Abstract
Acetylcholine binding proteins (AChBPs) are unique spatial homologs of the ligand-binding domains of nicotinic acetylcholine receptors (nAChRs), and they reproduce some pharmacological properties of nAChRs. X-ray crystal structures of AСhBP in complex with α-conotoxins provide important insights into the interactions of α-conotoxins with distinct nAChR subtypes. Although considerable efforts have been made to understand why α-conotoxin GIC is strongly selective for α3β2 nAChR, this question has not yet been solved. Here we present the structure of α-conotoxin GIC in complex with Aplysia californica AChBP (Ac-AChBP) at a resolution of 2.1 Å. Based on this co-crystal structure complemented with molecular docking data, we suggest the key residues of GIC in determining its high affinity and selectivity for human α3β2 vs α3β4 nAChRs. These suggestions were checked by radioligand and electrophysiology experiments, which confirmed the functional role of detected contacts for GIC interactions with Ac-AChBP and α3β2 nAChR subtypes. While GIC elements responsible for its high affinity binding with Ac-AChBP and α3β2 nAChR were identified, our study also showed the limitations of computer modelling in extending the data from the X-ray structures of the AChBP complexes to all nAChR subtypes.
The discovery and crystallization of the acetylcholine-binding protein (AChBP) from a fresh-water mollusc Lymnaea stagnalis1,2 was a great step in understanding the structure and function of both nicotinic acetylcholine receptors (nAChR) and other ligand-gated ion channels belonging to the family of Cys-loop receptors. The first X-ray structure2, based on the disposition of the bound buffer molecule, provided the first ideas about the localization of the binding sites at the interface between the AChBP subunits and also between those of true nAChRs. It was confirmed by the subsequent X-ray structure of the AChBP complex with nicotine3, a classical agonist for most of the nAChRs subtypes. Interestingly, this structure revealed that nicotine attachment is accompanied by considerable movement of the AChBP loop C from the periphery to the central axis, and finally, nicotine appears to be embraced by this loop. In contrast, one year later, the X-ray structures for AChBP complexes with antagonists such as α-cobratoxin4 or α-conotoxin PnIA (A10L, D14K)5 demonstrated that the loop C is shifted to the periphery by more than 10 Å. In fact, the first X-ray structures for AChBP itself and its complexes with nicotine and α-cobratoxin have been established for Lymnaea stagnalis protein (Ls-AChBP), while the majority of the presently known AChBP complexes were solved with AChBP from the sea-water mollusc Aplysia californica (Ac-AChBP) (see recent reviews6,7,8,9,10). These two proteins have remarkable differences in their affinity both to α-neurotoxins and different α-conotoxins and this is one of the factors that makes a step from the high-resolution X-ray structure of the AChBP complexes to the expected structures of the distinct nAChRs binding the same ligands more difficult. Other complications are due to the limitations of computer modelling methods at the stage of going from the coordinates of the AChBP complexes to the precise organization of the analysed particular nAChR subtype.
Our work focused on the Ac-AChBP interaction with α-conotoxins. The advantage of α-conotoxins is that, among other antagonists, they are the most selective tools that allow distinguishing distinct subtypes of nAChRs6,7,8,9,10. In spite of the availability of the X-ray structures of Ac-AChBPs complexes with different α-conotoxins5,11,12,13, further work is required to understand the specificity of interactions of α-conotoxins with distinct nAChR subtypes with complete details. Here we analysed the α-conotoxin GIC bound to the neuronal α3β2 nAChR. These receptors are present in the human brain, are involved in functions such as cognition and, among other neuronal nAChRs, are considered to be promising drug targets (see reviews14,15,16). α-Conotoxin GIC from Conus geographus venom is an extremely interesting peptide that potently and selectively blocks neuronal α3β2 nAChRs at very low concentrations (IC50 1.1 nM). This high selectivity for the human neuronal α3β2 receptor makes it one of the most attractive cholinergic ligands found in recent years. Although many efforts have been made to understand the high selectivity of GIC for the human α3β2 nAChR17,18,19, the answer is still not clear. In this report, we present the co-crystal structure at a resolution of 2.1 Å of the α-conotoxin GIC in complex with Ac-AChBP as a further step to shed light on the selectivity mechanisms in nAChRs research.
Results
Overall structure of the complex
The Ac-AChBP expressed in insect cells was purified and co-crystallized with synthesized α-conotoxin GIC using the vapour diffusion sitting drop method. The crystals of the protein-peptide complex belong to the P212121 space group with cell dimensions of a = 78.6 Å, b = 84.9 Å, and c = 208.6 Å (Supplementary Table 1). The crystal structure was determined by molecular replacement and refined to a resolution of 2.1 Å (Supplementary Table 1).
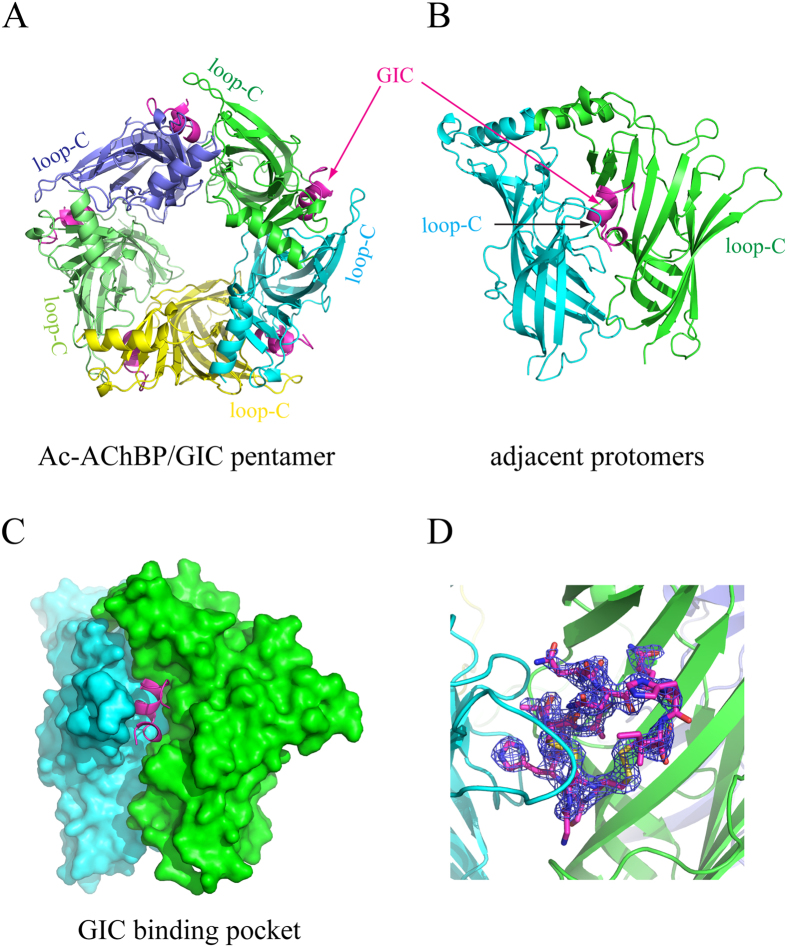
In the complex, the Ac-AChBP exists as a windmill-like pentamer bound with five α-conotoxin GIC peptides (Fig. 1A). Each ligand binding site in the Ac-AChBP is located between two adjacent protomers (Fig. 1B), and the five binding sites are structurally very similar due to the five-fold symmetry of the Ac-AChBP pentamer (Fig. 1A). In the complex, the bound α-conotoxins share a common orientation, with the central helix protruding into the ligand binding site of Ac-AChBP and the N and C termini of the bound α-conotoxin at the bottom and top parts of the ligand binding site, respectively (Fig. 1C,D).
Figure 1. The X-ray crystal structure of Ac-AChBP in complex with α-conotoxin GIC.
(A) The top view of the pentameric structure with five Ac-AChBP protomers, each in different colours and five α-conotoxin GIC molecules in magenta. (B) The side view of two adjacent protomers of the pentamer with a bound α-conotoxin GIC molecule (in magenta). (C) The side view of the surface model of two adjacent protomers with a bound α-conotoxin GIC molecule (in magenta) inside the binding pocket. (D) Fo-Fc electron density omit map contoured at 3.0 σ surrounding the GIC.
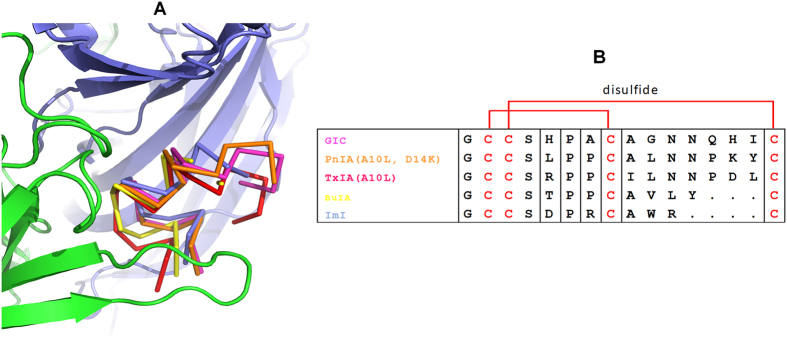
The pentameric Ac-AChBP/GIC complex is similar to previously reported complexes with other α-conotoxins. Upon structural superimposition, the Ac-AChBP/GIC complex structure had a RMSD of 0.71 Å for all paired Cα atoms compared with the Ac-AChBP/PnIA (A10L, D14K) complex (PDB code 2BR8)5, 0.54 Å with the Ac-AChBP/ImI complex (PDB code 2C9T and 2BYP)11,12, 0.58 Å with the Ac-AChBP/TxIA (A10L) complex (PDB code 2UZ6)13 and 0.60 Å with the Ac-AChBP/BuIA complex (PDB code 4EZ1). All five different α-conotoxins displayed a similar binding mode with the central helix of the peptide protruding into the binding site of Ac-AChBP (Fig. 2A), but the detailed interactions are different due to amino acid differences among the peptides (Fig. 2B). The structure of GIC alone has been previously determined by an NMR method (PDB code 1UL2)17. The backbone of the GIC (residues 2–16) was structurally conserved in the unbound and Ac-AChBP-bound states with a RMSD of 0.53 Å in this region.
Figure 2. Comparison of different α-conotoxins bound by Ac-AChBP.
(A) Backbone orientations observed in co-crystal structures of Ac-AChBP with five α-conotoxins. The backbone of GIC is shown in magenta, PnIA (A10L, D14K) in orange, ImI in light blue, TxIA (A10L) in red and BuIA in yellow. (B) Multiple sequence alignment of α-conotoxins GIC, PnIA (A10L, D14K), TxIA (A10L), ImI and BuIA. Disulfide bridges between Cys2-Cys8 and Cys3-Cys16 are shown in red.
Binding interface
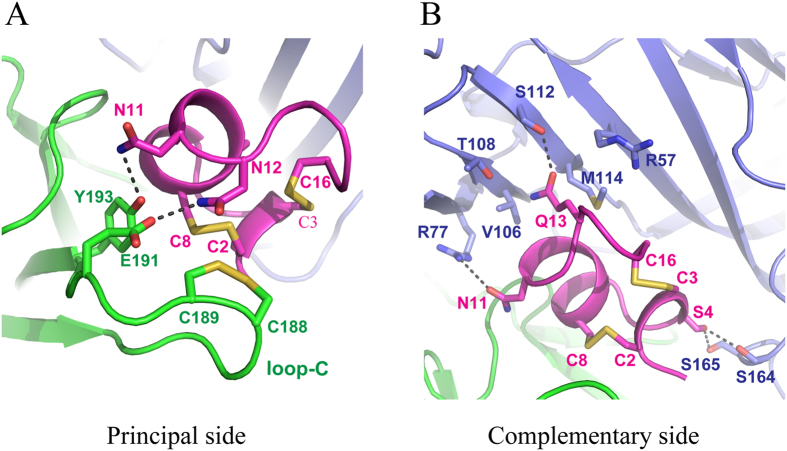
The GIC interacts with two adjacent Ac-AChBP protomers at the interface, forming the principal and complementary binding sides (Fig. 1B). Most interactions on the principal side were between the GIC and C loop (Gln-184 ~ Tyr-193) of one of the Ac-AChBP protomers (Fig. 1B and Table 1). The chemical nature of interactions at this side is a mixture of hydrophobic and hydrophilic interactions. The Cys-2/Cys-8 disulfide bridge of the GIC stacked onto the vicinal Cys-188/Cys-189 disulfide bond of the Ac-AChBP (Fig. 3A and Table 1). Other significant hydrophobic interactions included the contacts of Trp-145 and Tyr-193 of the Ac-AChBP with surrounding GIC residues including His-5, Pro-6, Ala-7, Cys-8, Asn-11 and Asn-12 (Table 1). Hydrogen-bonding interactions occurred between Asn-11 and Asn-12 of the GIC with Tyr-193 and Glu-191 of the Ac-AChBP, respectively (Fig. 3A). The complementary binding side was between the GIC and β-sheet of another Ac-AChBP protomer in the interface (Table 1). On this side, a notable interacting residue in the GIC was Gln-13, whose side chain resided in a pocket consisting of residues contacts with Arg-57, Val-106, Thr-108, Ser-112 and Met-114 of Ac-AChBP (Fig. 3B). Gln-13 also formed a hydrogen bond with Ser-112 of Ac-AChBP (Fig. 3B). On this side, additional hydrogen-bonding interactions included GIC Ser-4 to Ac-AChBP Ser-164 and Ser-165 and GIC Asn-11 to Ac-AChBP Arg-77 (Fig. 3B).
Table 1. Amino acid contacts with a distance cutoff 4.0 Å between α-conotoxin GIC and Ac-AChBP from the X-ray structure or α3β2 nAChR from modelling.
| GIC/Ac-AChBP crystal structure | GIC/α3β2-nAChR model | ||
|---|---|---|---|
| Principal side | |||
| GIC | Ac-AChBP | GIC | α3β2 |
| His-5 | Tyr-91 | His-5 | Tyr-93 |
| Pro-6, Ala-7 | Trp-145 | Pro-6, Ala-7 | Trp-149 |
| Ala-7 | Val-146 | Ala-7 | Ser-150 |
| Asn-11 | Ser-148 | Ala-7, Asn-11 | Asp-152 |
| Gly-1, Cys-2, His-5, Cys-8 | Tyr-186 | Gly-1, Cys-2, His-5, Cys-8 | Tyr-190 |
| Cys-2 | Cys-188 | Asn-12, Ile-15 | Cys-192 |
| Asn-12 | Cys-189 | Asn-12 | Cys-193 |
| Asn-11, Asn-12 | Glu-191 | Asn-11, Asn-12 | Glu-195 |
| His-5, Ala-7, Cys-8, Asn-11, Asn-12 | Tyr-193 | His-5, Cys-8, Asn12 | Tyr-197 |
| Complementary side | |||
| Ser-4 | Thr-34 | Ser-4 | Ser-38 |
| Ser-4, Pro-6 | Tyr-53 | Ser-4, His-5, Pro-6 | Trp-57 |
| Ala-9, Cys-16 | Gln-55 | ||
| Gln-13 | Arg-57 | Gln-13, Cys-16 | Glu-61 |
| Asn-11 | Arg-77 | Asn-11 | Arg-81 |
| Gly-10, Gln-13 | Val-106 | Gly-10, Gln-13 | Val-111 |
| Gln-13 | Thr-108 | ||
| Gln-13 | Ser-112 | Gln-13 | Ser-117 |
| Ala-9, Gly-10, Gln-13 | Met-114 | Ala-9, Gly-10, Gln-13 | Phe-119 |
| Pro-6, Ala-9 | IIe-116 | Leu-121 | |
| Cys-16 | Asp-157 | ||
| Ser-4 | Asp-162, Ser-164, Ser-165 | Ser-4 | Asp-171 |
Figure 3. Binding interface between GIC and Ac-AChBP.
(A) The disulfide bridge packing and hydrogen-bonding interactions (represented by black dashed line) on the principal side. Residues Asn-11 and Asn-12 of the GIC form hydrogen bonds with Tyr-193 and Glu-191 of the Ac-AChBP, respectively. Disulfide bond C2-C8 in the GIC closely packed together with C-188-C189 in the Ac-AChBP. (B) On the complementary side, the Gln-13 residue of GIC was located in a pocket surrounded by residues Arg-57, Val-106, Thr-108, Ser-112 and Met-114 of Ac-AChBP. Asn-11 of the GIC forms a hydrogen bond with Arg-77 of the Ac-AChBP. Ser-4 of the GIC also makes two hydrogen bonds with Ser-164 and Ser-165 of the Ac-AChBP.
Molecular docking and modelling of GIC complexes with the α3β2 nAChR subtype
GIC potently blocks the α3β2 subtype of human nAChR, showing the highest known selectivity (100,000-fold selectivity for the α3β2 subtype vs the muscle receptor of nAChR) for neuronal versus muscle subtypes of any nicotinic ligand characterized to date18. Using the co-crystal structure of Ac-AChBP/GIC as a template, we first modelled the structures of the α3 and β2 subunits and then generated a α3β2/GIC complex model. The interacting residues of the GIC and the α3β2 nAChR are listed in Table 1. The GIC-contacting residues in the α3β2 nAChR and Ac-AChBP are very similar in both position and chemical property of the side chain (Table 1). One notable change is the substitution of Ser-148 in the Ac-AChBP with Asp-152 in the α3β2 nAChR. In the α3β2/GIC model, the α3 subunit Asp-152 has more extensive interactions with GIC Asn-11 than the Ac-AChBP Ser-148 does in the Ac-AChBP/GIC crystal structure. Another notable change is the substitution of Arg-57 in the Ac-AChBP with Glu-61 in the α3β2 nAChR. The residue change would not significantly affect the GIC binding because Arg-57 in the Ac-AChBP and Glu-61 in the α3β2 nAChR are just involved in the formation of the pocket for GIC Gln-13, and both residues have no specific interactions with GIC in the Ac-AChBP/GIC crystal structure and α3β2/GIC model.
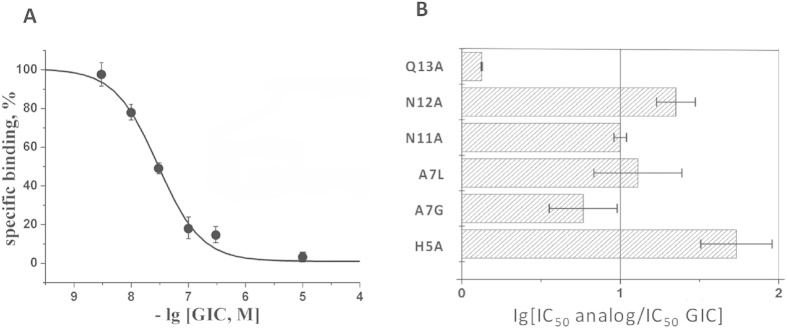
According to the structural and docking data, GIC His-5, Ala-7, Asn-11, Asn-12 and Gln-13 are residues that form the largest number of interactions with the Ac-AChBP or α3β2 nAChR (Table 1). Therefore, we chose these five sites and synthesized six GIC analogues (i.e., His5Ala, Ala7Gly, Ala7Leu, Asn11Ala, Asn12Ala and Gln13Ala) with single-site mutations in each analogue (Table 2). The wild-type GIC and these six analogues were synthesized by solid-phase peptide synthesis. The characterization of the synthetic GIC mutants by reverse-phase high performance liquid chromatography (HPLC) and mass spectrometry are shown in Supplementary Figure 1. All α-conotoxin GIC and its mutant peptides have CD spectra consistent with the CysI-III, CysII-IV disulphide isomer containing a helical character (See Supplementary Figure 2). We first studied the binding of these GIC analogues (in comparison with wild-type GIC) towards the Ac-AChBP by measuring their abilities to compete with [125I]-labelled α-bungarotoxin in a radioligand assay. In the dose response experiment, the wild-type GIC exhibited a complete inhibition of radioligand binding with IC50 = 29 ± 2 nM (Fig. 4A). Among the studied GIC mutants, only the Gln13Ala substitution essentially did not change the affinity for Ac-AChBP (Fig. 4B). The substitutions of Ala-7, Asn-11 and Asn-12 in GIC resulted in approximately one order drop in the binding affinity for Ac-AChBP (Fig. 4B). The most significant change was observed for the His5Ala mutation with an almost 100-fold increase in the IC50 value (Fig. 4B). We then tested the inhibition of the α3β2 nAChR by these GIC mutants in an electrophysiology experiment. All six GIC analogues except Gln13Ala showed a very significant decrease in the inhibition of the α3β2 nAChR compared with wild-type GIC (Table 2). The GIC analogues bearing the Ala7Leu or His5Ala mutation exhibited a more than 10,000-fold decrease in the inhibition (Table 2). These data further support our α3β2/GIC model built on the Ac-AChBP/GIC crystal structure.
Table 2. Amino acid sequences and blocking activities (in IC50s, nM) of α-conotoxin GIC and its analogues on expressed nAChR subtypes.
| Peptide | Sequence | Subtype | IC50 (nM) | Hill Slope | Subtype | IC50 (nM) |
|---|---|---|---|---|---|---|
| GIC | GCCSHPACAGNNQHIC* | hα3β2 | 1.13(1.10–1.16) | 0.764(0.559–0.969) | hα3β4 | 750(710–790) |
| GIC(Q13A) | GCCSHPACAGNNAHIC* | hα3β2 | 8.41(7.027–10.01) | 1.078(0.823–1.332) | hα3β4 | 660(630–690) |
| GIC(N12A) | GCCSHPACAGNAQHIC* | hα3β2 | 1980(840–4660) | 0.35(0.238–0.463) | hα3β4 | >10000 |
| GIC(N11A) | GCCSHPACAGANQHIC* | hα3β2 | 1600(545–4720) | 0.491(0.222–0.760) | hα3β4 | >10000 |
| GIC(A7L) | GCCSHPLCAGNNQHIC* | hα3β2 | >10000 | — | hα3β4 | >10000 |
| GIC(A7G) | GCCSHPGCAGNNQHIC* | hα3β2 | 4210(1566–11330) | 0.371(0.222–0.521) | hα3β4 | >10000 |
| GIC(H5A) | GCCSAPACAGNNQHIC* | hα3β2 | >10000 | — | hα3β4 | >10000 |
*C-terminal carboxamide.
Figure 4. The binding affinity of α-conotoxin GIC and its mutants to Ac-AChBP measured by a competitive radioligand assay.
(A) The inhibition curve for α-conotoxin GIC wild-type is shown. Each point is a mean ± s.e.m value of two measurements for each concentration of one experiment. The curve was calculated from the means ± s.e.m. using the ORIGIN 7.5 program. The calculated IC50 value is 29 ± 2 nM. (B) The bar diagram showing the differences in IC50 values for GIC mutants in comparison with wild-type peptides.
Potential structural basis for the high selectivity of GIC towards the α3β2 subtype
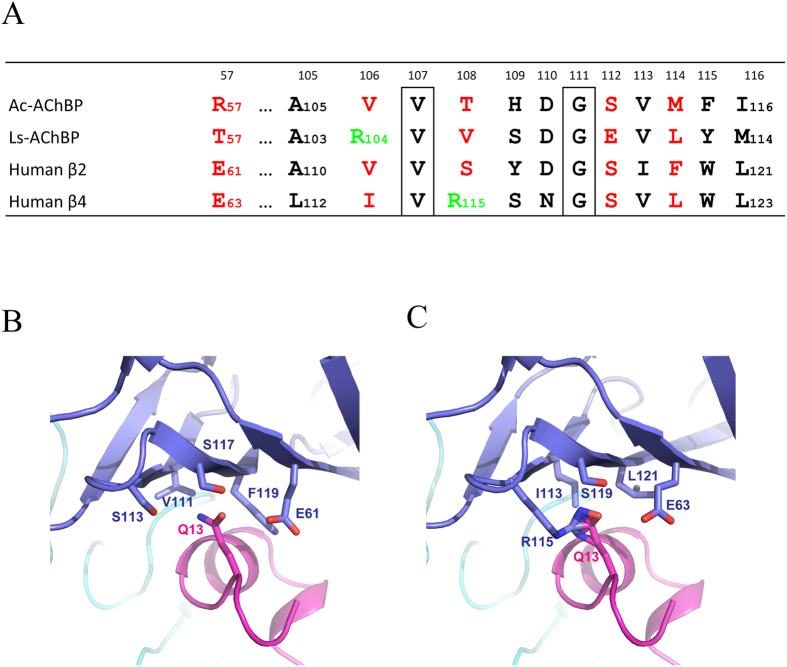
GIC exhibits a high affinity (~1 nM) to the α3β2 nAChR, but a much lower (~700 nM) affinity towards the α3β4 nAChR (Table 2). Sequence alignment of Ac-AChBP, AChBP from Lymnaea stagnalis (Ls-AChBP), as well as of human β2 and β4 nAChR subunits, are shown in Supplementary Fig. 3. The regions of nAChR subunits forming the binding sites are shown in Fig. 5A (summary from Supplementary Figure 3). With the structure of Ac-AChBP/GIC as a template, we also docked the GIC onto human α3β4 nAChR models. The comparison of the α3β2/GIC model, α3β4/GIC model and Ac-AChBP/GIC crystal structure allowed us to pinpoint the key amino acid residues on the complementary side, which appear to be responsible for selectivity of GIC for α3β2 vs α3β4. In the GIC/α3β2 model, the Gln-13 of GIC resides in a pocket surrounded by Glu-61, Val-111, Ser-113, Ser-117 and Phe-119 residues of the β2 subunit (Fig. 5B), which is very similar to the respective pocket formed by Arg-57, Val-106, Thr-108, Ser-112 and Met-114 residues on the complementary side of Ac-AChBP (Fig. 3B). No steric clashes were found between Gln-13 of GIC and the α3β2 receptor in the model, thus providing an explanation for a high affinity to both Ac-AChBP and the α3β2 receptor. Residues forming the pocket in the β4 subunit were changed to Glu-62, Ile-113, Arg-115, Ser-119 and Leu-121, which, except for Arg-115, were the same or similar to the respective residues in the β2 subunit (Fig. 5A). This position in the Ac-AChBP and Ls-AChBP are threonine and valine, respectively (Fig. 5A). Arg-115, which has a longer side chain compared to serine, threonine and valine, could bring steric hindrance in this pocket to accommodate Gln-13 of GIC and could even directly clash with it (Fig. 5C). The presence of arginine at this position would not favour the binding of the GIC with the α3β4 nAChR compared with the α3β2 nAChR, which is also represented by the decreased inhibition ability of GIC against the α3β4 receptor (Table 2).
Figure 5. Structure basis for the selectivity of GIC with α3β2 vs α3β4 nAChR.
(A) Primary sequence comparison of Ac-AChBP, Ls-AChBP, human β2 and human β4 residues forming the pocket on the complementary binding side to accommodate Gln-13 of the GIC. (B) The pocket in human β2 is able to accommodate Gln-13 of the GIC without steric clashes in the α3β2/GIC model. (C) Arg-115 in the pocket of human β4 could bring steric hindrance and even steric clashes with Gln-13 of the GIC and then disfavour the binding of α3β4 compared with α3β2 nAChR.
Discussion
Before this study, co-crystal structures of four different α-conotoxins in complexes with Ac-AChBP have been reported, namely for the PnIA variant (PDB code 2BR8)5, TxIA variant (PDB code 2UZ6)13, ImI (PDB code 2C9T and 2BYP)12 and BuIA (PDB code 4EZ1). All Ac-AChBP/α-conotoxin co-crystal structures are similar, but the residues interacting at the binding sites are different (Supplementary Table 2). As a result, different α-conotoxins exhibit a wide range in selectivity towards distinct nAChR subtypes, reflected in different pharmacological properties (Supplementary Table 3). The Ac-AChBP/TxIA (A10L) co-crystal structure showed that the Arg-5 of TxIA (A10L) forms a hydrogen bond with Tyr-186 and a salt bridge with Asp-195 residues of loop C, which account for the high binding affinity of this conotoxin variant to Ac-AChBP. A docking study of TxIA (A10L) onto α3β2 nAChR suggested that Arg-5 could have a salt bridge with Asp-197 and a hydrogen bond with Tyr-188 of the α3 subunit (corresponding to Asp-195 and Tyr-186 in Ac-AChBP)13. The Ac-AChBP/ImI co-crystal structure indicated that α-conotoxin Trp-10 residue could play a key role in binding to α3β2 nAChR. Substitution of Thr-57 in the β2 subunit by Lys-59 in the β4 subunit would result in steric clashes with the Trp-10 residue of ImI, which explains the decreased affinity for the α3β4 subtype7,12. α-Conotoxin GIC is one more α-conotoxin of strong selectivity for α3β2 nAChR. Compared with TxIA(A10L) and ImI, GIC does not contain the same respective Arg-5 or Trp-10 residues responsible for α3β2 binding and selectivity. Instead, it has His-5 and Gln-13 residues in these two positions.
Through the co-crystal structure of Ac-AChBP/GIC and the GIC docking to different nAChR subtypes, we came to the conclusion that His-5 and Gln-13 of GIC are important residues for its α3β2 nAChR binding and selectivity, respectively. These conclusions were further supported by the studies of GIC analogues in Ac-AChBP binding and α3β2 nAChR inhibition. The radioligand binding and electrophysiology experiments were different in sensitivity, but the results from both experiments revealed a common tendency: the His5Ala mutant showed the largest drop in ability for Ac-AChBP binding and for the α3β2 nAChR functional blocking, and the Gln13Ala substitution had the least influence on GIC activity in both experiments (Fig. 4B and Table 2). In accordance with the co-crystal structure, the His5Ala mutation led to a dramatic (by almost 2 orders of magnitude) decrease in affinity for Ac-AChBP. As shown in Table 1, the His-5 interacted extensively with Tyr-91, Tyr-186 and Tyr-193 of the Ac-AChBP and its mutation to Ala would lead to less interaction and decreased binding. In the α3β2/GIC model, His-5 of GIC also interacted with three tyrosines (i.e., Tyr-93, Tyr-190 and Tyr-197) of the α3 subunit. Therefore, its mutation to alanine would decrease its inhibitory effect by reducing the interactions with surrounding tyrosine residues in the α3 subunit. Similar to the His-5 position, substitutions of amino acid residues in Ala-7, Asn-11 and Asn-12 positions involved in the interactions on the principal side also had a significant impact on the activity of GIC in both assays (Fig. 4B and Table 2). Asn-11 and Asn-12 participate in the interaction with Ac-AChBP by forming hydrogen bonds with Glu-191 and Tyr-193 (Fig. 3A), and these two residues in Ac-AChBP were conserved in the α3 subunit. Ala-7 in GIC was different from the proline in PnIA, TxIA and BuIA and arginine in ImI. It formed hydrophobic interactions with Trp-145, Val-146 and Tyr-193 of Ac-AChBP. A change to leucine or glycine seems to break these interactions and thus reduce the activity of GIC (Fig. 4B and Table 2). These results, together with previous studies on other α3β2 nAChR selective α-conotoxins, suggested that the high affinity and specificity of α-conotoxin to the α3β2 nAChR subtype is determined basically by its interactions with the principal side of this receptor20.
In contrast, for Gln-13, which resides in a pocket formed by residues Arg-57, Val-106, Thr-108, Ser-112 and Met-114 on the complementary side of Ac-AChBP (Fig. 3B), its replacement for alanine did not significantly affect the activity in both assays (Fig. 4B and Table 2). Because the binding of GIC with Ac-AChBP or α3β2 nAChR is basically determined by multiple interactions on the principal side, a mutation of Gln-13, which is involved in the interactions on the complementary side, would not significantly affect the activity of GIC in Ac-AChBP binding and α3β2 nAChR inhibition. Previous studies have proposed that α-conotoxin residues on the complementary side, such as the Trp-10 in ImI, are important for the receptor selectivity of the toxin. Our docking studies with α3β2 vs α3β4 models also supported this suggestion. In the α3β2/GIC model, no steric clashes were found between Gln-13 of GIC and the β2 receptor subunit, which had a similar pocket consisting of Glu-61, Val-111, Ser-113, Ser-117 and Phe-119 (Fig. 5B). It provided an explanation for high affinity to both Ac-AChBP and α3β2 receptors by GIC. In contrast, the Arg-115 residue of the human β4 subunit, instead of threonine in Ac-AChBP and serine in the human β2 subunit (Fig. 5A), could have steric clashes with Gln-13 residues of GIC (Fig. 5C), which may explain the low affinity of GIC towards the α3β4 receptor subtype and its high selectivity of α3β2 vs α3β4. Our data from the co-crystal structure may also explain why Ls-AChBP shows a 10-fold decreased affinity for GIC in comparison with Ac-AChBP (Supplementary Table 3): the Arg-104 residue of Ls-AChBP (Val-106 Ac-AChBP) could also have steric clashes with the Gln-13 residue of GIC on the complementary side (Fig. 5A).
The models of α-conotoxin GIC complexes with α3β2 and α3β4 nAChRs were previously generated using methods of molecular docking 19. These models suggested that the GIC’s higher affinity for α3β2 vs α3β4 was due to the residues in the α3β2 subtype that are more closely located to the ligand than the respective residues in the α3β4 subtype. However, why the residues in the α3β2 subtype are situated more closely to GIC has not been explained. Our model based on the AChBP co-crystal structure allowed us to conclude that the major reason for α3β4 decreased affinity for GIC is probably the steric clashes between the Arg-115 residue of the β4 subunit and the peptide Gln-13 residue.
Notably, none of the substitutions led to an increase in the GIC affinity for α3β4 nAChR (Table 2). In general, our results demonstrated that more sophisticated computer methods are required to correctly transfer the data from the co-crystal structure of α-conotoxin or any other ligand, with a model like Ac-AChBP, to a true receptor. Even for the model as such, from the two most important GIC residues (His-5 and Gln-13), in our view, we experimentally could confirm only the role of His-5. This residue was also shown to be essential for recognition of α3β2 nAChR. It also should be noted that GIC has a high affinity both for Ac-AChBP and α3β2 nAChR, and the importance of one or another GIC residue was demonstrated by a decrease in the affinity of the chosen mutants. However, in the case of α3β4 nAChR, the task was the opposite: it was expected that the chosen GIC substitutions will raise the affinity to those subtypes. In general, increasing the affinity (or activity) is always a much more challenging task. Apparently, the co-crystal structure of GIC with Ac-AChBP is a good starting point and only extensive subsequent creation of substituted α-conotoxin libraries and a broad mutagenesis of the particular nAChR of interest will unravel the fine mechanisms of binding and might assist the design of more potent and selective antagonists and agonists.
In summary, we first determined the crystal structure of GIC with Ac-AChBP. Based on this structure, we obtained a more accurate model of α-conotoxin GIC complexed with α3β2 and also presented possible explanations for the high selectivity of GIC for α3β2. We believe that these results will help develop a better understanding of the α-conotoxin selectivity for distinct nAChR subtypes and may facilitate the design of α-conotoxin analogues for therapeutic purposes.
Materials and Methods
Chemical synthesis of GIC and its mutants
The linear peptides were assembled by solid-phase methodology on an ABI 433A peptide synthesizer (Applied Biosystems Inc., Foster City, CA, USA) using FastMoc [N-(9-fluorenyl)methoxycarbonyl] chemistry and standard side-chain protection, except for cysteine residues. A two-step oxidation protocol was used to fold the peptide selectively, as described previously20. Briefly, to form first disulfide bridge between Cys2 and Cys8, the peptides were added slowly to an equal volume of 20 mM potassium ferricyanide K3[Fe(CN)6] and 0.1 M Tris base, with the pH adjusted to 7.5 with acetic acid. The solution was mixed to react for 45 min, and the monocyclic peptide was purified by reverse-phase HPLC. Simultaneous removal of the S-acetamidomethyl groups and closure of the disulfide bridge between Cys3 and Cys16 was carried out by iodine oxidation, the bicyclic peptide was purified by HPLC on a reversed-phase C18 Vydac column (Hesperia, CA, USA) using a linear gradient of 10–40% B90 in 30 min. Solvent B was 90% ACN, 0.092% TFA, and H2O; Solvent A was 0.1% TFA in H2O. Analytical reversed phase-HPLC (Waters ACQUITY UPLC H-Class) and ESI -IT-TOF (Shimadzu, Kyoto, Japan) mass spectrometry confirmed the purity and molecular mass of the synthesized peptides. The purity and correctness of the structure of synthesized products were checked by HPLC re-chromatography and ESI mass-spectrometry (Fig. 1 Supplementary).
Protein expression and purification
The Ac-AChBP was expressed using the Bac-to-Bac baculovirus expression system (Invitrogen). Spodoptera frugiperda Sf9 insect cells were maintained in Insect-Xpress protein-free medium (Lonza) without serum. Full length Ac-AChBP with a C-terminal 6× His tag was cloned into the pFastBac-Dual vector (Invitrogen). The construct was transformed into bacterial DH10Bac competent cells, and the extracted bacmid was transfected into Sf9 cells using Cellfectin II Reagent (Invitrogen). The low-titre viruses (P0) were harvested after incubation of the transfected cells at 26 °C for 7 ~ 9 d and then amplified to generate a high-titre virus stock. The amplified high-titre viruses (P1) were used to infect 1 L of Sf9 cells at a density of 2 × 106 cells/ml. The supernatant of the cell culture containing soluble Ac-AChBP was harvested 48 h after infection and concentrated and buffer-exchanged to HBS (10 mM Hepes, pH 7.2, 150 mM NaCl). Ac-AChBP was captured by Nickel-charged resin (GE Healthcare) and eluted with 500 mM imidazole in HBS buffer (pH 7.2). Further purification was performed by gel-filtration chromatography using the Superdex 200 column (GE Healthcare).
Crystallization and data collection
Purified Ac-AChBP and synthesized α-conotoxin GIC were mixed at a molar ratio 1:1.5 at 4 °C for 2 h and then loaded to a Superdex 200 column. The complex was collected and concentrated to ~20 mg/ml in HBS buffer for crystallization. Crystals were successfully grown at 18 °C using the sitting drop vapour diffusion method by mixing equal volumes of protein and reservoir solution containing 1.5 M lithium sulfate monohydrate, 0.1 M Tris, PH 8.5. Crystals were cryocooled in liquid nitrogen with cryoprotectant (reservoir solution plus 20% (v/v) glycol) before data collection. Diffraction data were collected at the BL17U beam line of the Shanghai Synchrotron Research Facility (SSRF). Diffraction data were indexed, integrated and scaled with the program HKL200021.
Structural determination and refinement
The structure was determined by the molecular replacement method with PHASER22 in CCP4 suite23. The search model was Ac-AChBP–PnIA (D14K, A10L) (PDB code 2BR8). Iterative structural refinement was performed with the program PHENIX24 and the structure validation was performed by the program PROCHECK25. All structural figures were made with PyMol26.
Molecular modelling, docking and simulations
All the modelling, docking and simulations were performed in Discovery Studio Client 4.0 (Accelrys, San Diego, CA). The molecular models of extracellular ligand-binding domains of the human nAChRs such as (α3)2(β4)3, (α3)2(β2)3 were generated based on the template of Ac-AChBP structure using the homology modelling program Modeler version 9.0. The GIC docking was based on the reference model of the Ac-AChBP/GIC complex. The models were refined with a side-chain refinement and energy minimization process. The CHARMm forcefield was used for all simulations. All modelling and docking structures were verified by the program Profiles-3D in the Discovery Studio platform27, as well as by the MolProbity server28,29.
Competitive radioligand assay
For competitive radioligand assays we used 6× His tagged Ac-AChBP and Ls-AChBP, kindly provided by Prof. A.B. Smit (Free University of Amsterdam, the Netherlands), which were expressed and purified mainly as described above for Ac-AChBP. The competition experiments with [125I]-labelled α-bungarotoxin were carried out mainly as described in ref. 30. Shortly, α-conotoxin GIC or its mutants (in a concentration range of 0.03–100 μM) were pre-incubated 3 h at room temperature with the Ls- or Ac-AChBPs (the final protein concentration of 2.4 nM or 140 nM, respectively) in 50 μL of binding buffer (20 mM Tris-HCl buffer, 1 mg/mL of bovine serum albumin, pH 7.9). Next, [125I]- labelled α-bungarotoxin was added to final concentration 0.2 nM and the mixtures were additionally incubated for 5 min. Binding was stopped by rapid filtration on double DE-81 filters (Whatman) pre-soaked in binding buffer, unbound radioactivity being removed from the filters by washout (3 × 3 mL) with the binding buffer. Non-specific binding was determined in all cases using 3 h pre-incubation with 10 μM α-cobratoxin from Naja kaouthia venom.
The binding results were analysed using ORIGIN 7.5 (OriginLab Corporation, Northampton, MA, USA) fitting to a one-site dose-response curve by the equation: % response = 100/{1 + ([toxin]/IC50)n, where IC50 is the concentration at which 50% of the binding sites are inhibited and n is the Hill coefficient30.
Electrophysiology measurements and data analysis
The cRNAs of nAChR subunits (kindly provided by S. Heinemann, Salk Institute, San Diego, CA, USA) were obtained by in vitro transcription using the mMessage mMachine SP6 kit (Ambion, Austin, TX, USA). The MEGAclearTM kit (Ambion) was used to purify the cRNAs. Oocytes of Xenopus laevis were prepared and injected with capped RNA (cRNA) to express human α3β2, and human α3β4 nAChRs. Oocytes were injected within one day of harvesting and recordings were made 1–4 days post-injection, as described previously18. Briefly, oocytes were transferred to the recording chamber (~50 μL in volume) and gravity-perfused at 2 mL/min with ND-96 buffer containing 0.1 mg/mL bovine serum albumin (BSA). ACh-gated currents were obtained with a two-electrode voltage-clamp amplifier (Axoclamp 900A, Molecular Devices Corp., Sunnyvale, CA, USA). Oocytes were voltage clamped at −70 mV at room temperature. The continuous gravity perfused with standard ND-96 solution and stimulated with 2-s pulses of ACh once every minute. A total of 5 μL of different concentration toxins were placed in the chamber for 5 min, and then the perfusion system was applied during which 2-s pulses of 100 μM ACh were applied every minute until a constant level of blockage was achieved. The final results were acquired for at least 4 oocytes. The dose-response data were fitted to the equation: % Response = 100/{1 + ([toxin]/IC50)nH}, Each data point of the dose-response curve represents the average ± S.E. of 4 to 6 oocytes, where nH is the Hill coefficient, by nonlinear regression analysis using GraphPad Prism (GraphPad Software, San Diego, CA, USA)18.
PDB deposition
The coordinates and diffraction data have been deposited into the Protein Data Bank with accession code 5CO5.
Additional Information
How to cite this article: Lin, B. et al. From crystal structure of α-conotoxin GIC in complex with Ac-AChBP to molecular determinants of its high selectivity for α3β2 nAChR. Sci. Rep. 6, 22349; doi: 10.1038/srep22349 (2016).
Supplementary Material
Acknowledgments
This work was supported in part by the International Science & Technology Cooperation Program of China (2011DFR31210), National Natural Science Foundation of China (81420108028, 41366002, 31470751 and 31560243), State High-Tech Research and Development Project (863) of the Ministry of Science and Technology of China Grant 2012AA021706, and Changjiang Scholars and Innovative Research Team in University Grant IRT_15R15. Igor E. Kasheverov participation was supported by RFBR grant # 15-04-08338 and for Victor I. Tsetlin by RSF grant # 16-14-00215.
Footnotes
Author Contributions B.L., M.X., S.X., W.X. and S.L. designed research; B.L., M.X., X.Z., Y.W., D.Z., Y.H., I.K. and W.X. performed research; I.K., X.L., V.T., W.X. and S.L. analysed data; S.X. was responsible for the computer simulations. B.L., M.X., S.X., V.T., W.X. and S.L. wrote the paper. All authors contributed to reading and editing the manuscript.
References
- Smit A. B. et al. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature 411, 261–8 (2001). [DOI] [PubMed] [Google Scholar]
- Brejc K. et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–76 (2001). [DOI] [PubMed] [Google Scholar]
- Celie P. H. et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–14 (2004). [DOI] [PubMed] [Google Scholar]
- Bourne Y., Talley T. T., Hansen S. B., Taylor P. & Marchot P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J 24, 1512–22 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celie P. H. et al. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an α-conotoxin PnIA variant. Nat Struct Mol Biol 12, 582–588 (2005). [DOI] [PubMed] [Google Scholar]
- Kasheverov I. E., Utkin Y. N. & Tsetlin V. I. Naturally occurring and synthetic peptides acting on nicotinic acetylcholine receptors. Curr Pharm Des 15, 2430–2452 (2009). [DOI] [PubMed] [Google Scholar]
- Rucktooa P., Smit A. B. & Sixma T. K. Insight in nAChR subtype selectivity from AChBP crystal structures. Biochem Pharmacol 78, 777–787 (2009). [DOI] [PubMed] [Google Scholar]
- Tsetlin V., Utkin Y. & Kasheverov I. Polypeptide and peptide toxins, magnifying lenses for binding sites in nicotinic acetylcholine receptors. Biochem Pharmacol 78, 720–31 (2009). [DOI] [PubMed] [Google Scholar]
- Nys M., Kesters D. & Ulens C. Structural insights into Cys-loop receptor function and ligand recognition. Biochem Pharmacol 86, 1042–53 (2013). [DOI] [PubMed] [Google Scholar]
- Lebbe E. K., Peigneur S., Wijesekara I. & Tytgat J. Conotoxins targeting nicotinic acetylcholine receptors: an overview. Mar Drugs 12, 2970–3004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. B. et al. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J 24, 3635–3646 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulens C. et al. Structural determinants of selective α-conotoxin binding to a nicotinic acetylcholine receptor homolog AChBP. Proc Natl Acad Sci USA 103, 3615–3620 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S. et al. AChBP‐targeted α‐conotoxin correlates distinct binding orientations with nAChR subtype selectivity. EMBO J 26, 3858–3867 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst R., Rollema H. & Bertrand D. Nicotinic acetylcholine receptors: from basic science to therapeutics. Pharmacol Ther 137, 22–54 (2013). [DOI] [PubMed] [Google Scholar]
- Dineley K. T., Pandya A. A. & Yakel J. L. Nicotinic ACh receptors as therapeutic targets in CNS disorders. Trends Pharmacol Sci 36, 96–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoli M., Pistillo F. & Gotti C. Diversity of native nicotinic receptor subtypes in mammalian brain. Neuropharmacology 96, 302–11 (2015). [DOI] [PubMed] [Google Scholar]
- Chi S., Kim D., Olivera B., McINTOSH J. & Han K. Solution conformation of alpha-conotoxin GIC, a novel potent antagonist of alpha3beta2 nicotinic acetylcholine receptors. Biochem. J 380, 347–352 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. M. et al. Alpha-conotoxin GIC from Conus geographus, a novel peptide antagonist of nicotinic acetylcholine receptors. J Biol Chem 277, 33610–5 (2002). [DOI] [PubMed] [Google Scholar]
- Lee C., Lee S. H., Kim D. H. & Han K. H. Molecular docking study on the alpha3beta2 neuronal nicotinic acetylcholine receptor complexed with alpha-conotoxin GIC. BMB Rep 45, 275–80 (2012). [DOI] [PubMed] [Google Scholar]
- Hone A. J. et al. Positional Scanning Mutagenesis of α-Conotoxin PeIA Identifies Critical Residues That Confer Potency and Selectivity for α6/α3β2β3 and α3β2 Nicotinic Acetylcholine Receptors. J Biol Chem 288, 25428–25439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. & Minor W. Processing of X-ray diffraction data collected in oscillation mode. Vol. 276, 307–326 (Elsevier, 1997). [DOI] [PubMed]
- McCoy A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–3 (1994). [DOI] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58, 1948–54 (2002). [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., Macarthur M. W., Moss D. S. & Thornton J. M. {PROCHECK}: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993). [Google Scholar]
- DeLano W. L. Pymol Molecular Graphics System. DeLano Scientific, San Carlos, California, USA (2002). [Google Scholar]
- Eswar N. et al. Comparative protein structure modelling using Modeller. Curr Protoc Bioinformatics Chapter 5, Unit 5 6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I. W., Murray L. W., Richardson J. S. & Richardson D. C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32, W615–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35, W375–83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasheverov I. E., Zhmak M. N., Khruschov A. Y. & Tsetlin V. I. Design of new alpha-conotoxins: from computer modeling to synthesis of potent cholinergic compounds. Mar Drugs 9, 1698–714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.