Summary
Reason for performing study
Dysadhesion of the laminar basal epithelial cells (LBEC) from the underlying dermis is the central event leading to structural failure in equine laminitis. Although many studies of sepsis-related laminitis have reported multiple events occurring throughout the lamellar tissue, there is minimal information regarding signalling events occurring specifically in the LBEC.
Objectives
To determine the signalling events in the LBECs during the early stages of carbohydrate induced laminitis.
Study Design
Experimental study.
Methods
Eight horses were given an overload of carbohydrate (corn starch mixture, CHO) via nasogastric tube. Prior to administration of CHO, lamellar biopsies were taken from the left fore foot (CON). Biopsies were taken from the left hind foot at the onset of fever (DEV) and from the right fore foot at the onset of lameness (OG1). Laminar basal epithelial cells (LBECs) were isolated from cryosections using a LCM microscope. Next generation sequencing (RNA-Seq) was used to identify transcripts expressed in the LBECs for each time point and bioinformatic analysis was performed with thresholds for between group comparisons set at a greater than 2-fold change and p-value ≤0.05.
Results
Forty genes (22 increased/18 decreased) were significantly different from DEV time vs. CON and 107 genes (57 increased/50 decreased) were significantly different from OG1 time vs. CON. Significant increases in inflammatory genes were present in addition to significantly altered expression of genes related to extracellular matrix composition, stability and turnover.
Conclusions
Inflammatory response and extracellular matrix regulation signalling was strongly represented at the DEV and OG1 times. These results indicate that the LBEC is not only a casualty but also an active participant in lamellar events leading to structural failure of the digital lamellae in equine laminitis.
Keywords: horse, laminitis, laser capture microdissection, RNA-Seq, inflammation, extracellular matrix composition
Introduction
Lamellar failure occurs in laminitis due to dysadhesion of lamellar basal epithelial cells (LBECs) from the underlying dermis and subsequent displacement of the distal phalanx due to forces imposed on the lamellar structure [1]. Similar events occur in lamellar failure in sepsis-related laminitis as occur in organ failure in human sepsis including inflammatory mediator expression (i.e. cytokines and chemokines) [2–7], leukocyte emigration [8–11], oxidant stress [12, 13] and production of vasoactive molecules [14–18]. Epithelial injury/dysfunction is a central component of sepsis/SIRS related injury to organs such as the kidney, lung, and intestinal tract. Although inflammatory injury to epithelial cells is commonly reported in sepsis/SIRS [19–21], the epithelial cell itself can also amplify ongoing tissue inflammation in sepsis/SIRS by expressing numerous cytokines and chemokines [21, 22].
The majority of studies on cell signalling in laminitis use techniques such as real time- PCR and immunoblotting which assess homogenates of the entire lamellar tissue comprised of all cellular components of lamellar dermis and epidermis; thereby, not allowing assessment of signalling in a specific cell type. Techniques which allow visualisation of signal in the cells of interest (i.e. LBECs) such as immunohistochemistry [2, 23] and in situ hybridisation [4, 23] are are limited by the narrow availability of equine specific reagents and the qualitative nature of the results. Furthermore, there are only a few reports of successful isolation and culture of lamellar epithelial cells [24–26], which are historically difficult to isolate in pure culture and survive successfully through subsequent passages, and are not likely to respond identically in cell culture as in the lamellar milieu in the digital lamellar tissue. Since these cells are essential to determining mechanisms involved in lamellar failure, identification of a technique that would allow for the specific study of the LBEC was necessary.
Laser capture microdissection is a novel technique in which specific cells can be dissected from a tissue cryosection with a laser beam under microscopic guidance, allowing for the isolation of highly purified cell populations within a heterogeneous tissue section [27]. Collected samples can later be used for DNA, RNA or protein analysis. Laser capture microscopy was first described [28] in 1996 and since has been used for the study of a wide variety of disease states [29–34]. In specific regard to epithelial cells, LCM has also been used to determine differentially expressed genes produced in basal cell carcinoma [35], junctional epithelium in periodontal disease [36], ileal epithelium from neonates with necrotising enterocolitis [37], epidermal tissue from psoriasis lesions [38], and basal and granular cell involvement in epidermal differentiation [39].
The purpose of this study was to use LCM to isolate LBECs from lamellar biopsies collected from horses given an overload of carbohydrate to induce laminitis and to use a broad transcriptome analysis technique (RNA Sequencing; RNA-Seq) to identify signalling events occurring in the LBEC in the early stages of laminitis.
Materials and methods
Animals
Eight adult Standardbred horses with a median body weight of 444 kg (ranging from 407 kg to 476 kg) and a median age of 7.5 years (ranging from 4 to 14 years old) were used. Each horse was determined to be healthy and free of digital pathology by physical and lameness examination. Prior to the experiment, the horses were quarantined for 2 weeks, housed in stalls and fed a complete pelleted diet.
Carbohydrate Overload Model
A carbohydrate gruel consisting of 85% cornstarch and 15% wood flour (17.6 g/kg body weight) was administered to each horse via a nasogastric tube [7, 40, 41]. All horses received complete physical examinations, consisting of rectal temperature, heart rate, respiratory rate, abdominal sounds, digital pulses, evaluation with hoof testers, and gait evaluation immediately prior to nasogastric entubation, and at 2 h intervals following administration of carbohydrate. Control (CON) lamellar biopsies were collected from the left fore foot prior to carbohydrate administration. Lamellar biopsies were collected from the left hind foot for the developmental (DEV) time defined as at 24 h post CHO administration (n = 6) or the onset of fever (>38.6°C) if that occurred before 24 h (n = 1). Lamellar biopsies were collected from the right fore foot at the Obel Grade 1 (OG1) time defined as 48 h post CHO administration (n = 3) or the onset of OG1 lameness if that occurred prior to 48 h (n = 5). One horse did not develop a fever before the onset of OG1 which occurred at 24 h. Once the OG1 tissues were collected, each horse was subjected to euthanasia with pentobarbital sodium and phenytoin sodium (20 mg/kg of body weight i.v.).
Lamellar Biopsies
Horses were sedated and abaxial sesamoid perineural block with 2 ml mepivacaine (Carbocaine®)a over the medial and lateral palmar or plantar nerves were performed prior to biopsy collection. Once the horses were determined to have no sensation of the dorsal hoof/pastern region, 6 12-mm holes were drilled into the hard, keratinised dorsal hoof wall down to the softer stratum medium over the lamellae using a dremel tool. An 8-mm biopsy punch and a small curved bistoury knife were used to collect a lamellar biopsy from each hole (Fig 1). Biopsy of the lamellae contained both epidermal and dermal tissue. Four of the 6 biopsy sections were placed in OCT, immediately frozen in liquid nitrogen and later transferred to −80°C for storage until sectioned for LCM. Each biopsy site was covered with betadine soaked gauze and a cotton bandage was applied over the dorsal hoof surface and then secured to the foot with elasticon. Horses were monitored for any lameness associated with the biopsy sites, and when present, repeat perineural anaesthesia of that limb was performed using bupivacaineb.
Fig 1.
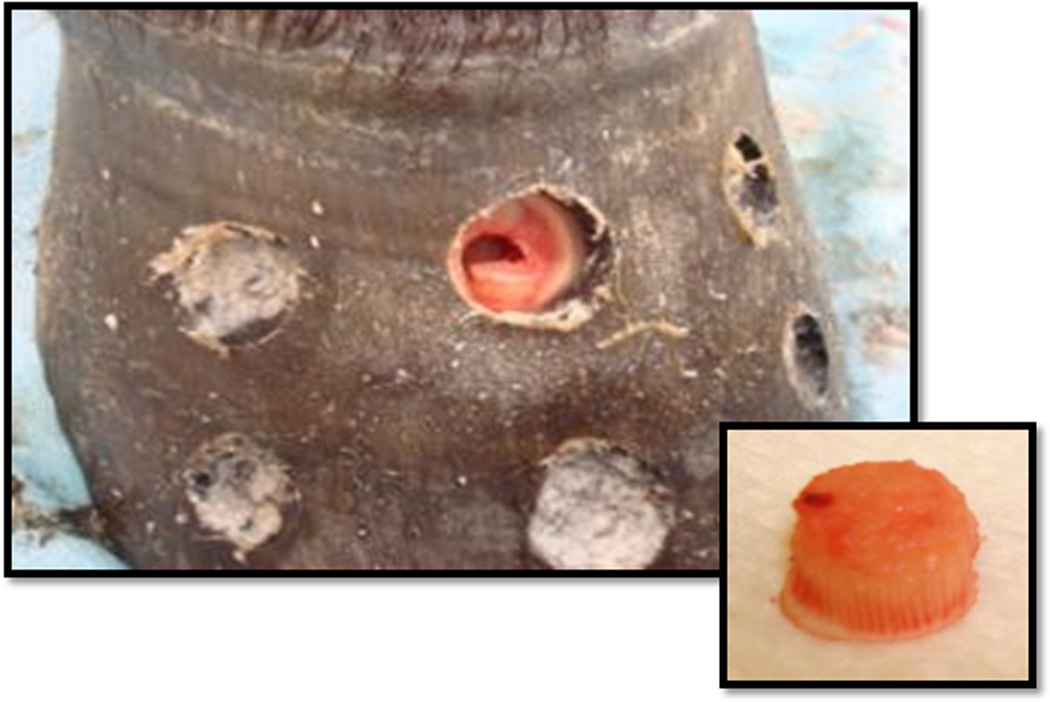
Six 8 mm biopsies were obtained from the dorsal hoof wall at each time point (CON, DEV, and OG1). Insert demonstrates biopsy at the epidermal and dermal interface.
Laser Capture Microdissection
Lamellar biopsies, frozen in OCT and stored at −80°C, were cut into 10 μm sections using a cryostatc. Three sections were mounted onto a RNase-Zapd treated polyethylene napthalate covered slidee and either used immediately or stored at −80°C in a 50 ml conical until used (within 48 h from sectioning).
Slides were stained with cresyl violetf in accordance to manufacturer’s instructions by first rehydrating with 95%, 70% and 50% ethanol followed by staining for 30 s and sequential dehydration with molecular grade ethanolg solutions. The xylene step was excluded from the protocol. After staining, slides were air dried and immediately used for LCM.
Slides were placed on the PALM LCMh for collection of tissue. LBEC were selected at 10X objective using the freehand drawing option on the PALM LCM software (Fig 2). Once selected the LBEC were cut and subsequently catapulted using an UV-energy of 50–60 and UV-focus of 80–82. Laser dissections of each slide were limited to 30–45 min to minimise RNA degradation. An average of 500,000 to 1,000,000 µm2 area of LBECs was collected from each section and a minimum of 2 different sections (biopsies) of lamellar tissue were used for each time point from each horse. Dissected tissue was captured into aqueous-free adhesive capsh. After collection, the caps were placed onto 200 µl RNase free tubes and stored at −80°C until RNA could be isolated.
Fig 2.
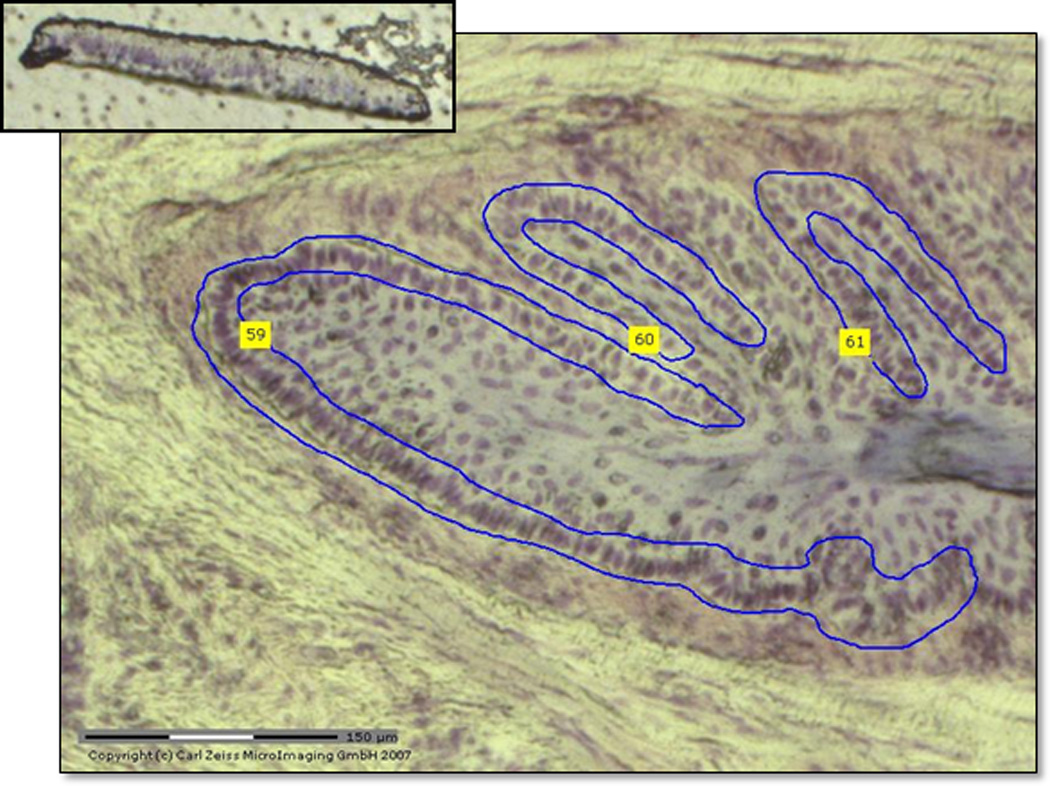
Selected section of lamellar basal epithelial cells (LBECs) stained with cresyl violet at 10X magnification from the PALM LCM microscope. Top insert demonstrates catapulted section of LBECs in adhesive cap.
RNA isolation and quality control
Total RNA was extracted from each sample using a kiti which included DNase treatment to remove genomic DNA contamination. Captured LBECs were pooled from each horse at each time point before isolation of total RNA. Total RNA from each individual sample was quantified using a NanoDrop spectrophotometer and the quality assessed using the pico chip on the Agilent Bioanalyzer 2100j. Total RNA ranged from 2 ng/µl to 14 ng/µl per sample. High quality samples for LCM isolation were demonstrated by having electropherograms with solid peaks at 18S and 28S and a RIN that was >7 (Fig 3).
Fig 3.
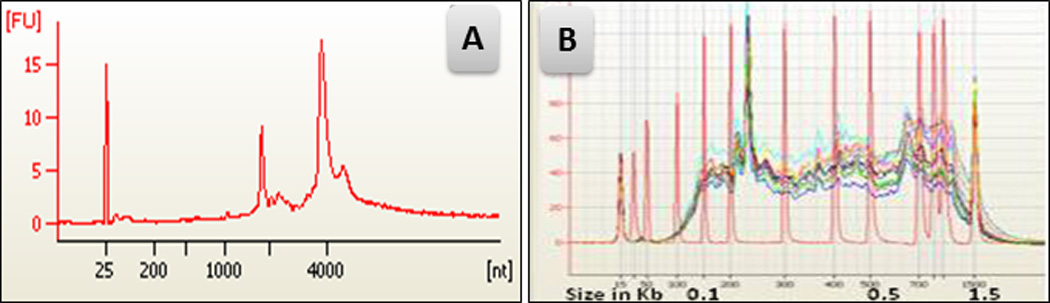
(A) Agilent bioanalyser data of RNA from captured LBECs demonstrating excellent RNA quality. (B) cDNA following RNA amplification (red DNA ladder overlaid to document high fidelity results).
RNA Sequencing (RNAseq)
A library was generated from rRNA-depleted total RNA using SOLiD Total RNA seq kit according to the manufacturer’s recommendationsk. Briefly, total RNA was fragmented with RNaseIII, a double-stranded RNA specific endoribonuclease. The fragmented RNA was purified by column chromatography, assessed by a bioanalyser and reverse transcribed. The cDNA was cleaned and size selected using Agencourt AMPure XP beadsl. The product was amplified with primers specific to the adapter sequences and then purified. Templated sequencing beads were produced via emulsion PCR with Applied Biosystems’ EZ Bead systemk. The templated sequencing beads were sequenced by ligation on an Applied Biosystems SOLiD 5500XL system. The xseq file was transferred to the Bioinformatics Shared Resource for further analysis.
NanoString nCounter profiling
In order to validate the RNA-seq results, we utilised the nCounter System from nanoString Technologiesm to identify the significant differences in lamellar gene expression profiles during laminitis. Several of the genes found to be regulated by RNA-Seq were included in a nanoString assay (custom Codeset) designed for assessment of 30 genes of interest for different ongoing laminitis studies. As the nanoString platform was used to evaluate samples from 2 different studies (the current study and a study of endocrinopathic laminitis), the genes of interest selected for evaluation were established from genes determined to be differentially regulated in both studies. Therefore, not all genes present in the assessment were taken from the RNAseq results of the current study. Sample numbers were limited to 6 horses per time point and were randomly selected. The nanoString platform is based on 2 sequence-specific nucleic acid probes for the mRNAs of interest, a biotin-labelled capture probe which allows attachment of all targets to a slide surface and a reporter probe in which a fluorescent barcode is attached to the nucleic acid probe to allow digital measurement of the prevalence and relative expression of each mRNA of interest within a given sample [42]. Due to the limited amount of RNA available, the samples were amplified using the nCounter Single Cell Expression Assay Protocolm in which 100 pg total RNA for each sample was reverse transcribedn and linearly amplifiedo for 12 cycles using Multiplex Target Enrichment primer pairsp specific to each gene of interest designed by nanoString to amplify the region of the mRNA to which the capture and reporter probes were designed to anneal. The amplified cDNA was then hybridised with the custom CodeSet (the capture and reporter probes used to assess mRNA concentrations for each gene of interest in the current study which were designed by nanoString from published equine sequences for the 30 genes of interest) on the nCounter Analysis System.
Data analysis
Samples from 8 horses at 3 different times, except for one horse at 2 different times have been sequenced using ABI SOLiD next generation sequencing platformk. cDNA fragments from all 23 samples are sequenced in one paired-end transcriptome sequencing run using F3 tag length of 50bp and F5 tag read length of 25bp for mate pairs. RNA-Seq produces short reads in the form of *.csfasta and *.qual files. Using SOLiD BioScope™ Software WTA workflow, these reads (14 million reads per sample on the average) were aligned to the UCSC equine reference genome assembly, Sep 2007 - EquCab2. Besides the genomic reference, WTA also uses gene annotations to improve the alignment. Gene annotations include the exons, genes and transcripts. The gene annotations file for the horse genome as refGene.txt is downloaded from the UCSC genome browser FTP site (http://hgdownload.cse.ucsc.edu/goldenPath/equCab2/). The alignment results were reported as separate *.bam files with around 8 million mapped reads per sample on the average.
Using publicly available open-source software Cufflinks [43] and the mapped RNA-Seq reads, the relative abundance of genes were measured. RNA-Seq fragment counts were properly normalised to calculate the transcript abundances in Fragments Per Kilobase of exon per Million fragments mapped (FPKM). The gene list was filtered based on the FPKM threshold value of 5 so that the gene was filtered out if the associated FPKM value is 5 or below for all the 23 samples resulting in 708 genes.
Using Cufflinks as the underlying quantification mechanism, Cuffdiff program calculates if the genes are differentially expressed in more than one condition and test for their significance. We performed group wise comparison between different time points (DEV vs. CON, OG1 vs. CON, OG1 vs. DEV) including all 8 horses for each time point (except for those comparisons with DEV times where n = 7 horses) using paired t-test to determine the significance of differentially expressed genes. We also performed pair-wise differential gene expression analysis for each horse to observe the differentially expressed genes across different times for each individual (DEV vs. CON = 7 comparisons, OG1vs. CON = 8 comparisons, OG1vs. DEV = 7 comparisons).
After differential expression analysis further filtering was performed based on the fold change value >2.0 and p-value <0.05. Based on the fold change/p-value cutoffs we compiled a differentially expressed gene list of 40, 107, 103 for each group of DEVvsCON, OG1vsCON and OG1vsDEV, respectively. These gene lists were further analysed using Ingenuity Pathway Analysisq (www.ingenuity.com) to determine the functional annotations and the canonical pathways involved in these most differentially expressed genes.
NanoString data were analysed using the nSolver Analysis Software analysis (nanoString Technologies). Briefly, the average of the house-keeping/reference genes geometric means across all lanes was used as the reference against which each lane was normalised. A normalisation factor was then calculated for each of the lanes based on the geometric mean of counts for the reference gene(s) in each lane relative to the average geometric mean of counts for the reference gene(s) across all lanes. This normalisation factor was then used to adjust the counts for each gene target and controls in the associated lane. A heat map was created (Fig 4) to show median-centered expression using centroid linkage and centered correlation metrics of each gene using Cluster 3.0 and JavaTreeView software algorithms applied to log2 transformed data.
Fig 4.
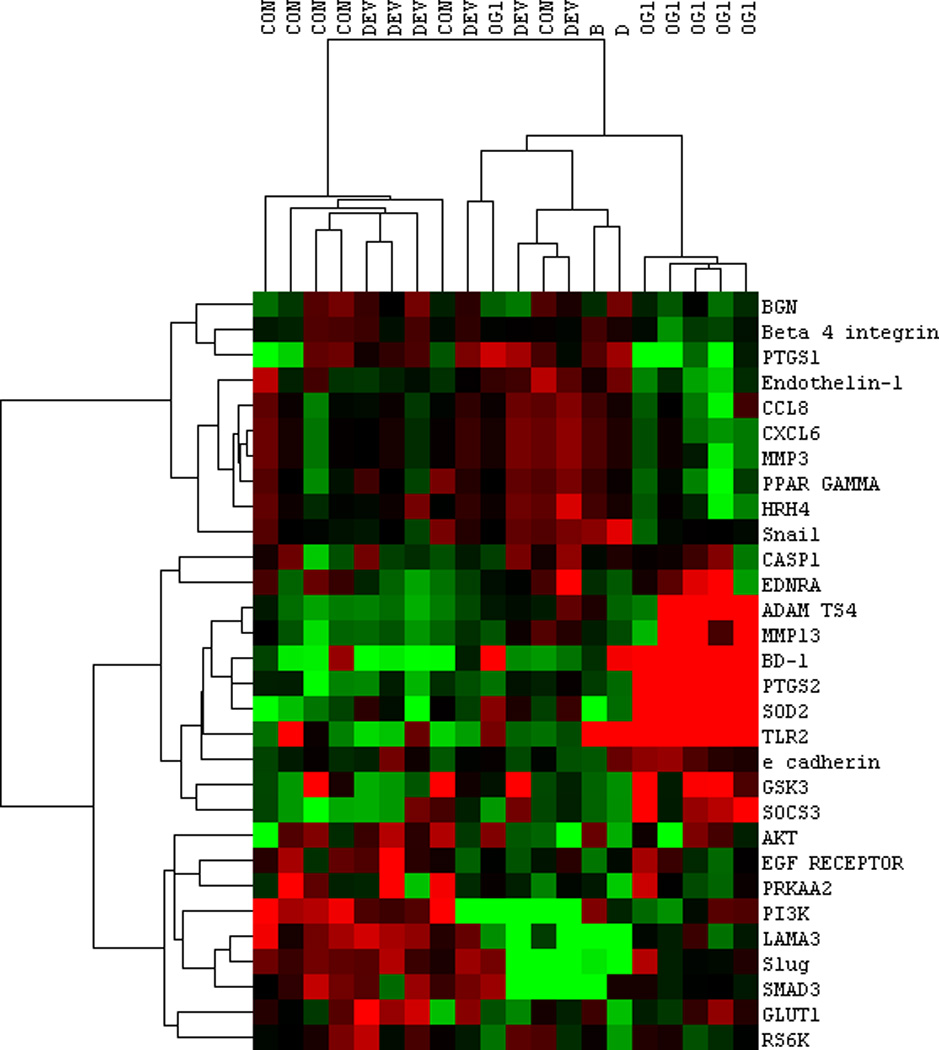
Heat map of nanoString results comparing LBEC selected gene expressions from horses at the CON, DEV and OG1 time points. Red, green, and black colours represent high, low, and mean expression levels, respectively. Note the clustering of highly expressed genes from RNA-seq data at OG1 time point. Genes selected for evaluation were determined by the differential expression from 2 different studies, therefore not all genes present on this heat map were differentially expressed in this study alone. Samples B and D are not related to this study.
Results
Clinical findings
Fever (>38.6°C) occurred in 7 of the 8 horses (one horse developed OG1 lameness at 24 h without developing a fever) at a median of 30 h post-CHO with the time of onset of fever ranging from 24 h-40 h. In the 5 out of 8 horses which exhibited obvious OG1 lameness, the lameness was detected at a median time of 42 h with a range between 24 h and 48 h post-CHO administration. In 3 OG1 horses, lamellar biopsies were taken at 48 h despite the lack of consistent signs of OG1 lameness (horse lifts feet incessantly, short, stilted gait at trot); the inability of perineural bupivacaine anaesthesia to entirely block the lameness in the limbs in which lamellar biopsies were performed was suspected to confound the detection of a mild lameness in these 3 animals at the OG1 time point.
DEV vs. CON
A total of 40 genes were differentially expressed at the DEV time point when compared to CON (Supplementary Item 1). Of these, 22 genes were up-regulated and 18 genes down-regulated. Ten percent of genes differentially expressed at the DEV time included those involved in extracellular matrix composition/regulation (Table 1) and 18% of genes were involved in inflammation (Table 2) or immune regulation (Fig 5). A small number of genes related to serotonin function (including up-regulation of serotonin receptor 4 and down-regulation of tryptophan hydroxylase 2), calcium regulation and selenoproteins were also differentially expressed (Fig 5 and Supplementary Item 1). Bioinformatic analysis (IPA software) identified 5 significant network functions, including cell morphology, carbohydrate metabolism, cell function and maintenance, and connective tissue development and function. The canonical pathways identified by bioinformatic analysis were primarily related to cytokine modulation of immune and epithelial cells (Fig 6).
Table 1.
Differentially expressed extracellular matrix genes by LBECs.
| DEV vs. CON | |||
|---|---|---|---|
| Gene ID | Gene Name | p-value | Regulation |
| LUM | lumican | 0.01378 | DOWN |
| HAPLN1 | hyaluronan and proteoglycan link protein 1 | 0.01618 | DOWN |
| ADAMTS4 | ADAM metallopeptidase with thrombospondin 1 motif 4 |
0.02168 | UP |
| MMP1 | matrix metallopeptidase 1 (interstitial collagenase) | 0.03815 | DOWN |
| OG1 vs. CON | |||
| Gene ID | Gene Name | p-value | Regulation |
| SDCBP | syndecan binding protein (syntenin) | 0.00001 | UP |
| BGN | biglycan | 0.00008 | DOWN |
| VIM | vimentin | 0.00023 | DOWN |
| SPARC | secreted protein, acidic, cysteine-rich (osteonectin) | 0.00119 | DOWN |
| THBS2 | thrombospondin 2 | 0.00217 | DOWN |
| FMOD | fibromodulin | 0.00519 | DOWN |
| DCN | decorin | 0.00788 | DOWN |
| MMP3 | matrix metallopeptidase 3 (stromelysin 1) | 0.00899 | UP |
| SCX | scleraxis | 0.01447 | DOWN |
| ADAMTS4 | ADAM metallopeptidase with thrombospondin 1 motif 4 |
0.02755 | UP |
| MMP13 | matrix metallopeptidase 13 (collagenase 3) | 0.0283 | UP |
| COL14A1 | collagen, type XIV, alpha 1 | 0.0342 | DOWN |
| TIMP1 | TIMP metallopeptidase inhibitor | 0.04343 | UP |
| OG1 vs. DEV | |||
| Gene ID | Gene Name | p-value | Regulation |
| SCX | scleraxis | 0.00058 | DOWN |
| SPARC | secreted protein, acidic, cysteine-rich (osteonectin) | 0.00075 | DOWN |
| MMP1 | matrix metallopeptidase 1 (interstitial collagenase) | 0.00193 | UP |
| SDCBP | syndecan binding protein (syntenin) | 0.00717 | UP |
| THBS2 | thrombospondin 2 | 0.00941 | DOWN |
| MMP3 | matrix metallopeptidase 3 (stromelysin 1) | 0.01159 | UP |
| BGN | biglycan | 0.01753 | DOWN |
| MMP13 | matrix metallopeptidase 13 (collagenase 3) | 0.03645 | UP |
| TIMP1 | TIMP metallopeptidase inhibitor | 0.04087 | UP |
Table 2.
Selected differentially expressed inflammatory genes by LBECs.
| CON vs. DEV | |||
|---|---|---|---|
| Gene ID | Gene Name | p-value | Regulation |
| HRH4 | histamine receptor H4 | 0.01384 | UP |
| LITAF | lipopolysaccharide-induced TNF factor | 0.02014 | UP |
| CON vs. OG1 | |||
| Gene ID | Gene Name | p-value | Regulation |
| IL4R | interleukin 4 receptor | 0.00013 | UP |
| PTGS1 | prostaglandin-endoperoxide synthase 1 (COX1) | 0.00068 | DOWN |
| PTGES | prostaglandin-E synthase | 0.00244 | UP |
| TLR2 | toll-like receptor 2 | 0.00360 | UP |
| BD-1 | beta-defensin-1 | 0.00417 | UP |
| PTGFR | prostaglandin F receptor | 0.00564 | UP |
| SOCS3 | suppressor of cytokine signalling 3 | 0.00598 | UP |
| C1QTNF7 | C1q and tumour necrosis factor related protein 7 | 0.00677 | DOWN |
| SOD2 | superoxide dismutase 2, mitochondrial | 0.01138 | UP |
| CXCL2 | chemokine (C-X-C motif) ligand 2; MIP-2a | 0.01543 | UP |
| IL1RN | interleukin 1 receptor antagonist | 0.01946 | UP |
| PLA2G4A | phospholipase A2, group IVA | 0.01957 | UP |
| CCL2 | chemokine (C-C motif) ligand 2; MCP-1 | 0.01962 | UP |
| CASP1 | caspase1; interleukin1 beta-converting enzyme | 0.02133 | UP |
| CCR2 | chemokine (C-C motif) receptor 2; MCP-1 receptor | 0.02309 | DOWN |
| IL12B | interleukin 12b; IL-12 p40 | 0.02400 | UP |
| TLR4 | toll-like receptor 4 | 0.02567 | UP |
| TNFSF12 | tumour necrosis factor (ligand) superfamily, member 12 | 0.02628 | DOWN |
| CXCL6 | chemokine (C-X-C motif) ligand 6 | 0.02632 | UP |
| SELE | E-selectin | 0.02935 | UP |
| TNFRSF13c | tumour necrosis factor receptor superfamily, member 13c | 0.03033 | DOWN |
| SPLA2 | secretory phopholipase A2 | 0.03126 | UP |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (COX-2) | 0.03191 | UP |
| CCL8 | chemokine (C-C motif) ligand 8; MCP-2 | 0.03455 | UP |
| SAA1 | serum amyloid A1 | 0.03895 | UP |
| CCL13 | chemokine (C-C motif) ligand 13 (MCP-4) | 0.03948 | UP |
| OG1 vs DEV | |||
| Gene ID | Gene Name | p-value | Regulation |
| iNOS | inducible nitic oxide synthase | 0.00009 | DOWN |
| PTGES | prostaglandin E synthase | 0.00291 | UP |
| TLR2 | toll-like receptor 2 | 0.00389 | UP |
| PTGFR | prostaglandin F receptor | 0.00429 | UP |
| BD-1 | beta-defensin-1 | 0.00644 | UP |
| C1QTNF7 | C1q and tumour necrosis factor related protein 7 | 0.01107 | DOWN |
| CASP1 | caspase1; interleukin1 beta-converting enzyme | 0.01287 | UP |
| IL1R1 | interleukin-1 receptor antagonist | 0.01288 | UP |
| CSF3 | colony stimulating factor 3 (granulocyte) | 0.01759 | DOWN |
| LITAF | lipopolysaccharide-induced TNF factor | 0.02018 | DOWN |
| SOD2 | superoxide dismutase 2, mitochondrial | 0.02046 | UP |
| IL-6 | interleukin-6 | 0.02774 | UP |
| SOCS3 | suppressor of cytokine signaling 3 | 0.03521 | UP |
| CCL13 | chemokine (C-C motif) ligand 13 (MCP-4) | 0.03576 | UP |
| SPLA2 | secretory phopholipase A2 | 0.03864 | UP |
| TLR4 | toll-like receptor 4 | 0.04020 | UP |
| CD14 | CD14 molecule | 0.04172 | UP |
| PTGS2 | prostaglandin-endoperoxide synthase 2 (COX2) | 0.04633 | UP |
| CCL2 | chemokine (C-C motif) ligand 2 (MCP-1) | 0.04951 | UP |
Fig 5.
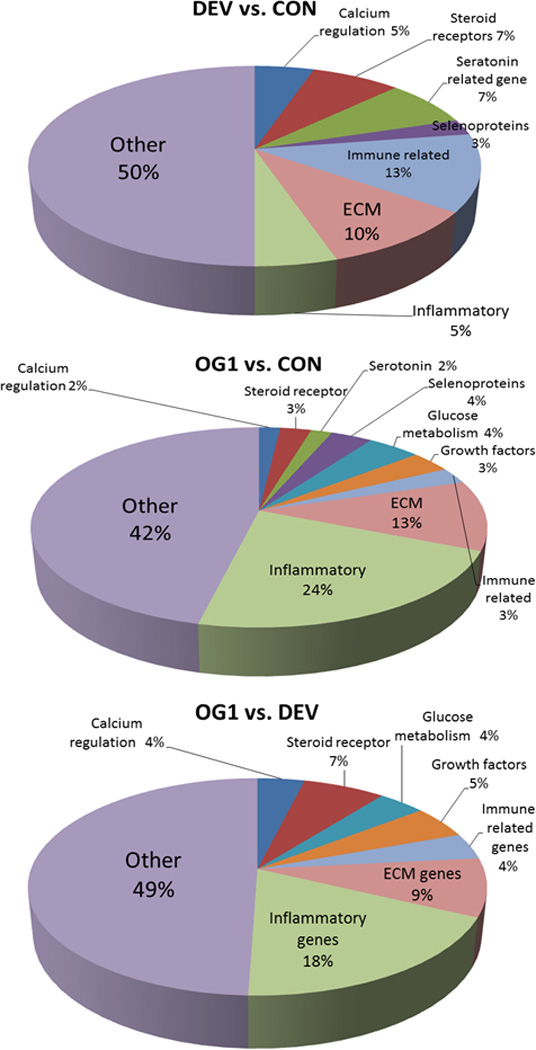
Pie charts representing the percent of differentially expressed genes in each of the compared groups (DEV vs. CON; OG1 vs. CON; and OG1 vs. DEV). While expressed genes could cross multiple categories, each gene was only assigned to one category. Other genes could not be grouped with sufficient numbers to include as individual categories but all differentially expressed genes are demonstrated in Supplementary Items 1–3 for review.
Fig 6.
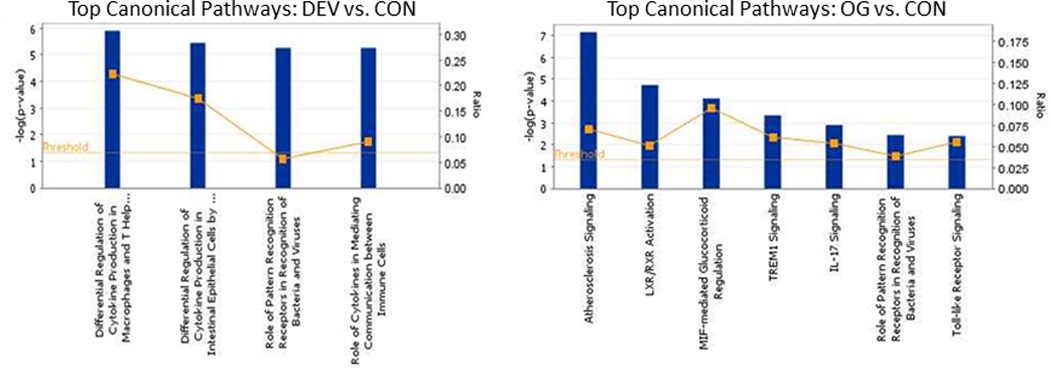
Top canonical pathways identified by IPA analysis of RNA in LBECs at the DEV time point compared to controls (left) and at the OG1 time point compared to controls (right).
OG1 vs. CON
A total of 107 genes were differentially expressed at the OG1 time vs. CON with 57 genes that were up-regulated and 50 genes down-regulated (Supplementary Item 2). Eleven significant network functions were identified with bioinformatic analysis of which included endocrine system function/small molecule biochemistry functions, cardiovascular system function/inflammatory response, and connective tissue disorders/inflammatory disease (Fig 7). Several canonical pathways were also identified by bioinformatic analysis including regulation of chemokines, roles in pattern recognition of bacteria and viruses, toll-like receptor signalling and arthrosclerosis signalling (Fig 6). Transcription factors that were predicted by bioinformatic analysis to be activated at the OG1 time point when compared to CON were RelA, NFκB complex, EGR1, HMGB1, NFκB-RelA, STAT3, STAT1 and MYC. Evaluation of the individual genes differentially expressed found that a high proportionate number of genes were associated with extra-cellular matrix composition/regulation (13%; Fig 5 and Table 1) and inflammatory response (24%; Fig 5 and Table 2). In addition to the ECM and inflammatory genes differentially expressed, those related to glucose/glycogen metabolism (4%; including the up-regulation of glucose-regulated protein and hexokinase), growth factors (3%; such as down-regulation of fibroblast growth factor 1 and bone morphogenetic protein 7) and variable regulation of selenoproteins (4%) were also present (Fig 5).
Fig 7.
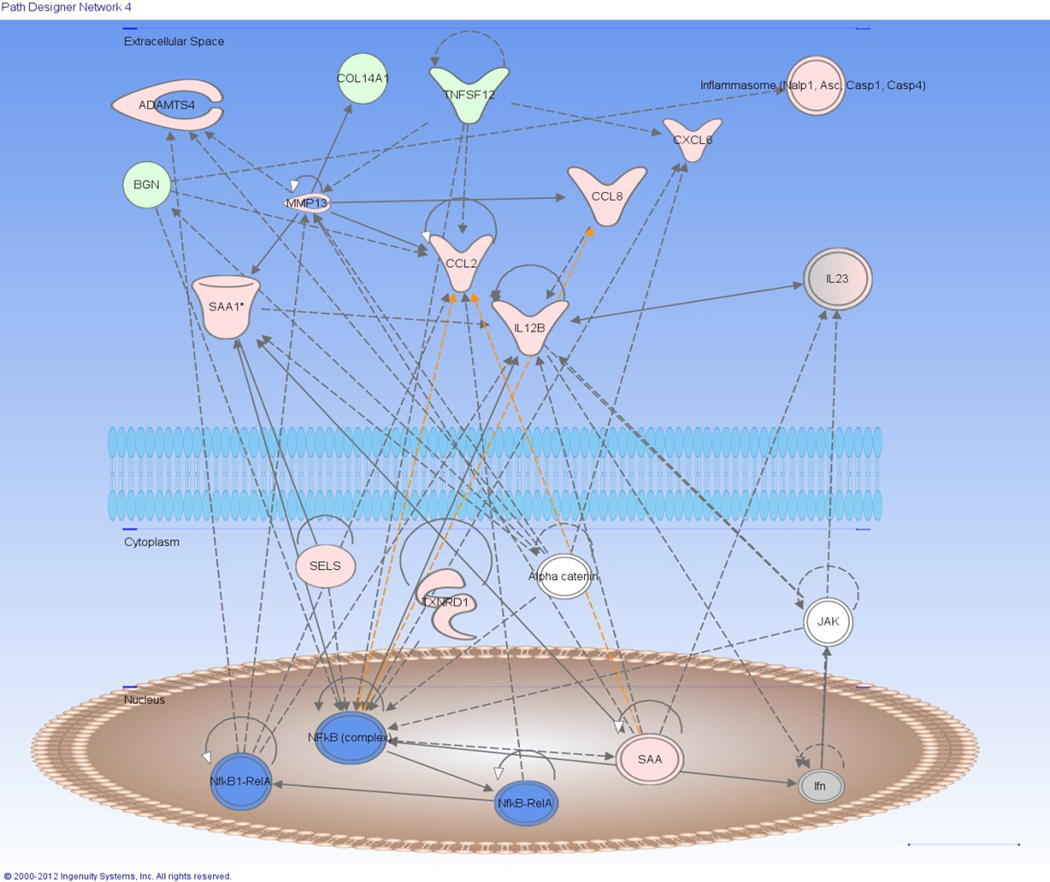
Differentially expressed genes from the selected network function “connective tissue disorders and inflammatory disease”. Diagram demonstrates interaction amongst numerous genes that were up-regulated (Pink) or down-regulated (Green) in the network and predicted to be activated (Blue) via IPA analysis. Dotted line = indirect relationship, straight line = direct relationship, White = non-regulated gene in OG1 horses.
OG1 vs. DEV
A total of 103 genes were differentially expressed at the OG1 time point compared to the DEV time point, with 59 genes up-regulated and 44 genes down-regulated (Supplementary Item 3). Top network functions identified with IPA analysis included lipid metabolism and small molecule biochemistry, cellular movement and connective tissue disorders, dermatological conditions and antimicrobial response. Several canonical pathways were also identified by bioinformatic analysis to be activated at OG1 compared to DEV including LXR/RXR activation, role of pattern recognition of bacteria and viruses, and role of macrophage and endothelial cells in rheumatoid arthritis. Transcription factors predicted to be activated at the OG1 time compared to DEV were HMGB1 and NFκB. As compared to controls, many of the genes differentially expressed in the OG1 vs. DEV time included those involved in inflammation (19%; Fig 5 and Table 2) and in extracellular matrix composition and turnover (9%; Fig 5 and Table 1). Differentially expressed genes related to calcium regulation, glucose/glycogen metabolism (including down-regulation of GLUT1), and growth factors (including as up-regulation of leptin) were present at the OG1 time compared to DEV (Fig 5). Many of the differentially expressed genes at the OG1 vs. CON were also differentially expressed when OG1 was compared to the DEV time point. For example, all 9 extracellular matrix components genes (Table 1) differentially expressed in OG1 vs. DEV were also expressed when compared to the CON time. Many of the inflammatory genes differentially expressed at the OG1 time were also similar between comparisons where only 5 out of 19 genes were not present when OG1 was compared to the CON time (Table 2).
NanoString
NanoString data were compared to the RNA-Seq results for selected genes. Results supported RNA-Seq findings overall, demonstrating a clustering of the OG1 animals, with similar increases in the majority of genes assessed that were increased in RNA-Seq (ADAMTS4, MMP13, beta defensin-1, COX-2/PTGS2, SOD2, TLR2) and decreased expression of COX-1/PTGS1 (Fig 4).
Discussion
To advance our understanding of lamellar pathophysiology, and therefore our treatment of laminitis, we must understand the events occurring at the level of the LBEC in the different types of laminitis. Numerous studies from multiple laboratories have documented which events are occurring in whole tissue extracts during the early stages of laminitis from several experimental models; however, evaluation of specific cell types within the lamellar tissue has been limited to histological evaluation. Utilising novel techniques we were able to successfully dissect the LBEC from cryosections and obtain high quality RNA from the captured LBECs to ascertain the differential regulation of genes. Furthermore, through networking analysis were able to identify genes in related signaling pathways most likely playing a role in LBEC dysfunction from sepsis-related laminitis. This report is the first to identify the differential expression of genes within a specific lamellar cell type, the LBECs. Interestingly, despite the fact that the Next Generation Sequencing techniques used were unbiased in regards to any specific cell signaling, the predominate mechanisms detected by our bioinformatic analysis of the RNA-Seq data performed on the LBEC appeared to be related to 2 events already of great interest in laminitis research, inflammatory and matrix signalling.
Epithelial cells, particularly keratinocytes, are an important part of the innate immune system, not only serving a structural/barrier function, but also in their ability to respond to bacterial ligands through pattern recognition receptors [44–47]. Human skin keratinocytes express several TLRs (including TLR2 and 4) and are able to respond to multiple pathogen associated molecular patterns (PAMPs) [45–48]; equine skin keratinocytes have been reported to respond to PAMPs more characteristic of gram negative bacteria (LPS and flagellin), resulting in the expression of proinflammatory cytokines and chemokines [44]. In this study, LBECs at the OG1 time demonstrated up-regulation of both TLR2 and 4 when compared to controls and IPA bioinformatics analysis revealed that the canonical pathways involving both TLR signalling and roles in pattern recognition of bacteria and viruses were activated at this time. These data combined with the fact that the primary transcription factors downstream of TLR signalling, NFkB and RelA, are predicted to be activated in these cells indicates that the inflammatory signalling occurring in the LBECs may not only be a response to extracellular events such as dermal inflammation associated with the influx of activated leukocytes in the lamellar interstitium [11], but may be due to the ligation of PAMPs (extravasated from the lamellar microvasculature) to the LBEC’s TLRs.
In addition to the TLRs, numerous additional inflammatory genes were differentially expressed in the LBECs, the majority of which were most prominent at the OG1 time point. Many of the inflammatory molecules that were up-regulated in the LBECs have been previously reported to be increased in sepsis-related laminitis, and include various chemokines (such as CXCL2 [MIP-2α], CCL2 [MCP-1] [6, 49, 50], CCL8 [MCP-2] [6], CCL13 [MCP-4] and CXCL6) [6], COX-2 [2, 7, 51], SOCS-3 [52], serum amyloid A [50], and beta defensin-1 [50]; most of these previously reported to be regulated in the CHO model of sepsis-related laminitis [6, 7, 52]. However, these previous findings were all from whole tissue homogenates and not from a single cell type such as the LBEC. As the epithelial cell of other organs is known to play an important role in the inflammatory processes of sepsis/SIRS through the overwhelming production of cytokines, chemokines, and other inflammatory mediators [22, 53] it is not surprising that the LBEC can respond in the same manner.
The up-regulation of monocytic chemokines in the LBEC at the OG1 time is consistent with previous reports of monocyte/macrophage migration into lamellar tissue at this time point after CHO administration [11] and is likely to be playing an important role in attracting this cell type toward the stressed LBEC in the septic horse. Stimulation of macrophages toward the LBEC can result in further production of inflammatory mediators, thereby promoting a continual overwhelming inflammatory response at the site of lamellar failure. An example of a perpetuating lamellar inflammatory response, possibly induced by extravasated monocytes, is demonstrated by the previously reported increased mRNA concentrations of IL-6 [7] and activation of STAT3 [52] (of which IL-6 is an important activator) in whole lamellar tissue homogenates at the OG1 time point in the CHO model. Furthermore, up-regulation of SOCS3, a STAT3 inhibitor, was present in the LBEC suggesting that these cells may also able to help modulate the inflammatory response within the lamellae. The expression pattern of the COX enzymes by LBECs in the current study also has similarities to previous reports from sepsis-related laminitis models in which whole lamellar tissue homogenates demonstrated increases in mRNA concentrations of COX-2 and decreases in COX-1 [2, 7]. This finding along with increased gene expression of other enzymes involved in prostanoid production (including phospholipase A2, secretory phospholipase A2, prostaglandin-E synthase) at the OG1 time point clearly indicates that the LBEC plays an active role in eicosanoid synthesis. Overall, the predominant up-regulation of inflammatory mediators by the LBEC, particularly at the OG1 time, combined with fact that the vast majority of predicted activated transcription factors from the bioinformatic analysis are central players in inflammatory signalling (i.e. NFkB, RelA, STAT1&3, HMGB1), indicate that inflammation is a critical event in the LBEC during the early stages of laminitis.
In addition to the numerous inflammatory genes that were differentially expressed in LBECs at OG1, there were also a significant number of regulated genes related to extracellular matrix/basement membrane composition and turnover. Enzymatic degradation of the extracellular matrix has been proposed to result in the detachment of the basal epithelial cells from the basement membrane and underlying dermis [23, 41, 54, 55]. Of these enzymes, the earliest and most studied are MMP2 [23] and MMP9 [54, 55]; however, these MMPs are now not thought to have an active a role as initially reported during the early stages of laminitis [56]. In the current study, increased mRNA concentrations of MMP3 (stomelysin) and MMP13 (collagenase) were detected in LBECs at the OG1 time compared to controls. MMP3 can degrade several components of the basement membrane (i.e. Collagen IV, laminin, proteoglycans) whereas MMP13 appears to primarily degrade collagens I-III. These collagens are components of the extracellular matrix rather than the basement membrane, although collagen III is a major component of the lamella reticularis, which provides the structural transition between the basal lamellae and the underlying dermis. The increased expression of these MMPs may not have a direct effect on basement membrane attachments, but can significantly affect the underlying dermis thereby weakening the overall lamellar dermal-epidermal interface. ADAMTS-4, an aggrecanase which has been immunolocalised to multiple lamellar cells including the lamellar epithelial cells [57] and reported to be increased in whole lamellar tissue extracts at the OG1 time in the CHO model, was found to be increased in LBECs in the current study at both DEV and OG1 time points [58]. Although ADAMTS-4 has primarily been discussed having an intracellular role in the LBEC in the breakdown of versican [57], it is possible that it plays a destructive role in the extracellular matrix affecting the epidermal-basement membrane attachments (S. Black, personal communication). Differences in the temporal response in ADAMTS-4 expression between our study and previous reports most likely is a result of specific increased expression of ADAMTS-4 in the LBECs at an earlier time, which may not be detectable in whole lamellar extracts of CHO horses [58]. The regulation of these matrix molecules indicate that the LBECs itself plays an early and active role in the regulation of matrix turnover in this model of laminitis.
A large number of matrix molecules were found to be down-regulated in the affected LBECs including a large number of small leucine-rich proteoglycans (SLRPs; biglycan, lumican, fibromodulin and decorin), the basement membrane glycoprotein osteonectin, and collagen 14α1. SLRPs are important ECM components regulating fibril organisation and stability of the ECM [59], and have also been shown to inhibit MMPs (including MMP13) [60]. The decreased expression of these SLRPs may impact the stability of the LBEC-dermal interface as deficiencies in type 1 (decorin and biglycan) [61] and type II (lumican and fibromodulin) [62]. SLRPs have been reported to lead to fragility and laxity of the skin. Similarly, a decrease in collagen 14α1, which was down-regulated in this study and documented to be expressed by basal epithelial cells in other species, also can lead to instability and separation of the epidermal/dermal interface [63]. Furthermore, as the absence of decorin results in abnormal collagen fibres in the corneal stroma, and the combined absence of both decorin and biglycan results in an even more severe phenotype with numerous abnormal fibres and disorganisation of the stroma [64], it is possible that, when multiple components of ECM assembly and organisation are decreased (as in this study), more severe ECM defects may result. The exact mechanism by which these ECM components are stimulated to be down-regulated has yet to be elucidated; however, it strongly suggests that aberrant regulation of the extracellular matrix plays an important role in dysadhesion of LBECs and subsequent failure of the lamella.
Smaller subsets of related genes, some of which have been previously evaluated in equine laminitis, were also found to be regulated in the LBEC at the DEV and OG1 times. Genes that were differentially expressed by the LBEC that have been previously examined in laminitis include those related to growth factor expression [65], serotonin signaling [66], and glucose metabolism [67, 68]. Other genes such as those involving calcium regulation, selenoproteins, and steroid receptor and metabolism have not been explored in equine laminitis and require further evaluation to determine the importance of their expression. Numerous other genes were differentially expressed which do not fall into discrete categories (see Supplementary Items ? for complete list of significantly expressed genes). Future studies are needed to determine which of these genes are of the greatest importance in LBEC dysfunction during the development of laminitis.
Although this study delineated a distinct cellular response in the LBEC layer which is likely to play a central role in lamellar failure during sepsis, the cellular components of the underlying dermal lamellae are also likely to play a role in the disease process both through interaction with the LBEC and by contributing directly to matrix dysregulation/injury. Tissue macrophages, recently documented to be present in the secondary dermal lamellae [8, 11], are a likely source of signalling molecules which may influence expression of the many genes noted to be regulated in the LBEC in the current study. Additionally, destructive enzymes which could be released from multiple cells types in the dermis and/or from the epidermis are likely playing a direct role in disruption of the laminar dermal architecture. However, unlike the dissection of one cell type which was possible with LCM of the LBEC layer, LCM of the lamellar dermis will result in dissection of multiple cell types (i.e. macrophages [8, 11], fibroblasts, cellular components of the dermal microvasculature, and lymphocytes [69]). Thus, delineation of regulated genes following laminar dermal LCM techniques will have to be incorporated with histologic techniques (i.e. in situ hybridisation or immunohistochemistry) of lamellar sections to determine the dermal cell types responsible for any dermal signalling events discovered in sepsis-related laminitis.
In conclusion, the use of LCM in this study allowed the isolation of the distinct target cell, the LBEC, along with the powerful method of RNA-Sequencing which enabled the identification of numerous differentially expressed genes in the LBECs in horses with CHO-induced laminitis. Using next generation sequencing (RNA-Seq) for the first time, genes from various signalling pathways were identified from a single cell population instead of from whole-tissue samples of lamina. Inflammatory response signalling and signalling related to extracellular matrix composition, stability, and turnover were strongly represented in the DEV and OG1 time points compared to control samples, demonstrating that these pathways are likely to play a critical role in the LBEC response in sepsis-related laminitis. Moreover, it is clear that the LBEC is not just a casualty of the events occurring within the lamina, but is likely to be an active participant in the events leading to structural failure. Further evaluation into regulation of the transcriptional networks linked to inflammatory and extracellular matrix signalling will hopefully result in the development of therapeutic targets thereby allowing for the prevention and treatment of LBEC injury and subsequent epidermal detachment in cases with sepsis/SIRS.
Supplementary Material
Acknowledgments
We thank Kirsteen H. Maclean (nanoString Technologies) for analysis of nanoString data, and Sarah Warner, OSU Comprehensive Cancer Center, for assistance with the RNA-Seq methodology.
Authorship
B. Leise contributed to study design, study execution, data interpretation, and preparation of the manuscript. M. Watts contributed to study execution. S. Roy contributed to study design. S. Yilmaz contributed to study execution, and data analysis and interpretation. H. Alder contributed to study execution, data analysis and interpretation, and preparation of the manuscript. J. Belknap contributed to study design, data analysis and interpretation, and preparation of the manuscript. All authors gave their final approval of the manuscript.
Source of funding
Supported by USDA-NIFA AFRI award 2009 01780 and grant UL1TR000090 from the National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Abbreviations
- ADAMTS-4
ADAM metallopeptidase with thrombospondin type 1 motif 4
- CHO
Carbohydrate overload
- CCL
Chemokine (C-C motif) ligand
- CON
Control
- COX
Cyclooxygenase
- CXCL
Chemokine (C-X-C motif) ligand
- DEV
Developmental
- ECM
Extracellular matrix
- FPKM
Fragments Per Kilobase of exon per Million fragments mapped
- HMGB1
High mobility group box-1
- IL-6
Interleukin-6
- LBECs
Laminar basal epithelial cells
- LCM
Laser capture microdissection
- MMP
Matrix metallopeptidase
- NF-κB
Nuclear factor kappa B
- OG1
Obel grade 1
- qRT-PCR
quantitative real-time polymerase chain reaction
- PAMP
Pathogen-associated molecular pattern
- RIN
RNA integrity number
- RNA-Seq
RNA Sequencing
- SIRS
Systemic inflammatory response syndrome
- SLRP
Small leucine-rich proteoglycans
- SOCS3
Suppressor of cytokine signalling 3
- SOD2
Superoxide dismutase 2
- STAT
Signal Transducing Activators of Transcription
- TLR
Toll–like receptor
- WTA
Whole transcriptome analysis
Footnotes
Manufacturers’ addresses
Carbocaine® Pharmcia & Upjohn; Bridgewater, NJ
Hospira, Inc; Lake Forest, IL
Leica Biosystems; Nussloch, Germany
Ambion; Austin, TX
PEN slides, Carl Zeiss MicroImaging GmbH; Munchen, Germany
LCM staining kit, Ambion; Austin, TX
Sigma-Aldrich; St. Louis, MO
PALM Technologies; Bernreid, Germany
RNeasy® Micro kit, Qiagen; Valencia, CA
Agilent; Wilmington, DE
Life Technologies; Carlsbad, CA
Agencourt Bioscience Corporation; Beckman Coulter, Inc; Indianapolis, IN
nanoString Technologies; Seattle, WA
Superscript VILO Mastermix; Invitrogen, Carlsbad, CA
TaqMan PreAmp Master Mix, 2x; Applied Biosystems, Foster City, CA
Integrated DNA Technologies; Coralville, IA
IPA, Ingenuity ® Systems; Redwood City, CA
Authors’ declaration of interests
No competing interests have been declared.
Ethical Animal Research
The experimental protocol was approved by the Institutional Animal Care and Use Committee at The Ohio State University.
Supplementary Items
Supplementary Item 1: Differentially expressed genes at the DEV vs. CON time.
Supplementary Item 2: Differentially expressed genes at the OG1 vs. CON time.
Supplementary Item 3: Differentially expressed genes at the OG1 vs. DEV time.
References
- 1.Pollitt CC. The anatomy and physiology of the suspensory apparatus of the distal phalanx. Vet. Clin. North Am. Equine Pract. 2010;26:29–49. doi: 10.1016/j.cveq.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Blikslager AT, Yin C, Cochran AM, Wooten JG, Pettigrew A, Belknap JK. Cyclooxygenase expression in the early stages of equine laminitis: a cytologic study. J. Vet. Intern. Med. 2006;20:1191–1196. doi: 10.1892/0891-6640(2006)20[1191:ceites]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 3.Belknap JK, Giguere S, Pettigrew A, Cochran AM, Van Eps AW, Pollitt CC. Lamellar pro-inflammatory cytokine expression patterns in laminitis at the developmental stage and at the onset of lameness: innate vs. adaptive immune response. Equine Vet. J. 2007;39:42–47. doi: 10.2746/042516407x155406. [DOI] [PubMed] [Google Scholar]
- 4.Faleiros RR, Leise BB, Westerman T, Yin C, Nuovo GJ, Belknap JK. In vivo and in vitro evidence of the involvement of CXCL1, a keratinocyte-derived chemokine, in equine laminitis. J. Vet. Intern. Med. 2009;23:1086–1096. doi: 10.1111/j.1939-1676.2009.0349.x. [DOI] [PubMed] [Google Scholar]
- 5.Fontaine GL, Belknap JK, Allen D, Moore JN, Kroll DL. Expression of interleukin-1beta in the digital laminae of horses in the prodromal stage of experimentally induced laminitis. Am. J. Vet. Res. 2001;62:714–720. doi: 10.2460/ajvr.2001.62.714. [DOI] [PubMed] [Google Scholar]
- 6.Faleiros RR, Leise BS, Watts M, Johnson PJ, Black SJ, Belknap JK. Laminar chemokine mRNA concentrations in horses with carbohydrate overload-induced laminitis. Vet. Immunol. Immunopathol. 2011;144:45–51. doi: 10.1016/j.vetimm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Leise BS, Faleiros RR, Watts M, Johnson PJ, Black SJ, Belknap JK. Laminar inflammatory gene expression in the carbohydrate overload model of equine laminitis. Equine Vet. J. 2011;43:54–61. doi: 10.1111/j.2042-3306.2010.00122.x. [DOI] [PubMed] [Google Scholar]
- 8.Faleiros RR, Nuovo GJ, Belknap JK. Calprotectin in myeloid and epithelial cells of laminae from horses with black walnut extract-induced laminitis. J. Vet. Intern. Med. 2009;23:174–181. doi: 10.1111/j.1939-1676.2008.0241.x. [DOI] [PubMed] [Google Scholar]
- 9.Loftus JP, Black SJ, Pettigrew A, Abrahamsen EJ, Belknap JK. Early laminar events involving endothelial activation in horses with black walnut- induced laminitis. Am. J. Vet. Res. 2007;68:1205–1211. doi: 10.2460/ajvr.68.11.1205. [DOI] [PubMed] [Google Scholar]
- 10.Riggs LM, Krunkosky TM, Noschka E, Boozer LA, Moore JN, Robertson TP. Comparison of characteristics and enzymatic products of leukocytes in the skin and laminar tissues of horses administered black walnut heartwood extract or lipopolysaccharide. Am. J. Vet. Res. 2009;70:1383–1390. doi: 10.2460/ajvr.70.11.1383. [DOI] [PubMed] [Google Scholar]
- 11.Faleiros RR, Johnson PJ, Nuovo GJ, Messer NT, Black SJ, Belknap JK. Laminar leukocyte accumulation in horses with carbohydrate overload-induced lamintis. J. Vet. Intern. Med. 2011;25:107–115. doi: 10.1111/j.1939-1676.2010.0650.x. [DOI] [PubMed] [Google Scholar]
- 12.Yin C, Pettigrew A, Loftus JP, Black SJ, Belknap JK. Tissue concentrations of 4-HNE in the black walnut extract model of laminitis: indication of oxidant stress in affected laminae. Vet. Immunol. Immunopathol. 2009;129:211–215. doi: 10.1016/j.vetimm.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Loftus JP, Belknap JK, Stankiewicz KM, Black SJ. Laminar xanthine oxidase, superoxide dismutase and catalase activities in the prodromal stage of black-walnut induced equine laminitis. Equine Vet. J. 2007;39:48–53. doi: 10.2746/042516406x151320. [DOI] [PubMed] [Google Scholar]
- 14.Eades SC, Stokes AM, Johnson PJ, LeBlanc CJ, Ganjam VK, Buff PR, Moore RM. Serial alterations in digital hemodynamics and endothelin-1 immunoreactivity, platelet-neutrophil aggregation, and concentrations of nitric oxide, insulin, and glucose in blood obtained from horses following carbohydrate overload. Am. J. Vet. Res. 2007;68:87–94. doi: 10.2460/ajvr.68.1.87. [DOI] [PubMed] [Google Scholar]
- 15.Hood DM, Grosenbaugh DA, Mostafa MB, Morgan SJ, Thomas BC. The role of vascular mechanisms in the development of acute equine laminitis. J. Vet. Intern. Med. 1993;7:228–234. doi: 10.1111/j.1939-1676.1993.tb01012.x. [DOI] [PubMed] [Google Scholar]
- 16.Moore RM, Eades SC, Stokes AM. Evidence for vascular and enzymatic events in the pathophysiology of acute laminitis: which pathway is responsible for initiation of this process in horses? Equine Vet. J. 2004;36:204–209. doi: 10.2746/0425164044877116. [DOI] [PubMed] [Google Scholar]
- 17.Bailey SR, Katz LM, Berhane Y, Samuels T, De Brauvere N, Marr CM, Elliott J. Seasonal changes in plasma concentrations of cecum-derived amines in clinically normal ponies and ponies predisposed to laminitis. Am. J. Vet. Res. 2003;64:1132–1138. doi: 10.2460/ajvr.2003.64.1132. [DOI] [PubMed] [Google Scholar]
- 18.Baxter GM, Laskey RE, Tackett RL, Moore JN, Allen D. In vitro reactivity of digital arteries and veins to vasoconstrictive mediators in healthy horses and in horses with early laminitis. Am. J. Vet. Res. 1989;50:508–517. [PubMed] [Google Scholar]
- 19.El-Achkar TM, Hosein M, Dagher PC. Pathways of renal injury in systemic gram-negative sepsis. Eur. J. Clin. Invest. 2008;38(Suppl. 2):39–44. doi: 10.1111/j.1365-2362.2008.02007.x. [DOI] [PubMed] [Google Scholar]
- 20.Shaykhiev R, Bals R. Interactions between epithelial cells and leukocytes in immunity and tissue homeostasis. J. Leukoc. Biol. 2007;82:1–15. doi: 10.1189/jlb.0207096. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Ma S. The cytokine storm and factors determining the sequence and severity of organ dysfunction in multiple organ dysfunction syndrome. Am. J. Emerg. Med. 2008;26:711–715. doi: 10.1016/j.ajem.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 22.Cavaillon JM, Annane D. Compartmentalization of the inflammatory response in sepsis and SIRS. J. Endotoxin Res. 2006;12:151–170. doi: 10.1179/096805106X102246. [DOI] [PubMed] [Google Scholar]
- 23.Kyaw-Tanner M, Pollitt CC. Equine laminitis: increased transcription of matrix metalloproteinase-2 (MMP-2) occurs during the developmental phase. Equine Vet. J. 2004;36:221–225. doi: 10.2746/0425164044877242. [DOI] [PubMed] [Google Scholar]
- 24.Ekfalck A, Rodriguez-Martinez H, Obel N. Cultivation of tissue from the matrix of the stratum medium of the equine and bovine hoof walls. Am. J. Vet. Res. 1990;51:1852–1856. [PubMed] [Google Scholar]
- 25.Wunn D, Wardrop KJ, Meyers K, Kramer J, Ragle C. Culture and characterization of equine terminal arch endothelial cells and hoof keratinocytes. Am. J. Vet. Res. 1999;60:128–132. [PubMed] [Google Scholar]
- 26.Visser MB, Pollitt CC. Characterization of extracellular matrix macromolecules in primary cultures of equine keratinocytes. BMC Vet. Res. 2010;6:16. doi: 10.1186/1746-6148-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, Geho DH, Petricoin EF, 3rd, Liotta LA. Laser-capture microdissection. Nat. Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 28.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 29.Roy S, Patel D, Khanna S, Gordillo GM, Biswas S, Friedman A, Sen CK. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc. Natl. Acad. Sci. USA. 2007;104:14472–14477. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn DE, Roy S, Radtke J, Gupta S, Sen CK. Laser microdissection and pressure-catapulting technique to study gene expression in the reoxygenated myocardium. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H2625–H2632. doi: 10.1152/ajpheart.01346.2005. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn DE, Roy S, Radtke J, Khanna S, Sen CK. Laser microdissection and capture of pure cardiomyocytes and fibroblasts from infarcted heart regions: perceived hyperoxia induces p21 in peri-infarct myocytes. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H1245–H1253. doi: 10.1152/ajpheart.01069.2006. [DOI] [PubMed] [Google Scholar]
- 32.Munshaw S, Hwang HS, Torbenson M, Quinn J, Hansen KD, Astemborski J, Mehta SH, Ray SC, Thomas DL, Balagopal A. Laser captured hepatocytes show association of butyrylcholinesterase gene loss and fibrosis progression in hepatitis C-infected drug users. Hepatology. 2012;56:544–554. doi: 10.1002/hep.25655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slevin M, Krupinski J, Rovira N, Turu M, Luque A, Baldellou M, Sanfeliu C, de Vera N, Badimon L. Identification of pro-angiogenic markers in blood vessels from stroked-affected brain tissue using laser-capture microdissection. BMC Genomics. 2009;10:113. doi: 10.1186/1471-2164-10-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu XS, Zhang ZG, Zhang RL, Gregg S, Morris DC, Wang Y, Chopp M. Stroke induces gene profile changes associated with neurogenesis and angiogenesis in adult subventricular zone progenitor cells. J. Cereb. Blood Flow Metab. 2007;27:564–574. doi: 10.1038/sj.jcbfm.9600371. [DOI] [PubMed] [Google Scholar]
- 35.Asplund A, Gry Bjorklund M, Sundquist C, Stromberg S, Edlund K, Ostman A, Nilsson P, Ponten F, Lundeberg J. Expression profiling of microdissected cell populations selected from basal cells in normal epidermis and basal cell carcinoma. Br. J. Dermatol. 2008;158:527–538. doi: 10.1111/j.1365-2133.2007.08418.x. [DOI] [PubMed] [Google Scholar]
- 36.Tsukamoto Y, Usui M, Yamamoto G, Takagi Y, Tachikawa T, Yamamoto M, Nakamura M. Role of the junctional epithelium in periodontal innate defense and homeostasis. J. Periodontal Res. 2012;47:750–757. doi: 10.1111/j.1600-0765.2012.01490.x. [DOI] [PubMed] [Google Scholar]
- 37.Nanthakumar N, Meng D, Goldstein AM, Zhu W, Lu L, Uauy R, Llanos A, Claud EC, Walker WA. The mechanism of excessive intestinal inflammation in necrotizing enterocolitis: an immature innate immune response. PLoS One. 2011;6:e17776. doi: 10.1371/journal.pone.0017776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsui H, Suarez-Farinas M, Belkin DA, Levenkova N, Fuentes-Duculan J, Coats I, Fujita H, Krueger JG. Combined use of laser capture microdissection and cDNA microarray analysis identifies locally expressed disease-related genes in focal regions of psoriasis vulgaris skin lesions. J. Invest. Dermatol. 2012;132:1615–1626. doi: 10.1038/jid.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Percoco G, Benard M, Ramdani Y, Lati E, Lefeuvre L, Driouich A, Follet-Gueye ML. Isolation of human epidermal layers by laser capture microdissection: application to the analysis of gene expression by quantitative real-time PCR. Exp. Dermatol. 2012;21:531–534. doi: 10.1111/j.1600-0625.2012.01509.x. [DOI] [PubMed] [Google Scholar]
- 40.Sprouse RF, Garner HE, Green EM. Plasma endotoxin levels in horses subjected to carbohydrate induced laminitis. Equine Vet. J. 1987;19:25–28. doi: 10.1111/j.2042-3306.1987.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 41.Johnson PJ, Tyagi SC, Katwa LC, Ganjam VK, Moore LA, Kreeger JM, Messer NT. Activation of extracellular matrix metalloproteinases in equine laminitis. Vet. Rec. 1998;142:392–396. doi: 10.1136/vr.142.15.392. [DOI] [PubMed] [Google Scholar]
- 42.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leise BS, Yin C, Pettigrew A, Belknap JK. Proinflammatory cytokine responses of cultured equine keratinocytes to bacterial pathogen-associated molecular pattern motifs. Equine Vet. J. 2010;42:294–303. doi: 10.2746/042516409X478523. [DOI] [PubMed] [Google Scholar]
- 45.Lebre MC, van der Aar AM, van Baarsen L, van Capel TM, Schuitemaker JH, Kapsenberg ML, de Jong EC. Human keratinocytes express functional Toll-like receptor 3, 4, 5, and 9. J. Invest. Dermatol. 2007;127:331–341. doi: 10.1038/sj.jid.5700530. [DOI] [PubMed] [Google Scholar]
- 46.Miller LS, Modlin RL. Toll-like receptors in the skin. Semin Immunopathol. 2007;29:15–26. doi: 10.1007/s00281-007-0061-8. [DOI] [PubMed] [Google Scholar]
- 47.Song PI, Park YM, Abraham T, Harten B, Zivony A, Neparidze N, Armstrong CA, Ansel JC. Human keratinocytes express functional CD14 and toll-like receptor 4. J. Invest. Dermatol. 2002;119:424–432. doi: 10.1046/j.1523-1747.2002.01847.x. [DOI] [PubMed] [Google Scholar]
- 48.Kollisch G, Kalali BN, Voelcker V, Wallich R, Behrendt H, Ring J, Bauer S, Jakob T, Mempel M, Ollert M. Various members of the Toll-like receptor family contribute to the innate immune response of human epidermal keratinocytes. Immunology. 2005;114:531–541. doi: 10.1111/j.1365-2567.2005.02122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budak MT, Orsini JA, Pollitt CC, Rubinstein NA. Gene expression in the lamellar dermis-epidermis during the developmental phase of carbohydrate overload-induced laminitis in the horse. Vet. Immunol. Immunopathol. 2009;131:86–96. doi: 10.1016/j.vetimm.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 50.Noschka E, Vandenplas ML, Hurley DJ, Moore JN. Temporal aspects of laminar gene expression during the developmental stages of equine laminitis. Vet. Immunol. Immunopathol. 2009;129:242–253. doi: 10.1016/j.vetimm.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 51.Waguespack RW, Cochran A, Belknap JK. Expression of the cyclooxygenase isoforms in the prodromal stage of black walnut-induced laminitis in horses. Am. J. Vet. Res. 2004;65:1724–1729. doi: 10.2460/ajvr.2004.65.1724. [DOI] [PubMed] [Google Scholar]
- 52.Leise BS, Watts M, Tanhoff E, Johnson PJ, Black SJ, Belknap JK. Laminar regulation of STAT1 and STAT3 in black walnut extract and carbohydrate overload induced models of laminitis. J. Vet. Intern. Med. 2012;26:996–1004. doi: 10.1111/j.1939-1676.2012.00944.x. [DOI] [PubMed] [Google Scholar]
- 53.Kinsey GR, Li L, Okusa MD. Inflammation in acute kidney injury. Nephron. Exp. Nephrol. 2008;109:e102–e107. doi: 10.1159/000142934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loftus JP, Belknap JK, Black SJ. Matrix metalloproteinase-9 in laminae of black walnut extract treated horses correlates with neutrophil abundance. Vet. Immunol. Immunopathol. 2006;113:267–276. doi: 10.1016/j.vetimm.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Mungall BA, Pollitt CC. Zymographic analysis of equine laminitis. Histochem. Cell Biol. 1999;112:467–472. doi: 10.1007/s004180050430. [DOI] [PubMed] [Google Scholar]
- 56.Visser MB, Pollitt CC. The timeline of metalloprotease events during oligofructose induced equine laminitis development. Equine Vet. J. 2012;44:88–93. doi: 10.1111/j.2042-3306.2011.00393.x. [DOI] [PubMed] [Google Scholar]
- 57.Pawlak E, Wang L, Johnson PJ, Nuovo G, Taye A, Belknap JK, Alfandari D, Black SJ. Distribution and processing of a disintegrin and metalloproteinase with thrombospondin motifs-4, aggrecan, versican, and hyaluronan in equine digital laminae. Am. J. Vet. Res. 2012;73:1035–1046. doi: 10.2460/ajvr.73.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Pawlak E, Johnson PJ, Belknap JK, Alfandari D, Black SJ. Effects of cleavage by a disintegrin and metalloproteinase with thrombospondin motifs-4 on gene expression and protein content of versican and aggrecan in the digital laminae of horses with starch gruel-induced laminitis. Am. J. Vet. Res. 2012;73:1047–1056. doi: 10.2460/ajvr.73.7.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S, Birk DE. The regulatory roles of small leucine-rich proteoglycans in extracellular assembly. FEBS J. 2013;280:2120–2137. doi: 10.1111/febs.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geng Y, McQuillan D, Roughley PJ. SLRP interaction can protect collagen fibrils from cleavage by collagenases. Matrix Biol. 2006;25:484–491. doi: 10.1016/j.matbio.2006.08.259. [DOI] [PubMed] [Google Scholar]
- 61.Corsi A, Xu T, Chen XD, Boyde A, Liang J, Mankani M, Sommer B, Iozzo RV, Eichstetter I, Robey PG, Bianco P, Young MF. Phenotypic effects of biglycan deficiency are linked to collagen fibril abnormalities, are synergized by decorin deficiency, and mimic Ehlers-Danlos-like changes in bone and other connective tissues. J. Bone Miner. Res. 2002;17:1180–1189. doi: 10.1359/jbmr.2002.17.7.1180. [DOI] [PubMed] [Google Scholar]
- 62.Jepsen KJ, Wu F, Peragallo JH, Paul J, Roberts L, Ezura Y, Oldberg A, Birk DE, Chakravarti S. A syndrome of joint laxity and impaired tendon integrity in lumican- and fibromodulin-deficient mice. J. Biol. Chem. 2002;277:35532–35540. doi: 10.1074/jbc.M205398200. [DOI] [PubMed] [Google Scholar]
- 63.Bader HL, Lambert E, Guiraud A, Malbouyres M, Driever W, Koch M, Ruggiero F. Zebrafish collagen XIV is transiently expressed in epithelia and is required for proper function of certain basement membranes. J. Biol. Chem. 2013;288:6777–6787. doi: 10.1074/jbc.M112.430637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang G, Chen S, Goldoni S, Calder BW, Simpson HC, Owens RT, McQuillan DJ, Young MF, Iozzo RV, Birk DE. Genetic evidence for the coordinated regulation of collagen fibrillogenesis in the cornea by decorin and biglycan. J. Biol. Chem. 2009;284:8888–8897. doi: 10.1074/jbc.M806590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carter RA, Treiber KH, Geor RJ, Douglass L, Harris PA. Prediction of incipient pasture-associated laminitis from hyperinsulinaemia, hyperleptinaemia and generalised and localised obesity in a cohort of ponies. Equine Vet. J. 2009;41:171–178. doi: 10.2746/042516408x342975. [DOI] [PubMed] [Google Scholar]
- 66.Bailey SR, Adair HS, Reinemeyer CR, Morgan SJ, Brooks AC, Longhofer SL, Elliott J. Plasma concentrations of endotoxin and platelet activation in the developmental stage of oligofructose-induced laminitis. Vet. Immunol. Immunopathol. 2009;129:167–173. doi: 10.1016/j.vetimm.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 67.Mobasheri A, Critchlow K, Clegg PD, Carter SD, Canessa CM. Chronic equine laminitis is characterised by loss of GLUT1, GLUT4 and ENaC positive laminar keratinocytes. Equine Vet. J. 2004;36:248–254. doi: 10.2746/0425164044877224. [DOI] [PubMed] [Google Scholar]
- 68.Toth F, Frank N, Chameroy KA, Bostont RC. Effects of endotoxaemia and carbohydrate overload on glucose and insulin dynamics and the development of laminitis in horses. Equine Vet. J. 2009;41:852–858. doi: 10.2746/042516409x479027. [DOI] [PubMed] [Google Scholar]
- 69.Faleiros RR, Nuovo GJ, Flechtner AD, Belknap JK. Presence of mononuclear cells in normal and affected laminae from the black walnut extract model of laminitis. Equine Vet. J. 2011;43:45–53. doi: 10.1111/j.2042-3306.2010.00121.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


