Abstract
Xylella fastidiosa is a xylem-limited phytopathogenic bacterium endemic to the Americas that has recently emerged in Asia and Europe. Although this bacterium is classified as a quarantine organism in the European Union, importation of plant material from contaminated areas and latent infection in asymptomatic plants have engendered its inevitable introduction. In 2012, four coffee plants (Coffea arabica and Coffea canephora) with leaf scorch symptoms growing in a confined greenhouse were detected and intercepted in France. After identification of the causal agent, this outbreak was eradicated. Three X. fastidiosa strains were isolated from these plants, confirming a preliminary identification based on immunology. The strains were characterized by multiplex PCR and by multilocus sequence analysis/typing (MLSA-MLST) based on seven housekeeping genes. One strain, CFBP 8073, isolated from C. canephora imported from Mexico, was assigned to X. fastidiosa subsp. fastidiosa/X. fastidiosa subsp. sandyi. This strain harbors a novel sequence type (ST) with novel alleles at two loci. The two other strains, CFBP 8072 and CFBP 8074, isolated from Coffea arabica imported from Ecuador, were allocated to X. fastidiosa subsp. pauca. These two strains shared a novel ST with novel alleles at two loci. These MLST profiles showed evidence of recombination events. We provide genome sequences for CFBP 8072 and CFBP 8073 strains. Comparative genomic analyses of these two genome sequences with publicly available X. fastidiosa genomes, including the Italian strain CoDiRO, confirmed these phylogenetic positions and provided candidate alleles for coffee plant adaptation. This study demonstrates the global diversity of X. fastidiosa and highlights the diversity of strains isolated from coffee plants.
INTRODUCTION
Xylella fastidiosa is a Gram-negative gammaproteobacterium limited to the xylem of host plants and transmitted by sap-feeding insects. This bacillus has been associated with economically disastrous diseases such as citrus variegated chlorosis (CVC) in orange trees, Pierce's disease (PD) in vineyards, and leaf scorch of olive trees. X. fastidiosa strains have been found to infect at least 309 plant species (1), mostly in the Americas. Nevertheless, X. fastidiosa strains have been isolated from nashi pear trees and grapevines in Taiwan (2, 3), from grapevines and almond trees in Iran (4), and recently from olive trees, almond trees, oleanders, and some other hosts in Italy (1, 5, 6). Detections of Xylella strains from plant species grown in other locations, such as Kosovo or Turkey, have not been not confirmed by strain isolation (1). It is important to note that most X. fastidiosa-colonized plants are asymptomatic (7).
X. fastidiosa is genetically diverse and has been divided into six subspecies (8–11), but only two subspecies (X. fastidiosa subsp. fastidiosa and X. fastidiosa subsp. multiplex) are currently taxonomically valid (12, 13). (i) X. fastidiosa subsp. fastidiosa causes PD and infects a large host range, including grapevine, almond trees, alfalfa, and maple (9). It has long been assumed that this subspecies originated from the United States, but recently it was proposed that a single genotype was introduced into the United States from Central America in the 1880s (14). (ii) X. fastidiosa subsp. multiplex is associated with scorch diseases of a range of trees, including almond, peach, and oak (1). This subspecies is thought to be native to temperate climates of northern America. X. fastidiosa subsp. multiplex causing plum leaf scald was first detected in 1935 in Argentina and then in Paraguay and Brazil. It was supposedly introduced from the United States (15). Once introduced into Brazil, these plum-infecting strains are suspected of recombining with native X. fastidiosa subsp. pauca strains, generating genetic variation which would have facilitated a switch from native hosts toward citrus and coffee (16). (iii) X. fastidiosa subsp. sandyi causes oleander leaf scorch (OLS). It is also supposed to have been introduced into the United States from Central America (9, 17). (iv) X. fastidiosa subsp. tashke was isolated from an ornamental tree (Chitalpa tashkentensis Elias and Wisura) in the United States (10) but has not been reported since that first study and remains poorly described. (v) X. fastidiosa subsp. morus infects mulberry (Morus spp.) and might have been generated by intersubspecific recombination events between X. fastidiosa subsp. fastidiosa and X. fastidiosa subsp. multiplex strains (11). (vi) Finally, X. fastidiosa subsp. pauca infects mostly Citrus spp. and Coffea spp. (18). Strains from this subspecies have thus far been isolated mainly from South America. Recently a variant of X. fastidiosa subsp. pauca was isolated from coffee and oleander plants in Costa Rica (19). Strains of X. fastidiosa recently isolated from olive trees in Argentina and in Italy are similar to this new variant of X. fastidiosa subsp. pauca (6, 20).
Coffee leaf scorch (CLS), due to X. fastidiosa, was first identified in 1995 in Brazil (21) and later in Latin American coffee-producing countries such as Costa Rica (22). Symptoms of CLS include drying of infected branches, shortening of internode regions, decreased fruit size, chlorosis, and early senescence of leaves. It affects plant productivity but rarely leads to plant death (23). Within the Rubiaceae family, the genus Coffea includes 124 described species according to Davis and colleagues (24, 25), but only two, Coffea arabica (65%) and Coffea canephora (35%), account essentially for the worldwide production of coffee (International Coffee Organization; http://www.ico.org/). C. canephora, the Robusta coffee, is a highly heterozygous diploid whose genome sequence has been recently deciphered (26). C. arabica, the Arabica coffee, is a tetraploid species issued from the hybridization of C. canephora and Coffea eugenioides (27). Both Robusta and Arabica species are subject to the propagation of somatic embryogenesis, through secondary embryogenesis from embryogenic suspensions (28). For propagation, the first step is to sample plant leaves from healthy plants in coffee plantations to establish explants. Testing primary plant material is hence essential to ensure safe propagation.
In April 2012, Coffea species plants grown in a containment facility in France were declared infected by X. fastidiosa, based on enzyme-linked immunosorbent assays (ELISAs) performed by a private laboratory in accordance with the grower's voluntary scheme of phytosanitary surveillance. These samples originated from plant cuttings imported from Central and Latin America. Coffee plants were asymptomatic in plantations. The cuttings have been rooted and cultivated in growth chambers since 2010. This outbreak was eradicated (29). The objectives of the present study were to isolate the pathogens, confirm their identification, and decipher their phylogenetic relationships with other X. fastidiosa strains infecting coffee plants as well as the recently reported CoDiRO strain isolated from olive trees in Italy. As the coffee plant-infecting strains are phylogenetically distant while having in common coffee plant infection abilities, our study focused on searching for determinants specific to coffee plant-infecting strains in genome sequences, including the two new genome sequences that are provided in this study.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A collection of 17 X. fastidiosa strains was established at the French Collection of Plant-Associated Bacteria (CIRM-CFBP; http://www6.inra.fr/cirm_eng/CFBP-Plant-Associated-Bacteria) (Table 1). These strains were grown on B-CYE medium (30), except that agar was replaced with Phytagel (Sigma; reference no. P8169) at 15 g liter−1. Incubation lasted up to 28 days at 25°C. Suspensions made from fresh cultures were stored in a 40% glycerol solution at −80°C.
TABLE 1.
List of X. fastidiosa strains used in the study
| CFBP strain code | Other code(s)a | X. fastidiosa subspeciesb (MLSA result) | Host of isolation | Place (yr) of isolationc |
|---|---|---|---|---|
| CFBP 7969 | LMG15553 | NA (fastidiosa) | Vitis rotundifolia cv. Carlos | NC, USA |
| CFBP 7970T | LMG17159, ATCC 35879, ICMP15197 | fastidiosa | Vitis vinifera | FL, USA |
| CFBP 8068 | LSV 00.54, ATCC 35873 | fastidiosa (multiplex) | Ulmus (elm) | DC, USA |
| CFBP 8069 | LNPV 00.56 PD 89.1 | NA (fastidiosa) | Vitis sp. | NA |
| CFBP 8070 | LSV 40.38 GA Plum | multiplex | Prunus spp. | GA, USA |
| CFBP 8071 | LSV 40.41, ATCC 35870, LMG15099 | NA (fastidiosa) | Prunus dulcis | CA, USA |
| CFBP 8072 | LSV41.03 | NA (pauca) | Coffea arabica | Ecuador (2012) |
| CFBP 8073 | LSV42.09 | NA (fastidiosa) | Coffea robusta | Mexico (2012) |
| CFBP 8074 | LSV42.10 | NA (pauca) | Coffea arabica | Ecuador (2012) |
| CFBP 8076 | LSV 42.31 2689 oak, ATCC 35874 | fastidiosa (multiplex) | Quercus rubra | DC, USA |
| CFBP 8077T | LSV 42.36, Ann-1, ATCC 700598 | multiplex/sandyi (sandyi) | Nerium oleander | CA, USA |
| CFBP 8078 | LSV 43.11, ATCC 35878 | fastidiosa (multiplex) | Vinca sp. | FL, USA |
| CFBP 8082 | LMG9064, ATCC 35876 | fastidiosa | Ambrosia artemifolia | FL, USA |
| CFBP 8083 | LMG15554, 13351 | NA (fastidiosa) | Vitis vinifera | NC, USA |
| CFBP 8084 | LMG15098, ATCC 35869 | NA (morus) | Morus alba | GA, USA |
For each strain, the first cited name refers to the collection that provided us the specimen, which is followed by the synonymous code in other collections. LSV, Laboratoire de la Santé des Végétaux, Anses, France, private collection; ATCC, American Type Culture Collection, USA; ICMP, International Collection of Microorganisms from Plants, New Zealand; LMG, BCCM/LMG, Belgium.
As originally indicated. If our MLSA results indicated a phylogenetic position different from the original, the subspecies derived from our MLSA is indicated in parentheses. NA, not available.
NA, not available.
Isolation from coffee samples.
Leaves were sampled from 40 symptomatic and asymptomatic coffee plants grown in containment facilities in France. The foliar symptoms were chlorotic blotches and reddish discoloration surrounding necrotic spots. Fragments of veins near the symptomatic blades, petioles, and midribs were dilacerated into a sterile phosphate-buffered saline (PBS) buffer (NaCl, 8 g liter−1; Na2HPO4·12H2O, 2.7 g liter−1; NaH2PO4·2H2O, 0.4 g liter−1) after surface sterilization with 70% alcohol. To isolate the pathogen, aliquots from leaf extracts were streaked on B-CYE and modified PWG media (31). Plates were incubated for up to 21 days at 28°C.
Immunological assays.
Plant extracts were subjected to an immunofluorescence analysis using a rabbit polyclonal antiserum. This antiserum was produced in collaboration with UR1268 BIA INRA, Angers-Nantes, France, and was validated in-house for specificity, inclusivity, and detection thresholds (data not shown). Plant extract aliquots of 40 μl and dilutions up to 1/1,000 were deposited on multiwell slides and fixed with 95% alcohol. Immunofluorescence was performed using EPPO standard PM7/97 (32).
DNA extraction.
Boiled bacterial suspensions were used for identification tests. For genomic sequencing, DNA was extracted using a DNeasy plant minikit (Qiagen) to reach a final concentration of 1 to 5 μg DNA in 50 μl.
PCR-based assays for identification of X. fastidiosa subspecies.
A multiprimer PCR test using ALM1/ALM2, XF2542-L/XF2542-R, and XF1968-L/XF1968-R primers was performed to differentiate strains from the three X. fastidiosa subspecies, fastidiosa, multiplex, and sandyi (33). Primers CVC-1 and 272-2-Int were used to specifically identify strains of X. fastidiosa subsp. pauca (34).
Housekeeping gene sequencing.
Primers and conditions for partial sequencing of seven housekeeping genes (cysG, gltT, holC, leuA, malF, nuoL, and petC) were provided as indicated on the X. fastidiosa MLST website (http://pubmlst.org/xfastidiosa/) (35) with the cysG-R primer sequence of Yuan et al. (17). PCR amplifications were performed with an Applied Biosystems thermocycler. The purity and yield of PCR products were checked by running an 8-μl reaction mixture in 1.2% agarose gels and poststaining with ethidium bromide. The remaining PCR products were sequenced with reverse and forward primers by Genoscreen (France).
Sequence acquisition, alignment, and analyses.
Forward and reverse nucleotide sequences were edited, assembled, translated, and aligned using Geneious Pro 4.8.5 software to obtain high-quality sequences (36). Sequences were concatenated by following the alphabetic order of the genes. Multilocus sequence analysis (MLSA) was conducted as described by Jacques et al. (37). Multilocus sequence typing (MLST) was performed according to the X. fastidiosa MLST website (http://pubmlst.org/xfastidiosa/). Table 2 presents a list of strains with MLST data available on the X. fastidiosa MLST website (http://pubmlst.org/xfastidiosa/) which were used in this study. Novel alleles and sequence types (STs) were submitted for inclusion in the X. fastidiosa MLST website website and were assigned numbers 28 and 29 for the two novel cysG alleles, 25 for the novel holC allele, 19 for the novel nuoL alleles, and ST74 and ST75 for the STs.
TABLE 2.
List of X. fastidiosa strains for which data available on the pubMLST website were used in this study
| Strain | X. fastidiosa subspecies | Country (yr) of isolation | Host of isolation |
|---|---|---|---|
| COF0209 | fastidiosa | Costa Rica (2000) | Coffea arabica |
| COF0222 | fastidiosa | Costa Rica (2000) | Coffea arabica |
| COF0245 | fastidiosa | Costa Rica (2000) | Coffea arabica |
| COF0246 | fastidiosa | Costa Rica (2000) | Coffea arabica |
| COF0394 | sandyi | Costa Rica (2000) | Coffea sp. |
| COF0400 | fastidiosa | Costa Rica (2000) | Coffea sp. |
| COF0402 | fastidiosa | Costa Rica (2000) | Coffea sp. |
| COF0404 | sandyi | Costa Rica (2000) | Coffea sp. |
| COF0405 | fastidiosa | Costa Rica (2000) | Coffea sp. |
| COF0406 | fastidiosa | Costa Rica (2000) | Coffea sp. |
| COF0412 | sandyi | Costa Rica (2009) | Coffea sp. |
| COF0413 | sandyi | Costa Rica (2009) | Coffea sp. |
| CVC0145 | pauca | Brazil (2000) | Citrus sp. |
Genome sequencing, assembling, and annotation.
The genome sequences of CFBP 8072 and CFBP 8073 were obtained with the Illumina HiSeq 2000 and HiSeq 2500 sequencing platforms (Genoscreen, France), respectively. Genome assembly was performed using a combination of Velvet (38), SOAPdenovo, and SOAPGapCloser (39). Annotation was conducted with EuGene-PP using similarities with known protein sequences (40).
Analyses of the Xylella genome sequences and their translated sequences.
Sequences of the CFBP 8072 and CFBP 8073 genomes were compared to 17 available genomes of X. fastidiosa (Table 3). The average nucleic identities based on blast (ANIb) (41) were calculated using JSpecies (http://imedea.uib-csic.es/jspecies/about.html#chap9 [42]). A list of orthologous genes shared between X. fastidiosa genomes was generated using the OrthoMCL companion tool (https://bbric-pipelines.toulouse.inra.fr/orthomcl-companion/web/index.html). Venn diagrams were obtained using jvenn software (43). The predicted proteome (translated genome) of each bacterial strain was used to generate a phylogenomic tree with CV-Tree (44). Prophage regions were identified using the PHAge Search Tool (PHAST [45]). Genes coding for putative virulence factors of X. fastidiosa were listed based on reports of Simpson et al. (46) and van Sluys et al. (47) and on the Carbohydrate-Active enZYmes (CAZy; http://www.cazy.org/ [48]) and KEGG (http://www.genome.jp/kegg/ [49]) databases and searched for in the genome sequences of strains CFBP 8072, CFBP 8073, 6c, and 32 based on tblastn.
TABLE 3.
Main characteristics of 19 X. fastidiosa genome sequences used in the study
| Strain designationa | X. fastidiosa subspecies | Accession no. | Host of isolation | Place (yr) of isolation | Genome size (Mb) | GC% | No. of proteins | No. of genes | No. of plasmids (size [kb]) | No. of contigs | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M23 | fastidiosa | NC_010577 | Prunus dulcis | CA, USA (2003) | 2.54 | 51.8 | 2,161 | 2,280 | 1 (38) | 2 | 69 |
| Temecula1 | fastidiosa | NC_004556 | Vitis vinifera | USA (NAa) | 2.52 | 51.8 | 2,034 | 2,123 | NA | 2 | 47 |
| GB514 | fastidiosa | NC_017562 | Vitis vinifera | TX, USA (NA) | 2.49 | 51.8 | 2,183 | 2,238 | 1 (26) | 2 | Unpublished |
| EB92.1 | fastidiosa | NZ_AFDJ00000000.1 | Sambucus nigra | USA (1992) | 2.48 | 51.5 | 2,337 | 2,392 | NA | 168 | 70 |
| CFBP 8073 | fastidiosa/sandyi | LKES00000000 | Coffea canephora | Mexico (2012) | 2.58 | 51.5 | 2,572 | 2,629 | NA | 328 | This study |
| Ann-1 | sandyi | CP006696 | Nerium oleander | USA (NA) | 2.75 | 52.1 | 2,598 | 2,728 | 1 (30) | 2 | 9 |
| Mul-MD | morus | AXDP00000000 | Morus alba | USA (NA) | 2.52 | 51.6 | 2,279 | 2,279 | NA | 101 | 71 |
| Mul0034 | morus | CP006740 | Morus alba | USA (NA) | 2.64 | 52.0 | 2,408 | 2,527 | 1 (24) | 2 | Unpublished |
| Dixon | multiplex | NZ_AAAL00000000 | Prunus dulcis | USA (NA) | 2.62 | 52.0 | 2,358 | 2,408 | NA | 32 | Unpublished |
| M12 | multiplex | NC_010513 | Prunus dulcis | CA, USA (2003) | 2.48 | 51.9 | 2,104 | 2,368 | NA | 1 | 69 |
| Griffin-1 | multiplex | AVGA01000000 | Quercus rubra | GA, USA (2006) | 2.39 | 51.7 | 2,053 | 2,240 | NA | 84 | 72 |
| Sy-VA | multiplex | JMHP00000000 | Platanus occidentalis | VA, USA (NA) | 2.48 | 51.6 | 2,226 | 2,226 | NA | 128 | 73 |
| ATCC 35871 | multiplex | NZ_AUAJ00000000 | Prunus salicina | GA, USA (NA) | 2.41 | 51.6 | 1,963 | 2,071 | NA | 66 | Unpublished |
| CFBP 8072 | pauca | LKDK00000000 | Coffea arabica | Ecuador (2012) | 2.50 | 51.9 | 2,545 | 2,599 | NA | 258 | This study |
| CoDiRO | pauca | JUJW00000000 | Periwinkle | Italy (2013) | 2.51 | 51.8 | 2,269 | 2,053 | 1 (35.3) | 12 | 74 |
| 6c | pauca | NZ_AXBS00000000 | Coffea sp. | Brazil (NA) | 2.61 | 52.4 | 2,336 | 2,506 | 1 (39.5) | 46 | 58 |
| 32 | pauca | NZ_AWYH00000000 | Coffea sp. | Brazil (NA) | 2.61 | 52.4 | 2,302 | 2,477 | NA | 56 | 58 |
| 9a5c | pauca | NC_002488 | Citrus sinensis | Brazil (1992) | 2.68 | 52.7 | 2,766 | 2,838 | 2 (51 and 1) | 3 | 46 |
| PLS 229b | JDSQ00000000 | Pyrus pyrifolia | Taiwan (NA) | 2.73 | 53.1 | 3,259 | NA | NA | 75 |
NA, not available.
As recognized after publication of the genome sequence, this strain does not belong to X. fastidiosa (63).
Identification of genomic fragments specific to X. fastidiosa strains isolated from coffee plants.
Genomic fragments specifically associated with strains isolated from coffee plants were highlighted by generating 20- and 25-bp-long k-mers in genome sequences of strains 32 and CFBP 8072. k-mers having perfect matches (as assessed with tblastn) with genome sequences of strains isolated from coffee plants (CFBP 8073, 6c, and CFBP 8072 or 32, respectively) were conserved. These k-mers were subsequently searched using blast against all X. fastidiosa genome sequences to remove aspecific fragments. All sequences were aligned using MUSCLE in Geneious Pro 4.8.5 software to visualize the specificity of the fragments and the effect of polymorphism at the protein level.
Nucleotide sequence accession numbers.
The CFBP 8072 and CFBP 8073 genome sequences reported here have been deposited in the NCBI genome database under accession numbers LKDK00000000 and LKES00000000, respectively.
RESULTS
Isolation and identification of X. fastidiosa strains from coffee samples.
The presence of X. fastidiosa was first assessed in 40 symptomatic and asymptomatic coffee plants based on immunofluorescence analyses, and four samples were detected as contaminated with X. fastidiosa. Three strains were isolated from three of these samples showing leaf scorches (see Fig. S1A and B in the supplemental material). Two strains, CFBP 8072 and CFBP 8074, were isolated from C. arabica plants originating from cuttings sampled on different trees in the same orchard in Ecuador, while the remaining strain, CFBP 8073, was isolated from C. canephora plants that originated from cuttings sampled in Mexico (see Fig. S1C to E).
According to multiple PCR identification tests (50–52), these three strains were identified as X. fastidiosa (data not shown). Moreover, a multiprimer PCR test (33) was performed for subspecies identification of these three X. fastidiosa strains. The CFBP 8073 strain presented a profile similar to that of X. fastidiosa subsp. sandyi CFBP 8077 (=strain Ann-1). Both strains presented a specific profile with one band at 638 bp (see Fig. S2A in the supplemental material). The strains CFBP 8072 and CFBP 8074 presented the same profile with two bands, one at 638 bp and one at 412 bp (see Fig. S2A). This unusual genetic profile was not previously described (33) and differed from all the profiles obtained with the 12 strains included in this study (see Fig. S2A). It should be noted, however, that no strains from X. fastidiosa subsp. pauca are available from any international culture collections and hence no strains from this subspecies could be included in this work as a control. The PCR identification test design by Pooler and Hartung (34) was performed on these two strains in order to test for identification as X. fastidiosa subsp. pauca. This test was designed to amplify a fragment specific to the X. fastidiosa subsp. pauca strains. The bands obtained for strains CFBP 8072 and CFBP 8074 were indeed at the expected size with this test (see Fig. S2B). As strains isolated from coffee plants from Brazil were already assigned to X. fastidiosa subsp. pauca (18), these results were considered primary indicators of identification evidence for these two strains. In summary, based on PCR identification tests, the CFBP 8073 strain isolated from the Mexican C. canephora plant was presumed to belong to X. fastidiosa subsp. sandyi, and strains CFBP 8072 and CFBP 8074 sampled from Ecuadorian C. arabica plants were putatively assigned to X. fastidiosa subsp. pauca.
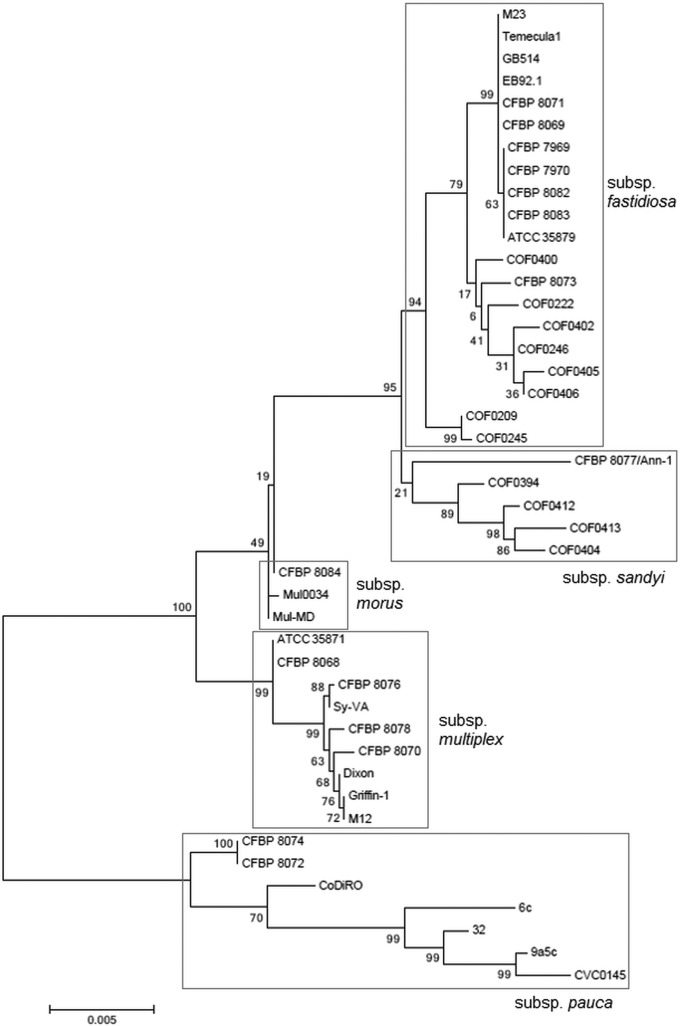
MLSA confirms that the newly isolated strains from coffee plants belong to two subspecies of X. fastidiosa.
Sequence analyses of seven housekeeping genes (cysG, gltT, holC, leuA, malF, nuoL, and petC) were performed, and the results were compared to the sequences of 41 isolates covering the X. fastidiosa subspecies fastidiosa, morus, multiplex, pauca, and sandyi by using a scheme dedicated to X. fastidiosa (http://pubmlst.org/xfastidiosa/). Maximum likelihood (ML) trees were constructed based on individual gene fragment sequences and on the concatenated data set (Fig. 1 and see Fig. S3 in the supplemental material). The three strains isolated from coffee plants originating from Mexico and Ecuador did not cluster on any of the phylogenetic trees. Strains CFBP 8072 and CFBP 8074 clustered on a branch close to the X. fastidiosa subsp. pauca clade. The strains CFBP 8072 and CFBP 8074 were identical based on their seven-housekeeping gene profiles; hence, only one strain, CFBP 8072, was kept for subsequent analyses.
FIG 1.
Maximum likelihood tree based on the concatenated partial sequences of cysG, gltT, holC, leuA, malF, nuoL, and petC. Bootstrap scores (1,000 replicates) are displayed at each node.
In contrast to the results obtained with the multiprimer identification PCR test, strain CFBP 8073, which was isolated from a Mexican coffee plant, clustered with other strains isolated from Costa Rican coffee plants (http://pubmlst.org/xfastidiosa/) (Fig. 1). This group of strains falls into X. fastidiosa subsp. fastidiosa. On the ML tree (Fig. 1), the Ann-1 strain of X. fastidiosa subsp. sandyi is close to a group of strains that was also isolated from coffee plants in Costa Rica (http://pubmlst.org/xfastidiosa/). Hence, coffee plant-colonizing strains are genetically diverse, not only in a comparison of strains from South America and Central America but also in a comparison of strains within Central America.
The phylogenetic trees built with an ML algorithm for each of the seven loci and the concatenated data set did not all depict the same phylogenetic history (Fig. 1; see Fig. S3 in the supplemental material). The Shimodaira-Hasegawa test (53) performed on individual gene sequences and on the data set of concatenated sequences (see Table S1 in the supplemental material) showed that all trees were significantly incongruent with each other but were not significantly different from the tree based on the data set of concatenated sequences, except for gltT. The seven genes used here presented a large range of GC content (from 50.6% for malF to 61.8% for cysG) (see Table S2 in the supplemental material), which embraces the mean genomic Xylella GC content (52%) (54). This large range of GC content is quite surprising for housekeeping genes, since these genes are supposed to evolve slowly and to code for basic metabolic functions. Nevertheless, neutrality estimates indicated that these genes were not positively or negatively selected (see Table S2). This was confirmed by the Ka/Ks ratios for the seven loci. The values ranged from 0.0631 (for gltT) to 0.2650 (for petC) (data not shown). All loci were polymorphic, and the number of polymorphic sites ranged from 19 for petC, the least polymorphic loci, to 48 for cysG, the most polymorphic locus. The number of alleles at each locus ranged from 9 for petC to 18 for cysG (see Table S2).
Allelic characterization at some loci highlights intersubspecific recombination events.
In MLST, the combination of the allele number at each of the seven loci gives rise to a sequence type (ST). To date, the PubMLST database describes 58 STs for X. fastidiosa, named ST1 to ST62, with ST12, ST36, ST59, and ST60 lacking sequences. While no one ST is shared between strains belonging to different subspecies, some allele sharing is observed between subspecies. This is the case for cysG (allele 18), gltT (allele 3), and petC (allele 3) for strains belonging to X. fastidiosa subsp. morus and X. fastidiosa subsp. multiplex (Table 4). X. fastidiosa subsp. fastidiosa and X. fastidiosa subsp. sandyi shared alleles 1 and 10 of gltT and allele 17 of holC.
TABLE 4.
Allele designations for each gene and ST determined from the concatenated data set for every strain of X. fastidiosa used in this studya
| Allele designationb for: |
ST | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| X. fastidiosa subspecies | Strain code | cysG | gltT | holC | leuA | malF | nuoL | petC | |
| fastidiosa | CFBP 8069 | 1 | 1* | 1 | 1 | 1 | 1 | 1 | 1 |
| CFBP 8071 | 1 | 1* | 1 | 1 | 1 | 1 | 1 | 1 | |
| EB92.1 | 1 | 1* | 1 | 1 | 1 | 1 | 1 | 1 | |
| GB594 | 1 | 1* | 1 | 1 | 1 | 1 | 1 | 1 | |
| M23 | 1 | 1* | 1 | 1 | 1 | 1 | 1 | 1 | |
| Temecula1 | 1 | 1* | 1 | 1 | 1 | 1 | 1 | 1 | |
| ATCC 35879 | 1 | 1* | 1 | 1 | 4 | 1 | 1 | 2 | |
| CFBP 7969 | 1 | 1* | 1 | 1 | 4 | 1 | 1 | 2 | |
| CFBP 7970 | 1 | 1* | 1 | 1 | 4 | 1 | 1 | 2 | |
| CFBP 8082 | 1 | 1* | 1 | 1 | 4 | 1 | 1 | 2 | |
| CFBP 8083 | 1 | 1* | 1 | 1 | 4 | 1 | 1 | 2 | |
| COF0246 | 12 | 1* | 18 | 1 | 10 | 10 | 1 | 17 | |
| COF0209 | 14 | 1* | 15 | 10 | 10 | 11 | 1 | 19 | |
| COF0222 | 12 | 11 | 17* | 1 | 10 | 11 | 1 | 20 | |
| COF0245 | 14 | 12 | 15 | 10 | 10 | 11 | 1 | 21 | |
| COF0400 | 23 | 1* | 20 | 13 | 10 | 5 | 1 | 47 | |
| COF0402 | 14 | 1* | 18 | 10 | 10 | 10 | 1 | 52 | |
| COF0406 | 12 | 10* | 18 | 1 | 10 | 10 | 1 | 55 | |
| COF0405 | 12 | 11 | 18 | 1 | 10 | 11 | 1 | 57 | |
| CFBP 8073 | 29 | 1* | 1 | 9 | 10 | 19 | 1 | 75 | |
| sandyi | Ann-1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 5 |
| COF0394 | 15 | 10* | 19 | 11 | 14 | 13 | 9 | 33 | |
| COF0412 | 25 | 1* | 19 | 11 | 11 | 12 | 9 | 54 | |
| COF0404 | 15 | 10 | 17* | 11 | 11 | 12 | 9 | 56 | |
| COF0413 | 15 | 10* | 16 | 11 | 11 | 12 | 9 | 61 | |
| morus | CFBP 8084 | 18* | 3* | 5 | 4 | 6 | 4 | 3* | 29 |
| Mul-MD | 18* | 3* | 5 | 4 | 6 | 4 | 3* | 29 | |
| Mul0034 | 8 | 3* | 5 | 4 | 6 | 4 | 5 | 30 | |
| multiplex | Dixon | 3 | 3* | 3 | 3 | 3 | 3 | 3* | 6 |
| Griffin-1 | 7 | 3* | 3 | 3 | 3 | 3 | 3* | 7 | |
| M12 | 7 | 3* | 3 | 3 | 3 | 3 | 3* | 7 | |
| Sy-VA | 5 | 7 | 4 | 3 | 5 | 3 | 3* | 8 | |
| CFBP 8076 | 5 | 4 | 4 | 3 | 5 | 3 | 3* | 9 | |
| CFBP 8070 | 3 | 5 | 6 | 5 | 3 | 3 | 4 | 10 | |
| ATCC 35871 | 18* | 3* | 9 | 3 | 5 | 3 | 3* | 41 | |
| CFBP 8068 | 18* | 3* | 9 | 3 | 5 | 3 | 3* | 41 | |
| CFBP 8078 | 3 | 3* | 4 | 3 | 5 | 15 | 3* | 51 | |
| pauca | CVC0145 | 9 | 8 | 10 | 7 | 7 | 8 | 7 | 11 |
| 9a5c | 9 | 8 | 10 | 7 | 7 | 7 | 6 | 13 | |
| 6c | 11 | 9 | 12 | 8 | 8 | 9 | 8 | 14 | |
| 32 | 10 | 8 | 11 | 7 | 8 | 8 | 6 | 16 | |
| CoDiRO | 24 | 14 | 10 | 7 | 16 | 16 | 6 | 53 | |
| CFBP 8072 | 28 | 8 | 25 | 7 | 8 | 16 | 6 | 74 | |
| CFBP 8074 | 28 | 8 | 25 | 7 | 8 | 16 | 6 | 74 | |
Allele numbers and STs are coded in agreement with the X. fastidiosa MLST website (http://pubmlst.org/xfastidiosa/).
For each locus, allele numbers marked with an asterisk are shared between different X. fastidiosa subspecies.
Strains CFBP 8072 and CFBP 8073 presented alleles already observed in X. fastidiosa strains, except that CFBP 8072 presented a yet undescribed cysG allele (allele 28) and a novel holC allele (allele 25), while CFBP 8073 presented a novel allele for cysG (allele 29) that was different from the new one of CFBP 8072 and a novel allele for nuoL (allele 19) (Table 4; see Fig. S4 in the supplemental material). Because both CFBP 8072 and CFBP 8073 presented yet undescribed alleles at some loci, these strains define two new STs. Strain CFBP 8072 is associated with ST74, while strain CFBP 8073 is affiliated with ST75.
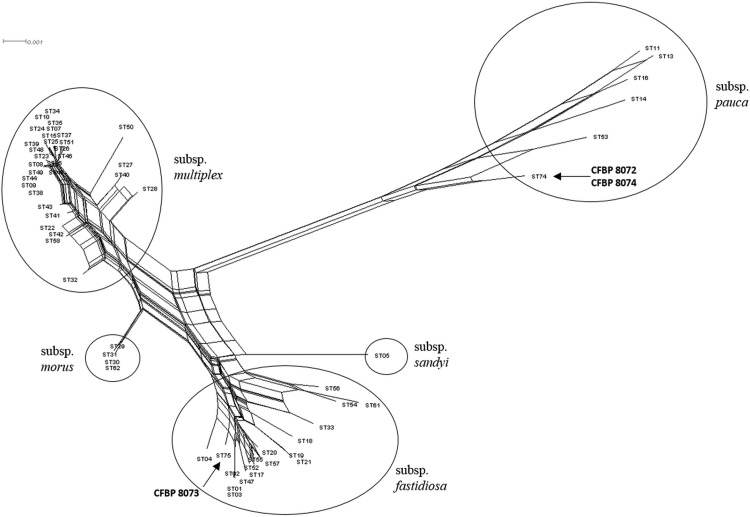
Phylogenetic networks highlight conflicting signals in the gene sequence data. Relevant reticulations were found mainly for cysG, gltT, holC, malF, and nuoL (Fig. 2; see Fig. S2 in the supplemental material). The position of the reticulations in a tree is a proxy of their time of occurrence, via the genetic distance. It is quite clear by these figures that recombination events occurred during evolution and even recently, at least for holC. It is also clear that reticulations are found at the base or along the branches that bear the two new STs defined for our strains.
FIG 2.
Split graph of multilocus sequence analysis of the X. fastidiosa strains of each sequence type (ST) for the data set of the concatenated sequences. The designation for each leaf indicates the ST number. See Tables 1 and 4 for strain designations and ST correlations, respectively.
Sequencing, assembly, and annotation of draft genomes from CFBP 8072 and CFBP 8073 strains.
As the MLSA and MLST analyses revealed that the two coffee-infecting strains were phylogenetically distant and distinct from previously described CLS-related strains (16, 55), we sequenced the genomes of X. fastidiosa strains CFBP 8072 and CFBP 8073. The shotgun sequencing yielded 17,551,806 and 20,038,038 paired-end reads with insert sizes of ca. 100 bp for CFBP 8072 and CFBP 8073, respectively. Assembling yielded 258 and 328 contigs larger than 250 bp (N50 = 87,332 and 67,596 bp), with the largest contig being 224,836 and 247,073 bp for a total assembly size of 2,496,737 and 2,582,171 bp for CFBP 8072 and CFBP 8073, respectively. The genome sizes and other general features of these genome sequences fell into the range of previously reported X. fastidiosa genome sequences (Table 3).
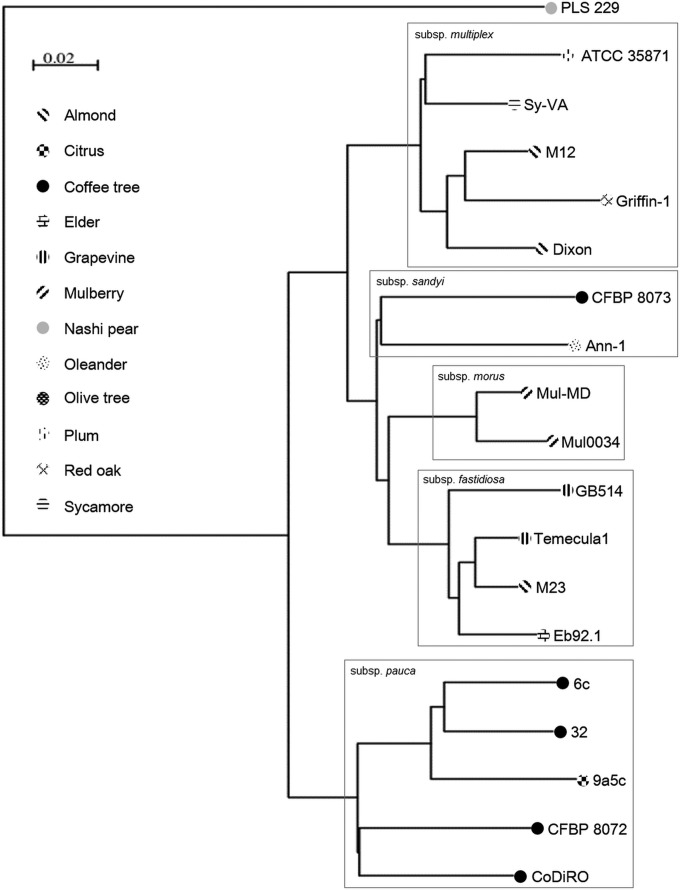
Phylogenetic analysis of CFBP 8072 and CFBP 8073 based on genomic data.
Based on these data, an entire genome-based phylogenetic tree was built using CV-Tree (44) by comparing protein sequences predicted from annotation analysis performed on CFBP 8072 and CFBP 8073 to 17 publicly available predicted proteomes (Fig. 3). The strain CFBP 8072 isolated from Arabica coffee originating from Ecuador clustered within X. fastidiosa subsp. pauca, yet in a different subclade. As found with multiprimer PCR but in contrast with MLSA, strain CFBP 8073 isolated from Robusta coffee originating from Mexico clustered with X. fastidiosa subsp. sandyi Ann-1 strain and not with the X. fastidiosa subsp. fastidiosa strains. The identities of CFBP 8072 and CFBP 8073 were also monitored with average nucleic identities (ANI) (see Table S3 in the supplemental material), which is an in silico substitution of the DNA-DNA hybridization method (42) (http://www.imedea.uib.es/jspecies/about.html). All strains share more than 95% identity, except strain PLS 229, for which ANIb values close to 83% were found for each pairwise comparison (see Table S3 in the supplemental material). Strain clustering at a threshold higher than 98% identity grouped strains into the previously defined subspecies: Temecula1, M23, GB514, and EB92-1 formed X. fastidiosa subsp. fastidiosa (ANIb values > 99.6%); Mul-MD and Mul0034 formed X. fastidiosa subsp. morus (ANIb values > 99.7%); ATCC 35871, Sy-VA, M12, Griffin-1 and Dixon formed X. fastidiosa subsp. multiplex (ANIb values > 99.3%); and strains 9a5c, 6c, 32, and CoDiRO formed X. fastidiosa subsp. pauca (ANIb values > 98.1%). The ANIb values between the strain Ann-1 genome and genomes of strains from X. fastidiosa subsp. morus and X. fastidiosa subsp. fastidiosa were between 97.8% and 98.1%, indicating close relationships but not as close as the ones existing within every other subspecies. Based on ANIb values, CFBP 8072 is similar to X. fastidiosa subsp. pauca but seems to present some divergence (ANIb values > 97.7% identity). In coherence with the MLSA results, strain CFBP 8073 was closer to the X. fastidiosa subsp. fastidiosa strains (ANIb values > 98.7%) than to the X. fastidiosa subsp. sandyi Ann-1 strain (ANIb values of 97.97 and 98.12) (see Table S3 in the supplemental material).
FIG 3.
CV-Tree results based on the entire genome sequences of 19 X. fastidiosa strains.
Gene content of CFBP 8072 and CFBP 8073 in relation to that of other X. fastidiosa strains.
In order to avoid bias due to different annotation methods, all the genome sequences (Table 3) were reannotated using EuGene-PP (40). We decided to exclude the PLS 229 genome from the comparative genomic analysis, since that strain does not belong to X. fastidiosa (56). According to gene predictions, the X. fastidiosa pan-genome was composed of 4,668 coding sequences (CDSs) (see Table S4 in the supplemental material); 3,529 of them (76%) were shared by at least two strains. Moreover, each strain harbored some single-copy specific proteins, ranging from 6 to 122, which represents 0.24 to 5.27% of their predicted proteomes.
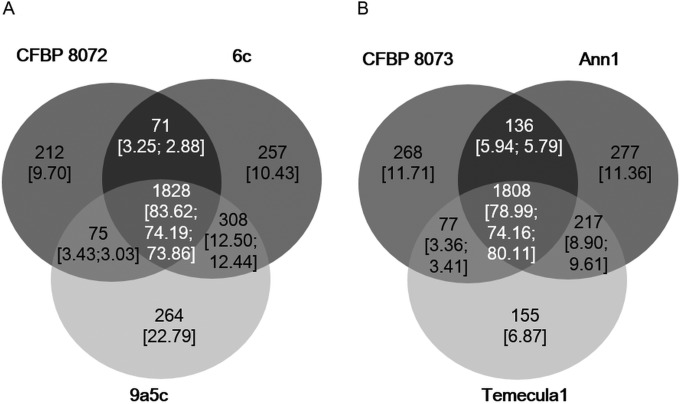
As strains CFBP 8072 and CFBP 8073 do not belong to the same subspecies, although they were both isolated from coffee plants, an OrthoMCL analysis was performed to identify the relative importance of the host plant versus the geographical location of strain isolation on the gene content (Fig. 4). The strain CFBP 8072 (C. arabica, Ecuador) genome was compared to those of X. fastidiosa subsp. pauca strains 6c (C. arabica, Brazil) and 9a5c (Citrus sinensis, Brazil). A smaller number of genes was shared by CFBP 8072 and 9a5c or 6c than between strains 6c and 9a5c together. As the size of the genomes in terms of groups of orthologs is variable among strains (see Table S4 in the supplemental material), values were reported for the total number of groups of orthologs per strain. Strain 6c shared 77.07% of its orthologs with CFBP 8072, while it shared 86.69% of its orthologs with 9a5c. This indicates that the geographical location (i.e., isolation from Brazil) had a stronger impact on gene content than the host plant itself (coffee plant for CFBP 8072 and 6c versus citrus plant for 9a5c) (Fig. 4).
FIG 4.
Venn diagrams illustrating results of OrthoMCL analyses. Values are the numbers of groups of orthologs, i.e., the CDSs present in single copy in each genome, while values in brackets are the percentages of ortholog groups relative to the total number of ortholog groups in each genome; values at intersections are indicated for listed strains clockwise from the left. (A) Comparison of gene contents of three X. fastidiosa subsp. pauca strains: CFBP 8072 isolated from coffee plants from Ecuador, 9a5c iolated from Brazilian citrus, and 6c isolated from the Brazilian coffee plant. (B) CFBP 8073 is compared with the X. fastidiosa subsp. fastidiosa strain Temecula1 and X. fastidiosa subsp. sandyi strain Ann-1 isolated from oleander.
In order to add an element to the positioning of CFBP 8073 in X. fastidiosa subsp. sandyi versus X. fastidiosa subsp. fastidiosa, the genome of strain CFBP 8073 (C. canephora, Mexico) was compared to those of the Temecula1 X. fastidiosa subsp. fastidiosa strain (V. vinifera, USA) and the X. fastidiosa subsp. sandyi Ann-1 strain (Nerium oleander, USA) (Fig. 4; see Table S1 in the supplemental material). These comparisons indicated that a higher number of orthologs was shared between CFBP 8073 and the Ann-1 strain (84.93% of total number of CFBP 8073 orthologs) than between CFBP 8073 and Temecula1 (82.79% of total number of CFBP 8073 orthologs), indicating a closer proximity of CFBP 8073 to X. fastidiosa subsp. sandyi than to X. fastidiosa subsp. fastidiosa.
Genes encoding potential virulence factors in the genome sequences of coffee plant-infecting X. fastidiosa.
Lists of candidate genes associated with secretion systems and with other virulence factors with a putative role in X. fastidiosa pathogenicity were established (see Table S5 in the supplemental material). Orthologs were searched for (tblastn) in the translated genome sequences of the four coffee plant-infecting X. fastidiosa strains (CFBP 8072, CFBP 8073, 6c, and 32). All candidates associated with the type I secretion system are conserved in the five strains. Most of the type II secretion system genes were also conserved, with few exceptions, such as a 1,4-beta-cellobiosidase gene that appeared to be poorly conserved in CFBP 8073. Genes associated with the type IV secretion system are poorly conserved in the coffee plant-infecting strain, and strain CFBP 8072 did not possess any of the searched candidates. The repertoire of genes coding for the type V secretion system appeared to be partially conserved, with the absence of several genes encoding hemolysins and hemagglutinins in both CFBP 8072 and CFBP 8073, as well as in the two other coffee plant-infecting strains. Finally, some other known virulence factors are conserved in CFBP 8072 and CFBP 8073; this is the case for the colicin V toxin. Four genes (hicA, hicB, higA, vapI) clustered in strain Temecula1 (PD1340 to PD1343) were missing from the CFBP 8072 genome sequence but were fully conserved in CFBP 8073.
A high prevalence of prophage sequences is a common feature of X. fastidiosa genomes.
Since evidence of homologous recombination was provided for both genomes, we checked the presence of prophages in a concatenated version of the genome sequences of CFBP 8072 and CFBP 8073 to assess the influence of horizontal gene transfer (HGT) on genome evolution. According to PHAST analysis (45), CFBP 8072 and CFBP 8073 contained 5 and 11 regions with sequences of phage origin, respectively (Table 5).
TABLE 5.
Identification of prophage sequences in the genome sequences of CFBP 8072 and CFBP 8073 based on PHAST analysisa
| Strain | Region length (kb) | No. of phage CDSs | Possible phage | No. of candidate phage CDSs | Completeness of phage sequence | GC% |
|---|---|---|---|---|---|---|
| CFBP 8072 | 17.2 | 14 | PHAGE_Haemop_Aaphi23_NC_004827 | 4 | Incomplete | 51.86 |
| 20.9 | 21 | PHAGE_Pseudo_PPpW_3_NC_023006 | 17 | Incomplete | 57.41 | |
| 41.4 | 29 | PHAGE_Haemop_Aaphi23_NC_004827 | 12 | Intact | 57.59 | |
| 25.7 | 23 | PHAGE_Xylell_Xfas53_NC_013599 | 8 | Incomplete | 52.74 | |
| 126.8 | 114 | PHAGE_Xylell_Xfas53_NC_013599 | 31 | Intact | 53.99 | |
| CFBP 8073 | 12.3 | 14 | PHAGE_Pseudo_PPpW_3_NC_023006 | 9 | Incomplete | 57.08 |
| 24 | 6 | PHAGE_Cellul_phiSM_NC_020860 | 1 | Incomplete | 54.78 | |
| 11.3 | 9 | PHAGE_Xantho_Cf1c_NC_001396 | 3 | Incomplete | 46.42 | |
| 31.9 | 15 | PHAGE_Xylell_Xfas53_NC_013599 | 14 | Questionable | 55.01 | |
| 25.9 | 23 | PHAGE_Xylell_Xfas53_NC_013599 | 13 | Intact | 56.16 | |
| 58.7 | 22 | PHAGE_Xylell_Xfas53_NC_013599 | 7 | Intact | 52.85 | |
| 31.1 | 7 | PHAGE_Ralsto_RSK1_NC_022915 | 2 | Incomplete | 52.72 | |
| 79.5 | 69 | PHAGE_Xylell_Xfas53_NC_013599 | 14 | Intact | 52.07 | |
| 23.7 | 16 | PHAGE_Haemop_Aaphi23_NC_004827 | 2 | Questionable | 54.95 | |
| 16.5 | 16 | PHAGE_Aggreg_S1249_NC_013597 | 6 | Incomplete | 54.42 | |
| 64.6 | 55 | PHAGE_Xylell_Xfas53_NC_013599 | 12 | Intact | 54.88 |
In each concatenated genome sequence, ordered from the largest to the smallest contig, PHAST identified regions of phage origin. Included are the size of each region, the number of predicted phage CDSs in that region, the predicted candidate phage with the highest number of CDSs, completeness of the phage sequence, and the GC%.
The sequences of 10 phages from 9a5c (XfP1 to -10) and eight from Temecula1 (Xpd1 to -8) were sought in the genomes of CFBP 8072 and CFBP 8073 due to the known positions of the phage regions in the 9a5c and Temecula1 genomes (57) (see Fig. S5 and S6 in the supplemental material). The data illustrated how phage regions were conserved in the whole genomes of the targeted strains and indicated that a given phage from strain 9a5c or Temecula1 was not necessarily found as a single region but could be found fragmented in several regions of the targeted genomes. Regarding the phages from strain 9a5c, both strains CFBP 8072 and CFBP 8073 possess phage regions with a high level of identity (see Fig. S5) with all but Xfp9 phages on at least 50% of the phage sequence length. The most relevant conservation was for phages Xfp6, Xfp7, Xfp8, and Xfp10 in CFBP 8072 and to a lower extent in CFBP 8073. The highest conservation of phage sequences in CFBP 8072 versus CFBP 8073 was not surprising, since both strains 9a5c and CFBP 8072 belong to the same subspecies, X. fastidiosa subsp. pauca, while CFBP 8073 does not. Considering the phages from strain Temecula1, both strains CFBP 8072 and CFBP 8073 possessed phage regions with a high level of identity (see Fig. S6) with all Xpd1 to -8 phages. The most relevant conservation was noted for Xpd1, Xpd2, Xpd6, and Xpd7.
Identification of allelic variants specific to X. fastidiosa isolated from coffee plants.
The genome sequences of the following four X. fastidiosa strains that were isolated from coffee plants are available: CFBP 8072 isolated from C. arabica originating from Ecuador, CFBP 8073 isolated from C. canephora originating from Mexico, and strains 6c and 32 (isolated from Coffea spp. from Brazil) (55, 58). To identify potential determinants of coffee plant adaptation, we compared these genome sequences to the 13 other available X. fastidiosa genome sequences representing X. fastidiosa strains which are not known to infect coffee plants (Table 3, excluding the PLS 229 strain that does not belong to X. fastidiosa). First, the core genome of coffee plant-infecting strains was compared to the pan-genome of non-coffee-plant-infecting strains. Unfortunately, we did not identify any orthologous group specific to coffee plant-infecting strains. Therefore, another approach based on the presence of specific k-mers associated with strains isolated from coffee plants was performed. A set of five fragments was obtained (see Fig. S7 in the supplemental material). These fragments are located in genes encoding a methionyl-tRNA synthetase (MetG), a von Willebrand factor type A, a cellobiosidase, a phage antirepressor, and a hypothetical protein. In regard to the methyl-tRNA synthetase, the k-mer region contained one single-nucleotide polymorphism (SNP) that had no effect on the amino acid translation. The k-mer fragment of the hypothetical protein contains six variable loci between coffee plant-infecting strains and non-coffee-plant-infecting strains; they were all synonymous. Two loci were variable in the coffee plant-specific k-mer region of the von Willebrand factor type A, one SNP had a nonsynonymous effect, the other SNP had a synonymous effect, and the combination of both loci was specific at the nucleotidic level, but the protein sequence was not specific. The specific k-mer fragment associated with a phage antirepressor was located either within the CDS or in an intergenic region approximately 250 bp upstream or 40 bp downstream of the CDS. Finally, the coffee plant-specific k-mer region was nonsynonymous for the cellobiosidase. In the latter case, this fragment was present five times in a serine/glycine-rich region of the gene in strain 6c but only twice in CFBP 8072 and once in strains 32 and CFBP 8073. These modifications did not localize in a functional domain but between a glycosyl hydrolase family 6 (GH6) domain (positions 33 to 401 in 9a5c protein) and a carbohydrate-binding module family 2 (CBM2) domain (positions 609 to 681 in 9a5c protein).
DISCUSSION
Several Xylella-contaminated coffee plants were recently intercepted in Europe (present study and reference 59). As coffee is not cropped in Europe, it is not submitted to any quarantine regulation. Nevertheless, since X. fastidiosa is listed in the A1 Annex of the EU Council Directive EC 2000/29, its occurrence in the European Union must be declared and contaminated plants should be eradicated. This work allows the identification of two novel STs for the X. fastidiosa strains isolated from coffee plant cuttings originating from Mexico and Ecuador. One strain fell into the X. fastidiosa subsp. fastidiosa-X. fastidiosa subsp. sandyi group, while the other fell into a genetic lineage close to X. fastidiosa subsp. pauca strains, being yet slightly divergent. Candidate alleles and/or ortholog coding elements associated with coffee plant adaptation are described.
Various assays are available to identify X. fastidiosa strains, but because of recombination events that affect the genomes of these pathogens, taxonomic assignation of strains can lead to conflicting results. Indeed, strain CFBP 8073, which was isolated from a Mexican coffee plant, is identified as X. fastidiosa subsp. sandyi based on a multiprimer PCR identification test (33), on a whole-genome-based phylogenetic tree using CV-Tree, and on ANI calculations, but based on MLSA, this strain clusters with other coffee plant-infecting strains isolated from Costa Rica (http://pubmlst.org/xfastidiosa/) into X. fastidiosa subsp. fastidiosa. The X. fastidiosa subspecies fastidiosa and sandyi are supposed to originate from Central America, and only a limited part of their original diversity was introduced into the United States (14, 17). Increasing the available genomic data in these two subspecies with strains isolated in the origin and/or diversification area would help to clarify this point.
Strain CFBP 8072 was assigned to X. fastidiosa subsp. pauca. This strain clusters on a branch close to, but divergent from, the X. fastidiosa subsp. pauca clade and especially in a divergent branch from the X. fastidiosa subsp. pauca strains that were isolated from Brazilian coffee plants (18). Interestingly, the CoDiRO strain, which is the causal agent of the current epidemics in olive trees in Italy (6), also belongs to X. fastidiosa subsp. pauca. It seems that first the clade clustering CFBP 8072 and CFBP 8074 diverged, and then the CoDiRO strain diverged from the clade containing the strains infecting coffee and citrus plants (6c, 32, and 9a5c). Adding genome sequences to represent the diversity of X. fastidiosa subsp. pauca strains is also needed to clarify the boundaries of this subspecies.
While an ANI threshold range (95 to 96%) for species demarcation had previously been suggested based on a comparative investigation between DNA-DNA hybridization and ANI values (42), no ANI threshold has been provided for subspecies delineations. Strain PLS 229, which was isolated from nashi in Taiwan (56), does not belong to the X. fastidiosa species, as indicated by ANIb values close to 83% for each pairwise comparison. This strain may form a novel species, tentatively named Xylella taiwanensis (56). ANIb results highlight that strain clustering at a threshold higher than 98% identity grouped strains into the previously defined subspecies. It is thus tempting to propose an ANIb value in the range of 98 to 99% as an indicative threshold for subspecies clustering; however, this would necessitate a much larger data set.
A first line of evidence of multiple recombination events at the origin of these coffee plant-infecting strains comes from the analysis of allele combinations. CFBP 8072 is an X. fastidiosa subsp. pauca strain showing alleles at gltT, leuA, and petC loci that are common among X. fastidiosa subsp. pauca strains (i.e., already identified in 9a5c, 32, and/or CoDiRO). The nuoL allele is of unknown origin and was recently reported in X. fastidiosa subsp. pauca strains isolated from oleander and coffee plants from Costa Rica (19) and in olive trees in Italy (6). The alleles of CFBP 8072 at cysG and holC loci are yet undescribed. Similarly, CFBP 8073 presented alleles at gltT, holC, malF, and petC that are common among X. fastidiosa subsp. fastidiosa strains (Table 4). In contrast, the leuA allele is quite rare and was previously identified in strain ALS12 with a totally different set of alleles at other loci that defined ST18 (35). Also, alleles of CFBP 8073 at cysG and nuoL loci were novel. These unusual allele combinations for X. fastidiosa strains are indications of recombination among strains from other subspecies. Donors may be of unknown origin, as is obviously the case for the novel alleles, or may have been previously described, which is the case for the leuA allele 9 already identified in X. fastidiosa subsp. multiplex ALS12 (9, 35).
Another line of indication for recombination events is brought forth by phylogenetic trees and networks. Conflicting signals in the gene sequence data suggest exchange or acquisition of genetic material among strains. In a phylogenetic network, alternative phylogenies are represented by parallelograms. The more reticulation there is in a network, the more conflicting signals exist in the sample, possibly due to exchange of genetic material. Relevant reticulations are found for five out of the seven housekeeping genes from the MLSA scheme (i.e., cysG, gltT, holC, malF, and nuoL). Recombination events occurred during evolution and even recently, especially at the base or along the branches that bear the two new STs defined for our strains. Lack of congruency among individual housekeeping genes and/or with the tree based on the concatenated data set was already observed with other sets of genes in a collection of Brazilian X. fastidiosa subsp. pauca strains (18). In this work, this observation is extended to the other X. fastidiosa subspecies.
From gene analysis, extensive evidence of intersubspecific recombination within X. fastidiosa has appeared to support genetic variation, potentially involved in host shifts. X. fastidiosa is naturally competent, at least in vitro, and recombination efficiencies are higher for attached cells (54), which is the case when Xylella cells are present in high numbers in insects and in xylem vessels (60). This natural competency could be a route through which horizontal gene transfer occurs for sympatric strains in natural environments. High recombination rates coupled with the uncontrolled movement of strains can have disastrous consequences. For example, X. fastidiosa subsp. multiplex causing plum leaf scald was first detected in South America, i.e., Brazil, Argentina, and Paraguay, in the mid-1930s (61). Strains isolated from infected plums in Brazil showed close genetic proximity with U.S. isolates of X. fastidiosa subsp. multiplex (62). The latter species is supposed to be native to the United States (14, 17). This pathogen, feasibly introduced into Brazil, could have evolved in sympatry with native X. fastidiosa subsp. pauca, whose host range is currently unknown. Then an intersubspecific recombination between representatives of these two subspecies may have led to a host shift from yet unknown native hosts of X. fastidiosa subsp. pauca to citrus and coffee (16). Recently, other cases of intersubspecific recombinations were documented in the United States as linked with the emergence of X. fastidiosa in mulberry, blueberry, and blackberry (11, 63).
The strains isolated from coffee plants grown in Central and South America are polymorphic at the seven MLST loci, with at least 17 STs (http://pubmlst.org/xfastidiosa/). At least four STs (ST14, ST16, ST53, and ST74) were identified for the coffee plant-infecting X. fastidiosa subsp. pauca strains. For these STs, no one locus presents a single allele, indicating that there is genetic variation at all seven loci. As the coffee plant species C. canephora and C. arabica are both grown in large but different areas of Latin America, it is not likely that bacterial genetic differentiation is linked to host genotypes. In addition, both coffee species were introduced less than 300 years ago in Latin America (64) via different introduction events. It has been suggested that in Brazil, coffee species had recruited X. fastidiosa subsp. pauca through intersubspecific recombination (16). Indeed, it was hypothesized that Brazilian X. fastidiosa subsp. pauca was originally unable to infect coffee plants, but that adaptation to this host plant became possible only after the introduction of novel genetic variation. The first description of CLS appears in 1995 (21). Even if the disease had been previously present but unidentified, it has been hypothesized that the recombination event leading to the host shift has been a relatively recent event (16). It should therefore be observed that either this original recombinant strain diversified quickly and invaded most equatorial coffee-producing areas from Central to South America or that coffee species recruited an already diversified but Xylella-adapted population.
Recombination is an important driving force of genome evolution in X. fastidiosa, including homologous recombination but also gene acquisition through HGT (15, 55). X. fastidiosa has the largest ratio of phage genes to genome size (7 to 9%) in a set of 37 phytopathogenic bacterial genomes (65). Various phage species, including Xfas53, were predicted in the sequenced genomes. The temperate phage Xfas53 has been propagated and purified from the X. fastidiosa strain Temecula (66). This lytic phage has a chimeric structure that combines characteristics of P2- and P22-type phages. It harbors a genome of 36.7 kb that contains 45 CDSs (66). In CFBP 8072, a large region of 126.8 kb is predicted to contain 31 CDSs from Xfas53, and 8 CDSs are located in another region. In CFBP 8073, five regions contained from seven to 14 Xfas53 CDSs. The other sequences identified by PHAST were similar to lytic prophages of various origin, some of which are found in other plant-pathogenic bacteria, such as Pseudomonas spp. (prophage F10) and Ralstonia solanacearum (prophage RSK1), or in bacteria associated with decaying plant material (prophage ΦSM from Cellulomonas spp.). Apart from lytic phages, sequences of filamentous phages have been identified. This is the case of the filamentous phage Cf1 that has been commonly found infecting Xanthomonas species. This phage has the ability to reduce the growth of infected cells (67). While several phages have been identified in X. fastidiosa strains 9a5c, 6c, and 32 (X. fastidiosa subsp. pauca), Temecula1 (X. fastidiosa subsp. fastidiosa), Dixon (X. fastidiosa subsp. multiplex), and Ann-1 (X. fastidiosa subsp. sandyi) (15, 46, 55, 57, 65), none of these were identified in CFBP 8072 or CFBP 8073 by PHAST but were identified, at least partially and/or fragmented, in the genome sequences of CFBP 8072 or CFBP 8073, based on a blast search. The prophage Xfp6 shared by the 9a5c and 6c strains (55) was also found in strains CFBP 8072 and CFBP 8073 with a very high level of conservation. Hence, phage activity is still in process in strain CFBP 8072 or CFBP 8073 and more generally in X. fastidiosa, being thus another mechanism contributing to the ongoing diversification of the pathogen.
Coffee plant-colonizing strains are genetically diverse, not only in a comparison of strains from South America and Central America but also in a comparison of those within Central America. Our results confirm data previously obtained by Montero-Astúa et al. (68). In order to identify potential determinants of coffee plant adaptation, genome sequences of coffee plant-infecting strains were compared. No groups of orthologs are specific to coffee plant-infecting strains. In contrast, the k-mer approach helped to identify SNPs or fragments that were specific to coffee plant-infecting strains. Most of them remained silent at the protein level and, hence, probably did not impact bacterial interactions with the plant. They may indicate ongoing differentiation of strains based on host constraint.
In this study, the presence of novel alleles together with previously described alleles in these STs show evidence of recombination events. This study emphasizes recombination as a factor of divergence among X. fastidiosa strains. As host shifting associated with the recombination of phylogenetically distant strains was already documented for X. fastidiosa, the current scenario of interception and emergence of X. fastidiosa strains in Europe highlight the importance of avoiding any further introductions in order to limit the risk of creating new genotypes or discovering new hosts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rodrigo P. P. Almeida for his advice concerning the isolation of X. fastidiosa, Leonardo de la Fuente for the gift of the Georgia plum strain (CFBP 8070), Jérôme Gouzy and Sébastien Carrère for annotating the genome sequences, and Matthieu Barret for carefully reading the manuscript.
Funding Statement
The 6-month salary of Stelly Mississipi to participate in this study was provided by Nestlé.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03299-15.
REFERENCES
- 1.EFSA Panel on Plant Health. 2015. Scientific opinion on the risks to plant health posed by Xylella fastidiosa in the EU territory, with the identification and evaluation of risk reduction options. EFSA J 13:3989. [Google Scholar]
- 2.Leu LS, Su CC. 1993. Isolation, cultivation, and pathogenicity of Xylella fastidiosa, the causal bacterium of pear leaf scorch disease in Taiwan. Plant Dis 77:642–646. doi: 10.1094/PD-77-0642. [DOI] [Google Scholar]
- 3.Su CC, Chang CJ, Chang CM, Shih HT, Tzeng KC, Jan FJ, Kao CW, Deng WL. 2013. Pierce's disease of grapevines in Taiwan: isolation, cultivation and pathogenicity of Xylella fastidiosa. J Phytopathol 161:389–396. doi: 10.1111/jph.12075. [DOI] [Google Scholar]
- 4.Amanifar N, Taghavi M, Izadpanah K, Babaei G. 2014. Isolation and pathogenicity of Xylella fastidiosa from grapevine and almond in Iran. Phytopathol Mediterr 53:318–327. [Google Scholar]
- 5.Cariddi C, Saponari M, Boscia D, de Stradis A, Loconsole G, Nigro F, Porcelli F, Potere O, Martelli GP. 2014. Isolation of a Xylella fastidiosa strain infecting olive and oleander in Apulia, Italy. J Plant Pathol 96:1–5. [Google Scholar]
- 6.Elbeaino T, Valentini F, Abou Kubaa R, Moubarak P, Yaseen T, Digiaro M. 2014. Multilocus sequence typing of Xylella fastidiosa isolated from olive affected by “olive quick decline syndrome” in Italy. Phytopathol Mediterr 53:533–542. [Google Scholar]
- 7.Janse JD, Valentini F, Purcell AH, Almeida RPP. 2012. Detection and identification methods and new tests as used and developed in the framework of COST873 for bacteria pathogenic to stone fruits and nuts. J Plant Pathol 94:S1.147-S1.154. [Google Scholar]
- 8.Schaad NW, Postnikova E, Lacy G, Fatmi M, Chang CJ. 2004. Xylella fastidiosa subspecies: X. fastidiosa subsp. piercei subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., and X. fastidiosa subsp. pauca subsp. nov. Syst Appl Microbiol 27:290–300. (Erratum, 27:763.) doi: 10.1078/0723-2020-00263. [DOI] [PubMed] [Google Scholar]
- 9.Schuenzel EL, Scally M, Stouthamer R, Nunney L. 2005. A multigene phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa. Appl Environ Microbiol 71:3832–3839. doi: 10.1128/AEM.71.7.3832-3839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randall JJ, Goldberg NP, Kemp JD, Radionenko M, French JM, Olsen MW, Hanson SF. 2009. Genetic analysis of a novel Xylella fastidiosa subspecies found in the southwestern United States. Appl Environ Microbiol 75:5631–5638. doi: 10.1128/AEM.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunney L, Schuenzel EL, Scally M, Bromley RE, Stouthamer R. 2014. Large-scale intersubspecific recombination in the plant-pathogenic bacterium Xylella fastidiosa is associated with the host shift to mulberry. Appl Environ Microbiol 80:3025–3033. doi: 10.1128/AEM.04112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bull CT De Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, Scortichini M, Stead DE, Takikawa Y. 2012. List of new names of plant pathogenic bacteria (2008-2010). J Plant Pathol 94:21–27. [Google Scholar]
- 13.Bull CT, Coutinho TA, Denny TP, Firrao G, Fischer-Le Saux M, Li X, Saddler GS, Scortichini M, Stead DE, Takikawa Y. 2014. List of new names of plant pathogenic bacteria (2011-2012). J Plant Pathol 96:223–226. [Google Scholar]
- 14.Nunney L, Yuan X, Bromley R, Hartung J, Montero-Astua M, Moreira L, Ortiz B, Stouthamer R. 2010. Population genomic analysis of a bacterial plant pathogen: novel insight into the origins of Pierce's disease of grapevine in the U.S. PLoS One 5:e15488. doi: 10.1371/journal.pone.0015488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nunes LR, Rosato YB, Muto NH, Yanai GM, da Silva VS, de Leite DB, Gonçalves ER, de Souza AA, Coletta-Filho HD, Machado MA, Lopes SA, de Oliveira RC. 2003. Microarray analyses of Xylella fastidiosa provide evidence of coordinated transcription control of laterally transferred elements. Genome Res 13:570–578. doi: 10.1101/gr.930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunney L, Yuan X, Bromley RE. 2012. Detecting genetic introgression: high levels of intersubspecific recombination found in Xylella fastidiosa in Brazil. Appl Environ Microbiol 78:4702–4714. doi: 10.1128/AEM.01126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan X, Morano L, Bromley R, Spring-Pearson S, Stouthamer R, Nunney L. 2010. Multilocus sequence typing of Xylella fastidiosa causing Pierce's disease and oleander leaf scorch in the United States. Phytopathology 100:601–611. doi: 10.1094/PHYTO-100-6-0601. [DOI] [PubMed] [Google Scholar]
- 18.Almeida RPP, Nascimento FE, Chau J, Prado SS, Tsai CW, Lopes SA, Lopes JRS. 2008. Genetic structure and biology of Xylella fastidiosa causing disease in citrus and coffee in Brazil. Appl Environ Microbiol 74:3690–3701. doi: 10.1128/AEM.02388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nunney L, Ortiz B, Russell SA, Ruiz Sánchez R, Stouthamer R. 2014. The complex biogeography of the plant pathogen Xylella fastidiosa: genetic evidence of introductions and subspecific introgression in Central America. PLoS One 9:e112463. doi: 10.1371/journal.pone.0112463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haelterman RM, Tolocka PE, Roca ME, Guzmán FA, Fernández FD, Otero ML. 2015. First presumptive diagnosis of Xylella fastidiosa causing olive scorch in Argentina. J Plant Pathol 97:391–403. [Google Scholar]
- 21.Beretta MJG, Harakava R, Chagas CM. 1996. First report of Xylella fastidiosa in coffee. Plant Dis 80:821. doi: 10.1094/PD-80-0821D. [DOI] [Google Scholar]
- 22.Rodríguez CM, Obando JJ, Villalobos W, Moreira L, Rivera C. 2001. First report of Xylella fastidiosa infecting coffee in Costa Rica. Plant Dis 85:1027. doi: 10.1094/PDIS.2001.85.9.1027A. [DOI] [PubMed] [Google Scholar]
- 23.de Lima JEO, Miranda VS, Hartung JS, Brlansky RH, Coutinho A, Roberto SR, Carlos EF. 1998. Coffee leaf scorch bacterium: axenic culture, pathogenicity, and comparison with Xylella fastidiosa of citrus. Plant Dis 2:94–97. [DOI] [PubMed] [Google Scholar]
- 24.Davis AP, Govaerts R, Bridson DM, Stoffelen P. 2006. An annotated taxonomic conspectus of the genus Coffea (Rubiaceae). Bot J Lin Soc 152:465–512. doi: 10.1111/j.1095-8339.2006.00584.x. [DOI] [Google Scholar]
- 25.Davis AP, Tosh J, Ruch N, Fay M. 2011. Growing coffee: Psilanthus (Rubiaceae) subsumed on the basis of molecular and morphological data; implications for the size, morphology, distribution and evolutionary history of Coffea. Bot J Lin Soc 167:357–377. doi: 10.1111/j.1095-8339.2011.01177.x. [DOI] [Google Scholar]
- 26.Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, Aury JM, Bento P, Bernard M, Bocs S, Campa C, Cenci A, Combes MC, Crouzillat D, da Silva C, Daddiego L, De Bellis F, Dussert S, Garsmeur O, Gayraud T, Guignon V, Jahn K, Jamilloux V, Joët T, Labadie K, Lan T, Leclercq J, Lepelley M, Leroy T, Li LT, Librado P, Lopez L, Muñoz A, Noel B, Pallavicini A, Perrotta G, Poncet V, Pot D, Priyono Rigoreau M, Rouard M, Rozas J, Tranchant-Dubreuil C, VanBuren R, Zhang Q, Andrade AC, Argout X, et al. 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345:1181–1184. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- 27.Lashermes P, Combes MC, Robert J, Trouslot P, D'Hont A, Anthony F, Charrier A. 1999. Molecular characterisation and origin of the Coffea arabica L. genome. Mol Gen Genet 261:259–266. doi: 10.1007/s004380050965. [DOI] [PubMed] [Google Scholar]
- 28.Quiroz-Figueroa FR, Fuentes-Cerda CFJ, Rojas-Herrera R, Loyola-Vargas VM. 2002. Histological studies on the developmental stages and differentiation of two different somatic embryogenesis systems of Coffea arabica. Plant Cell Rep 20:1141–1149. doi: 10.1007/s00299-002-0464-x. [DOI] [Google Scholar]
- 29.European and Mediterranean Plant Protection Organization. 2012. EPPO Reporting Service no. 8, Paris, 2012-08-01. 2012/165 - Xylella fastidiosa detected in a containment facility in France. http://archives.eppo.int/EPPOReporting/2012/Rse-1208.pdf.
- 30.Wells JM, Raju BC, Nyland G, Lowe SK. 1981. Medium for isolation and growth of bacteria associated with plum leaf scald and phony peach diseases. Appl Environ Microbiol 42:357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis MJ, Raju BC, Brlansky RH, Lee RF, Timmer LW, Norris RC, McCoy RE. 1983. Periwinkle wilt bacterium: axenic culture, pathogenicity, and relationships with other gram-negative, xylem-inhabiting bacteria. Phytopathology 73:1510–1515. doi: 10.1094/Phyto-73-1510. [DOI] [Google Scholar]
- 32.Anonymous. 2009. Indirect immunofluorescence test for plant pathogenic bacteria. EPPO standard PM7/97. EPPO Bull 39:413–416. [Google Scholar]
- 33.Hernandez-Martinez R, Costa HS, Dumenyo CK, Cooksey DA. 2006. Differentiation of strains of Xylella fastidiosa infecting grape, almonds and oleander using a multiprimer PCR assay. Plant Dis 90:1382–1388. doi: 10.1094/PD-90-1382. [DOI] [PubMed] [Google Scholar]
- 34.Pooler MR, Hartung JS. 1995. Specific PCR detection and identification of Xylella fastidiosa strains causing citrus variegated chlorosis. Curr Microbiol 31:377–381. doi: 10.1007/BF00294703. [DOI] [PubMed] [Google Scholar]
- 35.Scally M, Schuenzel EL, Stouthamer R, Nunney L. 2005. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl Environ Microbiol 71:8491–8499. doi: 10.1128/AEM.71.12.8491-8499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacques MA, Durand K, Orgeur G, Balidas S, Fricot C, Bonneau S, Quilévéré A, Audusseau C, Olivier V, Grimault V, Mathis R. 2012. Phylogenetic analysis and polyphasic characterization of Clavibacter michiganensis strains isolated from tomato seeds reveal that non-pathogenic strains are distinct from C. michiganensis subsp. michiganensis. Appl Environ Microbiol 78:8388–8402. doi: 10.1128/AEM.02158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo R, Liu B, Xie P, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Yu C, Wang B, Lu Y, Han C, Cheung D, Yiu S, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam T. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sallet E, Gouzy J, Schiex T. 2014. EuGene-PP: a next generation automated annotation pipeline for prokaryotic genomes. Bioinformatics 30:2659–2661. doi: 10.1093/bioinformatics/btu366. [DOI] [PubMed] [Google Scholar]
- 41.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 42.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. 2014. Jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Z, Hao B. 2009. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res 37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart D. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson AJ, Reinach FC, Arruda P, Abreu FA, Acencio M, Alvarenga R, Alves LM, Araya JE, Baia GS, Baptista CS, Barros MH, Bonaccorsi ED, Bordin S, Bové JM, Briones MR, Bueno MR, Camargo AA, Camargo LE, Carraro DM, Carrer H, Colauto NB, Colombo C, Costa FF, Costa MC, Costa-Neto CM, Coutinho LL, Cristofani M, Dias-Neto E, Docena C, El-Dorry H, Facincani AP, Ferreira AJ, Ferreira VC, Ferro JA, Fraga JS, França SC, Franco MC, Frohme M, Furlan LR, Garnier M, Goldman GH, Goldman MH, Gomes SL, Gruber A, Ho PL, Hoheisel JD, Junqueira ML, Kemper EL, Kitajima JP, Krieger JE, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151–159. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- 47.Van Sluys MA, de Oliveira MC, Monteiro-Vitorello CB, Miyaki CY, Furlan LR, Camargo LE, da Silva AC, Moon DH, Takita MA, Lemos EG, Machado MA, Ferro MI, da Silva FR, Goldman MH, Goldman GH, Lemos MV, El-Dorry H, Tsai SM, Carrer H, Carraro DM, de Oliveira RC, Nunes LR, Siqueira WJ, Coutinho LL, Kimura ET, Ferro ES, Harakava R, Kuramae EE, Marino CL, Giglioti E, Abreu IL, Alves LM, do Amaral AM, Baia GS, Blanco SR, Brito MS, Cannavan FS, Celestino AV, da Cunha AF, Fenille RC, Ferro JA, Formighieri EF, Kishi LT, Leoni SG, Oliveira AR, Rosa VE Jr, Sassaki FT, Sena JA, de Souza AA, Truffi D, et al. 2003. Comparative analyses of the complete genomes sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J Bacteriol 185:1018–1026. doi: 10.1128/JB.185.3.1018-1026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Firrao G, Bazzi C. 1994. Specific identification of Xylella fastidiosa using the polymerase chain reaction. Phytopathol Mediterr 33:90–92. [Google Scholar]
- 51.Minsavage GV, Thompson CM, Hopkins DL, Leite RMVBC, Stall RE. 1994. Development of a polymerase chain protocol for detection of Xylella fastidiosa in plant tissue. Phytopathology 84:456–461. doi: 10.1094/Phyto-84-456. [DOI] [Google Scholar]
- 52.Harper SJ, Ward LI, Clover GRG. 2010. Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology 100:1282–1288. doi: 10.1094/PHYTO-06-10-0168. [DOI] [PubMed] [Google Scholar]
- 53.Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log likelihoods with applications to phylogenetic inference. Mol Biol Evol 16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- 54.Kung SH, Almeida RPP. 2014. Biological and genetic factors regulating natural competence in a bacterial plant pathogen. Microbiology 160:37–46. doi: 10.1099/mic.0.070581-0. [DOI] [PubMed] [Google Scholar]
- 55.Barbosa D, Alencar VC, Souza Santos D, de Freitas Oliveira AC, de Souza AA, Coletta-Filho HD, Costa de Oliveira R, Nunes LR. 2015. Comparative genomic analysis of coffee-infecting Xylella fastidiosa strains isolated from Brazil. Microbiology 161:1018–1033. doi: 10.1099/mic.0.000068. [DOI] [PubMed] [Google Scholar]
- 56.Chen J, Su CC, Deng WL, Jan FJ, Chang CJ, Huang H. 2014. Analyses of Xylella whole genome sequences and proposal of Xylella taiwanensis sp. nov. Proceedings of the 2014 Pierce's Disease Research Symposium, 15 to 17 December 2014, Sacramento, CA, USA. [Google Scholar]
- 57.Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 58.Alencar VC, Barbosa D, Santos DS, Oliveira ACF, de Oliveira RC, Nunes LR. 2014. Genomic sequencing of two coffee-infecting strains of Xylella fastidiosa isolated from Brazil. Genome Announc 2:e01190-13. doi: 10.1128/genomeA.01190-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergsma-Vlami M, van de Bilt JLJ, Tjou-Tam-Sin NNA, van de Vossenberg BTLH, Westenberg M. 2015. Xylella fastidiosa in Coffea arabica ornamental plants imported from Costa Rica and Honduras in the Netherlands. J Plant Pathol 97:395. [Google Scholar]
- 60.Chatterjee S, Wistrom C, Lindow SE. 2008. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc Natl Acad Sci U S A 105:2670–2675. doi: 10.1073/pnas.0712236105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.French WJ, Kitajima EW. 1978. Occurrence of plum leaf scald in Brazil and Paraguay. Plant Dis Rep 62:1035–1038. [Google Scholar]
- 62.Mehta A, Leite RP, Rosato YB. 2001. Assessment of the genetic diversity of Xylella fastidiosa isolated from citrus in Brazil by PCR-RFLP of the 16SrDNA and 16S-23S intergenic spacer and rep-PCR fingerprinting. Antonie Van Leeuwenhoek 79:53–59. doi: 10.1023/A:1010219811555. [DOI] [PubMed] [Google Scholar]
- 63.Nunney L, Hopkins DL, Morano LD, Russell SE, Stouthamer R. 2014. Intersubspecific recombination in Xylella fastidiosa strains native to the United States: infection of novel hosts associated with an unsuccessful invasion. Appl Environ Microbiol 80:1159–1169. doi: 10.1128/AEM.02920-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pendergrast M. 2001. Uncommon grounds: the history of coffee and how it transformed our world. Texere, London, United Kingdom. [Google Scholar]
- 65.Varani AM, Monteiro-Vitorello CB, Nakaya HI, Van Sluys MA. 2013. The role of prophage in plant-pathogenic bacteria. Annu Rev Phytopathol 51:429–451. doi: 10.1146/annurev-phyto-081211-173010. [DOI] [PubMed] [Google Scholar]
- 66.Summer EJ, Enderle CJ, Ahern SJ, Gill JJ, Torres CP, Appel DN, Black MC, Young R, Gonzalez CF. 2010. Genomic and biological analysis of phage Xfas53 and related prophages of Xylella fastidiosa. J Bacteriol 192:179–190. doi: 10.1128/JB.01174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuo TT, Tan MS, Su MT, Yang MK. 1991. Complete nucleotide sequence of filamentous phage Cf1c from Xanthomonas campestris pv. citri. Nucleic Acids Res 19:2498. doi: 10.1093/nar/19.9.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montero-Astúa M, Hartung JS, Aguilar E, Chacón C, Li W, Albertazzi FJ, Rivera C. 2007. Genetic diversity of Xylella fastidiosa strains from Costa Rica, São Paulo, Brazil, and United States. Phytopathology 97:1338–1347. doi: 10.1094/PHYTO-97-10-1338. [DOI] [PubMed] [Google Scholar]
- 69.Chen J, Xie G, Han S, Chertkov O, Sims D, Civerolo EL. 2010. Whole-genome sequences of two Xylella fastidiosa strains (M12 and M23) causing almond leaf scorch disease in California. J Bacteriol 192:4534. doi: 10.1128/JB.00651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang S, Flores-Cruz Z, Kumar D, Chakrabarty P, Hopkins DL, Gabriel DW. 2011. The Xylella fastidiosa biocontrol strain EB92-1 genome is very similar and syntenic to Pierce's disease strains. J Bacteriol 193:5576–5577. doi: 10.1128/JB.05430-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guan W, Shao J, Zhao T, Huang Q. 2014. Genome sequence of a Xylella fastidiosa strain causing mulberry leaf scorch disease in Maryland. Genome Announc 2:e00916-13. doi: 10.1128/genomeA.00916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J, Huang H, Chang C-J, Stenger DC. 2013. Draft genome sequence of Xylella fastidiosa subsp. multiplex strain Griffin-1 from Quercus rubra in Georgia. Genome Announc 1:e00756-13. doi: 10.1128/genomeA.00756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guan W, Shao J, Davis RE, Zhao T, Huang Q. 2014. Genome sequence of a Xylella fastidiosa strain causing sycamore leaf scorch disease in Virginia. Genome Announc 2:e00773-14. doi: 10.1128/genomeA.00773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giampetruzzi A, Chiumenti M, Saponari M, Donvito G, Italiano A, Loconsole G, Boscia D, Cariddi C, Martelli GP, Saldarelli P. 2015. Draft genome sequence of the Xylella fastidiosa CoDiRO strain. Genome Announc 3:e01538-14. doi: 10.1128/genomeA.01538-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Su C-C, Deng W-L, Jan F-J, Chang C-J, Huang H, Chen J. 2014. Draft genome sequence of Xylella fastidiosa pear leaf scorch strain in Taiwan. Genome Announc 2:e00166-14. doi: 10.1128/genomeA.00166-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.