Abstract
Over the recent decade, the central importance of DNA supercoiling in chromosome organization and global gene regulation of bacteria became more and more visible. With a regulon comprising more than 2000 genes in Escherichia coli, DNA supercoiling is among the most influential regulators of gene expression found in bacteria so far. However, the mechanism creating thousands of diverse temporal gene expression patterns coordinated by DNA supercoiling remains unclear. In this study we show that a specific chromosomal arrangement of genes modulates the local levels of DNA supercoiling at gene promoters via transcription-coupled DNA supercoiling (TCDS) in the model organism E. coli. Our findings provide a consistent explanation for the strong positive coupling of temporal gene expression patterns of neighboring genes. Using comparative genomics we are furthermore able to provide evidence that TCDS is a driving force for the evolution of chromosomal gene arrangement patterns in other Enterobacteriaceae. With the currently available data of promoter supercoiling sensitivity we prove that the same principle is applicable also for the evolutionary distant gram-positive pathogenic bacterium Streptococcus pneumoniae. Moreover, our findings are fully consistent with recent investigations concerning the regulatory impact of TCDS on gene pairs in eukaryots underpinning the broad applicability of our analysis.
INTRODUCTION
In previous decades investigations of transcriptional regulation in bacteria and other kingdoms was strongly focused on DNA binding transcription factors forming large regulatory networks. However, even in the well-studied model organism Escherichia coli for the majority of genes no regulator is known although these genes exhibit complex expression patterns during the bacterial growth cycle. It is an open question whether their transcription factors or transcription factor binding sites are not yet identified or there are other mechanisms of regulatory control for at least a subset of these genes. Besides the classical transcription factor based gene regulation, DNA supercoiling has been shown to be essential for transcriptional regulation in bacteria. With more than half of the genes sensitive to DNA supercoiling (1) it is a central regulatory factor (2). DNA supercoiling is thought to affect transcription in multiple ways. Most importantly, it supports the local untwisting of DNA facilitating the transcription initiation by RNA polymerase (RNAP) (3). Moreover, it can affect the local DNA geometry (regulatory site phasing and DNA curvature) and thus modulate transcription factor binding affinities or the oligomeric state of the initiating RNAP (4–6).
In E. coli, DNA is negatively supercoiled. DNA supercoiling levels are under a tight homeostatic control (7–9). The homeostasis is preserved by the opposing activities of DNA gyrase and the topoisomerases I and IV. Free DNA superhelicity is furthermore buffered by abundant nucleoid-associated-proteins (NAPs) that wrap DNA and therewith constrain DNA supercoils (5,10,11). Although topoisomerases and NAPs balance the levels of global DNA supercoiling, the overall level of negative superhelicity continuously decreases from exponential to stationary growth phase (12). This effect is mainly mediated by changes in gyrase activity (13,14) and NAP composition (5,15,16). The alterations of global DNA supercoiling were shown to be directly pertinent to the growth phase-dependent gene regulation (1,17–19). Gene ontology analysis of supercoiling sensitive genes revealed basal anabolic and catabolic metabolism to be associated with genes requiring high and low levels of negative DNA supercoiling, respectively (1,20). It has been shown that during exponential phase, genes close to the origin of replication preferring high negative DNA supercoiling showed significantly higher expression levels than the average of genes in this region of the chromosome whereas genes preferring low negative DNA supercoiling levels showed significantly lower expression levels in the same region. This discrepancy of gene expression or supercoiling sensitive genes dropped toward the terminus region (21). It can be explained by a DNA supercoiling gradient along the oriC-ter axis (22). The gradient hypothesis was further supported by an asymmetric distribution of gyrase binding sites (21), namely a high binding frequency in the oriC-proximal half of the chromosome decreasing toward the terminus region. Although the temporal expression of supercoiling sensitive genes suggest regulation by a supercoiling gradient their chromosomal distribution shows no obvious pattern. This is surprising, since, taking the supercoiling gradient into account a clear separation of genes preferring high negative supercoiling (hyp genes) in the oriC proximal region and genes preferring relaxation of DNA (rel genes) in the terminus proximal region could be expected. One hypothesis to explain the discrepancy would be the differentiation of the expression patterns by placing genes with different supercoiling preference in the same local supercoiling context. For instance, in an origin-proximal region assumed to have high negative DNA supercoiling levels during exponential growth, the hyp genes would show a high activity whereas the rel gene activity would be low. Considering half of all genes being supercoiling sensitive it would results in an antagonistic expression of neighbors or alternating islands of genes with similar supercoiling sensitivity. However, there is no obvious pattern in the distribution of supercoiling sensitive genes or an antagonistic expression level of neighbors. Quite on the contrary, the expression levels of neighboring genes in general are positively correlated. According to our findings presented in this study and in full accordance with literature (23), gene expression of neighboring genes (in this study: operons) is highly similar throughout the bacterial growth cycle. The question remains how DNA supercoiling can coordinate DNA supercoiling sensitive genes taking their random chromosomal position into account? To uncouple expression of supercoiling sensitive genes from global DNA supercoiling levels, the modulation of local supercoiling levels would be necessary. The discrepancy between the DNA supercoiling provided globally and the promoter optimum is supplied locally. Such local adjustments of DNA supercoiling levels could be realized by transcription itself, owing to supercoiling generated by RNA-Polymerase (RNAP) during elongation dubbed transcription-coupled DNA supercoiling (TCDS). The underlying mechanism was first described in the twin supercoil domain model proposed by Liu and Wang in 1987 (24). In this model, RNAP creates a mirror-inverted shift of local DNA supercoiling during transcription with negative supercoils generated upstream and positive supercoils downstream of the translocating RNAP. The shift in DNA supercoiling is counteracted by the activity of Topoisomerase I relaxing negative supercoils upstream of the RNAP and gyrase transforming positive DNA supercoils into negative ones downstream of the RNAP. Despite the topoisomerase activity, remaining supercoils can freely diffuse within several kilobases (25–27) and affect local DNA supercoiling levels at proximal promoters. It has been shown for individual promoters in several bacterial species that TCDS affects the transcription of neighboring genes (28–31). A striking example of the regulatory effect of TCDS is the relay mechanisms first described for the ilvIH-leuO-leuABCD region in Salmonella typhimurium (32) and later also found in E. coli (33). It allows for the TCDS regulatory effect to bridge several kilobases on the chromosome without attenuation. Here, the upstream ilIH operon activates the downstream leuO promoter via its TCDS. leuO transcription subsequently activates the leuABCD operon via TCDS. This way, the TCDS signal is amplified on the way from ilvIH to leuABCD via LeuO mediating the coupling of gene expression of distant genes. However, DNA supercoil diffusion is limited by transient barriers creating supercoiling domains of about 10 kb (34). The size of the domains is however not uniform in literature. Earlier experiments reported domains with a size up to 100 kb (35,36). These domains are thought to be dependent on anchoring of the DNA in the membrane via the simultaneous transcription, translation and membrane insertion of membrane associated proteins (37), translocase activity e.g. FtsK (38) and by the NAP-dependent formation of topologically closed DNA domains. On the one hand, these domains may restrict the influence of TCDS to several kilobases, on the other hand they allow for a local accumulation of DNA supercoiling by blocked diffusion. But even without such supercoiling barriers transcription can produce sufficient TCDS to alter supercoiling levels significantly (39,40). Under the conservative assumption of 10 kb supercoiling domains, TCDS can still influence the expression of roughly five genes upstream and downstream of a transcribed gene due to the dense packing of genes on bacterial genomes. Taking into account that on average every second gene in E. coli is sensitive to DNA supercoiling, a systematic analysis of gene expression regulation by TCDS is required. In this study we analyze the effect of TCDS on gene expression and chromosome architecture. We show that the combination of gene orientation, promoter strength and supercoiling sensitivity are determinants of the local gene expression driving the arrangement of genes on the E. coli chromosome. Furthermore, we provide evidence that this rationale is conserved in other bacteria.
MATERIALS AND METHODS
Operon-based analysis
To avoid a bias by intra-operon similarities of expression, hence trivial coupling of gene expression or TCDS levels, we exclusively analyzed the data on an operon level. For this study we pooled the supercoiling sensitive genes found in wt, Δfis and Δhns Norfloxacin treatments (1) and called an operon hyp or rel if at least one gene was assigned to this category in Blot et al. No operon contained both types of genes. The expression level of the operon required for the TCDS model is an average over all genes in the operon taking into account the different contributions by gene length. Note that intra-operon expression levels were highly similar. Averaging should there not introduce any bias. To improve clarity we use the term gene instead of operon in the text, although all analysis was done on the operon level.
TCDS model
For the analysis of chromosomal TCDS we utilized the published temporal gene expression data of E. coli K12 (22). In our model each base within the annotated coding sequence is a source of positive and negative supercoiling according to the Liu–Wang model. The intensity of supercoil emission depends on the expression level of the gene. Therefore each base within a gene emits the same amount of negative TCDS upstream and positive TCDS downstream. The following formula mathematically describes the TCDS model.
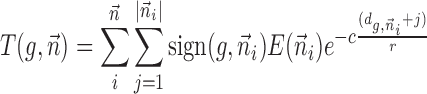 |
(1) |
TCDS T of the promoter of each gene g is calculated where n is the set of neighbors within range r; sign is the effect (positive or negative) dependent on the neighbor orientation and |ni| is the sequence length of the neighbor i. The formula describes an exponential decay of the TCDS effect with distance 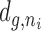 from its creation, to model diffusion and topoisomerase effects. The model realizes a decay to 1% of the TCDS at the border of the given range (constant c) and no effect beyond that range. This range is indicated for all results in the study. Furthermore, we assume that the TCDS of different neighbors is additive. To account for the length of the TCDS producing gene (a long gene may already span a considerable part of the TCDS range) we calculate the resulting TCDS from every base of the transcript to the target promoter (Equation 1 inner sum) and sum them up. The TCDS created at the start of transcription close to the promoter for example has a smaller impact on the downstream gene than the TCDS created at the end of transcription far more downstream and closer to the downstream gene. In order to avoid a bias by the incomplete and complex annotation of promoters, we approximated the promoter position of each gene by the 5′ end of the coding sequence. In bacteria, UTRs are rather short (≈100 bp) and homogeneous in length and should have no qualitative impact on the study. A similar reasoning holds for the approximation of transcript lengths. We approximated the mRNA length by the reliable and well documented lengths of coding sequences.
from its creation, to model diffusion and topoisomerase effects. The model realizes a decay to 1% of the TCDS at the border of the given range (constant c) and no effect beyond that range. This range is indicated for all results in the study. Furthermore, we assume that the TCDS of different neighbors is additive. To account for the length of the TCDS producing gene (a long gene may already span a considerable part of the TCDS range) we calculate the resulting TCDS from every base of the transcript to the target promoter (Equation 1 inner sum) and sum them up. The TCDS created at the start of transcription close to the promoter for example has a smaller impact on the downstream gene than the TCDS created at the end of transcription far more downstream and closer to the downstream gene. In order to avoid a bias by the incomplete and complex annotation of promoters, we approximated the promoter position of each gene by the 5′ end of the coding sequence. In bacteria, UTRs are rather short (≈100 bp) and homogeneous in length and should have no qualitative impact on the study. A similar reasoning holds for the approximation of transcript lengths. We approximated the mRNA length by the reliable and well documented lengths of coding sequences.
Chromosomal insert construction
In a first step we cloned a Gentamycin resistance gene and the green fluorescent protein derivative venus on a pUC18 plasmid. Overlap extension was used to join the gentamycin resistance and venus. The fragment was cloned into pUC18 via HindIII and EcoRI restriction. We replaced the native chromosomal open reading frame (ORF) of tufB with this construct in E. coli CSH50 using homologous recombination (RED/ET system). The strain showed no obvious deficiency in growth under the applied experimental conditions and the fluorescence signal matched the transcriptomics data of the wild-type. In a second step we cloned mCherry downstream of the tet promoter. For selection an Ampicillin resistance cassette was cloned downstream. Using homologous recombination we inserted this construct into the chromosome upstream of any reported tufB promoter without the disruption of any annotated sequence including repeats. Also the second construct showed no growth deficiencies under the applied experimental conditions. The activity pattern of tufBP was not altered under uninduced conditions compared to the precursor construct and the transcriptomics data. Primers used for each cloning step are listed in Table 1.
Table 1. Primer list for tufB chromosomal insertions.
| Primer forward | Primer reverse | Cloning step |
|---|---|---|
| 5′-CATGTTGCCTGGTTGATGTGGTGA TATCACCGATTTATCCGTGTCTTAGA GGGACAATCGATGGTGAGCAAGGGCG AGGAGC-3′ | 5′-GGGCATCAAATGATGCCCTTTTAGTGCG CATTGCGTCAAATGTTATCGGCAACATT ACCCTGTTATCCCTAGTTAC-3′ | venus primer with flanking tufB ORF homology sequences for RED E/T recombination |
| 5′-GTCTTCTTGCGCTAATTTTTTGCTAG GGATAACAGGGTAATTCTAGAGACGC ACACCGTGGAAAC-3′ | 5′-CATGGAATTCATTACCCTGTTATCCCTA TGGACTCACAAAGAAAAAACGCCCGGTG TGCAAGACCGAGCGTTCTGAACAACCCG GGGCGGCGTTGTGACAATTTAC-3′ | Gentamycin resistance gene primer for overlap extension with tetP-mCherry |
| 5′-CATGAAGCTTGTTGACACTCTATCAT TGATAGAGTTATTTTACCACTCGGAT CCAGGAGGAACAATATGGTGTCTATC ACTAAAGATCAAATCATGGTGAGCAA GGGCGAGGAG-3′ | 5′-ATTACCCTGTTATCCCTAGCAAAAAATT AGCGCAAGAAGACAAAAATCACCTTGCG CTAATGCTCTGTTACAGGGTACCTTACTTG TACAGCTCGTCCATG-3′ | tetP-mCherry primer for overlap extension with the Ampicillin resistance gene |
| 5′-GGGAGGAATAATAAGAAAAAATCT CTGCCAGGAAGCTATCGTTGAAAAGC AATGTGACGAGCGGATAACAATTTCA CACAGG-3′ | 5′-CACTTTTTCAACATCATTGTGCTCAACA ATGCGCTCCTGCTAAACCATAATTCTTTTT ATCAGAGGGTTTTCCCAGTCACGACGTT-3′ | primer for amplifying the tetP-mCherry-Ampicillin construct with homology for a divergent insertion in the upstream region of tufB |
Experimental conditions
All fluorescence measurements were conducted in LB medium at 37°C with continuous shaking using the Tecan infinite 200 pro reader. Measurements were taken every 5 min with a duration of ∼30 s. In order to repress the chromosomal tet promoter, we transformed a low copy plasmid (Pcatt-TRE18) harboring tetR. For induction of the repressed tet promoter we used Anhydrotetracycline a non-toxic Tetracyclin derivative. Final concentrations are given in the respective figure. The promoter activity was deduced from the fluorescence intensity of mCherry and venus in a Tecan infinite 200 pro fluorescence reader.
Experimental data selection of Streptococcus pneumoniae
For a proof of a general applicability of our approaches we investigated available gene expression data or another bacterium. For Streptococcus pneumoniae comprehensive transcriptomics data of Novobiocin treatment and multiple growth conditions were available at the GEO database. Different growth conditions were necessary to validate the stable bias of TCDS at supercoiling sensitive genes caused by the fixed order and orientation of genes under all conditions. For S. pneumoniae we chose the full set of supercoiling sensitive genes identified by the comparison of Novobiocin treated and untreated cells by Ferrandiz et al. 2004 (19). Gene expression data for TCDS calculations were taken from the Yadav et al. (41) biofilm dataset (GEO: GSM854585, GSM854586). The data consists of three replicates of planktonic and biofilm growth.
Comparative genomics approach
In order to investigate the conservation of TCDS effects we focused on the clade of Enterobacteriaceae. In this clade species haven't diverged to much to investigate a large set of conserved orthologous genes. Orthologous groups were determined using the software proteinortho (v5.07) with standard settings (E-value = 1e-5, blastp, α = 0.5, coverage=50%) and synteny option to separate co-orthologs. The set of Enterobacteriaceae was taken from the DoriC database (42) with each species being present once in the dataset. Protein sequences and annotation files were provided by the NCBI ftp site (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/). The resulting core genome of all NCBI and DoriC listed Enterobacteriaceae comprised more than half of the E. coli genome (see Supplementary list). With the limitation to the core genes equal amounts of investigated genes for all species were ensured. From the orthologous genes we determined triads consisting of a target gene, the neighbors with strongest positive and strongest negative TCDS contribution on the target gene TCDS level. To investigate conservation of TCDS dependent gene regulation we selected the triads with either a strongly positive correlation (Pearson coefficient ≥0.9) or a strongly negative correlation (Pearson coefficient ≤ −0.9) of TCDS and temporal gene expression of the target gene and an at least a moderate average TCDS bias in time (|TCDS| ≥ 0.1) of the target gene in E. coli. In addition, only triads were included in the statistic where all three genes remained within 10 kb distance to each other. This ensured a plausible TCDS impact on the target gene. For each conserved gene triad we derived the conservation frequency of the TCDS sign (positive or negative TCDS) on the target gene, so we tested whether the qualitative impact of the genes on the target remained the same. A positive TCDS impact can be realized by an upstream tandem or a downstream convergent orientation of the neighbor toward the target. In contrast, a neighbor with a divergent upstream or tandem downstream orientation has a negative TCDS contribution on the target gene TCDS level. Subsequently, for each triad we separately counted the number of conserved impact occurrences of the two other genes in the triad on the target gene within the Enterobacteria. The two groups of positive and negative TCDS contribution within the triad were further split in a positive and negative impact on target transcription in E. coli yielding four groups. The type of impact on target transcription depends on the type of TCDS impact on the target gene and the reaction of the target gene expression on the TCDS level (Figure 7B). For instance, the impact on transcription was called positive if the gene with a positive impact on target gene TCDS in E. coli (DNA relaxation) would belong to a target gene that prefers relaxation, hence showed a positive correlation of TCDS and transcription. If the majority (>50%) of orthologs showed a conservation in the TCDS impact we counted the gene as conserved for the respective group, otherwise as not conserved. Summing up these counts over all investigated gene triads, we obtained a statistic of the number of conserved TCDS effects on orthologous genes in Enterobacteriaceae.
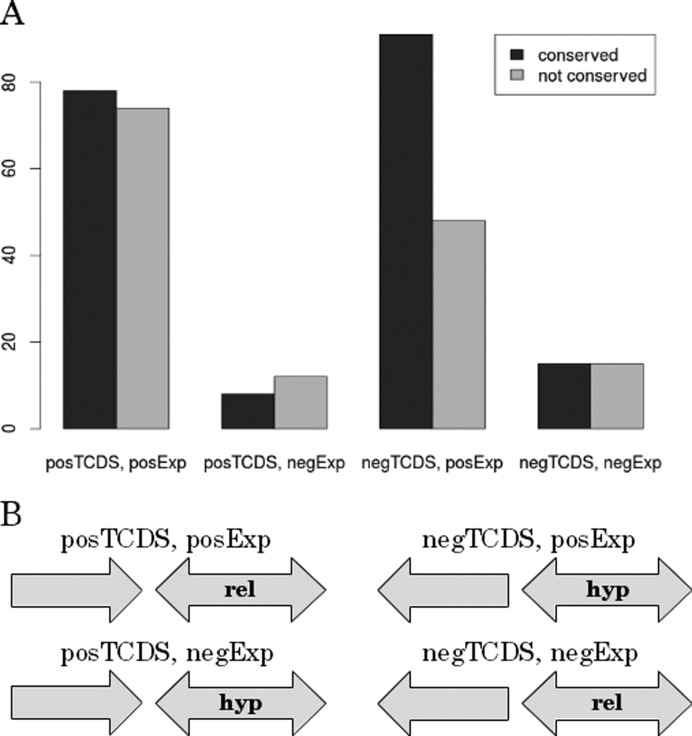
Figure 7.
Comparative genomics analysis of orthologs within the clade of Enterbacteriaceae. (A) Depicted is the conservation of effects of the neighboring genes with strongest positive and negative TCDS on the target gene. These two groups are further subdivided into the subset that contributes positively and negatively to target gene expression. We separately investigated four different groups: posTCDS, posExp = neighbor with strongest positive TCDS contribution on the target gene TCDS level and positive impact on gene expression of the target. posTCDS, negExp = neighbor with strongest positive TCDS contribution on the target gene TCDS level and negative impact on gene expression of the target. negTCDS, posExp = neighbor with strongest negative TCDS contribution on the target gene TCDS level and positive impact on gene expression of the target. negTCDS, negExp = neighbor with strongest negative TCDS contribution on the target gene TCDS level and negative impact on gene expression of the target. The abscissa depicts the total counts of conserved TCDS impact on the target gene. (B) Scheme of gene arrangement in the four separate groups. Hyp and rel indicate a preference for high negative and low negative supercoiling respectively inferred from the temporal correlation or anti-correlation of target gene TCDS and gene expression.
RESULTS
Distance-dependent similarity of temporal gene expression
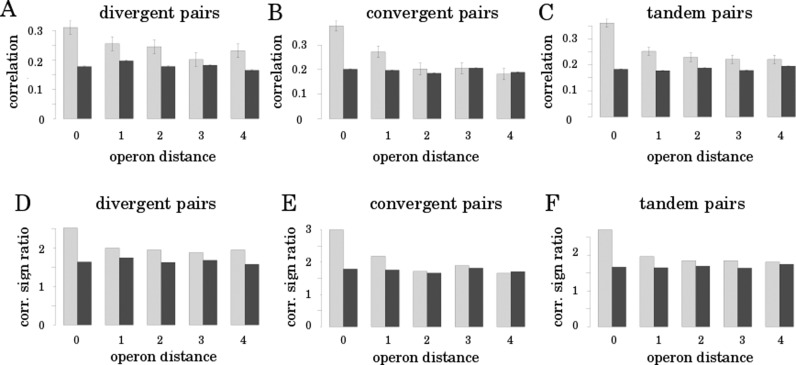
In order to study the global impact of TCDS on gene expression we first investigated the positive correlation of transcription of neighboring genes described in literature (23). For a solid foundation we correlated temporal gene expression curves sampled during the whole E. coli growth cycle from exponential to stationary phase (22). To avoid a bias by similarities in temporal gene expression due to cotranscription, we restricted our analysis to E. coli operons. Note that throughout the results section we analyze the data on the operon level and refer to genes for readability (see ‘Materials and Methods’ section). For a second control we split the pairs of operons in divergent, convergent and tandem operon pairs to identify biases by unknown operon structures or common regulatory sites. Such biases would be visible only in tandem or divergently oriented operons. Figure 1 depicts the average correlation coefficients of temporal expression patterns of operons at various distances and orientations. Our analysis yield two results. First of all, the correlation of temporal gene expression is independent of the relative orientation. This rules out unknown transcription units or operons as the major source of the similar gene expression of proximal genes. In this case only tandem pairs would show this effect. Furthermore, shared regulatory regions can be excluded as major contributor to the expression patterns. Here, mainly divergent genes which could share a regulatory region would show this effect. Common regulators, which could explain an independence of orientation can be excluded due to a wide scattering of regulatory sites on the chromosome and the different mode of regulation ( repression or activation ) of most regulators. Hence, neighboring operons with the same regulator and the same mode of regulation is rare. The remaining principle that could explain the effect is transcription activity of neighbors and the resulting local change of supercoiling via TCDS at the target gene promoter. This hypothesis is supported by the second observation, namely the decrease of expression similarity with increasing distance of the operons (Figure 1) matching roughly the 10 kb supercoiling domains.
Figure 1.
Spatially resolved correlation of temporal gene expression. The light-gray bars represent the mean Pearson correlation of the temporal expression of all operon pairs with a respective orientation. The black control bar indicates the mean correlation of randomly reassigned gene pairs from the same pool. The abscissa indicates the number of operons between the pair of investigated operons from 0 for a direct neighborhood up to 4. For the correlation of operon expression, the expression patterns of the closest genes of the pairs were chosen. Panels (A–C) depict the pairs with divergent, convergent and tandem orientation of operons respectively. Panels (D–F) depict the ratio of the numbers of positive and negative correlations incorporated in the mean correlation values of A–C. For the correlation values error bars are indicated (SEM).
Supercoiling sensitivity determines the orientation of neighboring genes
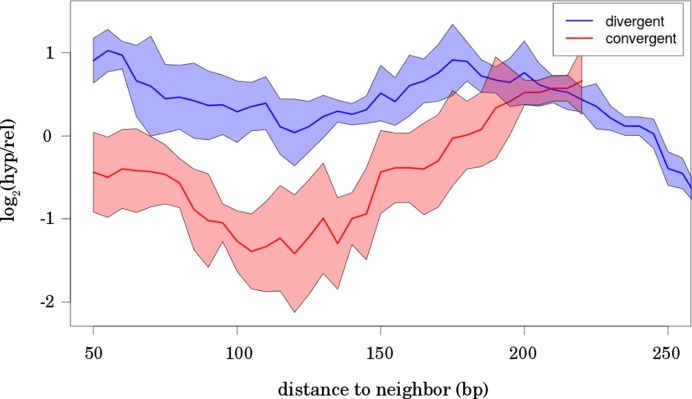
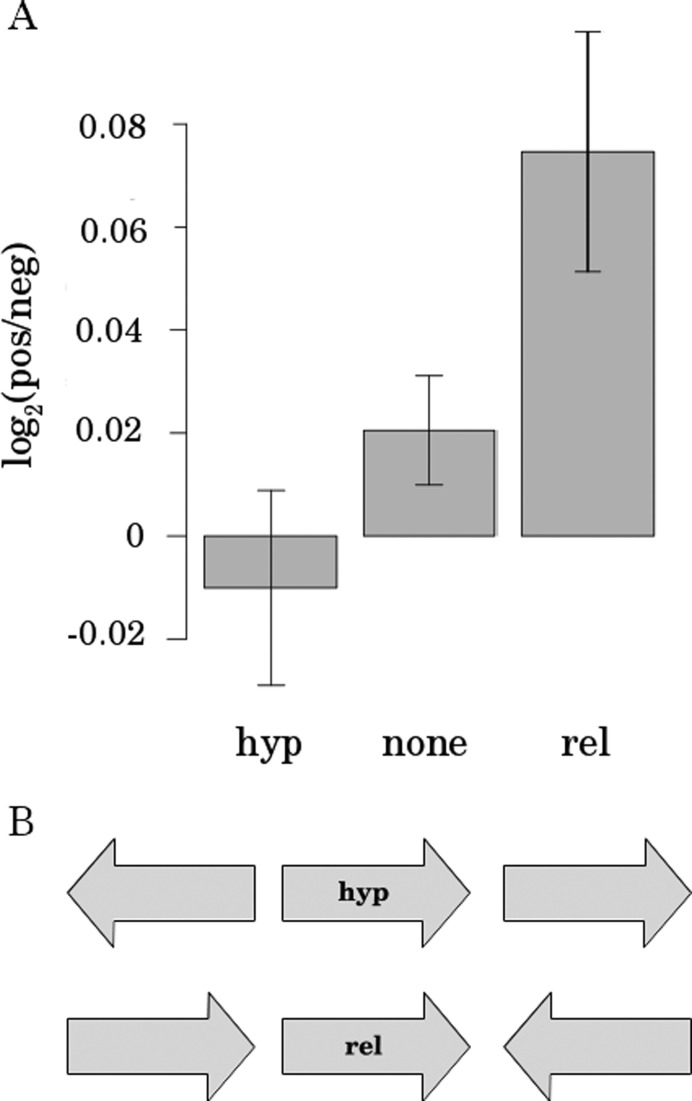
The directed supply of promoters with DNA supercoiling via TCDS is not trivial. According to the Liu and Wang twin-supercoiled-domain-model (24,43), transcription activity generates negative supercoiling upstream and positive supercoiling downstream of the transcribed gene. If TCDS plays a crucial role in the modulation of local DNA supercoiling, the orientation and arrangement of genes would be determinative for an optimal supply of supercoiling sensitive promoters with TCDS. We therefore investigated the relative orientation of direct neighbors of supercoiling sensitive genes and found that hyp genes were enriched in divergently oriented direct neighbors with small intergenic regions, whereas rel genes were enriched in arrangements with a close convergent direct neighbor (Figure 3). The close proximity of divergent neighbors for hyp genes and close convergent neighbors for rel genes indicates an efficient utilization of positive and negative TCDS by supercoiling sensitive genes. In the remaining part of the text the term ‘target gene’ will be used to describe the gene under investigation receiving TCDS, not meaning any particular class of genes. Our first results about gene expression similarity showed a coupling of indirect neighbors. We therefore extended the investigation of relative orientation to indirect neighbors by counting the number of genes pointing toward and away from the target gene within the range of supercoiling domains of 10 kb. The genes pointing toward the target gene provide positive TCDS to the promoter, whereas genes pointing away from the gene of interest provide negative TCDS. We found an increased frequency of genes pointing away from hyp genes and a predominant orientation of neighbors pointing toward rel genes (Figure 4). This finding suggests that TCDS is utilized as a regulatory mechanism operating via cooperative effects of spatial arrangement and orientation of genes.
Figure 3.
Neighbor distance resolved log2 ratio of the number of hyp and rel genes in sets of genes with divergent (blue) and convergent (red) neighbors. Distance between the genes is determined using ORF start positions for the divergent and ORF end positions for the convergent orientation. The abscissa indicates the distance to the neighbor ± 50 bps. Genes with distances falling in this range were taken for counting the hyp and rel genes. The different size of the sets (|hyp| > |rel|) was normalized in the counting. In the log ratio scale −1 and +1 indicates a 2-fold difference between hyp and rel gene counts. The analysis comprised the hyp and rel gene sets detected after the treatment of Escherichia coli wild-type, Δfis and Δhns cells with the topoisomerase inhibitor Norfloxacin (1). The curve depicts the mean log ratio of the three datasets. The trend is confirmed by all three datasets. Error bars (SD) are indicated by the colored area around the curve. In the case of divergent orientation of neighbors, short distances are preferred by the hyp genes, whereas rel genes prefer a longer distance to the neighbor. The opposite can be observed in the case of convergent orientation.
Figure 4.
TCDS disbalance in the supercoiling sensitive genes is due to an orientation bias of neighboring genes. (A) Mean log ratio of the number of genes with positive and negative TCDS contribution on target gene TCDS level within a range of 10 kb in the sets of hyp, rel and none (neither hyp, nor rel). Error bars (SEM) are indicated. (B) Scheme of the preferred orientations of hyp and rel gene neighbors.
A model for transcription-coupled DNA supercoiling
To quantitatively assess TCDS levels at supercoiling sensitive promoters we estimated the TCDS using time-resolved RNA-seq data. TCDS mainly depends on the strength of transcription (40), the length of the transcript (17,39,44) and the distance to its target. We developed a model assuming the TCDS level to be linearly correlated with transcription strength and gene length as well as an exponential decrease with distance to account for the supercoil diffusion and Topoisomerase activity. The attenuation of TCDS with distance is however not well understood. Hence, to estimate the robustness of the results presented in this study with respect to the model settings, we also tested a model with linear decay of TCDS down to 1% within the given range. The results were qualitatively identical to the results of the exponential decay model underpinning the robustness of the results (Supplementary Figure S1).
Supercoiling sensitivity determines the promoter TCDS level
In order to study the impact of TCDS on global transcription regulation, we investigated differences in TCDS received by supercoiling sensitive genes described above. For an outlier robust investigation of the differences of hyp and rel gene sets (e.g. by highly transcribed genes), we derived the median TCDS value for each set of genes. To rule out an expression strength bias between hyp and rel gene sets, we computed the promoter TCDS with and without the impact of the own gene expression. Without the TCDS of its own transcription, the promoters of rel genes showed positive TCDS level, whereas the hyp gene set showed a strongly negative TCDS level at the promoter region throughout all growth stages. Taking TCDS of its own transcription into account, the TCDS level got more negative (less positive for the rel gene set) at the promoter as the promoter is located upstream of the transcription complex. Hence, the negative TCDS of its own transcription shifts the TCDS at the promoter to a more negative value. Since hyp genes yield a slightly higher expression level on average than rel genes, the difference of TCDS between hyp and rel gene sets increased even more. If not mentioned otherwise, we excluded the effect of the own transcription in the following analyses to focus on the effects of neighboring genes. For a statistical evaluation of the difference, we randomly assigned genes to the hyp and rel gene sets and compared its median TCDS with the median TCDS derived for the correct hyp and rel gene sets. Compellingly, the differences between hyp and rel gene sets within the supercoiling domain range (10–15 kb; (34)) were highly significant throughout the growth cycle (Figure 2). These results suggest that promoters preferring high negative supercoiling are predominantly supplied with negative TCDS by transcription activity of their neighbors, whereas promoters preferring more relaxed DNA predominantly are supplied with positive TCDS by nearby transcription activity. Hence, supercoiling sensitive genes are supplied with TCDS matching the promoter supercoiling preference. In full accordance with the positive correlation of local gene expression patterns, the supplied TCDS supports the activity of the promoter and thereby positively couples gene expression patterns of neighboring genes.
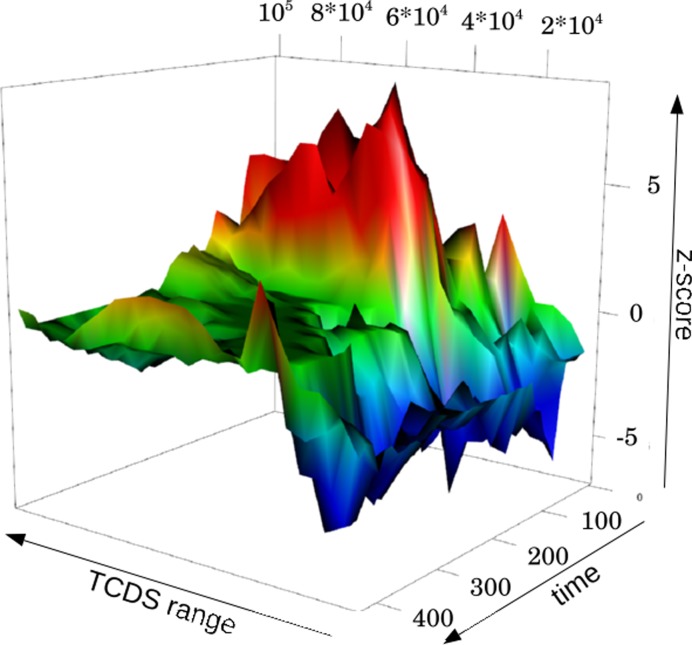
Figure 2.
Difference of TCDS in genes sensitive to high negative supercoiling (hyp) and DNA relaxation (rel) in Escherichia coli. The z-score of the difference of the median of TCDS in hyp and rel genes (medianTCDS(hyp)-medianTCDS(rel)) is plotted on the z-axis. z-scores are based on the TCDS bias between hyp and rel genes relative to the bias for random gene sets. The z-score is coded in color and height. A z-score larger than 2 or smaller than −2, indicating a significant deviation from randomness, is color-coded in red and blue respectively. Hence, a blue color indicates a significantly more negative TCDS at promoters preferring high negative supercoiling than at promoters preferring relaxation. The opposite holds for a red color. The time axis indicates the 43 different time steps spanning from inoculation (0) over exponential phase (≤120) to stationary phase (≥360) The TCDS axis shows the TCDS decay parameter of the mathematical model. The longer the TCDS range the slower the decay with distance. At the indicated range the TCDS reaches 1% of its initial strength. The blue cleft spans to ∼15 kb and has its minimum (highest significance) around 10 kb.
In vivo validation of model predictions
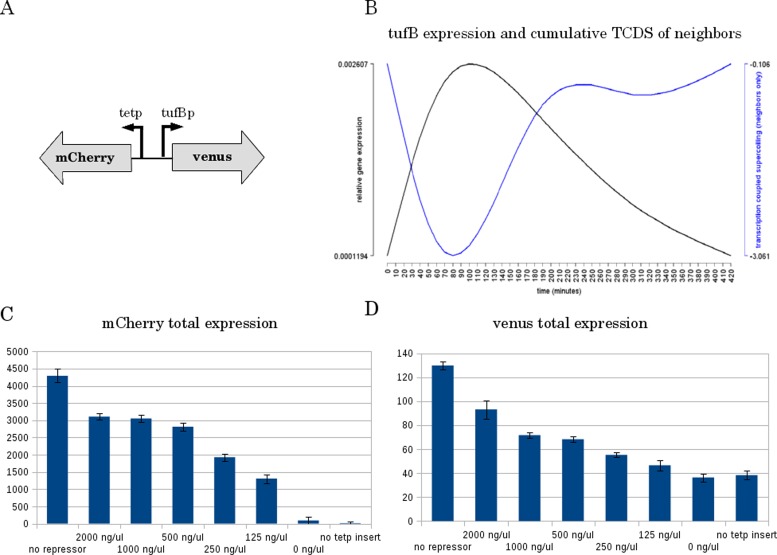
In order to experimentally validate our TCDS model predictions, we chose tufB, a gene with a clear correlation of TCDS (Figure 5B). The tufB promoter belongs to the stringent promoters assumed to be sensitive to high levels of negative superhelicity (45). However, expression of tufB has not been found to be supercoiling sensitive before. We inserted a divergently oriented tet promoter coupled to mCherry, a reporter gene encoding a fluorescent protein, upstream of the native locus of the tufB promoter that was itself coupled to venus (fluorescent reporter gene). The scheme of the construct is depicted in Figure 5A. The strain with the tufB coding sequence replace by venus showed no growth deficiency or any other noticeable phenotype in our experiments. Without induction we confirmed the wild-type expression pattern of tufB seen in the transcriptomics data (22). Under different levels of induction and without providing the repressor (fully induced tetp) we could modulate the expression of tufB gradually up to a 3-fold increase of expression during full induction. In addition to the exponential phase expression pattern we got an additional activation of tufB during stationary phase under induction/no repressor conditions due to the activity of the tetp in that phase. The result confirm that TCDS can change transcription of neighbors on the chromosome and that the effect depends on the level of transcription of neighboring genes. More compellingly, the results suggest that supercoiling sensitivity of a promoter can be predicted by applying our TCDS model on gene expression data and correlate TCDS and gene expression levels.
Figure 5.
Gene expression correlates with TCDS level of neighboring genes. (A) Scheme of the reporter gene arrangement. The tufB promoter was chosen due to its expression strength for reliable detection of the signal and the clear correlation of negative neighboring TCDS and gene expression. In the construct designed in the native locus of tufB the coding sequence of tufB is replaced by the reporter gene venus encoding a fluorescent protein. Upstream of the tufB promoter the tet promoter coupled to mCherry reporter gene is inserted. (B) Temporal expression of tufB in wild-type Escherichia coli (21) and TCDS derived from the expression and orientation of neighboring genes within 10 kb. (C) Total expression of tetP (mCherry) at 24 h after induction with different concentrations of Anhydrotetracyclin. Abscissa: no repressor (cells without repressor plasmid), induction with decreasing levels (from left to right) of Anhydrotetracyclin and no tetP insert (the construct without the tetP insert). Ordinate: total fluorescence of mCherry after 24 h. (D) The same as in (C) with venus total fluorescence measured 24 h after induction.
A clear TCDS bias at the target promoter is required for an impact on target promoter activity
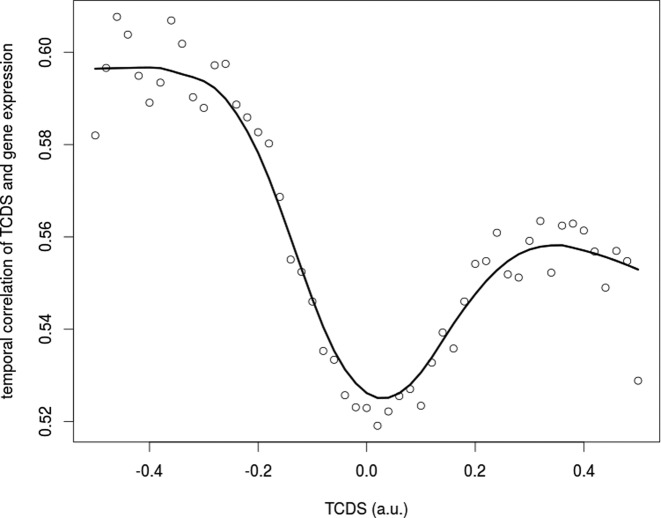
In order to check, whether the results obtained with tufB can be generalized, we investigated the link between the strength of the TCDS received by a gene promoter and the target gene expression. We computed a pearson correlation coefficient of the temporal curve of TCDS at the target promoter and the temporal gene expression curve of the target and plotted the temporal average TCDS of the target against the correlation coefficient. This allows for the analysis of expression correlation dependent on the general TCDS bias at the target promoter. It is conspicuous that at a certain threshold of the TCDS bias, correlation of expression of the gene under the control of the target promoter with the received TCDS steeply increases (Figure 6). Hence, only at a strong enough disproportion of neighboring positive and negative TCDS we observe an impact of TCDS on the expression pattern of the target gene underpinning the correctness and physiological relevance of our model. Moreover, the effect is not strictly symmetric for a positive and negative disproportion. In the case of a positive TCDS prevalence the effect is less pronounced. A plausible explanation is the general repressive nature of positive DNA supercoiling which can only be used for the activation of rel gene promoters, whereas negative supercoiling should have a positive effect on transcription for the majority of promoters.
Figure 6.
TCDS and gene expression pattern is strongly correlated at a threshold bias of TCDS at the promoter. On the abscissa the temporal average of TCDS is depicted. A deviation from 0 indicates a bias of TCDS. The ordinate shows the average temporal correlation of promoter TCDS and gene expression for genes with a TCDS denoted on the abscissa (±0.05 window). TCDS strength is computed in arbitrary units based on temporal RNA-seq data for a TCDS range of 10 kb excluding the TCDS generated by the expression of the investigated promoter. The pearson correlation coefficient is applied to correlate the temporal TCDS and expression curve.
The rules for TCDS-dependent gene arrangement are conserved in Enterobacteriaceae
To test the applicability of our findings on other bacteria, we asked whether the gene arrangement reflected TCDS-dependent regulation of gene expression in other bacteria. Lacking time-resolved transcriptomics data and supercoiling sensitive gene sets for related species we decided to investigate conservation of the qualitative TCDS impact (positive or negative) of neighboring genes on the target gene. The qualitative TCDS impact solely depends on the orientation of the neighbors toward their target gene, an information that is provided by NCBI gene annotation data. The clade of Enterobacteriaceae was chosen for the set of E. coli related bacteria. In this set the core genome (conserved genes in all investigated species) still comprises half of the E. coli genes. Hence, the core genome is rather large and allows for a TCDS impact analysis of many genes and their conserved neighbors. Furthermore, the genomes in this set have diverged enough to observe conservation of gene arrangements. For the analysis of conservation we defined ortholog triads that consist of the TCDS target gene and the two genes with highest positive and negative TCDS contribution on the target genes TCDS level in E. coli. For the clade of Enterobacteriaceae we investigated the conservation of the type of impact of the two neighbors on the target gene. The conservation of the neighbor with highest positive and highest negative impact was analyzed separately. We further split the groups in sets with positive and negative impact of the gene on the target gene expression. We therefore got four types of groups (Figure 7). The strongest negative TCDS contribution on the target gene TCDS level with a positive impact on transcription was highly conserved. This finding is consistent with earlier studies on conservation of divergent promoters (46). In this study we show that this is not bound to direct divergent neighbors. In fact the negative TCDS sign includes direct and distant (<10 kb) neighbors of a target in either an upstream divergent or downstream tandem arrangement. According to our data, the negative TCDS sign in the conserved triads is equally due to an upstream divergent and a downstream tandem orientation (data not shown). Hence, the TCDS sign was conserved in many cases even in the presence of a rearrangement of genes. Note, that there are generally low counts for negative impact of TCDS on transcription (negExp) showing the generally positive effect of TCDS on neighboring promoters irrespective of the TCDS sign.
Supercoiling sensitive genes of Streptococcus pneumoniae show a similar TCDS bias
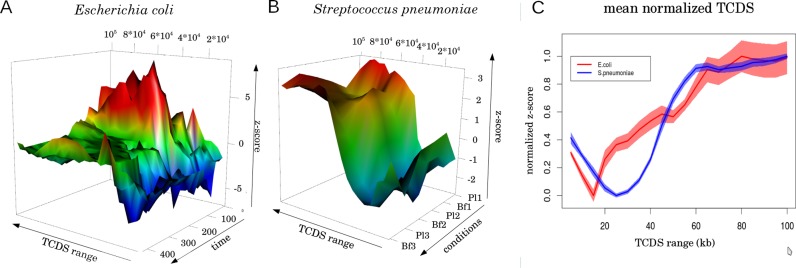
To further generalize our finding, we applied our analysis on the evolutionarily distant gram-positive bacterium S. pneumoniae. For this bacterium supercoiling sensitive genes were already determined (19) and several gene expression datasets available. In this study DNA supercoiling levels were altered by Novobiocin, a DNA relaxing agent. Novobiocin blocks the adenosine triphosphate (ATP) hydrolysis by the DNA gyrase B subunit and therefore stops the transformation of positive DNA supercoils into negative supercoils. This causes an imbalance in the homeostasis of DNA supercoiling and leads to a relaxation of DNA on a global level. In analogy to the E. coli analysis, we split these genes in hyp and rel where genes with a higher expression level in the untreated background were assigned to the hyp gene set and genes with higher expression in the Novobiocin background were assigned to the set of rel genes. Due to the lack of temporal gene expression data in these species we were not able to perform the time related analyses we performed for E. coli. However, using wild-type gene expression data of planktonic and biofilm growth conditions in S. pneumoniae we were able to test whether a stable TCDS bias could be observed between hyp and rel gene sets at physiological ranges of TCDS (Figure 8B). The results suggest a significant difference of TCDS at promoters with opposite DNA supercoiling preference indicated by the blue cleft at small TCDS ranges (Figure 8B). At about 20 kb TCDS range the (negative) difference of hyp and rel TCDS levels is maximal. The overall pattern resembles the pattern observed in the E. coli hyp/rel TCDS difference (Figure 8C).
Figure 8.
TCDS bias analysis of supercoiling sensitive genes for Escherichia coli and Streptococcus pneumoniae. In (A and B) the TCDS bias z-score for two bacterial species is depicted in depencence on the TCDS range and experimental conditions (see ‘Materials and Methods’ section). The z-score of the difference of the median of TCDS in hyp and rel genes (medianTCDS(hyp)-medianTCDS(rel)) is plotted on the z-axis. Z-scores are based on the TCDS bias between hyp and rel genes relative to the bias for random gene sets. The z-score is coded in color and height. A z-score larger than 2 or smaller than −2, indicating a significant deviation from randomness, is color-coded in red and blue respectively. Hence, a blue color indicates a significantly more negative TCDS at promoters preferring high negative supercoiling than at promoters preferring relaxation. The opposite holds for a red color. In (A) The time axis indicates the 43 different time steps spanning from inoculation (0) over exponential phase (≤120) to stationary phase (≥360) of E. coli In (B) the conditions axis indicates the three replicates of biofilm forming conditions (Bf1-Bf3) and the three replicates of planktonic growth (Pl1-Pl3) of S. pneumoniae. The TCDS range axis shows the TCDS decay parameter of the mathematical model. The longer the TCDS range the slower the decay with distance. At the indicated range the TCDS reaches 1% of its initial strength. (C) A summary of (A and B) for the spatial dependency of the TCDS bias. The mean values along the conditions axis are plotted and normalized by minimum and maximum of each plot.
DISCUSSION
In this study we have shown that local gene position and orientation is the result of selective pressure to maintain regulatory influence of DNA supercoiling generated by transcription of neighboring genes. In fact, genes arrangements are highly organized to cooperatively utilize neighboring TCDS. Local gene arrangement optimizes TCDS supply to support transcriptional activity of DNA supercoiling sensitive genes allover the chromosome. The cooperative nature of TCDS effects suggests a crosstalk between neighboring genes as a reciprocal control mechanism of gene expression. In the light of this study the chromosomal genes appear to be strategically positioned to utilize TCDS optimally. On an evolutionary scale, a gene neighborhood with a TCDS bias attracts genes preferring the respective level of supercoiling. Moreover, the distance to the TCDS source is minimized for these genes by short intergenic regions, whereas genes with an opposing supercoiling sensitivity show an increased distance to the source by larger intergenic regions. Several aspects of TCDS could favor its use in regulatory circuits. First of all, it links the expression of proximal genes without the need to organize continuous transcription units. Yet, in contrast to transcription units, the interaction can be weakened gradually by increasing the distance e.g by an insertion or rearrangement. This simple mechanism would greatly facilitate the modulation of the gene regulatory circuitry on an evolutionary scale. It has been shown that supercoiling sensitivity is highly dependent on the composition of DNA binding proteins (1). Several NAPs for instance can constrain DNA supercoils thereby buffering TCDS effects locally dependent on the presence of binding sites. Therefore, NAPs could separately influence the promoters to decouple the expression patterns if favorable. Furthermore, TCDS regulatory effects can be faster than those mediated by transcription factor interactions and also circumvent the necessity of the expensive synthesis of the latter. Thus, the spatial distribution of regulatory connections in the genetic system show a rather long range information transfer mediated by the TRN that potentially complements the short-range TCDS-mediated expression coupling (2). It seems likely that genes regulated by a classical regulator may regulate neighboring genes via TCDS and therewith locally spread the regulatory information. For this hypothesis, the promoter relay mechanism in the ilvIH-leuO-leuABCD (32,33) is a striking model. The ilvIH operon is induced under amino acid starvation by the binding of leucine-free Lrp, a classical regulatory mechanism. The resulting transcription activates the upstream gene leuO via negative TCDS which again via negative TCDS activates the divergently oriented leuABCD operon. In summary, an operon is activated by classical regulator binding and the regulatory information is subsequently passed on to the neighboring genes by changes in DNA supercoiling via TCDS. This would reduce the transcription factor binding sites and the corresponding regulator molecules needed for regulation and at the same time allow for a modulation of the transcription factor signal. More compellingly, the ilvIH-leuO-leuABCD region also offers a model for the the inverse case. The ilvIH operon activates the neighboring leuO gene via TCDS, whereas leuO in turn encodes a regulator. This implies that the classical regulatory network and the TCDS neighborhood regulatory communicate in both directions. The extent of this interplay remains to be investigated in future studies. One hint could be that for half of the genes in the well-studied model organism E. coli no regulator is known so far (according to regulondb). Another favorable effect of TCDS is the channeling of TCDS toward specific DNA regions potentially reducing ATP costs for the maintenance of high levels of global DNA supercoiling to achieve promoter activation. In addition, local supply of DNA supercoiling by TCDS allows for an optimal DNA supercoiling level at a gene promoter also in chromosomal regions that do not provide a sufficient level of global DNA supercoiling. This reduces the spatial contraints for a gene to be placed in chromosomal regions with a specific global supercoiling level, however, keeping it at the same time sensitive to global DNA supercoiling changes. Therefore, TCDS allows for a more homogeneous distribution of supercoiling sensitive genes on the chromosome as observed in E. coli, which increases the evolutionary flexibility of the spatial gene organization on the chromosome. Finally, we provide evidence that the same mechanisms also hold in other bacteria validating and generalizing previous studies on the conservation of gene arrangements. Although this study focuses on two bacteria and the clade of Enterobacteriaceae we are convinced that more transcriptomics data will show the generality of our results in bacteria and beyond. The awareness of TCDS impact on transcription has already inspired studies in Eukaryots (47) with similar conclusion on single gene levels. With TCDS as the cause and effect of DNA reading processes more astonishing results are expected in all kingdoms of life in the near future.
Supplementary Material
Acknowledgments
The author thanks Georgi Muskhelishvili for providing laboratory space and instruments as well as for valuable scientific discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
LOEWE program of the State of Hesse; Deutsche Forschungsgemeinschaft (DFG) [SO 1447/1-1]. Funding for open access charge: DFG [SO 1447/1-1].
Conflict of interest statement. None declared.
REFERENCES
- 1.Blot N., Mavathur R., Geertz M., Travers A., Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marr C., Geertz M., Hutt M.T., Muskhelishvili G. Dissecting the logical types of network control in gene expression profiles. BMC Syst. Biol. 2008;2:18. doi: 10.1186/1752-0509-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahms J.G., Dargouge O., Brahms S., Ohara Y., Vagner V. Activation and inhibition of transcription by supercoiling. J. Mol. Biol. 1985;181:455–465. doi: 10.1016/0022-2836(85)90419-x. [DOI] [PubMed] [Google Scholar]
- 4.Auner H., Buckle M., Deufel A., Kutateladze T., Lazarus L., Mavathur R., Muskhelishvili G., Pemberton I., Schneider R., Travers A. Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J. Mol. Biol. 2003;331:331–344. doi: 10.1016/s0022-2836(03)00727-7. [DOI] [PubMed] [Google Scholar]
- 5.Dorman C.J. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem. Soc. Trans. 2013;41:542–547. doi: 10.1042/BST20120222. [DOI] [PubMed] [Google Scholar]
- 6.Gerganova V., Maurer S., Stoliar L., Japaridze A., Dietler G., Nasser W., Kutateladze T., Travers A., Muskhelishvili G. Upstream binding of idling RNA polymerase modulates transcription initiation from a nearby promoter. J. Biol. Chem. 2015;290:8095–8109. doi: 10.1074/jbc.M114.628131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- 8.Neumann S., Quinones A. Discoordinate gene expression of gyrA and gyrB in response to DNA gyrase inhibition in Escherichia coli. J. Basic Microbiol. 1997;37:53–69. doi: 10.1002/jobm.3620370109. [DOI] [PubMed] [Google Scholar]
- 9.Snoep J.L., van der Weijden C.C., Andersen H.W., Westerhoff H.V., Jensen P.R. DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur. J. Biochem. 2002;269:1662–1669. doi: 10.1046/j.1432-1327.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 10.Berger M., Farcas A., Geertz M., Zhelyazkova P., Brix K., Travers A., Muskhelishvili G. Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep. 2010;11:59–64. doi: 10.1038/embor.2009.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouafa Z.A., Reverchon S., Lautier T., Muskhelishvili G., Nasser W. The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res. 2012;40:4306–4319. doi: 10.1093/nar/gks014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balke V.L., Gralla J.D. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J. Bacteriol. 1987;169:4499–4506. doi: 10.1128/jb.169.10.4499-4506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drlica K. Bacterial topoisomerases and the control of DNA supercoiling. Trends Genet. 1990;6:433–437. doi: 10.1016/0168-9525(90)90306-q. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez-Estrada A., Ramirez-Santos J., Gomez-Eichelmann M. d. e. l.C. Role of chaperones and ATP synthase in DNA gyrase reactivation in Escherichia coli stationary-phase cells after nutrient addition. Springerplus. 2014;3:656. doi: 10.1186/2193-1801-3-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen-Hughes T.A., Pavitt G.D., Santos D.S., Sidebotham J.M., Hulton C.S., Hinton J.C., Higgins C.F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 16.Schneider R., Travers A., Muskhelishvili G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol. Microbiol. 1997;26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 17.Peter B.J., Arsuaga J., Breier A.M., Khodursky A.B., Brown P.O., Cozzarelli N.R. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouzine F., Sanford S., Elisha-Feil Z., Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 19.Ferrandiz M.J., Arnanz C., Martin-Galiano A.J., Rodriguez-Martin C., de la Campa A.G. Role of global and local topology in the regulation of gene expression in Streptococcus pneumoniae. PLoS One. 2014;9:e101574. doi: 10.1371/journal.pone.0101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnenschein N., Geertz M., Muskhelishvili G., Hutt M.T. Analog regulation of metabolic demand. BMC Syst. Biol. 2011;5:40. doi: 10.1186/1752-0509-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobetzko P., Travers A., Muskhelishvili G. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. U.S.A. 2012;109:42–50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobetzko P., Glinkowska M., Travers A., Muskhelishvili G. DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle. Mol. Biosyst. 2013;9:1643–1651. doi: 10.1039/c3mb25515h. [DOI] [PubMed] [Google Scholar]
- 23.Jeong K.S., Ahn J., Khodursky A.B. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 2004;5:R86. doi: 10.1186/gb-2004-5-11-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L.F., Wang J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouviere-Yaniv J., Yaniv M., Germond J.E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka H., Yasuzawa K., Kohno K., Goshima N., Kano Y., Saiki T., Imamoto F. Role of HU proteins in forming and constraining supercoils of chromosomal DNA in Escherichia coli. Mol. Gen. Genet. 1995;248:518–526. doi: 10.1007/BF02423446. [DOI] [PubMed] [Google Scholar]
- 27.Malik M., Bensaid A., Rouviere-Yaniv J., Drlica K. Histone-like protein HU and bacterial DNA topology: suppression of an HU deficiency by gyrase mutations. J. Mol. Biol. 1996;256:66–76. doi: 10.1006/jmbi.1996.0068. [DOI] [PubMed] [Google Scholar]
- 28.Richet E., Raibaud O. Supercoiling is essential for the formation and stability of the initiation complex at the divergent malEp and malKp promoters. J. Mol. Biol. 1991;218:529–542. doi: 10.1016/0022-2836(91)90699-7. [DOI] [PubMed] [Google Scholar]
- 29.Burns H., Minchin S. Thermal energy requirement for strand separation during transcription initiation: the effect of supercoiling and extended protein DNA contacts. Nucleic Acids Res. 1994;22:3840–3845. doi: 10.1093/nar/22.19.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen D., Lilley D.M. Transcription-induced hypersupercoiling of plasmid DNA. J. Mol. Biol. 1999;285:443–448. doi: 10.1006/jmbi.1998.2358. [DOI] [PubMed] [Google Scholar]
- 31.Naughton C., Corless S., Gilbert N. Divergent RNA transcription: a role in promoter unwinding? Transcription. 2013;4:162–166. doi: 10.4161/trns.25554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C.C., Wu H.Y. Transcription-driven DNA supercoiling and gene expression control. Front. Biosci. 2003;8:d430–439. doi: 10.2741/968. [DOI] [PubMed] [Google Scholar]
- 33.Chen C.C., Chou M.Y., Huang C.H., Majumder A., Wu H.Y. A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J. Biol. Chem. 2005;280:5101–5112. doi: 10.1074/jbc.M411840200. [DOI] [PubMed] [Google Scholar]
- 34.Postow L., Hardy C.D., Arsuaga J., Cozzarelli N.R. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J. Mol. Biol. 1972;71:127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- 36.Sinden R.R., Pettijohn D.E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl. Acad. Sci. U.S.A. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynch A.S., Wang J.C. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J. Bacteriol. 1993;175:1645–1655. doi: 10.1128/jb.175.6.1645-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aussel L., Barre F.X., Aroyo M., Stasiak A., Stasiak A.Z., Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 39.Samul R., Leng F. Transcription-coupled hypernegative supercoiling of plasmid DNA by T7 RNA polymerase in Escherichia coli topoisomerase I-deficient strains. J. Mol. Biol. 2007;374:925–935. doi: 10.1016/j.jmb.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhi X., Leng F. Dependence of transcription-coupled DNA supercoiling on promoter strength in Escherichia coli topoisomerase I deficient strains. Gene. 2013;514:82–90. doi: 10.1016/j.gene.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yadav M.K., Kwon S.K., Cho C.G., Park S.W., Chae S.W., Song J.J. Gene expression profile of early in vitro biofilms of Streptococcus pneumoniae. Microbiol. Immunol. 2012;56:621–629. doi: 10.1111/j.1348-0421.2012.00483.x. [DOI] [PubMed] [Google Scholar]
- 42.Gao F., Luo H., Zhang C.T. DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 2013;41:D90–93. doi: 10.1093/nar/gks990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H.Y., Shyy S.H., Wang J.C., Liu L.F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 44.Sinden R.R., Pettijohn D.E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl. Acad. Sci. U.S.A. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Travers A., Muskhelishvili G. DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 46.Korbel J.O., Jensen L.J., von Mering C., Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat. Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 47.Meyer S., Beslon G. Torsion-mediated interaction between adjacent genes. PLoS Comput. Biol. 2014;10:e1003785. doi: 10.1371/journal.pcbi.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.