Abstract
Tumor spheroids are becoming an important tool for the investigation of cancer stem cell (CSC) function in tumors; thus, low-cost and high-throughput methods for drug screening of tumor spheroids are needed. Using neurospheres as non-adherent three-dimensional (3-D) cultures, we developed a simple, low-cost acridine orange (AO)–based method that allows for rapid analysis of live neurospheres by fluorescence microscopy in a 96-well format. This assay measures the cross-section area of a spheroid, which corresponds to cell viability. Our novel method allows rapid screening of a panel of anti-proliferative drugs to assess inhibitory effects on the growth of cancer stem cells in 3-D cultures.
Keywords: neurospheres, tumor spheroids, cancer stem cell, glioblastoma, acridine orange, microscopy
Solid tumors grow in a three-dimensional (3-D) spatial conformation, which is not mimicked by two-dimensional (2-D) monolayer cultures. Non-adherent tumor spheroids are commonly used as 3-D in vitro models in cancer research to provide an intermediate between conventional adherent cancer cell cultures and in vivo xenograft models (1). In addition to providing a 3-D model, tumor spheroids represent an important tool for studying and expanding cancer stem cell (CSC) populations derived from patient samples or established cancer cell lines. CSCs represent a challenge for cancer therapy, as they are often resistant to current therapies (2). Thus, CSCs grown as spheroids have become an important tool to investigate drugs for their potential to inhibit therapy-resistant CSC function.
Recently, novel high-throughput methodologies for studying tumor spheroids have been developed using luminescent, colorimetric, or fluorescent viability reagents to study a variety of tumor spheroid functions such as motility and invasion (3), effects of co-culture of different cell types (4,5), and hypoxia (6). However, most microscopic high-throughput analyses relying on fluorescent probes require removal of the probe from the supernatant before microscopy. For instance, when fluorescein diacetate (FDA) is used as a viability dye, the culture medium, which contains esterases from dead cells, needs to be removed because it can result in a high background signal (7). As tumor spheroids are non-adherent floating structures, removal of excess probe from the supernatant is difficult and may compromise tumor spheroid integrity. Additionally, common cell viability reagents can be costly (see Supplementary Table S1).
Here we present a convenient, low-cost method for spheroid analysis using fluorescent probes and microscopy. We used acridine orange (AO), a cell-permeable organic compound that emits light in the red and orange spectrums and has been used before to stain and analyze multicellular spheroids (8). When AO is combined with single-stranded RNA, AO dimers are created, and the AO emission maximum shifts to red (640 nm) (9). However, when it intercalates into double-stranded DNA, AO retains its monomeric properties, its fluorescence yield and lifetime increase more than 2-fold, and its emission maximum shifts to 525 nm (within the green spectrum) (9,10). As tumor spheroids are detected by DNA-bound AO in the green [fluorescein isothiocyanate (FITC)] channel (525 nm), removal of excess probe is not required, making AO an ideal tool for visualizing non-adherent, floating spheroids. Additionally, AO is very cost-effective compared to other dyes. Using our AO-based method, the staining cost for 1000 assays is $0.007, which is more than 5000 times lower than that of other dyes (for cost-comparison of dyes used for spheroid analysis, see Supplementary Table S1).
METHOD SUMMARY
Here we report a new low-cost and effective method for analysis of acridine orange–stained 3-D tumor spheroids by rapid-throughput fluorescence microscopy in a 96-well format.
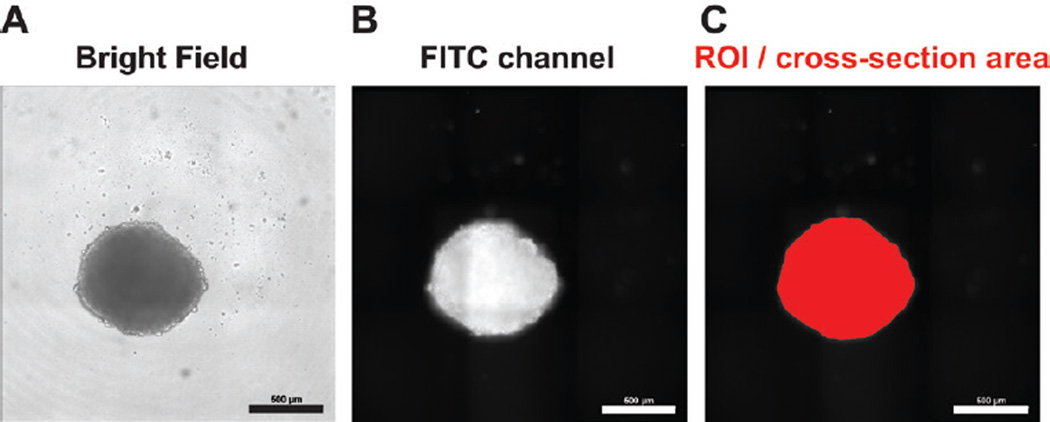
We used neurospheres derived from U87 glioblastoma cells, a well-established model system (11). A detailed protocol can be found in the Supplementary Materials. In brief, adherent U87 cells were dissociated with trypsin and seeded into low-adhesion flasks for suspension culture (4 × 106 cells per 75 cm2 flask) in cancer stem cell medium (CSC medium) comprised of serum-free DMEM/F12 medium supplemented with EGF (20 ng/mL), basic-FGF (20 ng/mL), heparin (5 µg/mL), B27 (2%), and gentamicin (0.1 mg/mL). The resulting primary neurospheres were cultured for up to eight passages. U87 neurospheres were then dissociated into single cells and subjected to flow cytometry using a BD FACSAria2 Special Order Research Product (SORP) instrument (BD Biosciences, San Jose, CA) in a biosafety cabinet. Cells were sorted by forward-scattered light (FSC) versus side-scattered light (SSC) and seeded into round-bottom 96-well plates (1000 cells per well in a 96-well suspension culture plate). Seeding cells by flow cytometry allows seeding of exact cell numbers per well while excluding debris or cells from the sub-G1 population, thereby ensuring uniformity at the beginning of the experiment. This is important because uniform spheroids and spheroid sizes are obtained by introducing defined numbers of viable cells to each well, and even small alterations in cell number or viability at seeding can result in substantial differences in neurosphere size after 14 days; however, manual counting and seeding of cells represents a viable alternative that has been used successfully by many laboratories (3–7). U87 neurospheres were allowed to grow for 2 weeks and then stained with 0.1 µg/ mL AO for 1 hour. Subsequently, neurospheres were imaged with a Nikon (Melville, NY) Eclipse Ti inverted microscope with an automated stage and a Nikon 10× air PlanApo objective (NA 0.45). Images were taken with an Andor Technology (Belfast, UK) Electron Multiplying Charge Coupled Device (EMCCD) camera (iXon3) with a 20 ms exposure. When using these settings, a 96-well plate is generally scanned in 5 min and 18 s, or less, which is faster than previously reported using a similar system (3). Analysis of neurosphere cross-section area was performed with Nikon Imaging Systems (NIS)-Elements high content analysis (HCA)/JOBS software as follows: The boundaries of neurospheres were defined by thresholding on fluorescence intensity and spheroid size (only objects ≥100 µm were defined as spheroids). The fill area (in µm2) within the region of interest (ROI) was defined as the neurosphere cross-section area. We confirmed that the measured area corresponds to the area detected in the FITC channel and to neurospheres visualized by bright-field microscopy (Figure 1).
Figure 1. Comparison of neurosphere images acquired by bright-field and fluorescence microscopy, and the final analyzed image.
U87 neurosphere cells (1000 cells per well) were seeded into round-bottom 96-well plates in 100 µl cancer stem cell (CSC) medium. After 2 weeks, neurospheres were stained with acridine orange (AO) by adding 1 µl CSC medium containing 10 µg/mL AO for 1 h. Subsequently, neurospheres were subjected to microscopy. The same neurosphere was visualized by bright-field microscopy (A) and using the FITC channel to detect AO-positive neurosphere cells (B). (C) The region of interest (ROI) in red identified by the HCA/JOBS software was used to determine neurosphere cross-section area. Scale bar: 500 µm.
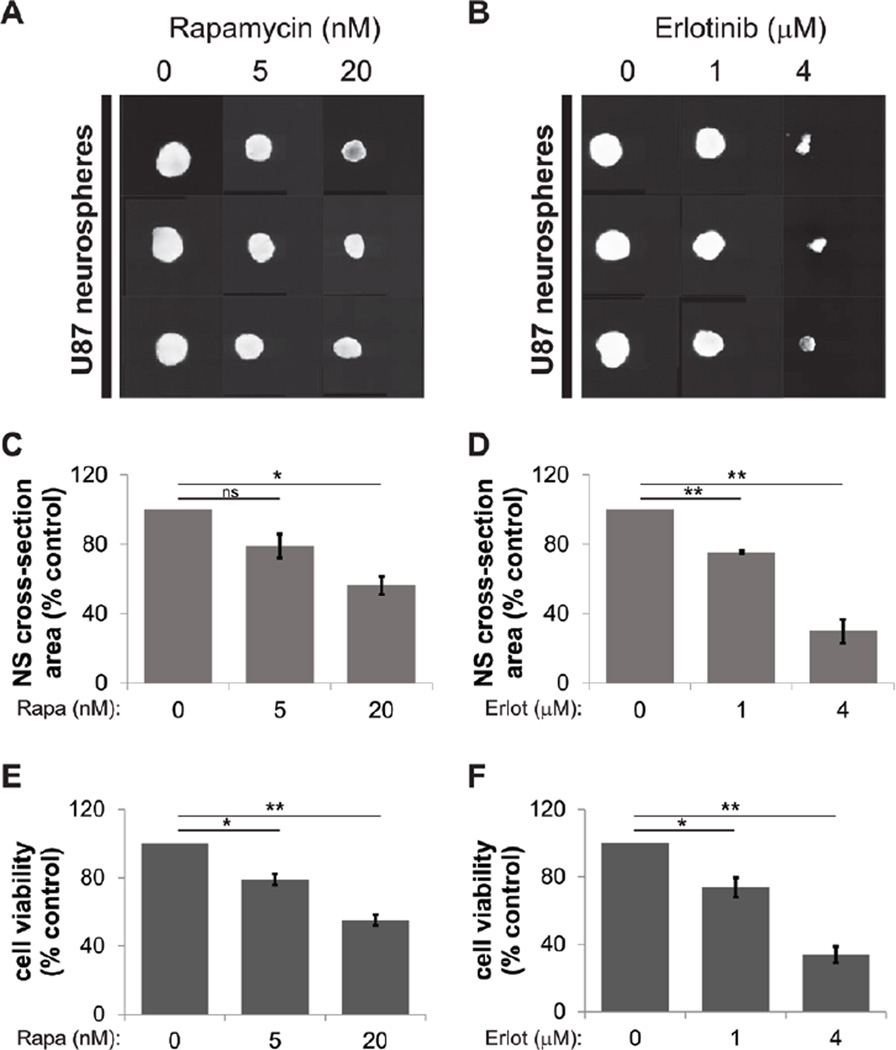
Next, we tested whether this protocol is suitable for monitoring the effect of anti-neoplastic agents on neurosphere formation. U87 neurosphere cells were seeded into 96-well round bottom plates and treated with vehicle (DMSO) alone or rapamycin, an inhibitor of the mammalian target of rapamycin complex 1 (mTORC1), or erlotinib, an epidermal growth factor receptor (EGFR) inhibitor that was previously shown to attenuate neurosphere size (12). To investigate whether the data obtained by our neurosphere assay correlate with cell viability, duplicate experiments were done in parallel for simultaneous analysis by the neurosphere assay and a water-soluble tetrazolium salt-1 (WST-1) proliferation assay as described previously (13). This assay measures cell metabolism by NAD(P)H-dependent reduction of tetrazolium dye and therefore provides a simple and indirect means of measuring spheroid viability. Increasing concentrations of rapamycin and erlotinib reduced neurosphere size as determined by fluorescence microscopy (Figure 2, A and B). Analysis of neurosphere images also revealed a dose-dependent decrease of neurosphere cross-section area after treatment with both antineoplastic drugs (Figure 2, C and D). Importantly, our methodology allows simultaneous collection of images (Figure 2, A and B) and cross-section area data (Figure 2, C and D) within 5 min, 18 s, and the data were markedly correlated with neurosphere cell viability as determined by the WST-1 assay (Figure 2, E and F).
Figure 2. Neurosphere size as measured by spheroid cross-section area corresponds with cell viability.
U87 neurosphere cells were seeded into round-bottom 96-well plates (1000 cells per well) and were treated with the indicated concentrations of rapamycin or erlotinib. After 2 weeks, neurospheres were stained with 0.1 µg/mL acridine orange (AO) for 1 h. Cells were analyzed by fluorescence microscopy (FITC channel), followed by measurement of neurosphere cross-section area using the HCA/JOBS software. (A and B) Tile-view of AO-stained neurospheres (in triplicate) detected in the FITC channel after treatment with DMSO as a control or the indicated concentrations of rapamycin (A) or erlotinib (B) both dissolved in DMSO. (C and D) Neurosphere images taken 2 weeks after incubation with DMSO and the indicated concentrations of rapamycin (C) or erlotinib (D) were analyzed for neurosphere cross-section area as described in the text. Data from 3 independent experiments are shown, each done in 10 technical replicates. Data are expressed as percentages of control DMSO-treated sample values. (E and F) Neurosphere viability as determined by WST-1 assay after 2 weeks of incubation with DMSO and the indicated concentrations of rapamycin (E) or erlotinib (F). Means ± sem are shown from the values of 3 independent experiments, each done in 10 technical replicates. Data are expressed as percentages of DMSO-treated samples (control). *, P < 0.05, **, P < 0.01 using a paired t-test.
Neurosphere cross-section area data collected by the automated HCA/JOBS system in a 96-well format allows for rapid analysis of neurosphere formation that correlates with neurosphere viability. Additionally, this assay is simple and provides greatly improved cost-efficiency compared with other staining methods used for tumor spheroid analysis (see Supplementary Table S1). Taken together, our neurosphere assay provides a novel tool to analyze spheroids of different origins for growth and size as measured by cross-section area, which we found correlates to viability, allowing rapid-throughput drug screening for tumor spheroid growth and thus CSC inhibition.
Supplementary Material
Acknowledgments
The work was supported by NIH grants R01 CA121192, CA155566, CA77816 and grant I01CX000916 from the Department of Veterans Affairs (LCP). JB is supported in part by a NIH/NCI training grant, T32 CA09560. This work was also supported by NIH grants R01 CA158911, R01 CA158911-S3, R01 NS093843, a Zell scholar award from Zell Family Foundation, Northwestern Brain Tumor Institute (SYC) and a Brain Cancer Research Award from James S. McDonnell Foundation (BH). This work was also supported by Northwestern University’s Center for Advanced Microscopy and the Flow Cytometry Facility, as well as Cancer Center Support Grant NCI CA060553. This paper is subject to the NIH Public Access Policy.
C.A. is supported in part by Nikon.
Footnotes
Supplementary material for this article is available at www.BioTechniques.com/article/114372.
Author contributions
F.E. designed the studies, performed the experiments, analyzed the data, and wrote the manuscript. A.A. designed the studies, performed experiments, analyzed data, and edited the manuscript. J.B., C.A., A.I., and A.D.A. performed experiments and analyzed data. B.H., S.-Y.C., and S.G. provided reagents, analyzed data, and edited the manuscript. L.C.P. designed studies, analyzed data, and edited the manuscript.
Competing interests
The other authors declare no competing interests.
References
- 1.Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francipane MG, Chandler J, Lagasse E. Cancer Stem Cells: A Moving Target. Curr Pathobiol Rep. 2013;1:111–118. doi: 10.1007/s40139-013-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, Lomas C, Mendiola M, et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann OI, Ilmberger C, Magosch S, Joka M, Jauch KW, Mayer B. Impact of the spheroid model complexity on drug response. J. Biotechnol. 2015;205:14–23. doi: 10.1016/j.jbiotec.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 5.Hsiao AY, Tung YC, Qu X, Patel LR, Pienta KJ, Takayama S. 384 hanging drop arrays give excellent Z-factors and allow versatile formation of co-culture spheroids. Biotechnol. Bioeng. 2012;109:1293–1304. doi: 10.1002/bit.24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senkowski W, Zhang X, Olofsson MH, Isacson R, Hoglund U, Gustafsson M, Nygren P, Linder S, et al. Three-Dimensional Cell Culture-Based Screening Identifies the Anthelmintic Drug Nitazoxanide as a Candidate for Treatment of Colorectal Cancer. Mol. Cancer Ther. 2015;14:1504–1516. doi: 10.1158/1535-7163.MCT-14-0792. [DOI] [PubMed] [Google Scholar]
- 7.Lindhagen E, Nygren P, Larsson R. The fluorometric microculture cytotoxicity assay. Nat. Protoc. 2008;3:1364–1369. doi: 10.1038/nprot.2008.114. [DOI] [PubMed] [Google Scholar]
- 8.Johnson TS, Bain E, Raju MR, Martin JC. Correlation of the growth of Chinese hamster V279-17lb multicellular spheroids with cytokinetic parameters. Cytometry. 1980;1:65–70. doi: 10.1002/cyto.990010113. [DOI] [PubMed] [Google Scholar]
- 9.Zelenin AV. Acridine Orange as a Probe for Cell and Molecular Biology. In: Mason WT, editor. Fluorescent and Luminescent Probes for Biological Activity: A Practical Guide to Technology for Quantitative Real-Time Analysis. 2nd. London, UK.: Academic Press; 1999. pp. 117–135. [Google Scholar]
- 10.Petit JM, Denis-Gay M, Ratinaud MH. Assessment of fluorochromes for cellular structure and function studies by flow cytometry. Biol. Cell. 1993;78:1–13. doi: 10.1016/0248-4900(93)90109-r. [DOI] [PubMed] [Google Scholar]
- 11.Yu SC, Ping YF, Yi L, Zhou ZH, Chen JH, Yao XH, Gao L, Wang JM, Bian XW. Isolation and characterization of cancer stem cells from a human glioblastoma cell line U87. Cancer Lett. 2008;265:124–134. doi: 10.1016/j.canlet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Feng H, Lopez GY, Kim CK, Alvarez A, Duncan CG, Nishikawa R, Nagane M, Su AJ, et al. EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J. Clin. Invest. 2014;124:3741–3756. doi: 10.1172/JCI73093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckerdt F, Beauchamp E, Bell J, Iqbal A, Su B, Fukunaga R, Lulla RR, Goldman S, Platanias LC. Regulatory effects of a Mnk2-eIF4E feedback loop during mTORC1 targeting of human medulloblastoma cells. Oncotarget. 2014;5:8442–8451. doi: 10.18632/oncotarget.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.