Abstract
During protein synthesis, ribosomes become stalled on polyproline-containing sequences, unless they are rescued in archaea and eukaryotes by the initiation factor 5A (a/eIF-5A) and in bacteria by the homologous protein EF-P. While a structure of EF-P bound to the 70S ribosome exists, structural insight into eIF-5A on the 80S ribosome has been lacking. Here we present a cryo-electron microscopy reconstruction of eIF-5A bound to the yeast 80S ribosome at 3.9 Å resolution. The structure reveals that the unique and functionally essential post-translational hypusine modification reaches toward the peptidyltransferase center of the ribosome, where the hypusine moiety contacts A76 of the CCA-end of the P-site tRNA. These findings would support a model whereby eIF-5A stimulates peptide bond formation on polyproline-stalled ribosomes by stabilizing and orienting the CCA-end of the P-tRNA, rather than by directly contributing to the catalysis.
INTRODUCTION
eIF-5A and EF-P were initially suggested to stimulate catalysis of the first peptide bond directly following translation initiation (1,2). Subsequently, both factors were shown to rescue ribosomes stalled during translation of polyproline-containing proteins, and that this activity required the presence of unique post-translational modifications (2–7). eIF-5A is post-translationally modified with hypusine (Nϵ-(4-amino-2-hydroxybutyl)lysine), a modification so far unique to IF-5A proteins (2,8,9). Hypusine is formed in two enzymatic reactions: the 4-aminobutyl moiety of spermidine is added to the ϵ-amino group of a conserved lysine residue (K51 in yeast) of eIF-5A by deoxyhypusine synthase (DHS), producing deoxyhypusine, which is converted in a second step to hypusine via hydroxylation by deoxyhypusine hydroxylase (DOHH) (2,9,10). The HYP2, DYS1 and LIA1 genes encoding IF-5A, DHS and DOHH, respectively, are strictly conserved across all eukaryotes. While the presence of genes encoding IF-5A and DHS are essential for viability, yeast strains lacking LIA1 are viable and grow only slightly slower than wild-type strains (2,9). Consistently, the deoxyhypusine-modified form of yeast eIF-5A is equally efficient at relieving ribosome stalled at polyproline stretches as the hypusine-modified eIF-5A (7). In contrast, loss of LIA1 is recessively lethal in Caenorhabditis elegans and Drosophila, suggesting that the final hydroxylation step plays an important role for eIF-5A activity in higher eukaryotes (2). Unlike eukaryotes and archaea, bacteria have developed diverse and unrelated pathways to modify the equivalent residue, including lysinylation (11–13) and rhamnosylation (14,15). While a crystal structure of an unmodified EF-P in complex with a bacterial 70S ribosome has been reported (16), to date no structures exist of the modified eIF-5A in complex with a eukaryotic 80S ribosome, and the molecular basis of its rescue activity, in particular of its unique hypusine modification, is not understood.
MATERIALS AND METHODS
Native pullouts of Ski complex bound ribosomal complexes using TAP-tagged Ski3
In vivo pullouts were performed using Dynabeads® M-270 Epoxy (Life Technologies) with yeast strains expressing C-terminally TAP-tagged Ski3 (strains obtained from Euroscarf). Ski3 is a subunit of the Ski complex which is involved in 3′ to 5′ mRNA degradation by the cytosolic exosome and mRNA quality control (17–19). Purifications were essentially performed as described before (20). Cultures were harvested at log phase immediately after the addition of 10 μg/ml cycloheximide and lysed by glass bead disruption. Incubation with IgG-coupled magnetic beads was done for 12 h at 4°C with slow tilt rotation followed by elution with AcTEV Protease for 180 min at 17°C.
Ribosome profiling of in vivo pullouts using TAP-tagged Ski3
Ribosome profiling experiments were performed as described previously (21), but with some modifications. Approximately five A260 units of the Ski-3-TAP tagged pullouts were used for ribosome profiling. The pullout fraction was digested with RNase I (Ambion) at 25°C for 45 min in a shaker at 500 rpm followed by 5 min incubation on ice with SUPERase-In (Ambion). Following nuclease digestion, ribosomes were pelleted through a sucrose cushion (20 mM HEPES pH 7.5, 100 mM KOAc, 10 mM MgCl2, 750 mM sucrose, 1 mM DTT, 0.5 mM PMSF, 10 μg/ml cycloheximide, Protease Inhibitor Cocktail (Roche)) by centrifugation at 312 000 × g for 45 min at 4°C. Ribosomal pellets were resuspended in ribosome-splitting buffer (20 mM Tris, 400 mM KCl, 2 mM MgCl2, 1 mM DTT and 1 mM puromycin) and incubated on ice for 30 min. Ribosomal subunits were pelleted again by centrifugation at 250 000 × g for 90 min at 4°C. The supernatant from this centrifugation step was used as the source for ribosome protected fragments (RPFs).
RPFs were further purified and size selected in a 15% denaturing sodium dodecyl sulphate-polyacrylamide gel electrophoresis gel for fragments between 26–62 nt using markers. Gel extracted fragments were precipitated and processed as given in the protocol (ARTseq™ Ribosome Profiling Kit, Epicentre, WI, USA) for library preparation and high-throughput sequencing. Sequencing was performed on an Illumina HiSeq 1500. Reads were adapter-trimmed using the software Cutadapt (version 1.2.1, EMBnet.journal, 2011). Reads mapping to ribosomal RNA, tRNA, small nuclear and nucleolar RNA were removed. Remaining reads were mapped to the yeast genome (R64–1–1, 19 July 2014) using Tophat (v2.0.8b) (22) with the following parameters: -a 4 –no-novel-juncs –GTF. Only uniquely mapped reads were used for further bioinformatic analysis. For identifying the P-site position within the footprints, a meta-gene analysis using 5′ end of the footprints around the start codon was performed. Based on this analysis, the first peak appeared 12 nt upstream of the start codon. A-, P- and E-site codons for all footprints were assigned by shifting 16, 13 and 10 nt, respectively. The number of footprints per amino acid in each site was calculated by summing up the shifted footprints over the corresponding codons. To normalize the occurrence, the codons in the second position downstream of the A-site were summed up and the occurrence for each amino acid was calculated. The values for the amino acids in A-, P- and E-site where then divided by the occurrence for each corresponding amino acid in this second position accordingly. For all the footprints mapping to ORFs, the normalized occurrence of A-, P- and E-site amino acids were then plotted in Supplementary Figure S1.
Cryo-electron microscopy and single particle reconstruction
The ribosomes from the TEV-eluate were adjusted to a concentration of 4 A260/ml (0.8 μM 80S ribosomes), applied to 2 nm pre-coated Quantifoil R3/3 holey carbon supported grids and vitrified using a Vitrobot Mark IV (FEI Company). Data collection was performed on a Titan Krios TEM (FEI Company) equipped with a Falcon II direct electron detector, operated at 300 keV using the EPU software (FEI). The magnification settings resulted in a pixel size of 1.084 Å/pixel. The dataset was provided with the semi-automatic software EPU (FEI Company) with a dose of 2.4 e−/Å−2 per frame for 13 frames in total. The frames were aligned using the Motion Correction software (23). Data were collected at a defocus range between −0.8 and −3.4 μm. Only micrographs that showed clearly visible Thon rings below 5.5 Å on the level of the rotationally averaged power spectra profiles were used for further analysis. Automatic particle detection was performed by the program SIGNATURE (24). Initial in silico sorting of the dataset consisting of 246.555 particles in total was performed using the SPIDER software package (25). Classes were obtained by competitive projection matching in SPIDER (26,27). The vast majority (>95%) of the particles we found were programmed with tRNAs. This dataset could be subdivided into two main classes, both containing A- and P-tRNAs and either with or without the Ski complex (76 816 and 88 640 particles, respectively). The large number of ribosomal particles without density for the Ski complex suggests that the Ski complex was not stably bound to these particles. To our surprise, both classes contained ribosome-bound eIF-5A. Nevertheless, only the class without Ski complex displayed a highly resolved density for eIF-5A that allowed model building. The density for eIF-5A in the Ski complex bound ribosome class was partially disordered and the density for the factor was fragmented. For high-resolution refinement, the dataset containing the eIF-5A particles was further cleaned by removing particles with low cross-correlation. The cleaned dataset (62 532 particles) was then processed further using RELION (28). To do this, the particle boxes were extracted using the coordinates obtained by SIGNATURE and normalized in RELION. The contrast transfer function (CTF) estimation was repeated using CTFFIND3 (29) and the dataset was subjected to auto-refinement in RELION using a ribosomal reference low-pass filtered to 70 Å. After auto-refinement, the dataset was subjected to movie processing and the particle-polishing feature in RELION. Here, only the first eight frames were used for the calculations, resulting in an accumulated dose of 24 e−/Å−2. Subsequent auto-refinement of ‘shiny’ particles resulted in a final reconstruction of 3.9 Å resolution according to a gold standard fourier shell correlation (FSC) cutoff of 0.143. This map was sharpened using automatic b-factor estimation in RELION and used for interpretation and model building. Local resolution was calculated using ResMap (30) and maps were visualized in UCSF Chimera (31). RELION data were processed on the Leibnitz-Rechenzentrum (LRZ) Munich.
Model building
For modeling the large ribosomal subunit (LSU), the crystal structure of the yeast ribosome (PDB ID: 4V88) (32) was taken as a template. Peptidyl A- and deacylated P-tRNA were modeled based on the crystal structure of the Thermus thermophilus 70S ribosome in the post-catalysis state of peptide bond formation (containing dipeptidyl-tRNA in the A site and deacylated tRNA in the P site, PDB ID: 1VY5) (33) and for eIF-5A a homology model was generated using HHPred (34). All structures were roughly fitted into the map using UCSF Chimera. Flexible fitting and, where necessary, de novo model building was done in Coot (35) followed by real space refinement in PHENIX (36). For the rRNA and the tRNAs, geometry restrictions were calculated using the ‘PDB to 3D restraints’ database prior to PHENIX refinement.
The eIF-5A homology model was obtained after a multiple alignment using HHPred. This model was subjected to geometry minimization using PHENIX and remodeled in Coot. The well-resolved hypusine-containing β3-β4 loop (residues 47–54) was modeled de novo and for the N-terminal extension (NTE; res 1–16) a poly-Ala model was generated. For uL16, the loop containing residues 103–111 (not present in the yeast ribosome X-ray structure (32)) was modeled de novo. The L1 stalk in the eIF-5A position was remodeled and a poly-alanine model for uL1 was generated using uL1 from the human 80S ribosome as a template (37) (PDB ID: 5AJ0). In a final step, all models were combined and subjected to real-space refinement using the PHENIX software.
RESULTS
Presence of eIF-5A on Ski complex-associated ribosomes
Translational stalling can also result when ribosomes translate mRNAs containing secondary structure, premature termination codons or aberrant mRNAs lacking stop codons, which in eukaryotes evokes specialized pathways of mRNA degradation, such as non-stop decay (NSD), nonsense-mediated decay (NMD) or No-Go decay (NGD), respectively (38,39). These stalled ribosomes are recognized by termination release factors (eRF1 and eRF3 in NMD), or their homologs (Dom34 and Hbs1 in NSD and NGD), which trigger recycling of the ribosome and degradation of the aberrant mRNAs (40). In yeast, the exosome-Ski (Ski2p, Ski3p, Ski8p) complex has been implicated in degradation of aberrant mRNA lacking a stop codon (18,19). We recently performed in vivo pullouts using a yeast strain expressing affinity-tagged Ski3p in order to investigate the interaction of the Ski complex with the ribosome that, to our surprise, also contained eIF-5A. Further biochemical analysis showed that the mRNAs in the A-, P- and E-sites contained a mixture of codons for all amino acids, with some enrichment for phenylalanine, histidine and proline codons at the A-site (Supplementary Figure S1). The lack of significant enrichment for proline and methionine within the P-site, suggests that the isolated complex does not represent a polyproline-stalled ribosomal state, nor an initiation complex, as might be expected for an eIF-5A substrate. Moreover, yeast eIF-5A has been implicated in mRNA degradation, with its inactivation leading to a stabilization of short-lived mRNAs (41,42) and especially nonsense-containing mRNAs (43). An alternative possibility is that the antibiotic cycloheximide, which was added to the cells before harvesting, promotes eIF-5A binding to the ribosome. Cycloheximide binds with the E-site of the eukaryotic ribosome overlapping the A76 position of the CCA-end of the E-tRNA (44,45) and prevents translocation of E-tRNA into the E-site (46,47). Indeed, eIF-5A could not be detected using mass spectrometry on the ribosomes obtained from replicate in vivo pullouts using the affinity-tagged Ski3p that were performed in the absence of cycloheximide. This suggests that cycloheximide generates a functional state of the ribosome, namely with a P-tRNA and free E-site, that is recognized and bound by eIF-5A.
Cryo-EM structure of eIF-5A on the ribosome
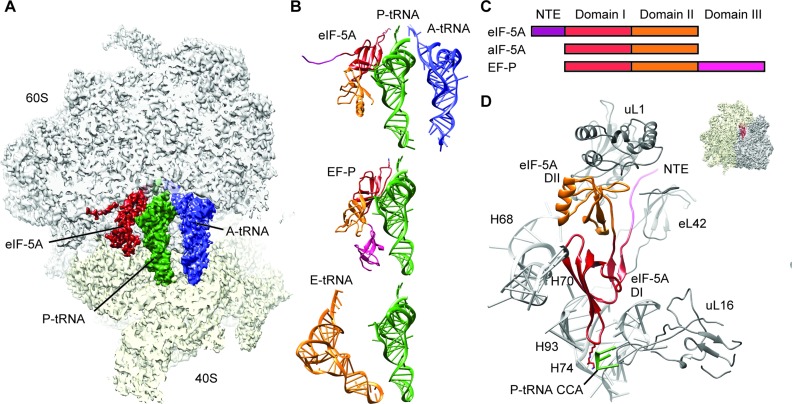
Cryo-EM analysis of this ribosome preparation and in silico sorting of this dataset yielded a subpopulation of ribosomal particles that contained stoichiometric occupancy of A-tRNA, P-tRNA and eIF-5A. The final reconstruction of the yeast eIF-5A–80S complex (Figure 1A) had an average resolution of 3.9 Å and local resolution extending to 3.5 Å in the core of the ribosome (Supplementary Figure S2A and B). The electron density for the ribosome, tRNAs, cycloheximide and the majority of eIF-5A was well-resolved (Supplementary Figure S2B–F), enabling us to present a complete molecular model of the yeast eIF-5A–80S complex. While the molecular model includes side chains for domains I and II of eIF-5A, the N-terminal residues 1–16 are less ordered and therefore only a polyalanine backbone trace was generated. On the ribosome, eIF-5A is located between the P- and E-sites, interacting with the E-site surface of P-tRNA (Figure 1A and B), analogous to the bacterial homolog EF-P, as well as overlapping the binding site of E-tRNA (Figure 1B) (48). The conformation of ribosome-bound eIF-5A is very similar to the unbound form observed in the crystal structures of yeast eIF-5A, as well as archaeal and eukaryotic IF-5As and bacterial EF-Ps (Supplementary Figure S3), with the exception that β3–β4 loop bearing the hypusine modification re-adjusts upon binding. In addition, we observe the NTE (Figures 1D and 2A; Supplementary Figure S2B) that was disordered in the eukaryotic crystal structures (49), and is absent from archaeal IF-5As and bacterial EF-Ps (Figure 1C; Supplementary Figure S3).
Figure 1.
Cryo-EM structure of eIF-5A bound to the yeast 80S ribosome. (A) Transverse section of the cryo-EM map of eIF-5A–80S complex, (40S, yellow; 60S, gray), revealing the binding site of eIF-5A (dark red), P-tRNA (green) and A-tRNA (blue). (B) Comparison of ribosome binding positions of eIF-5A, EF-P (16) and E-site tRNA(48), relative to A-tRNA (blue) and P-tRNA (green). The domains for eIF-5A and EF-P are colored according to (C). (C) Schematic representation of the domain structures of eIF-5A, aIF-5A and EF-P. (D) Molecular model for the interaction of domains I (DI, red) and II (DII, orange) as well as N-terminal extension (NTE, magenta) of eIF-5A with rRNA and ribosomal protein components of the ribosome (gray). Ribosomal insert shows the orientation of the view.
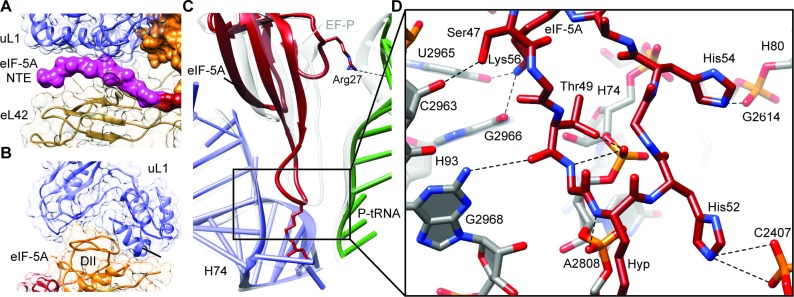
Figure 2.
Interaction of eIF-5A with the yeast 80S ribosome. (A) View of the NTE of eIF-5A sandwiched between ribosomal proteins uL1 (blue) and eL42 (tan). (B) Domain II (DII, orange) of eIF-5A inserts into the cleft between domains 1 and 2 of uL1 (blue). (C) Comparison of the ribosome binding position of domain I of eIF-5A (red), P-tRNA (green) and 25S rRNA helix 74 (H74, blue) relative to the EF-P, P-tRNA and H74 (gray) from the bacterial EF-P–70S complex (16). Arg27 of eIF-5A makes a potential hydrogen bond interaction (dashed line) with the P-tRNA (green). (D) Possible hydrogen bond interactions (dashed lines) between domain I of eIF-5A (red) with 25S rRNA nucleotides within helices H74, H80 and H93 of the ribosome (gray).
Interactions of eIF-5A on the ribosome
eIF-5A interacts with both rRNA and ribosomal protein components of the yeast 80S ribosome; Domain I of eIF-5A contacts predominantly 25S rRNA nucleotides located in helices H74 and H93 as well as to a lesser extent H68 and H70, whereas domain II and the NTE of eIF-5A contact ribosomal proteins uL1 and/or eL42 (Figure 1D). Domain II of eIF-5A is inserted between domains 1 and 2 of uL1 (Figure 2B), which induces a closed conformation of the L1 stalk, similar to that observed previously upon EF-P binding (16) (Supplementary Figure S4). The eIF-5A conformation of the L1 stalk results in a large inward movement toward the central protuberance, similar to that observed in the presence eEF2 (50) (Supplementary Figure S4). The eIF-5A conformation of the L1 stalk is stabilized by the eukaryotic-specific NTE of eIF-5A, which is sandwiched between uL1 and eL42 (Figure 2A). The absence of the NTE in bacterial EF-Ps may explain why the orientation of domain I differs when comparing the ribosome-bound EF-P and eIF-5A structures (Figure 2C), however, we cannot exclude that this results from differences in the species of tRNA in the P-site. On the bacterial ribosome, domain I of EF-P establishes a large interaction surface with acceptor stem of the P-tRNA (Figure 2C), comprising nine hydrogen bonds to the phosphate-oxygen backbone (16). In contrast, eIF-5A is shifted away from the P-tRNA and hydrogen bonding is observed from only Arg27 of eIF-5A to the backbone of nucleotides (G4-C5) of the P-tRNA (Figure 2C). Instead, contacts are observed from domain I of eIF-5A, in particular from the backbone and positively charged side chains of residues within the β3–β4 loop, which form a network of hydrogen bond interactions with 25S rRNA nucleotides, predominantly nucleotides 2806–2808 (Ec2437–2439) in H74 and 2963–2968 (Ec2594–2599) in H93 (Figure 2D and Supplementary Figure S5). Overall, the binding position of eIF-5A observed here is in excellent agreement with the hydroxyl-radical cleavages mapped onto the 25S rRNA and P-tRNA from iron tethers located in domains I and II of eIF-5A (7) (Supplementary Figure S6).
Hypusine of eIF-5A interacts with the CCA-end of the P-tRNA
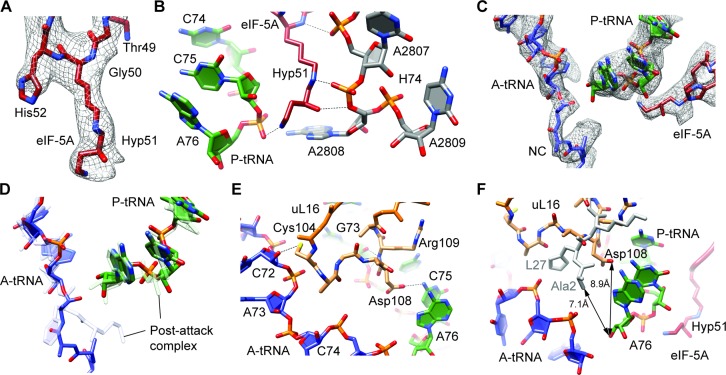
The electron density of eIF-5A enables the hypusine modification at the tip of the β3–β4 loop to be unambiguously modeled (Figure 3A). The hypusine moiety inserts into a pocket formed by the CCA-end of the P-tRNA and the backbone of nucleotides within H74, and is capped by nucleotide A2808 (EcA2439) that is flipped-out of H74 (Figure 3B). Two hydrogen bonds with 25S rRNA are possible from the K51 component of the hypusine moiety, namely, between the backbone amide (NH) and the ϵ-amino (NZ) group of K51, which are within 3.3 Å and 2.3 Å of the non-bridging phosphate-oxygens (OP1) of nucleotides U2807 and A2808 (Figure 3B). Although the amino-butyl component of the hypusine moiety extends the reach of eIF-5A toward the peptidyltransferase center, it is not sufficient to permit direct interaction with an amino acid or peptide attached to the P-tRNA (Figure 3B and C). Instead, the 4-amino group at the tip of the modification reaches only so far as to contact the backbone of the CCA-end of the P-tRNA, coming into hydrogen bond distance of the OP1 of A76 (Figure 3B). Our model suggests that the hydroxyl group would allow an additional hydrogen bond interaction with the O5’ bridging oxygen of A2808 (Figure 3B).
Figure 3.
Hypusine of eIF-5A at the peptidyltransferase center of the ribosome. (A) Electron density (gray mesh) and molecular model (red) for hypusine 51 (Hyp51) of eIF-5A. (B) Potential hydrogen bond interactions (dashed lines) between hypusine 51 (Hyp51) of eIF-5A (red) with the CCA-end of the P-tRNA (green) and 25S rRNA nucleotides within helix H74 (gray). (C) Electron density (gray mesh) and molecular models for the CCA-ends of P-tRNA (green) and A-tRNA with nascent chain (NC) (blue) as well as eIF-5A (red). (D) Comparison of A-tRNA (blue) and P-tRNA (green) from eIF-5A–80S complex with A- and P-tRNAs from post-attack complex (33) (gray). (E) Potential hydrogen bond interactions (dashed lines) between the loop of uL16 (orange), A-tRNA (blue) and P-tRNA (green). (F) Proximity of Asp108 of uL16 (orange) to the 3′ OH of A76 of the P-tRNA (8.9 Å) in the eIF-5A–80S complex, compared with the proximity of Ala2 of L27 (gray) to the 3′ OH of A76 of the P-tRNA (7.1 Å) in the pre-attack complex (33). The CCA-end of the A-tRNA (blue) and hypusine 51 (Hyp51) of eIF-5A (red) are shown for reference.
uL16 reaches into the PTC and stabilizes A- and P-tRNAs
Although electron density is observed for the nascent polypeptide chain within the ribosomal tunnel, it is not possible to determine the exact sequence of the nascent chain. This may be because the nascent chain is flexible but also because it represents a heterogeneous mixture of different sequences, as indicated by the biochemical analysis (Supplementary Figure S1). Nevertheless, the electron density clearly indicates that the nascent polypeptide chain is attached to the A-tRNA (Figure 3C), indicating that the eIF-5A complex is trapped in a pre-translocational state, but post-peptide bond formation. Similarly, the CCA-ends of the A- and P-tRNAs as well as 25S rRNA nucleotides within the PTC adopt conformations like those observed in recent X-ray structure of the bacterial ribosome in the post-peptide bond formation state (33) (Figure 3D). The eukaryotic-specific loop of uL16 (residues 104–110), which is poorly resolved in previous 80S structures (32,37), becomes ordered and establishes interactions with the CCA-ends of both the A- and P-tRNAs (Figure 3E). Specifically, the OD1 of Asp108 of uL16 is within hydrogen bonding distance of the N6 of A76 of the P-tRNA and the backbone amide and carboxyl of Arg109 are within 3.1–3.4 Å of the 2′ OH of G73 of the P-tRNA (Figure 3E). In addition, the SG of Cys 104 of uL16 can form a hydrogen bond with the 2′ OH of C72 of the P-tRNA (Figure 3E). This latter interaction of uL16 with the A-tRNA is likely to be important for loop stabilization since the loop remains disordered in the presence of P-tRNA but absence of A-tRNA (32,37). In bacteria with a shorter uL 16 loop, the N-terminus of ribosomal protein L27 occupies the equivalent position (Figure 3F), and is also ordered only upon A-tRNA binding (33,51,52). The N-terminus of L27 comes within 7.1 Å of the 3′ OH of A76 of the P-tRNA (Figure 3F) and has been proposed to play a role in a proton wire that couples aminoacyl-tRNA binding at the A-site to peptide bond formation (33). In the presence of A-tRNA, we now observe that Asp108 of uL16 comes within 8.9 Å of the 3′ OH of A76 of the P-tRNA (Figure 3F), raising the possibility that the uL16 loop could also play a more active role in peptide-bond formation. Consistently, mutations within the uL16 loop are lethal (53), possibly because they yield mutant ribosomes with altered A-tRNA binding and peptidyltransferase activities (54).
DISCUSSION
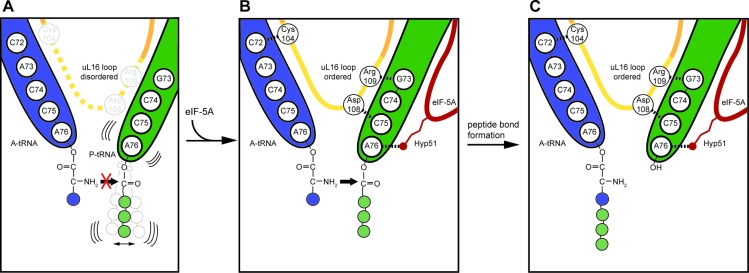
Although our structure of eIF-5A was not obtained on a polyproline-stalled ribosome, the results nevertheless allow us to suggest a model for how hypusinylated eIF-5A could contribute to rescuing ribosomes stalled during translation of geometrically challenging amino acid stretches in general and of polyproline-containing proteins in particular (Figure 4). Biochemical analysis have indicated that ribosomes stall on polyproline stretches because peptide-bond formation is slow between the peptidyl-Pro-Pro-tRNA in the P-site and the incoming Pro-tRNA in the A-site (4) (Figure 4A). In addition to proline being both a poor donor and acceptor for peptide bond formation (55–58), it has also been shown that polyproline sequences attached to the P-tRNA lead to a destabilization of the P-tRNA, and even peptidyl-tRNA drop-off (4). The polyproline-stalled ribosomes are thus trapped in a pre-translocational state with a free E-site, which is recognized by eIF-5A (Figure 4B). Our structure suggests that eIF-5A binding stabilizes the productive position of the P-tRNA, in particular, by establishing an interaction between the hypusine moiety of eIF-5A and the backbone of the A76 of the CCA-end of the P-tRNA (Figure 4B). We propose that the optimal geometry for peptide bond formation is facilitated together with the eukaryotic-specific loop of uL16, which becomes stabilized via interactions with the terminal ends of the A- and P-tRNAs (Figure 4B). Thus, eIF-5A facilitates the transfer of the nascent chain from the P-tRNA to the A-tRNA by buttressing the CCA-end of the P-tRNA between the hypusine modification and the loop of uL16 (Figure 4C). Moreover, our findings that cycloheximide promotes eIF-5A binding to the ribosome (Supplementary Figure S2F) suggest that ribosomes trapped with a free E-site are a substrate for eIF-5A, thus raising the possibility that eIF-5A may also play a more general role in rescue of stalled ribosomes.
Figure 4.
Model for eIF-5A action on the ribosome. (A) Certain nascent polypeptide chains, such as those containing polyproline stretches, destabilize the P-tRNA (green) and prevent peptide-bond formation with the incoming A-tRNA (blue). (B) The stalled ribosomes are recognized by eIF-5A, which binds such that the modified hypusine 51 (Hyp51) residue interacts with the A76 of the CCA-end of the P-tRNA. This interaction stabilizes the P-tRNA in the optimal geometry for peptide bond formation, leading to ordering of the loop of uL16, which in turn establishes interactions with both A- and P-tRNAs, facilitating efficient peptide bond formation. (C) Peptide bond formation leads to a deacylated tRNA in the P-site and A-tRNA bearing the nascent polypeptide chain extended by one amino acid.
ACCESSION NUMBERS
The cryo-EM map and associated atomic coordinates have been deposited in the EMDB and PDB with the accession codes EMDB-3227 and PDB ID 5GAK, respectively.
Supplementary Material
Acknowledgments
We thank Heidemarie Sieber, Susanne Rieder and Charlotte Ungewickell for technical assistance. We also thank Thomas Fröhlich for his assistance with MS analysis. We thank the Leibnitz-Rechenzentrum Munich for providing computational services.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB646 to R.B. and T.B.; WI3285/3-1 to D.N.W; GRK 1721 to R.B.; FOR1805 to R.B. and D.N.W.]; European Research Council [Advanced Grant CRYOTRANSLATION to R.B.]; Boehringer Ingelheim Fonds Fellowship to C.S.; DFG Fellowship through the Graduate School of Quantitative Biosciences Munich (QBM) to V.S. Funding for open access charge: Deutsche Forschungsgemeinschaft [SFB646 to R.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Glick B.R., Ganoza M.C. Identification of a soluble protein that stimulates peptide bond synthesis. Proc. Natl Acad. Sci. U.S.A. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dever T.E., Gutierrez E., Shin B.S. The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 2014;49:413–425. doi: 10.3109/10409238.2014.939608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ude S., Lassak J., Starosta A.L., Kraxenberger T., Wilson D.N., Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 4.Doerfel L.K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M.V. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 5.Peil L., Starosta A.L., Lassak J., Atkinson G.C., Virumae K., Spitzer M., Tenson T., Jung K., Remme J., Wilson D.N. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc. Natl. Acad. Sci. U.S.A. 2013;110:15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolstenhulme C.J., Parajuli S., Healey D.W., Valverde D.P., Petersen E.N., Starosta A.L., Guydosh N.R., Johnson W.E., Wilson D.N., Buskirk A.R. Nascent peptides that block protein synthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez E., Shin B.S., Woolstenhulme C.J., Kim J.R., Saini P., Buskirk A.R., Dever T.E. eIF5A promotes translation of polyproline motifs. Mol. Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park M.H., Cooper H.L., Folk J.E. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park M.H., Nishimura K., Zanelli C.F., Valentini S.R. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolff E.C., Park M.H., Folk J.E. Cleavage of spermidine as the first step in deoxyhypusine synthesis. The role of NAD. J. Biol. Chem. 1990;265:4793–4799. [PubMed] [Google Scholar]
- 11.Navarre W.W., Zou S.B., Roy H., Xie J.L., Savchenko A., Singer A., Edvokimova E., Prost L.R., Kumar R., Ibba M., et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanagisawa T., Sumida T., Ishii R., Takemoto C., Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 2010;17:1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- 13.Peil L., Starosta A.L., Virumae K., Atkinson G.C., Tenson T., Remme J., Wilson D.N. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat. Chem. Biol. 2012;8:695–697. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- 14.Lassak J., Keilhauer E.C., Furst M., Wuichet K., Godeke J., Starosta A.L., Chen J.M., Sogaard-Andersen L., Rohr J., Wilson D.N., et al. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat. Chem. Biol. 2015;11:266–270. doi: 10.1038/nchembio.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajkovic A., Erickson S., Witzky A., Branson O.E., Seo J., Gafken P.R., Frietas M.A., Whitelegge J.P., Faull K.F., Navarre W., et al. Cyclic rhamnosylated elongation factor P establishes antibiotic resistance in pseudomonas aeruginosa. MBio. 2015;6:e00823. doi: 10.1128/mBio.00823-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaha G., Stanley R.E., Steitz T.A. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 18.Frischmeyer P.A., van Hoof A., O'Donnell K., Guerrerio A.L., Parker R., Dietz H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 19.van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 20.Defenouillere Q., Yao Y., Mouaikel J., Namane A., Galopier A., Decourty L., Doyen A., Malabat C., Saveanu C., Jacquier A., et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J.Z., Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 26.Leidig C., Thoms M., Holdermann I., Bradatsch B., Berninghausen O., Bange G., Sinning I., Hurt E., Beckmann R. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 2014;5:1–8. doi: 10.1038/ncomms4491. [DOI] [PubMed] [Google Scholar]
- 27.Penczek P.A., Frank J., Spahn C.M. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J. Struct. Biol. 2006;154:184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mindell J.A., Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 30.Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 33.Polikanov Y.S., Steitz T.A., Innis C.A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behrmann E., Loerke J., Budkevich T.V., Yamamoto K., Schmidt A., Penczek P.A., Vos M.R., Burger J., Mielke T., Scheerer P., et al. Structural snapshots of actively translating human ribosomes. Cell. 2015;161:845–857. doi: 10.1016/j.cell.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoemaker C.J., Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson R.J., Hellen C.U., Pestova T.V. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 40.Franckenberg S., Becker T., Beckmann R. Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Curr. Opin. Struct. Biol. 2012;22:786–796. doi: 10.1016/j.sbi.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Zuk D., Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valentini S.R., Casolari J.M., Oliveira C.C., Silver P.A., McBride A.E. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160:393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrader R., Young C., Kozian D., Hoffmann R., Lottspeich F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J. Biol. Chem. 2006;281:35336–35346. doi: 10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- 44.Klinge S., Voigts-Hoffmann F., Leibundgut M., Arpagaus S., Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 45.Garreau de Loubresse N., Prokhorova I., Holtkamp W., Rodnina M.V., Yusupova G., Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513:517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 46.Pestova T.V., Hellen C.U. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selmer M., Dunham C.M., Murphy F.V. 4th, Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 49.Tong Y., Park I., Hong B.S., Nedyalkova L., Tempel W., Park H.W. Crystal structure of human eIF5A1: insight into functional similarity of human eIF5A1 and eIF5A2. Proteins. 2009;75:1040–1045. doi: 10.1002/prot.22378. [DOI] [PubMed] [Google Scholar]
- 50.Spahn C.M., Gomez-Lorenzo M.G., Grassucci R.A., Jorgensen R., Andersen G.R., Beckmann R., Penczek P.A., Ballesta J.P., Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voorhees R.M., Weixlbaumer A., Loakes D., Kelley A.C., Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenner L., Demeshkina N., Yusupova G., Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 2011;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 53.Hofer A., Bussiere C., Johnson A.W. Mutational analysis of the ribosomal protein Rpl10 from yeast. J. Biol. Chem. 2007;282:32630–32639. doi: 10.1074/jbc.M705057200. [DOI] [PubMed] [Google Scholar]
- 54.Sulima S.O., Gulay S.P., Anjos M., Patchett S., Meskauskas A., Johnson A.W., Dinman J.D. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res. 2014;42:2049–2063. doi: 10.1093/nar/gkt1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wohlgemuth I., Brenner S., Beringer M., Rodnina M.V. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J. Biol. Chem. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- 56.Pavlov M.Y., Watts R.E., Tan Z., Cornish V.W., Ehrenberg M., Forster A.C. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muto H., Ito K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem. Biophys. Res. Commun. 2008;366:1043–1047. doi: 10.1016/j.bbrc.2007.12.072. [DOI] [PubMed] [Google Scholar]
- 58.Johansson M., Ieong K.W., Trobro S., Strazewski P., Aqvist J., Pavlov M.Y., Ehrenberg M. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl Acad. Sci. U.S.A. 2011;108:79–84. doi: 10.1073/pnas.1012612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.