Abstract
The transition of Escherichia coli from the exponential into the stationary phase of growth induces the stringent response, which is mediated by the rapid accumulation of the alarmone nucleotide (p)ppGpp produced by the enzyme RelA. The significance of RelA’s functionality during the transition in the opposite direction, i.e. from the stationary phase into new exponential growth, is less well understood. Here we show that the relaxed strain, i.e. lacking the relA gene, displays a relative delay in regrowth during the new exponential growth phase in comparison with the isogenic wild type strain. The severity of the effect is a function of both the carbon source and amino acid composition of the outgrowth media. As a result, the loss of RelA functionality increases E. coli tolerance to the bactericidal antibiotic ampicillin during growth resumption in fresh media in a medium-specific way. Taken together, our data underscore the crucial role of medium composition and growth conditions for studies of the role of individual genes and regulatory networks in bacterial phenotypic tolerance to antibiotics.
Bacteria face rapid changes in nutrient availability to which they have to adapt: in periods of famine they need to slow down their metabolism and growth, and when the food source is abundant again they need to resume their rapid production of biomass. The simplest laboratory model of feast-to-famine transition is bacterial stationary phase liquid culture diluted into fresh media. The renewed availability of nutrients allows the starved bacteria to transition to exponential growth after an initial lag phase. To exercise this metabolic maneuver efficiently, both adequate responses to nutrient limitation during the stationary phase and to nutrient abundance upon re-dilution are of importance.
One of the key players coordinating bacterial metabolism is the intracellular alarmone (p)ppGpp (see several excellent recent reviews on the subject)1,2,3. In Escherichia coli two enzymes RelA and SpoT, the namesakes of the widely distributed RelA/SpoT Homolog (RSH) protein family4, control the intracellular concentration of this messenger nucleotide. RelA is a ribosome-associated factor that senses amino acid limitation by directly inspecting the aminoacylation status of the A-site tRNA5. Deacylated tRNA activates RelA’s strong (p)ppGpp synthesis activity6, and increased (p)ppGpp levels initiate a multilayered adaptation program. On the transcriptional level production of ribosomes is halted7 while expression of amino acid biosynthesis genes is induced8,9,10. At the same time diverse molecular targets are directly engaged by (p)ppGpp11, affecting protein synthesis, DNA replication and nucleotide biosynthesis1. While RelA is a one-trick pony, SpoT is a bifunctional enzyme capable of both (p)ppGpp synthesis12 and degradation13, which mediates (p)ppGpp accumulation during the response to various stimuli such as fatty acid14, iron15 and carbon source12 starvation. In addition to responding to nutritional downshifts, SpoT maintains basal (p)ppGpp levels during steady state growth16.
Rapid RelA-dependent accumulation of ppGpp is dubbed the stringent response17 and leads to cessation of stable RNA synthesis, inhibition of translation and growth arrest18. Loss of function relA mutants display a so-called relaxed phenotype characterized by a waste of cellular resources on continuous production of stable RNA during amino acid starvation18, diminished antibiotic tolerance19, and reduced production of glycogen20.
The classical growth curve of bacteria in batch culture contains a lag phase, an exponential phase and a stationary phase21. RelA mediates rapid accumulation of (p)ppGpp during the exit from exponential phase to entry into the stationary phase22, preparing the bacteria for starvation and cessation of growth. Interest in the physiology of relaxed (ΔrelA) strains has been reignited in the last decade, since the functionality of ribosome-dependent RSH enzymes i.e. RelA in Beta- and Gammaproteobacteria and Rel in the rest of bacterial clades4 has been linked to bacterial virulence23 and antibiotic tolerance19. Given the multiple roles played by (p)ppGpp during bacterial stationary phase physiology (for review see Navarro Llorens and colleagues24) we set out to systematically characterize how RelA functionality affects re-growth of E. coli from an overnight stationary culture in fresh media, a step involved in virtually all microbiological experiments, specifically focusing on the role of amino acids and carbon source composition of the outgrowth media.
Results
The growth resumption delay of a ΔrelA strain is dependent on the outgrowth medium and can be abolished by the addition of the complete set of 20 amino acids
We used two standard types of microbiology media: chemically defined minimal medium M925 and complex Lysogeny Broth (LB) medium26. LB is based on a mixture of nutrients originating from a pancreatic digest of casein from cow’s milk and autodigest of Saccharomyces cerevisiae, and as different nutrients are sequentially consumed, E. coli cultures undergo a succession of diauxic shifts along the growth curve27. M9 in its simplest formulation consists of a buffering system, a mixture of essential inorganic salts and a carbon source – usually glucose, as is used here – and it satisfies minimal nutrient requirements for growth of E. coli, while supplements such as amino acids and vitamins can be added separately.
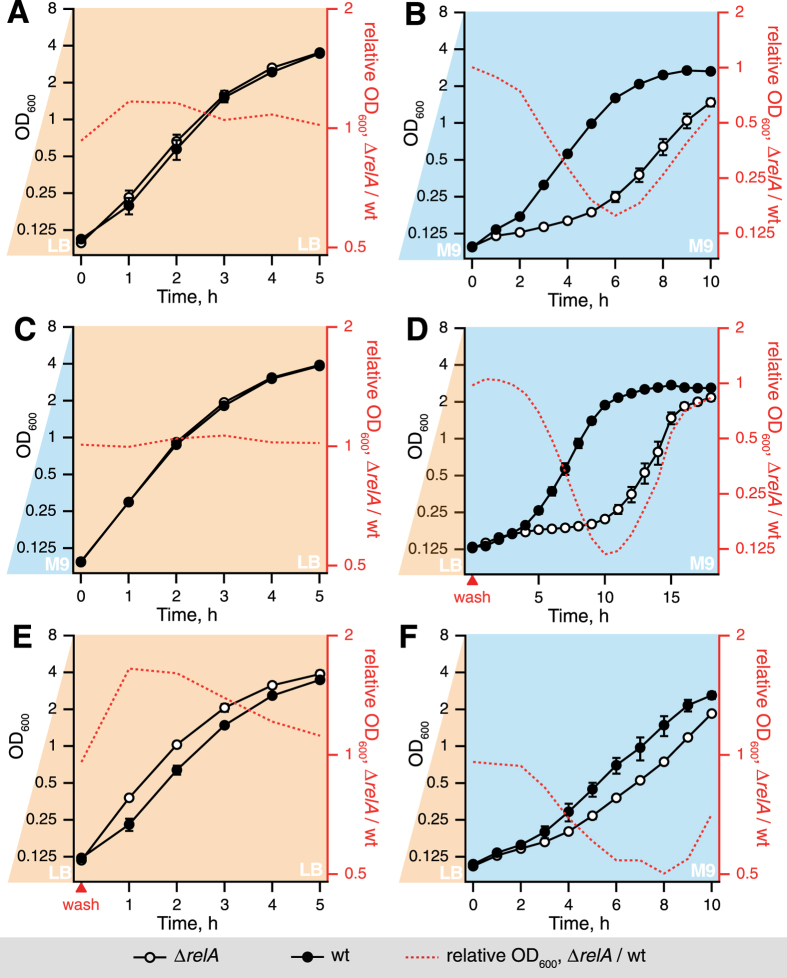
To test RelA’s role in growth resumption, K-12 E. coli wild type strain BW2511328 and isogenic relaxed ΔrelA were grown through exponential phase into stationary phase, kept in stationary phase (defined as less than 10% increase in OD600 within 1 hour) for 15 hours and diluted into fresh medium. The OD600 of cultures was followed throughout the time course. During the initial growth to stationary phase there is no substantial difference in the growth of the two strains, both in LB (designated with light beige shading) and M9 (designated with light blue shading) (Supplementary Figure 1), just as there is no difference in growth resumption of the wild type and the relaxed strain upon LB-to-LB transition (Fig. 1A, quantification of lag and doubling times is summarized in Table 1). At the same time, the ΔrelA strain showed a pronounced – around five hours – growth resumption delay during transition from LB to M9 medium supplemented with 0.4% glucose without additional supplements such as amino acids (Fig. 1D). As a simple numerical measure of the differences in growth resumption, we have plotted the ratio of OD600 for ΔrelA to wild type strain (Fig. 1A–F, red trace).
Figure 1. The outgrowth medium defines the growth resumption delay in ΔrelA E. coli strain.
The OD600 values of a wild type BW25113 strain (filled circles) and an isogenic ΔrelA strain (empty circles) were followed in LB (A, C and E, light beige shading) or M9 medium supplemented with 0.4% glucose (hereafter M9, light blue shading) (B,D,F). The ratio of OD600 for ΔrelA to OD600 of wild type strain (red dotted line) serves as a numerical measure of the difference in growth resumption kinetics between the two. Prior to inoculation, the seeder culture was kept for 15 hours in stationary phase in either LB (light beige shading) (A,D,E,F) or M9 (B,C) media. Cross-inoculation experiments M9-to-LB (C) and LB-to-M9 (D) demonstrate that the growth defect of ΔrelA is specific to the outgrowth medium, i.e. present only in M9. During the LB-to-M9 transition (D), cells were washed with M9 (indicated by the red triangle on the x axis) to reduce carry-over of medium. The washing procedure itself had only mild effect on cells, and if anything, favored growth resumption of ΔrelA cells (E). Results are shown as mean values of biological replicates (n ≥ 3) and error bars (too small to be seen for some of the points) indicate standard error of the mean.
Table 1. Quantification of the growth kinetics data of wild type and relaxed BW25113 E. coli.
| Figure | Initial | Wash | Outgrowth | Doubling time, min |
Lag phase, h |
||
|---|---|---|---|---|---|---|---|
| medium | step | medium | ΔrelA | wt | ΔrelA | wt | |
| S1A | LB | NA | NA | 26 ± 0.4 | 25 ± 0.3 | NA | NA |
| 1A | LB | − | LB | 39 ± 0.1 | 40 ± 1 | 0.2 ± 0.1 | 0.4 ± 0.1 |
| 1E | LB | + | LB | 36 ± 1 | 40 ± 1 | −0.1 ± 0.01 | 0.2 ± 0.1 |
| 1C | M9 | − | LB | 37 ± 1 | 37 ± 2 | 0.03 ± 0.02 | 0.04 ± 0.05 |
| S1B | M9 | NA | NA | 58 ± 1 | 67 ± 2 | NA | NA |
| 1B | M9 | − | M9 | 82 ± 7 | 70 ± 3 | 4.5 ± 0.2 | 1.1 ± 0.03 |
| 5A | M9 | + | M9 | 102 ± 0.5 | 68 ± 0.5 | 4.3 ± 0.3 | 1.0 ± 0.5 |
| 1F | LB | − | M9 | 91 ± 3 | 94 ± 7 | 3.6 ± 0.07 | 1.7 ± 0.3 |
| 1D | LB | + | M9 | 82 ± 2 | 87 ± 6 | 9.1 ± 0.3 | 3.4 ± 0.5 |
| 2B | M9 | − | M9 + AA | 43 ± 0.3 | 44 ± 1 | 0.4 ± 0.1 | 0.6 ± 0.06 |
| 5B | M9 | + | M9gly | 109 ± 5 | 123 ± 3 | 3.8 ± 0.3 | 2.9 ± 0.2 |
| S3 | M9 | − | M9gly + AA | 77 ± 3 | 73 ± 3 | 1.0 ± 0.2 | 1.2 ± 0.1 |
Lag phase is estimated by fitting the data points used to estimate the doubling time to an exponential growth model as per Monod21. M9glc corresponds to M9 medium supplemented with 0.4% glucose; AA indicates to the addition of the 20 amino acids set; gly indicates substitution of glucose for glycerol. Detailed description of the media composition and growth conditions is provided in the corresponding Figure legends. Bold letters indicate the step for which the parameters are quantified. NA signifies that corresponding parameter is not applicable for the experiment. SEM is rounded up to one significant digit. Results are reported as mean values of biological replicates (n ≥ 3), ± standard error of the mean.
The growth resumption delay of the ΔrelA strain could, in principle, stem from lower effective inoculum size as measurements of colony forming units (CFU) do show slightly lower cell count of the ΔrelA strain compared to the wild type during the stationary phase (Supplementary Figure 2A). However, cross-inoculation experiments LB-to-M9 and M9-to-LB show that the appearance of the growth resumption delay in the ΔrelA strain is specific to the nature of the outgrowth medium, specifically it is present in M9 but not LB (Fig. 1C,D), suggesting that reduction of the inoculum size is not the cause of the phenomenon. Washing the cells with M9 during the LB-to-M9 transition in order to remove traces of LB has a dramatic effect on the relative growth delay of ΔrelA strain: when this step is omitted the effect is considerably less pronounced (compare Fig. 1D,F). However, the wash per se is not responsible for the delay, since addition of the wash step during LB-to-LB transition, if anything, promotes an earlier regrowth of the ΔrelA strain (compare Fig. 1E,A).
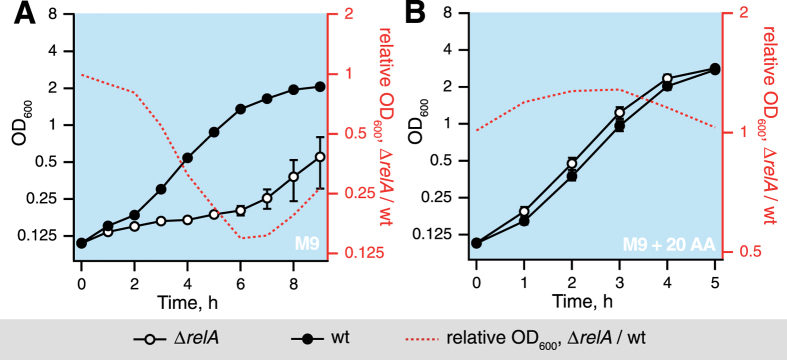
Eventual regrowth of the ΔrelA strain in M9 medium could, in principle, be mediated by a sub-population harboring compensatory mutations – a well-documented phenomenon for E. coli strains unable to produce (p)ppGpp due to a simultaneous disruption in both relA and spoT genes29. However, passage of the wild type and ΔrelA strain through a second regrowth phase faithfully replicated the growth delay effect (Fig. 2A), supporting the idea of composition of the outgrowth medium being responsible for the effect. Since the growth resumption lag was not apparent in LB medium, which has a high concentration of easily metabolizable amino acids27, we have tested whether amino acid supplementation of M9 rescues delayed outgrowth of the ΔrelA culture. Indeed, the growth resumption delay is rescued by addition of a full set of 20 amino acids (each at 100 μg/ml) to the outgrowth minimal medium (Fig. 2B), suggesting that amino acid limitation in M9 is, indeed, responsible for the effect. Measurements of CFUs are in good agreement with the OD600 trace (Supplementary Figure 2B).
Figure 2. Eventual growth resumption of the ΔrelA strain in M9 is not due to compensatory mutations and its regrowth delay can be relieved by the addition of the full set of 20 amino acids.
(A) After 15 hours in stationary phase in M9 supplemented with 0.4% glucose (hereafter M9), cells were diluted into fresh M9 and grown until the stationary phase. After 15 hours in the second stationary phase cells were diluted into fresh M9 and the OD600 of the second growth resumption of wild type (filled circles) and isogenic relaxed strain (empty circles) was followed. (B) The growth resumption delay of the ΔrelA culture disappeared upon addition of the full set of 20 amino acids (each at 100 μg/ml) to the outgrowth M9 medium. The ratio of OD600 for ΔrelA to OD600 of the wild type strain (red dotted line) serves as a numerical measure of the difference in growth resumption kinetics between the two. Results are shown as mean values of biological replicates (A, n = 2; B, n = 3) and error bars (too small to be seen for some of the points) indicate standard error of the mean.
Deprivation of methionine, valine and leucine in the outgrowth medium causes a relative delay in growth resumption of ΔrelA strain
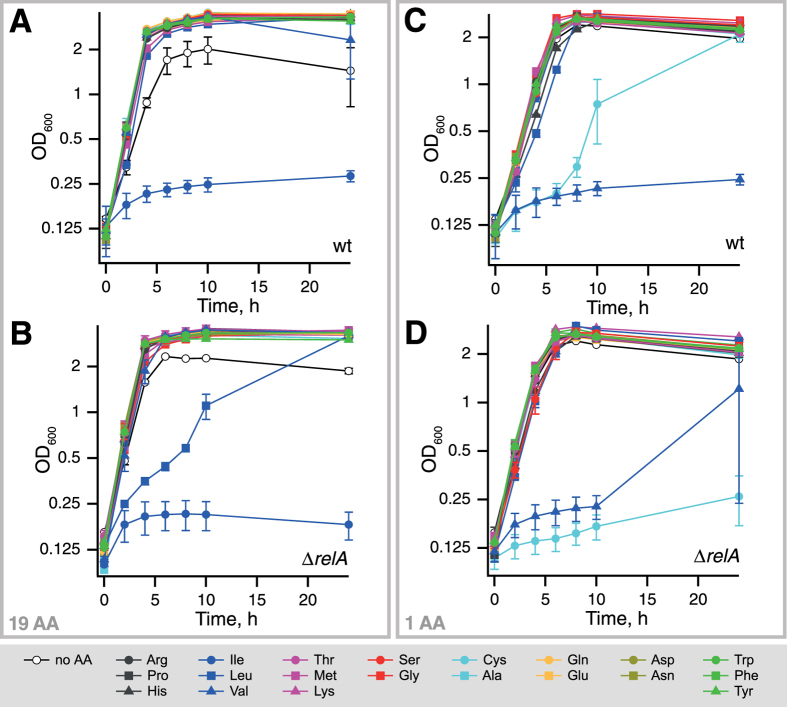
To test whether any specific amino acid is the limiting factor responsible for the delay in the resumption of the ΔrelA strain we tested growth recovery in M9 minimal media supplemented with single amino acid drop out sets, M9 supplemented with 0.4% glucose and 19 individual amino acids added at final concentration of 100 μg/ml. Deprivation of methionine, lysine or any of the branched-chain amino acids (BCAA) – isoleucine, leucine and valine – resulted in a growth resumption delay in both strains, although to a somewhat different degree in each case (Fig. 3A,B). The effect, however, was substantially stronger in the case of the ΔrelA strain (compare Fig. 3A,B,C).
Figure 3. The effects of amino acid composition on transition of wild type and relaxed BW25113 E. coli strains from stationary phase to new exponential growth.
After 15 hours in the stationary phase in M9 medium supplemented with 0.4% glucose, cells were gently pelleted, washed with M9 and diluted into fresh M9 medium supplemented with either 19 amino acids (A–C) or with one amino acid (D–F). Omitted (A–C) or added (D–F) amino acids are indicated by standard three-letter abbreviations with colours grouping amino acids to their biosynthesis pathways as per Keseler and colleagues55 with an exception of grey symbols for Arg, Pro and His which are synthesized via unrelated ad hoc pathways. Empty symbols designate the M9 medium without the addition of any amino acids. Growth resumption was followed for E. coli wild type (A,D) and ΔrelA (B,E) cultures. The ratio of OD600 for ΔrelA to OD600 of the wild type strain (C,F) serves as a numerical measure of the difference in growth resumption kinetics between the two. The results are shown as mean values of biological replicates (n ≥ 3). The error bars (too small to be seen for some of the points) indicate standard error of the mean and for the sake of clarity are omitted on traces lacking specific effects.
In order to separate amino acid dropout effects on bacterial growth per se from specific effects on growth resumption we have performed the same set of experiments using inoculum of E. coli cells from exponential, rather then stationary, phase – an approach that was used in the past to study auxotrophy of relA mutants30,31,32. When switched from minimal M9 medium lacking amino acid supplements into a 19 amino acid medium neither the wild type nor the ΔrelA strain were able to resume growth in isoleucine dropout media for 24 hours of observation: a well-known phenotype of the K-12 strains33,34,35 (Fig. 4A,B). This is in stark contrast with the stationary phase cultures, which did start regrowth after 3.1 ± 0.1 (wt), 3.4 ± 0.3 (ΔrelA) hours (Fig. 3A,B, Table 1). Additionally, the relaxed strain showed a specific growth delay when leucine is omitted. Tyrosine omission does not result in lower stationary phase OD600 when we use exponential phase culture inoculum, but does with the use of stationary phase inoculum (compare Fig. 3A,B and Fig. 4A,B).
Figure 4. The effects of amino acid composition on exponential growth of wild type and relaxed BW25113 E. coli strains.
Individual cultures in M9 medium were started from a single colony, grown up to OD600 of 0.8, diluted to OD600 of 0.1 and grown to 0.5. After that cells were gently pelleted, washed with M9 and resuspended in fresh M9 medium supplemented with either 19 amino acids (A,B) or with one amino acid (C,D). The results are shown as mean values of biological replicates (n = 2). The error bars (too small to be seen for some of the points) indicate standard error of the mean and for the sake of clarity are omitted on traces lacking specific effects.
Addition of individual amino acids does not rescue the growth resumption delay of the ΔrelA strain
Next, we set out to determine if the addition of any specific amino acid rescues the relative delay in growth resumption of the ΔrelA strain by testing the effects of addition of individual amino acids at final concentration of 100 μg/ml. None of the amino acids reversed the defect; conversely, several amino acids exacerbated it for both strains (Fig. 3D–F). Addition of valine and cysteine strongly inhibited the regrowth of both wild type and relaxed strain; serine completely inhibited the regrowth of ΔrelA, but not wild type (Fig. 3D,E). Growth inhibition by valine and cysteine is present when we use exponentially growing inoculum, suggesting that the effect is not specific for growth resumption but rather bacterial growth per se (Fig. 4). While the addition of histidine, serine, threonine, isoleucine and leucine caused a prolonged lag phase after stationary phase in both of the two strains (Fig. 3D,E), the severity of the effect was somewhat different, with serine causing a more pronounced growth resumption delay in the relaxed strain (Fig. 3E). The inhibitory effect of serine was absent in the case of exponentially growing cells (Fig. 4C,D).
Switching the carbon source of the outgrowth medium from glucose to glycerol abolishes the growth resumption delay of the relaxed strain
Rich LB and poor M9 minimal media dramatically differ in amino acid content: while in LB medium amino acids and peptides serve both as building blocks for protein as well as a source of carbon, ammonium and energy27, M9 usually lacks amino acids altogether and the most commonly used carbon source is glucose, as was used in the experiments described above (Figs 1, 2, 3, 4). As we have shown, the addition of 20 amino acids set to M9 supplemented with 0.4% glucose abolishes the delay in growth resumption of the ΔrelA strain (Fig. 2). Importantly, addition of amino acids also decreases the doubling time of both the wild type and the relaxed strain almost twice (from 70 ± 3 to 44 ± 1 and from 82 ± 7 to 43 ± 0.3 minutes, respectively, Table 1). One could argue the relative growth delay of the relaxed strain in the absence of amino acids is merely a consequence of the necessity of relA functionality during slow growth per se, rather then a specific effect of the lack of amino acids.
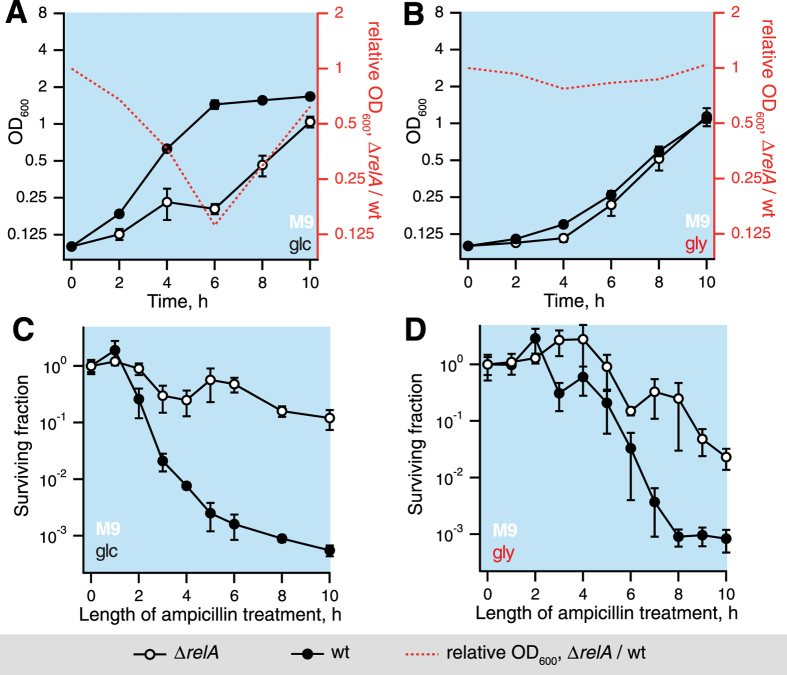
To probe this conjecture, we have performed the regrowth experiments while reducing the growth rate in M9 lacking amino acids by substituting the glucose, a preferred carbon source for E. coli, for less optimal carbon source, glycerol. This further reduction of the growth rate can be counteracted by the addition of 20 amino acid set, which allows us to probe the connection amongst amino acid and carbon source composition, growth rate and growth resumption delay in the ΔrelA strain. While the doubling time increases to 109 ± 5 (ΔrelA) and 123 ± 3 minutes (wt) in M9 supplemented with glycerol instead of glucose, the relaxed cells initiate the regrowth almost early as the wild type (Fig. 5A,B, Table 1). Addition of the 20 amino acids set to M9 medium supplemented with glycerol increases the growth rates to the levels similar to that in M9 supplemented with glucose. However, the growth resumption kinetics of the relaxed and wild type strains remain unchanged, i.e. ΔrelA and the wild type regrow similarly (Supplementary Figure 3, Table 1). Taken together, these results demonstrate that the relative growth delay of the relaxed strain is modulated by both carbon source and amino acid composition of the outgrowth media.
Figure 5. The effects of the carbon source composition of the outgrowth media on regrowth kinetics and ampicillin sensitivity upon transition of wild type and relaxed BW25113 E. coli strains from stationary phase to fresh M9 media.
After 15 hours in the stationary phase in M9 medium, cells were gently pelleted, washed with M9 and diluted into fresh M9 medium supplemented with 0.4% glucose (A) or glycerol (B), and the ratio of OD600 for ΔrelA to OD600 of wild type strain was plotted as a numerical measure of the differences in growth resumption between the two strains. To follow the ampicillin tolerance during E. coli regrowth in the presence of 0.4% glucose (C) or glycerol (D), the bacterial cultures were treated as described above but the regrowth medium was supplemented with ampicillin at 200 μg/ml and cell viability (colony forming units, CFU) was measured instead of OD600. Results are shown as mean values of biological replicates (n ≥ 3) and error bars indicate standard error of the mean.
Relaxed strain is killed by ampicillin considerably slower then the wild type during growth resumption in M9 supplemented with either glucose or glycerol
The bacterial growth rate is a key factor affecting antibiotic susceptibility. In the case of the antibiotic ampicillin the killing efficiency is believed to be directly proportional to the rate of growth36. Therefore, the effects of relA’s loss of functionality on growth resumption kinetics are expected to alter the antibiotic killing kinetics. To test this conjecture, we followed antibiotic killing by ampicillin after stationary phase cultures were diluted into M9 supplemented with either glucose (Fig. 5C) or glycerol (Fig. 5D). Surprisingly, the ΔrelA strain was killed considerably slower then the isogenic wild type under both conditions. In the case of the wild type strain there is a correlation between the regrowth and ampicillin killing kinetics, i.e. the earlier bacteria start regrowth, the more efficiently they are killed by ampicillin. At the same time the relaxed strain is killed by ampicillin considerably less efficient then the wild type even in if the growth kinetics are very similar in M9 supplemented with glycerol (compare Fig. 5B,D). As a result, the effect of relA disruption on ampicillin tolerance is heavily dependent on medium composition: while in the presence of glucose after 5 hours of incubation with ampicillin – time point that is often used for end-point persister measurements, e.g37,38. – the relaxed strain has approximately two orders of magnitude higher persister count, in M9 supplemented with glycerol persister frequencies for the two strains are nearly identical.
Discussion
E. coli growth, nutrient availability and RelA functionality
We have systematically analyzed the effects of amino acid and carbon source availability, and RelA functionality in K-12 BW25113 E. coli strains during their transition from stationary phase to new exponential growth. The RelA-specific effects during this transition are confounded by two aspects that one has to consider. First are the defects in amino acid metabolism that are specific to K-12 E. coli strains, the workhorse of microbiology for almost a century39. Due to a frameshift mutation in one of the central isoenzymes of acetohydroxy acid synthase (AHAS)34, addition of valine to minimal medium leads to cessation of growth that can be rescued by the addition of isoleucine, although the exact mechanism behind it is still matter of debate8,9,33. We clearly see the valine effect in our experiments (Figs 3 and 4). Second is the role of RelA and (p)ppGpp in amino acid biosynthesis. (p)ppGpp is crucial for amino acid synthesis as evidenced by both ppGpp0 (i.e. completely lacking the alarmone) E. coli12 and B. subtilis40 being auxotrophic for several amino acids including methionine and branched chain amino acids leucine, isoleucine and valine. The knock out strain used in the current work, while lacking RelA does have an intact copy of the second enzyme synthesizing (p)ppGpp in E. coli – SpoT12. While not directly causing auxotrophy, disruption of relA does lead to perturbed regulation of amino acid biosynthesis. Simultaneous addition of “one-carbon” amino acids (serine, glycine and methionine, SMG) suppresses bacterial growth, but while the wild type can overcome it, the relaxed can not30; and the effect is counteracted by addition of isoleucine31,41. The difference in the behaviors of wild type and relaxed strains is likely due the stringent response promoting biosynthesis of branched chain amino acids (BCAA), such as isoleucine8,9. We clearly see that omission of one of the BCAA results in RelA-specific retardation of growth resumption (Figs 3 and 4). Cysteine is known to cause transient amino acid starvation in the uropathogenic E. coli strain SP53642; the mechanism behind this phenomenon is not understood. We see manifestations of cysteine-induced starvation in our background: while inhibition of wild-type growth is transient, growth inhibition is near-complete in the course of 24 hours of observation of the relaxed strain (Fig. 4).
While the effects of amino acid composition on regrowth of the ΔrelA strain were expected, the effects of substitution of the carbon source in M9 media from glucose to glycerol were surprising (Fig. 5). In the presence of glucose ΔrelA strain regrows with a delay in comparison to the wild type, and in the presence of glycerol the two strains regrow equally well. The cause of this is not obvious, connections between (p)ppGpp and carbon metabolism are known; for example expression of the receptor protein of the global catabolic modulator cAMP (CRP) is under direct negative control of (p)ppGpp43. There are parallels between the effects on re-growth observed in this study and previous observations of the differential requirements for RelA in glycogen accumulation during amino acid starvation in the presence of different carbon sources44,45. The relA gene is needed when glucose is the carbon source, while the high cellular levels of cyclic AMP relieve the requirement for relA when glycerol is the carbon source20,45. Moreover, branched-chain amino acid biosynthesis is promoted by cAMP46. Since (p)ppGpp and amino acid metabolism are interconnected with carbon metabolism via many other pathways, such as tricarboxylic acid cycle47 the connections among carbon source, RelA functionality and re-growth are far from simple.
Bacterial regrowth kinetics is intimately connected with bacterial sensitivity to bactericidal antibiotics: the frequency of persisters is reflecting the awakening kinetics48. Increased cellular (p)ppGpp level was suggested to be the ultimate driver of persister formation49, and is implicated in antibiotic-specific tolerance mechanisms, i.e. protection from ampicillin acting via inhibition of cell wall biosynthesis50. Therefore, one could naively assume that the loss of RelA would result in, if anything, lower persister count, which is evidently not the case. Clearly, persistence is a multifaceted phenomenon, with media composition and growth conditions playing a major role via effects on metabolism51 and growth rate52.
Methods
Bacterial strains and plasmids
The relA deletion strain was constructed from strain BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) as described elsewhere28 using primers relAF (CGATTTCGGCAGGTCTGGTCCCTAAAGGAGAGGACGGTGTAGGCTGGAGCTGCTTC) and relAR (CAATCTACATTGTAGATACGAGCAAATTTCGGCCTAATTCCGGGGATCCGTCGACC) for template PCR. Kanamycin resistance cassette was removed and ΔrelA phenotype was confirmed on SMG plates30 (Supplementary Figure 4).
Media and growth conditions
Cells were grown with vigorous agitation (200–220 rpm) at 37 °C in LB (Becton, Dickinson and Company) and M9 minimal medium (48 mM Na2HPO4, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 0.1 mM CaCl2 and 2 mM MgSO4)25 or on LB agar plates (Becton, Dickinson and Company). M9 was supplemented with 0.4% (w/v) carbon source, which was glucose or glycerol. Amino acids were used at a concentration of 100 μg/ml, kanamycin at 25 μg/ml and ampicillin at 200 μg/ml. The data presented on Figs 3 and 4 were obtained using a 96-well plate reader Tecan Sunrise and the reset of the experiments were performed in flasks.
Growth recovery experiments
Bacterial cultures were started from single colonies on LB plate and grown until OD600 of 0.8. Resulting seeder culture was used to inoculate the experimental culture to starting OD600 of 0.1, which was grown aerobically into stationary phase (20 ml of medium in 125 ml flasks), kept in stationary phase for 15 h and directly diluted into fresh medium to OD600 of 0.1 or, during shift from LB to M9, harvested by centrifugation and, washed with M9 before transfer into fresh medium. Experiments with inclusion of 1 or 19 amino acids were conducted as follows: after 15 h in stationary phase, cells were harvested by centrifugation (in carbon source experiments washed with carbon source depleted M9), resuspended to OD600 of 0.1 and grown aerobically in fresh medium on 96-well plates in a volume of 80 μl per well. OD600 readings of the 96-well plates (plate reader Tecan Sunrise) were converted to values for 1 cm path length (spectrophotometer Thermo Helios β) (Supplementary Figure 5). The length of the lag phase was determined by an intercept between the initial inoculum density (OD600 = 0.1) and the tangent of fastest exponential part of the growth curve that determines the doubling time. Lag and doubling times were calculated separately for individual growth curves (n ≥ 3). Data analysis was performed in R53 and the code is provided in the Supplementary Information.
Antibiotic killing
15 h stationary phase cultures were prepared as described above for growth recovery experiments. The cells were then collected 10 min at 5000 g at room temperature, washed with M9 0.4% glucose or M9 0.4% glycerol, collected and resuspended again and diluted to OD600 of 0.1 in 20 ml medium in 125 ml flasks. The following ampicillin killing assays were performed essentially as described in54. A 10 μl aliquot was used for a CFU count at the zero hour time point, and then ampicillin was added to the remaining culture at 200 μg/ml. During following time course of ampicillin killing, flasks were incubated at 37 °C 200 rpm. Colony forming units were determined by series of tenfold dilutions out of which 5 μl was spotted on an LB plate. After overnight incubation of the plates at 37 °C, colonies were counted and CFU/ml was calculated.
Additional Information
How to cite this article: Varik, V. et al. Composition of the outgrowth medium modulates wake-up kinetics and ampicillin sensitivity of stringent and relaxed Escherichia coli. Sci. Rep. 6, 22308; doi: 10.1038/srep22308 (2016).
Supplementary Material
Acknowledgments
We are grateful to Gemma Atkinson and Mike Cashel for helpful discussions. This work was supported by the by grant IUT2-22 from the Estonian Research Council (TT); European Regional Development Fund through the Centre of Excellence in Chemical Biology (VH and TT); Estonian Science Foundation (grants ETF9012 and PUT37 to VH); Umeå University, the Swedish Research council Vetenskapsrådet (grant 2013-4680), Kempe and Ragnar Söderberg foundations (VH).
Footnotes
Author Contributions V.V. and T.T. conceived the project. V.V., V.H. and T.T. designed the experiments. V.V. and S.O. performed experiments. V.H. and T.T. coordinated the study. V.V. and V.H. wrote the paper with contributions from T.T. and S.O.
References
- Hauryliuk V., Atkinson G. C., Murakami K. S., Tenson T. & Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13, 298–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Bittner A. N. & Wang J. D. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol 24, 72–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A. O., Colomer-Winter C. & Lemos J. A. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 197, 1146–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson G. C., Tenson T. & Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6, e23479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X. et al. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep 14, 811–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W. A. & Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA 70, 1564–8 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B. J., Ross W., Gaal T. & Gourse R. L. rRNA transcription in Escherichia coli. Annu Rev Genet 38, 749–70 (2004). [DOI] [PubMed] [Google Scholar]
- Traxler M. F. et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68, 1128–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedin K. & Norel F. Comparison of DeltarelA strains of Escherichia coli and Salmonella enterica serovar Typhimurium suggests a role for ppGpp in attenuation regulation of branched-chain amino acid biosynthesis. J Bacteriol 183, 6184–96 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B. J., Berkmen M. B. & Gourse R. L. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA 102, 7823–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjee U., Ogata K. & Houry W. A. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol 85, 1029–43 (2012). [DOI] [PubMed] [Google Scholar]
- Xiao H. et al. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266, 5980–90 (1991). [PubMed] [Google Scholar]
- Sy J. In vitro degradation of guanosine 5′-diphosphate, 3′-diphosphate. Proc Natl Acad Sci USA 74, 5529–33 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfzadeh M., Keener J. & Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci USA 90, 11004–8 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D., Albrecht C., Cashel M. & D’Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol 56, 958–70 (2005). [DOI] [PubMed] [Google Scholar]
- Ryals J., Little R. & Bremer H. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol 151, 1261–8 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stent G. S. & Brenner S. A genetic locus for the regulation of ribonucleic acid synthesis. Proc Natl Acad Sci USA 47, 2005–14 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M., Gentry D., Hernandez V. & Vinella D. In In Escherichia coli Salmonella Cell Mol Biol (ed. Neidhart H.) 1458–1495 (Washington, D.C., 1996). [Google Scholar]
- Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother 67, 2069–89 (2012). [DOI] [PubMed] [Google Scholar]
- Leckie M. P., Tieber V. L., Porter S. E., Roth W. G. & Dietzler D. N. Independence of cyclic AMP and relA gene stimulation of glycogen synthesis in intact Escherichia coli cells. J Bacteriol 161, 133–40 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J. The growth of bacterial cultures. Annual Reviews in Microbiology 3, 371–394 (1949). [Google Scholar]
- Lazzarini R. A., Cashel M. & Gallant J. On the regulation of guanosine tetraphosphate levels in stringent and relaxed strains of Escherichia coli. J Biol Chem 246, 4381–5 (1971). [PubMed] [Google Scholar]
- Dalebroux Z. D. & Swanson M. S. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10, 203–12 (2012). [DOI] [PubMed] [Google Scholar]
- Navarro Llorens J. M., Tormo A. & Martinez-Garcia E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev 34, 476–95 (2010). [DOI] [PubMed] [Google Scholar]
- Sambrook J. & Russel D. Molecular Cloning: A Laboratory Manual (Cold Spring Harbour Laboratory Press, Cold Spring Harbour, New York, 2001). [Google Scholar]
- Luria S. E. & Burrous J. W. Hybridization between Escherichia coli and Shigella. J Bacteriol 74, 461–76 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezonov G., Joseleau-Petit D. & D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189, 8746–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A. & Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97, 6640–5 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy H. & Cashel M. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol 371, 596–601 (2003). [DOI] [PubMed] [Google Scholar]
- Uzan M. & Danchin A. A rapid test for the rel A mutation in E. coli. Biochem Biophys Res Commun 69, 751–8 (1976). [DOI] [PubMed] [Google Scholar]
- Uzan M. & Danchin A. Correlation between the serine sensitivity and the derepressibility of the ilv genes in Escherichia coli relA- mutants. Mol Gen Genet 165, 21–30 (1978). [DOI] [PubMed] [Google Scholar]
- Alföeldi L., Stent G. S., Hoogs M. & Hill R. Physiological Effects of the Rna Control (Rc) Gene in E. Coli. Z Vererbungsl 94, 285–302 (1963). [DOI] [PubMed] [Google Scholar]
- De Felice M., Levinthal M., Iaccarino M. & Guardiola J. Growth inhibition as a consequence of antagonism between related amino acids: effect of valine in Escherichia coli K-12. Microbiol Rev 43, 42–58 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P. et al. Molecular basis of valine resistance in Escherichia coli K-12. Proc Natl Acad Sci USA 78, 922–5 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. R., Shapiro B. E., Hung S. P., Mjolsness E. D. & Hatfield G. W. A mathematical model for the branched chain amino acid biosynthetic pathways of Escherichia coli K12. J Biol Chem 280, 11224–32 (2005). [DOI] [PubMed] [Google Scholar]
- Tuomanen E., Cozens R., Tosch W., Zak O. & Tomasz A. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132, 1297–304 (1986). [DOI] [PubMed] [Google Scholar]
- Maisonneuve E., Castro-Camargo M. & Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–50 (2013). [DOI] [PubMed] [Google Scholar]
- Orman M. A. & Brynildsen M. P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother 57, 3230–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36, 525–57 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriel A. et al. GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J Bacteriol 196, 189–201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfoeldi L. & Kerekes E. Neutralization of the Amino Acid Sensitivity of Rcrel Escherichia Coli. Biochim Biophys Acta 91, 155–7 (1964). [DOI] [PubMed] [Google Scholar]
- Sorensen M. A. & Pedersen S. Cysteine, even in low concentrations, induces transient amino acid starvation in Escherichia coli. J Bacteriol 173, 5244–6 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson J. et al. Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell 102, 475–85 (2000). [DOI] [PubMed] [Google Scholar]
- Taguchi M., Izui K. & Katsuki H. Augmentation of glycogen synthesis under stringent control in Escherichia coli. J Biochem 88, 379–87 (1980). [DOI] [PubMed] [Google Scholar]
- Leckie M. P., Tieber V. L., Porter S. E. & Dietzler D. N. The relA gene is not required for glycogen accumulation during NH4 + starvation of Escherichia coli. Biochem Biophys Res Commun 95, 924–31 (1980). [DOI] [PubMed] [Google Scholar]
- Sutton A. & Freundlich M. Regulation of cyclic AMP of the ilvB-encoded biosynthetic acetohydroxy acid synthase in Escherichia coli K-12. Mol Gen Genet 178, 179–83 (1980). [DOI] [PubMed] [Google Scholar]
- Hardiman T., Lemuth K., Keller M. A., Reuss M. & Siemann-Herzberg M. Topology of the global regulatory network of carbon limitation in Escherichia coli. J Biotechnol 132, 359–74 (2007). [DOI] [PubMed] [Google Scholar]
- Joers A., Kaldalu N. & Tenson T. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J Bacteriol 192, 3379–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K. & Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol 66, 103–23 (2012). [DOI] [PubMed] [Google Scholar]
- Rodionov D. G. & Ishiguro E. E. Direct correlation between overproduction of guanosine 3′,5′-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J Bacteriol 177, 4224–9 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato S. M. et al. The role of metabolism in bacterial persistence. Front Microbiol 5, 70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luidalepp H., Joers A., Kaldalu N. & Tenson T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J Bacteriol 193, 3598–605 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2013).
- Kaldalu N., Joers A., Ingelman H. & Tenson T. A General Method for Measuring Persister Levels in Escherichia coli Cultures. Methods Mol Biol 1333, 29–42 (2016). [DOI] [PubMed] [Google Scholar]
- Keseler I. M. et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res 37, D464–70 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.