A method to retrieve all major tumor-associated cell populations with high cell yield, while maintaining the expression of cell surface markers and functions of isolated cells.
Keywords: enzymatic digestion, collagenase, tumor-infiltrating immune cells
Abstract
Careful preparation of human tissues is the cornerstone of obtaining accurate data in immunologic studies. Despite the essential importance of tissue processing in tumor immunology and clinical medicine, current methods of tissue disaggregation have not been rigorously tested for data fidelity. Thus, we critically evaluated the current techniques available in the literature that are used to prepare human lung tumors for immunologic studies. We discovered that these approaches are successful at digesting cellular attachments and ECMs; however, these methods frequently alter the immune cell composition and/or expression of surface molecules. We thus developed a novel approach to prepare human lung tumors for immunologic studies by combining gentle mechanical manipulation with an optimized cocktail of enzymes used at low doses. This enzymatic digestion cocktail optimized cell yield and cell viability, retrieved all major tumor-associated cell populations, and maintained the expression of cell-surface markers for lineage definition and in vivo effector functions. To our knowledge, we present the first rigorously tested disaggregation method designed for human lung tumors.
Introduction
Detailed studies of solid human cancers face the challenge of converting the harvested tumor into a single-cell suspension for biologic analysis without altering the phenotype or functional activity of the target cell population. Ideally, this process, termed "disaggregation," should be gentle enough to maximize viability and minimize biologic alterations, yet robust enough to optimize cell yield and ensure that the final product accurately represents the in vivo cellular populations [1, 2]. Currently, no standardized techniques exist for the disaggregation of human tumor tissue
Traditionally, efforts have used matrix-degrading enzymes [2–10] and mechanical manipulation [3, 4, 11, 12] to disrupt cellular attachments, break down the ECM, and liberate individual cells [13]. Methods of mechanical disaggregation (mincing, slicing, homogenizing) are rapid and simple, but they tend to generate suspensions characterized by high cellular damage and low cell recovery [3, 4, 6]. Although these approaches do not alter biochemical cellular properties, they may not successfully retrieve the less-resilient immune cell populations in the tumor microenvironment and induce substantial cell death and damage [14]. The other classic approach, enzymatic digestion, has variable efficacy depending on the enzymes and the concentrations used [2]. Generally, enzymatic methods produce higher cell yields and viabilities compared with mechanical disaggregation [15–18], but they may have deleterious phenotypic effects through the cleavage of cell-surface markers and/or activation of the cells [1, 19].
Given the diversity of tumor ECMs and the importance of cellular attachments, it has been argued that tumor disaggregation techniques should be individualized for their tissue of interest. However, to date, most disaggregation techniques have not been developed for, and evaluated in, specific types of human tumors; rather, a wide variety of enzymes have been used to digest solid tumors with the assumption that this particular enzymatic dissociation technique will provide high cell yield without affecting functional activity of the cell populations being studied.
Our group is studying the tumor microenvironment of human lung cancer. Given the potential problems with each method, we chose to test rigorously and validate various techniques to process our human NSCLC specimens. The most important factors that we considered were the project-dependent endpoints, including high immune cell yield and high cell viability, along with maintenance of key surface markers and functional characteristics. We found that currently available methods caused alterations in the true immune cell profile and thus, can provide misleading results. We present an optimized approach for lung cancer that maintains cell yields and preserves surface markers. A similar strategy can be developed for other tumors of interest.
MATERIALS AND METHODS
Patients and specimen collection
NSCLC samples from patients who had undergone surgical resection were collected at the Hospital of the University of Pennsylvania (Philadelphia, PA, USA). All patients selected for entry into the study met the following criteria: 1) a new diagnosis of histologically confirmed NSCLC; 2) no prior treatment, including surgery, chemotherapy, or radiation for lung cancer; and 3) no other known malignancy. Peripheral blood samples were obtained from HDs. This study was approved by the Institutional Review Board. Informed consent was obtained from each donor or patient before collection of samples.
The combined mechanical and enzymatic approach to tumor disaggregation
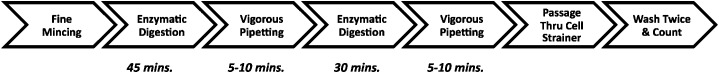
After the lung tumor was surgically removed, it was placed in wash media, i.e., Hyclone DMEM F12 media, supplemented with 2% FBS (Thermo Scientific, Waltham, MA, USA) and 1% penicillin-streptomycin (Life Technologies, Carlsbad, CA, USA) and brought immediately to the lab on ice. Samples were processed within 30–60 min of removal. The samples were then rinsed with serum-free Hyclone DMEM/F12 media, and under sterile conditions, the peritumoral, nonmalignant lung tissue and necrotic areas were removed with scissors. The remaining tumor tissue was split into equal parts (generally 0.5–1 g each) for disaggregation by the various methods. Each tumor fragment was then sliced into 1–2 mm3 pieces by sterile microdissection by use of curved scissors, fashioned with tungsten carbide inserts (Biomedical Research Instruments, Silver Spring, MD, USA). These pieces then underwent mechanical disaggregation (see Mechanical disaggregation below) or enzymatic digestion. For enzymatic digestion, tumor fragments were incubated in a sealed, 50 mL centrifuge tube with a final volume of 25 mL enzyme solution/0.5 g tissue and placed on a shaker with a speed of 85 rpm for 45 min at 37°C. After 45 min, tumor particles were pipetted vigorously by use of a 10 mL pipette to enhance disaggregation and then incubated further for another 30–50 min (based on the extent of digestion) under the same conditions. The sample was then pipetted vigorously one last time before being passed through a 70 μM nylon cell strainer (Fisher Scientific, Hampton, NH, USA). Gentle pressure was then applied with the black rubber bottom of a 5 mL syringe (Fisher Scientific) to any remaining, partially digested tissue on the cell strainer, which was then washed with 25 mL DPBS (Corning, Corning, NY, USA). Typically, <5% of the tissue (consisting of mainly acellular-connective tissue) remained on the strainer at the end of this step (see Scheme 1). The suspensions were centrifuged for 5 min at 300 g at room temperature, the supernatant aspirated, and the erythrocytes lysed by use of 1× RBC lysis buffer (Santa Cruz Biotechnology, Dallas, TX, USA). The remaining cells were washed twice in cell culture media with serum to deactivate any residual protease and collagenase. After the final wash, the cells were resuspended in cell-culture media (Hyclone DMEM/F12 media, supplemented with 10% FBS and 1% penicillin-streptomycin). Cells were counted on a hemocytometer. Viability was calculated by use of trypan blue (Sigma-Aldrich, St. Louis, MO, USA) exclusion or LIVE/DEAD cell staining by use of the Fixable Viability Dye eFluor 450 (eBioscience, San Diego, CA, USA).
Scheme 1. Combined M&E approach: a step-by-step description.
Enzymatic cocktails
Our custom-prepared LC-collagenase enzymatic cocktails all used Hyclone Leibovitz L-15 media, supplemented with 1% penicillin-streptomycin (Life Technologies) as the base. L-15 media are designed specifically for CO2-free culture conditions. The crude collagenases were all purchased from Worthington Biochemical (Lakewood, NJ, USA). Coll I has the original balance of collagenase, caseinase, clostripain, and tryptic activities; Coll II contains higher relative levels of protease activity, particularly clostripain; and Coll IV is designed to be especially low in tryptic activity. The concentrations of specific collagenases in all LC-collagenase cocktails were: Coll I (170 mg/L = 45–60 U/mL), Coll IV (170 mg/L = 45–60 U/mL), and Coll II (56 mg/L = 15–20 U/mL). The LC Collagenase cocktails differed only in their combinations of collagenases (see Fig. 1), not in the concentrations of Coll I, II, or IV. DNase I (25 mg/L = 50 K units/mL = 0.002%) and elastase (25 mg/L = 0.075 U/mL = 0.002%) were included in all LC collagenase cocktails. When used, the concentration of protease XIV (Sigma-Aldrich) was 250 mg/L (0.875 U/mL). The CAEC stock (Liberase, research grade; Roche Diagnostics, Indianapolis, IN, USA) was diluted to a concentration of 0.25 Wunsch U/mL in Leibovitz L-15 media. The HC Coll I cocktail was prepared by diluting Coll I in Leibovitz L-15 media to a concentration of 300 U/mL.
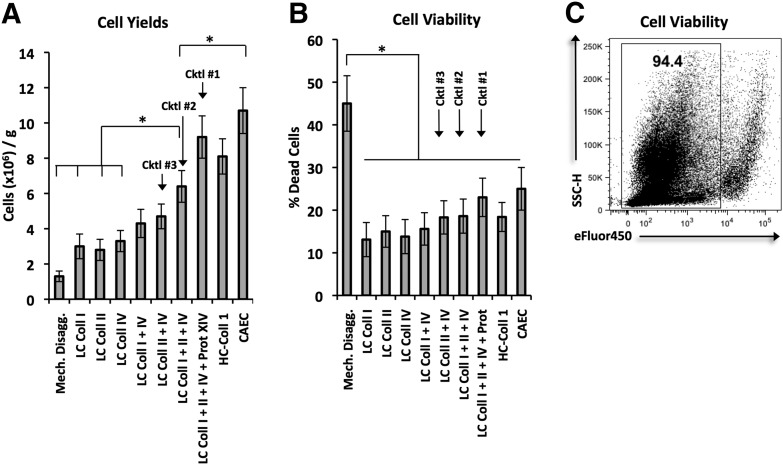
Figure 1. Cell yields and viabilities associated with various disaggregation methods in the combined M&E approach.
For bar graphs, each bar represents mean ± sem. Statistical analyses were performed with one-way ANOVA (*P < 0.05). Each method was tested on at least 4 tumor samples. (A) Average cell yields after disaggregation with multiple methods (millions/g). Tumors were divided into pieces of equal mass, disaggregated with different enzymatic cocktails, and the resulting single-cell suspensions counted via trypan blue exclusion, Cktl, Cocktail; Mech. disagg., mechanical disaggregation; Prot, protease. (B) Average percentage dead cells among disaggregation samples by use of trypan blue exclusion. (C) Cell viability after tumor digestion with Cocktail #2. Representative dot plots of disaggregated cells stained with Fixable Viability Dye eFluor 450 to detect live/dead cells. SSC-H, Side-scatter-height.
Mechanical disaggregation
Mechanical disaggregation was performed with the gentleMACS dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany), according to the “medium tumor” protocol set forth by the manufacturer. To replicate recent mechanical disaggregation results [4], the incubation steps with enzyme were omitted.
Peripheral blood processing, cell isolation, and enzymatic treatment
PBMCs and PBNs were isolated from the blood of HDs by use of Lymphoprep (AccuPrep, 1.077 g/ml; Nycomed, Oslo, Norway) density gradient centrifugation. In some experiments, CD11b+ PBMCs and CD15+ neutrophils were isolated by use of CD11b and CD15 MicroBeads, respectively, according to the manufacturer’s instructions (Miltenyi Biotec). T cells, from the PBMC fraction of HD whole blood, were isolated by use of T cell enrichment columns (R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s instructions.
Phagocytosis assay
Enzymatically treated or control CD15+ neutrophils (0.1 × 106) were incubated with pHrodo Escherichia coli BioParticles conjugates (Molecular Probes, Life Technologies). The cells were then incubated for 1 h at 37°C in 5% CO2. After incubation, the cells were washed twice with cell-culture media and resuspended in DPBS for flow cytometric analysis.
Cytotoxicity assay
One day before the experiment, 0.2 × 105 GFP-labeled A549 cells (a human lung carcinoma cell line) were plated per well in a sterile, tissue culture-treated Nunc F96 MicroWell black polystyrene plate (Thermo Scientific) in cell-culture media. On the day of the experiment, 0.2 × 105 enzymatically treated or control CD15+ neutrophils were added to the culture with PMA (10 ng/mL) or with PMA plus Apocynin (100 mM; both from Sigma-Aldrich). After a 24 h incubation, wells were washed to remove dead cells, 50 μL DPBS was added, and the remaining fluorescence was measured by use of the GloMax-Multi Detection System (Promega, Madison, WI, USA). Cytotoxicity was calculated according to the equation: [(GFP fluorescence of wells with tumor cells alone) – (GFP fluorescence of wells with added CD15+ neutrophils)]/(GFP fluorescence of wells with tumor cells alone) × 100.
Reactive oxygen species detection
The production of H2O2 by PBNs was measured by use of Amplex Red hydrogen peroxide/peroxidase assay kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions.
T Cell proliferation assay
Autologous PBMCs or T cells were isolated from the blood of HDs, as described above. CFSE-labeled T cells (0.1 × 106) or PBMCs were cocultured in cell-culture media with enzymatically treated or control CD11b+ PBMCs or CD15+ neutrophils at a 1:1 ratio in a 96-well U-bottom plate (Corning), coated with anti-CD3 antibody (1 µg/ml) and CD28 antibody (5 µg/ml). After 4 days, the T cells or PBMCs were collected and stained with monoclonal anti-CD3-allophycocyanin (BioLegend, San Diego, CA, USA). Proliferation was analyzed as the dilution of CFSE in CD3+-gated cells.
Flow cytometry
Flow cytometric analysis was performed according to standard protocols. Details about the antibodies used are listed in Supplemental Table 1. Matched isotype antibodies were used as controls. Data were acquired by use of the FACSCalibur or LSRFortessa flow cytometers (both from BD Biosciences, San Jose, CA, USA) and were analyzed by use of FlowJo software (Tree Star, Ashland, OR, USA).
Statistics
Unpaired Student’s t-tests were implemented to compare differences in continuous variables between two groups. ANOVA with post hoc testing was used for multiple comparisons. We considered differences statistically significant when P < 0.05. Graphical data were expressed as the mean ± sem. Statistical analysis was conducted by use of Stata Software (StataCorp LP, College Station, TX, USA).
RESULTS AND DISCUSSION
Comparison of current techniques in the disaggregation of human NSCLC
Multiple approaches have been reported previously to isolate immune cells from solid human tumors (Supplemental Table 2). However, unlike murine tumor models, no standardized techniques have been developed for the dissociation of human tumor tissue. We developed a panel of different methods to evaluate for the disaggregation of NSCLCs after conducting a literature search of the common techniques for solid human cancers (Supplemental Table 2). We included three standard techniques: HC-Coll I [4, 7, 20–23], a CAEC [24–27], and mechanical disaggregation alone [4]. We also hypothesized that combinations of enzymes in low concentration would synergize to optimize better cell yield, cell viability, and cell phenotype than would any enzyme alone or in HC. To test this hypothesis, we prepared 7 novel enzymatic cocktails (selected cocktail components outlined in Table 1) composed of different combinations of lLC-Coll I, II, and IV (composition outlined in Table 2) and protease XIV. In addition, we tailored these cocktails specifically for lung tissue by adding elastase to break down the significant amount of elastin in the lung.
TABLE 1.
Components of Cocktails #1–3, CAEC, and HC-Coll I
| Cocktail | Components |
|---|---|
| #1 | Coll I, II, IV; DNAse; elastase; protease XIV |
| #2 | Coll I, II, IV; DNAse; elastase |
| #3 | Coll II, IV; DNAse; elastase |
| CAEC | Coll I, II; neutral protease (thermolysin) |
| HC-Coll I | Coll I; DNAse; elastase |
TABLE 2.
Components of the crude collagenase preparations used in our custom-prepared LC-collagenase cocktails and HC-Coll I cocktail
| Collagenase | Components |
|---|---|
| I | Collagenase, caseinase, clostripain, and tryptic activities |
| II | Higher relative levels of protease activity (clostripain) |
| IV | Especially low in tryptic activity |
Our first objective was to evaluate these techniques based on maximum cell yield and cell viability. We found that cell yields increased as the number of enzymatic components increased (Fig. 1A). The addition of protease XIV to 3 collagenases (Cocktail #1) generated some of the highest cell yields, rivaling those of HC-Coll I and CAEC. Mechanical disaggregation and enzymatic cocktails composed of a single collagenase were associated with the lowest cell yields. In addition, mechanical disaggregation was associated with the lowest cell viability (Fig. 1B). Comparatively, every enzymatic cocktail resulted in significantly higher viabilities. Figure 1C demonstrates the high cell viability of single-cell suspension after tumor digestion with enzymatic cocktails. Based on the balance between high cell yield and high cell viability, we selected Cocktails #1–3 with multiple enzymes, HC-Coll I, and CAEC for further characterization of the effects on cell phenotype and function.
Cocktail #2 (LC-Coll I, II, IV) and Cocktail #3 (LC-Coll II, IV) preserve tumor-associated immune cell-surface markers
Our next objective was to determine the effect of tissue disaggregation on the expression of general tumor-associated immune cell-surface markers. We hypothesized that combinations of different collagenases at low concentrations would preserve these markers and that exposure to high levels of proteases with broad specificity (e.g., in Cocktail #1 and CAEC) would lead to the loss of these surface markers. For expression levels of surface markers, we used our results with mechanical disaggregation alone (by use of no enzymes) as our “gold standard,” as the mechanical disaggregation was not associated with any marker cleavage.
Lymphocyte markers
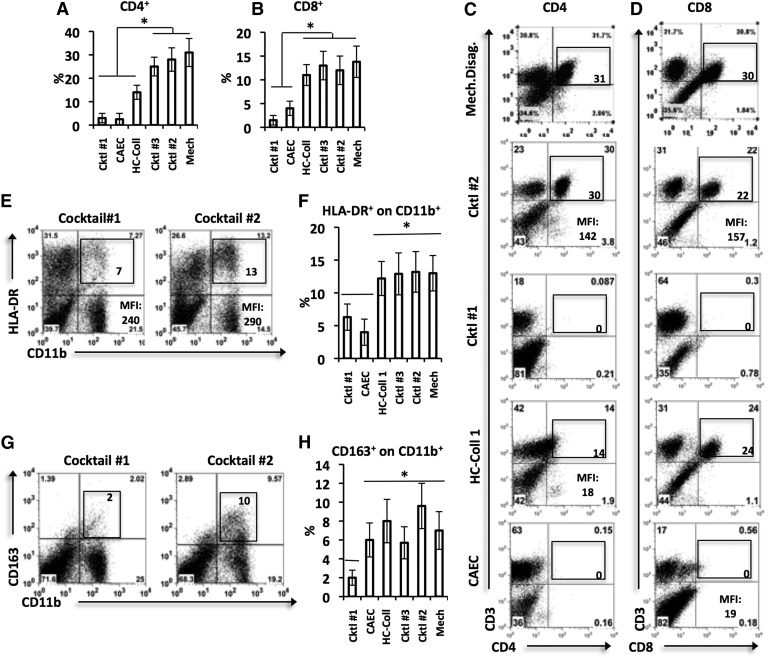
We investigated the lymphocytic markers CD3, CD4, and CD8 (T cells), CD45RO (antigen-experienced T cells), CD19 (B cells), and CD62L (marker of activation). Cocktail #1 (LC-Coll I, II, IV, protease XIV) and CAEC had the most negative impact on common lymphoid markers. Cocktail #1 and CAEC substantially cleaved CD4 (Fig. 2A and C; P < 0.05) and CD8 (Fig. 2B and D; P < 0.05). In addition, both Cocktail #1 and CAEC cleaved CD62L on CD3+ T cells (data not shown; P < 0.05). HC-Coll I had no effect on CD8 (Fig. 2B and D) but partially cleaved CD4 (Fig. 2A and C; P < 0.05). No significant differences existed between the CD4+ or CD8+ percentages or MFIs after disaggregation with Cocktail #2, Cocktail #3, or mechanical disaggregation. The expression of the CD45RO, CD3, and CD19 was not different among the groups (P > 0.05; data not shown).
Figure 2. The effects of multiple disaggregation methods on general tumor-associated immune cell-surface markers>.
Tumor tissue samples of equal mass underwent disaggregation with different enzymatic cocktails or mechanical disaggregation. The expression of the indicated surface markers within the disaggregation samples was then analyzed with flow cytometry on gated live CD45 cells. For bar graphs, each bar represents mean ± sem. Statistical analyses were performed with one-way ANOVA (*P < 0.05). Each method was tested on at least 3 tumor samples. (A and B) Graphical summaries of the proportion of CD4 and CD8 in the disaggregated tumor. (C and D) Representative dot plots displaying CD4 and CD8 expressions, respectively. One experiment of 4 is shown. Expression levels are marked in each gate, and MFIs are provided where appropriate. (E–H) Summary of the markers differentially cleaved by various disaggregation methods: HLA-DR on CD11b+ cells (E and F) and CD163+ on CD11b+ cells (G and H). Left panels show representative dot plots, and right panels summarize the data for all experiments.
Myeloid markers
We investigated the myeloid markers CD11b and CD33 (common myeloid lineage), CD15 and CD66b (neutrophils), CD14, HLA-DR (MHC class II), and CD163 (monocytes/macrophages). Cocktail #1 also had the broadest negative effects on the expression of common myeloid markers. Cocktail #1 partially cleaved HLA-DR (Fig. 2E and F) and CD163 (Fig. 2G and H) on CD11b+ cells, resulting in significantly decreased expression levels (P < 0.05). CAEC was milder than Cocktail #1, as its effects were limited to the partial cleavage of HLA-DR (Fig. 2F; P < 0.05). No significant differences existed among HLA-DR+, CD163+, or CD15+ percentages or MFIs after disaggregation with Cocktail #2, Cocktail #3, and mechanical disaggregation. The expression of the remaining myeloid markers, CD33, CD11b, and CD14, on CD11b cells was not different among the groups (P > 0.05; data not shown).
These findings suggest that enzymatic cocktails containing high levels of proteases with broad specificity protease XIV adversely affect the expression of cell-surface markers as a result of indiscriminate cleavage [18, 19]. Despite these effects, proteases with broad specificity, such as caseinase and clostripain, are included in crude collagenase preparations because of their ability to enhance the liberation of cell populations. Our results suggest that it should be a fine balance between collagenases or elastases with narrow specificity and proteases with broad specificity in an enzymatic cocktail; certain “milder” commercial collagenase preparations optimize this balance (Coll I and Coll IV), whereas other “harsher” preparations (Col II, CAEC, Cocktail #1) skew the balance, leading to inadvertent cell-marker damage. Whereas the 50–60 U/mL concentrations of Coll I and Coll IV that we used were 4- to 20-fold lower than levels reported in the literature [2–4, 6, 20, 28], we decreased the concentration of Col II by an additional factor of 3, as a result of its higher relative levels of proteases with broad specificity. In so doing, we intentionally limited the presence of proteases with broad specificity associated with this type of collagenase, which likely led to better maintenance of cell-surface marker expression associated with Cocktail #2.
In summary, Cocktail #2 and Cocktail #3 best preserved general immune cell markers. Given the slightly higher cell yield of Cocktail #2 compared with Cocktail #3, we moved forward with this cocktail and compared its phenotypic and functional effects more closely with those of CAEC and HC-Coll. Although mechanical disaggregation maintained the expression of lymphoid and myeloid markers, it consistently generated poor cell yields and cell viabilities (as demonstrated by Fig. 1A and B); thus, we did not proceed with further phenotypic and functional analysis by use of this method.
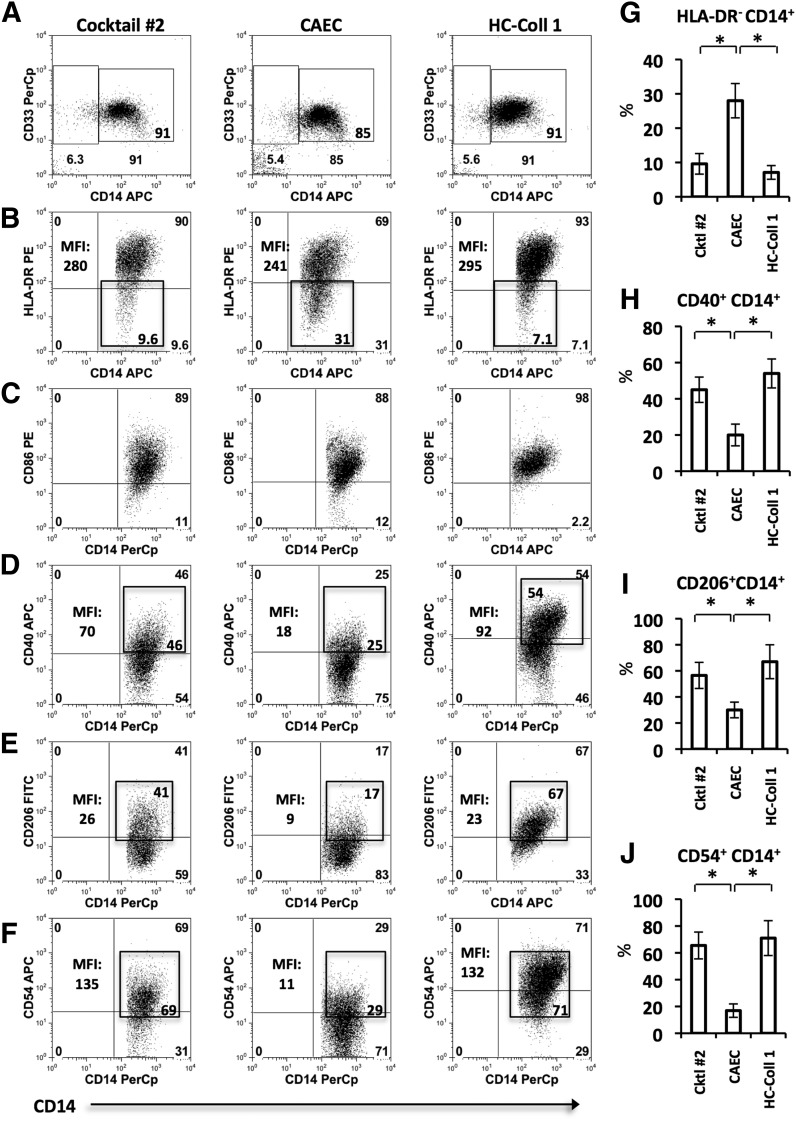
Given the recent interest in human tumor-associated APCs [29], defined as CD11b+CD15-CD14+CD33+ (Fig. 3A), we then compared the effects of CAEC, HC-Coll I, and Cocktail #2 on the expression of functionally significant surface markers in this population. We investigated the cell-surface proteins important in antigen presentation (HLA-DR), costimulation (CD86, CD40), adhesion (CD54), and mannose recognition (CD206). As demonstrated by the representative flow cytometry dot plots, CAEC partially cleaved HLA-DR+ (Fig. 3B and G), CD40+ (Fig. 3D and H), CD206+ (Fig. 3E and I), and CD54+ (Fig. 3F and J). There were no significant differences in marker expression between Cocktail #2 and HC-Coll I. Thus, relative to Cocktail #2 or HC-Coll I, CAEC had significant effects on key tumor-associated APC markers. Of particular importance is the artificial generation of a CD14+HLA-DR– cellular population (Fig, 3B). Recently isolated from the tumors of head and neck cancer patients, this population has been referred to as immunosuppressive monocytic myeloid-derived suppressor cells, based on its ability to inhibit T cell proliferation and IFN-γ production [27, 30].
Figure 3. The effect of multiple disaggregation methods on tumor-associated myeloid cell-surface markers.
Tumor tissue samples of equal mass underwent disaggregation with Cocktail #2 (LC-Coll I, II, IV), CAEC, or HC-Coll I, as described previously. The expression of indicated markers was investigated by use of flow cytometry. Each bar represents mean ± sem. Statistical analyses were performed with one-way ANOVA (*P < 0.05). (A) Allophycocyanin (APC in Fig. 3) gating strategy. Dot plots depict CD11b+CD15– cells; the CD14+CD33+ population was gated for further analysis. (B–F) Representative dot plots displaying the expression levels of HLA-DR, CD86, CD40, CD206, and CD54, respectively. One experiment of 4 is shown. Expression levels are marked in each gate, and MFIs are provided where appropriate. (G–J) Graphical summaries of the markers found to be differentially affected by disaggregation methods: HLA-DR-, CD40+, CD206+, and CD54+, respectively.
Our findings regarding the artificial cleavage of multiple cell-surface markers by certain disaggregation methods are significant. Not only may it help explain much of the lab-to-lab discrepancy present in the literature, but it’s also possible that such effects have led to false conclusions about the presence of specific cellular populations and their biologic characteristics. For example, the cell population characterized by CD14+HLA-DR– has been described recently as “immunosuppressive,” capable of inhibiting T cell proliferation and IFN-γ production [27, 31]. We found this population was artificially increased after treatment with CAEC as a result of reduced HLA-DR expression (CD14 expression was unchanged). On the other hand, we found another population, CD14+CD206+ macrophages, to be decreased after CAEC exposure as a result of partial cleavage of CD206 (a marker of M2-like tumor-associated macrophages).
Effects of digestion cocktails on the cell-surface marker expression profiles of peripheral blood leukocytes
Several groups have published data generated by the coincubation of PBMCs and blood neutrophils with enzymatic cocktails [4, 19, 32]. These studies provide a further test of the enzymes, as we could compare “digested” versus “nondigested” samples. Thus, our next goal was to investigate the effects of these enzymes on the cellular phenotypes of PBMCs and PBNs, which from 5 HDs, were treated with PBS (control), Cocktail #2, or CAEC for 45 min at 37°C before flow cytometric analysis.
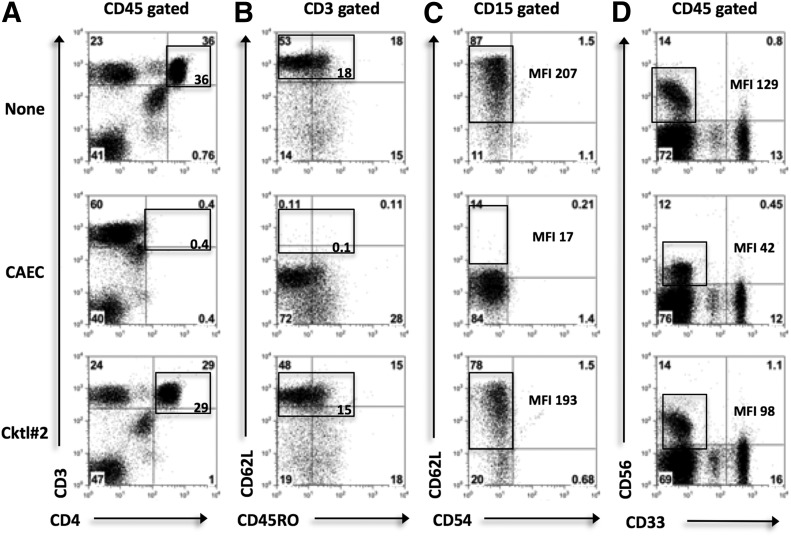
As shown in the representative dot plots, CAEC cleaved CD4 and CD62L (in the CD3+ fraction) on T cells (Fig. 4A and B) and CD62L on neutrophils (Fig. 4C), whereas Cocktail #2 had minimal effects on these markers. CAEC only partially cleaved the NK cell-surface marker CD56 (Fig. 4D). Whereas the CD56+ percentage was unchanged, as shown in the representative dot plots (Fig. 4D), the CAEC MFI was reduced significantly compared with DPBS and Cocktail #2 (P < 0.05). The expression levels of the other lymphoid markers analyzed were not significantly different among Cocktail #2, CAEC, and DPBS (Table 3). The only other myeloid marker cleaved solely by CAEC was CXCR2; its expression was reduced by 36% and was significantly lower than the expression levels after treatment with DPBS or Cocktail #2 (Table 4). CAEC and Cocktail #2 both partially cleaved CD40 and CXCR1.
Figure 4. The effect of enzymatic exposure on HD peripheral blood leukocyte surface markers.
PBMCs and PBNs were incubated with PBS [control (None)], Cocktail #2 (Coll I, II, IV), or CAEC for 30 min. Lymphocyte, myeloid, and NK cell-surface markers were then analyzed by use of flow cytometry. Statistical analysis was performed with one-way ANOVA. (A–D) Representative dot plots displaying the expression levels of CD4, CD62L on CD3+ cells, CD62L on CD15+ cells, and CD56, CD33 on CD45+ cells, respectively. One of 5 experiments is shown. Expression levels are marked in each gate, and MFIs are displayed where appropriate. Control marker expression was used as a baseline.
TABLE 3.
Tabular summary of the changes in expression levels of lymphoid markers
| Marker | CAEC | Cktl #2 | Marker | CAEC | Cktl #2 |
|---|---|---|---|---|---|
| CD3 | = | = | CD95/CD4 | = | = |
| CD4 | ↓↓ | = | CD95/CD8 | = | = |
| CD8 | ↓ | = | CD45RO/CD3 | = | = |
| CD62L/CD3 | ↓↓↓ | = | CCR5/CD3 | = | = |
| PD-1/CD4 | = | = | CCR7/CD3 | = | = |
| PD-1/CD8 | = | = | CD19 | = | = |
PD-1, Programmed death 1; =, 0-10%; ↓, 10-30%; ↓↓, 30-50%; ↓↓↓, >50%
TABLE 4.
Tabular summary of the changes in expression levels of myeloid markers
| Marker | CAEC | Cktl #2 |
|---|---|---|
| CD62L/CD15 | ↓↓↓ | = |
| CD62L/CD11b | N/A | = |
| HLA-DR/CD11b | = | = |
| CD11b | = | = |
| CD33 | = | = |
| CD14 | = | = |
| CD40/CD11b | ↓↓ | ↓ |
| CXCR1/CD15 | ↓↓↓ | ↓↓ |
| CXCR2/CD15 | ↓↓ | = |
=, 0-10%; ↓, 10-30%; ↓↓, 30-50%; ↓↓↓, >50%
Although both enzymatic cocktails demonstrated some degree of marker cleavage, our results confirm that Cocktail #2 has a less negative impact on the expression of cell-surface markers than CAEC, most likely as a result of reduced proteases with broad specificity and lower concentrations of collagenases.
Cocktail #2 has minimal effects on the functional activity of peripheral blood leukocytes
Together, our data have shown that the method of processing tissue has an impact on the surface marker-based phenotype of tumor-associated cellular populations. However, less clear is the effect of enzymatic protocols on the functional capacity of these populations. Given the results, suggesting that Cocktail #2 offers the best balance of cell yield, cell viability, and maintenance of cell-surface marker expression, we next evaluated the effect of enzymatic treatment with Cocktail #2 on immune cell functionality.
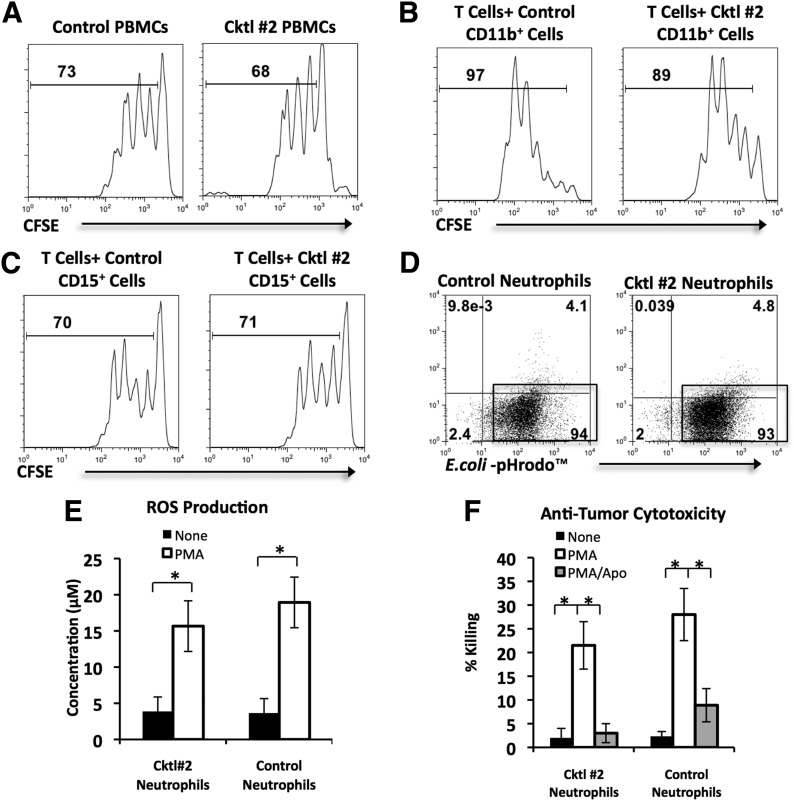
We compared enzymatically treated PBMCs and PBNs from HDs with PBS-treated cells. First, it has been reported that Coll I- and Coll IV-based enzymatic cocktails reduce the proliferative capacity of T cells in response to CD3 stimulation [4]. To prove that Cocktail #2 did not induce a similar effect on T cell proliferation, we cultured enzymatically treated and control CFSE-labeled PBMCs for 4 days in the presence of CD3 stimulation. There was no significant difference between the average proliferation of CD3+ T cells from enzymatically treated PBMCs and control PBMCs (Fig. 5A). Second, it is possible that enzymes may alter the ability of myeloid cells to regulate T cell responses. To address this issue, we cultured activated T cells with Cocktail #2-treated or control CD11b+ PBMCs, isolated from autologous whole PBMCs (Fig. 5B), or Cocktail #2-treated or control CD15+ neutrophils, isolated from whole blood (Fig. 5C). As demonstrated by the representative histograms (Fig. 5B and C), proliferation of T cells was unchanged when enzymatically digested CD11b+ PBMCs (P > 0.05) or CD15+ neutrophils (P > 0.05) were added in place of their respective controls, demonstrating that Cocktail #2 did not alter T cell proliferation or myeloid cell regulation of T cell proliferation. Furthermore, the enzymatic treatment of PBN did not affect the phagocytic activity of neutrophils and their ability to produce ROS after stimulation with PMA (Fig. 5D and E)). Neutrophils are known to be cytotoxic to tumor cells after stimulation with PMA. Figure 5F demonstrates that treatment of PBN with enzymatic cocktail did not change the ability of PMA-activated PBN to kill tumor cells (Fig. 5E). Thus, our results confirm that exposure to Cocktail #2 has minimal effects on the function of immune cells in multiple in vitro assays.
Figure 5. The effect of enzymatic exposure on the functional activity of HD peripheral blood leukocytes.
HD PBMCs and PBNs were incubated with PBS (control) or Cocktail #2 (Coll I, II, IV) for 30 min and then added to multiple in vitro functional assays. For bar graphs, statistical analysis was performed by Student’s t-tests (*P < 0.05). Each bar represents mean ± sem. (A) The effect of enzymatic exposure on T cell proliferation. CFSE-labeled control or Cocktail #2-treated PBMCs were stimulated with plate-bound anti-CD3 antibodies for 4 days. The proliferation of T cells was analyzed by CFSE dilution in the gated CD3+ cells. The histograms, representing 1 experiment of 5, show the percentage of dividing cells. (B and C) The effect of enzymatic exposure on myeloid cell regulation of T cell proliferation. CFSE-labeled T cells were stimulated with plate-bound anti-CD3/CD28 antibodies for 4 days in the presence of control or Cocktail #2-treated CD11b+ or CD15+ cells. The histograms, representing 1 experiment of 5, show the percentage of dividing T cells in the presence of CD11b+ cells or CD15+ cells. (D) Phagocytosis assay. Neutrophils were cocultured for 45 min with E. coli-pHrodo conjugates and then analyzed by flow cytometry. Representative flow cytometry dot plots from 1 of 5 experiments are shown. Percentages of cells that phagocytosed E. coli-pHrodo conjugates are displayed in gates. (E) ROS production by PMA-stimulated neutrophils, which were cultured in the presence or absence of PMA for 1 h before ROS concentration was measured in the supernatant by use of Amplex Red reagent. Graphical summary of 5 experiments is displayed. (F) Anti-tumor cytotoxicity of PMA-stimulated neutrophils. Neutrophils were cocultured in a 1:1 ratio with GFP-expressing A549 human NSCLC cells without PMA, in the presence of PMA, or the presence of PMA/apocynin (Apo) for 16 h. Graphical summary of 5 experiments is displayed.
Overall, our approach highlights the importance of careful analysis of the method of disaggregation to generate biologically valuable data. In this report, we present a rigorously tested disaggregation method for human NSCLCs. With the use of fresh human NSCLC samples, we showed that our optimal cocktail produces high cell yields without significantly compromising cell viability and maintains the expression of cell-surface markers and immune cell effector functions in the disaggregation of human lung tumor tissue. Unfortunately, no enzymatic cocktail is perfect with regard to every parameter. Although Cocktail #2 balanced cell yield and viability, its cell yield was not as high as CAEC, Cocktail #1, or HC-Coll I. The mild nature of this cocktail was also evident in the analysis of its effects on cell-surface markers, as it had some of the most minimal effects on the expression of common immune cell markers. Fortunately, this cocktail had no detectable impact on the performance of PBMCs and PBNs in multiple in vitro functional assays. We also processed 3 pleural mesotheliomas and 5 ovarian tumors to determine if our optimized cocktail for lung tumor could be used for other malignancies. Although our enzymatic cocktail was able to digest ovarian tumor tissue effectively (albeit with a longer incubation time), it was not effective in digesting mesotheliomas, which tend to be more fibrous. These findings indicate that different types of tumors will require a specific, empirically determined battery of enzymes to result in optimal tumor dissociation. Importantly, the analysis of the tumor dissociates after different combinations of enzymes will be necessary to ensure that this aspect of the experimental technique has been optimized for the investigated type of tumor.
In conclusion, we described a disaggregation method for human lung cancers that consists of a nontraumatic, gentle mechanical step, followed by enzymatic digestion with an optimized enzymatic cocktail composed of multiple low-concentration collagenases and elastase. This approach appears to minimize cleavage of cell-surface markers or changes in cell function, allowing more accurate analysis. Our data also suggest that a similar approach should be applied to any type of human tumors to be analyzed.
Supplementary Material
ACKNOWLEDGMENTS
J.G.Q. was supported by the U.S. National Institutes of Health (NIH) National Center for Research Resources (Grant TL1RR024133) and is now at the NIH National Center for Advancing Translational Sciences (Grant TL1R000138). S.S. was funded by the Lavin Family Foundation, CHEST Foundation, and NIH (R01 CA163256). Partial funding was provided by the Translational Center of Excellence in Lung Cancer, supported by the Penn Abramson Cancer Center. We thank Dr. Edmund Moon for providing the A549-GFP human lung carcinoma cell line.
Glossary
- CAEC
commercially available enzymatic cocktail
- CD62L
cluster of differentiation 62 ligand
- Coll I, II, IV
collagenases I, II, IV
- DPBS
Dulbecco’s PBS
- ECM
extracellular matrix
- HC
high concentration
- HD
healthy donor
- LC
low concentration
- M&E
mechanical and enzymatic
- MFI
mean fluorescence intensity
- NSCLC
nonsmall cell lung carcinoma
- PBN
peripheral blood neutrophil
- ROS
reactive oxygen species
Footnotes
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
AUTHORSHIP
E.E. and J.G.Q. conceived of the study and performed data analysis. Data acquisition was performed by P.B., J.G.Q., and S.S. Reagents were contributed by S.M.A., S.S., and W.W.H. Manuscript preparation was performed by J.G.Q. Manuscript revisions were performed by J.G.Q., E.E., S.S., S.M.A., and W.W.H. All authors read and approved the final manuscript.
DISCLOSURES
The authors declare no conflict of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
REFERENCES
- 1.Visscher D. W., Crissman J. D. (1994) Dissociation of intact cells from tumors and normal tissues. In Methods in Cell Biology (Darzynkiewicz Z., Robinson J. P., Crissman H. A., eds.), Academic Press, San Diego, CA, USA, 1–13. [DOI] [PubMed] [Google Scholar]
- 2.Allalunis-Turner M. J., Siemann D. W. (1986) Recovery of cell subpopulations from human tumour xenografts following dissociation with different enzymes. Br. J. Cancer 54, 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slocum H. K., Pavelic Z. P., Rustum Y. M., Creaven P. J., Karakousis C., Takita H., Greco W. R. (1981) Characterization of cells obtained by mechanical and enzymatic means from human melanoma, sarcoma, and lung tumors. Cancer Res. 41, 1428–1434. [PubMed] [Google Scholar]
- 4.Grange C., Létourneau J., Forget M. A., Godin-Ethier J., Martin J., Liberman M., Latour M., Widmer H., Lattouf J. B., Piccirillo C. A., Cailhier J. F., Lapointe R. (2011) Phenotypic characterization and functional analysis of human tumor immune infiltration after mechanical and enzymatic disaggregation. J. Immunol. Methods 372, 119–126. [DOI] [PubMed] [Google Scholar]
- 5.Petit V., Massonnet G., Maciorowski Z., Touhami J., Thuleau A., Némati F., Laval J., Château-Joubert S., Servely J. L., Vallerand D., Fontaine J. J., Taylor N., Battini J. L., Sitbon M., Decaudin D. (2013) Optimization of tumor xenograft dissociation for the profiling of cell surface markers and nutrient transporters. Lab. Invest. 93, 611–621. [DOI] [PubMed] [Google Scholar]
- 6.Novelli M., Savoia P., Cambieri I., Ponti R., Comessatti A., Lisa F., Bernengo M. G. (2000) Collagenase digestion and mechanical disaggregation as a method to extract and immunophenotype tumour lymphocytes in cutaneous T-cell lymphomas. Clin. Exp. Dermatol. 25, 423–431. [DOI] [PubMed] [Google Scholar]
- 7.Kedar E., Ikejiri B. L., Bonnard G. D., Herberman R. B. (1982) A rapid technique for isolation of viable tumor cells from solid tumors: use of the tumor cells for induction and measurement of cell-mediated cytotoxic responses. Eur. J. Cancer Clin. Oncol. 18, 991–1000. [DOI] [PubMed] [Google Scholar]
- 8.Langdon S. P. (2004) Isolation and culture of ovarian cancer cell lines. Methods Mol. Med. 88, 133–139. [DOI] [PubMed] [Google Scholar]
- 9.Daurkin I., Eruslanov E., Stoffs T., Perrin G. Q., Algood C., Gilbert S. M., Rosser C. J., Su L. M., Vieweg J., Kusmartsev S. (2011) Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res. 71, 6400–6409. [DOI] [PubMed] [Google Scholar]
- 10.Eruslanov E., Stoffs T., Kim W. J., Daurkin I., Gilbert S. M., Su L. M., Vieweg J., Daaka Y., Kusmartsev S. (2013) Expansion of CCR8(+) inflammatory myeloid cells in cancer patients with urothelial and renal carcinomas. Clin. Cancer Res. 19, 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamburger A. W., White C. P., Tencer K. (1982) Effect of enzymatic disaggregation on proliferation of human tumor cells in soft agar. J. Natl. Cancer Inst. 68, 945–949. [PubMed] [Google Scholar]
- 12.Von Hoff D. D., Casper J., Bradley E., Trent J. M., Hodach A., Reichert C., Makuch R., Altman A. (1980) Direct cloning of human neuroblastoma cells in soft agar culture. Cancer Res. 40, 3591–3597. [PubMed] [Google Scholar]
- 13.Cunningham R. E. (2010) Tissue disaggregation. Methods Mol. Biol. 588, 327–330. [DOI] [PubMed] [Google Scholar]
- 14.Costa A., Silvestrini R., Del Bino G., Motta R. (1987) Implications of disaggregation procedures on biological representation of human solid tumours. Cell Tissue Kinet. 20, 171–180. [DOI] [PubMed] [Google Scholar]
- 15.Rong G. H., Grimm E. A., Sindelar W. F. (1985) An enzymatic method for the consistent production of monodispersed viable cell suspensions from human solid tumors. J. Surg. Oncol. 28, 131–133. [DOI] [PubMed] [Google Scholar]
- 16.Besch G. J., Wolberg W. H., Gilchrist K. W., Voelkel J. G., Gould M. N. (1983) A comparison of methods for the production of monodispersed cell suspensions from human primary breast carcinomas. Breast Cancer Res. Treat. 3, 15–22. [DOI] [PubMed] [Google Scholar]
- 17.Pavelic Z. P., Slocum H. K., Rustum Y. M., Creaven P. J., Karakousis C., Takita H. (1980) Colony growth in soft agar of human melanoma, sarcoma, and lung carcinoma cells disaggregated by mechanical and enzymatic methods. Cancer Res. 40, 2160–2164. [PubMed] [Google Scholar]
- 18.Cerra R., Zarbo R. J., Crissman J. D. (1990) Dissociation of cells from solid tumors. Methods Cell Biol. 33, 1–12. [DOI] [PubMed] [Google Scholar]
- 19.Mulder W. M., Koenen H., van de Muysenberg A. J., Bloemena E., Wagstaff J., Scheper R. J. (1994) Reduced expression of distinct T-cell CD molecules by collagenase/DNase treatment. Cancer Immunol. Immunother. 38, 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt P. G., Robinson B. W., Reid M., Kees U. R., Warton A., Dawson V. H., Rose A., Schon-Hegrad M., Papadimitriou J. M. (1986) Extraction of immune and inflammatory cells from human lung parenchyma: evaluation of an enzymatic digestion procedure. Clin. Exp. Immunol. 66, 188–200. [PMC free article] [PubMed] [Google Scholar]
- 21.Takeo S., Yasumoto K., Nagashima A., Nakahashi H., Sugimachi K., Nomoto K. (1986) Role of tumor-associated macrophages in lung cancer. Cancer Res. 46, 3179–3182. [PubMed] [Google Scholar]
- 22.Perrot I., Blanchard D., Freymond N., Isaac S., Guibert B., Pachéco Y., Lebecque S. (2007) Dendritic cells infiltrating human non-small cell lung cancer are blocked at immature stage. J. Immunol. 178, 2763–2769. [DOI] [PubMed] [Google Scholar]
- 23.Chikamatsu K., Sakakura K., Toyoda M., Takahashi K., Yamamoto T., Masuyama K. (2012) Immunosuppressive activity of CD14+ HLA-DR- cells in squamous cell carcinoma of the head and neck. Cancer Sci. 103, 976–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Appelt A., Jany C., Heymer A. (2010) Cell isolation from lung tumor tissue. Euro. Biotechnol. News 9, 34–37. [Google Scholar]
- 25.Zhang B., Wang Z., Wu L., Zhang M., Li W., Ding J., Zhu J., Wei H., Zhao K. (2013) Circulating and tumor-infiltrating myeloid-derived suppressor cells in patients with colorectal carcinoma. PLoS ONE 8, e57114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akunuru S., Palumbo J., Zhai Q. J., Zheng Y. (2011) Rac1 targeting suppresses human non-small cell lung adenocarcinoma cancer stem cell activity. PLoS ONE 6, e16951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasquez-Dunddel D., Pan F., Zeng Q., Gorbounov M., Albesiano E., Fu J., Blosser R. L., Tam A. J., Bruno T., Zhang H., Pardoll D., Kim Y. (2013) STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J. Clin. Invest. 123, 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ensley J. F., Maciorowski Z., Pietraszkiewicz H., Klemic G., KuKuruga M., Sapareto S., Corbett T., Crissman J. (1987) Solid tumor preparation for flow cytometry using a standard murine model. Cytometry 8, 479–487. [DOI] [PubMed] [Google Scholar]
- 29.Quatromoni J. G., Eruslanov E. (2012) Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am. J .Transl. Res. 4, 376–389. [PMC free article] [PubMed] [Google Scholar]
- 30.Gabrilovich D. I., Nagaraj S. (2009) Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 9, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poschke I., Mougiakakos D., Hansson J., Masucci G. V., Kiessling R. (2010) Immature immunosuppressive CD14+HLA-DR-/low cells in melanoma patients are Stat3hi and overexpress CD80, CD83, and DC-sign. Cancer Res. 70, 4335–4345. [DOI] [PubMed] [Google Scholar]
- 32.Abuzakouk M., Feighery C., O’Farrelly C. (1996) Collagenase and dispase enzymes disrupt lymphocyte surface molecules. J. Immunol. Methods 194, 211–216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.