SUMMARY
The ubiquitous biological nanomotors were classified into two categories in the past: linear and rotation motors. In 2013, a third type of biomotor, revolution without rotation (http://rnanano.osu.edu/movie.html), was discovered and found to be widespread among bacteria, eukaryotic viruses, and double-stranded DNA (dsDNA) bacteriophages. This review focuses on recent findings about various aspects of motors, including chirality, stoichiometry, channel size, entropy, conformational change, and energy usage rate, in a variety of well-studied motors, including FoF1 ATPase, helicases, viral dsDNA-packaging motors, bacterial chromosome translocases, myosin, kinesin, and dynein. In particular, dsDNA translocases are used to illustrate how these features relate to the motion mechanism and how nature elegantly evolved a revolution mechanism to avoid coiling and tangling during lengthy dsDNA genome transportation in cell division. Motor chirality and channel size are two factors that distinguish rotation motors from revolution motors. Rotation motors use right-handed channels to drive the right-handed dsDNA, similar to the way a nut drives the bolt with threads in same orientation; revolution motors use left-handed motor channels to revolve the right-handed dsDNA. Rotation motors use small channels (<2 nm in diameter) for the close contact of the channel wall with single-stranded DNA (ssDNA) or the 2-nm dsDNA bolt; revolution motors use larger channels (>3 nm) with room for the bolt to revolve. Binding and hydrolysis of ATP are linked to different conformational entropy changes in the motor that lead to altered affinity for the substrate and allow work to be done, for example, helicase unwinding of DNA or translocase directional movement of DNA.
INTRODUCTION
Motors impact almost all aspects of daily life. For example, electric motors are machines that convert electrical energy into mechanical energy and further into kinetic energy to drive the operation of other devices, and these motors have been around since the 1740s, when Scottish monk Andrew Gordon introduced a simple electrostatic device (1). Similarly, bionanomotors are miniscule protein machines that produce mechanical motion by converting an energy source into work. These biological motors are responsible for most forms of motion in all life forms. Bionanomotors are essential in all aspects of crucial cellular processes critical to survival, such as mitosis, DNA replication, DNA repair, homologous recombination, Holliday junction resolution, RNA transcription, ATP synthesis, muscle contraction, viral genome packaging, and directional motility of cellular components to their destinations. Bionanomotors make possible the occurrence of otherwise thermodynamically unfavorable processes.
In the past, biological nanomotors were classified into two categories: linear and rotational motors. Recently, a third type of biological motor mechanism, revolution without rotation, was described and found to be widespread in bacteria, eukaryotic viruses, and dsDNA bacteriophages (http://rnanano.osu.edu/movie.html). Its impact is evident, as with the viral DNA-packaging motor, which was thought for decades to be rotational (2). Recent studies have revealed that none of the motor components rotates to any significant degree during genome packaging (3–7). The finding of revolution motion has now been applied to many mysteries regarding biological motor structures, functions, and mechanisms.
This review focuses on recent findings about various aspects of motor function, including chirality, stoichiometry, channel size, entropy and conformational change, and energy usage, in a wide variety of motor proteins: FoF1 ATPase, bacterial flagellar motors, helicases, viral double-stranded DNA (dsDNA)-packaging motors, bacterial chromosome translocases, myosin, kinesin, and dynein. Translocases of dsDNA are used to illustrate how these features relate to the motor mechanism in energy conversion, directional control, and sequential action. Nature has elegantly evolved a revolution mechanism to avoid coiling and tangling during lengthy dsDNA genome transportation.
CLASSIFICATION OF BIOMOTORS
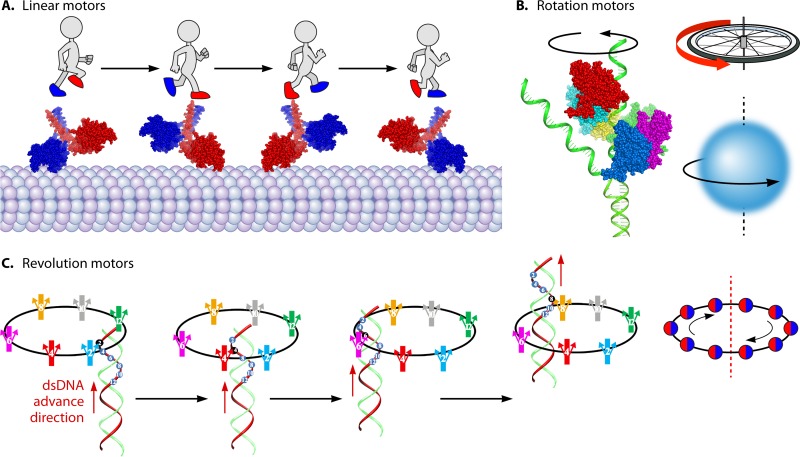
Although different biomotors may possess different structures and have distinct roles in cellular functions, they all need to undergo conformational changes to create motion. Based on their motion mechanisms, the biomotors are categorized into three classes: linear, rotary, and revolutional (Fig. 1) (8–16). Rotation is the circular movement of an object around its own axis, resembling the Earth rotating on its axis in a complete cycle every 24 h. Revolution is the turning of an object around a second object, resembling the action of the Earth revolving around the sun one circle per year (Fig. 1). Many motors assemble into hexamers and use ATP to trigger their conformational changes. Typical linear motors are myosin, kinesin, and dynein (17–19), typical rotation motors are FoF1 ATP synthase, helicases, and the bacterial flagellar motor (9, 20–24), and typical revolution motors are genomic dsDNA translocases for bacterial chromosome segregation and viral dsDNA packaging (12, 25). The representatives in the last category include the dsDNA translocases FtsK of Escherichia coli and SpoIIIE of Bacillus and the dsDNA-packaging motors of bacteriophages phi29, T3, T4, T7, P22, lambda, and SPP1 and the large animal dsDNA viruses (26).
FIG 1.
Illustration of different categories of motors. (A) Linear motors are like people walking (PDB code 3KIN). (B) Rotation motors are like a wheel and like Earth rotating on its own axis. (Adapted from reference 50.) (C) Revolution motors resemble Earth revolving around the sun without self-rotation. (Adapted from reference 14 with permission from Elsevier.)
Rotation Motors
FoF1 ATP synthase, helicases, and bacteria flagella are common representatives of rotation motors (20, 21, 27). Besides the flagellar motor, which is more intricate, many of these rotary ATPases are assembled into hexamers. Although many other nucleic acid-tracking ATPases are present in different oligomeric forms such as monomer and dimer, for those hexameric rotation motors that track along nucleic acids, most, if not all, rotate along nucleic acids with one strand of DNA or RNA passing through the channel. Most rotation dsDNA translocases share mechanisms that are distinct from those of the revolution dsDNA translocases. These rotation nanomotors include, but are not limited to, helicases (27–29), RNA polymerase (30), transcription termination factors (31), and others that participate in DNA recombination, repair, and Holliday junction resolution. Some RecA family ATPase monomers or dimers can assemble onto DNA as a filament (32, 33); whether the functional unit in the complex is a hexamer with a helical “open-washer” structure remains to be confirmed.
FoF1 complex.
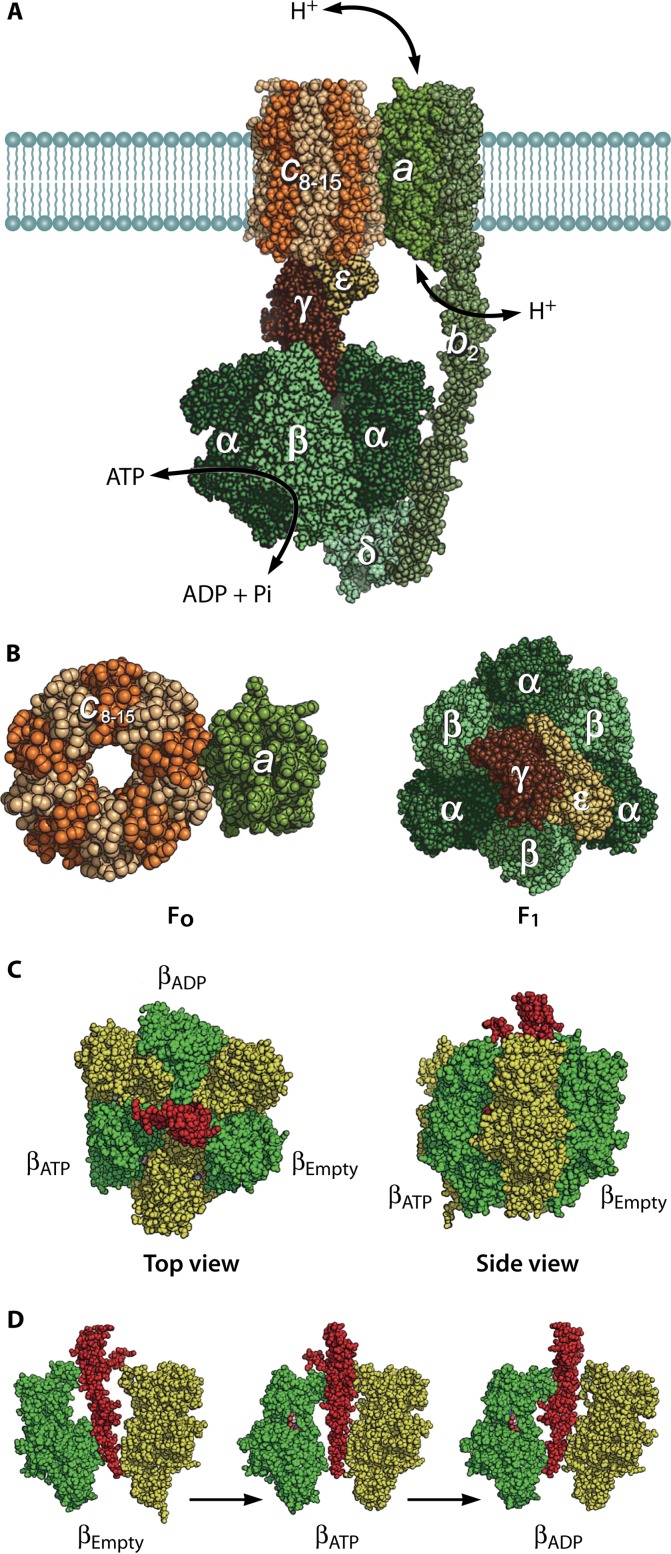
FoF1 ATP synthase is found in the inner membranes of mitochondria, the thylakoid membranes of chloroplasts, and the plasma membranes of bacteria (34, 35) and is responsible for ATP generation. This enzyme is composed of two rotary motors, Fo and F1. F1 is a membrane-protruding part and the catalytic domain of the ATP synthase. When isolated from the membrane, F1 acts as a rotary motor fueled by ATP hydrolysis. Fo is the membrane-spanning part of ATP synthase and conducts the proton translocation across the membrane down membrane potential. Upon proton translocation, Fo rotates the ring-shaped rotor against the stator complex composed of the a and b subunits (Fig. 2). Fo and F1 are connected by a common rotor axle and a peripheral stalk (Fig. 2 and 3). These connections allow interconversion of proton motive force (PMF) and free energy of ATP hydrolysis via mechanical rotation of the subunit complex (20).
FIG 2.
Structures of FoF1 ATP synthase and the α3β3γ subcomplex of F1. (A) Reconstituted structure of FoF1 ATP synthase from crystal structures of isolated subunit or subcomplexes: a3b3γε subcomplex (PDB code 3OAA), δ (PDB code 1ABV), b dimer (PDB codes 1B9U, 2KHK, and 1L2P), c ring (PDB code 3UD0), and putative structure of the a subunit (PDB code 1C17). Green parts represent the stator complex, including the peripheral stalk (δ-b2 subcomplex) that holds the a3b3 stator ring of F1 and ab2 stator of Fo. Brown parts represent the rotor complex (γε–c-ring subcomplex). (B) Fo and F1 (Fig. 1A), both viewed from the top. (C) Original crystal structure of F1 from bovine mitochondria (PDB code 1BMF). Sphere representations of the α, β, and γ subunits are shown in yellow, green, and red, respectively. Each β subunit carries either AMP-PNP, ADP, or neither and is designated βATP, βADP, or βEmpty, respectively. (D) Conformational states of 3 β subunits viewed from the side. α-β pairs are shown in green and yellow with the central γ subunit in red. α and β subunits are composed of the N-terminal domain, nucleotide binding domain, and C-terminal domain (from bottom to top). βEmpty has an open conformation in which the α-helical C-terminal domain rotates upward, opening the cleft of the nucleotide binding pocket. Both βATP and βADP have a closed conformation entrapping the nucleotide within the closed pocket. All α subunits represent the open conformation.
FIG 3.
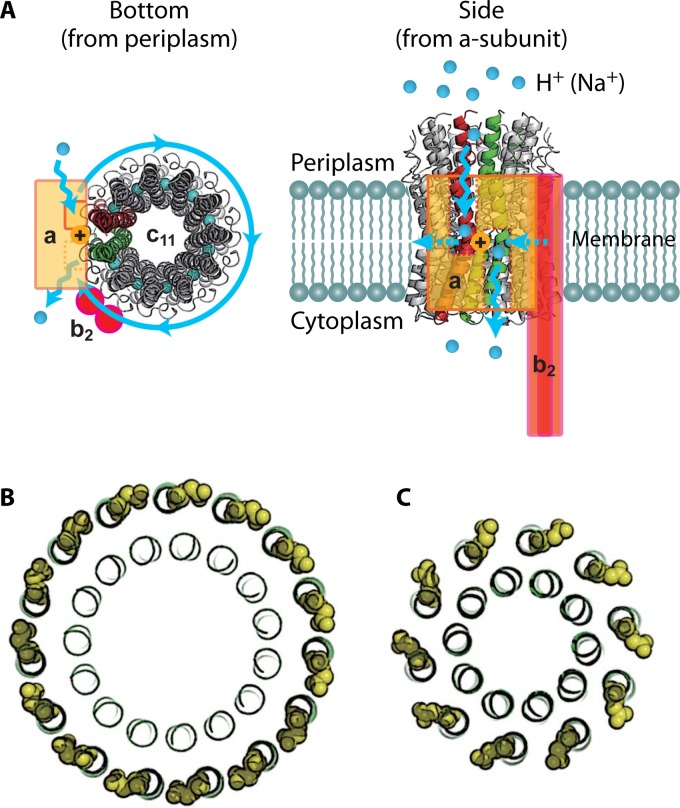
Structure of Fo. (A) Crystal structure of the c11 ring of Na+-transporting Fo from Ilyobacter tartaricus (PDB code 1YCE). The blue spheres in the middle of the c11 ring represent bound Na+ ions. The stator ab2 complex is shown in the schematic drawing. The a subunit has 2 hemichannels, each open to the periplasmic space or the cytoplasmic space. A proton transferring between the a and c subunits accompanies the rotation of the c ring. Two c-subunit monomers at the interface of the a subunit are shown in red and green, respectively. (B) “Ion-locked” conformation of cGlu62 (yellow sphere representation) in the crystal structure of the H+-transporting c15 ring from Spirulina platensis (PDB code 2WIE). (C) “Ion-unlocked” conformation of cGlu59 (yellow sphere representation) in the crystal structure of the H+-transporting c10 ring from yeast mitochondria (PDB code 3U2F).
Bacterial flagella.
Different species of bacteria use different motion methods in order to locate optimal growth conditions or avoid toxic substances. Most motile bacteria are driven by the rotation of flagellar filaments displaying a stiff helical structure approximately 10 μm in length and 20 μm in diameter. Each filament has its own individual rotation motor at its base (21, 36). These motors can rotate clockwise to produce forward movement or counterclockwise, resulting in tumbling and the change of direction. The flagellar motor is powered by a proton motive force. Many models, such as the “electrostatic proton turbine” model and the “turnstile” model, have been proposed to explain flagellar motor function (36, 37).
RNA polymerase.
The RNA polymerases are essential enzymes for the process of transcription. The RNA polymerase reads the DNA strand and transcribes it into RNA sequence. During this process, RNA polymerase rotates DNA through its channel; DNA rotation has been identified by direct observation by real-time optical microscopy measurements (38).
DNA helicase.
Nucleic acid helicases comprise a class of enzymes that convert chemical energy into mechanical work for unidirectional translocation along nucleic acids and separate the nucleic acid duplexes into transient single-stranded intermediates (39–42). Helicases are involved in many cellular processes, including DNA repair, replication, and recombination and RNA transcription, remodeling, splicing, and translation (39–42). There are numerous ways to classify nucleic acid helicases. Based on their sequence, helicases are divided into six main groups, SF1 to SF6 (40). The cores of SF1 and SF2 enzymes are structurally similar to each other (40). The most well studied SF3 helicase is the papillomavirus E1 helicase, and the most well studied SF4 helicase is T7 gp4 helicase (40). SF5 contains the bacterial Rho factor (40), which is an ATP-dependent hexameric RNA helicase involved in the termination of transcription in bacteria. SF6 contains some of the AAA+-like proteins, including the MCM proteins as well as RuvB (40).
Revolution Motors
DNA-packaging motor of dsDNA bacteriophages.
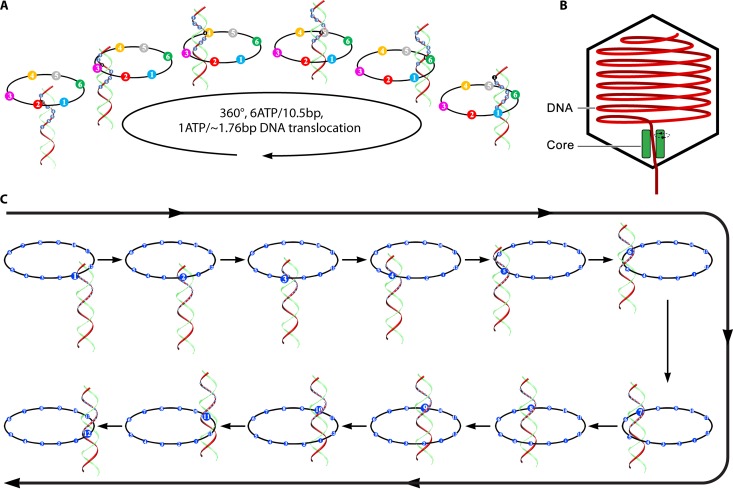
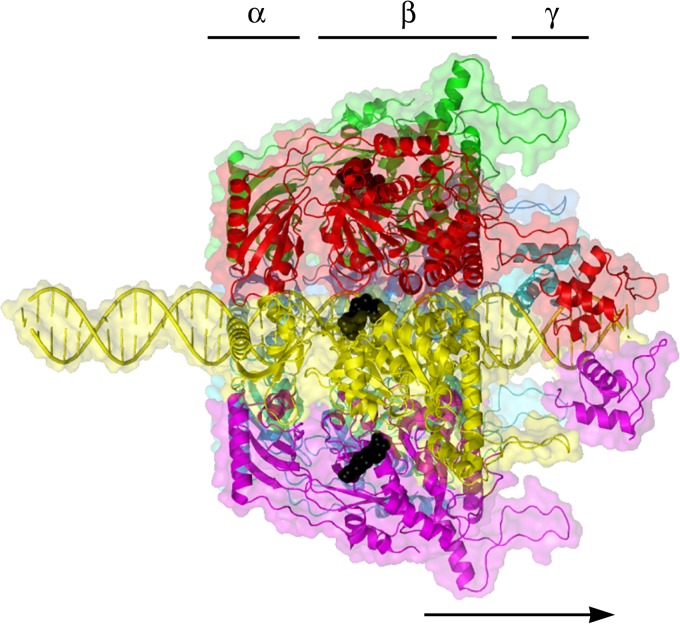
Packaging of a viral or bacteriophage genome into its capsid requires extensive translocation of the nucleic acid. The motor proteins employed by many DNA viruses and phages belong to the FtsK-HerA family of translocases (43, 44). DNA-packaging motors of dsDNA bacteriophages have been historically classified as rotation motors, while extensive investigations have revealed that none of the motor components rotate during active motor actions (Fig. 4) (3–5). Connector rotation has been excluded in both the phi29 motor, by using single-molecule force spectroscopy in combination with single-molecule fluorescence polarization spectroscopy (4), and the T4 motor, by tethering its connector in a packaging assay (3, 45) (Fig. 4). In addition, tethering of DNA ends to bead clusters showed active DNA translocation with no observation of the rotation of the beads (5, 7). DNA was found to twist by as little as 1.5 degrees per base pair translocated, confirming a nonrotation mechanism (6), since 1.5°/bp × 10.5 bp/helical turn = 15.7° is far below the 360° per complete helical turn. This puzzle of “rotation motors that do not rotate” was not solved until the breakthrough of the discovery of revolution motion in 2013 by Guo's group (12–16) (Fig. 5). Recent studies in bacteriophages suggested that the small twisting of the DNA is due to the conformational changes of DNA between the A form and the B form (46–48) and also to the conformational changes of the motor channel (49, 50). Such motors translocate DNA along the helix through unidirectional revolution, resulting in a thermodynamic edge over rotation motors involving dsDNA translocation.
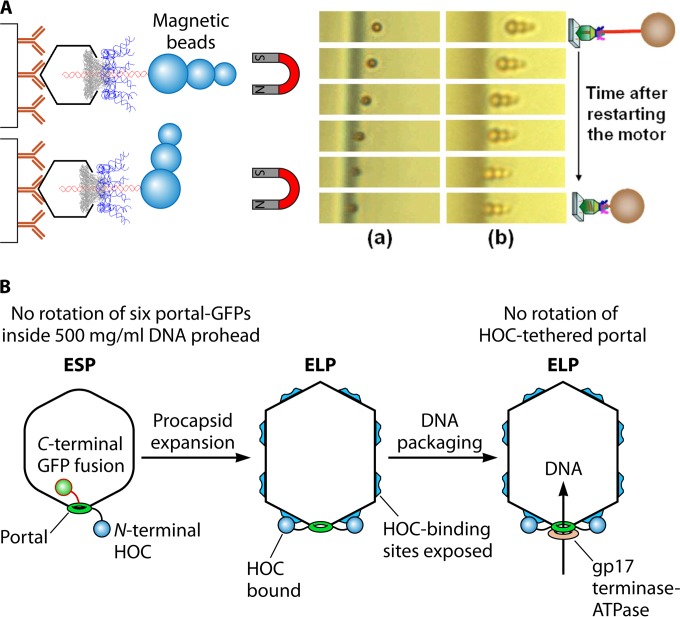
FIG 4.
Experiment with the phi29 and T4 motors revealing that neither connector nor dsDNA rotation is required for active DNA packaging. (A) Direct observation of DNA packaging horizontally using a dsDNA with its end linked to a cluster of magnetic beads for stretching the DNA. Panels a and b, real-time sequential images of DNA-magnetic bead complexes. (Adapted from reference 50 and reprinted from reference 5 with permission from AIP Publishing LLC.) (B) Experiment revealing that the T4 motor connector does not rotate during packaging. The packaging activity is not inhibited with the N terminus of the motor connector protein fused and tethered to its protease immune binding site on the capsid. GFP, green fluorescent protein; ESP, empty small particle; ELP, empty large particle; HOC, highly antigenic outer capsid protein. (Adapted from reference 16 and adapted from reference 3 with permission from John Wiley and Sons.)
FIG 5.
Schematic showing the sequential revolution motion in translocating dsDNA. (A) Revolution of dsDNA inside the ATPase hexameric ring. (Adapted from reference 12.) (B) Diagram of cryo-EM results showing offset of dsDNA in the channel of the bacteriophage T7 DNA-packaging motor. The dsDNA did not appear in the center of the channel; instead, the dsDNA tilted toward the wall of the motor channel. (Adapted from reference 131 with permission of the publisher.) (C) Revolution of dsDNA along the 12 subunits of the connector channel. (Adapted from reference 12.)
The revolution motion was first described in the well-studied phi29 dsDNA-packaging motor (12). Viral dsDNA-packaging motors consist of a protein portal channel and two packaging components for packaging the genome into the procapsid (Fig. 6). The phi29 motor contains a dodecameric connector channel (51, 52), a hexameric packaging RNA ring (7, 53–55), and an ATPase gp16 hexameric ring (56, 57) (Fig. 6). Many of the dsDNA viruses known so far use similar mechanisms during replication for their genome translocation into the procapsid (58–60). More-detailed descriptions and explanations about the motor structure characteristics and motion mechanisms are given in the sections below.
FIG 6.
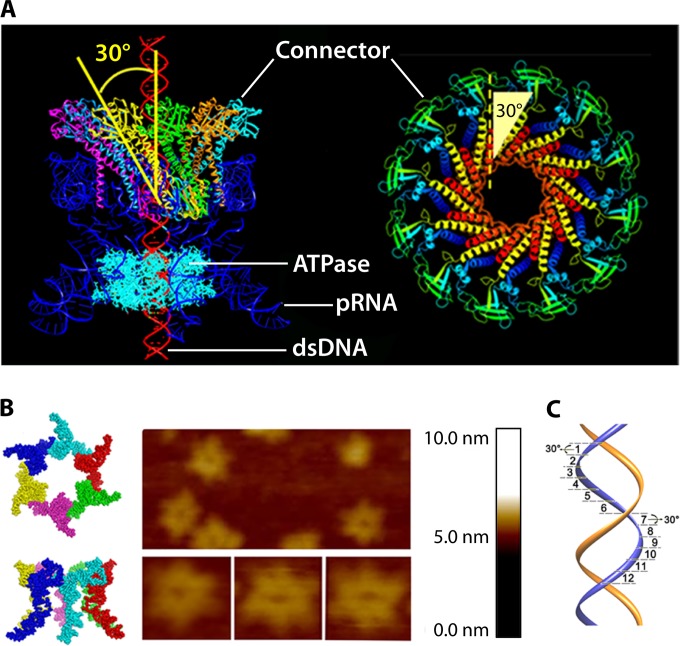
Depiction of the structure and function of the phi29 DNA-packaging motor. (A) Side view of phi29 dsDNA-packaging motor (left) and top view of phi29 connector (right). (B) Hexameric pRNA generated from crystal structures of its 3WJ core and AFM images of loop-extended hexameric pRNA. (C) DNA revolving inside the connector channel by contact with each connector subunit in a 30° transition step for each contact. (Adapted from references 13 and 56.)
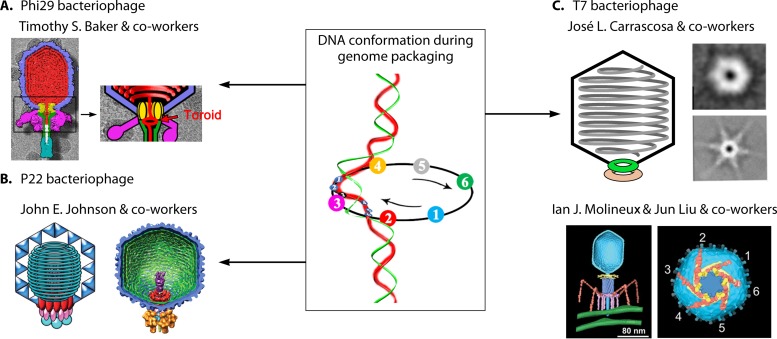
With inspiration from the revolution motion found in the phi29 dsDNA-packaging motor, studies in the other dsDNA motors were carried out, and it was found that the revolution mechanism is a common feature shared by dsDNA-packaging motors, as evidenced by the results from both crystal structure and biochemical studies. Crystal structure analysis of the motor channels of SPP1 (61), T7 (62), HK97 (63), P22 (64), and phi29 (52) revealed the existence of an antichiral arrangement between their channel subunits and the dsDNA helices. In addition, the packaged genome in many viruses has been found to spool free from rotation tangles inside the capsid (65–69). In phi29, the toroid of dsDNA has been shown by cryo-electron microscopy (cryo-EM) around the portal region (Fig. 7) (70–73), representing the accumulation of individual revolving DNAs processed by cryo-EM during revolution motion. A compression mechanism has also been found in the T4 DNA-packaging motor (47, 48), which agrees with the revolution mechanism. Overall, the lack of rotation of the DNA during packaging allows tight, ordered packaging of the DNA with little or no knotting or tangling, which as such does not impair subsequent DNA injection steps.
FIG 7.
Examples of spooling of DNA within capsids of phages to support the revolution mechanism. (A) Bacteriophage phi29. (Adapted from reference 71 with permission from Elsevier.) (B) Bacteriophage P22. (Adapted from reference 65 with permission from AAAS.) (C) Bacteriophage T7. (Adapted from reference 69 and adapted from reference 72 with permission from the American Society for Biochemistry and Molecular Biology.) The toroid formed at the phi29 portal position might be an accumulation of the images of the revolution motion during packaging, as shown in the image in the center. (Adapted from reference 15 with permission from Elsevier.)
dsDNA translocase FtsK/SpoIII E superfamily for bacterial chromosome segregation.
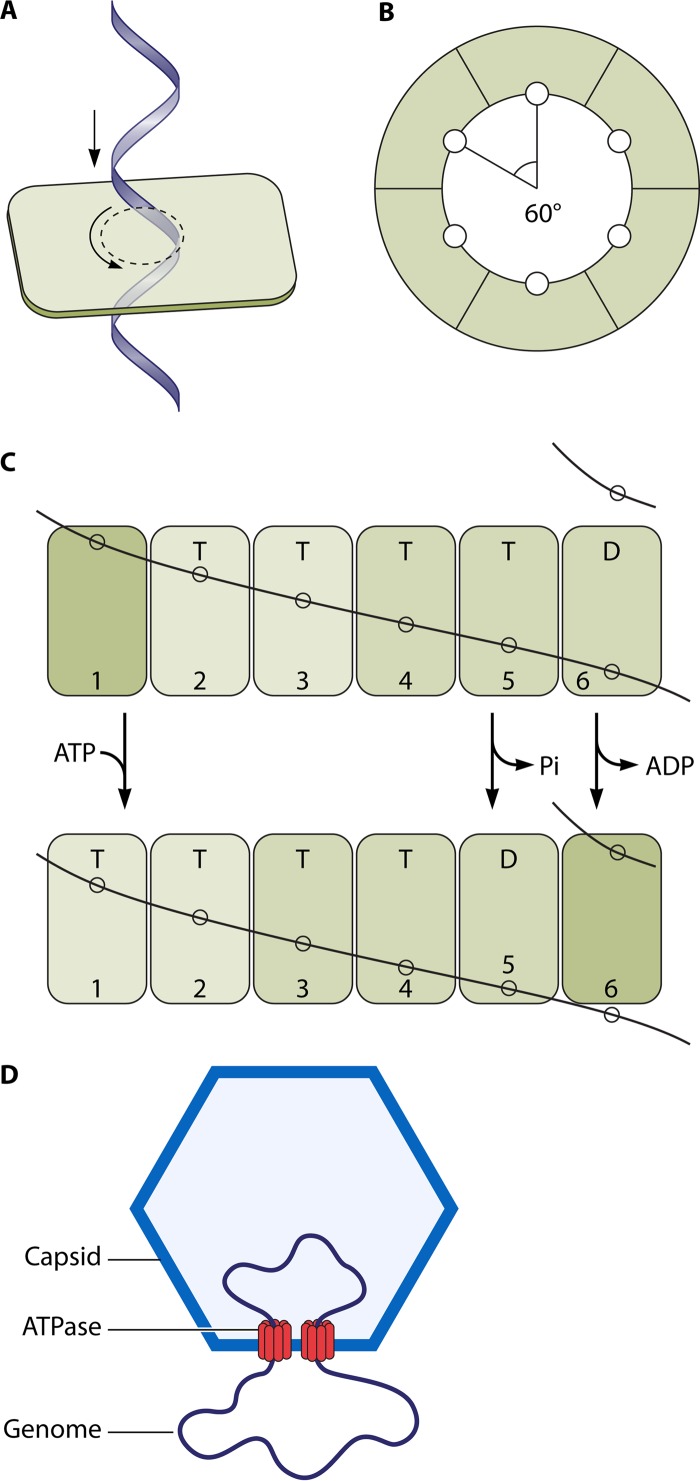
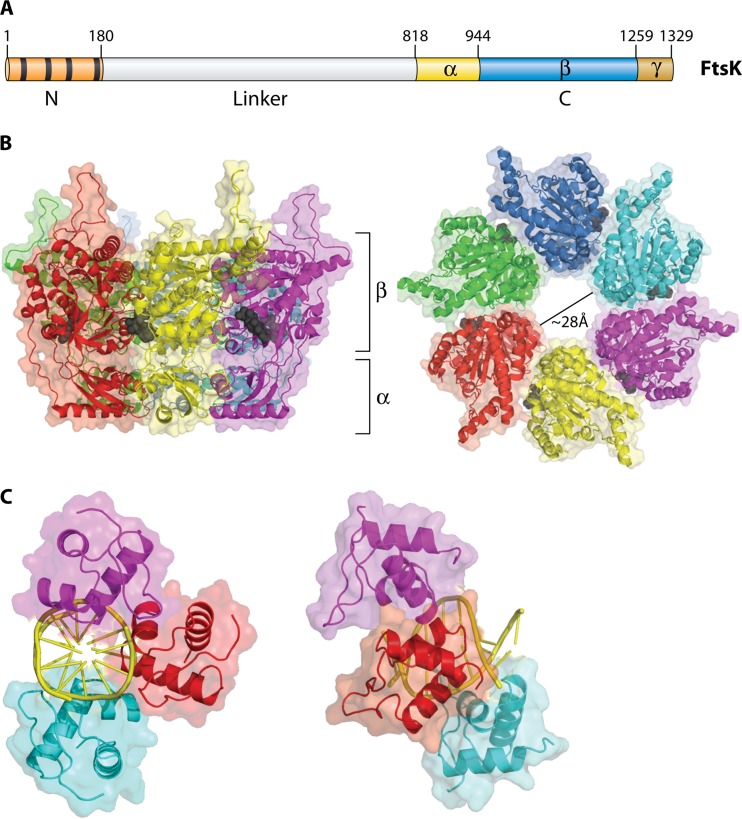
Members of the FtsK/SpoIIIE family of proteins are hexameric dsDNA translocases found in many bacterial species that also use a revolution mechanism for DNA translocation (Fig. 8). FtsK/SpoIIIE are part of the larger FtsK-HerA family, which is present throughout bacteria and archaea (43), and also contains the motor proteins of various conjugative plasmids and transposons. Based on a core with a RecA-like fold, this family has some added features that set it apart from other RecA-like proteins (43). FtsK is a dsDNA motor protein involved in the transportation of DNA and separation of intertwined chromosomes during cell division (74, 75). It functions to coordinate chromosome segregation, unlinking and recombining during cell division so that the closing division septum is free from DNA (76). It is one of the fastest and most powerful motors discovered to date, with a translocation speed of 17.5 kbp/s at 37°C (77) and a stalling force of 60 pN (78). The FtsK motor contains three functional components: one for DNA translocation, one for orienting the motor, and one for anchoring itself to the bacterial membrane (Fig. 9) (79). The DNA translocation motor is at the C terminus of FtsK and can be subdivided into three domains, α, β, and γ (80). While the α subdomain fold is unique to the FtsK family, the β subdomain, which contains residues for binding and hydrolyzing ATP, classifies FtsK as both a P-loop nucleoside triphosphatase (NTPase) and a hexameric translocase/helicase due to its RecA-like fold and sequence conservation common in these families (43). The third subdomain, γ, has two distinct roles: it acts as a protein-protein interaction domain to activate Xer-mediated recombination at dif (81) and also acts as a DNA binding domain to recognize and bind to specific 8-bp DNA sequences on the chromosome. FtsK-orienting polarized sequences (KOPS), with the sequence GGGNAGGG (82, 83), are highly skewed in their distribution so that on each chromosome arm the sequences are directed toward the terminus region, specifically switching orientation at the dif site. The role of these polarized sequences is to “determine” the directionality of translocation by acting as a recognition/preferential loading site for FtsK. Three γ domains bind to each 8-bp KOPS (Fig. 9C), leading to loading of an active hexameric motor to one side of the KOPS (84) (Fig. 10). This ensures that the motor is loaded correctly onto DNA in a specific orientation and that the subsequent translocation is toward the XerCD-dif site. Directional translocation of DNA is, therefore, sequence dependent at the motor-loading step; further, a translocating FtsK appears to ignore additional KOPS and reads through them (77, 84).
FIG 8.
Illustration showing how FtsK may undergo the revolution mechanism. (A) One strand of dsDNA contacts with the inner-channel wall of the hexameric ATPase. The continuous contact between DNA and ATPase does not require any rotation of the ATPase or DNA. (B) Each DNA contact is expected to be separated by 60° along the inner surface of the ATPase hexameric channel. (C) Sequential action of dsDNA translocation. DNA is shown as a line. T, ATP-bound; D, ADP-bound. (Adapted from reference 25 with permission of John Wiley and Sons [copyright 2010 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim].) (D) Speculation on the segregation and translocation of the mimivirus genome into the capsid via a revolution mechanism similar to that for FtsK. (Adapted from reference 15 with permission from Elsevier.)
FIG 9.
The FtsK motor protein. (A) E. coli FtsK protein domain structure. The N-terminal domain is in red, with each transmembrane domain represented by a black box. The numbers represent the FtsK amino acid numbers. The C terminus is subdivided into α, β, and γ domains. (B) Two views of the hexameric FtsK motor protein structure. On the left is a side on view emphasizing the α and β domains. Bound nucleotide (ADP) is shown in black in a space-filling model. On the right is a view down the symmetry axis, viewed from the β-domain side. (C) Structure of three γ domains bound to a KOPS DNA, seen along the DNA axis (left) and from the side (right).
FIG 10.
Model of the FtsK motor loaded at a KOPS site. The N termini of the γ domains are located on one side of the complex, where they would connect to the motor domains of FtsK. This leads to loading of the motor to one side of the KOPS site so that the motor is pointing in a defined direction. This gives the motor its subsequent directional translocation (the arrow denotes the direction the motor would move along the DNA).
SpoIIIE is an FtsK orthologue in B. subtilis that is vital for sporulation (85). Like FtsK, SpoIIIE shares a conserved C-terminal domain that harbors three subdomains, α, β, and γ (86, 87). The α and β subdomains translocate DNA through ATP binding and DNA-dependent hydrolysis (88), and the γ subdomain recognizes specific DNA sequences to guide DNA translocation (84, 89). These subdomains assemble into a hexameric ring that accommodates dsDNA in its central channel (80). During spore formation, an asymmetric cell division occurs, and a complete chromosome copy is pumped into the smaller spore compartment from a larger “mother cell” using the ATPase-driven motor of SpoIIIE (85, 90). Recent reports proposed that SpoIIIE motors also use a revolution mechanism for DNA translocation (50). Whether FtsK and SpoIIIE in fact respond to their respective directionality sequences differently or whether the two related proteins behave in a similar fashion remains to be studied.
DNA-packaging motors of large eukaryotic dsDNA viruses.
Comparative genomic studies of large eukaryotic dsDNA viruses, including mimivirus, vaccinia virus, and pandoravirus, showed remarkable similarity between their motor components and those of the FtsK-HerA superfamily (43, 44, 91), indicating that these viruses might also undergo revolution motion during genome packaging, allowing translocation of large genomes with minimal rotation of the DNA.
Linear Motors: Myosin, Kinesin, and Dynein
Myosin, kinesin, and dynein are linearly acting cytoskeletal motors. They are able to work in ensembles to generate large forces, such as during muscle contraction, or as single molecules to transport cargo along a specific cellular track. While myosins utilize actin filaments as tracks, kinesin and dynein are microtubule-based motors. The cytoskeletal motors are large protein families that are ubiquitously expressed in eukaryotic cells and are organized into classes based on their structure and function. Myosin motors are divided into 35 classes (92), while there are 14 classes of kinesin (93) and 9 classes of dynein (94). The “Mechanism of Linear Motors” section below focuses on the myosin mechanism as an example of linear motion, while the reader is referred to excellent reviews for the kinesin (95) and dynein (94) mechanisms.
Of the cytoskeletal linear motors, kinesin and myosin are more structurally related, as their nucleotide binding regions are similar to those of G proteins and other P-loop ATPases. The key features of the active site are the conserved switch I, switch II, and P loop (Walker A motif), which coordinate nucleotide binding and hydrolysis and also transmit key structural changes to the force-generating element of the motor (96) (Fig. 11). The members of the family of P-loop NTPases, G proteins, kinesins, and myosins are thought to have evolved from a common ancestor (97). Although the motor domains of the myosin and kinesin families are highly conserved, the N- and C-terminal domains are quite variable and allow dimerization, filament formation, protein-protein interactions, cargo binding, and light-chain binding.
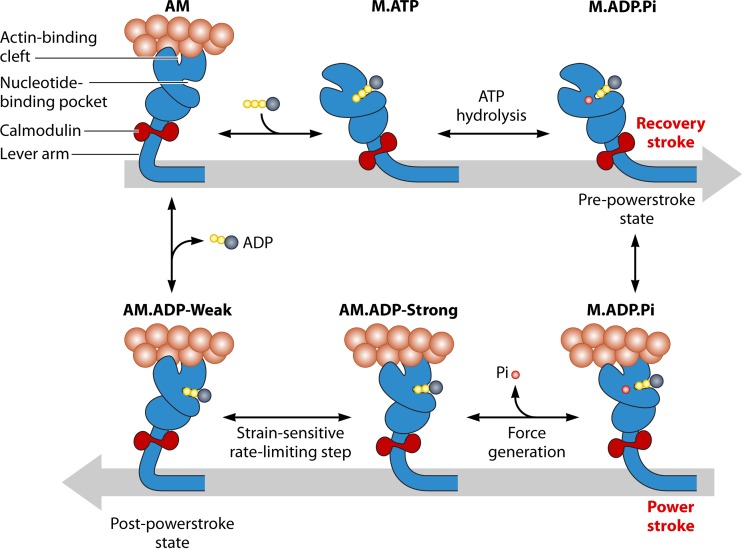
FIG 11.
Simplified schematic representation of the ATPase cycle of myosin, showing the proposed mechanism of how the motor is primed (recovery stroke) and generates force (power stroke) by coupling movement of the lever to key steps in the ATPase cycle. (Reprinted from reference 236 with permission.)
Dyneins are members of the AAA+ family of ATPases, and their structure consists of a ring of six AAA+ ATPase modules (94). The first AAA+ module is the key site of ATP hydrolysis, while sites 2 to 4 are also capable of hydrolyzing ATP, and sites 5 and 6 are not capable of hydrolyzing ATP. Dynein also contains a stalk and microtubule binding domain as well as a cargo binding tail that extends from the AAA+ ring.
STRUCTURE OF BIOMOTORS
Motor Structural Frame
Molecular motors exhibit wide variability in structure, while in general the structures consist of a mechanical frame containing both moving and static parts, along with an energy supply. In most molecular motors, the mechanical frame is formed by proteins, though the phi29 dsDNA-packaging motor also contains an RNA hexameric molecule as an essential component (53). The DNA or RNA polymerases are track-laying motors, and DNA and RNA can be considered components of these motors since they are required for the unwinding or polymerization process (98–101). The structure of the motor domain, which contains all the elements capable of converting chemical energy into mechanical work, is also highly conserved within various classes of motors (102, 103).
Channel, Pore, or Surrounding Ring
During cell segregation or binary fission, circular closed dsDNA can be translocated by the dsDNA translocases without breaking the covalently bonded backbones (79, 80, 90). In addition, concatemer dsDNA is the packaging substrate in many dsDNA viruses (104–106). This process can be finished by the dsDNA translocation motor, since the ATPase monomers can assemble into a hexamer on the DNA itself without a free 5′ or 3′ dsDNA end by utilizing the energy from ATP binding (50). This has been confirmed by electrophoretic mobility shift assay (EMSA), which showed the binding of ATPase onto single-ended or double-ended blocked DNA substrate (12), and by single-molecule fluorescent imaging and EM imaging, which showed that the first step in phi29 DNA packaging was the binding of multiple gp16s in a queue along the dsDNA, evidenced by the observation of a string of multiple Cy3-gp16 complexes on dsDNA chains in the presence of nonhydrolyzable ATPγS (12).
Stoichiometry of Motor Components
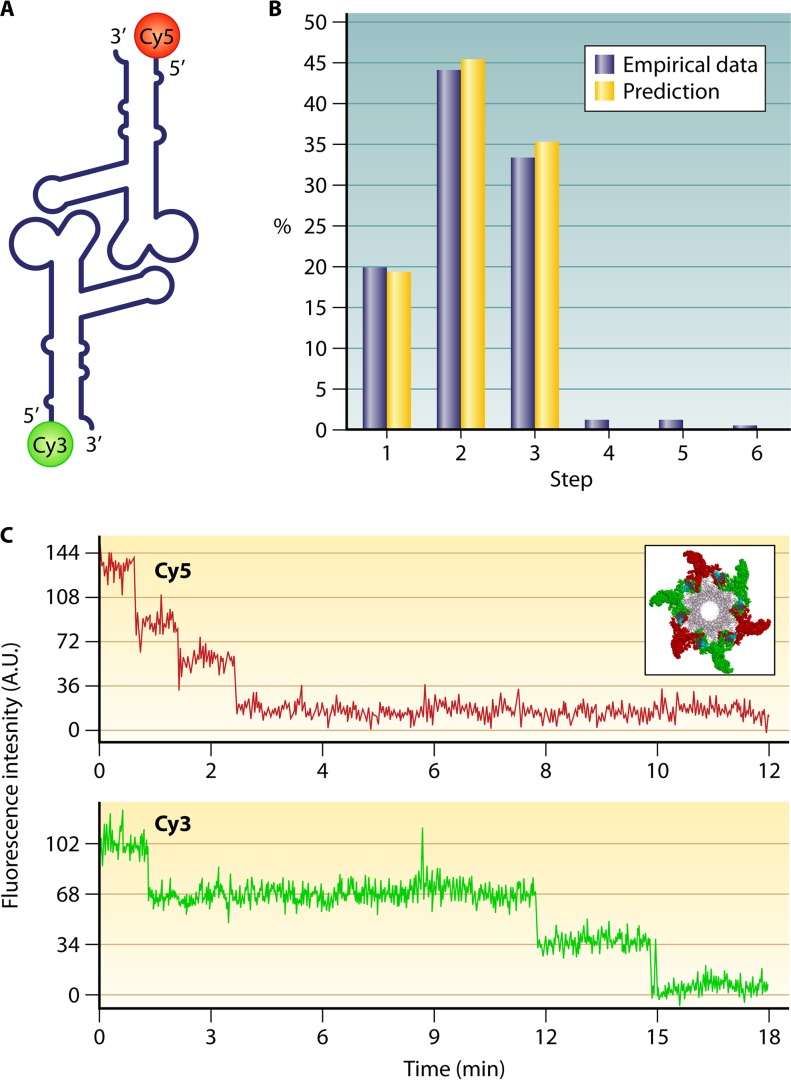
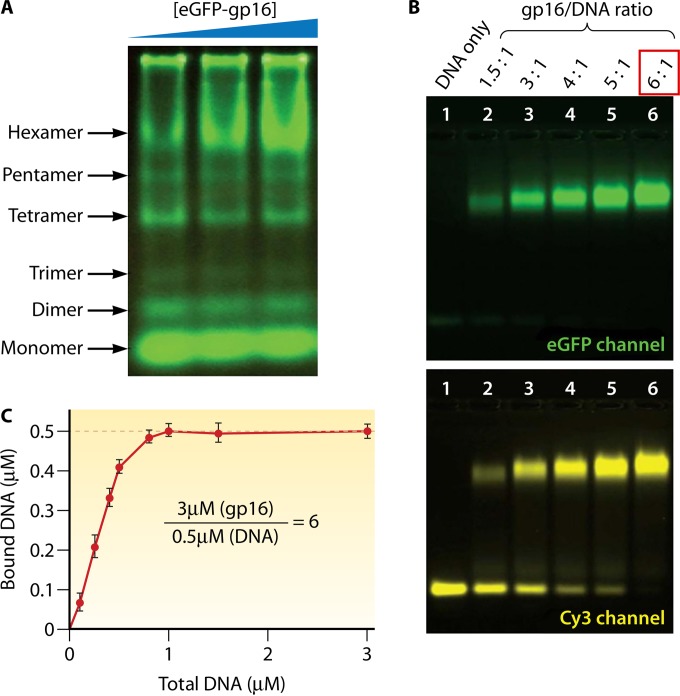
Structural studies of viruses in 1978 led to the definition of the popular 5-fold/6-fold mismatch mechanism (2), based on the findings that the viral icosahedron capsid is composed of many pentamers and hexamers (107) and that the DNA-packaging motor of dsDNA viruses resides within the pentameric vertex (108) with a dodecameric motor channel (62, 109–113). In 1998, the pRNA ring in the phi29 dsDNA-packaging motor was first determined to be a hexamer (54, 114, 115). Despite some researchers supporting a pentameric model (116–118), the hexameric pRNA formation was verified by cryo-electron microscopy (cryo-EM) (119), biochemical analysis (54, 114, 115), single-molecule photobleaching (7), gold labeling imaging by EM (120, 121), and RNA crystal structure studies (55). In fact, all six copies of pRNA were found to be retained on the active motor during packaging (7) (Fig. 12). Retention of six copies of pRNA upon packaging appears to dismiss the assumption that the motor components change from hexamers to pentamers (118, 122, 123). More recently, the ATPase gp16 of the phi29 dsDNA-packaging motor, which belongs to the ASCE superfamily, which has many hexameric members (124–126), has also been confirmed to be a hexamer in its final oligomeric state (Fig. 13) (56). Experimental evidence includes that from binomial distribution analysis, qualitative DNA binding assays, capillary electrophoresis (CE) assays, and EMSA (56, 127, 128). In addition, the description of biophysical studies identifying the four steps of motor motion (Fig. 14A) is related to the four relaying lysine layers embedded inside the phi29 connector channel inner wall (Fig. 14B and C) (see below) (13, 117, 129).
FIG 12.
Single-molecule photobleaching assay confirming the presence of six copies of phi29 motor pRNA. (A) pRNA dimer design constructed with Cy3- and Cy5-pRNA. (B) Comparison of empirical photobleaching steps with theoretical prediction of Cy3-pRNA in procapsids bound with dually labeled dimers. (C) Photobleaching steps for procapsids reconstituted with the dimer. (Adapted from reference 7 with permission of the publisher [copyright 2007 European Molecular Biology Organization].)
FIG 13.
Stoichiometric assays showing the formation of the phi29 ATPase hexamer. (A) A native gel reveals six oligomeric states of the ATPase; the hexamer formation increases as the concentration of protein is increased. (B) A slab gel showing the binding of ATPase to dsDNA in a 6:1 ratio, imaged in GFP (upper panel) and Cy3 (lower panel) channels for ATPase and dsDNA, respectively. (C) Quantification by varying the [ATPase]/[DNA] molar ratio. The concentration of bound DNA plateaus at a molar ratio of 6:1. (Adapted from reference 56.)
FIG 14.
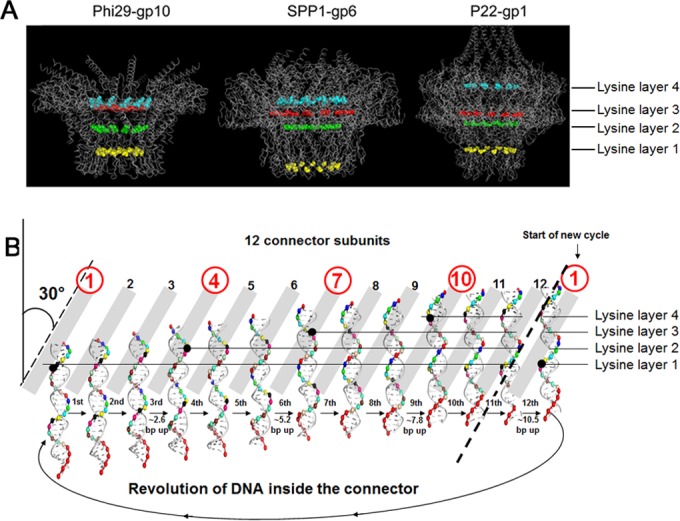
Four steps of pauses for each cycle during the packaging of phi29 dsDNA. (A) The presence of four lysine residues of motor channel protein leads to the formation of four positively charged rings within the negatively charged channels of different motors. (Adapted from reference 13 and adapted from reference 15 with permission from Elsevier.) (B) Diagram showing DNA revolution inside the phi29 connector channel with four steps of pauses due to the interaction of four positively charged lysine rings with the negatively charged dsDNA phosphate backbone. DNA revolution across the 12 connector channel subunits is shown. (Adapted from reference 15 with permission from Elsevier.) PDB codes: phi29 gp10, 1H5W; SPP1 gp6, 2JES; P22 gp1, 3LJ5.
Assembly of a hexamer appears to be crucial to the motor function, as demonstrated by the ATPase function of the FtsK (and SpoIIIE) DNA motors. The α and β domains hexamerize to form the DNA translocase motor, a ring-shaped multimer with a central channel through which the dsDNA substrate is threaded (80). Electron microscopy (EM) of FtsK(C) has revealed DNA-dependent hexamer formation and DNA passage through the hexameric ring. For the α and β domains, each forms a hexameric ring, and the two rings sit atop one another with an ∼10-Å cleft separating them and two strands connecting the rings (Fig. 9B).
Factors for Distinction of Revolution and Rotation Motors
Differentiation of the rotation motor from the revolution motor is challenging, especially as the dsDNA translocation motors involve both rotation and revolution motors. Below we introduce two simple ways to distinguish these two types of motors.
Chirality: left-handed for revolution motor and right-handed for rotation motor.
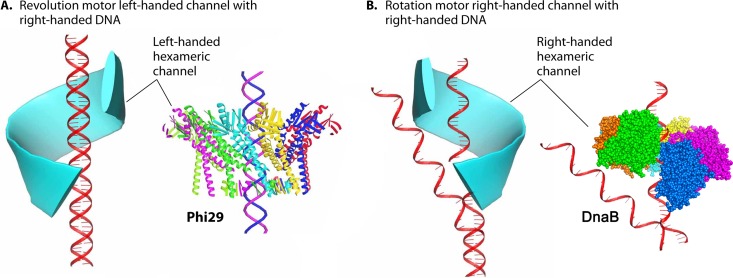
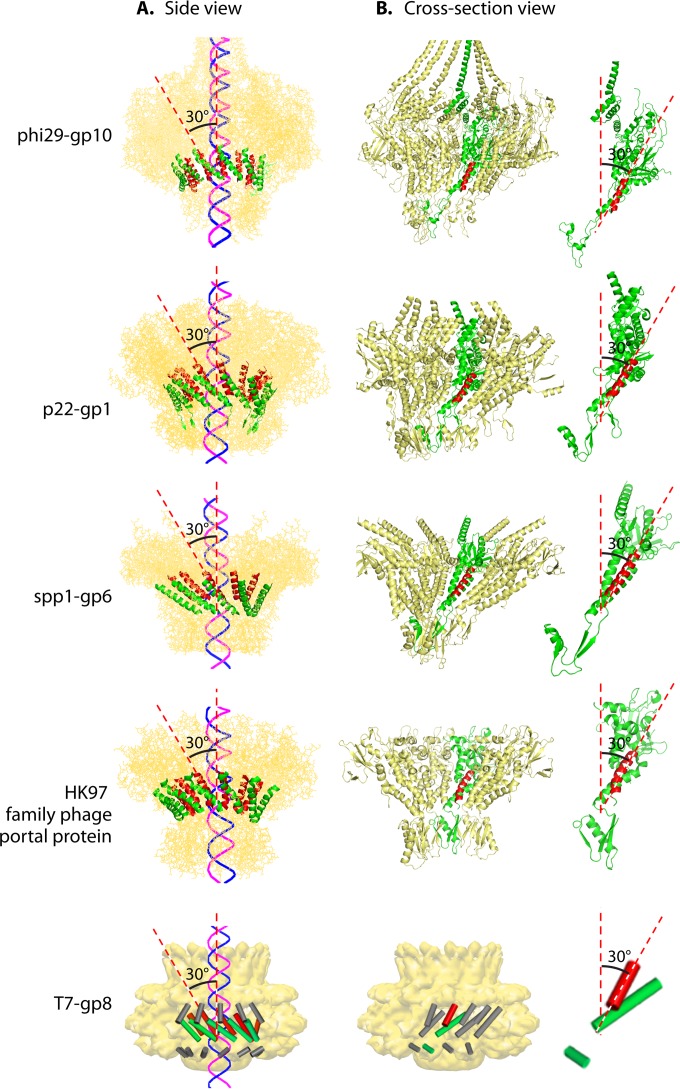
Chirality is one criterion to distinguish revolution motors from rotation motors. The motor channels (connectors) of SPP1 (61), T7 (62), HK97, P22 (64), and phi29 (52) all adopt an antichiral arrangement between the left-handed motor connector subunits and the right-handed DNA helices during packaging (Fig. 15) (12, 13). No apparent homology among these portal proteins has been shown by sequence alignment. However, they all share very similar three-dimensional structures, particularly a sequence of α-β-α-β-β-α stretches. The final α-helices of this structurally conserved stretch line the inside of the channel and are tilted at a 30° (left-handed) angle to the channel axis (Fig. 16). This remarkable conservation suggests a critical role of this conserved 30° antichiral arrangement in dsDNA packaging (50). The antichiral structure greatly facilitates the controlled one-way motion of dsDNA during packaging of the viral genome. As the DNA revolves around the inside of the channel, there is no rotation, coiling, or torsion force from the contact with any of the 12 connector subunits (12). On the other hand, rotation motors use right-handed channels for right-handed dsDNA translocation as a parallel thread.
FIG 15.
Use of channel chirality to distinguish revolution motors from rotation motors. (A) In revolution motors, the right-handed DNA revolves within a left-handed channel. (B) In rotation motors, the right-handed DNA rotates through a right-handed channel via the parallel thread, with DnaB shown as an example (130). (Adapted from reference 50.) PDB codes: phi29 gp10, 1H5W; DnaB, 4ESV.
FIG 16.
Quaternary structures showing the presence of the left-handed 30° tilting of the connector channels of different bacteriophages. External (A) and cross-sectional (B) views of the motor are shown, showing the antiparallel configuration between the left-handed connector subunits and the right-handed dsDNA helices. The 30° tilt of the helix (highlighted) relative to the vertical axis of the channel can be seen in a cross-sectional internal view of the connector channel and the view of its single subunit in panel B. (Adapted from reference 50 and adapted from reference 15 with permission from Elsevier.) PDB codes: phi29 gp10, 1H5W; HK97 family portal protein, 3KDR; SPP1 gp6, 2JES; P22 gp1, 3LJ5. T7 gp8 EM code: EMD-1231.
Channel size: larger than 3 nm for revolution motor and smaller than 2 nm for rotation motor.
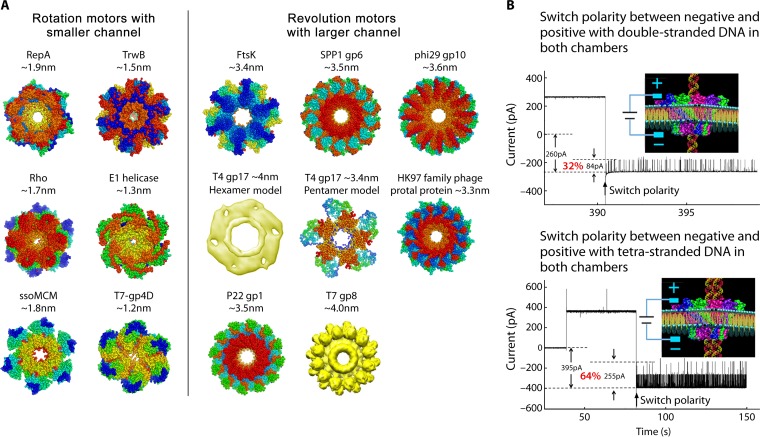
Hexameric rotation motors mostly process one of the nucleic acid strands that is threaded through the central channel of the motor, while the other is excluded during translocation (130). Rotation motors adopt a channel with close to full contact with nucleic acid during the translocation through the center of the channel, and as a result, the channel size has to be close to or smaller than the diameter of one strand of dsDNA, which is smaller than 2 nm. However, for revolution motors, both strands of the dsDNA advance within the channel through its contact with the channel wall from the side instead of proceeding through the center of the channel. This is in agreement with the cryo-EM images showing that the T7 dsDNA core tilts relative to its channel axis. A counterclockwise motion of the dsDNA in the T7 motor was observed when viewing from the N terminus of T7 connector (131) (Fig. 5B), in agreement with the direction of dsDNA revolution in the phi29 dsDNA-packaging motor (12, 13, 60, 132) (Fig. 5A). The revolution motor channels are thus larger than the dsDNA in order for there to be sufficient room for revolution. Crystal structures of different motors have confirmed the channel sizes, confirming that the diameters of the revolution motor channels are larger than 3 nm while those of the rotation motors are smaller than 2 nm (50). This appears to be true for the revolution motors of phi29, SPP1, T4, T7, HK97, and FtsK and rotation motors including Rho factor, E1 helicase, TrwB, ssoMCM, and RepA (50) (Fig. 17).
FIG 17.
Comparison of channel sizes of rotation and revolution motors. (A) Channel sizes of different biomotors that utilize the rotation mechanism (left panel) and the revolution mechanism (right panel). During DNA translocation, the rotation motors use smaller channels (<2 nm), while revolution motors use larger channels (>3 nm in diameter). (B) The larger size of revolution motors has also been proved by the single-pore conductance assay with the phi29 connector, showing the one-way traffic property of the channel with double-stranded or quadruple-stranded DNA. (Adapted from reference 50.) PDB codes: RepA, 1G8Y; TrwB, 1E9R; ssoMCM, 2VL6; Rho, 3ICE.; E1, 2GXA; T7 gp4D, 1E0J; FtsK, 2IUU; phi29 gp10, 1H5W; HK97 family portal protein, 3KDR; SPP1 gp6, 2JES; P22 gp1, 3LJ5. T7 gp8 EM ID: EMD-1231.
MOTION MECHANISM
Energy Conversion: Transition among Entropy, Randomness, Affinity, and Conformation Change as Driving Force
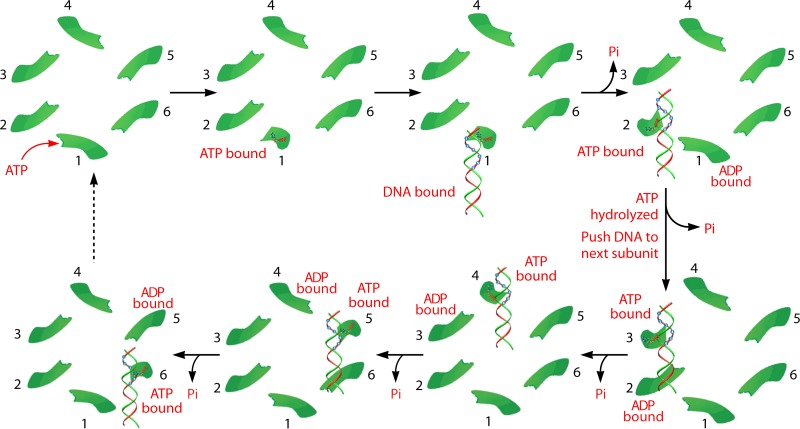
Several assays were used to study the ATP-ATPase interaction and elucidate how ATP energy translates to physical motion in phi29 DNA packaging. EMSA suggests that the ATPase undergoes conformational entropy changes upon ATP binding or ATP hydrolysis, leading to a high or low binding affinity of ATPase toward DNA, respectively (12, 56, 133). These changes in ATPase force the substrate DNA away from the ATPase and lead to physical motion of genomic DNA toward the second subunit, thereby moving the DNA toward the interior of the viral protein shell (Fig. 18). Six ATP molecules are used to complete one revolution cycle, with one ATP molecule to package 1.75 bp of dsDNA (10.5 bp/6ATP = 1.75 bp/ATP). Such conformational changes are abolished by site-directed mutation to the Walker A motif (56), which has been identified (57) and confirmed (12, 57, 134) to be responsible for ATP binding. Mutation to the gp16 Walker B motif, which is required for ATP hydrolysis, eliminates the catalytic force step (56). Thus, the conformational entropy change of the ATPase is a process that couples motion with the free energy changes and, most likely, an intrinsic property of the protein through evolution.
FIG 18.
Model of the sequential mechanism of the sequence action of the phi29 DNA-packaging motor. Binding of ATP to the conformationally disordered ATPase subunit stimulates an entropic and conformational change of the ATPase, thus fastening the ATPase at a less random configuration. This lower-entropy conformation enables the ATPase subunit to bind dsDNA and prime ATP hydrolysis. ATP hydrolysis triggers the second entropic and conformational change, which renders the ATPase low affinity for dsDNA, thus pushing the DNA to the next subunit that has already bound ATP. These sequential actions promote the movement and revolution of the dsDNA around the hexameric ATPase ring. (Adapted from reference 50 and adapted from reference 15 with permission from Elsevier.)
The P-loop lysine, catalytic glutamic acid, and arginine finger are well known to be critical to retain the catalytic power of ATPases. The P-loop lysine is the lysine in the phosphate binding loop (P loop), which is highly conserved among NTPases, with the general sequence GXXXXGKT/S. The glutamic acid, thought to be a general base for activating the hydrolysis of ATP, is also well conserved among ATPases and interacts with the ATP γ phosphate via a water molecule. The glutamic acid general base has been thought to activate the intervening water molecule to induce nucleophilic attack of the water molecule on the γ phosphate. Recent quantum chemical calculation and single-molecule analysis revealed that the role of general base may be not water activation but the enhancement of proton transfer from phosphoester to bulk solution (135). In some ATPases the glutamic acid also plays a role as a sensor for the ATP binding state and changes orientation drastically upon ATP binding (136). The arginine finger is highly conserved among G proteins, AAA proteins, and RecA-type ATPases. The arginine finger is located at an interface of nucleotide binding subunits; ATP binds the interface of two subunits, one of which possesses most of ATP binding residues while the other provides the arginine finger. The crystal structure of bovine mitochondrial F1 (MF1) with a chemical analogue of the γ phosphate suggests that the arginine finger stabilizes the transient state of the hydrolysis reaction, which was supported by biochemical (137), theoretical (138), and single-molecule (139) studies. The AAA+ family also contains another arginine, in a location structurally separate from the arginine finger, that acts to couple ATP binding and hydrolysis to conformational changes between subunits; this has been termed the sensor II motif (140, 141). This well-conserved core of catalytic residues both helps to hydrolyze nucleotides efficiently to provide power for the motor to work and allows the motor to couple ATP binding and hydrolysis to communication between subunits to allow either more complex motions or cooperative motion within multisubunit enzymes.
Mechanism of Rotation Motors
Step rotation of F1.
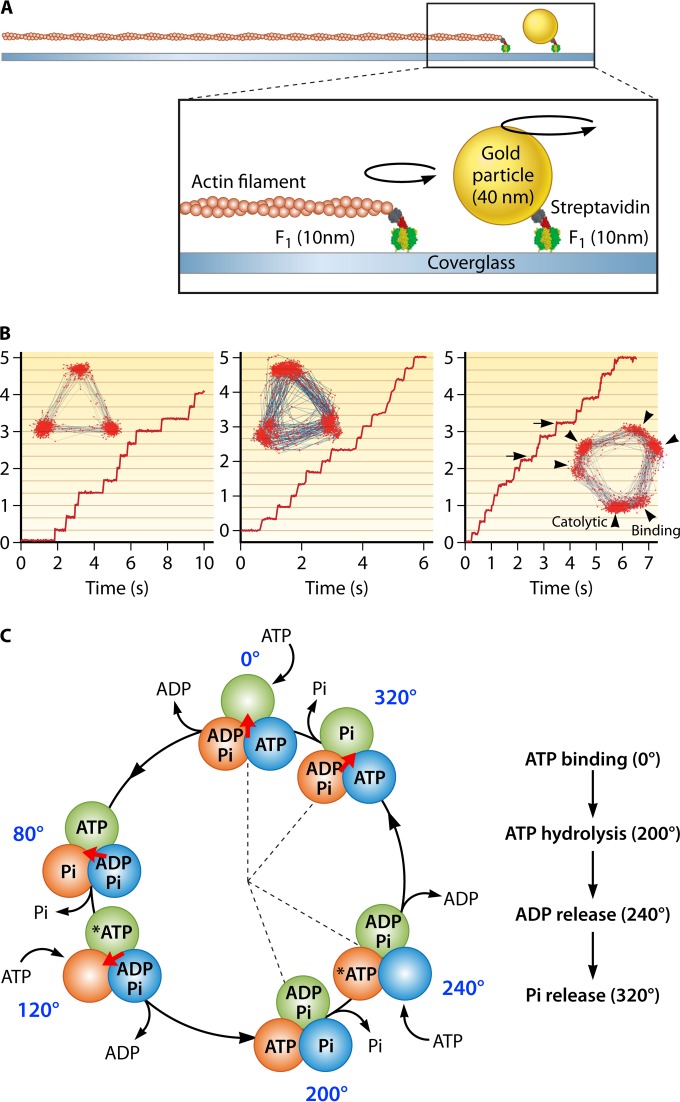
Rotation of the isolated F1 motor driven by ATP hydrolysis has been directly observed with an optical microscope (35, 142). Unidirectional rotation was first visualized for F1 from Bacillus sp. strain PS3 and has subsequently been observed in several kinds of rotary ATPases: the F1 proteins from Escherichia coli (143), spinach chloroplasts (143), and human mitochondria (144), as well as V-ATPases from Thermus thermophilus (145) and Enterococcus hirae (146). Among these, F1 from Bacillus PS3, termed TF1 (thermophilic F1), is the best characterized in terms of rotary dynamics. Therefore, the rotation features of F1 introduced here are based on the findings in the single-molecule rotation assay of TF1 unless otherwise mentioned. Consistent with the pseudo-3-fold symmetry of the α3β3 stator ring, F1 rotates γ in discrete 120° steps (147), each coupled with a single turnover of ATP hydrolysis (147, 148). An intermediate state has also been observed after ATP binding, with an 80° degree angle from the ATP waiting angle before hydrolysis (149). Thus, each 120° step can be resolved into 80° and 40° substeps (Fig. 19). A recent reaction scheme suggests that the 80° substep is driven by ATP binding and that the ADP release occurs on different β subunits. The 40° substep is initiated after hydrolysis of bound ATP, and Pi release also occurs on different β subunits (150) (Fig. 19C).
FIG 19.
Single-molecule rotation of F1. (A) Schematic image of the experimental setup. The α3β3 ring is fixed on the glass surface. A probe (fluorescently labeled actin filament or 40-nm colloidal gold) is attached to the γ subunit. (B) Left, rotation of F1 with 3 binding pauses separated 120°, which is caused by slow ATP binding at 200 nM. The inset shows the trajectory of the rotation. Center, rotation of a mutant F1 (βE190D) with 3 catalytic pauses at 2 mM ATP. Each pause is caused by the extremely slow ATP hydrolysis by the mutant. Right, rotation of mutant F1 (βE190D) at 2 μM ATP. Due to slow ATP binding and hydrolysis, 6 pauses are observed. The pauses before the 80° (arrowheads) and 40° (arrows) substeps correspond to binding and catalytic pauses, respectively. (C) Chemomechanical coupling scheme. Each circle indicates the chemical state of the catalytic sites. One catalytic site is highlighted in dark green. The central arrow (red) represents the angular position of the γ subunit. Each catalytic site retains the bound nucleotide as ATP until the γ subunit rotates 200° from the binding angle (0°). After a 200° rotation, the catalytic site executes the hydrolysis of ATP into ADP and Pi, each of which is released at 240° and 320°, respectively.
Single-molecule rotation assay of F1.
The dynamic behavior of ATP-driven rotation of F1 is well characterized in single-molecule rotation assay where the α3β3 stator ring is immobilized on a coverslip and the rotation probe is attached to the outwardly protruding part of the γ subunit of F1 (Fig. 19A). In earlier studies, a fluorescently labeled actin filament (0.5 to 5.0 m) was used (142, 147). In recent studies, nanoparticles with high scattering coefficients are often used, such as polystyrene beads (151), gold nanocolloids (151), and nanorods (152, 153), which enable imaging with high spatiotemporal resolution. In order to manipulate the rotation of F1, submicrometer-size magnetic beads are attached on the γ subunit as a rotation probe as well as a handle to control the orientation of the γ subunit with magnetic tweezers (148, 154, 155).
Conformational transition of the β subunit.
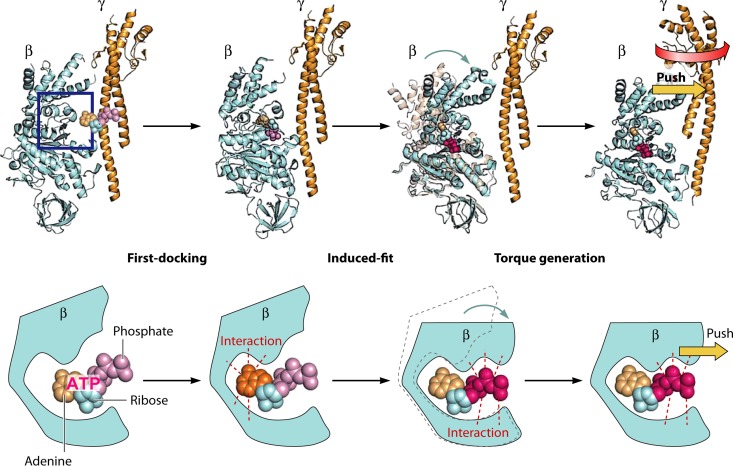
Many lines of experimental evidence showed that the catalytic β subunit undergoes a large conformational change (156–158). The clearest data came from the crystal structure of MF1, which showed that the β subunit changes conformational state upon nucleotide binding, rotating the C-terminal helical domain inwardly (156). This conformation is termed the closed form. On the other hand, the nucleotide-free β subunit assumes an open conformational state. When nucleotide-free β subunit undergoes the open-to-closed conformational transition upon nucleotide binding, it pushes the γ subunit. Therefore, of the steps of ATP binding, hydrolysis, and release of ADP and Pi, it is in fact ATP binding that is thought to be the major torque-generating step. Indeed, a nuclear magnetic resonance (NMR) study (157), single-fluorophore imaging (158), and recent high-speed atomic force microscopy (AFM) (159) confirmed that the β subunit does undergo the open-to-closed conformational transition upon ATP binding, as expected from the crystal structure of MF1.
It was proposed that hydrogen bond formation between the phosphate moiety of bound ATP and the catalytic residues is the main driving force of the induced-fit-type conformational change: the open-to-closed transition of the β subunit. The accompanying swing motion of the C-terminal domain of the β subunit would push the γ subunit to induce the rotation (Fig. 20).
FIG 20.
The open-to-closed transition of the β subunit of MF1. The accompanying swing motion of the C-terminal domain of the β subunit would push the γ subunit to induce the rotation.
PMF-driven rotation of Fo.
The most straightforward approach to address the dynamics of c-ring rotation is imaging of c-ring rotation under proton motive force (PMF)-driven conditions. Pioneering work on this was done by Börsch's group (160), where they employed single-molecule Förster resonance energy transfer (FRET) measurement to visualize c-ring rotation (161). Donor and acceptor fluorescent dyes were introduced to the a subunit and one of the c subunits that the stator and rotor subunits of EFoF1 reconstituted into liposomes. Under ATP synthesis conditions, the FRET signal showed multiple states that were attributed to pauses of the c ring every 36°, relating to the 10-fold symmetry of the c ring from Escherichia coli. Following on from the work of Börsch's group, a single-molecule rotation assay of FoF1 under ATP synthesis conditions was reported (162), where EFoF1 molecules were reconstituted into a supported membrane and PMF was charged by uncaging the caged proton between a coverslip and a supported membrane. In this experiment, EFoF1 molecules showed clear 120° steps, suggesting that the kinetic bottleneck was a catalytic event on F1. These experiments still have a critical experimental drawback in that the electrochemical potential is transient and not stable, which prevents elaborated analysis. Thus, the single-molecule analysis of the rotary dynamics of Fo still requires technical advancement. One remarkable technical achievement is the arrayed lipid bilayer chamber (ALBiC) system, which displays a million femtoliter chambers, each sealed with a lipid bilayer patch (163). This allows single-molecule measurement of transport activity of membrane proteins such as alpha-hemolysin and ATP synthase. One advantage of ALBiC system is hermetic sealing of chambers with lipid bilayers. Such new membrane technology would pave a way to the single-molecule study of rotary ATPase as well as other membrane protein machines by combination with single-molecule imaging techniques.
Rotation of helicases.
Several mechanisms of helicase translocation along nucleic acid lattices have been proposed. Currently the most popular models are the “inchworm stepping mechanism” and the “Brownian motor mechanism” (164, 165). Translocation begins with one binding site tightly bound to the substrate while the other one is bound weakly to it. “Inching forward” of the helicase is coupled with the NTP binding and hydrolysis cycle. Binding and coordination of the NTP-metal ion complex results in a large conformational change, which closes the cleft between the two nucleic acid binding sites. NTP hydrolysis and subsequent release of the NDP and inorganic phosphate are followed by the reversal of the conformational change, which results in the relative movement of the domain (or subunit) containing leading nucleic acid binding sites. One cycle of action is completed in six conformational changes. The Brownian mechanism requires only one nucleic acid binding site on the helicase, while two distinct conformational states, with high or low affinities toward its substrate depending on the different NTP ligation states, were proposed. Nucleic acids are translocated by the combination of Brownian motion and power stroke.
Mechanism of Revolution Motors
One-way traffic of dsDNA-packaging motor.
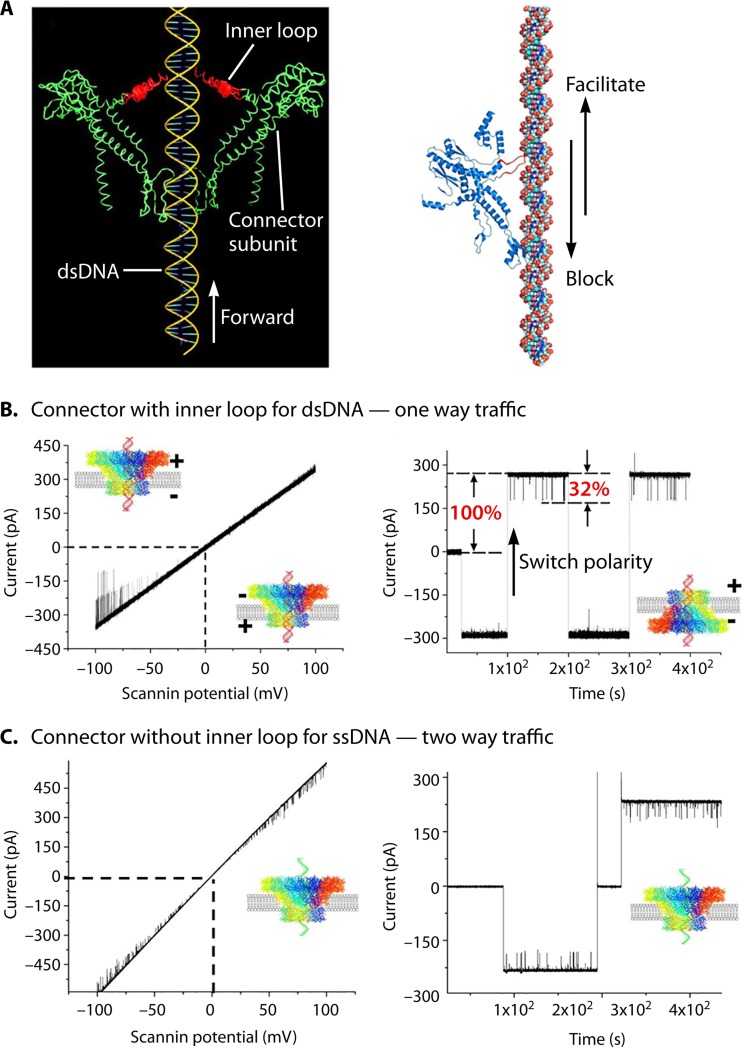
The special structure of the dsDNA-packaging motor generates several factors that coordinate a one-way traffic mechanism. As mentioned in the previous sections, first, ATP binding induces an entropic and conformational change of the ATPase giving an affinity for binding dsDNA. ATP hydrolysis induces a second entropic and conformational change resulting in a low affinity for dsDNA, thus pushing dsDNA toward the next subunit of the ATP hexamer. Second, the 30° left-handed twist of the channel wall produces an antiparallel arrangement between the channel wall and the right-handed helix of the dsDNA, facilitating the unidirectional translocation. Third, the one-orientation flow loops within the channel found in SPP1, phi29, and T4 generate a vector force for the one-way trafficking of dsDNA (Fig. 21); a mutant phi29 connector with a deletion of the internal loop N229-N246 failed to produce any virions (49, 166–168). The single-channel conductance assay has shown a one-way property for the wild-type connector (13, 132) and a two-way property for the loop-deleted connector (Fig. 21) (13, 169). These results suggest that the channel loops act as a ratchet during DNA translocation to prevent the DNA from leaving, in line with the “push-through one-way valve” model (13, 60, 132, 166). Analysis of the crystal structure and cryo-EM density map of SPP1 channel loops reveals that these loops are located in close proximity to dsDNA via nonionic interactions. The channel loops of phi29 and SPP1 might play similar roles in directional traffic of dsDNA, leading to entrance into the capsid through the connector channel (170).
FIG 21.
Role of the flexible inner channel loop of phage portal proteins in DNA one-way traffic. (A) Flexible loops within the phi29 (left) and SPP1 (right) connector channels function to interact with dsDNA, facilitating DNA to move forward but blocking reversal of DNA during DNA packaging. (B) Demonstration of one-way traffic of dsDNA through wild-type connectors using a ramping potential or a switching polarity (right). (C) ssDNA is translocated via two-way traffic with a loop-deleted connector. (Adapted from reference 13.)
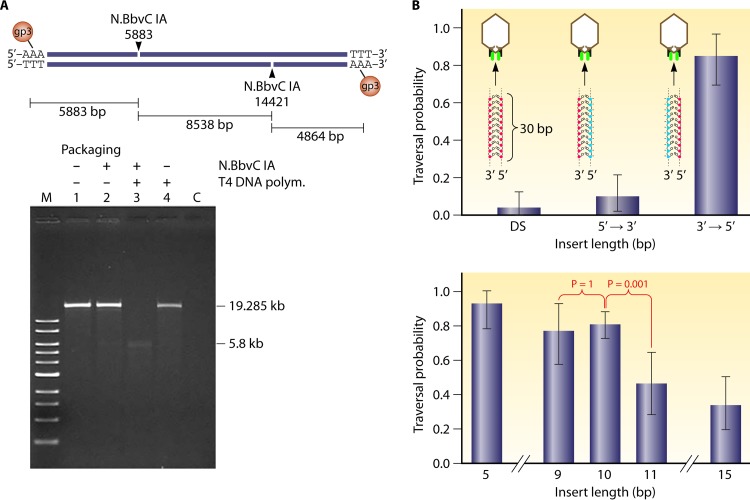
The 5′-to-3′ mode of revolution of one DNA strand along the channel wall to generate the vector force agrees with published data from T4 and phi29. During motor packaging, dsDNA is processed by contacting the connector with one strand of DNA in the 5′-to-3′ direction (171), and modification of the phi29 genome DNA in the 5′-to-3′ direction strand was found to stop dsDNA packaging (172) (Fig. 22). Extensions up to around 10 bases at the end of the DNA can be tolerated; however, extensions to 20 or more bases significantly blocked the DNA packaging of the phi29 or the T4 motor (Fig. 22) (172, 173).
FIG 22.
Effect of DNA chemistry and structure on its packaging of various bacteriophage dsDNA-packaging motors. (A) Design (upper panel) and results (lower panel) demonstrating the blockage of dsDNA packaging by single-stranded gaps. When a single-stranded gap is present, only the left-end fragment of phi29 genomic DNA is packaged. (Adapted from reference 171 with permission from Elsevier.) (B) Chemical modification of the negatively charged phosphate backbone on DNA packaging. Modification on the 3′→5′ strand does not block dsDNA packaging, but alteration on the other direction seriously affects DNA packaging, evidenced by its traversal probability. The insertion of the modified DNA with up to 10 bp can be tolerated, while further increasing the length will result in a 2-fold reduction of the traversal probability. The results support the finding of the revolution mechanism, showing that only one strand of the dsDNA interacts with the motor channel during revolution. (Adapted from reference 172 with permission from Macmillan Publishers Ltd.)
Additionally, the electropositive lysine layers present in many phage channels interact with one of the strands of the electronegative dsDNA phosphate backbone, resulting in a relaying contact that facilitates one-way motion and the generation of steps of transitional pausing during dsDNA translocation (12, 13, 116). The negatively charged phi29 connector interior surface is decorated with 48 lysine residues, resulting in the formation of four positively charged 12-lysine rings, as revealed by connector crystal analysis (52) (Fig. 14), which results in uneven speed alternations during the DNA translocation process, with four pauses (12, 13, 116), as previously reported for both phi29 (116, 129) and T4 (174). The lysine layers are nonessential for DNA packaging, while the interactions between lysine and the DNA backbone are involved in motor action. Similar patterns of four relaying electropositive lysine layers have been found in the SPP1, P22, and phi29 phages (52, 123). The effects of the lysine layers on genome translocation can be interpreted from the structural aspects. Taking the phi29 dsDNA-packaging motor as an example, its four lysine layers fall vertically within a 3.7-nm range (52) inside the 7-nm connector channel, spaced on average ∼0.9 nm apart. For B-type dsDNA, ∼2.6 bp will advance through each rise between two lysine layers (0.9 nm/0.34 nm · bp−1 = ∼2.6 bp). For every cycle of the DNA revolution (360°, 10.5 bp) through the channel of 12 subunits, a 0.875 mismatch occurs between the negative DNA phosphate base and the channel subunits with the positive lysine layer (10.5/12 = 0.875). To compensate for the distance variation due to this mismatch, the dsDNA phosphate backbone will interact with the positively charged lysine in the next subunit, leading to a slight pause in DNA advancement (Fig. 14). The continuation of the interactions between lysine layers and DNA backbones leads to the four pauses during packaging.
dsDNA translocases of the FtsK/SpoIIIE superfamily.
The crystal structure of the hexameric FtsK motor domain from Pseudomonas aeruginosa revealed a 6-fold symmetric ring with ADP bound in the active site of every subunit (80). There was very little structural information alone that could explain how the protein could convert the chemical energy from ATP hydrolysis into movement of the DNA substrate. However, the same study also presented a different crystal form of the motor domain, a monomeric form with ATP bound. A comparison of the two structures showed that when the β domains were superimposed, the α domains were shifted relative to each other in a hinge-like opening of α from β. The point in the α domain juxtaposed to the DNA substrate was able to move by 5.5 Å by this hinge-like mechanism (equivalent to 1.6 bp). If this movement were correlated to ATP hydrolysis, it would provide a mechanism by which the protein could pump DNA. The translocation of dsDNA with 1.6 bp per subunit of FtsK (25, 80) agrees well with the quantification for the phi29 DNA-packaging motor that each ATPase subunit uses one ATP to package 1.75 nucleotides (12, 13, 16, 56, 57) and for the bacteriophage T3 system with 1.8 bp per ATP (175). Based on these data, an inchworm mechanism has been proposed for FtsK (25, 80), with ATPase subunits acting in a sequential manner. In the model proposed by Massey et al. (80), for each ATP hydrolyzed, the DNA moves 2 bp (∼6.8 Å, slightly larger than the 5.5-Å difference between the two crystal forms) in an inchworm-like movement with contacts between both the α and β domains and the central DNA. The relative strength of the interaction between each domain and the DNA is dependent upon the ATP binding or hydrolysis state; upon hydrolysis of ATP, one DNA contact is lost while the other contact is strong but is shifted by ∼6.8 Å, resulting in net movement of DNA along the central channel of 2 bp. An integer number of bases was chosen, as it allows each monomer in the ring to contact the dsDNA in the same manner at every subunit, whether this is contact with the repeating sugar-phosphate backbone or with the bases themselves. Movement by a noninteger number of bases would mean that the protein-DNA contact necessary for movement of the substrate would be different in any two adjacent subunits. The movement of dsDNA by one subunit also functions to bring the next FtsK monomer in the ring to register with the DNA, and as such it hands the DNA substrate on to the adjacent monomer with minimal rotation (Fig. 8B). This second subunit then also translocates 2 bp of DNA upon hydrolyzing ATP, passing the DNA onto the next monomer, and so on around the ring. The result is 12 bp of DNA translocated per catalytic cycle of the hexamer if all 6 monomers hydrolyze ATP once. This figure is close to the 10.5 bp per helical turn in B-form DNA, with the extra 1.5 bp being manifested as a slight twisting. If the protein is anchored (both FtsK and SpoIIIE are membrane bound at their N termini), then this results in generation of DNA supercoiling, positive ahead of the motor and negative behind. Indeed, induction of supercoiling has been seen in bulk biochemical assays (176) and in single-molecule experiments (177). The observed supercoiling induction for FtsK translocation of one positive supercoil ahead of the motor per 150 bp translocated is in broad agreement with the proposed rotary inchworm model.
The inchworm model also proposed an obligatory handoff event between adjacent monomers within a single ring, such that the presence of a single catalytically inactive subunit would effectively inactivate the entire hexamer (80). This was supported by biochemical data. Mutants which were unable to bind ATP were mixed in different ratios with wild-type subunits, and the relative ATPase activity was compared to that of hexamers consisting of only wild-type subunits. With increasing proportions of mutant subunits, the ATPase activity decreased rapidly, following the predicted pattern for poisoning of the hexamer by an inactive subunit. This agrees with the finding that incorporation of one inactive subunit of the phage phi29 DNA-packaging motor completely blocks the function of the ATPase ring (12, 13, 15, 56).
Interestingly, a fusion protein in which three motor domains were joined to each other in a single polypeptide, a covalent trimer of FtsK, has been produced (178). This construct was found to be a very active DNA translocase motor. Within this trimeric construct, the Walker A and Walker B motifs, for nucleotide binding and hydrolysis, respectively, were selectively mutated. This design led to the surprising finding that a single active-site mutant or two nonadjacent mutants per hexamer did not cause a great decrease in ATPase activity and did not significantly decrease the speed of translocation along dsDNA (178). However, the presence of these mutations did reduce the ability of the hexamer to produce force as judged by the ability of the protein to displace either protein or DNA triplexes. It is important to consider that when the ATPase subunits were fused into a concatemer, there might have been unknown and unintended alterations either to the ATPase activity of individual monomers or to their ability to form higher-order multimers, which could mask the effect of mutant subunits. Conversely, a number of single-molecule studies have now shown that the linked trimeric FtsK proteins appear to translocate and respond to nucleotides similarly to unlinked hexamers (77, 179). Nevertheless, in order to explain these results, a new model in which more than one subunit within a hexameric ring would contact DNA concurrently was proposed. This model was based upon the escort or “spiral staircase” model of Rho and E1 helicases (27, 180). In these hexameric helicases, multiple subunits can contact the DNA/RNA substrate at the same time, with the single contact point for each monomer being at a different level around the ring, rather like the stairs in a spiral staircase. ATP hydrolysis moves one of the contact points downwards through the ring forcefully, and the other contacts move along passively. When the last contact point at the bottom of the ring is reached, the protein arm becomes free and can then move back up the top to reengage with the polynucleotide substrate and begin the cycle of movement down the staircase again. With a flexible and compressible single-stranded DNA/RNA substrate, the movement of the protein-DNA contacts is small enough that the protein can maintain with the DNA/RNA through a full catalytic cycle of every subunit in the ring. However, dsDNA is a much stiffer and noncompressible substrate than single-stranded DNA (ssDNA) or RNA, and as such a DNA translocase would have immense trouble utilizing an identical mechanism; the stiffer dsDNA substrate in a DNA translocase channel would mean that each single protein contact would have to move almost 30 Å to maintain contact with the DNA during a full catalytic cycle around the hexameric ring. This amount of movement of a contact point within a tight hexameric structure seems unlikely. Further, it would be energetically unfeasible to compress the double helix greatly yet still produce power from ATP hydrolysis. Therefore, an intermediate or “limited-escort” model was proposed for FtsK, whereby 3 adjacent monomers in a ring contact dsDNA simultaneously. This could allow for one inactive subunit to be skipped if the monomers on either side of it are proficient for translocation (25, 178). It would also imply that two adjacent mutant subunits would produce an inactive hexamer (178).
Both the rotary inchworm model and the partial-escort model are consistent with a mechanism that is largely that of a revolution motor. Both models propose that dsDNA touches the internal surface of the hexameric ring and that the contact point between the protein and the DNA revolves around the inner surface of the protein multimer with minimal rotation (12, 25, 84). Both models proposed that a defined number of bases would be moved per ATP hydrolyzed if each subunit contacts the DNA substrate in an identical fashion around the ring and the DNA must be moved by a defined length at each step to maintain these identical interactions. A slight twisting of the DNA at each step is then necessary to maintain the identity of the DNA and protein contacts around the ring; the angle between adjacent subunit active sites in a hexamer would be 360°/6 or 60°, whereas the angle between adjacent phosphates, around the dsDNA axis, would be 360°/10.5 (about 34°). If precisely 2 bp is translocated per protein subunit, then there is a requirement to twist the DNA an extra 8° to maintain the identity of each protein-DNA contact. DNA within cells is negatively supercoiled, with a supercoiling density in E. coli of around −0.05. If the in vivo negative supercoiling is accounted for, the amount of twisting required at each FtsK power step is reduced to around 5° per 2 bp translocated, which corresponds to one supercoil induced for every 144 bp translocated. This theoretical value is almost identical to the value of one supercoil per 150 bp observed using single-molecule experiments (177). If this is the case, a translocating FtsK will produce a small amount of positive supercoiling ahead of the protein. In the cell this might be removed either by the action of DNA gyrase or by the occasional slipping of the motor to release the torsional tension in the DNA.
With the close fit of the DNA into the central channel of FtsK, movement of the DNA relative to the protein would not require large amounts of rotational movement away from the central axis of the DNA. The point(s) of contact between FtsK and DNA and the concomitant wave of ATP binding and hydrolysis would revolve in a counterclockwise manner as viewed from the DNA entry side of the hexamer.
Complicated motors with multiple functional modules.
In cells, many motors, such as those involved in homologous recombination, DNA repair, Holliday junction resolution, or nuclear membrane-embedded pores or other membrane transporters, are composed of multiple functional modules present either as hexamers or as other oligomers. For example, the Holliday junction branch migration RuvB complex is made up of two hexamers that sit at two vertices of a RuvA tetramer (181, 182). Continuous translocation of the two dsDNA helixes with associated rotation has been proposed (183). Although the hexameric ATPase might participate in this motion process, the involvement of more than one functional module makes the classification of the motor as either rotation or revolution challenging. Chromosome or genomic DNA either in eukaryotic cells or in bacteria, as a very long string, would cause coiling or tangling if rotation of such a long double helix occurred, and resolution of the coiling requires the consumption of energy. Thus, it is speculated that some of the functional complexes within these complicated motors might belong to the revolution motor class, since the use of the revolution mechanism will avoid dsDNA coiling and tangling, as described in the above sections.
Mechanism of Linear Motors
Linear motors are highly efficient, with estimates that 60 to 70% of the energy from ATP hydrolysis is utilized for mechanical motion (183–185). In myosin, the actin filament is proposed to accelerate the structural changes in the force-generating element, which couples the mechanical and chemical cycles. The lever arm consists of the light-chain binding region. Variability in the length of the light-chain binding region in different myosin isoforms helped to prove the hypothesis that this region functions as a lever arm (189). In kinesin, the major difference is that the ATP binding step is associated with force generation, while the hydrolysis step occurs with kinesin bound to the microtubule (95). The neck linker or coiled-coil stalk has been demonstrated to be critical for movement. In dynein, the coiled coil of the stalk which connects the AAA ring and the microtubule binding domain is known to change conformation in a nucleotide-dependent manner and function as a force-generating element (94).
Conserved catalytic cycle of myosins.
The modified Lymn-Taylor cycle provides the minimal framework for explaining the conserved properties of the actomyosin ATPase cycle (193). The motor domain of myosin contains the sites for ATP binding (nucleotide binding pocket [NBP]) and actin binding (actin binding cleft) (17). These sites are coordinated with the reciprocal movement of the lever arm region during the recovery and power stroke states of the ATPase cycle (Fig. 11). In the absence of any nucleotide, myosin binds to actin tightly and forms a strongly bound complex. ATP binding to myosin causes the cross-bridge to detach from actin and enter the weak binding states. During the detached states, ATP is hydrolyzed by myosin, and the lever arm region of myosin primes itself into a pre-power stroke state (Fig. 11, recovery stroke) (194). Thereafter, myosin complexed with the hydrolysis products rebinds to actin in a weak binding state. Actin binding activates the release of phosphate from the active site, which is a key step in coupling the mechanical and chemical cycles, and then the release of ADP occurs when myosin is strongly bound to actin. During the actomyosin-bound state, myosin pulls on the actin filament, performing mechanical work which is produced by the swing of the lever arm (Fig. 11, power stroke).
Nucleotide binding region.
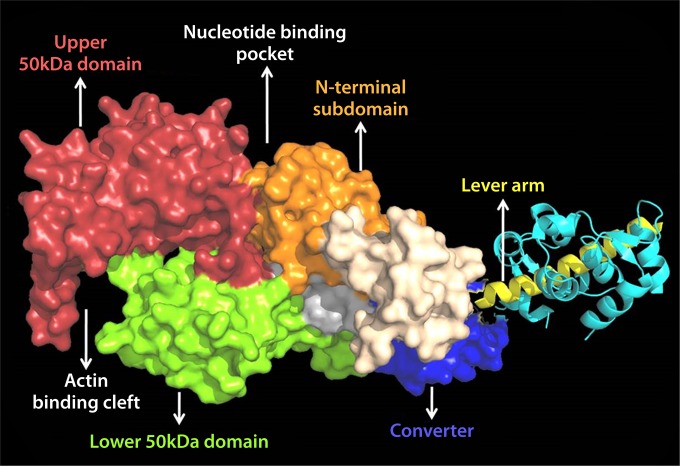
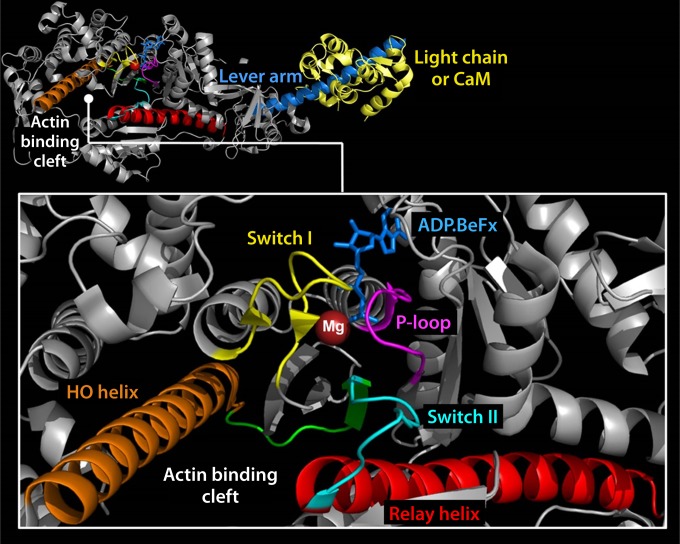
The coordination of ATP within the nucleotide binding pocket, cleavage of its phosphoanhydride bond, and the sequential release of products govern the mechanical cycle of myosins (193, 196, 197). Small structural changes in the conserved regions of the nucleotide binding pocket are communicated to the actin binding and lever arm regions (Fig. 23). Switch I and switch II coordinate the sequential release of products and transmit information from the NBP to the actin binding and lever arm regions (198). Switch II forms a salt bridge with switch I and also interacts with the γ phosphate of ATP (199, 200). The P loop is also involved in the coordination of the α and β phosphates of ATP. Additionally, magnesium (Mg) is coordinated to the oxygen on the β and γ phosphates and makes a direct or water-mediated contact with residues of switch I (Fig. 24). The switch elements undergo a conformational change to a “closed” state upon binding of ATP, which leads to a twisting of a seven-stranded β sheet (transducer), resulting in the opening of the actin binding cleft (201). Moreover, the twist of the transducer region also translates toward the C-terminal lever arm region via a highly conserved structural element called the relay helix (Fig. 24). The relay helix communication pathway induces the recovery stroke and formation of the pre-power stroke state of the lever arm. The actin binding cleft is a deep cleft between the upper (U50) and lower (L50) 50-kDa subdomains in the motor domain (Fig. 23). The binding of myosin to actin is proposed to cause movement of switch I, which induces a loss of Mg coordination, reducing its affinity and causing its eventual release (202–204). These and other rearrangements to the active site result in an isomerization to the weak-ADP state of the pocket and eventual release of ADP.
FIG 23.
Crystal structure showing the different subdomains of myosin along with the actin, nucleotide binding, and lever arm regions (PDB code 1W7J). (Reprinted from reference 236 with permission.)
FIG 24.
Crystal structure of myosin showing the key structural elements involved in the coordination of ATP and the energy transduction mechanism, as discussed in the text (PDB code 1W7J). (Reprinted from reference 236 with permission.)
Actin binding region.
The open-closed transition of switch I may be coupled to the closed-open equilibrium of the actin binding cleft. A 32-amino-acid-long alpha helix which traverses the upper 50-kDa domain of myosins, called the HO helix, and a related HG/HH helix have been demonstrated by molecular modeling studies to be strongly coupled to the open-closed transition of the cleft (205) (Fig. 24). Conformational changes in the HO helix during the myosin ATPase cycle correlate with ATP-induced dissociation and attachment to actin as demonstrated by intrinsic tryptophan fluorescence (206). The relay helix near the lower 50-kDa domain is a 4.7-nm-long α helix that has been well documented to be an essential feature of the force-generating region of myosin (194, 207, 208). It connects the nucleotide binding site to the lever arm region and goes from a kinked to a straight conformation during formation of the pre-power stroke state (194, 207, 208). The HO helix and the relay helix are connected via the switch II loop.
Lever arm region.
The lever arm movement during the power and recovery stroke stages of the catalytic cycle has been probed in a number of studies, including intrinsic tryptophan fluorescence (209, 210) polarization (211, 212), electron paramagnetic resonance (213), and more recently FRET (208) studies. There is still controversy about the timing of the power stroke in relationship to the product release steps of the ATPase cycle. Examining the kinetics of relay helix straightening (201) or lever arm rotation (216) by FRET during actin-activated phosphate release suggests that the power stroke occurs after actin binding and before phosphate release. A study that directly measured the movement of the lever arm by FRET in myosin V demonstrated two phases of the power stroke, a rapid phase that occurs before phosphate release and a slower phase that occurs after phosphate release but before ADP release (217). However, a recent crystal structure of myosin VI provides evidence that phosphate release gates the movement of lever associated with the power stroke (218). There is a good agreement on the details of movement of the lever arm during the recovery stroke. A FRET study reported that the reverse movement of the relay helix from a straight to a kinked conformation is associated with the reversal of the power stroke or the recovery stroke (208). Other studies also agree that the straight-to-kinked transition of the relay helix occurs after ATP binding and before hydrolysis (194, 207).
The structural mechanism of dynein-based motility is not well understood because of the lack of high-resolution structures. However, several recent reports have used a combination of electron microscopy and X-ray crystallography to shed light on the details of the key structural changes that drive microtubule binding and force generation (219). In the presence of an ATP analogue there is closure of the motor ring, which drives the movement of the linker domain, a structural element proposed to be crucial for force generation. The coiled-coil helix of the stalk domain also slides in a nucleotide-dependent manner, which alters the affinity for microtubules. Thus, dynein is similar to myosin in that the ATP binding and hydrolysis prime the motor for force generation, while binding to the track triggers the force-generating structural change.
Mechanism of Sequential Control and Coordination among Channel Subunits
Nucleic acid translocation and duplex unwinding by hexameric biomotors are coupled to the NTP binding and hydrolysis cycle. In nucleic acid translocation, there are six NTP binding sites and at least six nucleic acid binding sites in the hexamer. Individual subunits of a hexameric helicase may switch between several DNA binding states. The binding of NTP to a hydrolysis site, near the inner surface of the channel, induces a conformational change that exposes one pair of negatively and positively charged regions per nucleotide hydrolysis site. These charged regions are not of equal size, are oriented at an angle to the circumferential meridian, and are not constant, appearing and disappearing with the binding and hydrolysis of NTP, respectively. These charged regions can interact with the closest negatively charged DNA phosphate, which gives an “electrostatic push” in the direction of the charged-pair axis. The combined effect of the charged pairs produces a sustained torsional and axial thrust as NTPs are hydrolyzed sequentially around the motor ring.
Sequential action of the phi29 dsDNA-packaging motor was originally shown in phi29 pRNA (129, 220) and further in ATPase, evidenced by Hill constant determination and binomial distribution inhibition assays through the study of the effects of introducing mutant subunits on the oligomerization of gp16 ATPase (12, 50, 127, 128). The results also suggest that conformational changes occur within the gp16 subunits in different nucleotide states that enable them to communicate with each other.
POTENTIAL MOTOR APPLICATIONS
This review offers potential answers to the puzzles in nanotechnology that may shed light on inventions in the motion world. The system of riding along dsDNA provides a prototype for nanomechanical devices and for cargo transportation under control and along predefined paths at the nanoscale. The revolution mechanism offers a hint for the design of nanomolecular motors that can manipulate biomolecules in a controlled fashion. These nanomachines can also be applied for the construction of sophisticated nanodevices, including molecular sensors (221–223), bioreactors (224), chips (225), DNA-sequencing apparatuses (169, 221, 226–229), or other electronic and optical devices (230, 231). In addition, these multisubunit biocomplexes could serve as drug targets for therapeutics and have potential for diagnostic applications (222, 223, 232, 233), especially as the biomotors have an advantage of a high stoichiometry that could be applied to solve the drug resistance issue (234, 235).
CONCLUDING REMARKS AND PERSPECTIVES
Nanobiomotors are tiny machines that utilize a primary energy source to do mechanical work. They are crucial to the sustenance of living systems, since they provide for biological motion, help direct cellular components to proper destinations, package DNA, contract muscles, and perform a variety of other functions. Biomotors exhibit a diversity of complex structures. Most have the same basic components, including a mechanical frame (usually composed of proteins) with both moving and static parts, powered by an energy supply. This energy is typically derived from the binding and hydrolysis of ATP, which lead to conformational changes in the motor protein, resulting in movement. Some motors use energy produced from ion gradients. These motors are typically divided into categories based on the type of motion displayed: linear, rotary, and revolution motors. The action of revolution enables movement to be free of coiling and torque. Revolution motors have now solved many puzzles associated with viral DNA-packaging motor studies. They also have settled the discrepancies concerning the structure, stoichiometry, and functioning of DNA translocation motors. The rotation and revolution mechanisms can be distinguished by the size of channel: the channels of rotation motors are equal to or smaller than 2 nm, whereas channels of revolution motors are larger than 3 nm. Rotation motors use parallel threads that operate with a right-handed channel, while revolution motors use a left-handed channel to drive the right-handed DNA in an antiparallel arrangement.
ACKNOWLEDGMENTS
We thank Darshan Trivedi (Stanford University) for generating Fig. 11, 23, and 24 and for critical comments, Hui Li and Sijin Guo (University of Kentucky) for participating in figure preparation, and Richard N. Greenberg (University of Kentucky), Maria Spies (University of Iowa), and Dan Li for critical reading of the manuscript and helpful discussions.
This work was supported by NIH grants R01-EB012135, R01-EB019036, TR000875, and U01-CA151648 to P.G., an AHA grant to C.M.Y., and NHMRC APP1005697 and ARC Future Fellowship FT120100153 to I.G. Funding for P.G.'s Endowed Chair in Nanobiotechnology position is from the William Fairish Endowment Fund.
The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
P.G. is a cofounder of Biomotor and RNA Nanotech Development Co. Ltd.
Biographies

Peixuan Guo, Ph.D., is currently the First Sylvan G. Frank Endowed Chair in Pharmaceutics and Drug Delivery Systems at The Ohio State University College of Pharmacy and holds a joint appointment in the College of Medicine’s Department of Physiology & Cell Biology and in Ohio State’s Dorothy M. Davis Heart and Lung Research Institute (DHLRI). He held an Endowed Chair in Nanobiotechnology and was the Director of the Nanobiotechnology Center at the University of Kentucky before he moved to The Ohio State University in 2016. He received his Ph.D. in microbiology 1987 and was an NIH postdoctoral fellow in 1990, a Purdue University assistant professor in 1990, tenured in 1993, a full professor 1997, and honored as Purdue Faculty Scholar in 1998. He was Director of the NIH Nanomedicine Development Center from 2006 to 2011. He constructed the first viral DNA-packaging motor, discovered phi29 motor pRNA, pioneered RNA nanotechnology, incorporated a motor channel into lipid membrane, and discovered a third class of revolution biomotors without rotation. He received the Pfizer Distinguished Faculty Award in 1995 and a Lions Club Cancer Research Award in 2006 and was made a member of the Distinguished Alumni of University of Minnesota 2009 and the 100 Years Distinguished Chinese Alumni of the University of Minnesota in 2014. He is the editor of five nanotechnology journals, has had hundreds of reports on television (such as ABC, NBC, and the BBC), and has been featured on the websites of NIH, NSF, MSNBC, and NCI. He is a member of two prominent national nanotechnology initiatives sponsored by NIST/NIH/NSF and has been a member of the Examination Panel (Oversea Expert) of the Chinese Academy of Sciences since 2014.

Hiroyuki Noji, Ph.D., a Professor in the Department of Applied Chemistry, The University of Tokyo, specializes in single-molecule biophysics and has been studying the chemomechanical coupling mechanism of FoF1 ATP synthase by the use of single-molecule techniques. He is also known as an inventor of the femtoliter chamber array system for single-molecule enzymatic assays that is currently applied in single-molecule digital enzyme-linked immunosorbent assay (ELISA). Professor Noji was trained under the supervision of Professor Masasuke Yoshida and received his Ph.D. from Tokyo Institute of Technology in 1997. After a postdoctoral fellowship in the laboratory of Professor Kazuhiko Kinosita, Jr., he was appointed as an Associate Professor at the Institute of Industrial Science, The University of Tokyo, in 2001. In 2005, he moved to the Institute of Scientific and Industrial Research, Osaka University, as a full professor. Since 2010, he has been a Professor in the Department of Applied Chemistry, The University of Tokyo.

Christopher M. Yengo, Ph.D., received his doctoral degree in molecular physiology and biophysics from the University of Vermont. He was a postdoctoral fellow at the University of Pennsylvania. He was an Assistant Professor in the Biology Department at the University of North Carolina at Charlotte. He is currently an Associate Professor in the Department of Cellular and Molecular Physiology in the College of Medicine at Pennsylvania State University. He is interested in the fundamental mechanism of myosin-based force generation, the function of nonmuscle myosins, regulation of the actin cytoskeleton, and the role of myosin mutations in heart disease and deafness.

Zhengyi Zhao is currently a Ph.D. candidate in the College of Pharmacy, University of Kentucky. She graduated from Shenyang Pharmaceutical University in 2011 with a degree in pharmaceutical sciences and then joined Professor Peixuan Guo's lab in 2012. She has a broad training in molecular biology, nanobiotechnology, biophysics, and pharmaceutics. Her current research focuses on the study of the function, mechanism, and application of the bacteriophage phi29 dsDNA packaging nanomotor.

Ian Grainge, Ph.D., received his B.A. (1994) and M.A. (1995) from the University of Cambridge, United Kingdom, and his Ph.D. (1997) from the University of Oxford, United Kingdom. He did postdoctoral research at the University of Texas, Austin, at Cancer Research UK, Clare Hall Laboratories, and at the University of Oxford. He currently is a Lecturer at the University of Newcastle, Australia, and an Australian Research Council Future Fellow. His research focus is on genomic stability. His research interest is in the many mechanisms that organisms employ to keep their genomes intact and pass on that information to subsequent generations. This includes DNA replication, recombination, DNA repair, and chromosome partitioning and segregation. The goal is to understand these processes at the molecular level, and there is a particular fascination with molecular machines, such as helicases and DNA translocases, that underlie many of the vital processes in the cell. He has 20 years of experience in the area of recombination and 15 years of work on molecular motor proteins.
REFERENCES
- 1.McCluskey R. 2013. The Sixth Scottish University. The Scots colleges abroad: 1575 to 1799. Catholic Histor Rev 99:360–361. [Google Scholar]
- 2.Hendrix RW. 1978. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci U S A 75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann RG, Mullaney J, Black LW. 2006. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol 61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- 4.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Faik W, Anderson DL, Bustamante C. 2007. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol 5:558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang C, Zhang H, Shu D, Guo P, Savran C. 2008. Bright-field analysis of phi29 DNA packaging motor using a magnetomechanical system. Appl Phys Lett 93:153902–153903. doi: 10.1063/1.3000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, Schnitzbauer J, Grimes S, Jardine PJ, Bustamante C. 2014. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell 157:702–713. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shu D, Zhang H, Jin J, Guo P. 2007. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J 26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamoto RK, Ketchum CJ, al Shawi MK. 1999. Rotational coupling in the F0F1 ATP synthase. Annu Rev Biophys Biomol Struct 28:205–234. doi: 10.1146/annurev.biophys.28.1.205. [DOI] [PubMed] [Google Scholar]
- 9.Capaldi RA, Aggeler R. 2002. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem Sci 27:154–160. doi: 10.1016/S0968-0004(01)02051-5. [DOI] [PubMed] [Google Scholar]
- 10.Vale R. 1993. Motor proteins, p 175–211. In Kreis T, Vale R (ed), Guidebook to the cytoskeletal and motor proteins. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 11.Goldstein L, Vale R. 1991. Motor proteins. A brave new world for dynein. Nature 352:569–570. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz C, De Donatis GM, Zhang H, Fang H, Guo P. 2013. Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology 443:28–39. doi: 10.1016/j.virol.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Z, Khisamutdinov E, Schwartz C, Guo P. 2013. Mechanism of one-way traffic of hexameric phi29 DNA packaging motor with four electropositive relaying layers facilitating anti-parallel revolution. ACS Nano 7:4082–4092. doi: 10.1021/nn4002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo P. 2014. Biophysical studies reveal new evidence for one-way revolution mechanism of bacteriophage phi29 DNA packaging motor. Biophys J 106:1837–1838. doi: 10.1016/j.bpj.2014.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo P, Zhao Z, Haak J, Wang S, Wu D, Meng B, Weitao T. 2014. Common mechanisms of DNA translocation motors in bacteria and viruses using one-way revolution mechanism without rotation. Biotechnol Adv 32:853–872. doi: 10.1016/j.biotechadv.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo P, Schwartz C, Haak J, Zhao Z. 2013. Discovery of a new motion mechanism of biomotors similar to the earth revolving around the sun without rotation. Virology 446:133–143. doi: 10.1016/j.virol.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweeney HL, Houdusse A. 2010. Structural and functional insights into the myosin motor mechanism. Annu Rev Biophys 39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 18.Vale RD, Fletterick RJ. 1997. The design plan of kinesin motors. Annu Rev Cell Dev Biol 13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 19.Kikkawa M. 2013. Big steps toward understanding dynein. J Cell Biol 202:15–23. doi: 10.1083/jcb.201304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Junge W, Lill H, Engelbrecht S. 1997. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem Sci 22:420–423. doi: 10.1016/S0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 21.DeRosier DJ. 1998. The turn of the screw: the bacterial flagellar motor. Cell 93:17–20. doi: 10.1016/S0092-8674(00)81141-1. [DOI] [PubMed] [Google Scholar]
- 22.Trachtenberg S, Cohen-Krausz S. 2006. The archaeabacterial flagellar filament: a bacterial propeller with a pilus-like structure. J Mol Microbiol Biotechnol 11:208–220. doi: 10.1159/000094055. [DOI] [PubMed] [Google Scholar]
- 23.Holwill MEJ, Satir P. 1987. Generation of propulsive forces by cilia and flagella, p 120–130. In: Bereiter-Hahn J, Anderson OR, Reif WE (ed), Cytomechanics—the mechanical basis of cell form and structure. Springer, Berlin, Germany. [Google Scholar]
- 24.Jankowsky E, Gross CH, Shuman S, Pyle AM. 2000. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature 403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 25.Crozat E, Grainge I. 2010. FtsK DNA translocase: the fast motor that knows where it's going. Chembiochem 11:2232–2243. doi: 10.1002/cbic.201000347. [DOI] [PubMed] [Google Scholar]
- 26.Serwer P, Wang HY. 2005. Single-particle light microscopy of bacteriophages. J Nanosci Nanotechnol 5:2014–2028. doi: 10.1166/jnn.2005.447. [DOI] [PubMed] [Google Scholar]
- 27.Enemark EJ, Joshua-Tor L. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 28.Bailey S, Eliason WK, Steitz TA. 2007. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 29.Castella S, Burgin D, Sanders CM. 2006. Role of ATP hydrolysis in the DNA translocase activity of the bovine papillomavirus (BPV-1) E1 helicase. Nucleic Acids Res 34:3731–3741. doi: 10.1093/nar/gkl554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. 1998. Force and velocity measured for single molecules of RNA polymerase. Science 282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 31.Steinmetz EJ, Platt T. 1994. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc Natl Acad Sci U S A 91:1401–1405. doi: 10.1073/pnas.91.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing X, Bell CE. 2004. Crystal structures of Escherichia coli RecA in complex with MgADP and MnAMP-PNP. Biochemistry 43:16142–16152. doi: 10.1021/bi048165y. [DOI] [PubMed] [Google Scholar]
- 33.Cox MM. 2003. The bacterial RecA protein as a motor protein. Annu Rev Microbiol 57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida M, Muneyuki E, Hisabori T. 2001. ATP synthase—a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol 2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 35.Okuno D, Iino R, Noji H. 2011. Rotation and structure of FoF1-ATP synthase. J Biochem 149:655–664. doi: 10.1093/jb/mvr049. [DOI] [PubMed] [Google Scholar]
- 36.Berry RM. 19 April 2001. Bacterial flagella: flagellar motor. In Encyclopedia of life sciences. John Wiley & Sons Ltd., Chichester, United Kingdom. doi: 10.1038/npg.els.0000744. [DOI] [Google Scholar]
- 37.Berry RM. 2003. Theories of rotary motors. Philos Trans R Soc Lond B Biol 355:503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada Y, Ohara O, Takatsuki A, Itoh H, Shimamoto N, Kinosita K Jr. 2001. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature 409:113–115. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 39.Pyle AM. 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys 37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 40.Singleton MR, Dillingham MS, Wigley DB. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 41.Grigoriev DN, Moll W, Hall J, Guo P. 2004. Bionanomotor, p 361–374. In Nalwa HS. (ed), Encyclopedia of nanoscience and nanotechnology. American Scientific Publishers, Valencia, CA. [Google Scholar]
- 42.Kowalczykowski SC. 2015. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer LM, Makarova KS, Koonin EV, Aravind L. 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res 32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burroughs AM, Iyer LM, Aravind L. 2007. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn 3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 45.Black LW. 2015. Old, new, and widely true: the bacteriophage T4 DNA packaging mechanism. Virology 479–480:650–656. doi: 10.1016/j.virol.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey SC. 2015. The scrunchworm hypothesis: Transitions between A-DNA and B-DNA provide the driving force for genome packaging in double-stranded DNA bacteriophages. J Struct Biol 189:1–8. doi: 10.1016/j.jsb.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixit AB, Ray K, Black LW. 2012. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc Natl Acad Sci U S A 109:20419–20424. doi: 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray K, Sabanayagam CR, Lakowicz JR, Black LW. 2010. DNA crunching by a viral packaging motor: compression of a procapsid-portal stalled Y-DNA substrate. Virology 398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geng J, Fang H, Haque F, Zhang L, Guo P. 2011. Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials 32:8234–8242. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De-Donatis G, Zhao Z, Wang S, Huang PL, Schwartz C, Tsodikov VO, Zhang H, Haque F, Guo P. 2014. Finding of widespread viral and bacterial revolution dsDNA translocation motors distinct from rotation motors by channel chirality and size. Cell Biosci 4:30. doi: 10.1186/2045-3701-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jimenez J, Santisteban A, Carazo JM, Carrascosa JL. 1986. Computer graphic display method for visualizing three-dimensional biological structures. Science 232:1113–1115. doi: 10.1126/science.3754654. [DOI] [PubMed] [Google Scholar]
- 52.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol 315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 53.Guo P, Erickson S, Anderson D. 1987. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science 236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 54.Guo P, Zhang C, Chen C, Trottier M, Garver K. 1998. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 2:149–155. doi: 10.1016/S1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. 2013. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the phi29 hexameric motor pRNA. RNA 19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz C, De Donatis GM, Fang H, Guo P. 2013. The ATPase of the phi29 DNA-packaging motor is a member of the hexameric AAA+ superfamily. Virology 443:20–27. doi: 10.1016/j.virol.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo P, Peterson C, Anderson D. 1987. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage phi29. J Mol Biol 197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 58.Guo PX, Lee TJ. 2007. Viral nanomotors for packaging of dsDNA and dsRNA. Mol Microbiol 64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao VB, Feiss M. 2008. The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 60.Zhang H, Schwartz C, De Donatis GM, Guo P. 2012. “Push through one-way valve” mechanism of viral DNA packaging. Adv Virus Res 83:415–465. doi: 10.1016/B978-0-12-394438-2.00009-8. [DOI] [PubMed] [Google Scholar]
- 61.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. 2007. Structural framework for DNA translocation via the viral portal protein. EMBO J 26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. 2005. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol 347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol 299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 64.Olia AS, Prevelige PE, Johnson JE, Cingolani G. 2011. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol 18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. 2006. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 66.Molineux IJ, Panja D. 2013. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol 11:194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- 67.Petrov AS, Harvey SC. 2008. Packaging double-helical DNA into viral capsids: structures, forces, and energetics. Biophys J 95:497–502. doi: 10.1529/biophysj.108.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu B, Margolin W, Molineux IJ, Liu J. 2015. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci U S A 112:E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang JH, Olson N, Jardine PJ, Girimes S, Anderson DL, Baker TS. 2008. DNA poised for release in bacteriophage phi29. Structure 16:935–943. doi: 10.1016/j.str.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duda RL, Conway JF. 2008. Asymmetric EM reveals new twists in phage phi29 biology. Structure 16:831–832. doi: 10.1016/j.str.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 72.Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL. 2013. Structural characterization of the bacteriophage T7 tail machinery. J Biol Chem 288:26290–26299. doi: 10.1074/jbc.M113.491209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science 339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu XC, Weihe EK, Margolin W. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol 180:6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steiner W, Liu G, Donachie WD, Kuempel P. 1999. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol 31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 76.Grainge I. 2010. FtsK—a bacterial cell division checkpoint? Mol Microbiol 78:1055–1057. doi: 10.1111/j.1365-2958.2010.07411.x. [DOI] [PubMed] [Google Scholar]
- 77.Lee JY, Finkelstein IJ, Crozat E, Sherratt DJ, Greene EC. 2012. Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK. Proc Natl Acad Sci U S A 109:6531–6536. doi: 10.1073/pnas.1201613109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, Bustamante C, Cozzarelli NR. 2005. Sequence-directed DNA translocation by purified FtsK. Science 307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 79.Demarre G, Galli E, Barre FX. 2013. The FtsK family of DNA pumps. Adv Exp Med Biol 767:245–262. doi: 10.1007/978-1-4614-5037-5_12. [DOI] [PubMed] [Google Scholar]
- 80.Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. 2006. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell 23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 81.Grainge I, Lesterlin C, Sherratt DJ. 2011. Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res 39:5140–5148. doi: 10.1093/nar/gkr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bigot S, Saleh OA, Lesterlin C, Pages C, El KM, Dennis C, Grigoriev M, Allemand JF, Barre FX, Cornet F. 2005. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J 24:3770–3780. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levy O, Ptacin JL, Pease PJ, Gore J, Eisen MB, Bustamante C, Cozzarelli NR. 2005. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc Natl Acad Sci U S A 102:17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. 2008. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol Cell 31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu LJ, Errington J. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 86.Fiche JB, Cattoni DI, Diekmann N, Langerak JM, Clerte C, Royer CA, Margeat E, Doan T, Nollmann M. 2013. Recruitment, assembly, and molecular architecture of the SpoIIIE DNA pump revealed by superresolution microscopy. PLoS Biol 11:e1001557. doi: 10.1371/journal.pbio.1001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaimer C, Graumann PL. 2011. Players between the worlds: multifunctional DNA translocases. Curr Opin Microbiol 14:719–725. doi: 10.1016/j.mib.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 88.Barre FX. 2007. FtsK and SpoIIIE: the tale of the conserved tails. Mol Microbiol 66:1051–1055. doi: 10.1111/j.1365-2958.2007.05981.x. [DOI] [PubMed] [Google Scholar]
- 89.Sivanathan V, Allen MD, de BC, Baker R, Arciszewska LK, Freund SM, Bycroft M, Lowe J, Sherratt DJ. 2006. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat Struct Mol Biol 13:965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bath J, Wu LJ, Errington J, Wang JC. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- 91.Chelikani V, Ranjan T, Zade A, Shukla A, Kondabagil K. 2014. Genome segregation and packaging machinery in Acanthamoeba polyphaga mimivirus is reminiscent of bacterial apparatus. J Virol 88:6069–6075. doi: 10.1128/JVI.03199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Odronitz F, Kollmar M. 2007. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol 8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. 2004. A standardized kinesin nomenclature. J Cell Biol 167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. 2013. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol 14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kull FJ, Endow SA. 2013. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci 126:9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vale RD. 1996. Switches, latches, and amplifiers: common themes of G proteins and molecular motors. J Cell Biol 135:291–302. doi: 10.1083/jcb.135.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kull FJ, Vale RD, Fletterick RJ. 1998. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J Muscle Res Cell Motil 19:877–886. doi: 10.1023/A:1005489907021. [DOI] [PubMed] [Google Scholar]
- 98.Gelles J, Landick R. 1998. RNA polymerase as a molecular motor. Cell 93:13–16. doi: 10.1016/S0092-8674(00)81140-X. [DOI] [PubMed] [Google Scholar]
- 99.Lohman TM, Thorn K, Vale RD. 1998. Staying on track: common features of DNA helicases and microtubule motors. Cell 93:9–12. doi: 10.1016/S0092-8674(00)81139-3. [DOI] [PubMed] [Google Scholar]
- 100.West SC. 1996. DNA helicases: new breeds of translocating motors and molecular pumps. Cell 86:177–180. doi: 10.1016/S0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 101.Yu X, Hingorani MM, Patel SS, Egelman EH. 1996. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat Struct Biol 3:740–743. doi: 10.1038/nsb0996-740. [DOI] [PubMed] [Google Scholar]
- 102.Geeves MA, Holmes KC. 1999. Structural mechanism of muscle contraction. Annu Rev Biochem 68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- 103.Vale RD, Milligan RA. 2000. The way things move: looking under the hood of molecular motor proteins. Science 288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 104.Bravo A, Alonso JC. 1990. The generation of concatemeric plasmid DNA in Bacillus subtilis as a consequence of bacteriophage SPP1 infection. Nucleic Acids Res 18:4651–4657. doi: 10.1093/nar/18.16.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.White JH, Richardson CC. 1987. Processing of concatemers of bacteriophage T7 DNA in vitro. J Biol Chem 262:8854–8860. [PubMed] [Google Scholar]
- 106.Everett RD. 1981. DNA replication of bacteriophage T5. 3. Studies on the structure of concatemeric T5 DNA. J Gen Virol 52:25–38. [DOI] [PubMed] [Google Scholar]
- 107.Caspar DLD, Klug A. 1962. Physical principles in the construction of regular viruses. Cold Spr Harb Symp Quant Biol 27:1–24. doi: 10.1101/SQB.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 108.Bazinet C, King J. 1985. The DNA translocation vertex of dsDNA bacteriophages. Annu Rev Microbiol 39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- 109.Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, Rubinstein JL, Battaile KP, Chirgadze N, Maxwell KL, Davidson AR. 2010. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J Mol Biol 395:754–768. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- 110.Valpuesta JM, Fujisawa H, Marco S, Carazo JM, Carrascosa J. 1992. Three-dimensional structure of T3 connector purified from overexpressing bacteria. J Mol Biol 224:103–112. doi: 10.1016/0022-2836(92)90579-9. [DOI] [PubMed] [Google Scholar]
- 111.Kochan J, Carrascosa JL, Murialdo H. 1984. Bacteriophage lambda preconnectors: purification and structure. J Mol Biol 174:433–447. doi: 10.1016/0022-2836(84)90330-9. [DOI] [PubMed] [Google Scholar]
- 112.Doan DN, Dokland T. 2007. The gpQ portal protein of bacteriophage P2 forms dodecameric connectors in crystals. J Struct Biol 157:432–436. doi: 10.1016/j.jsb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 113.Carrascosa JL, Vinuela E, Garcia N, Santisteban A. 1982. Structure of the head-tail connector of bacteriophage phi 29. J Mol Biol 154:311–324. doi: 10.1016/0022-2836(82)90066-3. [DOI] [PubMed] [Google Scholar]
- 114.Zhang F, Lemieux S, Wu X, St Arnaud S, McMurray CT, Major F, Anderson D. 1998. Function of hexameric RNA in packaging of bacteriophage phi29 DNA in vitro. Mol Cell 2:141–147. doi: 10.1016/S1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 115.Hendrix RW. 1998. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell 94:147–150. doi: 10.1016/S0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- 116.Chistol G, Liu S, Hetherington CL, Moffitt JR, Grimes S, Jardine PJ, Bustamante C. 2012. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell 151:1017–1028. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G. 2010. Mechanochemistry of a viral DNA packaging motor. J Mol Biol 400:186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 118.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson D, Rossman MG. 2008. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure 16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ibarra B, Caston JR, Llorca O, Valle M, Valpuesta JM, Carrascosa JL. 2000. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J Mol Biol 298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- 120.Moll D, Guo P. 2007. Grouping of ferritin and gold nanoparticles conjugated to pRNA of the phage phi29 DNA-packaging motor. J Nanosci Nanotechnol 7:3257–3267. doi: 10.1166/jnn.2007.914. [DOI] [PubMed] [Google Scholar]
- 121.Xiao F, Zhang H, Guo P. 2008. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res 36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morais MC, Tao Y, Olsen NH, Grimes S, Jardine PJ, Anderson D, Baker TS, Rossmann MG. 2001. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J Struct Biol 135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA plus ATPases. J Struct Biol 146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 125.Wang F, Mei Z, Qi Y, Yan C, Hu Q, Wang J, Shi Y. 2011. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature 471:331–335. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- 126.Willows RD, Hansson A, Birch D, Al-Karadaghi S, Hansson M. 2004. EM single particle analysis of the ATP-dependent BchI complex of magnesium chelatase: an AAA(+) hexamer. J Struct Biol 146:227–233. doi: 10.1016/j.jsb.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 127.Chen C, Trottier M, Guo P. 1997. New approaches to stoichiometry determination and mechanism investigation on RNA involved in intermediate reactions. Nucleic Acids Symp Ser 36:190–193. [Google Scholar]
- 128.Trottier M, Guo P. 1997. Approaches to determine stoichiometry of viral assembly components. J Virol 71:487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. 2009. Intersubunit coordination in a homomeric ring ATPase. Nature 457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. 2012. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright ET, Serwer P, Jiang W. 2013. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc Natl Acad Sci U S A 110:6811–6816. doi: 10.1073/pnas.1215563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jing P, Haque F, Shu D, Montemagno C, Guo P. 2010. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett 10:3620–3627. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schwartz C, Fang H, Huang L, Guo P. 2012. Sequential action of ATPase, ATP, ADP, Pi and dsDNA in procapsid-free system to enlighten mechanism in viral dsDNA packaging. Nucleic Acids Res 40:2577–2586. doi: 10.1093/nar/gkr841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang LP, Guo P. 2003. Use of acetone to attain highly active and soluble DNA packaging protein gp16 of phi29 for ATPase assay. Virology 312:449–457. doi: 10.1016/S0042-6822(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 135.Hayashi S, Ueno H, Shaikh AR, Umemura M, Kamiya M, Ito Y, Ikeguchi M, Komoriya Y, Iino R, Noji H. 2012. Molecular mechanism of ATP hydrolysis in F1-ATPase revealed by molecular simulations and single-molecule observations. J Am Chem Soc 134:8447–8454. doi: 10.1021/ja211027m. [DOI] [PubMed] [Google Scholar]
- 136.Zhang X, Wigley DB. 2008. The ‘glutamate switch' provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat Struct Mol Biol 15:1223–1227. doi: 10.1038/nsmb.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nadanaciva S, Weber J, Wilke-Mounts S, Senior AE. 1999. Importance of F-1-ATPase residue alpha-Arg-376 for catalytic transition state stabilization. Biochemistry 38:15493–15499. doi: 10.1021/bi9917683. [DOI] [PubMed] [Google Scholar]
- 138.Dittrich M, Hayashi S, Schulten K. 2003. On the mechanism of ATP hydrolysis in F(1)-ATPase. Biophys J 85:2253–2266. doi: 10.1016/S0006-3495(03)74650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Komoriya Y, Ariga T, Iino R, Imamura H, Okuno D, Noji H. 2012. Principal role of the arginine finger in rotary catalysis of F-1-ATPase. J Biol Chem 287:15134–15142. doi: 10.1074/jbc.M111.328153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Guenther B, Onrust R, Sali A, O'Donnell M, Kuriyan J. 1997. Crystal structure of the delta′ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell 91:335–345. doi: 10.1016/S0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 141.Erzberger JP, Berger JM. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct 35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 142.Noji H, Yasuda R, Yoshida M, Kinosita K Jr. 1997. Direct observation of the rotation of F1-ATPase. Nature 386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 143.Hisabori T, Kondoh A, Yoshida M. 1999. The gamma subunit in chloroplast F-1-ATPase can rotate in a unidirectional and counter-clockwise manner. FEBS Lett 463:35–38. doi: 10.1016/S0014-5793(99)01602-6. [DOI] [PubMed] [Google Scholar]
- 144.Suzuki T, Tanaka K, Wakabayashi C, Saita E, Yoshida M. 2014. Chemomechanical coupling of human mitochondrial F1-ATPase motor. Nat Chem Biol 10:930–936. doi: 10.1038/nchembio.1635. [DOI] [PubMed] [Google Scholar]
- 145.Imamura H, Nakano M, Noji H, Muneyuki E, Ohkuma S, Yoshida M, Yokoyama K. 2003. Evidence for rotation of V-1-ATPase. Proc Natl Acad Sci U S A 100:2312–2315. doi: 10.1073/pnas.0436796100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Minagawa Y, Ueno H, Hara M, Ishizuka-Katsura Y, Ohsawa N, Terada T, Shirouzu M, Yokoyama S, Yamato I, Muneyuki E, Noji H, Murata T, Iino R. 2013. Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V-1-ATPase. J Biol Chem 288:32700–32707. doi: 10.1074/jbc.M113.506329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yasuda R, Noji H, Kinosita K Jr, Yoshida M. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93:1117–1124. doi: 10.1016/S0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 148.Rondelez Y, Tresset G, Nakashima T, Kato-Yamada Y, Fujita H, Takeuchi S, Noji H. 2005. Highly coupled ATP synthesis by F1-ATPase single molecules. Nature 433:773–777. doi: 10.1038/nature03277. [DOI] [PubMed] [Google Scholar]
- 149.Shimabukuro K, Yasuda R, Muneyuki E, Hara KY, Kinosita K, Yoshida M. 2003. Catalysis and rotation of F-1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40 degrees substep rotation. Proc Natl Acad Sci U S A 100:14731–14736. doi: 10.1073/pnas.2434983100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Watanabe R, Iino R, Noji H. 2010. Phosphate release in F-1-ATPase catalytic cycle follows ADP release. Nat Chem Biol 6:814–820. doi: 10.1038/nchembio.443. [DOI] [PubMed] [Google Scholar]
- 151.Yasuda R, Noji H, Yoshida M, Kinosita K Jr, Itoh H. 2001. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 152.Enoki S, Iino R, Niitani Y, Minagawa Y, Tomishige M, Noji H. 2015. High-speed angle-resolved imaging of a single gold nanorod with microsecond temporal resolution and one-degree angle precision. Anal Chem 87:2079–2086. doi: 10.1021/ac502408c. [DOI] [PubMed] [Google Scholar]
- 153.Spetzler D, York J, Daniel D, Fromme R, Lowry D, Frasch W. 2006. Microsecond time scale rotation measurements of single F-1-ATPase molecules. Biochemistry 45:3117–3124. doi: 10.1021/bi052363n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Watanabe R, Okuno D, Sakakihara S, Shimabukuro K, Iino R, Yoshida M, Noji H. 2012. Mechanical modulation of catalytic power on F-1-ATPase. Nat Chem Biol 8:86–92. doi: 10.1038/nchembio.715. [DOI] [PubMed] [Google Scholar]
- 155.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K. 2004. Mechanically driven ATP synthesis by F-1-ATPase. Nature 427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 156.Abrahams JP, Leslie AG, Lutter R, Walker JE. 1994. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 157.Kobayashi M, Akutsu H, Suzuki T, Yoshida M, Yagi H. 2010. Analysis of the open and closed conformations of the beta subunits in thermophilic F-1-ATPase by solution NMR. J Mol Biol 398:189–199. doi: 10.1016/j.jmb.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 158.Masaike T, Koyama-Horibe F, Oiwa K, Yoshida M, Nishizaka T. 2008. Cooperative three-step motions in catalytic subunits of F(1)-ATPase correlate with 80 degrees and 40 degrees substep rotations. Nat Struct Mol Biol 15:1326–1333. doi: 10.1038/nsmb.1510. [DOI] [PubMed] [Google Scholar]
- 159.Uchihashi T, Iino R, Ando T, Noji H. 2011. High-speed atomic force microscopy reveals rotary catalysis of rotorless F-1-ATPase. Science 333:755–758. doi: 10.1126/science.1205510. [DOI] [PubMed] [Google Scholar]
- 160.Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CAM, Graber P. 2004. Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat Struct Mol Biol 11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 161.Zimmermann B, Diez M, Zarrabi N, Graber P, Borsch M. 2005. Movements of the epsilon-subunit during catalysis and activation in single membrane-bound H+-ATP synthase. EMBO J 24:2053–2063. doi: 10.1038/sj.emboj.7600682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Watanabe R, Tabata KV, Iino R, Ueno H, Iwamoto M, Oiki S, Noji H. 2013. Biased Brownian stepping rotation of FoF1-ATP synthase driven by proton motive force. Nat Commun 4:1631. doi: 10.1038/ncomms2631. [DOI] [PubMed] [Google Scholar]
- 163.Watanabe R, Soga N, Fujita D, Tabata KV, Yamauchi L, Kim SH, Asanuma D, Kamiya M, Urano Y, Suga H, Noji H. 2014. Arrayed lipid bilayer chambers allow single-molecule analysis of membrane transporter activity. Nat Commun 5:4519. doi: 10.1038/ncomms5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Patel SS, Donmez I. 2006. Mechanisms of helicases. J Biol Chem 281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 165.Lohman TM, Tomko EJ, Wu CG. 2008. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol 9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 166.Fang H, Jing P, Haque F, Guo P. 2012. Role of channel lysines and “push through a one-way valve” mechanism of viral DNA packaging motor. Biophys J 102:127–135. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Grimes S, Ma S, Gao J, Atz R, Jardine PJ. 2011. Role of phi29 connector channel loops in late-stage DNA packaging. J Mol Biol 410:50–59. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Serwer P. 2010. A hypothesis for bacteriophage DNA packaging motors. Viruses 2:1821–1843. doi: 10.3390/v2091821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Geng J, Wang S, Fang H, Guo P. 2013. Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids. ACS Nano 7:3315–3323. doi: 10.1021/nn400020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, van HM, Tavares P. 2003. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J 22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Moll W-D, Guo P. 2005. Translocation of nicked but not gapped DNA by the packaging motor of bacteriophage phi29. J Mol Biol 351:100–107. doi: 10.1016/j.jmb.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 172.Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. 2009. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Oram M, Sabanayagam C, Black LW. 2008. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J Mol Biol 381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kottadiel VI, Rao VB, Chemla YR. 2012. The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4. Proc Natl Acad Sci U S A 109:20000–20005. doi: 10.1073/pnas.1209214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Morita M, Tasaka M, Fujisawa H. 1993. DNA packaging ATPase of bacteriophage T3. Virology 193:748–752. doi: 10.1006/viro.1993.1183. [DOI] [PubMed] [Google Scholar]
- 176.Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. 2002. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108:195–205. doi: 10.1016/S0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 177.Saleh OA, Bigot S, Barre FX, Allemand JF. 2005. Analysis of DNA supercoil induction by FtsK indicates translocation without groove-tracking. Nat Struct Mol Biol 12:436–440. doi: 10.1038/nsmb926. [DOI] [PubMed] [Google Scholar]
- 178.Crozat E, Meglio A, Allemand JF, Chivers CE, Howarth M, Venien-Bryan C, Grainge I, Sherratt DJ. 2010. Separating speed and ability to displace roadblocks during DNA translocation by FtsK. EMBO J 29:1423–1433. doi: 10.1038/emboj.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lee JY, Finkelstein IJ, Arciszewska LK, Sherratt DJ, Greene EC. 2014. Single-molecule imaging of FtsK translocation reveals mechanistic features of protein-protein collisions on DNA. Mol Cell 54:832–843. doi: 10.1016/j.molcel.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Thomsen ND, Berger JM. 2009. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rafferty JB, Sedelnikova SE, Hargreaves D, Artymiuk PJ, Baker PJ, Sharples GJ, Mahdi AA, Lloyd RG, Rice DW. 1996. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science 274:415–421. doi: 10.1126/science.274.5286.415. [DOI] [PubMed] [Google Scholar]
- 182.Han YW, Tani T, Hayashi M, Hishida T, Iwasaki H, Shinagawa H, Harada Y. 2006. Direct observation of DNA rotation during branch migration of Holliday junction DNA by Escherichia coli RuvA-RuvB protein complex. Proc Natl Acad Sci U S A 103:11544–11548. doi: 10.1073/pnas.0600753103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cooke R. 1997. Actomyosin interaction in striated muscle. Physiol Rev 77:671–697. [DOI] [PubMed] [Google Scholar]
- 184.Woledge RC, Curtin NA, Homsher E. 1985. Energetic aspects of muscle contraction. Monogr Physiol Soc 41:1–357. [PubMed] [Google Scholar]
- 185.Howard J. 2001. Mechanics of motor proteins and the cytoskeleton. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 186.Reference deleted.
- 187.Reference deleted.
- 188.Reference deleted.
- 189.Tyska MJ, Warshaw DM. 2002. The myosin power stroke. Cell Motil Cytoskeleton 51:1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- 190.Reference deleted.
- 191.Reference deleted.
- 192.Reference deleted.
- 193.Lymn RW, Taylor EW. 1971. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry 10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 194.Nesmelov YE, Agafonov RV, Negrashov IV, Blakely SE, Titus MA, Thomas DD. 2011. Structural kinetics of myosin by transient time-resolved FRET. Proc Natl Acad Sci U S A 108:1891–1896. doi: 10.1073/pnas.1012320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Reference deleted.
- 196.De la Cruz EM, Ostap EM. 2004. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol 16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 197.Eisenberg E, Greene LE. 1980. The relation of muscle biochemistry to muscle physiology. Annu Rev Physiol 42:293–309. doi: 10.1146/annurev.ph.42.030180.001453. [DOI] [PubMed] [Google Scholar]
- 198.Kintses B, Gyimesi M, Pearson DS, Geeves MA, Zeng W, Bagshaw CR, Malnasi-Csizmadia A. 2007. Reversible movement of switch 1 loop of myosin determines actin interaction. EMBO J 26:265–274. doi: 10.1038/sj.emboj.7601482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sasaki N, Shimada T, Sutoh K. 1998. Mutational analysis of the switch II loop of Dictyostelium myosin II. J Biol Chem 273:20334–20340. doi: 10.1074/jbc.273.32.20334. [DOI] [PubMed] [Google Scholar]
- 200.Onishi H, Ohki T, Mochizuki N, Morales MF. 2002. Early stages of energy transduction by myosin: roles of Arg in switch I, of Glu in switch II, and of the salt-bridge between them. Proc Natl Acad Sci U S A 99:15339–15344. doi: 10.1073/pnas.242604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Coureux PD, Sweeney HL, Houdusse A. 2004. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J 23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rosenfeld SS, Houdusse A, Sweeney HL. 2005. Magnesium regulates ADP dissociation from myosin V. J Biol Chem 280:6072–6079. doi: 10.1074/jbc.M412717200. [DOI] [PubMed] [Google Scholar]
- 203.Hannemann DE, Cao W, Olivares AO, Robblee JP, De la Cruz EM. 2005. Magnesium, ADP, and actin binding linkage of myosin V: evidence for multiple myosin V-ADP and actomyosin V-ADP states. Biochemistry 44:8826–8840. doi: 10.1021/bi0473509. [DOI] [PubMed] [Google Scholar]
- 204.Trivedi DV, Muretta JM, Swenson AM, Thomas DD, Yengo CM. 2013. Magnesium impacts myosin V motor activity by altering key conformational changes in the mechanochemical cycle. Biochemistry 52:4710–4722. doi: 10.1021/bi4004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ovchinnikov V, Trout BL, Karplus M. 2010. Mechanical coupling in myosin V: a simulation study. J Mol Biol 395:815–833. doi: 10.1016/j.jmb.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yengo CM, De la Cruz EM, Chrin LR, Gaffney DP, Berger CL. 2002. Actin-induced closure of the actin-binding cleft of smooth muscle myosin. J Biol Chem 277:24114–24119. doi: 10.1074/jbc.M111253200. [DOI] [PubMed] [Google Scholar]
- 207.Agafonov RV, Negrashov IV, Tkachev YV, Blakely SE, Titus MA, Thomas DD, Nesmelov YE. 2009. Structural dynamics of the myosin relay helix by time-resolved EPR and FRET. Proc Natl Acad Sci U S A 106:21625–21630. doi: 10.1073/pnas.0909757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Muretta JM, Petersen KJ, Thomas DD. 2013. Direct real-time detection of the actin-activated power stroke within the myosin catalytic domain. Proc Natl Acad Sci U S A 110:7211–7216. doi: 10.1073/pnas.1222257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Conibear PB, Malnasi-Csizmadia A, Bagshaw CR. 2004. The effect of F-actin on the relay helix position of myosin II, as revealed by tryptophan fluorescence, and its implications for mechanochemical coupling. Biochemistry 43:15404–15417. doi: 10.1021/bi048338j. [DOI] [PubMed] [Google Scholar]
- 210.Malnasi-Csizmadia A, Woolley RJ, Bagshaw CR. 2000. Resolution of conformational states of Dictyostelium myosin II motor domain using tryptophan (W501) mutants: implications for the open-closed transition identified by crystallography. Biochemistry 39:16135–16146. doi: 10.1021/bi001125j. [DOI] [PubMed] [Google Scholar]
- 211.Corrie JE, Brandmeier BD, Ferguson RE, Trentham DR, Kendrick-Jones J, Hopkins SC, van der Heide UA, Goldman YE, Sabido-David C, Dale RE, Criddle S, Irving M. 1999. Dynamic measurement of myosin light-chain-domain tilt and twist in muscle contraction. Nature 400:425–430. doi: 10.1038/22704. [DOI] [PubMed] [Google Scholar]
- 212.Forkey JN, Quinlan ME, Shaw MA, Corrie JE, Goldman YE. 2003. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature 422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 213.Baker JE, Brust-Mascher I, Ramachandran S, LaConte LE, Thomas DD. 1998. A large and distinct rotation of the myosin light chain domain occurs upon muscle contraction. Proc Natl Acad Sci U S A 95:2944–2949. doi: 10.1073/pnas.95.6.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Reference deleted.
- 215.Reference deleted.
- 216.Muretta JM, Rohde JA, Johnsrud DO, Cornea S, Thomas DD. 2015. Direct real-time detection of the structural and biochemical events in the myosin power stroke. Proc Natl Acad Sci U S A 112:14272–14277. doi: 10.1073/pnas.1514859112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Trivedi DV, Muretta JR, Swenson AM, Davis JP, Thomas DD, Yengo CM. 2015. Direct measurements of the coordination of lever arm swing and the catalytic cycle in myosin V. Proc Natl Acad Sci U S A 112:14593–14598. doi: 10.1073/pnas.1517566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Llinas P, Isabet T, Song L, Ropars V, Zong B, Benisty H, Sirigu S, Morris C, Kikuti C, Safer D, Sweeney HL, Houdusse A. 2015. How actin initiates the motor activity of myosin. Dev Cell 33:401–412. doi: 10.1016/j.devcel.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schmidt H, Zalyte R, Urnavicius L, Carter AP. 2015. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518:435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chen C, Guo P. 1997. Sequential action of six virus-encoded DNA-packaging RNAs during phage phi29 genomic DNA translocation. J Virol 71:3864–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Haque F, Li J, Wu H-C, Liang X-J, Guo P. 2013. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today 8:56–74. doi: 10.1016/j.nantod.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Haque F, Lunn J, Fang H, Smithrud D, Guo P. 2012. Real-time sensing and discrimination of single chemicals using the channel of phi29 DNA packaging nanomotor. ACS Nano 6:3251–3261. doi: 10.1021/nn3001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang S, Haque F, Rychahou PG, Evers BM, Guo P. 2013. Engineered nanopore of phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano 7:9814–9822. doi: 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Noireaux V, Libchaber A. 2004. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci U S A 101:17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Steuerwald D, Fruh SM, Griss R, Lovchik RD, Vogel V. 2014. Nanoshuttles propelled by motor proteins sequentially assemble molecular cargo in a microfluidic device. Lab Chip 14:3729–3738. doi: 10.1039/C4LC00385C. [DOI] [PubMed] [Google Scholar]
- 226.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. 1996. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A 93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. 2008. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci U S A 105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Soong RK, Bachand GD, Neves HP, Olkhovets AG, Craighead HG, Montemagno CD. 2000. Powering an inorganic nanodevice with a biomolecular motor. Science 290:1555–1558. doi: 10.1126/science.290.5496.1555. [DOI] [PubMed] [Google Scholar]
- 229.Gust D. 2015. Supramolecular photochemistry applied to artificial photosynthesis and molecular logic devices. Faraday Discuss doi: 10.1039/C5FD00142K. [DOI] [PubMed] [Google Scholar]
- 230.McNally B, Singer A, Yu Z, Sun Y, Weng Z, Meller A. 2010. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett 10:2237–2244. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chandler EL, Smith AL, Burden LM, Kasianowicz JJ, Burden DL. 2004. Membrane surface dynamics of DNA-threaded nanopores revealed by simultaneous single-molecule optical and ensemble electrical recording. Langmuir 20:898–905. doi: 10.1021/la035728i. [DOI] [PubMed] [Google Scholar]
- 232.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. 2011. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P. 2012. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Fang H, Zhang P, Huang LP, Zhao Z, Pi F, Montemagno C, Guo P. 2014. Binomial distribution for quantification of protein subunits in biological nanoassemblies and functional nanomachines. Nanomedicine 10:1433–1440. doi: 10.1016/j.nano.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shu D, Pi F, Wang C, Zhang P, Guo P. 2015. New approach to develop ultra-high inhibitory drug using the power-function of the stoichiometry of the targeted nanomachine or biocomplex. Nanomedicine 10:1881–1897. doi: 10.2217/nnm.15.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Trivedi DV. 2014. Allosteric communication and force generation in myosin motors. Doctoral dissertation. Pennsylvania State University, University Park, PA. [Google Scholar]