Abstract
Forming biofilms may be a survival strategy of Shiga toxin-producing Escherichia coli to enable it to persist in the environment and the food industry. Here, we evaluate and characterize the biofilm-forming ability of 39 isolates of Shiga toxin-producing Escherichia coli isolates recovered from human infection and belonging to seropathotypes A, B, or C. The presence and/or production of biofilm factors such as curli, cellulose, autotransporter, and fimbriae were investigated. The polymeric matrix of these biofilms was analyzed by confocal microscopy and by enzymatic digestion. Cell viability and matrix integrity were examined after sanitizer treatments. Isolates of the seropathotype A (O157:H7 and O157:NM), which have the highest relative incidence of human infection, had a greater ability to form biofilms than isolates of seropathotype B or C. Seropathotype A isolates were unique in their ability to produce cellulose and poly-N-acetylglucosamine. The integrity of the biofilms was dependent on proteins. Two autotransporter genes, ehaB and espP, and two fimbrial genes, z1538 and lpf2, were identified as potential genetic determinants for biofilm formation. Interestingly, the ability of several isolates from seropathotype A to form biofilms was associated with their ability to agglutinate yeast in a mannose-independent manner. We consider this an unidentified biofilm-associated factor produced by those isolates. Treatment with sanitizers reduced the viability of Shiga toxin-producing Escherichia coli but did not completely remove the biofilm matrix. Overall, our data indicate that biofilm formation could contribute to the persistence of Shiga toxin-producing Escherichia coli and specifically seropathotype A isolates in the environment.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) strains, including enterohemorrhagic E. coli (EHEC), are food-borne and waterborne human enteric pathogens responsible for infections. They are associated with important public health concerns in developed countries. Symptoms associated with STEC infections range from abdominal cramps and bloody diarrhea to postinfectious complications, such as hemolytic-uremic syndrome (HUS). HUS, a life-threatening complication of STEC infections, is a consequence of Shiga toxin production. According to the Public Health Agency of Canada (PHAC), more than 30,000 cases of STEC infections occur each year in Canada (1). Environmentally, the main reservoir for STEC is cattle. Transmission of STEC to humans typically is associated with the consumption of contaminated food such as undercooked beef, fresh produce, unpasteurized milk, or contaminated drinking water (2), but these pathogens also can be transmitted from person to person or via direct contact with animals or their feces.
The 2003 Karmali seropathotype model classifies STEC into seropathotypes based on their reported incidence in human disease, outbreaks, and/or association with the development of severe symptoms in humans (3). Serotypes frequently responsible for hemorrhagic colitis (HC) and HUS, O157:H7 and O157:NM, are assigned to seropathotype A. Seropathotype B strains are associated with outbreaks and HUS, but less commonly than those of seropathotype A, and they include serotypes O26:H11, O103:H2, O111:NM, O121:H19, O45:H2, and O145:NM. These serotypes belong to six serogroups (O26, O45, O103, O111, O121, and O145) that have been described by the CDC as the cause of most non-O157 STEC infections and are known as the big 6 (4). Seropathotype C serotypes are associated with sporadic HUS cases but no epidemics. The serotypes in group C are O91:H21, O104:H21, O113:H21, O5:NM, O121:NM, and O165:H25. Seropathotype D serotypes are associated with diarrhea but not with outbreaks or HUS cases. Seropathotype E serotypes include STEC serotypes with no reported implication in human infection (3).
Biofilms are multicellular communities of bacteria that attach to abiotic or biotic surfaces and produce an extracellular polymeric matrix (5). The biofilm-forming ability of STEC has been investigated under different conditions, using different STEC isolates representing different serotypes and origins (6–8). Biofilm formation by STEC may be regarded as a survival strategy for this bacterium (9–12). Indeed, bacteria within a biofilm are protected against several stresses, such as a nutrient-limited environment and sanitizers (13). This biofilm-mediated protection against sanitizers impedes contamination control, especially in food-processing plants (7). This allows biofilms to increase their survival and persistence in hostile environments such as water, a nutrient-limited environment, and food-processing plants, an environment where sanitizers are applied (11, 13, 14). In general, studies concerning STEC biofilms highlighted that their formation is heterogeneous and mostly dependent on the strains and the conditions used (7). However, there is limited information about the composition of the STEC biofilm matrix. Moreover, non-O157 isolates continue to gain importance as pathogens of concern (15). Their biofilm-forming abilities were shown to be different from those of O157 isolates (8). This gives importance to characterizing STEC biofilms and to understanding how these bacteria travel through the food supply chain from preharvest to consumer goods. The production of many adhesins has been associated with biofilm formation. Fimbriae such as type 1 fimbriae, curli, long polar fimbriae, and F9 fimbriae have been hypothesized to be required largely during the attachment step (16). The maturation step of STEC biofilm is thought to involve mostly autotransporters such as EhaA, EhaB, Agn43, Cah, and EspP (8, 17–20) and exopolysaccharides such as poly-N-acetylglucosamine (PGA), cellulose, and colanic acid (21). We hypothesize that its ability to form biofilms contributes to STEC persistence and that this consequently could influence their incidence. Therefore, the aim of this study was to evaluate and characterize the biofilm-forming ability of STEC isolates recovered from human infection that represent the most important seropathotypes. Furthermore, we investigated the putative link between adhesins or surface molecules/structures and biofilm formation.
We also characterized the composition of matrix and tolerance to sanitizers of the strongest biofilm formers.
MATERIALS AND METHODS
STEC isolates, storage, and culture conditions.
The collection of 39 STEC isolates associated with human infections that represent 14 different serotypes used in this study are described in Table 1 (3, 22–25). French isolates were provided by Christine Martin (INRA Clermont-Ferrand Theix), and North American isolates were provided by Mohamed Karmali from the Laboratory for Food-Borne Zoonoses of the Public Health Agency of Canada. Isolates were stored at −80°C in lysogeny broth (LB) containing 15% (vol/vol) glycerol. Each test was performed in triplicate and in three separate experiments. For each experiment, STEC isolates first were streaked onto LB agar and incubated overnight at 37°C.
TABLE 1.
List of STEC isolates used in this study and biofilm formation under static and dynamic conditions
| Serotype and name | Reference or sourced | Seropathotype | Biofilm quantificationc |
|
|---|---|---|---|---|
| Statica | Dynamicb | |||
| O157:H7 | ||||
| EDL933 | 22 | A | 0.86 (±0.29) | 12 (±0.6) |
| Sakai | 23 | A | 1.81 (±0.48) | 35 (±2) |
| 86-24 | 24 | A | 0.63 (±0.06) | 88 (±2) |
| AJH-4 | 3 | A | 0.25 (±0.11) | 90 (±1) |
| AJH-5 | 3 | A | 0.65 (±0.19) | 76 (±12) |
| AJH-6 | 3 | A | 0.38 (±0.06) | 67 (±6) |
| AJH-7 | 3 | A | 0.44 (±0.11) | 78(±2) |
| AJH-8 | 3 | A | 0.50 (±0.31) | 82 (±2) |
| AJH-9 | 3 | A | 0.31 (±0.06) | 97 (±2) |
| AJH-10 | 3 | A | 0.65 (±0.18) | 76 (±3) |
| AJH-11 | 25 | A | 1.21 (±0.39) | 28 (±4) |
| AJH-12 | 25 | A | 0.61 (±0.06) | 80 (±9) |
| AJH-13 | 25 | A | 0.37 (±0.02) | 24 (±5) |
| AJH-14 | 25 | A | 0.70 (±0.15) | 97 (±1) |
| O157:NM | ||||
| AJH-15 | 3 | A | 1.00 (±0.17) | 23 (±5) |
| O157:H26 | ||||
| CJH-1 | 25 | C | 0.34 (±0.16) | 99 (±0.3) |
| O26:H11 | ||||
| BJH-1 | 3 | B | 0.18 (±0.06) | 86 (±4) |
| BJH-2 | 3 | B | 0.35 (±0.07) | 65 (±33) |
| BJH-3 | 3 | B | 0.38 (±0.13) | 9 (±2) |
| BJH-4 | PHAC | B | 0.21 (±0.067) | 44 (±3) |
| O45:H2 | ||||
| BJH-5 | PHAC | B | 0.22 (±0.04) | 44 (±3) |
| BJH-6 | PHAC | B | 0.13 (±0.05) | 27 (±3) |
| BJH-7 | PHAC | B | 0.13 (±0.03) | 31 (±1) |
| BJH-8 | PHAC | B | 0.16 (±0.04) | 43 (±1) |
| O103:H2 | ||||
| BJH-9 | 25 | B | 0.30 (±0.13) | 37 (±5) |
| O111:H8 | ||||
| BJH-10 | PHAC | B | 0.61 (±0.3) | 67 (±6) |
| BJH-11 | PHAC | B | 0.25 (±0.1) | 53 (±6) |
| O111:NM | ||||
| BJH-12 | 3 | B | 0.43 (±0.25) | 11 (±1) |
| BJH-13 | 3 | B | 0.41 (±0.11) | 61 (±2) |
| BJH-14 | 3 | B | 0.28 (±0.09) | 19 (±1) |
| O121:H19 | ||||
| BJH-15 | PHAC | B | 0.68 (±0.31) | 93 (±3) |
| BJH-16 | PHAC | B | 0.65 (±0.30) | 92 (±10) |
| BJH-17 | PHAC | B | 0.65 (±0.19) | 75 (±4) |
| BJH-18 | 3 | B | 0.80 (±0.30) | 87 (±4) |
| O145:H25 | ||||
| CJH-2 | PHAC | C | 0.37 (±0.12) | 82 (±2) |
| O145:NM | ||||
| BJH-19 | PHAC | B | 0.15 (±0.12) | 99 (±1) |
| BJH-20 | PHAC | B | 0.51 (±0.30) | 96 (±1) |
| BJH-21 | PHAC | B | 0.08 (±0.09) | 82 (±4) |
| O113:H21 | ||||
| CJH-3 | 3 | C | 1.57 (±0.32) | 84 (±4) |
Absorbance read at 595 nm after crystal violet staining.
Percentage of area covered by the biofilm or cells determined by ImageJ analysis.
Values are averages (± standard errors) from at least three independent biological replicates.
PHAC, Public Health Agency of Canada.
Static biofilm formation assay.
The biofilm formation assay was performed as previously described (26). Briefly, overnight cultures at 37°C in LB media were diluted (1:100) in 5 ml of M9 medium with glucose (0.4%, wt/vol) and minerals (1.16 mM MgSO4, 2 μM FeCl3, 8 μM CaCl2, and 16 μM MnCl2) and incubated for 24 h at 37°C. These cultures were diluted (1:100) in M9 medium supplemented with glucose and minerals and were inoculated in triplicates into the microtiter plates (Costar 3370; Corning, NY). For all biofilm assays, the biofilm-positive STEC strain EDL 933 (26) and the biofilm-negative STEC isolate BJH-21 were used as positive and negative biofilm controls, respectively. Moreover, wells filled with sterile media were used as the blank value. After a 24-h incubation at 30°C, unattached cells were removed by washing three times with phosphate-buffered saline (PBS; 1.76 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4). Plates were dried at 37°C for 15 min, and biofilms were stained with crystal violet (0.1% [wt/vol]) for 2 min. The crystal violet solution was removed, and the biofilms were washed three times with PBS and then dried at 37°C for 15 min. The stain was released with 150 μl of 80% (vol/vol) ethanol and 20% (vol/vol) acetone. Biofilms were quantified by measuring the absorbance at 595 nm with a microplate reader (Powerwave; BioTek Instruments, Winooski, VT). According to their absorbance, isolates were categorized as strong (A595 > 0.6), medium (0.6 > A595 > 0.3), and weak (0.3 > A595 > 0.1) biofilm formers or as non-biofilm formers (A595 < 0.1).
Biofilms in the BioFlux flowthrough device.
Growth of biofilms in the BioFlux 200 device (Fluxion Biosciences, South San Francisco, CA) was performed as described before (26). Briefly, E. coli cultures were prepared as described above, and 1 ml from the 24-h culture was pelleted and resuspended in fresh prewarmed (30°C) M9 medium with glucose (0.4%, wt/vol) to an optical density at 600 nm (OD600) of ∼1. The microfluidic channels then were inoculated and the plate was incubated for 2 h at 30°C to allow bacteria to bind to the surface. Fresh M9 medium with glucose (0.4%, wt/vol) was added to the input reservoir, and the flow of fresh medium then was initiated at 1.0 dyne/cm2 for 5 h, followed by a decrease of the flow to 0.5 dyne/cm2 for an additional 17 h. Once the incubation was completed, biofilms were washed by injecting PBS, stained by injecting crystal violet, and washed by injecting PBS from the input reservoir for 20 min at 0.5 dyne/cm2. Images of BioFlux biofilms were obtained using an inverted fluorescence microscope (Olympus CKX41; Markham, ON, Canada), a digital camera (Retiga EX; Q Imaging Surrey, BC, Canada), and the software provided with the BioFlux 200 device. Images of biofilms and cell attachment obtained from three independent replicates were analyzed using ImageJ (National Institutes of Health, Bethesda, MD). The thresholds of the 16-bit greyscale images were adjusted to fit the bacterial clusters, and these modified images were analyzed using the “Analyze Particles” function. The percentage of area covered represents the surface of the picture covered by the biofilms or the adhered cells.
CR and calcofluor phenotype assays.
Congo red (CR) or calcofluor white (CF) assays were performed as described previously (27). Briefly, 2 μl of an overnight LB culture grown at 37°C was spotted onto M9 agar supplemented with 0.4% glucose and minerals containing 0.004% (vol/vol) CR and 0.002% (vol/vol) brilliant blue or containing 0.02% (vol/vol) CF (F-3543; Sigma) and 1 mM HEPES. Once the drops were dry, the plates were incubated for 24 h or 48 h at 30°C or 37°C. The binding of CR was evaluated based on the color of the colony (red or pink colonies were considered positive and white colonies were considered negative), and binding of CF was evaluated by the emission of fluorescence by colonies exposed to a UVA light (400 to 315 nm). Statistical analysis was performed by using a Mann-Whitney test.
PCR detection of autotransporter and fimbria-encoding genes.
The presence of autotransporter and fimbria-encoding genes was detected by PCR on bacterial lysates using primers described in the supplemental material (see Table S1). The primers were designed on conserved regions of genes based on alignment analysis of homologous genes. Two sets of primers were used to detect autotransporter-encoding genes ehaA, ehaB, ehaC, ehaD, and ehaG. The first set of primers amplified the passenger-encoding domain sequence (named ehaα) where nucleotide divergences have been described. The other set of primers amplified the translocation-encoding domain sequence (named ehaβ), which is highly conserved (8, 18, 28).
Detection of type 1 fimbriae.
The capacity of bacterial isolates to express a d-mannose-binding phenotype was measured by the ability to agglutinate Saccharomyces cerevisiae cells as described previously (29). Briefly, the 24-h M9 cultures prepared as described above were diluted (1:100) in 20 ml M9-0.4% glucose and incubated at 30°C without agitation for 24 h. An initial suspension of approximately 2 × 1011 CFU/ml in PBS was diluted by 2-fold serial dilution in a microtiter plate (Corning Costar no. 2797), and an equal volume of a commercial yeast suspension in PBS (final concentration, 1.5% [wt/vol]) was added to each well. After 30 min of incubation at 4°C, yeast agglutination was monitored visually by agglutination and precipitation of cells. The agglutination titer was established as the lowest bacterial dilution at which agglutination was observed. If α-d-mannopyranose (5% [wt/vol]) (Sigma) inhibited agglutination, yeast agglutination was considered to be due to type 1 fimbriae.
CLSM.
Biofilms were prepared as described above, and after 24 h of incubation at 30°C, biofilms were washed and stained with FilmTracer FM1-43 Green biofilm cell stain (Molecular Probes, Eugene, OR) according to the manufacturer's instructions as described previously (30). To determine the composition of the biofilm matrix, biofilms were stained with wheat germ agglutinin (WGA; Oregon Green 488; binds to N-acetyl-d-glucosamine and N-acetylneuraminic acid residues; Molecular Probes), FilmTracer SYPRO Ruby biofilm matrix stain (labels most classes of proteins; Molecular Probes), calcofluor white [binds to (1-3)-β- and (1-4)-β-d-glucopyranoside; Sigma], or BOBO-3 iodide (stains extracellular DNA; Molecular Probes) according to the manufacturer's instructions. After a 30-min incubation at room temperature protected from light, the fluorescent marker solution was removed, biofilms were washed with water, and the wells were filled with 100 μl of water or PBS for WGA-stained biofilms. Stained biofilms were visualized by confocal laser scanning microscopy (CLSM; FV1000 IX81; Olympus, Markham, ON, Canada), and images were acquired using FluoView software (Olympus).
Biofilm dispersion assay.
Enzymatic dispersion of established biofilms was performed as described previously (31, 32). Briefly, biofilms were grown as described above, and after 24 h of incubation, 37.5 μl of solution containing DNase I (100 μg/ml in 150 mM NaCl, 1 mM CaCl2), dispersin B (20 μg/ml in PBS; Kane Biotech Inc., Winnipeg, MB, Canada), cellulase (100 μg/ml in 50 mM sodium citrate, pH 5.0), or proteinase K (100 μg/ml in 50 mM Tris-HCl, pH 7.5, 1 mM CaCl2) was added directly to the biofilms. Control wells were treated with 37.5 μl of the buffer without enzymes. Wells were treated for 1 h at 37°C, and biofilms were washed, stained, and quantified as described above.
Sanitizer treatments.
Five sanitizers commonly used on the farm and in food-processing industries were used to test the ability of STEC biofilm cells and matrix to survive sanitizer treatments. The sanitizers were Virkon, a peroxide compound; Aseptol 2000, a quaternary ammonium chloride-based sanitizer; bleach, a chlorine compound; and hydrogen peroxide (H2O2; Sigma) and ethanol. Sanitizers were diluted with sterile distilled water to concentrations that did not require rinsing after their use as indicated by the manufacturer. Biofilms were formed as described above, and after 24 h of incubation at 30°C, media were removed and wells were filled with 200 μl of PBS (nontreated control) or sanitizers. Five different sanitizers were used and prepared as follows. A bleach solution containing 200 ppm of sodium hypochlorite; a 1% (vol/vol) Aseptol 2000 solution (Laboratoire Meriel, Pleumeleuc, France) containing 0.127% glutaraldehyde and quaternary ammonium compounds, including 0.014% (vol/vol) dodecyl dimethyl ammonium chloride, 0.06% (vol/vol) benzalkonium chloride, and 0.007% (vol/vol) alkyl dimethyl ethyl benzyl ammonium chloride; a 1% (wt/vol) Virkon solution (Vetoquinol, Lavaltrie, QC, Canada) containing 0.214% (wt/wt) potassium monopersulfate; a 5% (vol/vol) H2O2 solution (Sigma); or a 70% (vol/vol) ethanol solution. After 10 min of incubation at 25°C, wells were washed three times with 200 μl of neutralizing broth (Difco Laboratories). To evaluate the viability of STEC cells after the sanitation treatments, cells were harvested by scraping the surface with sterile pipette tips and rinsing the well with 200 μl of PBS. Bacterial cells were pooled from four replicate wells that received the same sanitization treatment. The volume of PBS was adjusted to 1 ml, vigorously vortexed, serially diluted in fresh PBS, and plated onto LB agar. The plates were incubated overnight at 37°C, and colonies on the plates then were counted to determine the number of STEC cells present. In addition to CFU, the metabolic activity of the treated biofilms also was evaluated using the CellTiter-Blue reagent (Promega Corporation) as described previously (33). CellTiter-Blue reagent is a resazurin solution that is reduced by viable cells into resorufin, which emits fluorescence. Briefly, treated biofilms were incubated at 37°C in LB containing 40 μl of CellTiter-Blue reagent. After 3 h of incubation, the level of fluorescence (excitation wavelength [λex], 570 nm; emission wavelength, [λem], 600 nm) was measured using a microplate reader. The percentage of remaining metabolic activity allowed us to evaluate the number of surviving bacteria. Percent resazurin reduction was calculated using the following formula: (experimental well absorbance − negative-control absorbance)/positive-control absorbance × 100. To evaluate if the biofilm matrix was removed, crystal violet was added to sanitizer-treated biofilms as described for the biofilm formation assay.
Statistical analysis.
Statistical analyses were performed using GraphPad Prism, version 4.02 (GraphPad Software, San Diego, CA). The results for static biofilms at the serogroup level and the results for enzymatic digestion were analyzed using a nonparametric one-way analysis of variance (ANOVA) with Dunn's multiple-comparison posttest. A Mann-Whitney test with two-tailed distribution was performed to compare biofilm formation and seropathotype, the ability to bind CR and CF, or the presence of autotransporter and fimbria-encoding genes.
RESULTS
Seropathotype A isolates (O157:H7) have greater capacity to form biofilms.
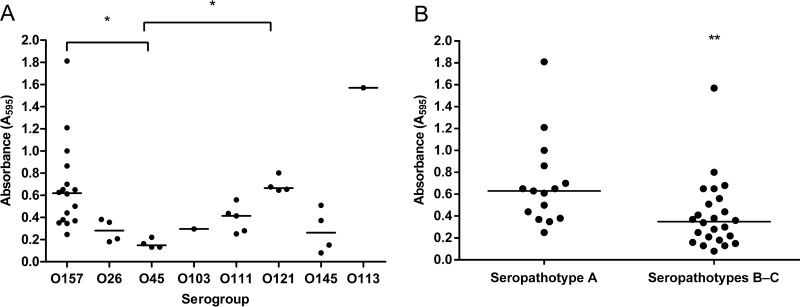
STEC biofilm production was characterized using the previously determined static and dynamic conditions (26). STEC organisms were grown in M9 supplemented with 0.4% glucose media in polystyrene microplates incubated at 30°C for 24 h, and biofilms were evaluated by crystal violet staining. Biofilm formation was tested after 24 h, 48 h, and 72 h of incubation. The 24-h time point was selected because the OD values of crystal violet staining were the highest at this time and had decreased after 48 h (data not shown). The capacity of isolates to form biofilms under static conditions was highly variable (OD of 0.08 for the lowest, OD of 1.8 for the highest). Depending on their capacity, isolates were classified as strong (A595 > 0.6), medium (0.6 > A595 > 0.3), or weak (0.3 > A595 > 0.1) biofilm formers or as non-biofilm formers (A595 < 0.1). Of the 39 isolates, 14 (35.9%) formed a strong biofilm, 13 (33.3%) formed a medium biofilm, 11 (28.2%) formed a weak biofilm, and 1 isolate of serotype O145:NM (2.5%) was not able to form biofilm (Fig. 1A). Interestingly, we noticed that under static conditions, the majority of seropathotype A isolates formed significantly stronger biofilms (P < 0.01) than isolates of other seropathotypes (Fig. 1B). At the serogroup level, isolates of serogroups O157 and O121 were strong biofilm formers, while O45 and O26 isolates were weak biofilm formers (Fig. 1A). When biofilms were grown under dynamic conditions, the capacity for STEC isolates to form biofilms was characterized by the coverage area. Under these conditions, 64.1% of STEC isolates were able to cover more than 50% of the area (Table 1). Under dynamic conditions, the majority (66.7%) of seropathotype A isolates formed strong biofilm covering more than 65% of the area. In addition, most isolates from serogroups O157 and O121 that formed strong biofilms under static conditions also were able to cover 69% and 89% of the area, respectively, under dynamic conditions. Similar observations were made for isolates from serogroup O45 that formed weak biofilms under static conditions and cover only 36% of the area in microfluidics. However, biofilms formed by O145:NM isolates were able to cover 92% of the area under dynamic conditions, whereas their biofilms were weak or absent when grown statically (Fig. 1A).
FIG 1.
Biofilm formation by STEC isolates grouped by serotype (A) and by seropathotype (B). Biofilms were formed in M9 medium plus glucose (0.4%, wt/vol) under static conditions in microtiter plates at 30°C and stained with crystal violet. The absorbance at 595 nm was measured. Results are averages from at least 3 biological replicates. The horizontal bars represent the medians. Statistical analysis was performed by using a nonparametric one-way ANOVA with Dunn's multiple-comparison posttest (A) and a t test for seropathotype comparison (B). *, P < 0.05; **, P < 0.01.
Production of curli and cellulose among STEC isolates is heterogeneous.
The occurrence of curli and cellulose or cellulose-like extracellular material production was analyzed. We grew STEC isolates at 30°C and 37°C for 24 h and 48 h on minimal agar plates containing Congo red dye (CR), indicative of curli and cellulose production, as well as on plates containing calcofluor white (CF), a fluorochrome that binds to polysaccharides with β-1,3 and β-1,4 linkages, such as cellulose, chitin, and succinoglycans (27). CR binding was a common characteristic in STEC isolates, since 37/39 (94.8%) of the tested isolates turned red on CR agar plates under at least one of the tested conditions (minimal medium; 30°C or 37°C; 24 h or 48 h) (see Table S2 in the supplemental material). The ability to bind CF was less frequent than that of CR (15/39; 38.5%), but the number of CF-positive isolates was more significant at 30°C (12/39; 30.7%) than at 37°C (9/39; 23.1%) (see Table S2). However, these CR and CF phenotypes do not strictly correlate with the capacity to form a biofilm under our experimental conditions (see Fig. S1).
Seropathotype A isolates (O157:H7) aggregate to yeast in a mannose-independent manner.
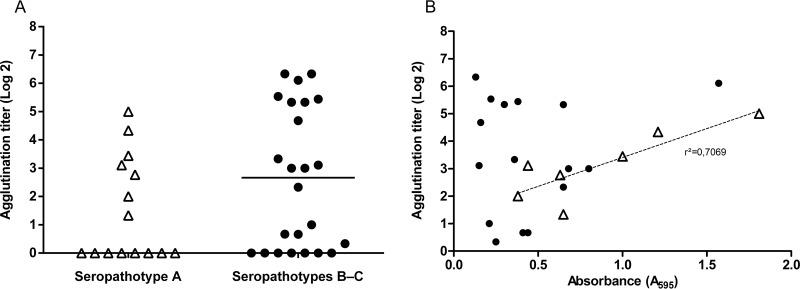
Among STEC isolates, 17 non-O157 isolates produced type 1 fimbriae as they were able to agglutinate yeast in a mannose-dependent manner (Fig. 2A); no correlation between biofilm formation and production of type 1 fimbriae was observed (Fig. 2B). Interestingly, 7 seropathotype A (O157) isolates were able to agglutinate yeast in a mannose-independent manner (Fig. 2A), and this agglutination phenotype correlated with biofilm formation (r2 = 0.7) (Fig. 2B).
FIG 2.
Yeast agglutination of seropathotype A and seropathotype B or C isolates (A) and mannose-dependent and -independent yeast agglutination as a function of biofilm formation (B). Yeast agglutination was determined with 24-h static culture of isolates grown in M9 supplemented with 0.4% glucose. (A) Seropathotype A isolates are represented by filled or open triangles, and seropathotypes B and C are represented by solid dots. Some seropathotype A and seropathotype B and C isolates were unable to agglutinate to yeast. The horizontal bars represent the median. (B) A linear regression analysis was made between the yeast agglutination titer and the amount of biofilm formed under static conditions. There was no correlation between mannose-dependent agglutination titer (indicative of type 1 fimbria production) and biofilm formation (correlation coefficient [r2] = 0.01) represented by solid dots, whereas there was a correlation between mannose-independent agglutination isolates and biofilm formation (r2 = 0.70) (both represented by open triangles). Results are averages from at least 3 biological replicates.
Association between the presence of fimbrial and autotransporter genes and biofilm formation.
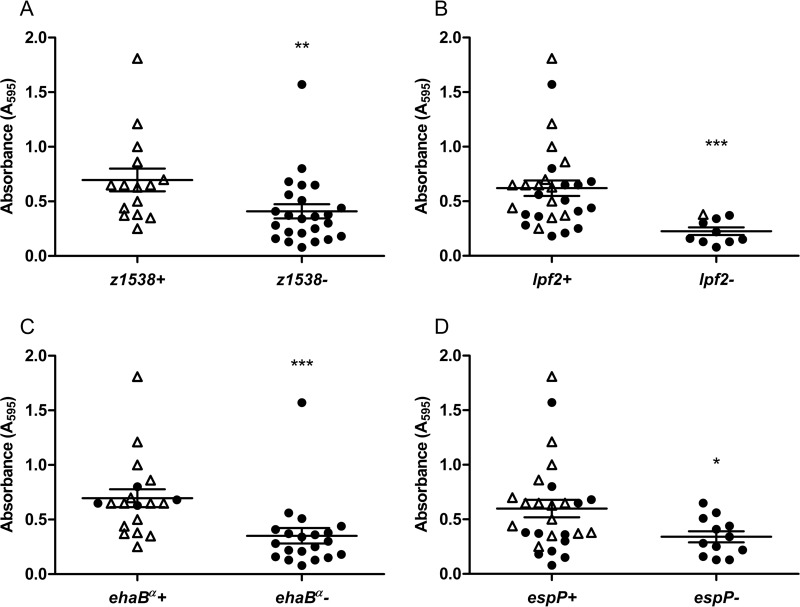
The presence of genes coding for factors that are putatively involved in STEC biofilm formation (8, 18, 34–36) was examined by PCR using primers designed in conserved regions based on sequence alignment (see Table S1 in the supplemental material). We observed an association between the presence of fimbrial genes z1538 and lpf2 and autotransporter gene ehaBβ or espP and biofilm formation (Fig. 3). As revealed by crystal violet staining, z1538-positive isolates and lpf2-positive isolates formed significantly (P < 0.01 and P < 0.001, respectively) more biofilms than z1538-negative or lpf2-negative isolates (Fig. 3A and B). For genes ehaA, ehaB, ehaC, ehaD, and ehaG, we used primers designed to amplify the variable passenger-encoding domain, named α, and the more conserved translocating-encoding domain, named β. Interestingly, the β domain sequence, ehaBβ, was detected in most isolates (38/39), while the α passenger domain sequence, ehaBα, was detected only in O157:H7, O157:NM, and O121:H19 isolates (see Table S3). Furthermore, ehaBα-positive isolates formed significantly more (P < 0.001) biofilms than ehaBα-negative ones (Fig. 3C). This suggests that the passenger-encoding domain of the gene for EhaB in STEC plays a role in biofilm formation. Moreover, espP-positive isolates formed significantly (P < 0.05) more biofilms than espP-negative isolates (Fig. 3D). These data indicate that isolates with genes z1538, lpf2, ehaBβ, and/or espP were able to form significantly more biofilms than isolates without those genes.
FIG 3.
Biofilm formation and presence of fimbrial genes z1538 (A) and lpf2 (B) and autotransporter genes ehaBβ (C) and espP (D). Open triangles represent seropathotype A isolates, and solid dots represent seropathotype B and C isolates. The longer horizontal bars represent the means, and the shorter error bars represent the standard errors of the means. Statistical analysis was performed by using a Mann-Whitney test with two-tailed distribution. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
STEC biofilm characterization.
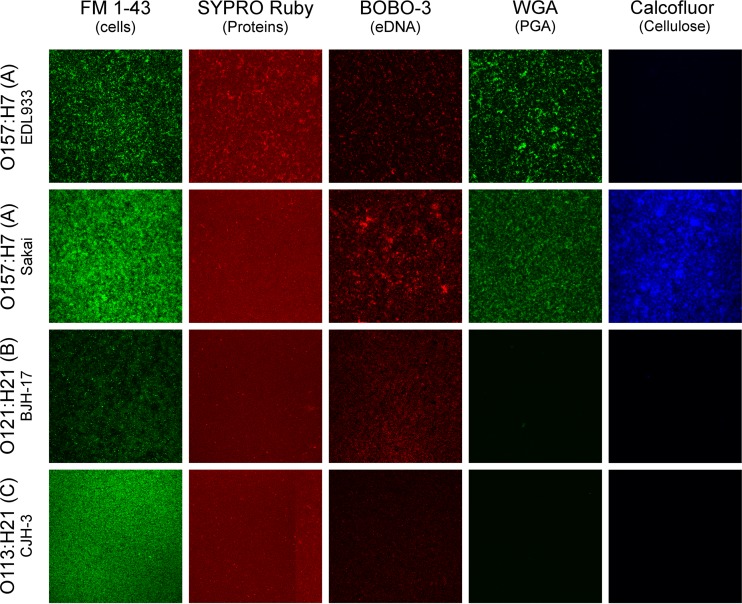
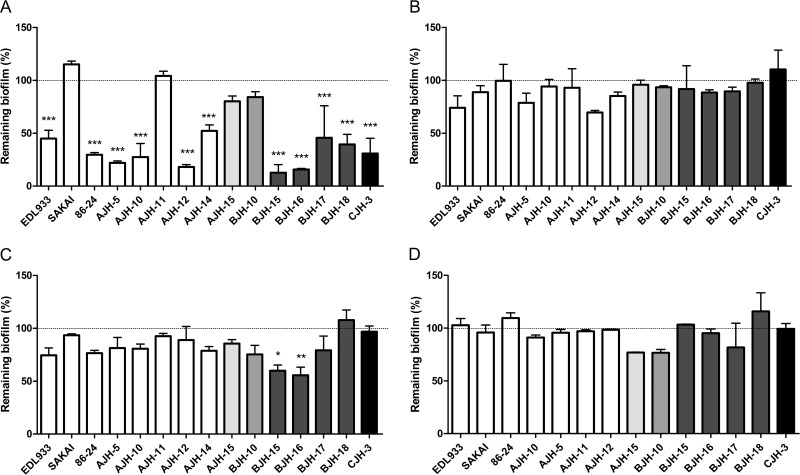
Confocal laser scanning microscopy (CLSM) using different fluorescent probes was performed with 15 high-biofilm-producing isolates (9 seropathotype A [O157], 5 seropathotype B, and 1 seropathotype C). The four isolates shown in Fig. 4 were selected to represent results obtained for the other 11 isolates. The data obtained with FM1-43, a membrane stain, highlighted the morphological heterogeneity of STEC biofilms (Fig. 4). In general, protein and DNA, stained with SYPRO Ruby and BOBO-3, respectively, were weakly detected. Polysaccharides were observed only in biofilms produced by seropathotype A (O157:H7/O157:NM) isolates. Among the 9 seropathotype A (O157:H7/O157:NM) isolates, 7 were stained with WGA, suggesting the presence of PGA or at least the presence of N-acetyl-d-glucosamine and N-acetylneuraminic acid residues in the biofilm matrix. Finally, cellulose was detected in the biofilm matrix of 2 isolates of seropathotype A (Fig. 4). The biofilms also were digested with enzymes to further characterize the composition of the matrix. Among the 15 selected isolates, 11 isolates were significantly sensitive to the proteinase K treatment (Fig. 5A). Two O121:H19 isolates were sensitive to cellulase treatment (Fig. 5C). Isolates were not affected by DNase I (Fig. 5B). Although PGA was observed in some isolates by CLSM, no isolates were affected by dispersin B treatments (Fig. 5D). This suggested that proteins and, in some isolates, cellulose play a larger role than extracellular DNA or PGA in STEC biofilm integrity.
FIG 4.
Images of 4 STEC biofilms obtained by CLSM. Biofilms were formed under static conditions in wells of microtiter plates and stained with FilmTracer FM1-43, SYPRO Ruby, BOBO-3, wheat-germ agglutinin (WGA)-Oregon green 488, and calcofluor. The four isolates shown were selected to represent results obtained for the other 11 isolates. A field-of-view representative of 3 biological replicates is shown for each isolate.
FIG 5.
Effect of enzymatic treatments on STEC biofilms. Dispersion of STEC biofilms formed under static conditions in microtiter plates by proteinase K (A), DNase I (B), cellulose (C), and dispersin B (D) is shown. Results are averages from at least 3 biological replicates. The horizontal bars represent the means, and the shorter error bars represent the standard errors of the means. Statistical analysis was performed by using a nonparametric one-way ANOVA with a Dunn's multiple-comparison posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Efficacy of sanitizers on STEC biofilms.
To determine the influence of biofilms on STEC sensitivity toward sanitizers commonly used to decontaminate farm or food production surfaces, four types of disinfectant, chlorine (200 ppm bleach), quaternary ammonium compound (1% [vol/vol] Aseptol 2000), alcohol (70% [vol/vol] ethanol), and peroxide compounds (1% [vol/vol] Virkon and 5% [vol/vol] H2O2) were applied on 10 STEC isolates selected based on their capacity to form strong biofilms (5 seropathotype A, 4 seropathotype B, and 1 seropathotype C). The impact of sanitizers on STEC biofilm was determined by enumerating viable cells and, with the resazurin-based CellTiter-Blue solution, was used to measure the metabolic activity of cells remaining after the treatment. After a 10-min exposure, the five sanitizers decreased the number of viable cells by 99.2 to 99.7% and the metabolic activity was reduced by 97.9 to 98.9%. There was no significant difference between seropathotypes or isolates (Table 2). Moreover, a protective effect of biofilm was observed, since planktonic cells were more sensitive than biofilm cells to sanitizer treatments (data not shown). None of the sanitizers were able to totally remove STEC biofilm matrix, as evaluated by crystal violet (Table 2). Bleach (43% or 22% of remaining matrix) was the most effective sanitizer, followed by H2O2 (76% of remaining matrix) and ethanol (91% or 85% of remaining matrix). Aseptol 2000 was not able to remove the matrix of any seropathotype biofilms, and Virkon only affects seropathotype B biofilm matrix. In brief, sanitizers effectively reduce the viability of STEC cells within biofilms but are not able to completely remove the biofilm matrix (Table 2).
TABLE 2.
Bacterial viability, metabolic activity, and remaining biofilm matrix of seropathotype A or seropathotypes B and C microtiter biofilms after sanitizer treatments
| Biofilm characterization after treatment | Value for seropathotype(s)a: |
|||||
|---|---|---|---|---|---|---|
| A (n = 5) |
B-C (n = 5) |
|||||
| Viabilityb | Metabolic activityc | Biofilm matrixd | Viability | Metabolic activity | Biofilm matrix | |
| Chlorine (bleach; 200 ppm) | 0.6 (±0.2) | 0.8 (±0.7) | 53 (±14) | 0.8 (±0.4) | 1.7 (±0.6) | 22 (±8) |
| QACe (Aseptol 2000; 1%) | 0.3 (±0.2) | 1.2 (±0.7) | 100 (±0) | 0.4 (±0.3) | 2.1 (±1.1) | 100 (±3) |
| Peroxide compounds | ||||||
| Virkon (1%) | 0.3 (±0.1) | 0.4 (±0.1) | 100 (±2) | 0.6 (±0.3) | 1.9 (±1.1) | 65 (±9) |
| H2O2 (5%) | 0.3 (±0.1) | 0.7 (±0.4) | 72 (±16) | 0.7 (±0.3) | 1.7 (±0.8) | 56 (±16) |
| Alcohol (ethanol; 70%) | 0.3 (±0.1) | 2.4 (±0.9) | 92 (±10) | 0.4 (±0.2) | 2.8 (±0.9) | 71 (±17) |
Biofilms were formed under static conditions and were treated with sanitizers for 10 min. The impact of treatments was compared to that of control PBS treatment. Results are expressed in percentages. The data represent the means from three independent biological replicates.
Viable cells were determined by bacterial cell enumeration (CFU).
The metabolic activity was determined by fluorescence of CellTiter-Blue solution (λex, 570 nm; λem, 600 nm).
The remaining biofilm matrix was evaluated by absorbance (595 nm) of crystal violet staining.
QAC, quaternary ammonium chloride.
DISCUSSION
Biofilm-forming abilities of STEC isolates recovered from human.
STEC, an important food-borne and waterborne pathogen, and STEC isolates of different origins (cattle, water, food, and human) have been shown to be able to form biofilms (6–8). Biofilms are thought to increase STEC survival and persistence in hostile environments such as water- or food-processing plants (37). Therefore, we hypothesized that biofilms could be involved in the transmission of STEC from the environment to humans. Consequently, for this study, we selected STEC isolates associated with human infections. Biofilm formation of 39 STEC isolates belonging to seropathotypes A, B, and C was investigated. As shown in other studies, biofilm formation on polystyrene surfaces appeared to be a strain-dependent phenomenon (7, 8, 38, 39). An association between STEC serotypes and biofilm-forming abilities was suggested (40); however, in other studies this was not observed (7, 8). In our study, because of the low number of isolates per serotype, this association could not be determined.
As previously observed, static or dynamic conditions used to form biofilms can influence the biofilm-forming capacity of isolates (26). This is especially observed for O145 isolates that formed stronger biofilms under dynamic conditions than under static conditions. Under static conditions, metabolic waste and dispersing signals can accumulate, and this may negatively affect the biofilm-forming ability. Moreover, biofilm architecture can dramatically change when biofilms are grown under dynamic conditions (41, 42) and could explain the difference in the biofilm-forming abilities of some STEC isolates (Table 1). Nevertheless, this difference was not observed for all STEC isolates, which indicates that the mechanism behind STEC biofilm formation is heterogeneous. Using the seropathotype classification, we found that under static conditions, seropathotype A isolates formed significantly stronger biofilms than those of seropathotypes B and C (Fig. 1B), and that under dynamic conditions the majority of seropathotype A isolates were able to form strong biofilms. This biofilm-forming capacity of seropathotype A isolates could contribute to their persistence in the environment and influence their high relative incidence and their frequent involvement in outbreaks (3). It is reasonable to assume that an STEC strain coexists with several other STEC isolates as well as with other bacterial species (43–47). Therefore, mixed STEC biofilms should be investigated in future work.
Factors potentially involved in STEC biofilm formation.
Biofilm formation has been shown to be mediated by bacterial surface structures that are regulated by environmental conditions. Here, we show that the production of curli as measured with Congo red binding was dependent on the isolates and growth conditions despite all STEC strains carrying the curli gene csgA (7). CF results indicative of curli and/or cellulose production also were variable and are similar to those of other studies (7, 8, 48). While curli or cellulose production in some STEC isolates was shown to be associated with biofilm formation (6, 8, 34, 49–51), these were shown to be nonessential for biofilm formation (7). In our study, no correlation was found between Congo red or calcofluor-positive isolates and biofilm formation or seropathotype (see Fig. S1 in supplemental material). This highlights a high variability in the mechanisms regulating the production of factors associated with biofilm formation and suggests that STEC strains adapt differently to various environments.
Although all isolates were PCR positive for type 1 fimbriae (see Table S3 in supplemental material), only seropathotype B and C isolates produced type 1 fimbriae. It has been shown that strains of seropathotype A isolates are unable to express type 1 fimbriae due to a 16-bp deletion in the regulatory switch region of fimA (52–54). Interestingly, 7 seropathotype A isolates were able to agglutinate yeast in a mannose-independent manner and were able to autoagglutinate (Fig. 2A and data not shown). This phenotype correlated with biofilm formation (r2 = 0.70) for those 7 isolates, suggesting that the production of this unidentified factor could contribute to biofilm formation (Fig. 2B).
Fimbriae and autotransporters are adhesion factors associated with biofilm formation in STEC (55). Fimbria-encoding genes z1538 and lpf2, the passenger domain of the autotransporter gene ehaB, and the autotransporter gene espP are present in isolates producing significantly more biofilms (Fig. 3). As previously observed, all seropathotype A isolates were positive for z1538, a gene present in the seropathotype A-specific genomic O island 47 (56) while absent from other isolates. Given that seropathotype A isolates (z1538 positive) formed significantly more biofilm than seropathotype B and C isolates (z1538-negative isolates) (Fig. 3A), it is possible that fimbrial adhesin encoded by O island 47 play a role in biofilm formation. To support this, Low et al. observed that z1538 fimbrial gene cluster expression was increased under conditions similar to those used for biofilm formation (35). In addition, isolates positive for lpf2 formed significantly more biofilm than lpf2-negative isolates (Fig. 3B). This suggests a biofilm formation role for the long polar fimbria of Lpf2. Expression of lpf2 in E. coli O157:H7 has been shown to be increased under conditions similar to those used for biofilm formation (57) and when grown at 28°C rather than at 37°C (35). Lpf2 has been suggested to contribute to STEC adhesion (58). The precise role of both the z1538 gene cluster and lpf2 during biofilm formation needs to be confirmed. The α passenger domain of ehaB was detected in isolates that formed strong biofilms at significant levels, such as those belonging to O157:H7, O157:NM, and O121:H19 serotypes (Fig. 3C). ehaB of reference strain EDL 933 (O157:H7) was shown to be involved in biofilm formation when overexpressed in E. coli K-12 (17). Therefore, it is possible that these EhaB variants found in O157:H7 and O121:H19 contribute to the establishment of biofilm. The plasmid-encoding gene espP was detected in most STEC isolates (Fig. 3D). Using an espP mutant, this gene already was shown to be associated with biofilm formation in O157:H7 (19). Seropathotype A isolates are equipped with fimbrial and autotransporter genes that could contribute to their biofilm capacity and their adaptation to different environments. Moreover, lpf2, ehaB, and espP also were found in the genome of environmental STEC strains, suggesting that those genes contribute to the STEC environmental lifestyle (59).
STEC biofilm matrix composition.
Biofilm matrix compositions of E. coli isolates were shown to be highly heterogeneous (20, 60, 61). The use of CLSM and fluorescent probes showed the presence of PGA and cellulose in the biofilm of seropathotype A isolates that are strong biofilm producers (Fig. 4). These were shown to be important in the development and architecture of biofilms in E. coli (20). To our knowledge, this is the first description of the matrix composition of STEC biofilms. Furthermore, a previous study showed that O157:H7 mutants lacking genes encoding PGA or cellulose synthesis were unable to adhere to alfalfa sprouts (21). Therefore, it is possible that our biofilm conditions stimulated the production of cellulose and PGA in seropathotype A isolates but not in seropathotype B and C isolates. Genome sequence database and PCR results indicate that O26:H11, O103:H2, O111:NM, O111H:8, O121:H19, and O157:H26 isolates are negative for pgaA, coding for PGA export from the periplasm, while it is present in seropathotype A isolates (data not shown) (62). This could explain why seropathotype A isolates have a greater potential to form biofilm than others. Alternatively, some other polysaccharide structure could be produced in seropathotype B and C isolates (63, 64). Nevertheless, biofilms were resistant to dispersin B treatment that targets PGA, but they were sensitive to a proteinase K treatment (Fig. 5). These findings suggest that proteins play an important role in biofilm integrity, while PGA seems to be less necessary for biofilm integrity. Previous studies suggested that in E. coli, proteins such as adhesins (fimbriae and curli) and autotransporters participate in the structure of the biofilm by establishing cell-surface, cell-cell, and cell-matrix interactions (18, 34, 50, 65), while polysaccharides may have a protective role (66).
Sanitizer treatments.
Food-processing and farm environments provide a variety of conditions which might favor the formation of biofilm (67). Standard cleaning and sanitizing practices are used to reduce sources of microbial contamination on their products. As previously described, strong attachment of biofilms on surfaces may affect the efficiency of sanitizers and protect bacteria against sanitation protocols that are used to reduce contamination (7, 68, 69). In the present study, we examined if sanitizers commonly used in good sanitation practices were effective at both reducing STEC viability in biofilms and eliminating the biofilm matrix. Although sanitizer treatments were effective at reducing significantly the viability and the metabolism of STEC in biofilms, the biofilm matrix either remained or was partly removed depending on the sanitizer (Table 2). These results agree with previous reports (6, 7, 68, 69). Moreover, no significant differences in viability or metabolic activity were seen between seropathotypes or isolates (7). Additionally, matrix composition, such as the presence of cellulose or PGA, had no protective effect on the cells. Sanitizers cannot completely remove biofilm matrix on food-processing surfaces left after a sanitation protocol, which could lead to a faster recolonization of the surface (70). Interestingly, some sanitizers, such as bleach or H2O2, were more effective at removing the biofilm matrix of STEC. Our biofilm study is restricted to specific conditions and represents to some degree biofilm formation on food-processing equipment design and surface materials; nevertheless, our results are indicative of biofilm characteristics of STEC that caused human infections. Thus, STEC biofilm formation is a potential hazard in food hygiene and may become a source of cross-contamination in farm and food-processing environments. Because detached biofilms may serve as a continuous contamination source, prevention, removal, and inactivation of STEC biofilms are critical for improving hygiene and contamination control and enhancing food safety.
Conclusions.
We show that STEC isolates recovered from human infections formed biofilms in a seropathotype-dependent manner. Specifically, seropathotype A isolates clustering as serotypes O157:H7 and O157:NM, with the highest relative incidence and frequently involved in outbreaks, had a greater ability to form biofilms than seropathotype B or C isolates. Seropathotype A isolates were the only ones able to produce cellulose and PGA. Additionally, biofilm integrity was dependent on proteins, but they were not overly abundant in the biofilm matrix. Furthermore, two autotransporters, EhaB and EspP, and two fimbriae, one encoded by genes found on genomic O island 47 and Lpf2, were identified as potential genetic determinants for biofilm formation. Interestingly, some seropathotype A isolates were able to agglutinate yeast in a mannose-independent manner. This phenotype correlated with biofilm formation, suggesting that an unidentified biofilm-associated factor is produced by seropathotype A isolates. Treatment with sanitizers reduced STEC viability but did not completely remove the biofilm matrix. Overall, our data indicate that biofilm formation could contribute to the persistence of seropathotype A isolates in the environment and to the transmission and, consequently, the high incidence of serotype O157:H7 and O157:NM infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. A. Karmali, K. Ziebell, and R. P. Johnson at the Laboratory for Food-Borne Zoonoses of the Public Health Agency of Canada for supplying North American STEC isolates and C. Martin from INRA Clermont-Ferrand Theix for supplying French STEC isolates. We are grateful to Judith Kashul for editing the manuscript. We thank Frederic Berthiaume for his help with confocal laser scanning microscopy assays.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02983-15.
REFERENCES
- 1.Thomas MK, Murray R, Flockhart L, Pintar K, Pollari F, Fazil A, Nesbitt A, Marshall B. 2013. Estimates of the burden of foodborne illness in Canada for 30 specified pathogens and unspecified agents, circa 2006. Foodborne Pathog Dis 10:639–648. doi: 10.1089/fpd.2012.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferens WA, Hovde CJ. 2011. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog Dis 8:465–487. doi: 10.1089/fpd.2010.0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol 41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis 192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 5.Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 112:1466–1477. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park YJ, Chen J. 2015. Control of the biofilms formed by curli- and cellulose-expressing Shiga toxin-producing Escherichia coli using treatments with organic acids and commercial sanitizers. J Food Prot 78:990–995. doi: 10.4315/0362-028X.JFP-14-382. [DOI] [PubMed] [Google Scholar]
- 7.Wang R, Bono JL, Kalchayanand N, Shackelford S, Harhay DM. 2012. Biofilm formation by Shiga toxin-producing Escherichia coli O157:H7 and non-O157 strains and their tolerance to sanitizers commonly used in the food processing environment. J Food Prot 75:1418–1428. doi: 10.4315/0362-028X.JFP-11-427. [DOI] [PubMed] [Google Scholar]
- 8.Biscola FT, Abe CM, Guth BE. 2011. Determination of adhesin gene sequences in, and biofilm formation by, O157 and non-O157 Shiga toxin-producing Escherichia coli strains isolated from different sources. Appl Environ Microbiol 77:2201–2208. doi: 10.1128/AEM.01920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villegas NA, Baronetti J, Albesa I, Polifroni R, Parma A, Etcheverria A, Becerra M, Padola N, Paraje M. 2013. Relevance of biofilms in the pathogenesis of Shiga toxin-producing Escherichia coli infection. ScientificWorldJournal 2013:607258. doi: 10.1155/2013/607258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu JH, Kim H, Beuchat LR. 2004. Attachment and biofilm formation by Escherichia coli O157:H7 on stainless steel as influenced by exopolysaccharide production, nutrient availability, and temperature. J Food Prot 67:2123–2131. [DOI] [PubMed] [Google Scholar]
- 11.Ryu JH, Beuchat LR. 2005. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and curli production on its resistance to chlorine. Appl Environ Microbiol 71:247–254. doi: 10.1128/AEM.71.1.247-254.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos Mendonça RC, Morelli AMF, Pereira JAM, de Carvalho MM, de Souza NL. 2012. Prediction of Escherichia coli O157:H7 adhesion and potential to form biofilm under experimental conditions. Food Control 23:389–396. doi: 10.1016/j.foodcont.2011.08.004. [DOI] [Google Scholar]
- 13.Marouani-Gadri N, Augier G, Carpentier B. 2009. Characterization of bacterial strains isolated from a beef-processing plant following cleaning and disinfection—influence of isolated strains on biofilm formation by Sakai and EDL 933 Escherichia coli O157:H7. Int J Food Microbiol 133:62–67. doi: 10.1016/j.ijfoodmicro.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 14.Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. 2006. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J Bacteriol 188:3572–3581. doi: 10.1128/JB.188.10.3572-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crim SM, Griffin PM, Tauxe R, Marder EP, Gilliss D, Cronquist AB, Cartter M, Tobin-D'Angelo M, Blythe D, Smith K, Lathrop S, Zansky S, Cieslak PR, Dunn J, Holt KG, Wolpert B, Henao OL. 2015. Preliminary incidence and trends of infection with pathogens transmitted commonly through food-foodborne diseases active surveillance network, 10 U.S. sites, 2006-2014. MMWR Morb Mortal Wkly Rep 64:495–499. [PMC free article] [PubMed] [Google Scholar]
- 16.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JA, Molin S, Prensier G, Arbeille B, Ghigo JM. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51:659–674. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- 17.Wells TJ, McNeilly TN, Totsika M, Mahajan A, Gally DL, Schembri MA. 2009. The Escherichia coli O157:H7 EhaB autotransporter protein binds to laminin and collagen I and induces a serum IgA response in O157:H7 challenged cattle. Environ Microbiol 11:1803–1814. doi: 10.1111/j.1462-2920.2009.01905.x. [DOI] [PubMed] [Google Scholar]
- 18.Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol 10:589–604. doi: 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- 19.Puttamreddy S, Cornick NA, Minion FC. 2010. Genome-wide transposon mutagenesis reveals a role for pO157 genes in biofilm development in Escherichia coli O157:H7 EDL 933. Infect Immun 78:2377–2384. doi: 10.1128/IAI.00156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beloin C, Roux A, Ghigo JM. 2008. Escherichia coli biofilms. Curr Top Microbiol Immunol 322:249–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthysse AG, Deora R, Mishra M, Torres AG. 2008. Polysaccharides cellulose, poly-beta-1,6-N-acetyl-d-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl Environ Microbiol 74:2384–2390. doi: 10.1128/AEM.01854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 23.Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, Ono A, Yanagawa H. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am J Epidemiol 150:787–796. doi: 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- 24.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med 109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 25.Pradel N, Bertin Y, Martin C, Livrelli V. 2008. Molecular analysis of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients and dairy samples in France. Appl Environ Microbiol 74:2118–2128. doi: 10.1128/aem.02688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tremblay YD, Vogeleer P, Jacques M, Harel J. 2015. High-throughput microfluidic method to study biofilm formation and host-pathogen interactions in pathogenic Escherichia coli. Appl Environ Microbiol 81:2827–2840. doi: 10.1128/AEM.04208-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Da Re S, Ghigo JM. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol 188:3073–3087. doi: 10.1128/JB.188.8.3073-3087.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Totsika M, Wells TJ, Beloin C, Valle J, Allsopp LP, King NP, Ghigo JM, Schembri MA. 2012. Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl Environ Microbiol 78:2179–2189. doi: 10.1128/AEM.06680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crepin S, Houle S, Charbonneau ME, Mourez M, Harel J, Dozois CM. 2012. Decreased expression of type 1 fimbriae by a pst mutant of uropathogenic Escherichia coli reduces urinary tract infection. Infect Immun 80:2802–2815. doi: 10.1128/IAI.00162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bello-Orti B, Deslandes V, Tremblay YD, Labrie J, Howell KJ, Tucker AW, Maskell DJ, Aragon V, Jacques M. 2014. Biofilm formation by virulent and non-virulent strains of Haemophilus parasuis. Vet Res 45:104. doi: 10.1186/s13567-014-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tremblay YD, Lamarche D, Chever P, Haine D, Messier S, Jacques M. 2013. Characterization of the ability of coagulase-negative staphylococci isolated from the milk of Canadian farms to form biofilms. J Dairy Sci 96:234–246. doi: 10.3168/jds.2012-5795. [DOI] [PubMed] [Google Scholar]
- 32.Orgaz B, Kives J, Pedregosa AM, Monistrol IF, Laborda F, SanJosé C. 2006. Bacterial biofilm removal using fungal enzymes. Enzyme Microb Technol 40:51–56. doi: 10.1016/j.enzmictec.2005.10.037. [DOI] [Google Scholar]
- 33.Tremblay YD, Caron V, Blondeau A, Messier S, Jacques M. 2014. Biofilm formation by coagulase-negative staphylococci: impact on the efficacy of antimicrobials and disinfectants commonly used on dairy farms. Vet Microbiol 172:511–518. doi: 10.1016/j.vetmic.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Cookson AL, Cooley WA, Woodward MJ. 2002. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int J Med Microbiol 292:195–205. doi: 10.1078/1438-4221-00203. [DOI] [PubMed] [Google Scholar]
- 35.Low AS, Holden N, Rosser T, Roe AJ, Constantinidou C, Hobman JL, Smith DG, Low JC, Gally DL. 2006. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ Microbiol 8:1033–1047. doi: 10.1111/j.1462-2920.2006.00995.x. [DOI] [PubMed] [Google Scholar]
- 36.Paton AW, Srimanote P, Woodrow MC, Paton JC. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun 69:6999–7009. doi: 10.1128/IAI.69.11.6999-7009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogeleer P, Tremblay YD, Mafu AA, Jacques M, Harel J. 2014. Life on the outside: role of biofilms in environmental persistence of Shiga-toxin producing Escherichia coli. Front Microbiol 5:317. doi: 10.3389/fmicb.2014.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Ordonez A, Alvseike O, Omer MK, Heir E, Axelsson L, Holck A, Prieto M. 2013. Heterogeneity in resistance to food-related stresses and biofilm formation ability among verocytotoxigenic Escherichia coli strains. Int J Food Microbiol 161:220–230. doi: 10.1016/j.ijfoodmicro.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Dourou D, Beauchamp CS, Yoon Y, Geornaras I, Belk KE, Smith GC, Nychas GJ, Sofos JN. 2011. Attachment and biofilm formation by Escherichia coli O157:H7 at different temperatures, on various food-contact surfaces encountered in beef processing. Int J Food Microbiol 149:262–268. doi: 10.1016/j.ijfoodmicro.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Nesse LL, Sekse C, Berg K, Johannesen KC, Solheim H, Vestby LK, Urdahl AM. 2013. Potentially pathogenic Escherichia coli can produce biofilm under conditions relevant for the food production chain. Appl Environ Microbiol doi: 10.1128/AEM.03331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purevdorj B, Costerton JW, Stoodley P. 2002. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 68:4457–4464. doi: 10.1128/AEM.68.9.4457-4464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vejborg RM, Klemm P. 2009. Cellular chain formation in Escherichia coli biofilms. Microbiology 155:1407–1417. doi: 10.1099/mic.0.026419-0. [DOI] [PubMed] [Google Scholar]
- 43.Carter MQ, Xue K, Brandl MT, Liu F, Wu L, Louie JW, Mandrell RE, Zhou J. 2012. Functional metagenomics of Escherichia coli O157:H7 interactions with spinach indigenous microorganisms during biofilm formation. PLoS One 7:e44186. doi: 10.1371/journal.pone.0044186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu NT, Nou X, Bauchan GR, Murphy C, Lefcourt AM, Shelton DR, Lo YM. 2015. Effects of environmental parameters on the dual-species biofilms formed by Escherichia coli O157:H7 and Ralstonia insidiosa, a strong biofilm producer isolated from a fresh-cut produce processing plant. J Food Prot 78:121–127. doi: 10.4315/0362-028X.JFP-14-302. [DOI] [PubMed] [Google Scholar]
- 45.Liu NT, Nou X, Lefcourt AM, Shelton DR, Lo YM. 2014. Dual-species biofilm formation by Escherichia coli O157:H7 and environmental bacteria isolated from fresh-cut processing facilities. Int J Food Microbiol 171:15–20. doi: 10.1016/j.ijfoodmicro.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Wang R, Kalchayanand N, Bono JL. 2015. Sequence of colonization determines the composition of mixed biofilms by Escherichia coli O157:H7 and O111:H8 strains. J Food Prot 78:1554–1559. doi: 10.4315/0362-028X.JFP-15-009. [DOI] [PubMed] [Google Scholar]
- 47.Wang R, Kalchayanand N, Bono JL, Schmidt JW, Bosilevac JM. 2012. Dual-serotype biofilm formation by Shiga toxin-producing Escherichia coli O157:H7 and O26:H11 strains. Appl Environ Microbiol 78:6341–6344. doi: 10.1128/aem.01137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlich GA, Chen CY, Cottrell BJ, Nguyen LH. 2014. Growth media and temperature effects on biofilm formation by serotype O157:H7 and non-O157 Shiga toxin-producing Escherichia coli. FEMS Microbiol Lett 354:133–141. doi: 10.1111/1574-6968.12439. [DOI] [PubMed] [Google Scholar]
- 49.Ryu JH, Kim H, Frank JF, Beuchat LR. 2004. Attachment and biofilm formation on stainless steel by Escherichia coli O157:H7 as affected by curli production. Lett Appl Microbiol 39:359–362. doi: 10.1111/j.1472-765X.2004.01591.x. [DOI] [PubMed] [Google Scholar]
- 50.Uhlich GA, Cooke PH, Solomon EB. 2006. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol 72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JH, Kim YG, Cho MH, Wood TK, Lee J. 2011. Transcriptomic analysis for genetic mechanisms of the factors related to biofilm formation in Escherichia coli O157:H7. Curr Microbiol 62:1321–1330. doi: 10.1007/s00284-010-9862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enami M, Nakasone N, Honma Y, Kakinohana S, Kudaka J, Iwanaga M. 1999. Expression of type I pili is abolished in verotoxin-producing Escherichia coli O157. FEMS Microbiol Lett 179:467–472. doi: 10.1111/j.1574-6968.1999.tb08764.x. [DOI] [PubMed] [Google Scholar]
- 53.Roe AJ, Currie C, Smith DG, Gally DL. 2001. Analysis of type 1 fimbriae expression in verotoxigenic Escherichia coli: a comparison between serotypes O157 and O26. Microbiology 147:145–152. doi: 10.1099/00221287-147-1-145. [DOI] [PubMed] [Google Scholar]
- 54.Li B, Koch WH, Cebula TA. 1997. Detection and characterization of the fimA gene of Escherichia coli O157:H7. Mol Cell Probes 11:397–406. doi: 10.1006/mcpr.1997.0132. [DOI] [PubMed] [Google Scholar]
- 55.McWilliams BD, Torres AG. 2015. Enterohemorrhagic Escherichia coli adhesins, p 157–174. In Sperandio V, Hovde C (ed), Enterohemorrhagic Escherichia coli and other Shiga toxin-producing E. coli. ASM Press, Washington, DC. doi: 10.1128/microbiolspec.EHEC-0003-2013. [DOI] [Google Scholar]
- 56.Shen S, Mascarenhas M, Morgan R, Rahn K, Karmali MA. 2005. Identification of four fimbria-encoding genomic islands that are highly specific for verocytotoxin-producing Escherichia coli serotype O157 strains. J Clin Microbiol 43:3840–3850. doi: 10.1128/JCM.43.8.3840-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torres AG, Milflores-Flores L, Garcia-Gallegos JG, Patel SD, Best A, La Ragione RM, Martinez-Laguna Y, Woodward MJ. 2007. Environmental regulation and colonization attributes of the long polar fimbriae (LPF) of Escherichia coli O157:H7. Int J Med Microbiol 297:177–185. doi: 10.1016/j.ijmm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Newton HJ, Sloan J, Bennett-Wood V, Adams LM, Robins-Browne RM, Hartland EL. 2004. Contribution of long polar fimbriae to the virulence of rabbit-specific enteropathogenic Escherichia coli. Infect Immun 72:1230–1239. doi: 10.1128/IAI.72.3.1230-1239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balière C, Rincé A, Blanco J, Dahbi G, Harel J, Vogeleer P, Giard J-C, Mariani-Kurdjian P, Gourmelon M. 2015. Prevalence and characterization of Shiga toxin-producing and enteropathogenic Escherichia coli in shellfish-harvesting areas and their watersheds. Front Microbiol 6:1356. doi: 10.3389/fmicb.2015.01356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sutherland IW. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol 9:222–227. doi: 10.1016/S0966-842X(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 61.Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, Chapman MR, Hadjifrangiskou M, Henderson JP, Hultgren SJ. 2013. Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio 4:e00645-13. doi: 10.1128/mBio.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF III, Romeo T. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-d-glucosamine. J Bacteriol 190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danese PN, Pratt LA, Kolter R. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol 182:3593–3596. doi: 10.1128/JB.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol 2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- 65.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo JM, Beloin C. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J Bacteriol 190:4147–4161. doi: 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeh JY, Chen J. 2004. Production of slime polysaccharide by EHEC and STEC as well as the influence of culture conditions on slime production in Escherichia coli O157:H7. Lett Appl Microbiol 38:488–492. doi: 10.1111/j.1472-765X.2004.01523.x. [DOI] [PubMed] [Google Scholar]
- 67.Giaouris E, Heir E, Hebraud M, Chorianopoulos N, Langsrud S, Moretro T, Habimana O, Desvaux M, Renier S, Nychas GJ. 2013. Attachment and biofilm formation by foodborne bacteria in meat processing environments: causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci 97:298–309. doi: 10.1016/j.meatsci.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 68.Fouladkhah A, Geornaras I, Sofos JN. 2013. Biofilm formation of O157 and non-O157 Shiga toxin-producing Escherichia coli and multidrug-resistant and susceptible Salmonella typhimurium and Newport and their inactivation by sanitizers. J Food Sci 78:M880–M886. doi: 10.1111/1750-3841.12123. [DOI] [PubMed] [Google Scholar]
- 69.Marouani-Gadri N, Firmesse O, Chassaing D, Sandris-Nielsen D, Arneborg N, Carpentier B. 2010. Potential of Escherichia coli O157:H7 to persist and form viable but non-culturable cells on a food-contact surface subjected to cycles of soiling and chemical treatment. Int J Food Microbiol 144:96–103. doi: 10.1016/j.ijfoodmicro.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 70.Larsen P, Olesen BH, Nielsen PH, Nielsen JL. 2008. Quantification of lipids and protein in thin biofilms by fluorescence staining. Biofouling 24:241–250. doi: 10.1080/08927010802040255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.