Abstract
Global proteomic analyses of complex protein samples in nanogram quantities require a fastidious approach to achieve in-depth protein coverage and quantitative reproducibility. Biological samples are often severely mass limited and can preclude the application of more robust bulk sample processing workflows. In this study, we present a system that minimizes sample handling by using online immobilized trypsin digestion and solid phase extraction to create a simple, sensitive, robust, and reproducible platform for the analysis of nanogram-size proteomic samples. To demonstrate the effectiveness of our simplified nanoproteomics platform, we used the system to analyze preimplantation blastocysts collected on day 4 of pregnancy by flushing the uterine horns with saline. For each of our three sample groups, blastocysts were pooled from three mice resulting in 22, 22, and 25 blastocysts, respectively. The resulting proteomic data provide novel insight into mouse blastocyst protein expression on day 4 of normal pregnancy because we characterized 348 proteins that were identified in at least two sample groups, including 59 enzymes and blastocyst specific proteins (eg, zona pellucida proteins). This technology represents an important advance in which future studies could perform global proteomic analyses of blastocysts obtained from an individual mouse, thereby enabling researchers to investigate interindividual variation as well as increase the statistical power without increasing animal numbers. This approach is also easily adaptable to other mass-limited sample types.
Global proteomic analyses are now widely applied across biological research to study changes in an organism(s) proteome correlated with a perturbation, phenotype and/or time series of interest (1). Furthermore, it has been broadly established that efficient and reproducible sample preparation workflows are crucial to successful quantitative proteome comparisons, especially when applying label free methods (2, 3). However, biological samples are often severely limited in quantity, which prohibits the application of more robust bulk sample processing workflows due to, for example, contamination, carryover, or sample losses (4). This has limited the effective application of global proteomics for many sample types of great interest, eg, laser capture microdissection of single cells from complex tissues, fluorescence-activated cell sorting of unique cell populations from heterogeneous cell mixtures, circulating tumor cells, and blastocysts. In a typical proteomics experiment, bulk homogenization is applied to generate sufficient protein for processing (>10 μg protein) and can blend the proteomes from many different cell types and disparate tissue regions. The resulting average proteome can effectively render unobservable proteome changes of interest and prohibit important applications. Even standard isobaric labeling protocols require multiple manual sample handling steps, resulting in significant protein losses and precludes their application to samples less than 1 μg (5). Consequently, there is considerable interest in the development of a simple, robust platform for analyzing nanogram quantities of protein obtained from cell type-specific samples.
Due to the desire for cell type-specific proteomic analyses, a number of approaches have been developed and applied to samples as small as 500 mammalian cells (∼150 ng protein) (5–9). These protocols aim to reduce sample losses incurred during handling through two main strategies. The first is to replace the traditional detergents and chaotropes with cleavable detergents or organic solvents that are easily removed without solid-phase extraction (SPE) (7, 9). However, without SPE, these methods suffer from a lack of flexibility due to incompatibility with salts and other reagents that many samples contain. The second approach is the so-called proteomic reactor approach. With this strategy all sample manipulations are carried out in a single vessel and the necessary chemistry and sample washing is facilitated by ultrafiltration devices or immobilizing proteins and/or peptides on a solid-phase support (5, 6, 8). These protocols offer far greater reagent flexibility but presently require many manual sample handling steps and long processing times, and results can vary considerably (10, 11).
Immobilized enzyme reactors (IMERs) have also been shown to be of potential use for the analysis of nanogram quantities of protein, either in a single-pot method or integrated into online sample handling systems (12–14). The advantages of the IMER approach include a larger enzyme to substrate ratio, rapid digestions, long-term stability, good reusability, and facile incorporation into integrated handling systems (13, 15). We have found that high enzyme to substrate ratios are particularly advantageous when handling nanogram sample quantities because they provide favorable digestion kinetics in highly dilute solutions without excess trypsin contamination of the resulting peptide solution. Although online handling systems offer great potential for reproducible sample handling and full automation, their inherent complexity makes robust operation challenging and impedes application to biological studies in which larger sample numbers are required.
Herein we designed a system that combines minimal sample handling with the advantages of IMER digestion and online solid phase extraction to create a simple (requires only fluidic components commonly found in most proteomic laboratories), sensitive, robust, and reproducible platform for the analysis of nanogram-size proteomic samples. We used our system to analyze mouse blastocysts; in this study, we were able to detect and quantify cell type-specific proteins in addition to providing novel insight into mouse blastocyst protein expression on day 4 of normal pregnancy.
Materials and Methods
Mice
All mice used in this investigation were housed in the Cincinnati Children's Hospital Medical Center Animal Care Facility according to National Institutes of Health and institutional guidelines for the use of laboratory animals. All protocols of the present study were reviewed and approved by the Cincinnati Children's Hospital Research Foundation Institutional Animal Care and Use Committee. Mice were provided with autoclaved rodent LabDiet 5010 (Purina) and UV light-sterilized reverse osmosis/deionization constant circulation water ad libitum and were housed under a constant 12-hour light, 12-hour dark cycle. Wild-type female mice were mated with wild-type males (d 1 of pregnancy = vaginal plug) (16). Uteri were flushed with physiological saline to recover blastocysts prior to their attachment with the uterine lining; zona-encased blastocysts were collected at 3:00 pm on day 4 of pregnancy. For each of our three samples analyzed by simplified nanoproteomics platform (SNaPP), blastocysts were pooled from three mice, resulting in 22, 22, and 25 blastocysts, respectively.
Simplified nanoproteomics platform
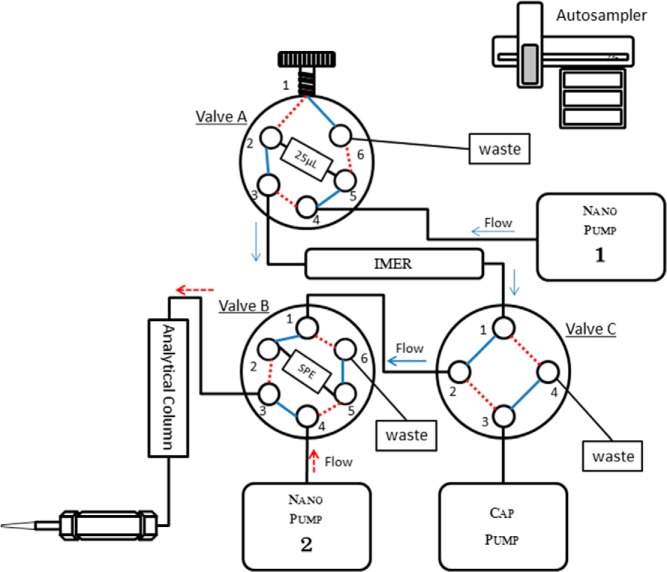
Protein extraction, denaturation, and reduction is achieved by adding 10 μL of homogenization buffer (8 M urea, 5 mM dithiothreitol in 50 mM ammonium bicarbonate at pH 8) and sonicating in a bath sonicator for 1 minute, followed by 30 minutes incubation at 37°C. The resulting extract is diluted with 50 mM ammonium bicarbonate to bring the final urea concentration below 3 M for injection on the SNaPP system. The SNaPP is achieved using the configuration illustrated in Figure 1. The system includes the following fluidic components: two Agilent Nanoflow 1200 pumps (Agilent Technologies), one capillary pump, a six-port injection valve with 25 μL sample loop, two six-port valves (VICI Valco), and a PAL autosampler (Leap Technologies). The trypsin-based immobilized enzyme reactor consists of Poroszyme immobilized trypsin beads packed into a 10-cm long, 150-μm inner diameter capillary. The IMER column is incubated in a butterfly portfolio heater set at 37°C. A typical digestion time is 75 minutes, and the flow rate is set at 200 nL/min of 5% acetonitrile in 95% 50 mM Tris (pH 8) and 5 mM CalCl2 at pressure less than 70 bars (Figure 1; nanopump 1). Each digestion is followed by 5 μL wash of the digestion column with 50% acetonitrile and 50% 50 mM Tris. Prior to analysis, the digestion column is conditioned with 15 injections of 250 ng Shewanella oneidensis cell lysate to remove excess trypsin peptides from autolysis and passivate nonspecific binding sites. After online digestion, the peptides are trapped and desalted via on-line SPE consisting of 4 cm, 150 μm inner diameter 5-μm Jupiter C18 minutes (Phenomenex).
Figure 1.
Schematic of the SNaPP for global proteomics of nanogram quantities. Autosampler injects protein sample to 25 μL sample loop at valve A. After injection, valve A position is then switched, and nanopump 1 carries out immobilized trypsin digestion at 200 nL/min with 5% acetonitrile in 95% 50 mM Tris (pH 8) for 75 minutes at approximately 70 bars. Digested peptides are loaded directly on SPE after immobilized trypsin digestion. Valve C then switches position and the capillary pump carries out desalting with nanopure water with 0.1% formic acid at 2.5 μL/min flow rate for 20 minutes. After desalting, valve C then switches position and nanopump 2 carries out peptide separation with a 100-minute reversed-phase gradient from 8% to 75% mobile phase B. Mobile phase A consists of nanopure water with 0.1% formic acid, and mobile phase B consists of acetonitrile with 0.1% formic acid.
Desalting is carried out by washing SPE with nanopure water with 0.1% formic acid at 2.5 μL/min flow rate for 20 minutes. Nanopump 2 (Figure 1) carries out the liquid chromatography separation using a flow rate of 300 nL/min. Mobile phase A consists of nanopure water with 0.1% formic acid. Mobile phase B consists of acetonitrile with 0.1% formic acid. The analytical column is 40 cm long 75 μm inner diameter packed in-house with 3-μm-diameter C18 bonded particles (PhenomenexA). A 100-minute, reversed-phase gradient from 8% to 75% B is used to elute the peptides. In this study, mass spectrometry analysis is performed using either a LTQ-Orbitrap or Q-Exactive mass spectrometer (Thermo Scientific) outfitted with a custom, electrospray ionization interface. For the LTQ-Orbitrap, the heated capillary temperature and spray voltage were 275°C and 2.2 kV, respectively. The LTQ-Orbitrap mass spectra were collected from 400–2000 mass to charge ratio at a resolution of 60 000 followed by a data-dependent ion trap collision-induced dissociation tandem mass spectrometry (MS/MS) of the six most abundant ions. For Q-Exactive Plus, the heated capillary temperature and spray voltage were 325°C and 2.3 kV, respectively. Mass spectra were collected from 400–2000 mass to charge ratio at a resolution of 70 000 followed by a data-dependent ion trap higher-energy collisional dissociation MS/MS at a resolution of 17.5 000 of the 10 most abundant ions.
Results and Discussion
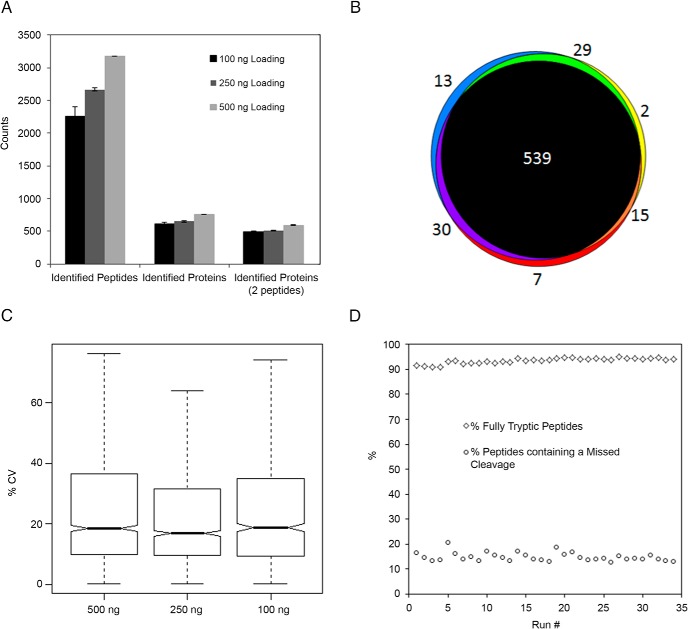
We characterized the performance of the SNaPP system by coupling it to an LTQ-orbitrap XL (Thermo Scientific) and analyzing serially diluted aliquots of a Shewanella oneidensis whole-cell lysate. Initial protein concentration was determined by Coomassie protein assay kit (Thermo Scientific). To determine the performance of SNaPP system handling small sample loadings, three replicate injections at total protein loadings of 100, 250, and 500 ng were obtained. To limit the impact of column carryover on our measurements, two full system blanks were carried out immediately preceding the 100-ng injections, and loading masses were analyzed in ascending order. The average number of protein identifications per replicate is displayed in Figure 2A. The data showed an essentially linear decrease in identifications with decreases in sample loading but still confidently identified more than 2000 peptides and 600 proteins for the 100-ng protein loadings. We observed very high reproducibility in the proteins identified (Figure 2B) and a median peptide intensity coefficient of variance (CV) of less than 20% for all three sample loadings (Figure 2C). This high level of reproducibility for label-free quantification is very encouraging because precise handling is increasingly challenging as sample amounts are reduced. Intriguingly, we saw no statistically significant difference in reproducibility of peptide intensities with decreased protein loading. We evaluated the stability of our immobilized trypsin column by carrying out 35 replicate injections of 500 ng of our cell lysate sample. The digestion quality is illustrated in Figure 2D by the percentage of identified peptides that contain a missed cleavage and the percentage of fully tryptic peptide identifications. These data also demonstrate consistent column performance over the 35 digestions.
Figure 2.
A, Number of confident (FDR < 1%) protein and peptide identifications from three replicate analyses at each protein loading. Error bars represent SD. B, Overlap of protein identifications at 500 ng protein loading. C, Box plot of peptide CV distributions at the different protein loadings. D, Stability of immobilized trypsin column demonstrated by performing 35 replicate injections on the same column.
To evaluate the capability to detect sample type-specific protein signatures and novel biological insight from small sample concentrations, we coupled our system to a Q-Exactive mass spectrometer (Thermo Scientific) for the analysis of mouse blastocysts. Based on the size and number of blastocysts in our samples, it was estimated that each sample contained between 440 and 500 ng (∼20 ng/blastocyst of total protein [17]; 22, 22, and 25 blastocyst were collected from samples 1, 2, and 3, respectively).
Before analyzing our blastocyst samples, we first validated the performance of our system in combination with the Q-Exactive mass spectrometer (Thermo Scientific) by running three replicates of our quality control sample at a protein loading of 200 ng. As expected, protein coverage was significantly increased, yielding an approximately 3-fold increase in both protein and peptide identifications compared with the Orbitrap XL (Thermo Scientific). Furthermore, median peptide CV decreased by 5%. We believe the increased sensitivity allows for more accurate determination of peak areas, resulting in the observed improvement in reproducibility.
SNaPP characterization of our three blastocyst samples identified 348 proteins (Supplemental Table 1), which ranged in length from 44 (thymosin-β-10) to 1675 (clathrin heavy chain 1) amino acids. The Gene Ontology classification (18) of these proteins (networks and bar graphs in Supplemental Figures 1–3) characterized them as mainly localizing to the nucleus and cytoplasm and having essential roles associated with ribosomes, RNA binding, ATP binding, and DNA binding, all of which are essential to blastocyst growth and survival. Protein identifications were made using MS-GF+ scoring to generate a spectra level confidence value (19), and the resulting spectral identifications were filtered to less than a 1% peptide level false discovery rate (based on decoy searches) using MS-GF+ MS/MS spectral scores and parent ion mass errors as described previously (20). Each reported protein was identified by at least one unique peptide (peptides that are not shared between multiple proteins) and quantified in at least two of our three blastocyst samples. Table 1 contains 22 proteins with both localization and biological functions significant to blastocysts; the biological significance of these proteins is detailed below and in Supplemental Table 1. Table 1 depicts the relative normalized log2 abundance values of each protein and the percentage coverage (number of peptides identified) across the three blastocyst samples. The protein abundance values in Table 1 highlight the high degree of correlation across the three samples; consequently, future studies could accurately quantitate significant blastocyst protein abundance changes in perturbed systems (ie, on time vs delayed implantation), even with variable numbers of peptides (percentage coverage) identified in each analysis.
Table 1.
Proteins Identified in Preimplantation Blastocysts
| Uniprot Gene Name UniProt Protein | Protein Name | Abundancea/Sequence Coverage, %b |
|||||
|---|---|---|---|---|---|---|---|
| Blastocyst Sample 1 (n = 22) | Blastocyst Sample 2 (n = 22) | Blastocyst Sample 3 (n = 25) | |||||
| Astl ASTL_MOUSE | Astacin-like metalloendopeptidase | 25.17 | 4% | 25.04 | 6% | 26.41 | 2% |
| Zp1 ZP1_MOUSE | Zona pellucida sperm-binding protein 1 | 29.51 | 20% | 29.01 | 22% | 28.35 | 20% |
| Zp2 ZP2_MOUSE | Zona pellucida sperm-binding protein 2 | 35.01 | 48% | 35.08 | 46% | 34.92 | 43% |
| Zp3 ZP3_MOUSE | Zona pellucida sperm-binding protein 3 | 33.82 | 28% | 33.24 | 25% | 32.51 | 28% |
| Ovgp1 OVGP1_MOUSE | Oviduct-specific glycoprotein | 26.82 | 23% | 27.06 | 27% | 27.43 | 27% |
| Cct8 TCPQ_MOUSE | T-complex protein 1 subunit theta | 23.70 | 2% | 24.09 | 2% | ||
| Hmga2 HMGA2_MOUSE | High-mobility group protein HMGI-C | 27.23 | 21% | 22.00 | 21% | 26.14 | 21% |
| Mea1 MEA1_MOUSE | Male-enhanced antigen 1 | 22.74 | 10% | 23.84 | 10% | 23.61 | 10% |
| Nasp NASP_MOUSE | Nuclear autoantigenic sperm protein | 22.19 | 3% | 24.83 | 3% | 21.63 | 3% |
| Nlrp14 NAL14_MOUSE | Germ cell specific leucine-rich repeat NTPase | 26.13 | 13% | 26.05 | 12% | 25.69 | 5% |
| Cops8 CSN8_MOUSE | COP9 signalosome complex subunit 8 | 23.62 | 17% | 23.44 | 17% | ||
| Dppa5a DPA5A_MOUSE | Developmental pluripotency-associated protein 5A | 24.59 | 54% | 24.23 | 53% | 23.08 | 54% |
| Dppa3 DPPA3_MOUSE | Developmental pluripotency-associated protein 3 | 25.75 | 8% | 26.10 | 8% | 25.95 | 8% |
| Khdc3 KHDC3_MOUSE | KH domain-containing protein 3 | 26.38 | 16% | 25.94 | 17% | 26.75 | 22% |
| Nlrp5 NALP5_MOUSE | Maternal antigen that embryos require | 25.36 | 11% | 26.10 | 12% | 25.41 | 7% |
| Npm2 NPM2_MOUSE | Nucleoplasmin-2 | 29.75 | 48% | 30.59 | 48% | 29.88 | 43% |
| Oosp1 OOSP1_MOUSE | Oocyte-secreted protein 1 | 20.61 | 9% | 22.05 | 9% | ||
| Padi6 PADI6_MOUSE | Protein-arginine deiminase type-6 | 31.02 | 78% | 31.40 | 78% | 30.76 | 65% |
| Alppl2 PPBN_MOUSE | Alkaline phosphatase, placental-like | 25.48 | 5% | 24.21 | 7% | ||
| Spin1 SPIN1_MOUSE | Spindlin-1 | 25.49 | 23% | 25.44 | 18% | 25.08 | 23% |
| Psmc3 PRS6A_MOUSE | 26S protease regulatory subunit 6A | 23.84 | 11% | 21.95 | 11% | 24.27 | 11% |
| Sftpd SFTPD_MOUSE | Pulmonary surfactant-associated protein D | 25.36 | 3% | 25.48 | 3% | ||
Abbreviations: n, the number of blastocysts collected per mouse. Blank values indicate where unique peptides were not detected for that protein in that blastocyst sample.
In DAnTE software, the area under the curve for the unique peptide peaks detected in the MS1 scans were log2 scaled and central tendency normalized and assessed at the protein level using Rrollup (reference peptide based scaling) (42).
Percentage coverage values were calculated from unique and nonunique peptides.
Our system characterized numerous proteins found in the zona pellucida (ZP), which is a thick transparent membrane surrounding mammalian ova before fertilization and fertilized embryos before implantation. Zona pellucida glycoproteins create a sperm-egg interface regulating species-specific binding of sperm to unfertilized eggs and preventing sperm from binding to fertilized eggs (blockade of polyspermy) (21). The mouse zona pellucida is composed of three glycoproteins (Zp1, Zp2, and Zp3), of which Zp2 is proteolytically cleaved by ovastacin (Astl), an oocyte-specific member of the astacin family of metalloendoproteases, after gamete fusion to prevent polyspermy (22). The molecular chaperone, Cct8, is involved in mediating sperm-oocyte interactions (23). We also characterized oviduct-specific glycoprotein (Ovgp1) (24), sperm proteins (Hmga2, Mea1, Nasp, Nlrp14), oocyte/embryo proteins (Cops8, Dppa5a, Khdc3, Nlrp5 [25, 26], Npm2, Oosp1 [27], Padi6, Alppl2, Spin1 [28]), proteasomal ATPase Psmc3 and pulmonary surfactant-associated protein D (Sftpd). Previous studies have shown that Psmc3-deficient mice died before implantation, displaying defective blastocyst development (29), and fetal Sftpd Met31Thr polymorphism plays a significant role in the genetic predisposition to spontaneous preterm birth in humans (30).
Our system also characterized 59 enzymes classified as oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases (Supplemental Table 1). Identified oxidoreductases included dehydrogenases with diverse metabolic roles (Impdh2, Ldhb, Mdh2, Gapdh, Aldh2, Aldh7a1, Pdhb, Paox), five peroxiredoxins (Prdx1, Prdx2, Prdx3, Prdx4, and Prdx6), and superoxide dismutase (Sod1). Peroxiredoxin and superoxide antioxidants perhaps protect blastocysts from oxidative stress (31, 32). The dehydrogenase Aldh7a1 also protects cells from oxidative stress by metabolizing a number of lipid peroxidation-derived aldehydes (33). Subcategorization of identified transferases include a methyltransferase (Bhmt), acetyltransferases (Acat1 and Ggct), a glycosyltransferase (Gyg1), and phosphotransferases (Pkm and Ckb [34]). Identification of spermidine aminopropyltransferase (Sms) and polyamine oxidase (Paox) supports the importance of polyamines to preimplantation mouse embryos (35). The 28 identified hydrolases include Astl (Table 1), Alppl2 (Table 1), Hint1, Park7, Asna1, and hydrolases acting on ester bonds (Rgn, Mtmr14, Acp1), peptide bonds (Ctsa, Apeh, deubiquitinating enzymes [Eif3f, Otub1, and ubiquitin carboxyl-terminal hydrolase isozyme L1, Uchl1] [36, 37], Prss2, cathepsin [Ctsb and Ctsd], proteasome subunits [Psma5 and Psmb6]), carbon-nitrogen bonds (Padi6 [Table 1], and Gda), and acid anhydrides (Dctpp1, Nudt5, Atp5b, Atp6v1a, Ddx21, Ddx5, Eif4a1, and Vcp). We also identified diverse isomerases (Rpe, Ppia, Bpgm, and intramolecular oxidoreductases [Mif, Pdia3, Pdia6, Ptges3]), a lyase (Ddt), and ligases (Trim28 and Uhrf1).
To demonstrate the ability of SNaPP to detect posttranslational modifications in global digests, we searched our blastocyst data for peptides containing diglycine adducted (114.04 Da) lysine residues, and these peptides are referred to as ubiquitin remnant-containing peptides (38) or ubiquitin signature peptides (39). If a peptide is modified by polyubiquitination, monoubiquitination, or an ubiquitin-like modifier (eg, NEDD8 or ISG15), trypsin digestion results in a diglycine tag. We characterized six modified proteins across all three blastocyst samples (Supplemental Table 1) including ATP synthase-coupling factor 6 (Atp5j), calmodulin (Calm1), elongation factor 1δ (Eef1d), 40S ribosomal protein S12 (Rps12), thymosin β-10 (Tmsb10), and ubiquitin carboxyl-terminal hydrolase isozyme L1 (Uchl1) (37). A previous study suggested that ubiquitin-dependent proteolysis occurs in the trophoblast (vs inner cell mass) of mouse blastocysts (40).
In summary, our online sample handling platform allows for robust semiautomated proteomic analysis of conventionally problematic samples containing between 100 and 500 ng protein content (5–25 blastocysts). The proteomic results from this study provided novel insights into the proteomic landscape of blastocysts that would likely be obscured by bulk proteomic measurements. The ability to do future global proteomic analyses on, for example, blastocysts from a single mouse, represents an important advance for developmental research because it allows for the investigation of interindividual variation and greatly increases the statistical power attainable while decreasing the number of animals needed, as opposed to a large number of animals previously reported for blastocyst proteomic analysis (41). This approach should be easily adaptable to other cell-specific sample types.
Acknowledgments
Portions of the experimental work described herein were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy and located at the Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the Department of Energy.
This work was supported in part by the Laboratory Directed Research and Development Program at the Pacific Northwest National Laboratory, National Institutes of Health Grant P41GM103493 (to R.D.S.) and Grant R01HD068524 from the National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development and March of Dimes (to S.K.D.). The Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the Department of Energy under Contract DE-AC05-76RL0 1830.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CV
- coefficient of variance
- IMER
- immobilized enzyme reactor
- MS/MS
- tandem mass spectrometry
- SnaPP
- simplified nanoproteomics platform
- SPE
- solid-phase extraction.
References
- 1. Nilsson T, Mann M, Aebersold R, Yates JR, Bairoch A, Bergeron JJM. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. [DOI] [PubMed] [Google Scholar]
- 2. Piehowski PD, Petyuk VA, Orton DJ, et al. Sources of technical variability in quantitative LC-MS proteomics: human brain tissue sample analysis. J Proteome Res. 2013;12:2128–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 2010;28:710–721. [DOI] [PubMed] [Google Scholar]
- 4. Altelaar AFM, Heck AJR. Trends in ultrasensitive proteomics. Curr Opin Chem Biol. 2012;16:206–213. [DOI] [PubMed] [Google Scholar]
- 5. Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. [DOI] [PubMed] [Google Scholar]
- 7. Wang HX, Qian WJ, Mottaz HM, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4:2397–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wisniewski JR, Ostasiewicz P, Mann M. High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J Proteome Res. 2011;10:3040–3049. [DOI] [PubMed] [Google Scholar]
- 9. Braakman RBH, Tilanus-Linthorst MMA, Liu NQ, et al. Optimized nLC-MS workflow for laser capture microdissected breast cancer tissue. J Proteomics. 2012;75:2844–2854. [DOI] [PubMed] [Google Scholar]
- 10. Liebler DC, Ham AJL. Spin filter-based sample preparation for shotgun proteomics. Nat Methods. 2009;6:785. [DOI] [PubMed] [Google Scholar]
- 11. Nel AJ, Garnett S, Blackburn JM, Soares NC. Comparative reevaluation of FASP and enhanced FASP methods by LC-MS/MS. J Proteome Res. 2015;14(3):1637–1642. [DOI] [PubMed] [Google Scholar]
- 12. Hustoft HK, Vehus T, Brandtzaeg OK, et al. Open tubular lab-on-column/mass spectrometry for targeted proteomics of nanogram sample amounts. PLoS One. 2014;9:e106881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun L, Zhu G, Dovichi NJ. Integrated capillary zone electrophoresis-electrospray ionization tandem mass spectrometry system with an immobilized trypsin microreactor for online digestion and analysis of picogram amounts of RAW 264.7 cell lysate. Anal Chem. 2013;85:4187–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Slysz GW, Lewis DF, Schriemer DC. Detection and identification of sub-nanogram levels of protein in a nanoLC-trypsin-MS system. J Proteome Res. 2006;5:1959–1966. [DOI] [PubMed] [Google Scholar]
- 15. Girelli AM, Mattei E. Application of immobilized enzyme reactor in on-line high performance liquid chromatography: a review. J Chromatogr B Anal Technol Biomed Life Sci. 2005;819:3–16. [DOI] [PubMed] [Google Scholar]
- 16. Xie H, Sun X, Piao Y, et al. Silencing or amplification of endocannabinoid signaling in blastocysts via CB1 compromises trophoblast cell migration. J Biol Chem. 2012;287:32288–32297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weitlauf HM. Changes in the protein content of blastocysts from normal and delayed implanting mice. Anat Rec. 1973;176:121–123. [DOI] [PubMed] [Google Scholar]
- 18. Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7:3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Piehowski PD, Petyuk VA, Sandoval JD, et al. STEPS: a grid search methodology for optimized peptide identification filtering of MS/MS database search results. Proteomics. 2013;13:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wassarman PM. Zona pellucida glycoproteins. J Biol Chem. 2008;283:24285–24289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burkart AD, Xiong B, Baibakov B, Jimenez-Movilla M, Dean J. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J Cell Biol. 2012;197:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dun MD, Smith ND, Baker MA, Lin M, Aitken RJ, Nixon B. The chaperonin containing TCP1 complex (CCT/TRiC) is involved in mediating sperm-oocyte interaction. J Biol Chem. 2011;286:36875–36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sendai Y, Komiya H, Suzuki K, et al. Molecular cloning and characterization of a mouse oviduct-specific glycoprotein. Biol Reprod. 1995;53:285–294. [DOI] [PubMed] [Google Scholar]
- 25. Tong ZB, Gold L, De Pol A, et al. Developmental expression and subcellular localization of mouse MATER, an oocyte-specific protein essential for early development. Endocrinology. 2004;145:1427–1434. [DOI] [PubMed] [Google Scholar]
- 26. Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mano H, Nakatani S, Aoyagi R, et al. IF3, a novel cell-differentiation factor, highly expressed in murine liver and ovary. Biochem Biophys Res Commun. 2002;297:323–328. [DOI] [PubMed] [Google Scholar]
- 28. Oh B, Hwang SY, Solter D, Knowles BB. Spindlin, a major maternal transcript expressed in the mouse during the transition from oocyte to embryo. Development. 1997;124:493–503. [DOI] [PubMed] [Google Scholar]
- 29. Sakao Y, Kawai T, Takeuchi O, et al. Mouse proteasomal ATPases Psmc3 and Psmc4: genomic organization and gene targeting. Genomics. 2000;67:1–7. [DOI] [PubMed] [Google Scholar]
- 30. Karjalainen MK, Huusko JM, Tuohimaa A, Luukkonen A, Haataja R, Hallman M. A study of collectin genes in spontaneous preterm birth reveals an association with a common surfactant protein D gene polymorphism. Pediatr Res. 2012;71:93–99. [DOI] [PubMed] [Google Scholar]
- 31. Hirota Y, Acar N, Tranguch S, et al. Uterine FK506-binding protein 52 (FKBP52)-peroxiredoxin-6 (PRDX6) signaling protects pregnancy from overt oxidative stress. Proc Natl Acad Sci USA. 2010;107:15577–15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho YS, Gargano M, Cao J, Bronson RT, Heimler I, Hutz RJ. Reduced fertility in female mice lacking copper-zinc superoxide dismutase. J Biol Chem. 1998;273:7765–7769. [DOI] [PubMed] [Google Scholar]
- 33. Brocker C, Lassen N, Estey T, et al. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Forsey KE, Ellis PJ, Sargent CA, Sturmey RG, Leese HJ. Expression and localization of creatine kinase in the preimplantation embryo. Mol Reprod Dev. 2013;80:185–192. [DOI] [PubMed] [Google Scholar]
- 35. Zwierzchowski L, Czlonkowska M, Guszkiewicz A. Effect of polyamine limitation on DNA synthesis and development of mouse preimplantation embryos in vitro. J Reprod Fertil. 1986;76:115–121. [DOI] [PubMed] [Google Scholar]
- 36. Sekiguchi S, Kwon J, Yoshida E, et al. Localization of ubiquitin C-terminal hydrolase L1 in mouse ova and its function in the plasma membrane to block polyspermy. Am J Pathol. 2006;169:1722–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mtango NR, Sutovsky M, Susor A, Zhong Z, Latham KE, Sutovsky P. Essential role of maternal UCHL1 and UCHL3 in fertilization and preimplantation embryo development. J Cell Physiol. 2012;227:1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng J, Schwartz D, Elias JE, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. [DOI] [PubMed] [Google Scholar]
- 40. Sutovsky P, Motlik J, Neuber E, et al. Accumulation of the proteolytic marker peptide ubiquitin in the trophoblast of mammalian blastocysts. Cloning Stem Cells. 2001;3:157–161. [DOI] [PubMed] [Google Scholar]
- 41. Fu Z, Wang B, Wang S, et al. Integral proteomic analysis of blastocysts reveals key molecular machinery governing embryonic diapause and reactivation for implantation in mice. Biol Reprod. 2014;90:52. [DOI] [PubMed] [Google Scholar]
- 42. Polpitiya AD, Qian W-J, Jaitly N, et al. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics (Oxford, England). 2008;24:1556–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]