Abstract
For many genomic loci, there are more than one potential cleavage and polyadenylation site, resulting in the generation of multiple distinct transcripts. When the proximal polyadenylation site is present within the coding region of the transcript, alternative polyadenylation can result in proteins with distinct amino acid sequences and potentially distinct functions. In most cases, the different possible polyadenylation sites are all present within the 3′ untranslated regions (UTRs), and the amino acid sequence of the encoded proteins are not affected by polyadenylation site selection. In individual instances, the selection of the proximal versus distal polyadenylation site in the 3′UTR can dramatically affect transcript stability and translatability. In some instances, UTR alternative polyadenylation generates RNA isoforms that have distinct subcellular localization patterns, and that can regulate the location of the encoded protein in an RNA-guided manner. In a recent paper, the laboratory of Christine Mayr demonstrated that alternative polyadenylation of the transmembrane protein CD47 results in transcripts with the same localization pattern, but the encoded protein localizes to the endoplasmic reticulum when it is encoded by the transcript generated by using the proximal polyadenylation site in 3′UTR, and the identical protein localizes to the plasma membrane when the transcript is encoded by the distal polyadenylation site, also in the 3′ UTR. Unlike previous studies, the mechanism of localization does not rely on differential trafficking of the mRNA and is instead, based on RNA-mediated recruitment of proteins to the cytoplasmic side of CD47 that support its plasma membrane localization. Other transmembrane proteins were discovered to be regulated similarly. The results demonstrate that the choice of polyadenylation site can affect protein localization and function, even when the sequence of the protein is unaffected. Further, the transcript encoding a protein can serve as a scaffold to recruit additional proteins that affect the protein’s fate.
Keywords: alternative polyadenylation, plasma membrane, HuR, SET, RAC1, CD47
INTRODUCTION
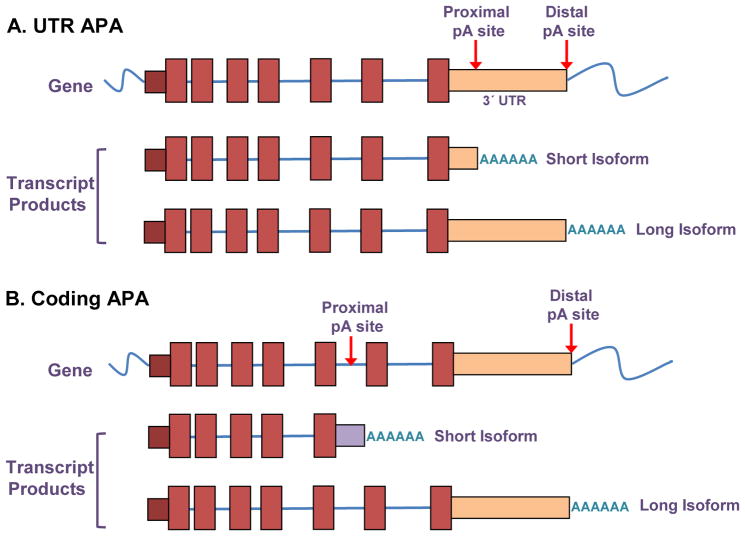
For many of the genes within the human genome, their end is in question. That is, there are different sites within the genomic locus at which the transcript can be cleaved and a poly(A) tail added, thus initiating the process that will generate the transcript end [1]. As a result of the multiple possibilities for cleavage and polyadenylation, a process called alternative polyadenylation, a single genetic locus can result in the production of multiple different transcripts with different ending positions. If the proximal and distal polyadenylation sites are both in the 3′ untranslated regions (UTRs), then this is considered UTR alternative polyadenylation, while if the proximal site is within the coding region, this is considered coding alternative polyadenylation (Figure 1).
Figure 1.
Schematic of alternative polyadenylation. In UTR APA (top), a gene has two potential alternative polyadenylation sites that can result in two distinct transcripts. Both the proximal and distal polyadenylation sites are present in the 3′ UTR. For coding APA (bottom), the proximal polyadenylation site is within the coding region of the gene, and two different isoforms result in the expression of distinct proteins.
Prevalence of alternative polyadenylation
While individual examples of alternative polyadenylation have been reported since the 1980s [2, 3], the prevalence of alternative polyadenylation genome-wide was not easily determinable until cDNA and expressed sequence tag sequencing (EST) data became available. In 2004, Ben Tian, Carol Lutz and colleagues analyzed cDNA and EST data and learned that alternative polyadenylation is prevalent: 54% of human genes and 32% of mouse genes were discovered to have multiple polyadenylation sites [4].
In another important study published in 2008, Sandberg and colleagues, inferring the levels of different isoforms of a transcript based on the extent of hybridization to each of the probes for a single gene on microarrays [5], discovered that when T cells are activated to proliferate, their 3′ UTRs become shorter. Thirteen genes shifted to the use of a more distal polyadenylation site and 86 genes shifted to the use of a more proximal polyadenylation site with activation. Further analysis by these authors revealed that cell lines and more proliferative cells tended to use proximal polyadenylation sites, while differentiated tissue used more distal sites.
With the advent of methods for high-throughput sequencing of RNA, multiple labs have developed methods to specifically sequence the 3′ ends of genes [1, 6–10]. The availability of these methods has made it possible to define more precisely the selection of polyadenylation sites on a genome-wide scale [11, 12]. These studies have revealed that 69–79% of mammalian genes [1, 7] and about half of genes in flies [13], worms [14] and zebrafish [15] have the potential to generate transcripts with different 3′ UTRs [12]. Polyadenylation site selection has been found to vary with proliferation [5, 16], among tissues [7], and with transformation to cancer [11]. The lengths of 3′ UTRs tend to increase upon differentiation [8, 17, 18], and shorten with proliferation [5, 16]. For instance, transcripts isolated from stem cells and testes are more likely to use proximal polyadenylation sites in 3′UTRs [13, 17], while transcripts isolated from differentiated neurons are more likely to terminate at distal polyadenylation sites [13, 18, 19]. Further, reprogramming of differentiated cells into induced pluripotent stem cells results in a shortening of 3′ UTRs [20]. Transcripts present in cancer cells, both cancer cell lines and primary tumors, are more likely to terminate at proximal polyadenylation sites [11, 21, 22]. The findings thus demonstrate that UTR alternative polyadenylation is widespread. These studies also demonstrate that the selection of polyadenylation sites is altered in proliferation, differentiation, activation, reprogramming, and carcinogenesis.
Functional role for coding region alternative polyadenylation
In cases in which the proximal polyadenylation site is within the coding region of the gene, alternative polyadenylation can result in two or more distinct proteins. Elegant studies in the 1980s demonstrated that a change in the selection of the polyadenylation site for the immunoglobulin M heavy chain is critically important for the immune response. The long isoform of the IgM heavy chain, expressed in resting lymphocytes, contains a transmembrane domain and its incorporation results in membrane-bound antibody [2, 3]. During lymphocyte activation, there is a shift toward increased utilization of a more proximal polyadenylation site. This results in a shorter, secreted protein that terminates prior to the transmembrane domain.
In addition to these findings demonstrating clear genome-wide patterns of alternative polyadenylation, complementary studies have articulated models for the functional importance of coding APA. Screening EST databases, Vorlova and colleagues discovered a coding region alternative polyadenylation event in 31 different genes in which the same genetic locus produces a transcript that encodes a receptor tyrosine kinase and a different, shorter transcript that encodes a secreted protein [23]. These secreted forms were discovered to act as soluble decoys that interact with the ligand for the receptor expressed by the full-length protein. Switching expression from the full-length vascular endothelial receptor 2 to a shorter, soluble antagonistic isoform in human vascular endothelial cells resulted in a strong antiangiogenic effect.
As another example, alternative cleavage and polyadenylation factors have been discovered to autoregulate themselves. CstF-77 is a protein partner in the CstF complex that contributes to cleavage and polyadenylation reactions. A polyadenylation site within exon 3 of CstF-77 creates a truncated and inactive transcript [24]. When cleavage and polyadenylation activity is high, the creation of this shorter, inactive CstF-77 transcript may serve as a negative regulator of cleavage and polyadenylation activity.
Functional role of UTR alternative polyadenylation in transcript stability
Emerging data has supported a functional role for UTR alternative polyadenylation. As 3′ UTRs can serve as recognition sites for molecules that affect the fate of a transcript, for instance, microRNAs or RNA-binding proteins [25, 26], changes in 3′ UTR length have the potential to substantially affect a transcript’s decay rate and its abundance. In 2009, Christine Mayr and David Bartel demonstrated that short mRNA isoforms for cyclin D2 and insulin-like growth factor 2 that lack recognition sites for the microRNAs let-7 or miR-15/16 are more stable [11]. The shorter isoforms of these transcripts were shown to be abundant in cancer cells and, because they lack microRNA recognition sites, more stable. Further, overexpression of IGF2BP1/IMP-1 expressed from the short isoform was sufficient to transform cells.
As additional examples, the likelihood of developing systemic lupus erythromatosis is associated with a genetic polymorphism in a proximal polyadenylation site of human interferon regulatory factor 5 (IRF5) that causes a mutation in the polyadenylation signal. This mutation causes differential expression of two isoforms of IRF5 [27]. The long isoform was shown to be less stable than the short isoform. This mutation affects IRF5 levels and, together with other mutations in the same gene, affects the risk of systemic lupus erythromatosis. In human glioblastomas, the O6-methylguanine-DNA methyltransferase (MGMT) gene is thought to perform disadvantageous repair of damage induced by chemotherapy [28]. Silencing of the gene is associated with a survival advantage after treatment with radiation and the alkylating agent temozolomide (TMZ) [29, 30]. In addition to methylation of the MGMT promoter leading to its silencing, recent data demonstrates that there are two different isoforms for MGMT, and that expression of the long MGMT isoform renders it susceptible to targeting by microRNAs that have recognition sites in the sequences present in the long, but not the short, MGMT transcript [31]. Expression of the long MGMT transcript in gliomas correlated with low MGMT expression, which would sensitize the cells to alkylating agents. Alternative polyadenylation has also been implicated in circadian rhythms, as a recent study demonstrated that two cold-induced RNA-binding proteins regulate genes that are part of the circadian response in mouse embryonic fibroblasts by affecting alternative polyadenylation of targeted transcripts [32]. Finally, an RNA-binding protein that regulates flowering in plants has been demonstrated to regulate the expression of alternatively processed antisense RNAs at the locus of a key flowering transcription repressor [33].
Taken together, these reports and others create a compelling argument that alternative polyadenylation plays a critically important role in controlling transcript decay and abundance for specific genes that control cell functionality. These findings are further supported by two recent studies in yeast, both of which demonstrated that the same genomic locus can be expressed as a series of transcripts with varying levels of stability [34, 35]. A difference of even a single nucleotide in the 3′ UTR was found by both groups to have a large effect on a transcript’s decay rate, thus supporting the importance of the specific nucleotide at which a transcript terminates in controlling its fate.
However, genome-wide analyses of the importance of alternative polyadenylation in transcript decay and abundance in mammalian cells have clarified that most instances in which there is a change in the use of a polyadenylation site are likely to have little effect on the specific transcript. These studies show that instances of large, functional effects may be more the exception than the rule. Genome-wide analyses have also shown that the longer isoform of a transcript may not always exhibit more rapid degradation than the shorter isoform of the transcript, indicating that the portion of the 3′ UTR that is present in the longer, but not the shorter isoform, may contain either destabilizing or stabilizing motifs. Spies, Burges, and Bartel determined genome-wide measurements of decay rates for mRNAs with alternative 3′ UTRs in murine 3T3 cells [25]. Isoforms generated from more distal polyadenylation sites had slightly lower mRNA stability, on average, than isoforms generated from proximal polyadenylation sites, consistent with the potential importance of destabilizing elements in 3′ UTRs. However, for most transcripts, the effect was small. Overall, the authors concluded that the selection of polyadenylation site had little effect on a transcript’s expression level or decay rate. In another study, mice were engineered to overexpress poly(A) binding protein nuclear 1, a protein important for polyadenylation [36]. RNA isolated from the quadruceps of these mice exhibited changes in polyadenylation site usage compared with controls. Among 2012 genes with changes in polyadenylation site usage, 916 also exhibited differential expression between the engineered and control mice, which was more than expected by chance. Overall, the use of more proximal polyadenylation sites was associated with an upregulation of the transcript level of the shorter isoform in mice overexpressing the poly(A) binding protein compared with control mice. This trend is consistent with the loss of destabilizing elements from the 3′ UTRs, but the preference for upregulation compared with downregulation was modest, making it less clear that the expectation should be the loss of microRNA target sites when 3′ UTRs shorten. Studies such as these have led some to question the extent to which alternative polyadenylation events are important drivers of cellular processes [37].
Alternative polyadenylation and translatability
In addition to effects on transcript degradation rates, alternative polyadenylation can also impact a transcript’s translatability, that is, the extent to which a transcript is converted to protein. Indeed, because mRNAs are circular, the 3′ UTR is located adjacent to the sites within the transcript that control the recruitment of translation initiation factors to the message’s 5′ end [25]. Using luciferase reporter assays as a readout, Mayr and Bartel demonstrated that translation from the shorter isoforms for cyclin D2, IMP-1 and Dicer resulted in higher protein production than translation from the longer isoform [11], thus establishing a potentially important role for alternative polyadenylation in the expression of these transcripts, and possibly more broadly as a regulator of translation rate, which could reflect a role for microRNAs or RNA binding proteins that target the transcript in its translatability. A study of the localization of transcripts to the polyribosomal versus cytoplasmic fractions of human embryonic kidney 293T cells revealed higher enrichment in the polyribosomal fraction for shorter isoforms, which would support a model in which short isoforms lacking microRNA recognition sites are more actively translated [38]. However, subsequent studies have yielded a less consistent picture. In the paper by Spies, Burges and Bartell, the different isoforms for a single gene had very similar distributions of the number of bound ribosomes [25]. To the extent that there was a difference, shorter transcripts were, surprisingly, less likely to be translated than longer transcripts on a genome-wide basis, the opposite of the expectation based on the previous studies. In another study, while changes in polyadenylation site selection were observed in naïve versus activated T cells, they were not associated with corresponding changes in mRNA or protein abundance [39]. Other examples in which the isoform with the longer 3′ UTR is preferentially translated have also been reported. For instance, the serotonin receptor, a major regulator of anxiety-based behaviors, undergoes alternative polyadenylation [40], and polymorphisms that increase the use of the proximal polyadenylation site result in increased fear and heightened anxiety and depressive symptoms [41]. The long, but not the short, isoform of the serotonin receptor contains binding sites for the RNA-binding protein hnRNPK, which increases serotonin receptor expression levels [42]. Similarly, for the cell cycle gene polo in Drosophila, the longer isoform is translated at three times the rate of the shorter isoform [43]. Drosophila lacking the distal polyadenylation signal cannot express the more highly expressed longer isoform and die due to lack of proliferation of abdominal cells. Thus, while in some instances, alternative polyadenylation clearly generates transcripts with different translatability, the effect is not always in a consistent direction, and the global importance of genome-wide alternative polyadenylation on translation remains unclear.
Conservation of alternative polyadenylation
To better understand the functional importance of alternative polyadenylation in gene function, the extent to which alternative polyadenylation events are conserved among species has been analyzed to determine whether there is selective pressure to retain individual polyadenylation sites. In one such analysis, of 4800 genes, the specific locations of the polyadenylation sites were conserved between mice and humans in approximately 500 genes [44]. The authors concluded that these genes may be under selection to preserve the specific location of these alternative polyadenylation sites. As conservation of the specific site was a relatively rare event, the authors concluded that the gain and loss of polyadenylation sites is a common occurrence in mammalian genomes. Other studies have discovered a lack of correlation between the specific location of polyadenylation sites in the mouse and human genomes [39]. As proximal sites tend to be weaker than distal sites, a model has been presented in which new, weaker 3′ sites are being consistently formed upstream to stronger 5′ sites [44]. These are lost quickly if they are weaker than the 5′ site and do not confer any advantage. This process would tend to produce the observed result: weaker 3′ sites and stronger 5′ sites.
Alternative polyadenylation and transcript localization
Some functionally-important, evolutionarily-conserved instances of alternative polyadenylation may not affect a transcript’s abundance, decay rate or translatability, but rather, its localization. Some transcripts are actively transported to specific subcellular locations. For instance, the process of learning and memory requires that specific transcripts are actively transported to neuron projections such as axons and dendrites [45–47]. Translation of these transcripts at synapses provides proteins required for learning-related plasticity [45]. These actively-transported transcripts contain cis-acting motifs in their 3′ UTRs, or less commonly 5′ UTRs, that recruit RNA-binding proteins [48]. Through these RNA-binding proteins, the transcripts become recognized by molecular motors that transport the messenger RNAs to positions within the neuron such as the axon or dendrite [49]. Alternative polyadenylation can result in the production of transcripts with and without the motifs critical for localization. An and colleagues demonstrated that for brain-derived neurotrophic factor (BDNF), transcripts expressed as the long isoform, but not the short isoform, are present not only in the cell soma, but also in the dendrites [50]. Mice that expressed the short, but not the long, isoform of BDNF exhibited impairment in long-term potentiation of dendrites, without the same effect on the soma. In another example, Duchene and colleagues examined subcellular localization to the mitochondria. They discovered that in Arabidopsis, alternative polyadenylation generates two isoforms of the voltage-dependent anion channel 3 (VDAC3) of the outer mitochondrial membrane. The 3′ UTR sequence present in the longer, but not the shorter, isoform of VDAC3 is necessary and sufficient to target VDAC3 mRNA to the mitochondria [51]. Thus, in these examples, sequences present in the alternatively present 3′ UTR were responsible for targeting the transcript to specific cellular locations, resulting in preferential localization of the encoded proteins.
Alternative polyadenylation regulates CD47 localization to the plasma membrane
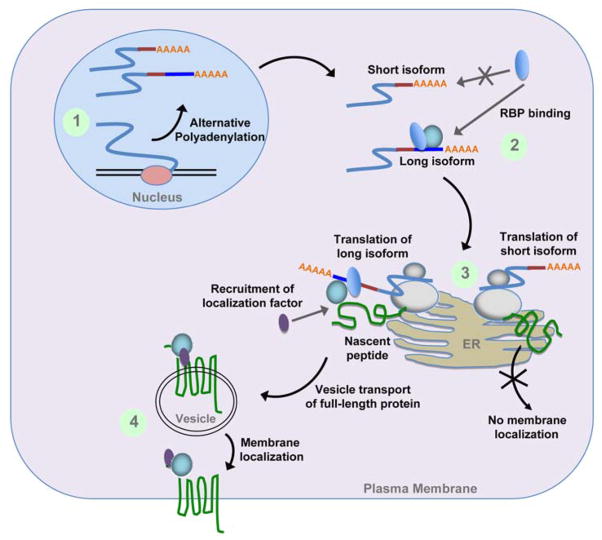
This summer 2015, Binyamin Berkovits and Christine Mayr published a paper in Nature entitled “Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization” [52]. In this report, Berkovits and Mayr focus on the CD47 membrane protein, a widely-expressed cell surface marker that allows cells to label themselves as ‘self’ and protect themselves from phagocytosis by macrophages. They discovered that when CD47 is encoded by a transcript with a short 3′ UTR, more of the protein is localized at the endoplasmic reticulum (ER), while if CD47 is encoded by a transcript with a long 3′ UTR, CD47 localizes mostly to the cell surface (Figure 2). This did not reflect a difference in the localization of the mRNA itself, as both the shorter and the longer transcripts were localized similarly near the perinuclear ER. Thus, this differential localization represents a new mechanism of control.
Figure 2.
Schematic of CD47 processing. CD47 is encoded by a short isoform and a long isoform through UTR alternative polyadenylation. RNA-binding proteins bind to the long, but not the short, isoform of CD47. After translation in the endoplasmic reticulum, localization factors are recruited to the long isoform that facilitate the transport of the encoded protein into vesicles and to the plasma membrane.
The alternatively present CD47 3′ UTR sequence contained tracts of uridines, and the HuR RNA-binding protein is known to associate with transcripts through uridine-rich regions [53–56]. Knockdown of HuR did not affect the total levels of CD47 mRNA or protein, but HuR knockdown did reduce the localization of the transcript to the cell surface. They hypothesized that HuR mediates its effects on the cell surface expression of CD47 through protein-protein interactions. Previous studies have shown that HuR interacts with SET [57], that SET interacts with RAC1 [58], and that active RAC1 translocates SET to the plasma membrane [58]. Berkovits and Mayr discovered that shRNAs directed against either SET or RAC1 resulted in no change in the total protein levels of CD47, but reduced the level of CD47 on the plasma membrane. Further, several other genes encoding transmembrane proteins that also contain HuR binding sites in their 3′ UTRs (CD44, ITGA1 and TNFRSF13C) also exhibited reduced surface expression, but similar overall expression, when HuR was knocked down.
Functionally, expression of CD47 from the long transcript resulted in protection against phagocytosis by macrophages, while expression of CD47 from the shorter transcript, even if expression levels were similar, resulted in less than complete protection against phagocytosis. Further, overexpression of CD47 from the long, but not the short, isoform resulted in strong co-localization with RAC1 at cellular ruffles called lamellipodia, and promoted the formation of lamellipodia at the leading edge. Thus, the functional effects of expression from the shorter versus the longer transcript were different, even though the sequence of the coding region of CD47 was unchanged. The findings, taken together, elucidate a mechanism whereby alternative polyadenylation can affect the localization of the encoded protein independent of an effect on the localization of the transcript itself. The results also define a new pathway for HuR-mediated translocation of proteins to the plasma membrane that affects at least four, and possibly more, transmembrane proteins.
A new method for targeting of proteins to subcellular components
Our understanding of how proteins are targeted to different subcellular compartments was significantly advanced by Gunter Blobel in 1975 when he discovered that proteins are localized to different subcellular compartments based on the presence of signal sequences present in the protein [59]. In the model he put forth, Blobel defined a series of amino acids located immediately after the initiation codon that are present only on those mRNAs whose translation products will be synthesized by ER-bound ribosomes. These proteins are destined to reside within the ER, the Golgi, the plasma membrane or other cellular compartments, but not the cytoplasm. According to this model, the amino acid sequence of a protein contains the necessary information to determine the protein’s final subcellular localization.
Since the initial publication of this model, additional amino acid sequences that direct protein sorting have been identified. For instance, a mannose-6-phosphate modification is a signal for a protein to be targeted to the lysosome [60]. Amino acid sequences indicating that a specific protein should be retained within the ER have also been identified. These proteins often, but not always, contain a KDEL amino acid sequence near the C terminus [61]. The KDEL receptor actively transports proteins containing the KDEL sequence from the Golgi to the ER. Thus, the prevailing model for protein targeting has involved the recognition of amino acid motifs within the protein to actively transport proteins to their proper subcellular compartment, mechanisms by which proteins interact with other proteins in that particular compartment, and/or mechanisms to rescue proteins that have escaped their appropriate compartment and reposition them [62].
Proteins that are integral to membranes represent an important category of proteins that must be actively sorted to the proper cellular compartment [62]. The amino acid sequence KKXX or RRXX at the cytoplasmic side of the C terminus represents a signal for transmembrane proteins to be retained in the ER [63–65]. This sequence is thought to function by allowing the protein to interact with coat protein I of the COP I complex [66], and thereby facilitate the retrograde transport of membrane proteins that escape the ER and enter the Golgi apparatus. In addition, an internally positioned, also cytoplasmically localized, RXR motif has been discovered to be important for the retention of membrane proteins destined for the ER [67]. The RXR motif is also used to retrieve resident ER proteins that mistakenly enter the Golgi apparatus. Amino acid sequences that are important for promoting export of membrane proteins from the ER have also been identified. A motif containing DXE [68] and neighboring residues [69] accelerates the ER export of a viral membrane protein. Further, studies of the trafficking of the Kir family revealed an export signal, FCYENE [70]. Thus, for many membrane proteins, information on whether the protein is intended to be retained within the ER or processed through the Golgi apparatus and vesicles to be inserted into the plasma membrane is encoded within the amino acid sequence of the protein itself.
How does the CD47 protein maintain localization both at the endoplasmic reticulum and plasma membrane?
Against this background, the findings of Berkovits and Mayr that CD47 can be either retained within the endoplasmic reticulum or appear at the surface of the cell, with exactly the same amino acid sequence, may be considered surprising. Their elegant findings explain that CD47 encoded by the long isoform is translocated to the plasma membrane through a mechanism that depends on HuR, SET and Rac1. Another interesting question is “How does CD47 encoded by the short transcript maintain its localization within the ER?” For instance, CD47 does not contain a KKXX or RRXX sequence at its C terminus, nor does it contain an RXR motif. One possibility could be that CD47 contains a previously undefined ER retention signal; indeed, a recent study discovered a new amino acid-based ER retention signal in the mTORC protein [71]. However, if there were a retrieval signal encoded in the CD47 coding region, then how does the same protein progress in an anterograde manner through the Golgi, enter vesicles and localize to the plasma membrane?
One possible consideration is whether CD47 in the ER has folded properly. Indeed, when the CD47 extracellular domain was replaced by the sequence for GFP, localization to the ER was particularly strong. Perhaps CD47 expressed by the short transcript is more likely to be incompletely folded, and ‘stuck’ in the endoplasmic reticulum, than CD47 expressed by the long transcript. One possible explanation for the difference in localization for the protein encoded by the different isoforms could be that the encoded mRNA itself facilitates the folding process. The possibility that RNA can serve as a chaperone and help a nascent protein strand avoid self-aggregation has been proposed [72, 73], and specific examples have been identified. The 23S rRNA and the V domain of the 23S rRNA can function as molecular chaperones in vitro [74, 75], and the ribosomal protein S3a can act as a molecular chaperone for the hepatitis B virus X protein [76]. In the case of CD47, the topology makes a role for the transcript itself in protein folding less likely: the mRNA is in the cytoplasm and the portion of the protein that is swapped for GFP in the engineered version is in the ER.
Taken together, the data in the recent paper by Berkovits and Mayr demonstrate a potential for an important cellular function mediated by alternative polyadenylation events in which both polyadenylation sites are present in the 3′ UTR. Further, they demonstrate that there may be an important function for alternative polyadenylation events that do not alter the decay rate or translatability of the encoded isoforms, and do not affect the overall level of the encoded protein. Their data also show that the localization of a protein can be determined by the message from which it is transcribed, and that the same amino acid sequence can localize to more than one of the multiple compartments within a cell, demonstrating that the information needed for protein targeting is not completely encoded within the coding sequence. Finally, they discovered a mechanism whereby the RNA-binding protein HuR has the capacity to regulate the localization of proteins to the plasma membrane. Further studies will allow for a better understanding of how widespread the HuR-mediated mechanism for protein localization is, and whether CD47 alternative polyadenylation is important, for instance, in the identification of transformed cells as no longer ‘self’.
Acknowledgments
HAC was the Milton E. Cassel scholar of the Rita Allen Foundation (http://www.ritaallenfoundation.org). ELJ was supported in part by a National Science Foundation (http://www.nsf.gov) Graduate Research Fellowship DGE-0646086. This work was funded by grants to HAC from the National Institute of General Medical Sciences (http://www.nigms.nih.gov), PhRMA Foundation grant 2007RSGl9572, National Science Foundation (http://www.nsf.gov) Grant OCI-1047879 to David August, National Institute of General Medical Sciences (http://www.nigms.nih.gov) R01 GM081686, National Institute of General Medical Sciences (http://www.nigms.nih.gov), and the Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research (https://www.stemcell.ucla.edu). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ABBREVIATIONS
- 3′ UTR
3′ untranslated region
- APA
alternative polyadenylation
- BDNF
brain-derived neurotrophic factor
- ER
endoplasmic reticulum
- VDAC3
voltage- dependent anion channel 3
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
References
- 1.Tian B, Manley JL. Trends Biochem Sci. 2013;38:312. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. Cell. 1996;87:941. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 3.Alt FW, Bothwell AL, Knapp M, Siden E, Mather E, Koshland M, Baltimore D. Cell. 1980;20:293. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- 4.Tian B, Hu J, Zhang H, Lutz CS. Nucleic Acids Res. 2005;33:201. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Science. 2008;320:1643. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, Attie O, Chen K, Salehi-Ashtiani K, Vidal M, Harkins TT, Bouffard P, Suzuki Y, Sugano S, Kohara Y, Rajewsky N, Piano F, Gunsalus KC, Kim JK. Science. 2010;329:432. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. Genome Res. 2012;22:1173. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Nat Methods. 2013;10:133. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Cell. 2010;143:1018. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon OK, Brem RB. RNA. 2010;16:1256. doi: 10.1261/rna.2038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayr C, Bartel DP. Cell. 2009;138:673. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Genes Dev. 2013;27:2380. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, Eisman RC, Andrews J, Kaufman T, Cherbas P, Celniker SE, Graveley BR, Lai EC. Cell Rep. 2012;1:277. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jan CH, Friedman RC, Ruby JG, Bartel DP. Nature. 2011;469:97. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. Genome Res. 2012;22:2054. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Oude Vrielink JA, Agami R. Genome Biol. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Proc Natl Acad Sci USA. 2009;106:7028. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. RNA. 2011;17:761. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B. Proc Natl Acad Sci USA. 2011;108:15864. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji Z, Tian B. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Cancer Res. 2009;69:9422. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vorlova S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L. Mol Cell. 2011;43:927. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo W, Ji Z, Pan Z, You B, Hoque M, Li W, Gunderson SI, Tian B. PLoS Genet. 2013;9:e1003613. doi: 10.1371/journal.pgen.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spies N, Burge CB, Bartel DP. Genome Res. 2013;23:2078. doi: 10.1101/gr.156919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Mol Cell. 2007;27:91. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G, Bauer JW, Gillett C, Burtt N, Cunninghame Graham DS, Onofrio R, Petri M, Gunnarsson I, Svenungsson E, Ronnblom L, Nordmark G, Gregersen PK, Moser K, Gaffney PM, Criswell LA, Vyse TJ, Syvanen AC, Bohjanen PR, Daly MJ, Behrens TW, Altshuler D. Proc Natl Acad Sci USA. 2007;104:6758. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaina B, Christmann M, Naumann S, Roos WP. DNA Repair (Amst) 2007;6:1079. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. N Engl J Med. 2005;352:997. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 30.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO European Organisation for Research Treatment of Cancer Brain Tumor Radiation Oncology Groups National Cancer Institute of Canada Clinical Trials Group. Lancet Oncol. 2009;10:459. [Google Scholar]
- 31.Kreth S, Limbeck E, Hinske LC, Schutz SV, Thon N, Hoefig K, Egensperger R, Kreth FW. Acta Neuropathol. 2013;125:671. doi: 10.1007/s00401-013-1081-1. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Hu W, Murakawa Y, Yin J, Wang G, Landthaler M, Yan J. Sci Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornyik C, Terzi LC, Simpson GG. Dev Cell. 2010;18:203. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Gupta I, Clauder-Munster S, Klaus B, Jarvelin AI, Aiyar RS, Benes V, Wilkening S, Huber W, Pelechano V, Steinmetz LM. Mol Syst Biol. 2014;10:719. doi: 10.1002/msb.135068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Cell. 2014;156:812. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Klerk E, Venema A, Anvar SY, Goeman JJ, Hu O, Trollet C, Dickson G, den Dunnen JT, van der Maarel SM, Raz V, ‘t Hoen PA. Nucleic Acids Res. 2012;40:9089. doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neve J, Furger A. Biochem Soc Trans. 2014;42:1190. doi: 10.1042/BST20140054. [DOI] [PubMed] [Google Scholar]
- 38.Sterne-Weiler T, Martinez-Nunez RT, Howard JM, Cvitovik I, Katzman S, Tariq MA, Pourmand N, Sanford JR. Genome Res. 2013;23:1615. doi: 10.1101/gr.148585.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gruber AR, Martin G, Muller P, Schmidt A, Gruber AJ, Gumienny R, Mittal N, Jayachandran R, Pieters J, Keller W, van Nimwegen E, Zavolan M. Nat Commun. 2014;5:5465. doi: 10.1038/ncomms6465. [DOI] [PubMed] [Google Scholar]
- 40.Battersby S, Ogilvie AD, Blackwood DH, Shen S, Muqit MM, Muir WJ, Teague P, Goodwin GM, Harmar AJ. J Neurochem. 1999;72:1384. doi: 10.1046/j.1471-4159.1999.721384.x. [DOI] [PubMed] [Google Scholar]
- 41.Hartley CA, McKenna MC, Salman R, Holmes A, Casey BJ, Phelps EA, Glatt CE. Proc Natl Acad Sci USA. 2012;109:5493. doi: 10.1073/pnas.1202044109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon Y, McKenna MC, Rollins DA, Song M, Nuriel T, Gross SS, Xu G, Glatt CE. Proc Natl Acad Sci USA. 2013;110:11624. doi: 10.1073/pnas.1301485110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, Coelho PA, Carmo AM, Sunkel CE, Proudfoot NJ, Moreira A. EMBO J. 2011;30:2431. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ara T, Lopez F, Ritchie W, Benech P, Gautheret D. BMC Genomics. 2006;7:189. doi: 10.1186/1471-2164-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Cell. 1997;91:927. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 46.Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Neurochem Res. 2002;27:1065. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- 47.Zhong J, Zhang T, Bloch LM. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meer EJ, Wang DO, Kim S, Barr I, Guo F, Martin KC. Proc Natl Acad Sci USA. 2012;109:4639. doi: 10.1073/pnas.1116269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirokawa N. J Neurosci. 2006;26:7139. doi: 10.1523/JNEUROSCI.1821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Cell. 2008;134:175. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michaud M, Ubrig E, Filleur S, Erhardt M, Ephritikhine G, Marechal-Drouard L, Duchene AM. Proc Natl Acad Sci USA. 2014;111:8991. doi: 10.1073/pnas.1402588111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berkovits BD, Mayr C. Nature. 2015;522:363. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. Nat Methods. 2011;8:559. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 54.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Mol Cell. 2011;43:340. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 55.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Mol Cell. 2011;43:327. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uren PJ, Burns SC, Ruan J, Singh KK, Smith AD, Penalva LO. J Biol Chem. 2011;286:37063. doi: 10.1074/jbc.C111.266882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brennan CM, Gallouzi IE, Steitz JA. J Cell Biol. 2000;151:1. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.ten Klooster JP, Leeuwen I, Scheres N, Anthony EC, Hordijk PL. EMBO J. 2007;26:336. doi: 10.1038/sj.emboj.7601518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blobel G, Dobberstein B. J Cell Biol. 1975;67:835. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coutinho MF, Prata MJ, Alves S. Mol Genet Metab. 2012;105:542. doi: 10.1016/j.ymgme.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 61.Alberts B, Johnson A, Lewis J. Molecular Biology of the Cell. New York: Garland Science; 2002. [Google Scholar]
- 62.Saraste J, Kuismanen E. Semin Cell Biol. 1992;3:343. doi: 10.1016/1043-4682(92)90020-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vincent MJ, Martin AS, Compans RW. J Biol Chem. 1998;273:950. doi: 10.1074/jbc.273.2.950. [DOI] [PubMed] [Google Scholar]
- 64.Nilsson T, Jackson M, Peterson PA. Cell. 1989;58:707. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 65.Jackson MR, Nilsson T, Peterson PA. EMBO J. 1990;9:3153. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dancourt J, Barlowe C. Annu Rev Biochem. 2010;79:777. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 67.Zerangue N, Schwappach B, Jan YN, Jan LY. Neuron. 1999;22:537. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 68.Nishimura N, Balch WE. Science. 1997;277:556. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 69.Sevier CS, Weisz OA, Davis M, Machamer CE. Mol Biol Cell. 2000;11:13. doi: 10.1091/mbc.11.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma D, Zerangue N, Lin YF, Collins A, Yu M, Jan YN, Jan LY. Science. 2001;291:316. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 71.Liu X, Zheng XF. Mol Biol Cell. 2007;18:1073. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi SI, Lim KH, Seong BL. Int J Mol Sci. 2011;12:1979. doi: 10.3390/ijms12031979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi SI, Han KS, Kim CW, Ryu KS, Kim BH, Kim KH, Kim SI, Kang TH, Shin HC, Lim KH, Kim HK, Hyun JM, Seong BL. PLoS One. 2008;3:e2677. doi: 10.1371/journal.pone.0002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Das B, Chattopadhyay S, Bera AK, Dasgupta C. Eur J Biochem. 1996;235:613. doi: 10.1111/j.1432-1033.1996.00613.x. [DOI] [PubMed] [Google Scholar]
- 75.Chattopadhyay S, Das B, Dasgupta C. Proc Natl Acad Sci USA. 1996;93:8284. doi: 10.1073/pnas.93.16.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim KH, Kim KH, Choi SI, Park ES, Park SH, Ryu K, Park YK, Kwon SY, Yang SI, Lee HC, Sung IK, Seong BL. PLoS One. 2011;6:e22258. doi: 10.1371/journal.pone.0022258. [DOI] [PMC free article] [PubMed] [Google Scholar]