Abstract
The phosphoenolpyruvate phosphotransferase system (PEP-PTS) and adenylate cyclase (AC) IV (encoded by BB0723 [cyaB]) are well conserved in different species of Borrelia. However, the functional roles of PEP-PTS and AC in the infectious cycle of Borrelia have not been characterized previously. We examined 12 PEP-PTS transporter component mutants by needle inoculation of mice to assess their ability to cause mouse infection. Transposon mutants with mutations in the EIIBC components (ptsG) (BB0645, thought to be involved in glucose-specific transport) were unable to cause infection in mice, while all other tested PEP-PTS mutants retained infectivity. Infectivity was partially restored in an in trans-complemented strain of the ptsG mutant. While the ptsG mutant survived normally in unfed as well as fed ticks, it was unable to cause infection in mice by tick transmission, suggesting that the function of ptsG is essential to establish infection by either needle inoculation or tick transmission. In Gram-negative organisms, the regulatory effects of the PEP-PTS are mediated by adenylate cyclase and cyclic AMP (cAMP) levels. A recombinant protein encoded by B. burgdorferi BB0723 (a putative cyaB homolog) was shown to have adenylate cyclase activity in vitro; however, mutants with mutations in this gene were fully infectious in the tick-mouse infection cycle, indicating that its function is not required in this process. By transcriptome analysis, we demonstrated that the ptsG gene may directly or indirectly modulate gene expression of Borrelia burgdorferi. Overall, the PEP-PTS glucose transporter PtsG appears to play important roles in the pathogenesis of B. burgdorferi that extend beyond its transport functions.
INTRODUCTION
Lyme disease, a bacterial infection transmitted to humans by the bite of Ixodes ticks infected with spirochetes of the genus Borrelia, is the most common vector-borne infection in United States. According to a recent estimate by the Centers for Disease Control and Prevention, approximately 300,000 new human cases of Lyme disease are diagnosed in the United States each year (1, 2). An early symptom of Lyme disease is the development of a skin rash known as erythema migrans; later manifestations of infection may include arthritis, carditis, and neurologic effects (3).
Borrelia burgdorferi, the causative agent of Lyme disease in North America, possesses a genome consisting of a small, 910-kbp linear chromosome along with numerous linear and circular plasmids (4, 5). Genome annotation of B. burgdorferi B31 revealed that the functions of two-thirds of the putative open reading frames (ORFs) are not known. Unlike many other pathogenic bacteria, B. burgdorferi lacks genes encoding known toxins or secretion systems (4, 5). Borrelial plasmids contain a large number of genes important in either infectivity in mammals or survival in the tick vector (6).
Gene regulation in B. burgdorferi is a complex process that involves interplay between several regulatory factors, including the two-component signal transduction systems Hk1-Rrp1 and Hk2-Rrp2, the alternative sigma factors RpoN (σ54) and RpoS (σs), BosR, an unorthodox DNA-binding protein, the small noncoding RNA DsrABb, and Hfq and CsrA, two RNA-binding proteins (reviewed in reference 7). The Hk1-Rrp1 pathway plays regulatory roles by producing the second messenger cyclic di-GMP (c-di-GMP) and is required for the survival of B. burgdorferi in Ixodes ticks (8–10). Conversely, Hk2-Rrp2 activates the RpoN-RpoS pathway, which is essential for this pathogen to successfully accomplish tick-mouse transmission and establish mammalian infection (11–13). Recent studies demonstrated that a c-di-GMP-binding protein, PlzA, connects these two signal transduction pathways (14). Environmental stimuli such as temperature, pH, oxygen, carbon dioxide, and undefined mammalian host cell signals have been shown to modulate gene expression in Borrelia (15–19).
The spirochete maintains an enzootic cycle through transmission back and forth between its arthropod vector and mammalian vertebrate hosts. Since Borrelia species lack most of the biosynthetic genes found in other bacteria, these organisms face additional challenges when adapting to the different nutrient conditions in these divergent environments. Although genome sequence analysis indicated the presence of numerous homologs of carbohydrate transporters, B. burgdorferi actually uses very few carbohydrates to support its growth, including glucose, mannose, N-acetylglucosamine, maltose, chitobiose, and glycerol (20). In many bacteria, the highly conserved phosphoenolpyruvate phosphotransferase system (PEP-PTS) plays an important dual role in both carbohydrate transport and the regulation of gene expression based on carbohydrate availability (21–24). The PEP-PTS consists of multiple components, including the common cytoplasmic components enzyme I (EI) and histidine protein (HPr), the sugar-specific cytoplasmic components enzyme IIA (EIIA) and enzyme IIB (EIIB), and the sugar specific membrane-associated component(s) enzyme IIC (EIIC). The PTS monitors the availability of consumable carbohydrates and the resulting energy state of the cell by sensing the intracellular PEP-to-pyruvate ratio. Subsequently, this information is transmitted to other cellular targets, hence modulating various cellular functions in response to carbohydrate availability (21, 24). In Gram-negative bacteria, EIIAGlc (glucose transporter) serves as a global regulator which, coupled with cyclic AMP (cAMP) signaling (synthesized by adenylate cyclase), regulates carbohydrate metabolism and gene regulation, whereas HPr (phosphorylation at serine 46) coupled with catabolite control protein A (CcpA) serves as a global regulator in Gram-positive bacteria (25).
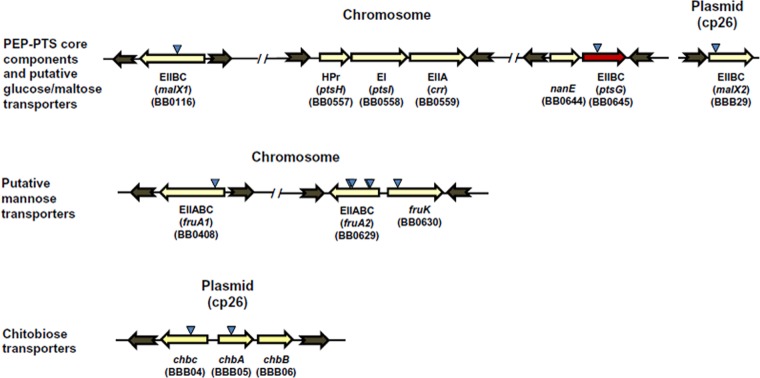
Genome sequence analyses indicate that, like other bacteria, Borrelia species possess PEP-PTS core components (EI and HPr) along with several sugar-specific EII components encoded by paralogous genes on both the chromosome and plasmids (Fig. 1). Additionally, a putative class IV adenylate cyclase encoded by the gene BB0723 (cyaB) is present in the B. burgdorferi genome. These PEP-PTS components and cyaB are well conserved in both Lyme disease and relapsing fever Borrelia strains. However, the potential role(s) that the PEP-PTS and cAMP signaling may play in gene regulation and pathogenesis of Borrelia species has not been determined.
FIG 1.
Arrangement of putative PEP-PTS component genes of B. burgdorferi B31. Triangles indicate the locations of transposon insertions. The red arrow indicates that ptsG is required for mammalian infectivity. The chitobiose transporter locus (chbC, chbA, and chbB) and the putative glucose/maltose EIIBC gene malX2 are located on cp26 plasmid, while the remainder of the PEP-PTS-encoding genes are located on the chromosome.
Recent signature-tagged mutagenesis (STM) analyses indicated that mutations in PTS carbohydrate transporter genes of B. burgdorferi exhibited a low- to no-infectivity phenotype (26). In the present study, we have analyzed in greater detail the mouse infectivity of mutants of PEP-PTS-associated carbohydrate transporters by needle and tick inoculation. Also, the role of cyaB in mouse infectivity and in the tick survivability and transmission of infection from tick to mice was assessed. Transcriptome analyses further indicated that ptsG of B. burgdorferi has important roles in the transcriptional regulation of multiple genes, including several involved in virulence of this pathogen.
MATERIALS AND METHODS
Bacterial strains and growth media.
The PEP-PTS and cyaB mutants were inactivated by transposon-mediated mutagenesis as part of an STM study in our laboratory in which 4,479 Himar1 mutants of B. burgdorferi 5A18NP1 were generated (26). All mutant clones were confirmed by PCR analysis using primers flanking the insertion site determined previously; the primers are listed in Table S1 in the supplemental material. In some STM mutants, the initial culture contained a second transposon mutant as a coisolate (see Fig. S1 in the supplemental material); in these cases, the clone containing the desired mutation was separated from the contaminant by replating in BSKII agarose medium (27, 28) supplemented with appropriate antibiotics. Each mutant and parental strains of B. burgdorferi were grown at 37°C in 5% CO2 in BSKII medium (29) supplemented with appropriate antibiotics for a maximum of 3 in vitro passages prior to use in experiments. The plasmid content of each clone was determined as described previously (30). The parental B. burgdorferi 5A18NP1strain and all transposon mutant progeny lack lp28-4 and lp56 plasmids (26).
Complementation of the ptsG gene using the pKFSS1 shuttle vector.
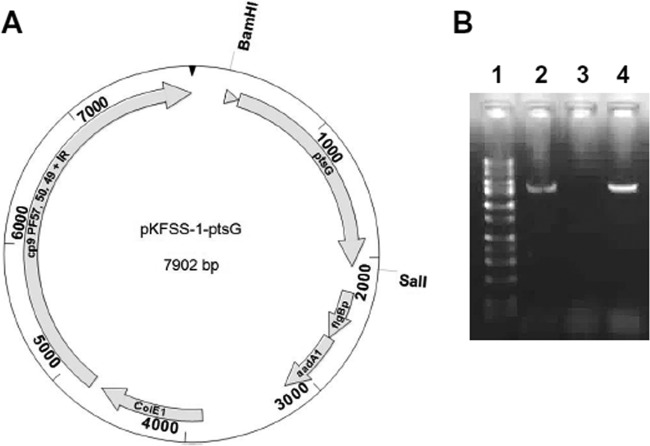
The ptsG mutant clone T10TC291 was complemented with pKFSS1 carrying a wild-type copy (WT) of ptsG and with upstream putative native promoter sequence. The pKFSS1 shuttle vector (31) contains a streptomycin/spectinomycin resistance marker encoded by aadA1 gene, which is under the control of the flgB promoter (flgBp), the ColE1 origin of replication, and required genetic elements (cp9, PF 57, 50, 49, and inverted repeat [IR]) that require replicating the shuttle plasmid in B. burgdorferi. An 1,817-bp DNA fragment including its potential promoter region (251 bp) was PCR amplified using the two primers BamHIptsGN and SalIptsGC (see Table S1 in the supplemental material) and genomic DNA from the parental B. burgdorferi 5A18NP1 strain as the template. The DNA fragment was cloned into the pKFSS1 shuttle vector using its unique BamHI and SalI restriction sites. The resulting recombinant plasmid pKFSS1::ptsG (Fig. 2A) was transformed into Escherichia coli TOP10 cells. The properties of pKFSS1::ptsG were confirmed by restriction analysis, PCR, and sequencing. pKFSS1::ptsG (20 to 25 μg) was electroporated into the ptsG mutant as described previously (32). pKFSS1 was also electroporated into parental B. burgdorferi 5A18NP1 and the ptsG mutant as controls. Transformants were isolated by streptomycin selection (50 μg/ml) in BSKII agarose plates, and transformation was confirmed by PCR using primers specific to the pKFSS1 shuttle vector. The plasmid contents of the complemented strains were determined by a Luminex procedure (30) to aid in the selection of clones for further experimentation. A similar approach was performed to develop the cyaB::pKFSS1-cyaB complemented plasmid and strain, using the indicated primers in Table S1 in the supplemental material.
FIG 2.
In trans complementation of ptsG mutant T10TC291. (A) Schematic diagram of the development of the ptsG complemented construct using the shuttle vector pKFSS1. ptsG with upstream putative native promoter sequence (triangle) was PCR amplified from parental 5A18NP1 B. burgdorferi and cloned into the BamHI-SalI sites of shuttle vector pKFSS1 (31). (B) RT-PCR analysis for the detection of transcripts of the ptsG gene is shown for the 5A18NP1 parent strain (lane 2), the ptsG mutant (lane 3), and the complemented strain (lane 4). Lane 1, DNA size standards.
RNA extraction, cDNA library preparation, and sequencing for transcriptome sequencing (RNA-seq).
Parental 5A18NP1 and ptsG mutant strains of B. burgdorferi were grown on BSKII medium. RNA was isolated from 1 × 109 cells collected at mid-logarithmic phase using a commercial RNA isolation kit (Ambion) according to the instructions provided by the manufacturer. Genomic DNA was removed by treatment with DNase I. RNA concentration was measured using a NanoDrop spectrometer (Thermo Scientific). cDNA library preparations, sequencing reactions, and initial bioinformatics analysis were conducted at Genewiz, Inc. (South Plainfield, NJ, USA). Illumina TruSeq RNA library preparation, clustering, and sequencing reagents were used throughout the process following the manufacturer's recommendations (Illumina, San Diego, CA, USA). The samples were sequenced using a 2 × 100 paired-end (PE) configuration. Image analysis and base calling were conducted with the HiSeq control software on the HiSeq2000 instrument. These base calls were used to generate bcl files using Illumina's CASAVA 1.8.2 program. The resulting bcl files were subsequently converted to fastq files, and sequence reads that passed Illumina's standard filtering were retained for further analysis using Illumina's CASAVA 1.8.2 program.
Raw sequence data generated from Illumina HiSeq2000 was converted into fastq files and demultiplexed using Illumina CASSAVA 1.8.2 program. fastq files from each sample were imported into the CLC Genomics Workbench (version 7.0.4.). Sequence reads were trimmed to remove bases with low quality at the ends (error rate of <0.05), and then the sequence reads were mapped to the reference genome of B. burgdorferi B31. Sequence hit count and reads per kilobase per million (RPKM) values were calculated for individual genes. Comparison of gene expression between two samples was conducted, and fold changes of the ptsG mutant compared to the parental 5A18NP1 strain were calculated (see Table S3 in the supplemental material). For unsupervised hierarchical clustering and principal-component analysis (PCA), the RPKM values for the two samples were quantile normalized, followed by log2 transformation. PCA and hierarchical clustering analysis were performed to examine sample separation between these two groups.
qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) was performed to validate the transcript level findings obtained using RNA-seq. qRT-PCR was performed on selected genes using the same RNA sample preparation as in the RNA-seq studies but different cDNA preparations. Additionally, vlsE expression in WT, ptsG mutant, and ptsG complemented (ptsGC) strains was determined from independent experiments using mid-logarithmic-phase cultures of the indicated strains. Primers were designed using AlleleID software (Primer Biosoft) and are listed in the Table S2 in the supplemental material. cDNA was generated from 1 μg RNA using iScript cDNA synthesis kit (Bio-Rad) according to the instruction manual. Assays were optimized as needed; PCR efficiency and product melting curves were analyzed and were within acceptable ranges. qRT-PCR was performed using iQ SYBR green supermix on the CFX touch real-time PCR detection system (Bio-Rad). flaB and/or enolase genes of B. burgdorferi B31 were used as the endogenous controls to normalize the expression. We also used no-reverse-transcriptase (NRT) controls and no-template controls (NTC) to monitor possible genomic DNA contamination of the RNA samples. Differential gene expression and fold differences were calculated by the relative quantification method (ΔΔ threshold cycle [ΔΔCT]) using Bio-Rad CFX manager software.
Mouse infection studies.
All experiments involving mice were performed according to a protocol approved by the Animal Welfare Committee of the University of Texas Health Science Center at Houston. PEP-PTS and cyaB mutant clones and the parental strain 5A18NP1 were examined individually for infectivity in 4-week-old C3H/HeNHsd mice by needle inoculation. B. burgdorferi was cultured in BSKII medium to mid-log phase, and groups of 3 mice were inoculated subcutaneously at the base of the tail with 5 × 105 organisms as described previously (27). The mice were sacrificed at 14 and/or 28 days postinoculation, and skin (at the inoculation site), tibiotarsal joint, heart, and urinary bladder were collected aseptically and were cultured in 6 ml of BSKII medium supplemented with appropriate antibiotics at 37°C in 5% CO2. The cultures were examined for the presence of spirochetes by dark-field microscopy every week up to 4 weeks.
qPCR.
The bacterial loads in skin and heart tissues from mice infected with either the 5A18NP1 WT strain or different PEP-PTS mutants of B. burgdorferi were determined using quantitative PCR (qPCR) as described previously (33). Briefly, total DNA (bacterial and mammalian) was extracted from ∼10 to 50 mg of skin and heart tissues using the DNeasy blood and tissue kit (Qiagen). The DNA concentration was measured using a NanoDrop spectrometer (Thermo Scientific), and 100 ng of DNA of each sample was used as the template for qPCR assay. qPCR was performed using iQ SYBR green supermix on the CFX touch real-time PCR detection system (Bio-Rad). DNA fragments of the flaB and mouse β-actin (34) genes were PCR amplified and cloned into pCR2.1Topo and pCR8/GW/Topo, respectively. B. burgdorferi and mammalian genomic copy numbers were determined separately in each tissue sample by using standard curves generated with known 10 to 106 copies of pCR2.1-flaB and pCR8/GW-β-actin using primer sets listed in Table S2 in the supplemental material. All mouse tissue samples were examined in triplicate technical replicates, and the standard curve for flaB and β-actin of each plate had a linear regression coefficient of determination (r2) of ≥0.98 (Fig. 3; see Fig. S2 in the supplemental material). No-template controls (NTC) and DNA extracted from uninfected mouse tissues were used as controls to optimize the assay conditions of qPCR.
FIG 3.
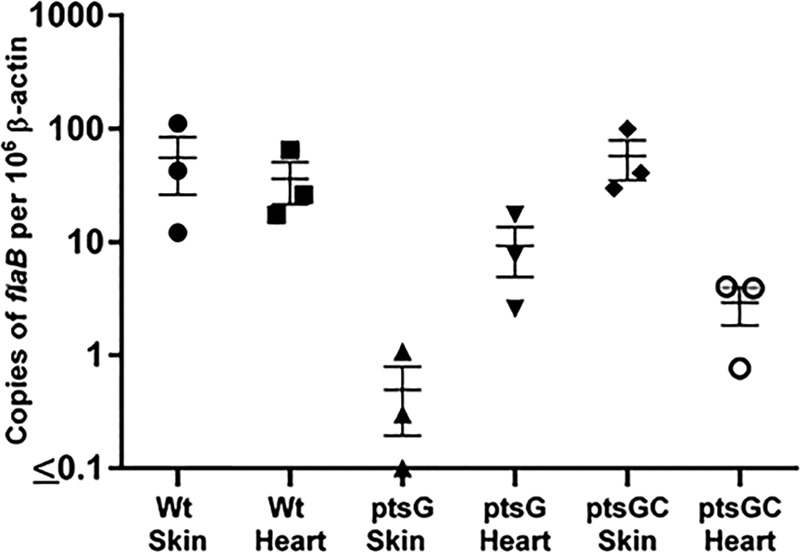
B. burgdorferi burdens in mouse tissues (skin and heart) of WT, ptsG mutant, and complemented ptsG mutant (ptsGC) strains. flaB copy numbers were determined by qPCR, representing the number of B. burgdorferi genomes. β-Actin copy numbers in the same samples were also determined to normalize the flaB copy number to the host tissue DNA. The data represent numbers of copies of B. burgdorferi genomes per 106 copies of β-actin ± standard errors of the means (SEM) for three biological replicates for each condition. The Student t test was not significant (P > 0.05) for comparisons of values for the ptsG mutant with those of either WT or ptsGC organisms.
Tick inoculation studies.
All mouse studies involving tick inoculation were performed at the Tulane National Primate Research Center and were reviewed and approved by its Institutional Animal Care and Use Committee (IACUC). Inoculation of Ixodes scapularis nymphs by capillary feeding, followed by mouse inoculation via tick feeding, was carried out as described previously (32, 35). Briefly, B. burgdorferi strains were cultured in BSKII medium containing appropriate antibiotics for parental, ptsG mutant, and complemented strains of B. burgdorferi. Capillary feeding with cultures containing 5.2 to 5.8 × 107 organisms per ml was utilized to inoculate I. scapularis ticks, and the ticks were rested for 21 to 25 days prior to (i) assessment of B. burgdorferi infection by culture and direct immunofluorescence and (ii) feeding of 10 to 12 ticks each on three 8- to 10-week-old female C3H/HeN mice. Ticks were allowed to feed to repletion and were collected for analysis by culture and immunofluorescence. Mice were bled and euthanized 4 weeks after tick feeding was completed. Serum was analyzed for anti-VlsE-C6 peptide reactivity by enzyme-linked immunosorbent assay (ELISA), and heart, bladder, ear, and tibiotarsal joint tissues were placed in BSKII medium for up to 8 weeks to assess the presence of B. burgdorferi by dark-field microscopy.
Recombinant CyaB expression and purification.
CyaB of B. burgdorferi was expressed and purified in E. coli using expression vector pQE30-Xa (Qiagen), which contains a sequence encoding an N-terminal His tag. Briefly, cyaB was amplified using primers cyaBFBamHIN and cyaBRSalIC (see Table S1 in the supplemental material). The resultant PCR fragment contained a BamHI site at the 5′ end and a SalI site at the 3′ end. After digestion with BamHI and SalI, the PCR fragment was ligated into the corresponding restriction sites of pQE30-Xa, generating the plasmid construct pQE30-Xa-CyaB. The plasmid was utilized to transform Top10 or KRX E. coli strains by electroporation. Expression was induced in KRX/pQE30-Xa-CyaB cells in LB medium at 37°C for 2 to 3 h following addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). An E. coli lysate was produced using a French press as described previously (36). CyaB was purified using HisLink protein purification resin (Promega, Madison, WI) according to standard methods, and the purified protein preparation was examined by SDS-PAGE followed by Coomassie blue R staining or Western blotting using anti-His antibody.
AC functional assay.
The adenylate cyclase (AC) functional assay was performed as described previously with some modifications (37). Briefly, ∼3 μg of purified CyaB protein was added to the 50-μl reaction mixture containing 20 nM Tris-HCl, 100 mM NaCl, 5 mM ATP, and 10 mM MnCl2. Triplicate samples were incubated at different temperatures (37°C, 50°C, and 65°C) and time intervals (30 min, 2 h, and 4 h), and cAMP concentrations were measured using cAMP Biotrak enzyme immunoassay (EIA) system (Amersham, GE Health Care) according to the manufacturer's instructions.
RESULTS
Components of the putative glucose PEP-PTS carbohydrate transporter of B. burgdorferi are required to establish mouse infection.
B. burgdorferi possesses multiple genes encoding PTS membrane transporters: three EIIBC components of glucose/maltose transporters, two encoded on the chromosome (ptsG and malX1) and one encoded on plasmid cp26 (malX2); two chromosomally encoded mannose-specific EIIABCs (fruA1 and fruA2) (20); and separate chitobiose-specific EIIA, EIIB, and EIIC components (chbA, chbB, and chbC) encoded on cp26 (Fig. 1). Previous studies in our laboratory generated transposon mutants with mutations in these carbohydrate transporters (26). Individual PEP-PTS and cyaB STM mutant clones were examined by PCR using primers flanking the insertion sites to confirm the transposon insertion and thus ascertain that each clone represented a pure culture. Some of the isolates appeared to be mixed cultures of two or more transposon mutant clones, based on the presence of faint WT-sized bands in the PCR products (see Fig. S1 in the supplemental material); subsequently these clones were separated as pure cultures by replating. PCR analyses showed that two of the mutant clones previously determined (26) to have transposon insertions in ptsH or chbB did not have inserts in these regions (see Fig. S1 in the supplemental material); these clones were hence excluded from further experimentation. Plasmid analyses further determined that the transposon mutants in this group had retained the plasmids known to be required for infectivity (lp25, lp28-1, lp36, and cp26) and lacked only plasmids that do not have a detectable influence on mouse infection (e.g., lp5 and cp9). Following these preliminary studies, we examined 10 different mutants with mutations in PEP-PTS-encoding genes and cyaB for mouse infectivity by needle inoculation. The results showed that two mutants with mutations in the putative glucose specific EIIBC (ptsG) (T10TC291 and T09TC107) were noninfectious in mice (Table 1). Three of the mutants with mutations in the putative mannose-specific EIIABC gene (fruA2) were infectious, while the clone T11P02A09 was noninfectious in this model. The other PEP-PTS mutants (including those with mutations in fruA1, malX1, malX2, fruK, cbhA, and cbhC) and the two cyaB mutants tested retained infectivity (Table 1). Some of the mutants (ptsG and cyaB) were examined for mouse infectivity after both 14 and 28 days postinfection (p.i.); the infectivity results were identical at the different time points, with none of the ptsG mutant cultures being positive and all of the cyaB mutant cultures being positive. The noninfectivity of the ptsG mutant T10TC291 and the fruA2 mutant T11P02A09 was confirmed in additional mouse needle inoculation studies (data not shown). To examine the potential effects of these mutations on in vitro growth, the growth rates of T10TC291 and T11P02A09 were examined during culture in BSKII medium and BSK-Lite (20), which lacks d-glucose other than what is present in complex medium additives (rabbit serum and yeast extract). During in vitro growth in BSKII medium, both of these mutants showed similar growth kinetics, as did the parental 5A18NP1 strain of B. burgdorferi (see Fig. S3 in the supplemental material). At later time points, the ptsG and fruA2 mutants exhibited slightly reduced concentrations in BSK-Lite relative to BSKII medium; however, these differences were not statistically significant (P > 0.05) (see Fig. S3 in the supplemental material). Because the other three fruA2 mutants were infectious (including one with an adjacent insertion site), further study of this gene has not been pursued; it is possible that T11P02A09 has secondary genetic changes that have rendered it noninfectious. Therefore, further characterization was focused on the effects of ptsG mutation on the infectivity of B. burgdorferi.
TABLE 1.
Mouse infectivity of B. burgdorferi clonal isolates of cyaB and PEP-PTS mutants
| Strain or clone | Mutated gene |
Insertion siteb | Insertion ratioc | No. of positive cultures/totala |
% positive sites | Additional plasmid(s) absentd | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene no. | Designation | Ear | Skin | Joint | Heart | Bladder | All sites | |||||
| 5A18NP1 (WT) | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | |||||
| T11TC373 | BB0723 | cyaB | 762143 | 0.02 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | lp5, lp38 |
| T08TC498e | BB0723 | cyaB | 761661 | 0.93 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | cp32-4 |
| T10TC291 | BB0645 | ptsG | 684111 | 0.45 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/15 | 0 | lp5, lp28-2 |
| T09TC107 | BB0645 | ptsG | 684634 | 0.78 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/15 | 0 | lp5, lp38 |
| T07TC473e | BB0408 | fruA1 | 421164 | 0.12 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | lp5 |
| T11P02A09e,f | BB0629 | fruA2 | 661152 | 0.20 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/15 | 0 | cp9, lp5 |
| T05TC025e,f | BB0629 | fruA2 | 661156 | 0.20 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | lp5 |
| T05TC436e,f | BB0629 | fruA2 | 660210 | 0.70 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | cp9, lp5 |
| T06TC053e,f | BB0629 | fruA2 | 660192 | 0.71 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | cp9, lp5 |
| T08P02E04 | BB0116 | malX1 | 113341 | 0.52 | 3/3 | 3/3 | 3/3 | 2/3 | 1/3 | 12/15 | 80 | lp5, lp28-2 |
| T04TC008 | BBB29 | malX2 | 25103 | 0.17 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | lp5 |
| T05TC295 | BBB05 | chbA | cp26, 4229 | 0.42 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | cp9, lp5 |
| T08P01C04 | BBB04 | chbC | cp26, 3501 | 0.23 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | lp5, lp21 |
| T09TC250 | BB0630 | fruK | 661801 | 0.21 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 | cp9, lp5 |
Groups of 3 C3H/HeN mice were each inoculated subcutaneously with 5 × 105 organisms of the strains shown. The mice were euthanized on day 28 postinoculation, and cultures were inoculated with the indicated aseptically collected tissues. Cultures were monitored for presence of B. burgdorferi by dark-field microscopy as described in Materials and Methods.
Transposon insertion site in the chromosome sequence (GenBank accession no. AE000783.1) or cp26 (GenBank accession no. AE000792.1) for chbA and chbC. 5A18NP1 is the parental strain.
Number of nucleotides from the 5′ end of the gene to the insertion site divided by the total number of nucleotides in the gene.
5A18NP1 and all transposon mutant progeny lack lp28-4 and lp56. Plasmids absent in 5A18NP1 and the mutants are not required for mouse infectivity (60).
The initial culture of the clone contained a second transposon mutant, as determined by PCR (see Materials and Methods). Each clone was separated from the coisolate by replating, and its purity was confirmed by a second PCR.
One fruA2 mutant clone (T11P02A09) was noninfectious, whereas the remaining three were infectious.
Additionally, we determined bacterial loads in skin and heart tissues for representative mutants using qPCR. The samples utilized were from the same tissue sites used for day 28 cultures (Table 1), and the results were expressed as the number of B. burgdorferi flaB copies relative to the number of mouse β-actin gene copies. Overall, qPCR results showed infectivity patterns consistent with the culture results, except that in some cases Borrelia DNA was detected at low levels in culture-negative samples (Fig. 3; see Fig. S2 in the supplemental material). Skin and heart tissues from mice (n = 3) infected with the parental strain 5A18NP1 yielded values of 38.8 ± 35.9 and 30.9 ± 17.8 flaB copies per 106 β-actin copies (geometric mean ± standard error [SE]), indicating variability in the infection levels in different animals. In comparison, the ptsG mutant T10TC291 gave skin and heart values of 0.32 ± 0.38 and 7.1 ± 5.3, respectively (Fig. 2). The bacterial load in the inoculation site skin of mice infected with the ptsG complemented strain (ptsGC) was similar to that in mice infected with the WT strain; however, as expected based on the culture results, the qPCR values were much lower in the heart (Fig. 3). The low-infectivity fruA2 mutant T11P02A09 also exhibited reduced numbers of organisms by qPCR (see Fig. S2 in the supplemental material). Although there was variability in the flaB copy numbers between the biological replicates of each tissue, a similar range of bacterial loads was observed in tissues of mice inoculated with WT and the infectious mutants examined (fruA1, fruA2 [T05TC025], and malX2) (see Fig. S2 in the supplemental material). Although there were notable reductions in the bacterial loads in mice inoculated with the ptsG mutant T10TC291 relative to those in mice inoculated with WT organisms and (for skin samples) the complemented mutant, these differences were not statistically significant (P > 0.05 by Student's t test).
The ptsG mutant clone (T10TC291) was complemented in trans on the shuttle vector pKFSS1 (pKFSS1::ptsG) using 251 bp of upstream sequences to include a potential ptsG promoter; promoter sequences could not be clearly delineated by sequence homology. The ptsG transcript was restored in the complemented strain (Fig. 2B). The initial mouse infection experiment with the complemented strain was performed without antibiotic selection in the inoculated mice. In this experiment, we were unable to culture B. burgdorferi from the mice inoculated with the ptsG complemented strain at either 14 days or 28 days postinoculation (data not shown). One possible reason that we did not observe restoration of mouse infectivity in the complemented strain may be loss of the shuttle vector during the infection process. To test this hypothesis, we first examined the retention of the complementing plasmid during 10 sequential in vitro passages in BSKII medium with and without streptomycin for the selection of the shuttle vector. Our results show that after 4 to 5 passages, the ptsG complemented strain lost the shuttle vector when cultured without streptomycin, while in the presence of streptomycin, the strain retained the shuttle vector throughout the course of the experiment (see Fig. S4 in the supplemental material). These in vitro results prompted us to repeat the mouse experiment with streptomycin selection during the course of infection. We observed that all mice (n = 4) were culture positive at the inoculation site (skin) after 14 days postinfection and that the spirochete disseminated to additional tissue sites in two of the mice (Table 2). Spirochetes recovered from tissue specimens had retained the shuttle vector, as confirmed by PCR (data not shown).These complementation experiments provided further evidence that the ptsG gene is required for the establishment of infection in mice, although the restoration of infectivity was not complete.
TABLE 2.
Mouse infectivity of the ptsG complemented strain of B. burgdorferia
| Strain | No. of positive cultures/total |
% positive | |||||
|---|---|---|---|---|---|---|---|
| Ear | Skin | Joint | Heart | Bladder | All sites | ||
| 5A18NP1 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 15/15 | 100 |
| ptsG::pKFSS1 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/15 | 0 |
| ptsG::pKFSS1-ptsG | 1/4 | 4/4 | 2/4 | 1/4 | 0/4 | 8/20 | 40 |
Methods were as described in footnote a of Table 1. ptsG, ptsG transposon mutant T10TC291. pKFSS1, empty vector control. pKFSS1-ptsG, vector with ptsG plus 200 bp of upstream sequence.
ptsG is not required for tick colonization but is essential for transmission of B. burgdorferi from ticks to mice.
We also examined the roles of ptsG and cyaB in tick infection and transmission of B. burgdorferi. Capillary feeding (35) was utilized to inoculate groups of nymphal I. scapularis ticks with the ptsG mutant, the complemented ptsG mutant (ptsG::pKFSS1-ptsG), two cyaB mutants, and the parental strain 5A18NP1. All of the B. burgdorferi strains were retained in unfed ticks and proliferated after the ticks obtained a blood meal. cyaB mutants were able to establish mouse infection after tick transmission (Table 3); however, none of the mice were culture positive or exhibited seroconversion following the feeding of ticks infected with ptsG mutant T10TC291 (Table 3). The ptsG complemented strain (ptsG::pKFSS1-ptsG) infected one of three mice as determined by positive culture and serologic assays (Table 3). The spirochetes recovered from 4 different tissues retained the same genotype as the infecting strain, as confirmed by PCR. Taken together, these data indicate that ptsG is essential for establishment of infection in mice either by needle inoculation or by tick transmission. In contrast, cyaB gene functions were not required for the maintenance of the enzootic cycle of B. burgdorferi under the conditions used in these studies.
TABLE 3.
Roles of ptsG and cyaB in tick colonization and transmission of B. burgdorferi from ticks to mice
| Strain (mutated gene) | Insertion ratio | Unfed ticks |
Fed ticks |
Mouse infection by tick transmission |
|||
|---|---|---|---|---|---|---|---|
| No. of positive cultures/total | Mean no. of spirochetes per field ± SD | No. of positive cultures/total | Mean no. of spirochetes per field ± SD | No. of positive cultures/total (no. of positive mice/total) | No. of mice with positive serology/total | ||
| 5A18NP1 | 7/10 | 0.05 ± 0.06 | 15/15 | 4.5 ± 4.1 | 12/12 (3/3) | 3/3 | |
| T11TC373 (cyaB) | 0.02 | 10/10 | 0.14 ± 0.15 | 17/17 | 3.3 ± 1.7 | 12/12 (3/3) | 3/3 |
| T08TC498 (cyaB) | 0.93 | 7/10 | 0.04 ± 0.04 | 20/22 | 1.2 ± 0.9 | 12/12 (3/3) | 3/3 |
| T10TC291 (ptsG) | 0.45 | 10/10 | 0.13 ± 0.12 | 22/22 | 3.9 ± 3.8 | 0/12 (0/3) | 0/3 |
| ptsG::pKFSS1-ptsG | 9/10 | 0.08 ± 0.07 | 23/23 | 6.3 ± 6.7 | 4/12 (1/3) | 1/3 | |
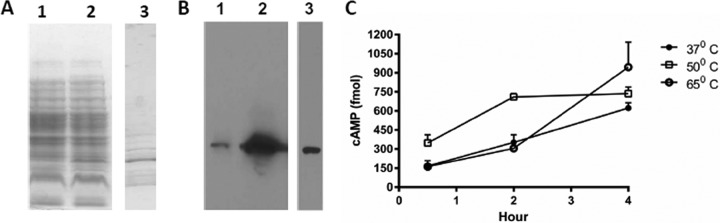
Recombinant B. burgdorferi CyaB possesses adenylate cyclase activity.
A polyhistidine-tagged form of B. burgdorferi CyaB was overexpressed and purified from E. coli using Ni affinity binding (Fig. 4A and B). Purified B. burgdorferi CyaB protein was incubated with substrate ATP at different temperatures and time intervals, and cAMP production was measured using an ELISA procedure. cAMP levels gradually accumulated with increased incubation time, and the highest activity was observed at higher temperatures (Fig. 4C). This result is consistent with the similarity of the B. burgdorferi CyaB predicted amino acid sequence with class IV adenylate cyclases identified in other bacteria, which are more active at higher temperatures (38, 39).
FIG 4.
The B. burgdorferi CyaB homolog encoded by BB0723 has adenylate cyclase activity. (A and B) SDS-PAGE (A) and Western blotting with anti-His tag antibody (B) were used to analyze the expression and purification of recombinant CyaB. Noninduced (lanes 1) and IPTG-induced (lanes 2) E. coli cell lysates and purified recombinant B. burgdorferi CyaB (lanes 3) are shown. (C) cAMP production by purified recombinant B. burgdorferi CyaB was higher at elevated temperatures, as measured using an enzyme immunoassay.
Additionally, we showed that parental 5A18NP1 B. burgdorferi and the complemented cyaB mutant strain produce cyaB transcripts, whereas neither of the cyaB mutants examined produces transcripts as determined by RT-PCR when grown in the BSKII medium at 37°C and 5% CO2 (see Fig. S5 in the supplemental material). Attempts to detect the CyaB protein in borrelial lysates using anti-CyaB antibody against either the recombinant protein or a conjugate of a predicted antigenic peptide (KEIDNIINQFGLKENIETRP) were unsuccessful (data not shown).
ptsG is involved in modulation of transcript levels of virulence-associated genes of B. burgdorferi.
Since the ptsG mutant was unable to cause infection in mice, we hypothesized that (similar to PEP-PTS components of other bacterial pathogens) ptsG in B. burgdorferi may be involved in the regulation of B. burgdorferi virulence factors essential for the establishment of infection in mice. Hence, we performed transcriptome profiling using RNA-seq to compare the ptsG mutant to the parental 5A18NP1 B. burgdorferi B31 strain at mid-logarithmic phase (∼5 × 107 cells/ml), grown on BSKII medium. Hierarchical clustering and principal-component analysis (PCA) of the RNA-seq data revealed that a number of genes were differentially expressed in the ptsG mutant (T10TC291) compared with the gene expression profile of 5A18NP1 parental strain. Overall RNA-seq analysis revealed that a number of genes were upregulated in the ptsG mutant; most of these genes encode lipoproteins (Table 4; see Table S3 in the supplemental material). A smaller number of genes were downregulated in this mutant compared to in the parental B. burgdorferi strain (Table 5; see Table S3 in the supplemental material). The trends in transcript level differences between the ptsG mutant and the WT control were generally confirmed for selected genes by qRT-PCR (Table 6). However, the increased transcript levels of ospC and dbpB and decreased levels of chbC transcripts in the ptsG mutant observed by RNA-seq were not confirmed by qRT-PCR (Table 6). Some of the genes that exhibited altered transcript levels in the ptsG mutant, such as vlsE, arp, ospC, dbpB, and dbpA, are known virulence-associated genes of B. burgdorferi that are required for the establishment of mammalian infection. Expression of fruA1 was upregulated in the ptsG mutant (>5-fold) when examined by qRT-PCR, but RNA-seq analysis revealed similar levels of fruA1 transcript for the parental and ptsG mutant strains. We also investigated expression of vlsE in the ptsG complemented strain by qRT-PCR. vlsE expression was moderately reduced in the complemented strain compared with the ptsG mutant but was not restored to wild-type levels (Fig. 5). Although our data revealed trends in vlsE expression levels in these strains (Fig. 5), the differences between strains (WT versus ptsG and WT versus ptsGC) were not statistically significant (P > 0.05). Overall, these results indicate that ptsG may modulate gene transcription in B. burgdorferi by either direct or indirect mechanisms.
TABLE 4.
Genes with increased transcript levels in the ptsG mutant T10TC291 relative to the parental control (WT) as determined by RNA-seq
| Locus | Gene product | Replicon | No. of reads (ptsG/WT) | Fold difference (ptsG/WT) |
|---|---|---|---|---|
| BB_F0041 | VlsE | lp28-1 | 1,087/63 | 17.3 |
| BB_F01 | Arthritis-related protein (Arp) | lp28-1 | 722/54 | 13.4 |
| BB_0844 | Hypothetical protein | Chromosome | 8,251/682 | 12.1 |
| BBC05 | Hypothetical protein | cp9 | 13,007/1,174 | 11.1 |
| BB_O02 | Hypothetical protein | cp32-7 | 11/1 | 11.0 |
| BB_K07 | Hypothetical protein | lp36 | 2,776/275 | 10.1 |
| BB_F14 | Hypothetical protein | lp28-1 | 9/1 | 9.0 |
| BB_H41 | Hypothetical protein | lp28-3 | 532/67 | 7.9 |
| BB_H02 | Hypothetical protein | lp28-3 | 15/2 | 7.5 |
| BB_A36 | Lipoprotein | lp54 | 24,140/3,334 | 7.2 |
| BB_K50 | Protein P37 | lp36 | 6,396/1,058 | 6.1 |
| BB_K32 | Fibronectin-binding protein | lp36 | 5,600/1,018 | 5.5 |
| BB_M37 | BppC | cp32-6 | 5/1 | 5.0 |
| BB_A25 | DbpB | lp54 | 13,855/2,804 | 4.9 |
| BB_K52 | Protein p23 | lp36 | 305/62 | 4.9 |
| BB_M28 | MlpF | cp32-6 | 932/204 | 4.6 |
| BB_A24 | DbpA | lp54 | 16,087/3,694 | 4.4 |
| BB_A34 | Solute-binding protein | lp54 | 2,535/636 | 4.0 |
| BB_B19 | OspC | cp26 | 379,664/95,325 | 4.0 |
TABLE 5.
Genes downregulated in the BB_0645/ptsG mutant in RNA-seq assay
| Locus | Gene product | Replicon | No. of reads (ptsG/WT) | Fold difference (ptsG/WT) |
|---|---|---|---|---|
| BB_U12 | Hypothetical protein | lp21 | 1/25 | −25.0 |
| BB_F32 | vls2-vls16 (silent cassettes) | 28-1 | 20/129 | −6.5 |
| BB_S21 | Hypothetical protein | cp32-3 | 10/25 | −2.5 |
| BB_H30 | Pseudogene | lp28-3 | 7/17 | −2.4 |
| BB_B04 | Chitobiose transporter ChbC | cp26 | 44,261/96,928 | −2.2 |
TABLE 6.
Validation of differential gene expression of selected genes in the ptsG mutant using qRT-PCR
| Gene no. | Gene name | Fold difference (ptsG/WT) determined by: |
|
|---|---|---|---|
| RNA-seq | qRT-PCRa | ||
| BB_F0041 | vlsE | 17.3 | 4.4 |
| BB_0844 | Hypothetical gene | 13.4 | 18.6 |
| BB_A24 | dbpA | 4.4 | 6.1 |
| BB_B25 | dbpB | 5.0 | 1.8 |
| BB_B19 | ospC | 4.0 | 1.1 |
| BB_B04 | chbC | −2.2 | 1.5 |
| BB_0408 | fruA1 | 1.3 | 5.3 |
| BB_0407 | manA | 1.3 | −1.04 |
| BB_A74 | osm28 | 2.2 | 1.9 |
| BB_0733 | plzA | 1.6 | 1.7 |
| BB_0771 | rpoS | 2.1 | 1.7 |
| BB_0450 | rpoN | 1.2 | −1.5 |
| BB_0448 | hpr | 1.2 | −1.6 |
Results for the fold difference determined by qRT-PCR analysis are representative of one experiment performed on 3 or 4 technical replicates of the same RNA samples used for RNA-seq.
FIG 5.
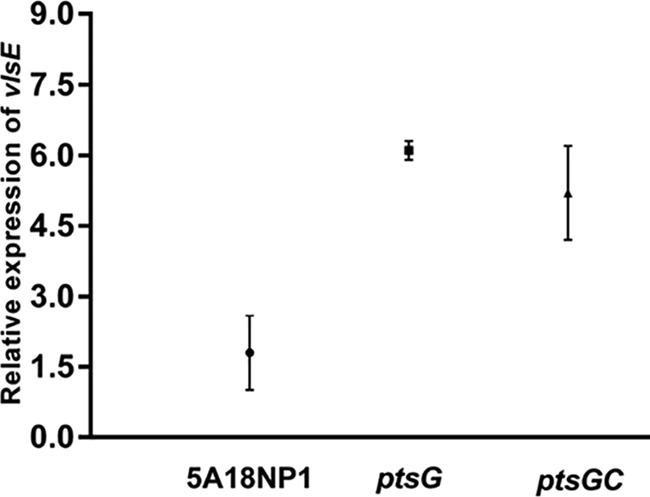
vlsE transcript levels in ptsG mutant and ptsG complemented (ptsGC) strains relative to those in the parental 5A18NP1 B. burgdorferi strain, as determined using qRT-PCR. The results obtained with two biological replicates for each strain from a representative experiment are shown. The Student t test was not significant (P > 0.05) when the data for the WT versus the ptsG mutant and the WT versus the complemented strain were compared. Transcript levels for the enolase gene (BB_0337) of B. burgdorferi B31 were used as an endogenous control to normalize vlsE expression.
DISCUSSION
To ensure successful transmission and acquisition between tick vector and susceptible hosts, Lyme disease Borrelia organisms tightly regulate gene expression in these two diverse environments. Although several studies have demonstrated that the alternative sigma factors RpoN and RpoS and other factors play key roles in the control of gene expression in Borrelia (13, 40, 41), additional, as-yet-unidentified regulatory factors are likely to be involved in this complex regulatory network. In many Gram-positive and Gram-negative organisms, the PEP-PTS acts as an important signal transduction system that controls gene expression depending on carbohydrate availability. Besides carbohydrate utilization, the PEP-PTS regulates several important virulence mechanisms, including biofilm formation, the generation and uptake of quorum-sensing signals, motility, and chemotaxis, in different bacteria (22, 23, 42, 43). Deletion of PEP-PTS genes results in an attenuated phenotype in different pathogens. Disruption of EI alters carbohydrate metabolism or nitrogen metabolism of Legionella pneumophila (44), Pseudomonas aeruginosa (45, 46), and group A Streptococcus (47), thereby reducing their virulence in animal models. Inactivation of EIIA or HPr in Brucella melitensis also leads to an attenuated phenotype (48, 49). In this study, we demonstrated that the PEP-PTS component PtsG modulates virulence of B. burgdorferi, possibly by regulating expression of genes that contribute to virulence.
Genome sequence analysis indicates that B. burgdorferi possesses six putative PEP-PTS carbohydrate transporters (Fig. 1) (5). Prior studies demonstrated that sugars that typically utilize PEP-PTS-mediated transport and phosphorylation (including glucose, mannose, and chitobiose) support the growth of B. burgdorferi (20). In addition, it was hypothesized that FruA1 and FruA2, previously annotated as fructose transporters, most likely represent mannose transporters in Borrelia, because fructose does not support the growth of B. burgdorferi (20). However, the sugar specificities of the PEP-PTS transporters, except for the chitobiose transporter (50), of B. burgdorferi have not been determined experimentally.
In the present study, we examined the infectivities of 12 PEP-PTS transporter component mutants using a mouse model in order to determine their role in establishment of infection in vertebrate host. Our data showed that ptsG (putative glucose transporter) mutants were unable to establish infection in mice, while strains with mutations in other PEP-PTS transporter genes were infectious (Table 1 and Fig. 3; see Fig. S2 in the supplemental material). In addition, one fruA2 mutant (T11P02A09) was noninfectious in mice, while the other three fruA2 mutants were infectious (Table 1; see Fig. S2 in the supplemental material). This discrepancy could be due to secondary genetic changes in the noninfectious fruA2 mutant, although plasmid analyses indicate that plasmids required for mouse infectivity are not missing. Overall these results indicate that PtsG plays a crucial role in establishing infection in the vertebrate host, consistent with the likelihood that d-glucose is the principal carbohydrate utilized by B. burgdorferi during mammalian infection (20). Given that infectivity using a high dose of organisms (5 × 105) is a qualitative and relatively insensitive measure, it is likely that some of the other genes tested are also required for optimal infection of mammals; this possibility will be examined in future studies.
Chitobiose, the dimer of chitin, is a component of tick cuticle. A previous study demonstrated that the ChbC component of the chitobiose transporter of B. burgdorferi is required for chitobiose utilization, but the chbC mutant did not show a defect in any stage of the mouse-tick infectious cycle (50). Our results support previous findings that the chitobiose transporter genes are not essential for mouse infection (50).
The roles of the ptsG and cyaB genes in the tick cycle were examined. We speculate that some of the other genes that do not play role in mammalian infection still may have role in the tick cycle. Determination of role of a specific gene in the tick cycle is laborious and expensive; therefore, it was not possible to investigate the roles of several genes in the survivability within the tick and transmission of the pathogen by tick bite in the current study. Nonetheless, in future studies, we will determine the roles of additional genes involved in PEP-PTS-mediated carbohydrate transport in the tick cycle of B. burgdorferi.
Infectivity was partially restored by in trans complementation of the ptsG mutant, as examined by both needle inoculation and tick transmission (Tables 2 and 3). Treatment of the needle-inoculated mice with streptomycin was utilized to select for the retention of the pKFSS-1::ptsG construct, but similar infectivity results were obtained with tick-inoculated mice without streptomycin treatment. It is interesting to note that in needle inoculation, all the mice (n = 4) were culture positive at the dermal inoculation site, but only two mice exhibited dissemination to other sites (heart, joint, and ear) (Table 2 and Fig. 3). The failure of ptsG complementation to restore full infectivity may be due to several possible reasons: (i) instability of the shuttle vector during infection, resulting in loss of the complementing plasmid; (ii) reduced or aberrant ptsG expression from the shuttle vector; (iii) secondary genetic changes in the transposon mutant; or (iv) lack of 5′ sequences required for optimal transcription. Upstream of ptsG, the B. burgdorferi genome contains a homolog of nanE (BB0644), encoding an epimerase that is involved in the amino and nucleotide sugar utilization pathway (51). The putative nanE and ptsG ORFs are separated by only 45 nucleotides; it is therefore possible that these genes are cotranscribed. In future studies, we will further examine these possibilities through the generation of a ptsG deletion mutant and the exploration of alternative complementation approaches.
Purified B. burgdorferi CyaB protein showed highest activity at higher temperatures, which is in agreement with previous studies performed with class IV adenylate cyclases isolated from Aeromonas hydrophila and Yersinia pestis (38, 39). A conserved motif, EXEXK, initially detected in class IV ACs from other bacteria, was also present in the active site of B. burgdorferi AC. The infectivity data (Tables 1 and 3) showed that the functions of CyaB are not essential for maintaining the enzootic cycle of B. burgdorferi in our model system; however, the high degree of conservation of cyaB among both Lyme disease and relapsing fever Borrelia strains indicates that the gene product does play a role in the survival of these organisms in their natural environments. Combining these data and the fact that the B. burgdorferi genome does not contain any homolog of cAMP receptor protein (CRP) (52), we speculate that unlike in Gram-negative bacteria, cAMP signaling may not couple with PEP-PTS to regulate virulence genes of B. burgdorferi.
Transcriptome analysis of the ptsG mutant showed that transcription of a number of virulence-associated genes is upregulated in the ptsG mutant compared with the parental B. burgdorferi strain (Tables 4 and 6). These data indicate that components of PEP-PTS transporters may directly or indirectly regulate gene transcription of B. burgdorferi. Transcript levels of several lipoprotein genes, including vlsE, were elevated in the ptsG mutant, indicating that PtsG or a PtsG-dependent factor(s) may repress the expression of lipoprotein genes. Previous studies indicated that vlsE may be regulated at transcriptional level through DNA-binding proteins (53). A B. burgdorferi homolog of SpoVG has been shown to bind to a sequence within the vlsE open reading frame (54). In a separate study the same group showed that EbfC (BB0462), a bacterial nucleoid-associated protein, binds to the coding region of ptsG (55). Therefore, expression of ptsG may be directly regulated by EbfC. However, the mechanism by which ptsG regulates gene transcription of vlsE and other virulence factors has yet to be determined.
The loss of regulation of the expression of lipoprotein genes could be responsible for the noninfectious nature of the ptsG mutant in mammalian infection. Previous studies suggested that increased expression of dbpA and vlsE impaired dissemination, which resulted in quick clearance of spirochete from mice (56, 57). We speculate that the repression of gene transcription through ptsG may have physiological importance under certain environmental conditions.
In other bacteria, ptsG may be indirectly associated with the regulation of gene expression and virulence. For instance, in Vibrio cholerae, Richard et al. (58) demonstrated that a small regulatory RNA, TarA, regulates expression of ptsG mRNA, which in turn may regulate other genes and the virulence of this pathogen. In E. coli, the small regulatory RNA SgrS controls ptsG transcript levels (59). It is possible that ptsG transcripts of B. burgdorferi are also regulated by a small regulatory RNA(s).
It is possible that the ptsG mutant of B. burgdorferi may simply be unable to transport sufficient carbohydrates for energy production, resulting in a noninfectivity phenotype. Currently, we do not know the substrate specificity of the ptsG transporter in B. burgdorferi; however, based on homology, it is likely involved in transporting glucose and/or a related sugar. In a separate study, E. B. Troy et al. (personal communication) examined in vitro carbohydrate utilization of selected B. burgdorferi mutants in the STM library by using transposon sequencing (Tn-seq) in a BSK basal medium containing low levels of glucose (BSK-Lite) supplemented with different carbohydrates. However, their study did not indicate differential growth of ptsG or fruA2 mutants in the presence of glucose versus other carbohydrate substrates (e.g., maltose); thus, no information was revealed regarding carbohydrate specificity of the encoded transporter proteins. Since the B. burgdorferi genome contains genes encoding several potential glucose-specific EIIBC components (ptsG, malX1, and malX2), loss of one transporter may be compensated for by other EII sugar transporters. Generating strains with multiple mutations in the ptsG, malX1, and malX2 genes as well as development of a better defined basal medium may be helpful in determining the substrate specificities of ptsG and other putative EIIBC components. At this point, it is unclear whether the profound reduction of B. burgdorferi infectivity by ptsG mutation is due to the disruption of transport activities, effects on gene expression, or indirect influences. Additional experimental analysis is needed to clarify how ptsG is involved in the pathogenicity of B. burgdorferi.
In summary, our studies indicate that a component(s) of the PEP-PTS may modulate the ability of B. burgdorferi to infect mammals by regulating transcription of different virulence-associated genes. We hypothesize that the phosphorylation states of different PEP-PTS proteins are altered in response to carbohydrate availability in the arthropod and/or mammalian host environments, which in turn facilitates differential gene expression in B. burgdorferi. Therefore, it will be of interest to investigate in detail the mechanisms of PEP-PTS gene regulation in B. burgdorferi, which will in turn promote a better understanding of the enzootic cycle and pathogenesis of the organisms that cause Lyme disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erin B. Troy and Linden Hu (Tufts University School of Medicine) for sharing unpublished information, Doug Litwin (UT Health Graduate School of Biomedical Science) for assisting with primer design for qRT-PCR, and Diane Edmondson and Sabitha Prabhakaran for their assistance in protein extraction and anti-CyaB antibody development in mice.
B.K.K. is the recipient of the Jeane B. Kempner postdoctoral fellowship award from the University of Texas Medical Branch, Galveston. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01 AI059048.
Funding Statement
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01 AI059048. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00917-15.
REFERENCES
- 1.Kuehn BM. 2013. CDC estimates 300,000 US cases of Lyme disease annually. JAMA 310:1110. doi: 10.1001/jama.2013.278331. [DOI] [PubMed] [Google Scholar]
- 2.Mead PS. 2015. Epidemiology of Lyme disease. Infect Dis Clin North Am 29:187–210. doi: 10.1016/j.idc.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Marques AR. 2010. Lyme disease: a review. Curr Allergy Asthma Rep 10:13–20. doi: 10.1007/s11882-009-0077-3. [DOI] [PubMed] [Google Scholar]
- 4.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35:490–516. [DOI] [PubMed] [Google Scholar]
- 5.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 6.Chaconas G, Norris SJ. 2013. Peaceful coexistence amongst Borrelia plasmids: getting by with a little help from their friends? Plasmid 70:161–167. doi: 10.1016/j.plasmid.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuels DS. 2011. Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- 8.Kostick JL, Szkotnicki LT, Rogers EA, Bocci P, Raffaelli N, Marconi RT. 2011. The diguanylate cyclase, Rrp1, regulates critical steps in the enzootic cycle of the Lyme disease spirochetes. Mol Microbiol 81:219–231. doi: 10.1111/j.1365-2958.2011.07687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caimano MJ, Kenedy MR, Kairu T, Desrosiers DC, Harman M, Dunham-Ems S, Akins DR, Pal U, Radolf JD. 2011. The hybrid histidine kinase Hk1 is part of a two-component system that is essential for survival of Borrelia burgdorferi in feeding Ixodes scapularis ticks. Infect Immun 79:3117–3130. doi: 10.1128/IAI.05136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He M, Ouyang Z, Troxell B, Xu H, Moh A, Piesman J, Norgard MV, Gomelsky M, Yang XF. 2011. Cyclic di-GMP is essential for the survival of the Lyme disease spirochete in ticks. PLoS Pathog 7:e1002133. doi: 10.1371/journal.ppat.1002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boardman BK, He M, Ouyang Z, Xu H, Pang X, Yang XF. 2008. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun 76:3844–3853. doi: 10.1128/IAI.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. 2005. Borrelia burgdorferi Sigma54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A 102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caimano MJ, Eggers CH, Hazlett KR, Radolf JD. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect Immun 72:6433–6445. doi: 10.1128/IAI.72.11.6433-6445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He M, Zhang JJ, Ye M, Lou Y, Yang XF. 2014. Cyclic di-GMP receptor PlzA controls virulence gene expression through RpoS in Borrelia burgdorferi. Infect Immun 82:445–452. doi: 10.1128/IAI.01238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, Caimano M, Skare J, Swingle K, Akins D, Schwartz I. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun 71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll JA, Garon CF, Schwan TG. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun 67:3181–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seshu J, Boylan JA, Gherardini FC, Skare JT. 2004. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect Immun 72:1580–1586. doi: 10.1128/IAI.72.3.1580-1586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyde JA, Trzeciakowski JP, Skare JT. 2007. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J Bacteriol 189:437–445. doi: 10.1128/JB.01109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks CS, Hefty PS, Jolliff SE, Akins DR. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun 71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Lackum K, Stevenson B. 2005. Carbohydrate utilization by the Lyme borreliosis spirochete, Borrelia burgdorferi. FEMS Microbiol Lett 243:173–179. doi: 10.1016/j.femsle.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Lengeler JW, Jahreis K. 2009. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib Microbiol 16:65–87. doi: 10.1159/000219373. [DOI] [PubMed] [Google Scholar]
- 22.Pereira CS, Santos AJ, Bejerano-Sagie M, Correia PB, Marques JC, Xavier KB. 2012. Phosphoenolpyruvate phosphotransferase system regulates detection and processing of the quorum sensing signal autoinducer-2. Mol Microbiol 84:93–104. doi: 10.1111/j.1365-2958.2012.08010.x. [DOI] [PubMed] [Google Scholar]
- 23.Houot L, Chang S, Absalon C, Watnick PI. 2010. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect Immun 78:1482–1494. doi: 10.1128/IAI.01356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poncet S, Milohanic E, Maze A, Nait Abdallah J, Ake F, Larribe M, Deghmane AE, Taha MK, Dozot M, De Bolle X, Letesson JJ, Deutscher J. 2009. Correlations between carbon metabolism and virulence in bacteria. Contrib Microbiol 16:88–102. doi: 10.1159/000219374. [DOI] [PubMed] [Google Scholar]
- 25.Saier MH Jr, Chauvaux S, Deutscher J, Reizer J, Ye JJ. 1995. Protein phosphorylation and regulation of carbon metabolism in gram-negative versus gram-positive bacteria. Trends Biochem Sci 20:267–271. doi: 10.1016/S0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 26.Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, Chaconas G, Philipp MT, Norris SJ. 2012. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS One 7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norris SJ, Howell JK, Garza SA, Ferdows MS, Barbour AG. 1995. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect Immun 63:2206–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dever LL, Jorgensen JH, Barbour AG. 1992. In vitro antimicrobial susceptibility testing of Borrelia burgdorferi: a microdilution MIC method and time-kill studies. J Clin Microbiol 30:2692–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 30.Norris SJ, Howell JK, Odeh EA, Lin T, Gao L, Edmondson DG. 2011. High-throughput plasmid content analysis of Borrelia burgdorferi B31 by using Luminex multiplex technology. Appl Environ Microbiol 77:1483–1492. doi: 10.1128/AEM.01877-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol 185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin T, Gao L, Edmondson DG, Jacobs MB, Philipp MT, Norris SJ. 2009. Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi. PLoS Pathog 5:e1000679. doi: 10.1371/journal.ppat.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wager B, Shaw DK, Groshong AM, Blevins JS, Skare JT. 2015. BB0744 affects tissue tropism and spatial distribution of Borrelia burgdorferi. Infect Immun 83:3693–3703. doi: 10.1128/IAI.00828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maruskova M, Esteve-Gassent MD, Sexton VL, Seshu J. 2008. Role of the BBA64 locus of Borrelia burgdorferi in early stages of infectivity in a murine model of Lyme disease. Infect Immun 76:391–402. doi: 10.1128/IAI.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Indest KJ, Howell JK, Jacobs MB, Scholl-Meeker D, Norris SJ, Philipp MT. 2001. Analysis of Borrelia burgdorferi vlsE gene expression and recombination in the tick vector. Infect Immun 69:7083–7090. doi: 10.1128/IAI.69.11.7083-7090.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrenz MB, Hardham JM, Owens RT, Nowakowski J, Steere AC, Wormser GP, Norris SJ. 1999. Human antibody responses to VlsE antigenic variation protein of Borrelia burgdorferi. J Clin Microbiol 37:3997–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Topal H, Fulcher NB, Bitterman J, Salazar E, Buck J, Levin LR, Cann MJ, Wolfgang MC, Steegborn C. 2012. Crystal structure and regulation mechanisms of the CyaB adenylyl cyclase from the human pathogen Pseudomonas aeruginosa. J Mol Biol 416:271–286. doi: 10.1016/j.jmb.2011.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith N, Kim SK, Reddy PT, Gallagher DT. 2006. Crystallization of the class IV adenylyl cyclase from Yersinia pestis. Acta Crystallogr Sect F Struct Biol Cryst Commun 62:200–204. doi: 10.1107/S1744309106002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sismeiro O, Trotot P, Biville F, Vivares C, Danchin A. 1998. Aeromonas hydrophila adenylyl cyclase 2: a new class of adenylyl cyclases with thermophilic properties and sequence similarities to proteins from hyperthermophilic archaebacteria. J Bacteriol 180:3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol 65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dulebohn DP, Hayes BM, Rosa PA. 2014. Global repression of host-associated genes of the Lyme disease spirochete through post-transcriptional modulation of the alternative sigma factor RpoS. PLoS One 9:e93141. doi: 10.1371/journal.pone.0093141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Black RA, Hobson AC, Adler J. 1983. Adenylate cyclase is required for chemotaxis to phosphotransferase system sugars by Escherichia coli. J Bacteriol 153:1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang W, Pascual-Montano A, Silva AJ, Benitez JA. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 44.Higa F, Edelstein PH. 2001. Potential virulence role of the Legionella pneumophila PtsP ortholog. Infect Immun 69:4782–4789. doi: 10.1128/IAI.69.8.4782-4789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan MW, Mahajan-Miklos S, Ausubel FM. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gera K, Le T, Jamin R, Eichenbaum Z, McIver KS. 2014. The phosphoenolpyruvate phosphotransferase system in group A Streptococcus acts to reduce streptolysin S activity and lesion severity during soft tissue infection. Infect Immun 82:1192–1204. doi: 10.1128/IAI.01271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delrue RM, Lestrate P, Tibor A, Letesson JJ, De Bolle X. 2004. Brucella pathogenesis, genes identified from random large-scale screens. FEMS Microbiol Lett 231:1–12. doi: 10.1016/S0378-1097(03)00963-7. [DOI] [PubMed] [Google Scholar]
- 49.Wu Q, Pei J, Turse C, Ficht TA. 2006. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol 6:102. doi: 10.1186/1471-2180-6-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tilly K, Grimm D, Bueschel DM, Krum JG, Rosa P. 2004. Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component. Vector Borne Zoonotic Dis 4:159–168. doi: 10.1089/1530366041210738. [DOI] [PubMed] [Google Scholar]
- 51.Plumbridge J, Vimr E. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J Bacteriol 181:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian G, Koonin EV, Aravind L. 2000. Comparative genome analysis of the pathogenic spirochetes Borrelia burgdorferi and Treponema pallidum. Infect Immun 68:1633–1648. doi: 10.1128/IAI.68.3.1633-1648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bykowski T, Babb K, von Lackum K, Riley SP, Norris SJ, Stevenson B. 2006. Transcriptional regulation of the Borrelia burgdorferi antigenically variable VlsE surface protein. J Bacteriol 188:4879–4889. doi: 10.1128/JB.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jutras BL, Chenail AM, Rowland CL, Carroll D, Miller MC, Bykowski T, Stevenson B. 2013. Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins. PLoS One 8:e66683. doi: 10.1371/journal.pone.0066683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jutras BL, Bowman A, Brissette CA, Adams CA, Verma A, Chenail AM, Stevenson B. 2012. EbfC (YbaB) Is a new type of bacterial nucleoid-associated protein and a global regulator of gene expression in the Lyme disease spirochete. J Bacteriol 194:3395–3406. doi: 10.1128/JB.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Q, Seemanaplli SV, McShan K, Liang FT. 2007. Increasing the interaction of Borrelia burgdorferi with decorin significantly reduces the 50 percent infectious dose and severely impairs dissemination. Infect Immun 75:4272–4281. doi: 10.1128/IAI.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Q, McShan K, Liang FT. 2008. Modification of Borrelia burgdorferi to overproduce OspA or VlsE alters its infectious behaviour. Microbiology 154:3420–3429. doi: 10.1099/mic.0.2008/019737-0. [DOI] [PubMed] [Google Scholar]
- 58.Richard AL, Withey JH, Beyhan S, Yildiz F, DiRita VJ. 2010. The Vibrio cholerae virulence regulatory cascade controls glucose uptake through activation of TarA, a small regulatory RNA. Mol Microbiol 78:1171–1181. doi: 10.1111/j.1365-2958.2010.07397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wadler CS, Vanderpool CK. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A 104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purser JE, Norris SJ. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.