Abstract
The gut microbiota plays important roles in its host. However, how each microbiota member contributes to the behavior of the whole population is not known. In this study, we therefore determined protein expression in the cecal microbiota in chickens of selected ages and in 7-day-old chickens inoculated with different cecal extracts on the day of hatching. Campylobacter, Helicobacter, Mucispirillum, and Megamonas overgrew in the ceca of 7-day-old chickens inoculated with cecal extracts from donor hens. Firmicutes were characterized by ABC and phosphotransferase system (PTS) transporters, extensive acyl coenzyme A (acyl-CoA) metabolism, and expression of l-fucose isomerase. Anaerostipes, Anaerotruncus, Pseudoflavonifractor, Dorea, Blautia, and Subdoligranulum expressed spore proteins. Firmicutes (Faecalibacterium, Butyrivibrio, Megasphaera, Subdoligranulum, Oscillibacter, Anaerostipes, and Anaerotruncus) expressed enzymes required for butyrate production. Megamonas, Phascolarctobacterium, and Blautia (exceptions from the phylum Firmicutes) and all Bacteroidetes expressed enzymes for propionate production pathways. Representatives of Bacteroidetes also expressed xylose isomerase, enzymes required for polysaccharide degradation, and ExbBD, TonB, and outer membrane receptors likely to be involved in oligosaccharide transport. Based on our data, Anaerostipes, Anaerotruncus, and Subdoligranulum might be optimal probiotic strains, since these represent spore-forming butyrate producers. However, certain care should be taken during microbiota transplantation because the microbiota may behave differently in the intestinal tract of a recipient depending on how well the existing communities are established.
INTRODUCTION
The intestinal tracts of warm-blooded animals represent a unique ecological niche. This niche is characterized by a constant temperature, availability of nutrients, and the absence of oxygen. It is not surprising that it is occupied with billions of microorganisms, which collectively comprise the gut microbiota (1). The gut microbiota plays an important role in its vertebrate host. The microbiota facilitates the digestion of food or feed components which may otherwise remain unavailable for the host (2). By-products of metabolism of certain microbiota members provide epithelial cells with butyrate, i.e., the most preferred energy source (3), and simultaneously suppress the expression of virulence factors of pathogens (4). Moreover, continuous interactions between gut microbiota and host provide stimuli for the host's immune system, keeping it activated and ready for an immediate and appropriate response to bacterial, viral, or protozoan pathogens (5). Some microbiota members may produce biologically active molecules such as γ-aminobutyric acid (6). It is therefore not surprising that the combined metabolism of the entire host microbiota considerably affects the behavior of its host up to its emotional behavior and psychology (7, 8).
The composition of gut microbiota is nowadays commonly determined by next-generation sequencing. Most frequently this is achieved by sequencing PCR products originating from the amplification of 16S rRNA genes (9–11). Less frequently it is performed by shotgun sequencing of randomly fragmented metagenomic DNA purified from intestinal contents (12, 13). Unfortunately, neither of these approaches allows for understanding protein expression of individual microbiota members in vivo. However, this information would be critical if one wants to actively modify the microbiota composition either by direct inoculation of vertebrates with probiotic cultures or by use of feed or food additives positively selecting for desired bacteria.
The correct development of normal gut microbiota is important for young individuals, which are always more susceptible to infections than adults. Colonization of the intestinal tract begins immediately after birth or hatching, with parents being the first and most important source of beneficial microbiota. However, the development of gut microbiota in chickens in commercial production is quite specific, as newly hatched chicks never come into contact with adult hens. The absence of contact with adult hens as sources of gut microbiota has therefore been overcome by the application of microbiota cultured from the fecal material of adult hens (14). However, one can never exclude that transferred gut microbiota will be free of both known and yet-unknown pathogens or will cause overgrowth of certain microbiota members in a new niche. This may result in a shift in the overall metabolism of the microbial population with negative effects on the health status of the recipient.
Recently we described gut microbiota development in the cecum throughout the life of egg-laying hens (11). The ceca of newly hatched chickens are first colonized by members of family Enterobacteriaceae (phylum Proteobacteria), followed by Lachnospiraceae and Ruminococcaceae from the phylum Firmicutes. Representatives of families Rikenellaceae, Bacteroidaceae, Prevotellaceae, and Porphyromonadaceae, all from phylum Bacteroidetes, and of family Veillonellaceae from phylum Firmicutes, appear as the last ones. The reasons for such development are not clear. It is not clear whether such development can be sped up by inoculating newly hatched chickens with gut microbiota from donors of different ages, whether all microbiota from adult birds can colonize the ceca of newly hatched chickens, or whether some microbiota members which are restricted by competing microbiota in adult hens may overgrow in the ceca of newly hatched chickens. Furthermore, the roles of individual microbiota members are far from being understood.
To begin understanding the roles of individual cecal microbiota members, we purified and identified proteins from the cecal microbiota of chickens of different ages and of 1-week-old chickens inoculated by microbiota from hens of different ages. Analysis of the proteins expressed by the gut microbiome enabled (i) characterization of the composition of the microbial community in the chicken cecum, (ii) identification of the microbiota members capable of overgrowth in recipient chickens, (iii) determination of the most important metabolic pathways operating in total microbiota, (iv) determination of the most important metabolic pathways specific for selected bacterial genera, and (v) identification of bacteria with specific biological processes such as spore formation. This knowledge allows for identification of bacteria with a high probiotic potential defined by a simple mode of administration and desired metabolic activities.
MATERIALS AND METHODS
Donor and recipient chickens.
The metaproteome was analyzed in the cecal microbiota of 1-, 3-, 16-, 28-, and 42-week-old donor birds and 7-day-old recipient chickens of the ISA Brown egg-laying hybrid (Hendrix Genetics, Boxmeer, Netherlands), which were inoculated with the cecal extracts of the donors on the day of hatching and sacrificed 7 days later. Each group consisted of 3 chickens, though we failed with protein purification and analysis in one 42-week-old donor. Proteomic data were therefore obtained for 29 chickens or hens. The cecal extracts used for the inoculation of recipient chickens were prepared by pooling an equal amount of cecal contents of 3 donors of the same age, resuspending in phosphate-buffered saline (PBS) with 0.05% cysteine, and centrifuging at 500 × g for 1 min. The supernatant was diluted 4 times in PBS with cysteine, and 200 μl was used for inoculation of newly hatched chickens. The chickens were allowed unlimited access to water and common mashed/granulated MINI feed from a local producer for chickens up to the age of 6 weeks.
Sequencing of the V3/V4 variable region of 16S rRNA genes.
Cecal content samples were homogenized using zirconia silica beads (BioSpec Products) in a MagNALyzer (Roche Diagnostics). Following homogenization, the DNA was extracted using the QIAamp DNA stool minikit according to the manufacturer's instructions (Qiagen). The DNA concentration and quality were determined spectrophotometrically, and the DNA was stored at −20°C until use. Prior to PCR, DNA samples were diluted to 5 ng/μl and used as a template in PCR with forward primer 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-MID-GT-CCTACGGGNGGCWGCAG-3′ and reverse primer 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-MID-GT-GACTACHVGGGTATCTAATCC-3′. The sequences in italic served as an index and adapter ligation, while underlined sequences allowed for amplification over the V3/V4 region of 16S rRNA genes. MID represents different sequences of 5, 6, 9, or 12 bp in length, and these were designed to differentiate samples into groups. PCR amplification and cleanup were performed using KAPA Taq HotStart PCR kit (Kapa Biosystems). In the next step, the DNA concentration was determined fluorometrically and the DNA was diluted to 100 ng/μl. Groups of 14 PCR products with different MID sequences were pooled and indexed with a Nextera XT index kit following the manufacturer's instructions (Illumina). Prior to sequencing, the concentrations of differently indexed samples were determined using a KAPA Library Quantification Complete kit (Kapa Biosystems). All indexed samples were diluted to 4 ng/μl, and 20% of phiX DNA was added. Sequencing was performed using the MiSeq reagent kit v3 and MiSEQ 2000 apparatus according to the manufacturer's instructions (Illumina).
The fastq files generated as an Illumina sequencing output were uploaded into Qiime software. Reverse reads from pair end sequencing were shortened to a length of 250 bp, and pair ends were joined. Quality trimming criteria were set to a value of 19 and no mismatch in the MID sequences. In the next step, chimeric sequences were predicted and excluded by the slayer algorithm from the analysis. The resulting sequences were then classified with RDP Seqmatch with an operational taxonomic unit (OTU) discrimination level set to 97% and de novo clustering of OTUs.
Protein purification.
Cecal contents (50 to 100 mg) were resuspended in 2 ml of 0.1% polysorbate 80, homogenized, and centrifuged for 1 min at 50 × g. Supernatant was transferred to a new tube and centrifuged at 4,000 × g for 10 min. The washing step with 0.1% polysorbate 80 was repeated 5 times. After the last washing step, the pellet was resuspended in 100 μl of 1% SDS and incubated at 100°C for 1 h. Subsequently, the protein lysate was mixed with 1.5 ml of TRI reagent and processed according to the manufacturer's recommendations (MRC).
Mass spectrometry.
Purified proteins were dissolved in 300 μl of 8 M urea and processed according to the FASP protocol (15) using a 10,000-molecular-weight-cutoff (MWCO) Vivacon 500 filtration device (Sartorius Stedim Biotech). Initial washing of proteins was performed with 8 M urea followed by centrifugation for 12 min at 12,000 × g. The reduction of the disulfide bonds was performed with 10 mM dithiothreitol for 15 min at room temperature, and acetylation was done with 50 mM iodoacetamide for 15 min at room temperature. After 3 washings with 10 mM triethylammonium bicarbonate, trypsin (Promega) was added at a 1:50 ratio, and the digestion proceeded for 16 h at 30°C.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of tryptic peptides was performed using a Dionex UltiMate 3000 RSLC liquid chromatograph connected to a LTQ-Orbitrap Velos Pro hybrid mass spectrometer (Thermo Scientific). For each analysis, 5 μg of peptide sample was used. Each sample was separated on an Easy Spray C18 column (length, 25 cm; inner diameter, 75 μm; particles, 3 μm) using a mixture of solvent A (0.1% formic acid) and solvent B (0.1% formic acid in 20:80 [vol/vol] H2O-acetonitrile [ACN]) at a flow rate of 300 nl/min and a 150-min reverse-phase gradient of 4% to 40% of solvent B. The mass spectrometer was operated in a data-dependent mode. From the mass spectra (Orbitrap analyzer; resolution 30,000 FWHM [full width at half-maximum]; mass range, 390 to 1,700 m/z), the 10 most intense peptides were fragmented using collision-induced dissociation (CID) fragmentation (normalized collision energy, 35) followed by an MS/MS scan (LTQ analyzer). Dynamic exclusion was enabled (30-s duration).
Raw LC-MS/MS data were analyzed using Proteome Discoverer v1.4. MS/MS identification was performed by the SEQUEST algorithm using the custom protein sequence database (see below). For each search, precursor and fragment mass tolerances were 10 ppm and 0.5 Da, respectively. Cysteine carbamidomethylation was set as static modification, and methionine oxidation was set as potential modification. Only peptides with a false-discovery rate of ≤5% were considered. Label-free protein quantification was based on peptide spectral match counts (PSMs).
Database construction and protein sequence annotation.
The main source of protein sequences and functional annotations for this study was the Clusters of Orthologous Groups (COG) database. However, as COG is currently lacking genera which were identified as commonly present in the chicken cecum, e.g., Dorea or Blautia (11), the COG protein database was enriched for the proteins of genera present in the STRING database but absent in the COG database. All the protein sequences were stored in FASTA format and were used as a database for SEQUEST algorithm. An additional SQLite database was created to store relations between protein identification and COG functional and NCBI taxonomy annotations. The proteins of individual samples identified and quantified using Proteome Discoverer were then annotated with both taxonomy and COG functional annotation using in-house-written R scripts. To estimate the microbial composition of samples at any taxonomic level, all spectral counts for specific taxa were summed and normalized to the spectral count of the total sample. For characterization of the metabolism of individual genera, spectral counts for particular enzymes and genera were summed from all available samples.
Identification of genera with similar biochemical pathways.
The order of individual enzymes and the order of genera were permuted based on results from hierarchical clustering. The dissimilarity of individual points was calculated as one minus Spearman's correlation of the points. To avoid false and random correlations originating from bacterial species of low abundance, only genera with at least 200 PSMs in COG metabolism category were included in this analysis. Moreover, out of all proteins present in the selected genera, only proteins/enzymes forming at least 1% of all spectral counts in at least one genus were analyzed. However, the complete data set, including the proteins detected in only a single sample and at a single PSM, can be found in Table S1 in the supplemental material.
Identification of genera with different abundances in donor and recipient chickens.
Both DNA sequencing and proteomic data were used to identify differentiating genera of donor and recipient chickens. This comparison was performed between all donor and recipient chickens; however, to correct for different microbiota composition in donors of different ages, the following filtering criteria were adopted. We analyzed separately donors or recipients of a particular age and removed all genera which were not detected in at least 3 donor or 3 recipient chickens of a particular group. For example, negative selection was applied for a genus which was detected twice in three 3-week-old donor chickens and once in 3 corresponding recipients. This was done to avoid inclusion of underrepresented genera at the detection limit of the protocols used. The percent representation of each genus in all donors or recipients was determined, and significant differences were calculated using Student's t test. Finally we excluded genera which did not exceed a 0.25% representation and did not pass a 3-fold difference in abundance in donor and recipient chickens.
Ethics statement.
The handling of animals in the study was performed in accordance with current Czech legislation (Animal Protection and Welfare Act no. 246/1992 Collection of the Government of the Czech Republic). The specific experiments were approved by the Ethics Committee of the Veterinary Research Institute and then by the Committee for Animal Welfare of the Ministry of Agriculture of the Czech Republic (permit no. MZe1449).
RESULTS
Complete proteome in the cecal contents of donor chickens and hens and in 1-week-old recipient chickens.
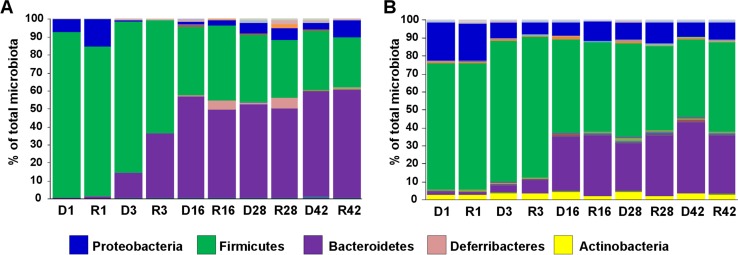
Altogether, 14,039 proteins were detected and assigned to bacterial species and COG categories (see Table S1 in the supplemental material). When these data were used for characterization of the composition of chicken cecal microbiota, the same developmental pattern as recorded by sequencing of the 16S rRNA genes was observed (Fig. 1).
FIG 1.
Microbiota composition in the ceca of donor chickens and hens and recipient chickens. Recipient chickens were inoculated on the day of hatching with the cecal extracts of donor hens (ages are indicated) and sacrificed 7 days later. (A) Cecal microbiota composition determined by sequencing of the V3/V4 variable regions of 16S rRNA genes. (B) Cecal microbiota composition determined by protein mass spectrometry. Blue, Proteobacteria; green, Firmicutes; purple, Bacteroidetes; pink, Deferribacteres; yellow, Actinobacteria. Deferribacteres were represented mainly by the genus Mucispirillum. However, since the complete proteome of this genus is not known, its proteins were not present in the protein database, and its overgrowth in 7-day-old chickens inoculated with cecal microbiota of 16- and 28-day-old donors could not be confirmed using protein mass spectrometry. D, donor birds aged 1, 3, 16, 28, and 42 weeks. R, recipient chickens receiving microbiota from birds aged 1, 3, 16, 28, and 42 weeks.
Genera represented differently in donor and recipient chickens.
Comparison of the microbiota compositions in donor and recipient chickens showed that the ceca of 7-day-old chickens could be colonized by most bacterial species, with the following exceptions. Genera which were difficult to transfer from donors to recipients included Fusobacterium, Methanobrevibacter, Paraprevotella, and Rikenella, since these bacteria were detected significantly less frequently in the microbiota of the recipients than in the donors. On the other hand, Flexispira, Campylobacter, Mucispirillum, Nitratiruptor, Ornithinibacillus, Helicobacter, Megamonas, Wolinella, Solibacillus, and Caldicellulosiruptor were detected in the microbiota of the recipients in a significantly higher abundance than in the donors (Table 1).
TABLE 1.
Genera with different abundances in donor and recipient chickens
| Genus | % (mean ± SD) in: |
Fold difference | Protocola | |
|---|---|---|---|---|
| Donors | Recipients | |||
| Flexispira | 0.03 ± 0.03 | 1.61 ± 1.53 | 53.94 | DNA |
| Campylobacter | 0.01 ± 0.01 | 0.35 ± 0.21 | 51.02 | DNA |
| Mucispirillum | 0.36 ± 0.37 | 6.59 ± 5.28 | 18.13 | DNA |
| Nitratiruptor | 0.04 ± 0.08 | 0.29 ± 0.15 | 7.17 | Protein |
| Ornithinibacillus | 0.07 ± 0.04 | 0.45 ± 0.38 | 6.70 | Protein |
| Helicobacter | 0.69 ± 0.73 | 3.71 ± 5.14 | 5.38 | DNA |
| Megamonas | 1.84 ± 1.99 | 7.24 ± 7.24 | 3.92 | Protein |
| Wolinella | 0.13 ± 0.15 | 0.49 ± 0.28 | 3.84 | Protein |
| Solibacillus | 0.09 ± 0.06 | 0.33 ± 0.01 | 3.82 | Protein |
| Caldicellulosiruptor | 0.08 ± 0.10 | 0.27 ± 0.26 | 3.19 | Protein |
| Fusobacterium | 0.45 ± 0.40 | 0.10 ± 0.10 | 0.22 | Protein |
| Methanobrevibacter | 0.77 ± 0.29 | 0.05 ± 0.10 | 0.07 | Protein |
| Paraprevotella | 0.54 ± 0.56 | 0.03 ± 0.09 | 0.06 | DNA |
| Rikenella | 0.79 ± 0.04 | 0.00 ± 0.00 | 0.00 | DNA |
Indicates whether the significant difference was recorded using 16S rRNA sequencing or protein mass spectrometry.
Identification of gut microbiota genera with similar metabolism.
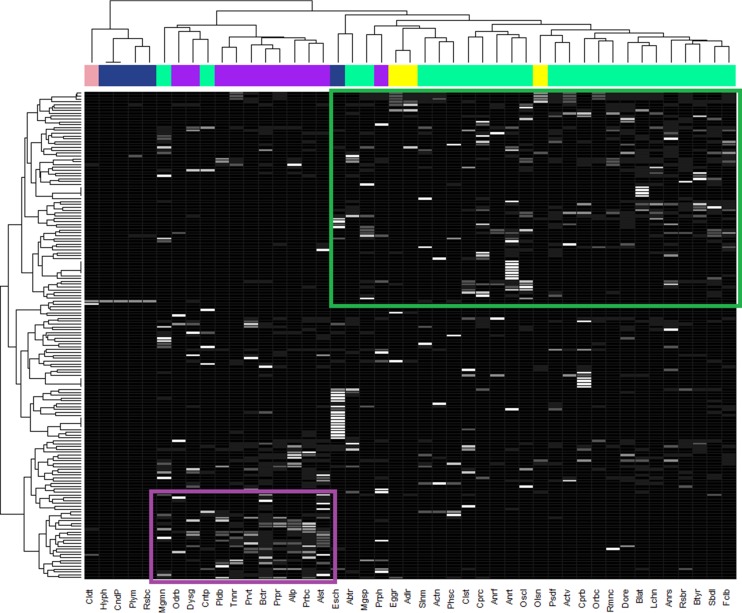
To address the reasons explaining differences in microbiota composition in donor and recipient chickens following microbiota transplantation, we analyzed expressed metabolic pathways in major genera. Hierarchical clustering showed that genera belonging to the same phyla clustered close to each other; i.e., they exhibited similar protein expression. Separate clusters were formed by (i) Firmicutes and Actinobacteria, (ii) Bacteroidetes, and (iii) Proteobacteria excluding E. coli, which clustered within Firmicutes. Two genera from the phylum Firmicutes, i.e., Megamonas and Centipeda, clustered with Bacteroidetes (Fig. 2).
FIG 2.
Bacterial genera and expressed enzymes involved in metabolism. The heat map was generated using total PSMs detected for each genus and particular protein. The heat map therefore integrates both relative expression within a given species and its numerical presence. color scaling is over the rows. Green box, enzymes characteristic of colonizers from the phylum Firmicutes. Purple box, enzyme characteristic of colonizers from the phylum Bacteroidetes. Color coding at the top: green, Firmicutes; purple, Bacteroidetes; blue, Proteobacteria; yellow, Actinobacteria; pink, Deferribacteres. Abtr, Abiotrophia; Actn, Acetonema; Actv, Acetivibrio; Adlr, Adlercreutzia; Allp, Alloprevotella; Alst, Alistipes; Anrf, Anaerofustis; Anrs, Anaerostipes; Anrt, Anaerotruncus; Bctr, Bacteroides; Blat, Blautia; Btyr, Butyrivibrio; Cldt, Calditerrivibrio; Clst, Clostridium; CndP, “Candidatus Pelagibacter”; Cntp, Centipeda; Cprb, Coprobacillus; Cprc, Coprococcus; Dore, Dorea; Dysg, Dysgonomonas; Eggr, Eggerthella; Esch, Escherichia; Fclb, Faecalibacterium; Hyph, Hyphomicrobium; Lchn, Lachnoanaerobaculum; Mgmn, Megamonas; Mgsp, Megasphaera; Odrb, Odoribacter; Olsn, Olsenella; Orbc, Oribacterium; Oscl, Oscillibacter; Phsc, Phascolarctobacterium; Pldb, Paludibacter; Plym, Polymorphum; Prbc, Parabacteroides; Prph, Porphyromonas; Prpr, Paraprevotella; Prvt, Prevotella; Psdf, Pseudoflavonifractor; Rmnc, Ruminococcus; Rsbc, Roseobacter; Rsbr, Roseburia; Sbdl, Subdoligranulum; Slnm, Selenomonas; Tnnr, Tannerella. For protein identification, see Fig. S1 in the supplemental material.
Major colonizers and dominant biological processes.
For the next analysis, we reduced the number of analyzed genera to those belonging to the 10 most abundant genera in at least one group of donor or recipient chickens. The classification of detected proteins into major biological processes showed that Campylobacter, Bacillus, Anaerotruncus, and Oscillibacter expressed flagellar or chemotaxis proteins. Tfp pilus proteins were detected in Parabacteroides, Prevotella, and Subdoligranulum, and proteins of the type II, Sec-dependent secretion system were detected in the majority of representatives from the phyla Bacteroidetes and Firmicutes. Small, acid-soluble spore protein was detected in Anaerostipes, Anaerotruncus, Pseudoflavonifractor, and Dorea. Spore germination protein YaaH was detected in Bacillus, uncharacterized spore protein YtfJ was detected in Blautia, Subdoligranulum, and Anaerotruncus, and spore coat protein CotF was expressed in Anaerostipes and Blautia.
Major colonizers and dominant metabolic processes.
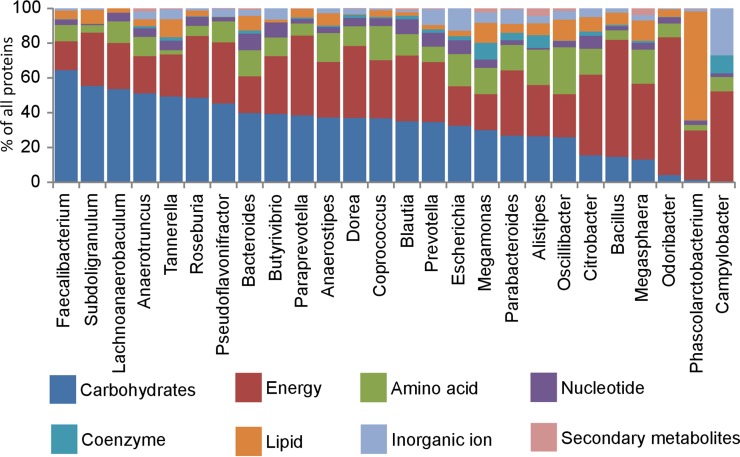
Classification of detected proteins into major metabolic categories showed that Odoribacter, Phascolarctobacterium, and Campylobacter nearly did not express enzymes for carbohydrate metabolism. Odoribacter complemented the absence of carbohydrate metabolism by high expression of proteins involved in energy metabolism, with ATP phosphoenolpyruvate carboxykinase being the most abundant protein of Odoribacter in this category. Phascolarctobacterium complemented the absence of carbohydrate metabolism by an increase in lipid metabolism, with methylmalonyl coenzyme A (methylmalonyl-CoA) mutase and methylmalonyl-CoA carboxyltransferase being the most abundant lipid metabolism proteins. Campylobacter complemented the absence of carbohydrate metabolism by an increase in metabolic processes associated with energy production and transport of inorganic ions across membranes (Fig. 3).
FIG 3.
Classification of expressed proteins in the major colonizers of the chicken cecum within the category “metabolism.” Individual genera are ordered according to decreasing preference for carbohydrate metabolism.
Metabolic characteristics of Firmicutes.
Firmicutes were characterized by the acquisition of Fe2+ by the FeoAB transport system and lipid metabolism using acyl-CoA intermediates. Firmicutes used the ATP-binding cassette (ABC) and phosphotransferase system (PTS) transporters for the uptake of carbohydrates, glycerol, and phosphate (Fig. 2, green box). The expression of l-fucose isomerase was also quite characteristic of Firmicutes, since it was found in Anaerostipes, Blautia, Butyrivibrio, Coprococcus, Dorea, Pseudoflavonifractor, Subdoligranulum, and Megamonas. Some unique proteins expressed by certain representatives of Firmicutes included the SudA and SudB subunits of sulfide dehydrogenase in Roseburia or CO dehydrogenase/acetyl-CoA synthase in Blautia.
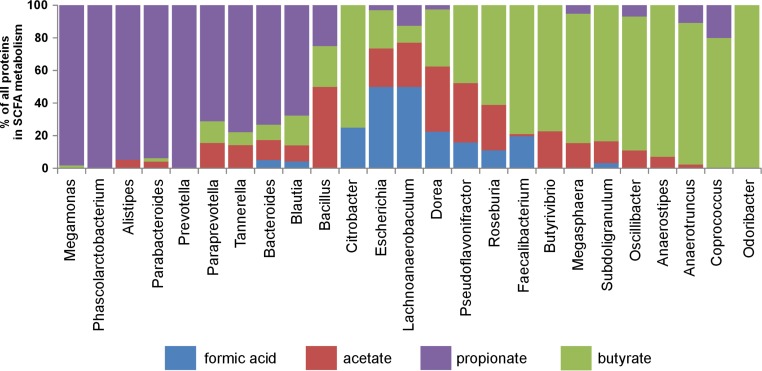
Since short-chain fatty acids (SCFA) play an important role in gut physiology (3), we specifically analyzed pathways leading to butyrate and propionate synthesis. There were 3 potential pathways leading to butyrate production. The first and major one was based on acetyl-CoA acetyltransferase. The second required acetyl/propionyl-CoA carboxylase, and the last one was dependent on butanol dehydrogenase.
Anaerotruncus, Faecalibacterium, Megasphaera, Oscillibacter, Subdoligranulum, and Butyrivibrio expressed acetyl-CoA acetyltransferase and at least one of two additional enzymes necessary for butyrate production, i.e., 3-hydroxyacyl-CoA dehydrogenase or enoyl-CoA hydratase. These three enzymes also clustered close to each other, indicating their tight coexpression (Fig. 2). In Butyrivibrio and Subdoligranulum, we also detected expression of butyrate kinase, an enzyme involved in the last step of one of the four potential butyrate production pathways (16).
Butyrate could also be produced by acetyl/propionyl-CoA carboxylase, which was detected in Faecalibacterium, Phascolarctobacterium, and Subdoligranulum. Interestingly, in genomic sequences of Phascolarctobacterium and Subdoligranulum proteins of this COG were designated glutaconyl-CoA decarboxylase, a different enzyme also known to be involved in butyrate production from glutarate (16).
The final butyrate production pathway was dependent on class IV alcohol dehydrogenase. This protein was moderately expressed in Anaerostipes, Anaerotruncus, Blautia, Dorea, Oscillibacter, Pseudoflavonifractor, and Subdoligranulum. Except for Subdoligranulum, in genomic sequences of the remaining genera this protein was designated NADPH-dependent butanol dehydrogenase.
Megamonas, Phascolarctobacterium, Blautia, and Lachnoanaerobaculum were the likely major propionate producers in Firmicutes. However, while propionate production in Megamonas and Phascolarctobacterium was via methylmalonyl-CoA mutase, epimerase, and decarboxylase, Blautia and Lachnoanaerobaculum expressed acetyl-CoA carboxylase.
Megamonas is one of the few members of phylum Firmicutes which is present mainly in the cecal microbiota of adult hens (11). Since it was also capable of overgrowth in recipient chickens, we analyzed its metabolism in greater detail. Megamonas uniquely expressed sugar alcohol permeases specific for sorbitol and galactitol and trihydroxycyclohexane-1,2-dione dehydratase involved in inositol metabolism. Megamonas expressed the Na+/melibiose symporter and α-galactosidase required for the digestion of the α-1,6 linkage between galactose and glucose present in melibiose. Megamonas expressed alanine dehydrogenase, which converts alanine into pyruvate and ammonia. Megamonas expressed the flavoprotein subunit of succinate dehydrogenase in an operon with a fumarase, but not the Fe-S subunit as in representatives of Bacteroidetes. Megamonas also highly expressed Ni-Fe hydrogenase, cobalamin-binding methylmalonyl-CoA mutase, and outer membrane cobalamin receptor.
Metabolic characteristics of Bacteroidetes.
Bacteroidetes were characterized by the expression of lipid metabolism dependent on acyl carrier proteins, expression of the pentose cycle, and respiration of fumarate. Of some rather unusual proteins, Parabacteroides expressed 3′-phosphoadenosine 5′-phosphosulfate sulfotransferase and Bacteroides expressed sulfate adenylyltransferase, which are both involved in sulfate assimilation. Alistipes expressed glutamate decarboxylase, metabolizing glutamate into γ-aminobutyric acid (GABA).
Representatives of phylum Bacteroidetes exhibited extended enzymatic activities in polysaccharide degradation, since α-amylase, α-1,2-mannosidase, endo-1,4-β-mannosidase, or glycogen-debranching α-1,6-glucosidase was specifically expressed in Bacteroidetes and not in Firmicutes (Fig. 2, purple box). Bacteroides, Alistipes, Paraprevotella, and Tannerella were characterized by high expression of xylose isomerase. In addition, Bacteroides expressed sucrose-6-phosphate hydrolase, α-l-fucosidase, α-glucosidase, glycosyl hydrolase, and d-arabinose 5-phosphate isomerase.
Representatives of the phylum Bacteroidetes were mainly propionate producers. For propionate production, Alistipes, Bacteroides, Parabacteroides, Paraprevotella, Prevotella, and Tannerella expressed cobalamin-binding methylmalonyl-CoA mutase and/or methylmalonyl-CoA epimerase. In Alistipes and Tannerella, the methylmalonyl-CoA epimerase gene formed an operon with the acetyl-CoA carboxylase gene. Although the COG annotation of the last protein was acetyl-CoA carboxylase, in Parabacteroides this gene was annotated as methylmalonyl-CoA decarboxylase, indicating its alternative function.
Despite high expression of enzymes leading to propionate production, representatives of Bacteroidetes could also produce butyrate. Paraprevotella expressed acetyl-CoA acetyltransferase. Bacteroides and Tannerella expressed acetyl/propionyl-CoA carboxylase (glutaconyl-CoA decarboxylase), and Bacteroides, Parabacteroides, and Tannerella expressed alcohol dehydrogenase, designated butanol dehydrogenase in the Tannerella genome. However, all these enzymes were expressed at a much lower level in Bacteroidetes than enzymes catalyzing propionate biosynthesis (Fig. 4).
FIG 4.
Expression of enzymes involved in SCFA biosynthesis. PSMs of proteins belonging to the COGs specified below were summarized for the major cecum colonizers and main SCFA (butyrate, propionate, acetate, and formic acid) biosynthesis pathways. For butyrate biosynthesis, PSMs of proteins belonging to the following COGs were summarized: COG4770, acetyl/propionyl-CoA carboxylase; COG0183, acetyl-CoA acetyltransferase; COG1979, alcohol dehydrogenase YqhD; COG1454, class IV alcohol dehydrogenase; COG3426, butyrate kinase; COG1250, 3-hydroxyacyl-CoA dehydrogenase; and COG1024, enoyl-CoA hydratase. For propionate biosynthesis, PSMs of proteins belonging to the following COGs were summarized: COG0777, acetyl-CoA carboxylase; COG4799, acetyl-CoA carboxylase; COG0439, biotin carboxylase; COG0346, catechol 2,3-dioxygenase; COG2185, methylmalonyl-CoA mutase; COG1884, methylmalonyl-CoA mutase; and COG4869, propanediol utilization protein. For acetate biosynthesis, PSMs of proteins belonging to the following COGs were summarized: COG0282, acetate kinase; COG0280, phosphotransacetylase; and COG1012, NAD-dependent aldehyde dehydrogenase. For formic acid production, PSMs of proteins belonging to the following COG were summarized: COG1882, pyruvate-formate lyase.
Reduced NAD(P)H from fermentation could be oxidized using fumarate as an electron acceptor, since Alistipes, Bacteroides, Parabacteroides, Paraprevotella, and Prevotella expressed both Fe-S and flavoprotein subunits of fumarate reductase. Apart from the proton gradient, Alistipes, Bacteroides, Odoribacter, Paraprevotella, Parabacteroides, Prevotella, and Tannerella also expressed the Nqr system of Na+-translocating NADH:ubiquinone oxidoreductase, thus generating an Na+ gradient across the cytoplasmic membrane.
The last group of proteins specific to Bacteroidetes included ExbBD, TonB, and multiple outer membrane receptors. Outer membrane receptor proteins belonging to COG1629 were detected 105 times, and 103 of them were expressed by different representatives of Bacteroidetes, including Alistipes (5 different receptors of this COG), Bacteroides (10 different receptors of this COG), Odoribacter (n = 1), Parabacteroides (n = 13), Prevotella (n = 6), and Tannerella (n = 6). Outer membrane cobalamin receptors (COG4206) were detected 41 times, and 39 of them were expressed by different representatives of Bacteroidetes, including Alistipes (n = 1), Bacteroides (n = 6), Odoribacter (n = 1), Parabacteroides (n = 1), Paraprevotella (n = 1), Prevotella (n = 1), and Tannerella (n = 1). The outer membrane receptors for ferrienterochelin and colicins (COG4771) were detected 79 times, and 68 of them were expressed by different representatives of Bacteroidetes, including Alistipes (n = 4), Bacteroides (n = 7), Parabacteroides (n = 7), Prevotella (n = 7), and Tannerella (n = 1).
The above-mentioned receptors in the outer membrane are commonly associated with TonB, which spans the periplasmic space, and ExbBD proteins present in the cytoplasmic membrane (17, 18). Not surprisingly, biopolymer transport protein ExbB/TolQ (COG0811), biopolymer transport protein ExbD (COG0848), and TonB (COG0810) were all detected in Alistipes, Bacteroides, Parabacteroides, and Tannerella.
DISCUSSION
In this study, we characterized the gut microbiome following microbiota transplantation and protein expression of the main gut colonizers. This information was used to explain colonization patterns and to predict the most promising probiotic genera suitable for colonization of the ceca of newly hatched chickens.
Flexispira, Campylobacter, Mucispirillum, Nitratiruptor, Ornithinibacillus, Helicobacter, Megamonas, Wolinella, Solibacillus, and Caldicellulosiruptor were capable of efficient colonization of newly hatched chickens, while Fusobacterium, Methanobrevibacter, Paraprevotella, and Rikenella were difficult to transfer from donors to recipients. The fact that we failed to transfer members of the latter group from donors to recipients could have been caused either by the inability of these genera to colonize the environment of ceca of the youngest chickens or by their extreme sensitivity to oxygen and loss of viability during cecal extract preparation. Reasons for the overgrowth of the former genera are unclear. However, Flexispira, Campylobacter, Helicobacter, and Wolinella all belong to the order Campylobacterales, which has carbohydrate-independent metabolism (Fig. 3) (19, 20). Since the carbohydrate supply in newly hatched chickens is limited and chickens up to the age of 3 or 4 days live from resorption of egg yolk rich in fat and protein (21), the carbohydrate-independent metabolic pathway may allow Campylobacterales to overgrow.
Overgrowth of Megamonas in inoculated chickens should be also considered with concern due to its high-level expression of cobalamin receptor and cobalamin-dependent methylmalonyl-CoA mutase. High demands for cobalamin may lead to reduced availability of cobalamin for young chickens. In addition, Megamonas expressed alanine dehydrogenase, resulting in ammonia production, which may increase the pH of the cecal contents and affect metabolism of both host epithelial cells and other microbiota members (22). This means that care should be taken during microbiota transplantation because the microbiota may behave one way in a well-established community and differently when repopulating the poorly colonized intestinal tract of a recipient.
Similar to findings in previous studies, GroEl/GroES, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), glutamate dehydrogenase, electron transfer proteins, rubrerythrin, or NifU homologs were detected as proteins highly expressed by gut microbiota (23–26). However, in this study we attempted to characterize protein expression at least in the major gut colonizers to a deeper extent, as 29 processed samples from donor and recipient chickens were used.
Alistipes, Bacteroides, Parabacteroides, and Tannerella expressed ExbBD, TonB, and multiple receptors classified as required for cobalamin, iron, or enterochelin transport. Interestingly, one such receptor identified in Caulobacter has been shown to be involved in transport of maltotetraose or larger maltodextrins in an ExbBD- and TonB-dependent fashion (18). Since Bacteroides is known to metabolize oligosaccharides of host origin derived from sulfated glycoproteins, e.g., mucins (27), we predict that at least some of these receptors together with ExbBD and TonB are used for the transport of oligosaccharides across the outer membrane. The Na+ gradient generated by the Nqr system may power the transport of oligosaccharides. Moreover, Bacteroides can remove sulfate residues from sulfated glycoproteins (28), and consistent with this, we detected enzymes enabling sulfate assimilation in Bacteroides and Parabacteroides.
Rather unexpectedly, Alistipes expressed glutamate decarboxylase, resulting in the production of GABA from glutamate. GABA, a neurotransmitter, can be produced from glutamate by a rather limited number of isolates of the genera Lactobacillus and Bifidobacterium in vitro (6). Another Lactobacillus strain affected the behavior of mice by activating the transcription of GABA receptors (7). Studies in humans showed that the presence of Alistipes correlated with depression (8), and Alistipes was increased in the fecal microbiota of frail residents of long-term care facilities (10). This might be associated with GABA production, though it is clear that the mere presence of the enzyme does not guarantee that GABA is released into the gut lumen.
Short-chain fatty acids (SCFA) play an important role in gut physiology. SCFA, and butyrate in particular, serve as a source of energy for colonic epithelial cells (3). In addition, SCFA negatively affect expression of virulence factors of bacterial pathogens (4). This is why we focused on the expression of enzymes leading to the production of propionate or butyrate in greater detail. Although additional organic acids may be released as terminal products of bacterial fermentation, enzymes involved in propionate metabolism dominated over those involved in butyrate metabolism in all representatives of Bacteroidetes. Megamonas and Phascolarctobacterium, the two genera from Firmicutes associated with the microbiota of adult hens (11), also expressed enzymes mainly required for propionate production (Fig. 4). Of the bacteria characteristic of young chickens, Blautia preferentially expressed enzymes of propionate rather than butyrate biosynthesis. Bacteroides was reported to produce both propionate and succinate as terminal products of its metabolism (29, 30), and the availability of cobalamin was shown to affect the propionate/succinate ratio in Prevotella. In the presence of cobalamin, propionate was a major terminal metabolic by-product, while succinate accumulated with decreasing cobalamin availability (31). This indicates that cobalamin availability is central for the conversion of succinate into propionate by cobalamin-dependent methylmalonyl-CoA mutase in vivo.
Roseburia, Faecalibacterium, Butyrivibrio, Megasphaera, Subdoligranulum, Oscillibacter, Anaerostipes, Anaerotruncus, Coprococcus, and Odoribacter expressed enzymes favoring the production of butyrate over propionate. Butyrate production by Firmicutes might at least partially explain their dominance in young, quickly growing chickens, which have a high butyrate requirement in order to supply the cells of the enlarging intestine. However, once the final body weight at around week 18 of life is achieved (11), maximal butyrate production is no longer required. Instead, digestion of complex polysaccharides by representatives of Bacteroidetes results in the production of both propionate and butyrate, which might represent an advantageous balance between sustained existence and acquisition of enough energy from available nutrients. This would explain the gradual succession of Firmicutes from the ceca of approximately 4-week-old chickens and establishment of a balance in the ratio of Firmicutes and Bacteroides coinciding with sexual maturity (11).
However, there might be at least two additional factors affecting the colonization of the ceca of newly hatched chickens. First, all Firmicutes expressed l-fucose isomerase, while Bacteroides expressed xylose isomerase. Fucosylated residues can be found as part of mucins with a different spatial distribution along the intestinal tract (32). Moreover, fucose residues are used as receptors of many bacterial adhesins (33). On the other hand, xylose is the first sugar attaching, e.g., chondroitin, to serine residues in proteoglycans (34). A different spatial distribution or age-dependent expression of fucosylated or xylosylated chicken glycoproteins then may positively select for particular microbiota members.
The last factor explaining the gradual colonization of the chicken cecum is that of spore formation. Anaerostipes, Anaerotruncus, Bacillus, Blautia, Dorea, Pseudoflavonifractor, and Subdoligranulum expressed spore-forming proteins. This means that although these genera are strict anaerobes, they can survive in feed or water as spores and enter the digestive tract immediately after the start of independent foraging. Finally, bacterial genera which are capable of both butyrate production and spore formation may have characteristics expected in probiotic strains. Specifically, based on data in this study, Anaerostipes, Anaerotruncus, and Subdoligranulum might therefore be suitable probiotic candidates for oral inoculation of newly hatched chickens.
Supplementary Material
ACKNOWLEDGMENT
We thank Peter Eggenhuizen for his English language corrections.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03473-15.
REFERENCES
- 1.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.Fleming SE, Fitch MD, DeVries S, Liu ML, Kight C. 1991. Nutrient utilization by cells isolated from rat jejunum, cecum and colon. J Nutr 121:869–878. [DOI] [PubMed] [Google Scholar]
- 4.Boyen F, Haesebrouck F, Vanparys A, Volf J, Mahu M, Van Immerseel F, Rychlik I, Dewulf J, Ducatelle R, Pasmans F. 2008. Coated fatty acids alter virulence properties of Salmonella Typhimurium and decrease intestinal colonization of pigs. Vet Microbiol 132:319–327. doi: 10.1016/j.vetmic.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 5.O'Hara AM, Shanahan F. 2006. The gut flora as a forgotten organ. EMBO Rep 7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett E, Ross RP, O'Toole PW, Fitzgerald GF, Stanton C. 2012. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol 113:411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- 7.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. 2011. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A 108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naseribafrouei A, Hestad K, Avershina E, Sekelja M, Linlokken A, Wilson R, Rudi K. 2014. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil 26:1155–1162. doi: 10.1111/nmo.12378. [DOI] [PubMed] [Google Scholar]
- 9.Li K, Bihan M, Yooseph S, Methe BA. 2012. Analyses of the microbial diversity across the human microbiome. PLoS One 7:e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, Fitzgerald GF, Deane J, O'Connor M, Harnedy N, O'Connor K, O'Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O'Toole PW. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 11.Videnska P, Sedlar K, Lukac M, Faldynova M, Gerzova L, Cejkova D, Sisak F, Rychlik I. 2014. Succession and replacement of bacterial populations in the caecum of egg laying hens over their whole life. PLoS One 9:e115142. doi: 10.1371/journal.pone.0115142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sergeant MJ, Constantinidou C, Cogan TA, Bedford MR, Penn CW, Pallen MJ. 2014. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS One 9:e91941. doi: 10.1371/journal.pone.0091941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Methner U, Barrow PA, Martin G, Meyer H. 1997. Comparative study of the protective effect against Salmonella colonisation in newly hatched SPF chickens using live, attenuated Salmonella vaccine strains, wild-type Salmonella strains or a competitive exclusion product. Int J Food Microbiol 35:223–230. doi: 10.1016/S0168-1605(96)01236-6. [DOI] [PubMed] [Google Scholar]
- 15.Wisniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 16.Vital M, Howe AC, Tiedje JM. 2014. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5:e00889. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Held KG, Postle K. 2002. ExbB and ExbD do not function independently in TonB-dependent energy transduction. J Bacteriol 184:5170–5173. doi: 10.1128/JB.184.18.5170-5173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neugebauer H, Herrmann C, Kammer W, Schwarz G, Nordheim A, Braun V. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J Bacteriol 187:8300–8311. doi: 10.1128/JB.187.24.8300-8311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Line JE, Hiett KL, Guard-Bouldin J, Seal BS. 2010. Differential carbon source utilization by Campylobacter jejuni 11168 in response to growth temperature variation. J Microbiol Methods 80:198–202. doi: 10.1016/j.mimet.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Mohammed KA, Miles RJ, Halablab MA. 2004. The pattern and kinetics of substrate metabolism of Campylobacter jejuni and Campylobacter coli. Lett Appl Microbiol 39:261–266. doi: 10.1111/j.1472-765X.2004.01574.x. [DOI] [PubMed] [Google Scholar]
- 21.Noy Y, Sklan D. 1998. Yolk utilisation in the newly hatched poult. Br Poult Sci 39:446–451. doi: 10.1080/00071669889042. [DOI] [PubMed] [Google Scholar]
- 22.Davila AM, Blachier F, Gotteland M, Andriamihaja M, Benetti PH, Sanz Y, Tome D. 2013. Re-print of “Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host.” Pharmacol Res 69:114–126. doi: 10.1016/j.phrs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, Jansson JK. 2009. Shotgun metaproteomics of the human distal gut microbiota. ISME J 3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 24.Gosalbes MJ, Durban A, Pignatelli M, Abellan JJ, Jimenez-Hernandez N, Perez-Cobas AE, Latorre A, Moya A. 2011. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One 6:e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolmeder CA, de Been M, Nikkila J, Ritamo I, Matto J, Valmu L, Salojarvi J, Palva A, Salonen A, De Vos WM. 2012. Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS One 7:e29913. doi: 10.1371/journal.pone.0029913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, Underwood A, Gielbert A, Woodward MJ, Petrovska L. 2014. Metaproteomics analysis reveals the adaptation process for the chicken gut microbiota. Appl Environ Microbiol 80:478–485. doi: 10.1128/AEM.02472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berry D, Mader E, Lee TK, Woebken D, Wang Y, Zhu D, Palatinszky M, Schintlmeister A, Schmid MC, Hanson BT, Shterzer N, Mizrahi I, Rauch I, Decker T, Bocklitz T, Popp J, Gibson CM, Fowler PW, Huang WE, Wagner M. 2015. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci U S A 112:E194–E203. doi: 10.1073/pnas.1420406112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjdia A, Martens EC, Gordon JI, Berteau O. 2011. Sulfatases and a radical S-adenosyl-l-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem 286:25973–25982. doi: 10.1074/jbc.M111.228841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adamberg S, Tomson K, Vija H, Puurand M, Kabanova N, Visnapuu T, Jogi E, Alamae T, Adamberg K. 2014. Degradation of fructans and production of propionic acid by Bacteroides thetaiotaomicron are enhanced by the shortage of amino acids. Front Nutr 1:21. doi: 10.3389/fnut.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isar J, Agarwal L, Saran S, Saxena RK. 2006. Succinic acid production from Bacteroides fragilis: process optimization and scale up in a bioreactor. Anaerobe 12:231–237. doi: 10.1016/j.anaerobe.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Strobel HJ. 1992. Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl Environ Microbiol 58:2331–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmen Larsson JM, Thomsson KA, Rodriguez-Pineiro AM, Karlsson H, Hansson GC. 2013. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastrointest Liver Physiol 305:G357–G363. doi: 10.1152/ajpgi.00048.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juge N. 2012. Microbial adhesins to gastrointestinal mucus. Trends Microbiol 20:30–39. doi: 10.1016/j.tim.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Mikami T, Kitagawa H. 2013. Biosynthesis and function of chondroitin sulfate. Biochim Biophys Acta 1830:4719–4733. doi: 10.1016/j.bbagen.2013.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.