution were also investigated (Fig. 3(g)). As shown in Figs. 3(h) and 3(i), NaCl does not elicit any chemotactic response of C. reinhardtii, indicating NaCl is a chemotactically neutral chemical. This result also supports that chemotaxis to NH4Cl and CoCl2 was elicited by NH4+ and Co2+, respectively, not by Cl− ion.
C. Chemotactic response to HCO3−
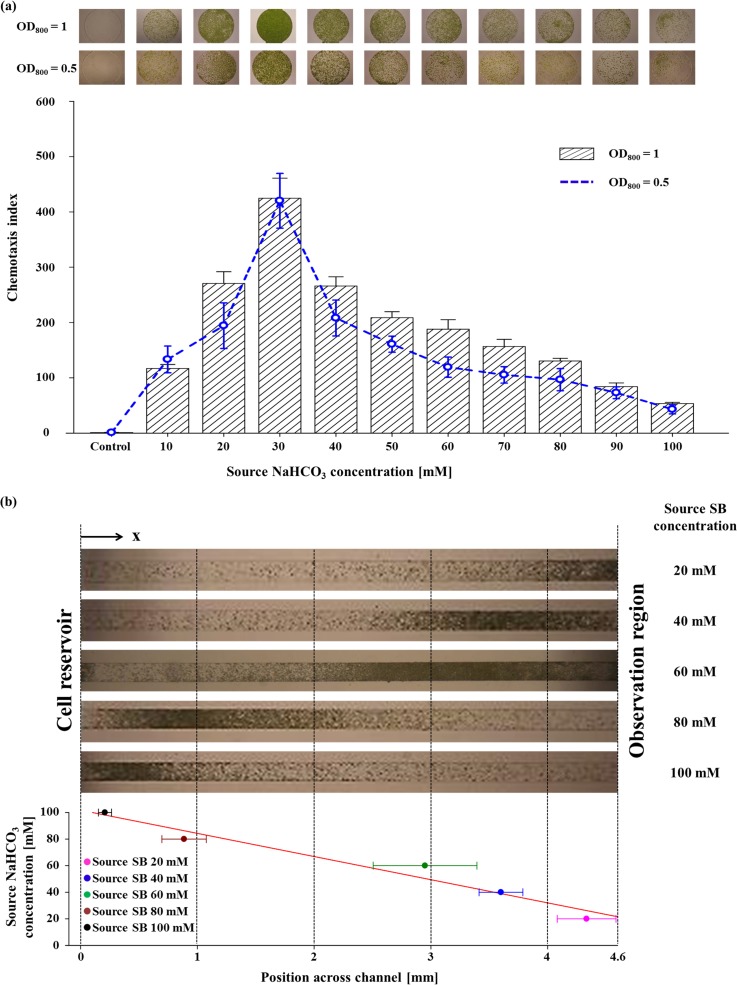
When dissolved in water, CO2 exists in forms of dissolved inorganic carbon. Among them, HCO3− is the most abundant inorganic carbon source for the aquatic photosynthesis15 in moderate conditions of aquatic ecosystem.15 To adapt CO2-limiting environment, the microalgae induce CCM for facilitating HCO3− uptake and elevating internal inorganic carbon concentration for better carbon fixation efficiency with CAs and HCO3− transporters.14,16 Since motile microorganisms are capable of detecting and finding the optimal position for a stable supplement of nutrient sources by means of chemotaxis,23 we thought that HCO3− elicits the chemotaxis of a motile microalga, C. reinhardtii. The chemotactic responses of cells to various NaHCO3 concentrations (from 10 mM to 100 mM at an interval of 10 mM) were analyzed under darkness to preclude the phototactic response. As expected, we observed a strong chemotactic response of C. reinhardtii towards HCO3−, as shown in the upper images of Fig. 4(a). We obtained the CIs at each concentration of HCO3− (the bottom of Fig. 4(a)). We found that the CIs gradually increase as the concentration of HCO3− increases with a maximum value of 424.6 at 30 mM (equivalent to 26.0 mM at the observation region), followed by a gradual decrease to 53.4 at 100 mM of HCO3−. We also analyzed the chemotactic response with a decreased cell density by half (OD800 of 0.5) to investigate whether the initial cell number in the cell reservoir affects the chemotaxis index. As shown in Fig. 4(a), the CIs showed no significant changes in the magnitude and the profile of chemotaxis index, although the cell number in the observation region decreased. This is because the number of cells reached the observation region by random motility was also reduced. This result indicates that the initial cell density is not a factor of the CI. Under the same cell density, notably, the chemotaxis index of HCO3− was 2.34-fold higher than NH4+ (CI: 181.2), supporting that HCO3− is a more favorable chemoattractant and implying that it is an important nutrient source to C. reinhardtii. Since the cells used in this experiment were cultivated in TP medium containing 7 mM of NH4Cl under ambient CO2 condition, the most limiting nutrient may be the inorganic carbon. Therefore, it seems that HCO3− triggered a stronger chemotactic response of the cells than NH4+. In addition, we found that the chemotactic response of C. reinhardtii to HCO3− is light-independent, considering that the assay was carried out under a long-term dark condition.
FIG. 4.
Chemotactic response of C. reinhardtii towards HCO3−. All the experiments were carried out in triplicates (n = 3). Error bars are the standard deviations. (a) The chemotaxis index (CI) of the wild-type cells (OD800 = 1) in response to HCO3− at various source NaHCO3 (sodium bicarbonate; SB) concentrations (from 10 mM to 100 mM at an interval of 10 mM). The blue dashed line indicates the chemotactic response of the wild-type cells at an OD800 of 0.5. The upper images show the representative microscopy images of the accumulated cells at each source concentration. The cell behavior assay in the absence of HCO3− gradient was performed as a control experiment. (b) Cell population band formation in response to the different source NaHCO3 concentrations (from 20 mM to 100 mM at an interval of 20 mM) across the cell migration channel. The top panel shows the representative cell bands formed at different position of the cell migration channel, while the bottom panel shows the center positions of the cell bands. The red line indicates the linear regression of the center position. The linearity of the center positions (r2 = 0.956) indicates the existence of the most preferred HCO3− concentration in the chemotaxis of C. reinhardtii.
Subsequently, we checked the cell distribution in the cell migration channel for further analysis. Interestingly, the dense population bands appeared at different positions in the cell migration channel (the top panel of Fig. 4(b)) according to the different concentrations of HCO3−. The dense population bands migrated toward the chemo-effector source as the concentration of source became lower, indicating that cells move to find the optimal concentration of HCO3− according to different source concentrations. For accurate analysis of the band location, we estimated the center position of the dense population band by densitometry. The bottom panel of Fig. 4(b) shows a linear distribution of the center positions according to the source concentration, which also supports the existence of the most preferred HCO3− concentration for the chemotaxis of C. reinhardtii, since the concentration profile across the channels shows near linearity after sufficient diffusion, as shown in Fig. 2.
D. Circadian rhythm of chemotaxis to HCO3−
The circadian rhythm of C. reinhardtii is an internal biological process, which controls various physiological, metabolic phenomena to occur with the diurnal cycle.47 Till date, several biological phenomena such as phototaxis,9 chemotaxis,8,48 and UV sensitivity49 in C. reinhardtii have been shown to follow a circadian rhythm. Although the effect of circadian rhythms on chemotaxis has been studied previously, the studies have been limited to only nitrogen sources, such as ammonium and nitrate.8,48
To test the circadian rhythm of chemotactic response towards HCO3−, the wild-type cells were cultured under the low CO2 condition and acclimated to a photoperiod of 12 h-light and 12 h-dark (LD 12:12) to synchronize circadian clock. Each experiment was performed after HCO3− concentration gradient was generated for 4 h using 30 mM of NaHCO3. Since the chemotactic response of cells under circadian rhythm rapidly changes according to time, we analyzed the chemotactic response during a short span of time (1 h) with the cells sampled every 6 h to minimize the effect of time on the chemotactic response. Fig. 5 shows that the intensity of the chemotaxis that varied according to the time of sampling. As a result, the chemotaxis to HCO3− was maximal at the middle of the dark period while it was lowest at the middle of the light period. This rhythmic pattern is consistent with a previous work, which revealed that the chemotaxis to NH4+ under circadian rhythm shows the maximum during the dark period and minimum during the light period.8 C. reinhardtii is able to uptake inorganic carbon (HCO3− and CO2) and efficiently convert them into organic compounds in the light phase by the means of CCM and photosynthesis.50 Thus, they can successfully cover their carbon requirement. In the dark phase, however, the light-dependent carbon assimilation process would stop, and the dark respiration starts to increase the internal production of CO2.40 As a consequence, the efflux of CO2 from the cells takes place. To compensate for the loss, a necessity of the carbon sources and the intensity of chemotactic response (CI) also might increase to allow the cells to find a nutrient-rich environment.8
FIG. 5.
The chemotaxis index of the cells synchronized to Light (on): Dark (off) (12 h:12 h) cycle. The chemotactic response was assayed for 1 h using the cells sampled every 6 h. Chemotaxis to HCO3− showed the maximum level at the middle of the dark period (18 h) and the minimum level during the day phase (6 h). The upper images show the representative images of the attracted cells at each sampling time. Data are the mean (bar graph) and standard deviation (error bars) of three biological replicates (n = 3).
E. Correlation of chemotaxis with carbon dioxide concentrating mechanism
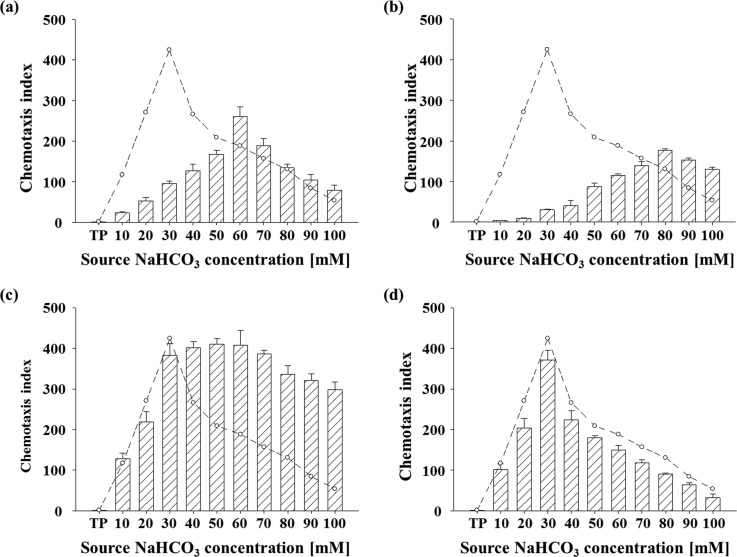
C. reinhardtii developed the ability to cope with variable CO2 concentrations.14 We investigated the effect of various factors that involved in CCM and inorganic carbon utilization, including transcriptional regulation of CCM, bicarbonate transporters, and external carbonic anhydrases. It was known that C. reinhardtii exhibits a relatively low affinity to inorganic carbon when grown under the high CO2 concentration (5% (v/v)).51 As shown in Fig. 6(a), the distribution of CIs of wild-type cells (CC-125) acclimated to the high CO2 (5%) showed a pattern with a peak shifted to higher concentration of NaHCO3 (skewness: −0.137 ± 0.023) compared to the pattern of wild-type cells acclimated to low CO2 (ambient) (skewness: 0.551 ± 0.033), but showed lower peak chemotactic index, indicating that cells grown in the high CO2 prefer higher concentration of HCO3− with lower intensity of chemotactic response. This result supports the previous report as mentioned above51 and shows that the chemotaxis to HCO3− of C. reinhardtii is highly correlated with CO2 utilization and acclimation.
FIG. 6.
The distribution of CI in response to several factors related to CCM according to different NaHCO3 concentrations. Data and error bars are the means and standard deviations of triplicate experiments (n = 3). Dashed line indicates the chemotactic response of the wild-type cells (OD800 1) acclimated to the CO2 (ambient) condition. CI distributions of (a) the wild-type cells acclimated to 5% CO2-enriched air. (b) CC-2702 (cia5), a CCM-deficient mutant. (c) The wild-type cells treated with DIDS, inhibitor of putative HCO3−/Cl− transporter. (d) The wild-type cells treated with AZ, inhibitor of external carbonic anhydrase. TP: TP medium used as a negative control.
We also analyzed the chemotactic response of a mutant (CC-2702, cia5) of CC-125 with a point mutation in CIA5, a transcription regulator that induce CCM.14 Therefore, this mutant is unable to acclimate to the low CO2 condition due to the impaired component of CCM including putative bicarbonate transporters.14 The distribution of CIs of the mutant showed a pattern with a more shifted peak to high concentration of NaHCO3 (skewness: −0.618 ± 0.043), indicating that the mutant lacking induction of CCM requires higher concentration of inorganic carbon source to compensate the loss of CCM induction. Because it was known that putative bicarbonate transporters are impaired in the cia5 mutant, we further analyzed the effect of inhibition of putative bicarbonate transporters on the chemotaxis to HCO3− using a specific inhibitor of putative HCO3−/Cl− transporters, DIDS. Interestingly, the wild-type cells treated with DIDS showed significantly enhanced chemotaxis index at higher concentrations of NaHCO3− than 30 mM, compared to the wild-type cells acclimated to low CO2 (Fig. 6(c)), which suggests that transport of bicarbonate is correlated with chemotaxis to bicarbonate. However, the wild-type cells that were treated with a specific inhibitor of external CAs, AZ, showed no noticeable difference with the untreated cells (Fig. 6(d)), indicating that external CAs are not directly involved in the chemotaxis to HCO3−.
IV. CONCLUSION
Chemotaxis is the most crucial behavioral response to chemical stimuli, which allows microorganisms to find nutrients or avoid toxic compounds for their survival. Microalgae, the aquatic photosynthetic microorganisms, require inorganic carbon sources for photosynthesis and HCO3−, the most abundant form of inorganic carbon in the water15 is an important carbon source of microalgae. In this study, we demonstrated for the first time the chemotactic response of C. reinhardtii to HCO3− using an agarose gel-based flow-free microfluidic platform under various conditions. For a more accurate and quantitative analysis of the chemotaxis of C. reinhardtii, we developed a modified diffusion-based microfluidic platform based on the previous agarose gel-based microfluidic device,26 which can provide long-term stable gradient with unique features including a separated observation region useful for quantitative analysis of chemotaxis at the single-cell level and single-layer device that is easy to fabricate without a complicated two-layer lithography process. Furthermore, a long cell migration channel provides an additional advantage in monitoring the profile of population band according to chemical concentrations and exposure time. The simple design and fabrication process make it easier to perform chemotaxis assays practically in general biology laboratories. We confirmed that the microfluidic platform is suitable for long-term chemotaxis assays using previously known chemoattractants and a chemotactically neutral molecule. Using the agarose gel-based microfluidic system, we found that C. reinhardtii showed the strongest chemotactic response at 26 mM of HCO3−, which is 2-fold higher than NH4+. We also observed that the cell population band migrates to preferred concentration in the microchannel with chemical gradient according to the source concentration. In addition, it was revealed that the chemotaxis of C. reinhardtii to HCO3− also displayed circadian rhythmicity with a peak during the dark period. Even though the sensing mechanism, such as the chemoreceptor and the signaling pathways, is not identified yet, this study suggests that there is a clear link between the chemotactic behavior and the inorganic carbon metabolism. Furthermore, it promotes understanding the physiological role of the chemotaxis of C. reinhardtii to HCO3− for efficient inorganic carbon uptake and metabolism.
ACKNOWLEDGMENTS
This study was supported by Grant No. 2014M1A8A1049278 from Korea CCS R&D Center of the NRF funded by the Ministry of Science, ICT, and Future Planning of Korea, the National Research Foundation of Korea (NRF) (Grant No. NRF-2013R1A2A1A01015644/2010-0027955), the Korea Institute of Energy Technology Evaluation and Planning and Ministry of Trade, Industry and Energy of in “Energy Efficiency and Resources Technology R&D” project Korea (Grant No. 20152010201900), and University-Institute Cooperation Program (2013).
References
- 1. Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., and Darzins A., Plant J. 54, 621 (2008). 10.1111/j.1365-313X.2008.03492.x [DOI] [PubMed] [Google Scholar]
- 2. Lim H. S., Kim J. Y. H., Kwak H. S., and Sim S. J., Anal. Chem. 86, 8585 (2014). 10.1021/ac502324c [DOI] [PubMed] [Google Scholar]
- 3. Borowitzka M. A., J. Appl. Phycol. 25, 743 (2013). 10.1007/s10811-013-9983-9 [DOI] [Google Scholar]
- 4. Kim J. Y. H., Kwak H. S., Sung Y. J., Choi H. I., Hong M. E., Lim H. S., Lee J., Lee S. Y., and Sim S. J., Sci. Rep. 6, 21155 (2016). 10.1038/srep21155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barra L., Chandrasekaran R., Corato F., and Brunet C., Mar. Drugs 12, 1641 (2014). 10.3390/md12031641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stephenson P. G., Moore C. M., Terry M. J., Zubkov M. V., and Bibby T. S., Trends Biotechnol. 29, 615 (2011). 10.1016/j.tibtech.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 7. Polle J. E. W., Kanakagiri S. D., and Melis A., Planta 217, 49 (2003). 10.1007/s00425-002-0968-1 [DOI] [PubMed] [Google Scholar]
- 8. Byrne T. E., Wells M. R., and Johnson C. H., Plant Physiol. 98, 879 (1992). 10.1104/pp.98.3.879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stavis R. L. and Hirschberg R., J. Cell Biol. 59, 367 (1973). 10.1083/jcb.59.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sjoblad R. D. and Frederikse P. H., Mol. Cell. Biol. 1, 1057 (1981). 10.1128/MCB.1.12.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hirschberg R. and Rodgers S., J. Bacteriol. 134, 671 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Govorunova E. G. and Sineshchekov O. A., Biochem.-Moscow 70, 717 (2005). 10.1007/s10541-005-0176-2 [DOI] [PubMed] [Google Scholar]
- 13. Spalding M. H., J. Exp. Bot. 59, 1463 (2008). 10.1093/jxb/erm128 [DOI] [PubMed] [Google Scholar]
- 14. Fang W., Si Y., Douglass S., Casero D., Merchant S. S., Pellegrini M., Ladunga I., Liu P., and Spalding M. H., Plant Cell 24, 1876 (2012). 10.1105/tpc.112.097949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y., Stessman D. J., and Spalding M. H., Plant J. 82, 429 (2015). 10.1111/tpj.12829 [DOI] [PubMed] [Google Scholar]
- 16. Yamano T., Sato E., Iguchi H., Fukuda Y., and Fukuzawa H., Proc. Natl. Acad. Sci. U.S.A. 112, 7315 (2015). 10.1073/pnas.1501659112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adler J., Hazelbauer G. L., and Dahl M. M., J. Bacteriol. 115, 824 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Young L. Y. and Mitchell R., Appl. Microbiol. 25, 972 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stocker R. and Seymour J. R., Microbiol. Mol. Biol. Rev. 76, 792 (2012). 10.1128/MMBR.00029-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adler J., Science 153, 708 (1966). 10.1126/science.153.3737.708 [DOI] [PubMed] [Google Scholar]
- 21. Adler J., Science 166, 1588 (1969). 10.1126/science.166.3913.1588 [DOI] [PubMed] [Google Scholar]
- 22. Mesibov R. and Adler J., J. Bacteriol. 112, 315 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagy K., Sipos O., Valkai S., Gombai É., Hodula O., Kerényi Á., Ormos P., and Galajda P., Biomicrofluidics 9, 044105 (2015). 10.1063/1.4926981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stern D. B., Witman G. B., and Harris E. H., The Chlamydomonas Sourcebook: Organellar and Metabolic Processes, 2nd ed. ( Elsevier, Amsterdam, 2008), p. 265. [Google Scholar]
- 25. Englert D. L., Manson M. D., and Jayaraman A., Nat. Protoc. 5, 864 (2010). 10.1038/nprot.2010.18 [DOI] [PubMed] [Google Scholar]
- 26. Si G., Yang W., Bi S., Luo C., and Ouyang Q., Lab Chip 12, 1389 (2012). 10.1039/c2lc21219f [DOI] [PubMed] [Google Scholar]
- 27. Ahmed T., Shimizu T., and Stocker R., Integr. Biol. 2, 604 (2010). 10.1039/c0ib00049c [DOI] [PubMed] [Google Scholar]
- 28. Long T. and Ford R. M., Environ. Sci. Technol. 43, 1546 (2009). 10.1021/es802558j [DOI] [PubMed] [Google Scholar]
- 29. Mao H., Cremer P. S., and Manson M. D., Proc. Natl. Acad. Sci. U.S.A. 100, 5449 (2003). 10.1073/pnas.0931258100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Amarie D., Glazier J. A., and Jacobson S. C., Anal. Chem. 79, 9471 (2007). 10.1021/ac0714967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim T., Pinelis M., and Maharbiz M. M., Biomed. Microdevices 11, 65 (2009). 10.1007/s10544-008-9210-7 [DOI] [PubMed] [Google Scholar]
- 32. Cheng S. Y., Heilman S., Wasserman M., Archer S., Shuler M. L., and Wu M., Lab Chip 7, 763 (2007). 10.1039/b618463d [DOI] [PubMed] [Google Scholar]
- 33. Ahmed T., Shimizu T. S., and Stocker R., Nano Lett. 10, 3379 (2010). 10.1021/nl101204e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim M. and Kim T., Anal. Chem. 82, 9401 (2010). 10.1021/ac102022q [DOI] [PubMed] [Google Scholar]
- 35. Moroney J. V., Husic H. D., Tolbert N. E., Kitayama M., Manuel L. J., and Togasaki R. K., Plant Physiol. 89, 897 (1989). 10.1104/pp.89.3.897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gorman D. S. and Levine R. P., Proc. Natl. Acad. Sci. U.S.A. 54, 1665 (1965). 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qin D., Xia Y., and Whitesides G. M., Nat. Protoc. 5, 491 (2010). 10.1038/nprot.2009.234 [DOI] [PubMed] [Google Scholar]
- 38. Williams T. G. and Turpin D. H., Plant Physiol. 83, 92 (1987). 10.1104/pp.83.1.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moroney J. V., Husic H. D., and Tolbert N. E., Plant Physiol. 79, 177 (1985). 10.1104/pp.79.1.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hanawa Y., Watanabe M., Karatsu Y., Fukuzawa H., and Shiraiwa Y., Plant Cell Physiol. 48, 299 (2007). 10.1093/pcp/pcl066 [DOI] [PubMed] [Google Scholar]
- 41. Merrett M. J., Nimer N. A., and Dong L. F., Plant Cell Environ. 19, 478 (1996). 10.1111/j.1365-3040.1996.tb00340.x [DOI] [Google Scholar]
- 42. Zhao D., Zhu Z., Li D., Xu R., Wang T., and Wu K., Biochemistry 54, 6806 (2015). 10.1021/acs.biochem.5b00847 [DOI] [PubMed] [Google Scholar]
- 43. Iellem A., Mariani M., Lang R., Recalde H., Panina-Bordignon P., Sinigaglia F., and D′Ambrosio D., J. Exp. Med. 194, 847 (2001). 10.1084/jem.194.6.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rani S. A., Pitts B., and Stewart P. S., Antimicrob. Agents Chemother. 49, 728 (2005). 10.1128/AAC.49.2.728-732.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zeebe R. E., Geochim. Cosmochim. Acta 75, 2483 (2011). 10.1016/j.gca.2011.02.010 [DOI] [Google Scholar]
- 46. Macfie S. M., Tarmohamed Y., and Welbourn P. M., Arch. Environ. Contam. Toxicol. 27, 454 (1994). 10.1007/BF00214835 [DOI] [Google Scholar]
- 47. Mittag M., Kiaulehn S., and Johnson C. H., Plant Physiol. 137, 399 (2005). 10.1104/pp.104.052415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ermilova E. and Zalutskaya Z., Plants 3, 113 (2014). 10.3390/plants3010113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nikado S. S. and Johnson C. H., Photochem. Photobiol. 71, 758 (2000). [DOI] [PubMed] [Google Scholar]
- 50. Singh H., Shukla M. R., Chary K. V. R., and Rao B. J., PLoS One 9, e106457 (2014). 10.1371/journal.pone.0106457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Badger M. R., Kaplan A., and Berry J. A., Plant Physiol. 66, 407 (1980). 10.1104/pp.66.3.407 [DOI] [PMC free article] [PubMed] [Google Scholar]