Abstract
In order to identify new cancer-associated metabolites that may be useful for early detection of lung cancer, we performed a global metabolite profiling of a non-small cell lung cancer (NSCLC) line and immortalized normal lung epithelial cells from the same patient. Among several metabolites with significant cancer/normal differences, we identified a unique metabolic compound, N-acetylaspartate (NAA) in cancer cells — undetectable in normal lung epithelium. NAA’s cancer-specific detection was validated in additional cancer and control lung cells as well as selected NSCLC patient tumors and control tissues. NAA’s cancer-specificity was further supported in our analysis of NAA synthetase (gene symbol: NAT8L) gene expression levels in The Cancer Genome Atlas: elevated NAT8L expression in approximately 40% of adenocarcinoma and squamous cell carcinoma cases (N=577), with minimal expression in all non-malignant lung tissues (N=74). We then showed that NAT8L is functionally involved in NAA production of NSCLC cells through siRNA-mediated suppression of NAT8L, which caused selective reduction of intracellular and secreted NAA. Our cell culture experiments also indicated that NAA biosynthesis in NSCLC cells depends on glutamine availability. For preliminary evaluation of NAA’s clinical potential as a circulating biomarker, we developed a sensitive NAA blood assay and found that NAA blood levels were elevated in 46% of NSCLC patients (N=13) in comparison with age-matched healthy controls (N=21) among individuals aged 55 years or younger. Taken together, these results indicate that NAA is produced specifically in NSCLC tumors through NAT8L overexpression and its extracellular secretion can be detected in blood.
Keywords: lung cancer, N-acetylaspartate, NAT8L, blood, biomarker
Introduction
Lung cancer is the leading cause of cancer death worldwide, leading to 1.6 million deaths every year (1). The majority of lung cancer cases are diagnosed in late stages, and early-stage detection and treatment are now known to reduce mortality rates, as recently reported for non-invasive screening with low-dose CT (LDCT) scan (2). Currently, LDCT screening is recommended only for the high-risk population of smokers over 55 years of age. This limitation is due to high false positive rates (96.4%) as well as risks of radiation exposure in LDCT. For better screening methods, recent studies have attempted to use diverse biological fluid samples from patients for finding new lung cancer biomarkers (3-5). Unlike diagnostic biomarkers that are required to have high sensitivity (i.e. high true positive rates) for clinical application, screening biomarkers must have high specificity (i.e. low false positive rates) in order to avoid a large number of people without lung cancer from undergoing invasive or costly procedures for confirmation (6). For example, specificity should be at least 99.6% (false positive rate < 0.4%) for screening tests on ovarian cancer among postmenopausal women to be clinically beneficial (7). Among recent studies on new lung cancer biomarkers, only two small-scale studies identified blood markers showing cancer-specificity higher than 99% (8, 9).
In order to discover new biomarker molecules for detecting cancer cases with high specificity, a small group of recent biomarker discovery studies have paid special attention to finding unique metabolites (small metabolic compounds) produced at levels significantly higher in tumors and minimal in most non-malignant cells and tissues. These efforts are based on new insights revealed in distinct metabolism of cancer cells and the fact that metabolites offer more possibilities of non-invasive tumor detection — such as imaging — than DNA, RNA, or proteins. A few such studies showed promising results for gliomas (10, 11) and prostate cancers (12, 13). For lung cancer, previous metabolic profiling studies on cancer cell lines or tumors did not report new cancer-specific metabolites, presumably due to their focus on characterizing cancer-selective metabolic fluxes and pathways of common metabolites (14-22).
In this study, we used a metabolite profiling approach with special focus on finding uncommon metabolites produced by non-small cell lung cancer (NSCLC) cells, but not by most healthy or non-malignant cells. This approach allowed us to identify a unique metabolite, N-acetylaspartate (NAA). We then examined NAA’s cancer-specificity and the mechanistic basis of its production in cancer cells. We also conducted proof-of-principle experiments with selected blood samples from lung cancer patients and controls as the first attempt to evaluate the feasibility of using NAA as one of the circulating biomarkers for lung cancer.
Materials and Methods
Metabolite extraction and mass spectrometry analysis of cell lines, media and tissues
All reagents were purchased from Sigma-Aldrich unless noted otherwise. All non-small cell lung cancer (NSCLC) cell lines have been authenticated, i.e. DNA fingerprinted for provenance with the Power-Plex 1.2wkit (Promega) and confirmed to be identical to the DNA fingerprint library maintained by ATCC and the Minna/Gazdar laboratory, and confirmed to be free of mycoplasma by e-Myco kit (Boca Scientific) (23). All NSCLC cell lines except HCC4017 were cultured with RPMI1640 (Invitrogen) supplemented with 5% fetal bovine serum (FBS)(Atlanta Biologicals). HCC4017 and patient-matched immortalized lung epithelial cells (HBEC30KT) were cultured in ACL4 medium (24) with 2% FBS, which was developed for the two lines to grow with reasonable rates under the same culture conditions. For isotope labeling experiments, RPMI1640 medium without glucose or glutamine (Invitrogen) was supplemented with glucose or glutamine whose carbons were either unlabeled or uniformly labeled with 13C ([U-13C]). From confluent cells, polar metabolites were extracted with methanol/water (1:1) (25) and derivatized with methoxyamine hydrochloride and N-methyl-N-(trimethylsilyl)trifluoroacetamide after adding 2H27-myristic acid as internal standard.
Snap-frozen tissues (resected NSCLC tumors from University of Texas Southwestern Medical Center (UTSW) Tissue Resource collected under IRB approved protocols; non-malignant lung tissues of healthy individuals from Biochain) were homogenized in ice-cold methanol/phosphate buffer. After centrifugation, supernatants were collected and dried. Metabolites in cell culture media were extracted with methanol and dried (11). Tumor and media samples were then derivatized as with cells above.
For Gas Chromatography – Mass Spectrometry (GC-MS) analysis of cell, media and tumor samples, derivatized metabolites were analyzed with Agilent 7890A/5975C using can mode at m/z 50-550. GC-MS datasets were deconvoluted with AMDIS (26) and metabolites were identified with NIST Spectral Library. Quantitative comparisons in different sample sets were made through SpectConnect (27) and were manually confirmed with Agilent’s ChemStation software. In this setting, the detection limit for NAA in tissues was 2 μM.
Protein extraction and Western blot
Proteins were extracted with two cycles of freeze-thaw method in phosphate-buffered saline, and Western blot was performed on 30 μg protein extracts per sample using commercial antibodies (anti-NAT8L from Abcore and anti-β-actin from EMD Millipore). See Supplementary Info for more details on protein extraction and Western blot.
NAT8L knockdown with siRNA
100,000 cells were plated per well in six-well plates and were transiently transfected with 40 nM synthetic small interfering RNA (siRNA) targeting NAT8L or scrambled negative control siRNA (OriGene) using DharmaFECT transfection reagent (ThermoFisher) according to the manufacturer’s protocol. The sequences for si(NAT8L) (human) were CGGACAUCGAGCAGUACUACAUGAA (#1), GCACUCUGCAUGACUUUAAUUCUTG (#2), and GGAAUAUACAGACAGACGUAAAGTG (#3). After 48 hrs, transfected cells were collected for further analyses.
RNA-seq data analysis of TCGA data for NAT8L
RNA-seq data in gene-level normalized read counts were downloaded from The Cancer Genome Atlas (TCGA) data portal (28). The data were logarithm transformed (base 2) and were quantile-normalized with R package for the same distribution of gene expression levels between samples.
NAA analysis of blood samples
Pooled and individual healthy plasma samples were obtained from Innovative Research (IR). Patient and age-matched control plasma samples were from the University of Texas Southwestern Medical Center(UTSW) (Dallas, TX) and the Sungkyunkwan University School of Medicine (SKK) (Seoul, Korea) under IRB-approved protocols. 2H3-Methylmalonic acid (D3-MMA) was purchased from Cerilliant (Round Rock, TX). Strong anion exchange columns (SAX, 100 mg sorbent mass and 3 mL reservoir volume) were procured from Biotage USA (Charlotte, NC). All solvents including water, methanol and acetonitrile were of HPLC grade (ThermoFisher).
Blood plasma samples were initially cleaned up with pretreatment methods as modified from those developed by Simon-Manso et al. in NIST (29). An aliquot of 200 μL blood plasma sample, with 1 μg D3-MMA added as internal standard, was centrifuged (5000g, 15 min). The supernatant was pre-treated with methanol (800 μL, vortex for 30 seconds, and two cycles of -20°C storage and vortexing). Then, the mixture was centrifuged at 19,600g for 10 min at room temperature. The supernatant was dried under vacuum, which was reconstituted in HPLC grade 0.6 mL water, vortexed and centrifuged at 425g for 5 minutes. The sample was then loaded onto the SAX columns, which were previously conditioned with 1 mL methanol and 1 mL HPLC grade water. The columns were washed sequentially with 1 mL HPLC grade water, 1 mL acetonitrile and 0.5 mL MTBE (Methyl-tert-butyl-ether). The analytes were eluted with 3 mL of 3% formic acid in MTBE and was dried under vacuum. The residue was reconstituted in 25 μL ethyl acetate, derivatized with 25 μL N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide, at 70 °C for 20 minutes.
GC-MS analysis was carried out on the same Agilent GC-MS system as above, using 2 °C/min ramping from 70 °C to 180 °C, 1°C/min upto 185°C and at 40 °C/min till 325 °C with a final hold for 15 min. N-acetylaspartate and internal standards were targeted on selected ion monitoring mode, wherein the ions targeted for derivatives were: m/z 346 (quantifier) and 287 (qualifier) for NAA; m/z 312 (quantifier) and 354 (qualifier) for 2H27-myrisitic acid and m/z 292 (quantifier) and 189 (qualifier) for D3-MMA. NAA concentration was estimated through fitting the NAA peak area of m/z 346 (normalized with peak area of D3-MMA (m/z 292)) against standard curves constructed with pooled commercial plasma containing supplemented NAA at 0 to 1 μM. In order to confirm the identities of these targets in sample GC-MS data, the ratios between peak intensities of quantifier and qualifier fragments and the retention times of these peaks were compared between standards and samples. Using these methods, the detection limit for NAA in blood plasma was 38 nM.
Statistical Analysis
The data are reported as the mean ± standard error of the mean from at least three data points generated from independent treatments. Data were analyzed by a two-tailed student’s t-test, and p values < 0.05 were considered statistically significant using Microsoft Excel.
Results
Discovery of N-acetylaspartate (NAA) in lung cancer cells
In order to expedite the discovery of candidates for cancer-specific metabolites in lung cancer, we exploited a unique system of a non-small cell lung cancer (NSCLC) cell line and a line of immortalized bronchial epithelial cells derived from the same patient, HCC4017 and HBEC30KT, for the initial discovery. After molecular characterization, we validated the selected candidate’s cancer specificity in additional NSCLC cell lines and NSCLC tumors. The mechanistic basis of this cancer specificity was further investigated with NSCLC cell lines, and its clinical potential as a circulating biomarker of lung cancer was evaluated with selected blood samples from lung cancer patients (Fig. 1).
Figure 1. Schematic of the workflow in this study.
In global metabolite profiling of HCC4017 and HBEC30KT using gas chromatography – mass spectrometry (GC-MS) methods modified from our previous work (30), we identified 194 non-redundant compounds and found that 47 of them were not detected in HBEC30KT cells but only in HCC4017 cells. We were able to positively match 17 of 47 to known compounds in the NIST spectral library, but not the remaining 30 compounds due to incompleteness of this library (see Supplementary Table S1 for the full list of these 194 compounds and their fold differences in HCC4017 vs. HBEC30KT cells). The 17 known compounds were all common metabolites that can be detected in many healthy or non-malignant cells and tissues, which was inconsistent with our objective to identify candidates of cancer-specific lung cancer markers. Thus, we focused on characterizing the identities of the 30 unidentified compounds.
While most of these 30 compounds showed mass spectra significantly different from those of the closest hits from the spectral library (see Supplementary Table S1 for the closest hits), one compound’s mass spectrum was very closely matched to that of an uncommon metabolite, methylmalonylglycine (thus was tentatively named as “methylmalonylglycine-like (MMG-L)”) (see Fig. 2A for MMG-L’s chromatographic peak and Fig. 2B for its mass spectrum and library search result; Peak #105 in Supplementary Table S1). This similarity allowed us to determine MMG-L’s exact identity, with a combination of chemical synthesis of methylmalonylglycine and high-accuracy mass spectrometry coupled with liquid chromatography, as N-acetylaspartate (NAA) (see Supplementary Info and Supplementary Figs. S1, S2, and S3 for details). This study is the first to report its detection in lung cancer cells, which is supported by recent reports of NAA detection in ovarian and prostate cancers (31, 32).
Figure 2. Discovery of NAA in lung cancer cells.
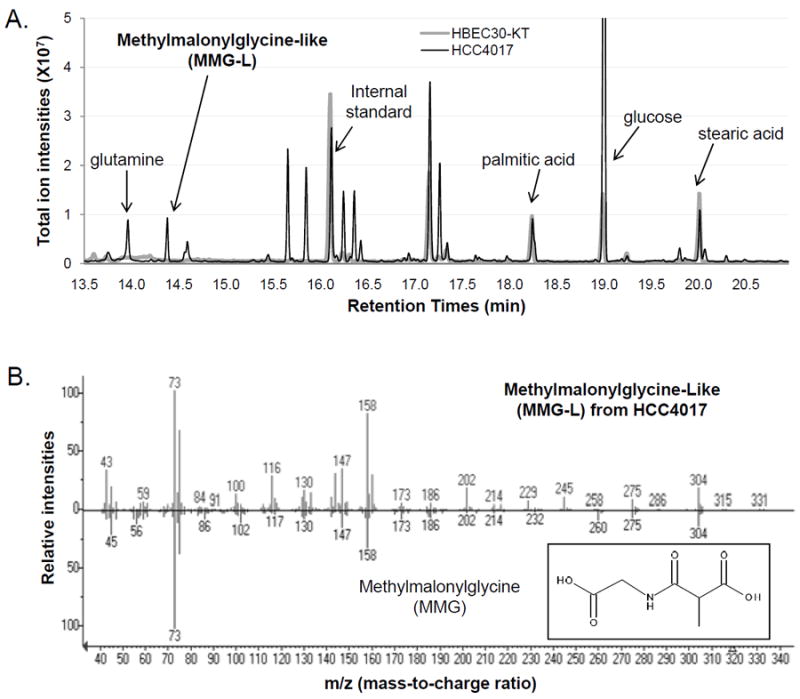
(A) In global metabolic profiling on lung cancer cells (HCC4017) and patient-matched control lung cells (HBEC30KT), an unidentified metabolite MMG-L (later characterized as NAA) was detected only in HCC4017 lung cancer cells, and is shown along with a few common metabolites of high abundance and the internal standard (2H27-myristic acid). Only a selected range of chromatographic retention times is shown on the X-axis for clarity. (B) The GC-MS mass spectrum of MMG-L from HCC4017 lung cancer cells (top panel) is compared with that of the top hit in search against NIST spectral library, methylmalonylglycine (MMG) (bottom panel). The fragment at m/z=245 is detected only for MMG-L while the fragment at m/z=260 is detected only for MMG.
Validation of cancer specificity of NAA and its biosynthetic enzyme NAT8L in lung cancer
In order to characterize the prevalence and the dynamic range of NAA production in lung cancer cells with diverse genetic backgrounds, NAA levels were then examined in eight additional NSCLC cell lines (see Supplementary Table S2 for clinical information and common oncogenic mutations for all cell lines used in this study). All examined cancer cell lines produced NAA but HBEC30KT or another immortalized normal lung cells (HBEC34KT) did not (Fig. 3A). We then investigated NAA’s potential as a secreted marker of lung tumors with culture media used in growing NSCLC cell lines. NAA levels in conditioned media from confluent cells were compared across 11 NSCLC cell lines and HBEC30KT. These lines included four from above and eight additional NSCLC lines (HCC2450, H522, H2347, H1819, H2126, H1568, H1993, and HCC1195), which allowed us to expand the range of intracellular and secreted NAA levels in NSCLC cells. The results show that intracellular NAA levels were reasonably well correlated with secreted NAA levels (R2=0.62) (Fig. 3B), indicating that NAA is produced and secreted into extracellular space by lung cancer cells.
Figure 3. Validation of NAA’s cancer specificity in lung cancer.
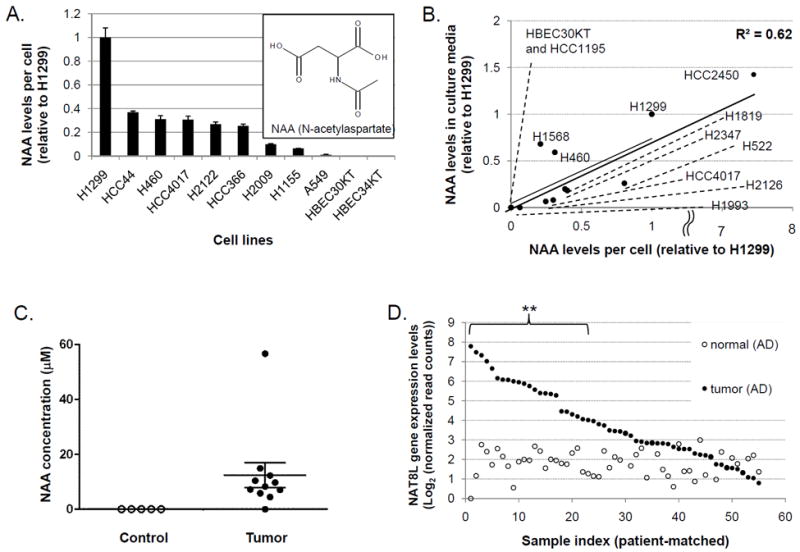
(A) NAA levels in nine NSCLC cell lines, HBEC30KT, and HBEC34KT relative to its average level in H1299 cells. Error bars indicate standard errors of normalized NAA levels (N=3).
(B) Intracellular and extracellular NAA levels in 11 NSCLC cell lines and HBEC30KT. Data shown are averages of triplicate data. Error bars are omitted for clarity.
(C) NAA concentrations in control lung tissues (“Control”, N=5) and NSCLC tumors (“Tumor”, N=11). Horizontal lines and error bars indicate means and standard errors of tissue NAA concentrations.
(D) Gene expression levels of NAT8L, the key enzyme for NAA biosynthesis, in 55 adenocarcinoma (AD) lung tumors as compared with patient-matched non-malignant lung tissues.
To evaluate the cancer selectivity of NAA in NSCLC tumors, we first analyzed NAA levels in 11 resected lung tumor samples from NSCLC patients and 5 lung tissues from non-cancer individuals with GC-MS (See Supplementary Table S3 for clinical information of these lung tissue samples). NAA was detected in 10 of 11 samples between 4.5 and 56.7 μM (12.6 ± 4.6 μM), but not in any of 5 non-malignant lung tissues (Fig. 3C). Despite the small sample sizes, this preliminary result was consistent with the cell line results above in terms of NAA’s cancer-selective detection in NSCLC.
To further examine the cancer-specificity of NAA in a larger number of lung tumors, we took advantage of the RNA-seq data in TCGA (The Cancer Genome Atlas) database (28) and the known dependence of NAA biosynthesis on NAA synthetase (gene symbol: NAT8L) (33). NAT8L’s gene expression levels were analyzed for 577 NSCLC tumors and 74 non-malignant lung tissues in TCGA database. Among 55 lung adenocarcinoma (AD) tumors with patient-matched non-malignant lung tissues, 24 tumors (44%) showed significant elevation of NAT8L expression above baseline (Fig. 3D: ** = greater than two-fold elevation over all non-malignant lung tissues and p<0.01:). Similar results were obtained from 16 lung squamous cell carcinoma tumors (SC) vs. patient-matched control tissues (elevated expression in 6 tumors (38%)) and all 577 NSCLC tumors (overexpression in 154 of 355 AD tumors (43%) and 89 of 222 SC tumors (40%)) (Supplementary Figs. S4 and S5). Taken together, these data suggest that NAA production in lung cancer may be not only selective but also specific to cancer cells, as compared with non-malignant lung cells.
Functional involvement of NAT8L in NAA production of lung cancer cells
Our TCGA data analysis above indicated the cancer-specific involvement of NAT8L in NAA production of lung tumors, which prompted us to examine NAT8L’s functional roles in NAA biosynthesis at the cellular level. NAT8L protein expression levels in most of eight selected NSCLC cell lines were significantly higher than in HBEC30KT cells (Supplementary Fig. S6). NAT8L’s functional involvement in NAA production of NSCLC cells was examined through siRNA-mediated knockdown experiments. Suppression of NAT8L with si(NAT8L)#1 led to selective reduction of intracellular NAA levels (by 72%) as compared with direct precursors of NAA synthesis (pyruvate, as a surrogate for acetyl-CoA, and aspartate)(34) and their directly-related metabolites (Fig. 4A). The si(NAT8L) #1 was chosen for this experiment because it showed the most efficient protein-level knockdown in H1299 cells (by 68%) out of three independent siRNAs for NAT8L (Fig. 4B). This dependence of NAA on NAT8L was also observed in another NSCLC cell line (HCC4017) (Supplementary Fig. S7). NAT8L suppression also caused selective decreases in extracellular NAA levels in comparison with two major nutrients in culture medium (glutamine and glucose) and two secreted metabolites ( lactate and alanine) related to NAA (Fig. 4C).
Figure 4. Functional involvement of NAT8L enzyme in NAA biosynthesis of lung cancer cells.
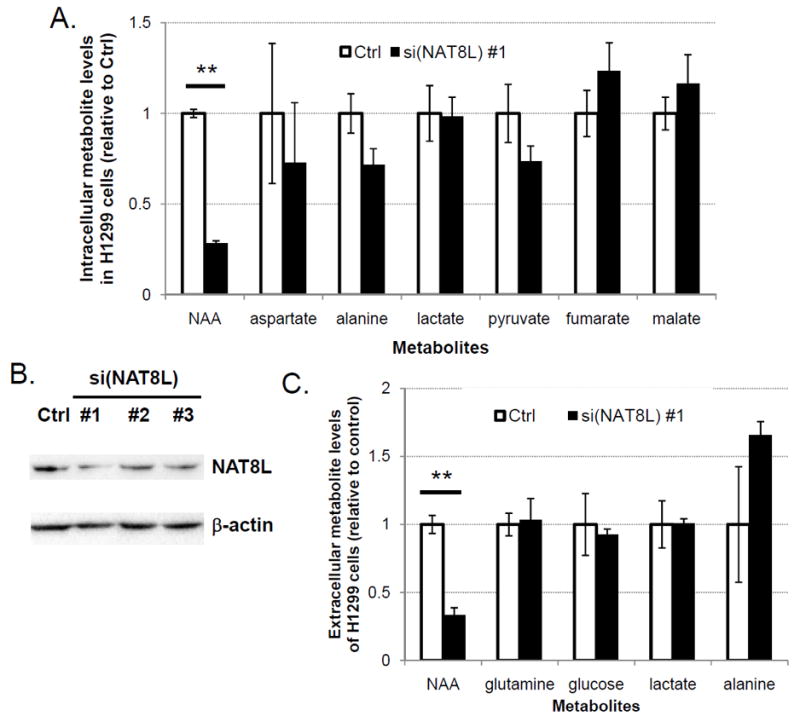
(A) Relative levels of NAA, its precursors and related metabolites in H1299 cells under non-targeting control (Ctrl) or si(NAT8L) #1 conditions. Error bars indicate standard errors (N=3) (** = p<0.01).
(B) NAT8L protein levels in H1299 cells treated with the control siRNA (Ctrl) or three independent NAT8L siRNAs.
(C) Relative Levels of NAA, glutamine, glucose, lactic acid, and alanine in culture medium of H1299 cells under the control (Ctrl) or si(NAT8L) #1 conditions. Error bars indicate standard errors (N=3) (** = p<0.01).
Glutamine dependence of NAA production in lung cancer cells
In order to further characterize the mechanism of NAA biosynthesis in lung cancer cells, we examined which major nutrients provide carbon sources for NAA through 13C-labeling experiments (35). When uniformly 13C-labeled ([U-13C]) glucose or glutamine were used in culture medium for H1299 lung cancer cells, acetyl group of NAA was labeled primarily with 13C from glucose (as indicated by relative enrichment of “M+2” peak) while aspartate group of NAA was labeled mainly with 13C from glutamine (as indicated by relative enrichment of “M+4” peak) (Fig. 5A). This result is consistent with glutamine’s contribution to aspartate through glutaminolysis, commonly observed in many cancer cells including lung cancer (36).
Figure 5. Glutamine dependence of NAA production in lung cancer cells.
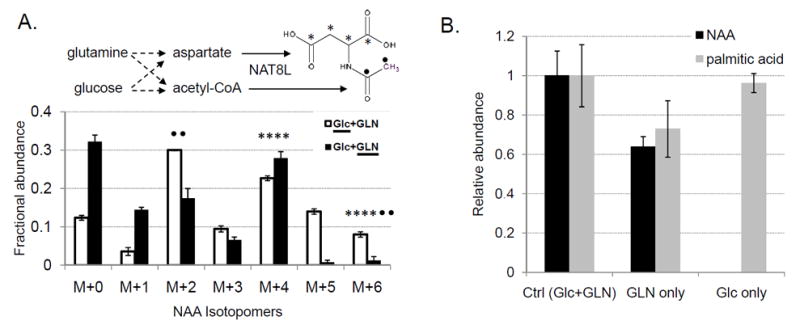
(A) Isotope labeling patterns of NAA in H1299 cells grown in medium containing [U-13C] glucose and unlabeled glutamine (Glc+GLN), or unlabeled glucose and [U-13C] glutamine (Glc+GLN). 13C-labeled aspartate and acetyl-CoA in NAA are marked with asterisks and closed circles, respectively. M+0 indicates unlabeled isotopomer of NAA (m/z=304) while M+X indicates NAA labeled with X carbons from 13C-labeled precursors. Error bars indicate standard errors of normalized metabolite levels (N=3).
(B) Levels of NAA and palmitic acid in H1299 cells under control, glutamine (GLN)-only, or glucose (Glc)-only conditions. Error bars indicate standard errors (N=3).
Then, we investigated whether glucose or glutamine is required in NAA production in NSCLC cells. When glucose was removed from the culture medium, intracellular NAA levels were decreased by approximately 40%, concomitant with 30% decrease in free palmitic acid (Fig. 5B). Interestingly, NAA production was completely abolished when glutamine was removed from the medium whereas little change was observed in free palmitic acid levels. Moreover, NAA levels in selected NSCLC cell lines showed significantly better correlation with intracellular glutamine levels (R2=0.68) than glucose levels or NAT8L protein amounts (R2<0.1) (Supplementary Fig. S8).
Since NAA can also be a degradation product of another unique metabolite, N-acetylaspartylglutamate (NAAG)(34), we further examined whether NAAG is present in lung cancer cells. NAAG level in HCC4017 cells was below our detection limit by LC-MS (1,000 fold less than NAA: see Supplementary Fig. S9), indicating that NAAG is not likely to be an alternative precursor for NAA in lung cancer cells. Therefore, the enzyme for producing NAA from NAAG (gene symbol: FOLH1) was not examined for its involvement in NAA metabolism of lung cancer cells.
Preliminary evaluation of NAA’s potential as a circulating marker of lung cancer
We then investigated the feasibility of using NAA as a blood biomarker for NSCLC tumors. NAA has been undetectable in blood of healthy individuals, but it is detected in blood of patients suffering from a rare neurological disease called Canavan disease (37), which is thought to be caused by excessive accumulation of NAA in brain and cerebrospinal fluid (CSF) of these patients due to their genetic defects of NAA-degradation enzyme, aspartoacylase. By roughly comparing 17 μM NAA in blood (37) with 380 μM NAA in CSF of these patients (38) (0.8 μM in CSF of healthy individuals (39)) and NAA concentration of lung tumors in our data (4.5 ~ 14.9 μM in 9 of 10 samples: Fig. 3A), we suspected that the majority of NAA blood levels in lung cancer patients may be approximately 0.2 μM or lower.
Since previously reported methods for NAA blood assay (detection limit: 0.35 μM) (37) were not sensitive enough to cover this concentration range, we developed a sensitive method by taking advantage of NAA’s chemical property and optimizing the mass spectrometry condition. NAA is negatively charged in blood (pKa = 3.1), so it can be selectively enriched with strong anion exchange columns out of complex plasma samples. In addition, GC-MS methods used above can be modified for maximum sensitivity and minimum background noise for NAA detection through adjustment of gas chromatography program and application of ‘selected ion monitoring’ mode in mass spectrometry. Combining these two approaches allowed us to improve the limit of detection for blood NAA by nearly 10-fold — to approximately 0.038 μM (= 38 nM) — with 12% coefficient of variation (Supplementary Fig. S10).
To evaluate the cancer-specificity of the NAA blood assay, which depends heavily on their levels in individuals without cancer, we first examined NAA levels of blood plasma samples from 40 healthy individuals (ages 31-73 years) (see Supplementary Table S4 for clinical information of all plasma samples: in addition to 29 samples from the University of Texas Southwestern Medical Center (UTSW) (Dallas, TX) (N=15) and the Sungkyunkwan University (SKK) (N=14), 11 samples from a commercial source (Innovative Research (IR)) were included in order to bridge the age-diversity gap in control samples). In agreement with previous reports, 21 samples from individuals aged 55 years or younger showed low to no detectable NAA blood levels (less than 50 nM) (Fig. 6). Importantly, three of the 21 control samples were from individuals with non-cancer lung abnormalities (benign pulmonary nodule, pulmonary fibrosis, and inactive tuberculosis). However, unexpectedly, the majority of 19 plasma samples from older individuals (56-73 years) showed increased NAA blood levels: 62 ± 19 nM (Supplementary Fig. S11).
Figure 6. Preliminary evaluation of NAA as a circulating biomarker for lung cancer.
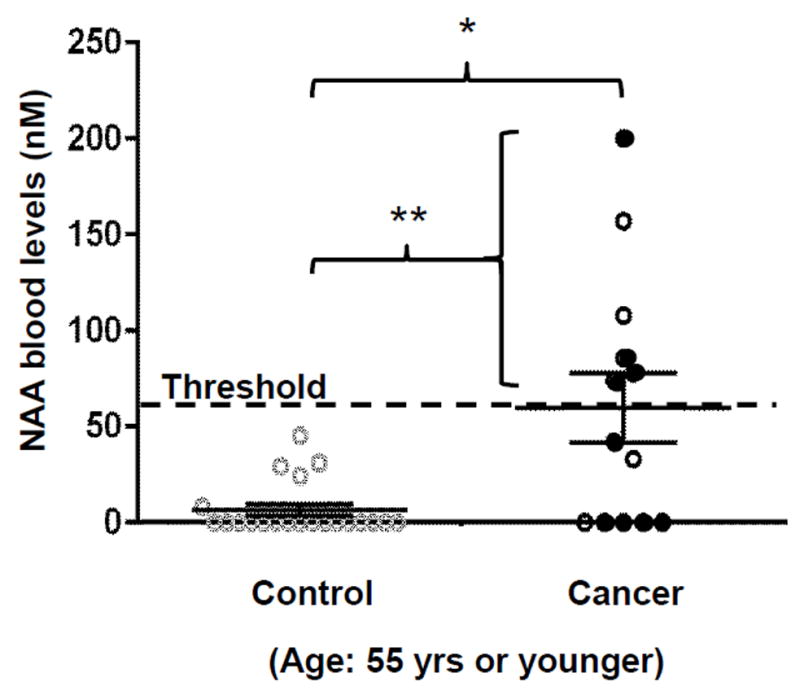
NAA blood levels in healthy controls (“Control”) vs. NSCLC patients (“Cancer”) at 55 years of age or younger (** = p<0.01, * = p<0.05). Filled circles indicate nine NSCLC patients diagnosed as stage 1A. Horizontal lines and error bars indicate means and standard errors of NAA blood levels.
Thus, our efforts to evaluate the clinical feasibility of the NAA blood assay were focused on samples from lung cancer patients at 55 years or younger. Six of 13 NSCLC patients (46%) showed NAA levels over 60 nM threshold (120 ± 20 nM, p<0.01) including four of nine NSCLC patients at stage 1A, compared with 21 age -matched controls (Fig. 6). None of the lung cancer patients in this age group were treated with chemo- or radio-therapy except the patient for “CA6” blood sample (collected after chemo and radiotherapy: NAA blood level=157 nM). These results are consistent with the highly cancer-specific overexpression of NAT8L in lung tumors (approximately 40% of all lung tumors) observed in the TCGA data analysis shown above. In contrast, NAA blood levels in the older age group (over 55 years: N=21) did not show significant differences from age-matched controls discussed above (N=19): 47 ± 14 nM (up to 190 nM; p=0.5).
Discussion
In this study, we used a metabolic profiling approach based on gas chromatography – mass spectrometry (GC-MS) with the specific objective to identify candidates of cancer-specific markers. GC-MS was chosen as the analytical method because of the richness of the available spectral libraries (approximately 200,000 unique compounds in the NIST spectral library (Electron Ionization))(40) despite the limitation of requiring derivatization of extracted metabolites before analysis. Among 194 non-redundant compounds identified in the GC-MS analysis of the isogenic pair of lung cancer and non-malignant cells (HCC4017 and HBEC30KT), we identified a new candidate of cancer-specific metabolite in lung cancer by focusing our efforts on characterizing the exact identities of 30 compounds that were detected only in cancer cells but could not be positively identified with library search.
To characterize these 30 unidentified compounds, we followed a commonly used approach: (1) come up with candidate identities, (2) obtain the corresponding synthetic compounds, and (3) compare their chromatographic retention times and mass spectra with those of compounds from original samples. Because this process is laborious and time-consuming, we prioritized the 30 unidentified cancer-selective compounds based on their similarities to the top hits from the library searches. The high spectral similarity of MMG-L to methylmalonylglycine in the NIST library allowed us to successfully characterize MMG-L as N-acetylaspartate (NAA).
NAA has been detected primarily in brains and cerebrospinal fluids, but not in other tissues or blood of healthy individuals (34, 37, 39, 41). Although NAA was recently reported as one of the potential markers of ovarian or prostate cancers (31, 32), our study is the first to report its production in lung cancer cells and lung tumors and elucidate the mechanistic basis — cancer-specific overexpression of NAA’s biosynthetic enzyme, NAA synthetase (gene symbol: NAT8L) (33). We demonstrated that NAT8L plays a direct and specific role in NAA biosynthesis and extracellular secretion by lung cancer cells, using glucose and glutamine as major carbon sources. NAT8L has not been reported in previous proteomic studies of any cancer, including lung cancer cells or lung tumors (42-44).
The results in this study indicate that NAA’s roles in lung cancer cells may be regulatory rather than metabolic. In brain, NAA’s major role is to provide acetate in the synthesis of fatty acids and other lipids of myelin (45, 46), after being synthesized from acetyl-CoA and aspartate via NAT8L in neuronal cells and transported to oligodendrocytes for cleavage to acetate and aspartate via aspartoacylase (34). However, in lung cancer cells, the synthesis of palmitic acid (the most abundant fatty acid) was not affected significantly upon reduction of NAA synthesis after NAT8L suppression. This discrepancy can be explained by the low NAA levels in lung tumors (average 14 μM, approximately 500-fold lower than those in brains). Also, we report for the first time that NAA’s biosynthesis in lung cancer cells may depend on glutamine as the major carbon source for the aspartate moiety of NAA. Glutamine’s contribution to aspartate through the glutaminolysis pathway is well established in cancer cells, where glutamine is successively converted to glutamate (via glutaminase), α-ketoglutarate (via glutamate dehydrogenase or alanine/aspartate transaminases), oxaloacetate (via citric acid cycle), and to aspartate (via aspartate transaminase) (47, 48). Thus, our data suggest that NAA’s role in these cells may be related to regulation of utilizing glutamine and other related nutrients. More questions about the specific roles that NAT8L or NAA play in proliferation or survival of cancer cells and the underlying mechanisms should be addressed in future studies.
We also explored the clinical feasibility of using NAA as one of the circulating biomarkers for lung tumors. Our data support that NAA molecules are produced by a significant subset of lung cancer cells and tumors in a cancer-specific manner, are secreted into extracellular space, and can be detected in blood with the sensitive methods that we developed. Our preliminary evaluation suggests that NAA alone may be a blood biomarker for a limited subset of NSCLC tumors within the age group of 55 years old or younger who do not have conditions that can cause excessive accumulation of NAA (such as Canavan Disease). Although this age group represents only 11% of total lung cancer cases, their cases appear to behave more aggressively, with fewer stage I cases and more stage IV cases at diagnosis (49, 50). It is also noteworthy that, although NAA’s capability to distinguish individuals with benign lung conditions from patients with malignant lung tumors were not thoroughly investigated in this study, 21 non-cancer individuals at 55 years of age or younger (all of which showed negative NAA blood assay results) included three with non-cancer lung abnormalities.
The most significant limitation of this preliminary evaluation is that it is not clear how much of NAA in lung cancer patients’ blood is derived from lung tumors. One of the ways to examine this issue is to analyze NAA levels in blood and tumor samples of these patients collected before and after surgical removal of their lung tumors. As an initial effort, we analyzed NAA levels in six pairs of such blood samples from lung adenocarcinoma patients (before vs. after surgery). In partial support of the argument that lung tumors contribute to increasing blood NAA levels, two of six pre-surgery samples showed positive NAA levels while the two corresponding post-surgery samples (as well as four other post-surgery samples) had no detectable NAA (data not shown).
An important consideration in potential clinical application of our findings is that NAA blood levels in the healthy individuals older than 55 years seemed to be comparable to those in lung cancer patients. To our understanding, blood NAA in these old healthy individuals is most likely derived from brain and/or cerebrospinal fluid (CSF) through blood-brain barrier (BBB) leakage. For younger healthy individuals, NAA concentrations decrease dramatically from brains (5 to 8 mM) to CSF (0.8 μM) (19–58 years) (39) and no NAA has been detected in blood samples (27–50 years). These stark differences are thought to be caused by tight barriers between brain and CSF as well as between CSF and blood in healthy individuals under 50 years old. On the other hand, increased permeability of blood-brain barrier (in particular, blood-CSF barrier) has been reported in old healthy individuals by three independent studies. Healthy 60–87 year-olds have shown increased BBB permeability as compared with healthy 21–50 year-olds approximately by two to three-fold (51, 52), which has been recently confirmed in a high-resolution MRI study on healthy 55–91 year-olds vs. healthy 23–47 year-olds (53). These findings indicate that, in order to further examine how much of the NAA from lung tumors contribute to the NAA detected in blood samples of target population, future studies should include control samples from individuals with neurological conditions associated with BBB leakage.
Also, an important clinical question that our study started to address but has not fully answered is how many lung cancer patients within the 55-and-under age group are likely to produce NAA in their lung tumors and show detectable NAA levels in blood. Our large-scale analysis on 577 NSCLC tumors in the TCGA database indicated that approximately 40% of patient tumors may overexpress NAT8L. In fairly good agreement, this percentage was close to 46% (6 of 13 total) or 44% (4 of 9 in stage 1A) of lung cancer patients at 55 years or younger showing NAA blood levels above threshold. In order to determine NAA’s sensitivity in detecting lung tumors with statistical reliability, a significantly larger number of patient blood samples collected before cancer treatment should be analyzed, based on recommendations by Pepe et al.(6, 54). In addition, control samples properly matched for source, age, sex, and smoking history should be used for estimating the cancer-specificity of the NAA blood assay more accurately. For example, the samples from the age group of 55 years or younger in our proof-of-principle experiments were not properly matched for source, sex and smoking history (sources - UTSW and SKK for Cancer, UTSW, SKK, and IR for Control; female percentages - 61% in Cancer and 43% in Control; smoker percentages - 46% in Cancer and 38% in Control).
Finally, two technical limitations of our study are that the reproducibility of the NAA blood assay and the effect of blood samples’ storage time on the assay results were not completely addressed. We showed the intra-assay variability to be 12% in terms of coefficient of variation, but intra-subject, inter-assay and inter-laboratory variabilities need to be evaluated in future studies. We observed that the NAA blood assay can be applied to archived samples stored at -80°C for up to 7 years because NAA blood levels in patient samples stored for 0~3 years (53 ± 28 nM: from SKK) were not significantly different from those stored for 3~7 years (75 ± 36 nM: from UTSW, p=0.63).
In conclusion, we discovered NAA’s cancer-specific production in NSCLC tumor cells and characterized NAA’s dependence on the elevated expression of its biosynthetic enzyme (NAT8L) and the availability of glutamine. Our preliminary evaluation of NAA’s clinical potential as a circulating marker for lung tumors suggests that it may be useful in identifying approximately 40% of lung cancer patients among a narrowly-defined target population: those under 55 years of age without neurological conditions associated with NAA accumulation or blood-brain barrier leakage. Future work should include (1) in-depth investigation into elucidating the physiological roles of NAA molecule and NAT8L enzyme in lung tumors and (2) rigorous evaluation of the clinical potential and benefit of using NAA as one of the circulating biomarkers of lung tumors in a defined target population.
Supplementary Material
Acknowledgments
We thank UTSW Tissue Shared Resource, Luc Girard, Michael Peyton, and Adwait Sathe for sharing samples, data, and technical expertise.
Financial Support: This study was supported by University of Texas at Dallas Startup funds and NCI Lung Cancer SPORE Career Development Award and Developmental Research Award (P50CA70907) for H. Yoo, NCI Lung Cancer SPORE (P50CA70907) and UTSW CCSG (P30-CA142543) for J.D. Minna, Welch Foundation (I-1414) for M.A. White, CPRIT (RP130272) and Welch Foundation (I-1733) for R.J. DeBerardinis, and Welch Foundation (AT-1595) for J.M. Ahn.
Footnotes
Conflicts of Interest: R.J. DeBerardinis is a member of advisory boards for Agios Pharmaceuticals and Peloton Therapeutics.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassanein M, Callison JC, Callaway-Lane C, Aldrich MC, Grogan EL, Massion PP. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila) 2012;5:992–1006. doi: 10.1158/1940-6207.CAPR-11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsay JC, DeCotiis C, Greenberg AK, Rom WN. Current readings: blood-based biomarkers for lung cancer. Semin Thorac Cardiovasc Surg. 2013;25:328–34. doi: 10.1053/j.semtcvs.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascaux C, Peled N, Garg K, Kato Y, Wynes MW, Hirsch FR. Early detection and screening of lung cancer. Expert Rev Mol Diagn. 2010;10:799–815. doi: 10.1586/erm.10.60. [DOI] [PubMed] [Google Scholar]
- 6.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–8. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu KH, Skates S, Hernandez MA, Bedi D, Bevers T, Leeds L, et al. A 2-stage ovarian cancer screening strategy using the Risk of Ovarian Cancer Algorithm (ROCA) identifies early-stage incident cancers and demonstrates high positive predictive value. Cancer. 2013;119:3454–61. doi: 10.1002/cncr.28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108:3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ajona D, Pajares MJ, Corrales L, Perez-Gracia JL, Agorreta J, Lozano MD, et al. Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. Journal of the National Cancer Institute. 2013;105:1385–93. doi: 10.1093/jnci/djt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–9. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan AP, Rajendiran TM, Ateeq B, Asangani IA, Athanikar JN, Yocum AK, et al. The role of sarcosine metabolism in prostate cancer progression. Neoplasia. 2013;15:491–501. doi: 10.1593/neo.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 14.Hori S, Nishiumi S, Kobayashi K, Shinohara M, Hatakeyama Y, Kotani Y, et al. A metabolomic approach to lung cancer. Lung Cancer. 2011;74:284–92. doi: 10.1016/j.lungcan.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Duarte IF, Rocha CM, Gil AM. Metabolic profiling of biofluids: potential in lung cancer screening and diagnosis. Expert Rev Mol Diagn. 2013;13:737–48. doi: 10.1586/14737159.2013.835570. [DOI] [PubMed] [Google Scholar]
- 16.Fan TW, Lane AN, Higashi RM, Bousamra M, 2nd, Kloecker G, Miller DM. Metabolic profiling identifies lung tumor responsiveness to erlotinib. Exp Mol Pathol. 2009;87:83–6. doi: 10.1016/j.yexmp.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, et al. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhri VK, Salzler GG, Dick SA, Buckman MS, Sordella R, Karoly ED, et al. Metabolic alterations in lung cancer-associated fibroblasts correlated with increased glycolytic metabolism of the tumor. Mol Cancer Res. 2013;11:579–92. doi: 10.1158/1541-7786.MCR-12-0437-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kami K, Fujimori T, Sato H, Sato M, Yamamoto H, Ohashi Y, et al. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics. 2013;9:444–53. doi: 10.1007/s11306-012-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen T, Gao L, Wen Z, Wu C, Tan CS, Toh WZ, et al. Exploratory investigation of plasma metabolomics in human lung adenocarcinoma. Mol Biosyst. 2013;9:2370–8. doi: 10.1039/c3mb70138g. [DOI] [PubMed] [Google Scholar]
- 21.Lam CW, Law CY. Untargeted Mass Spectrometry-Based Metabolomic Profiling of Pleural Effusions: Fatty Acids as Novel Cancer Biomarkers for Malignant Pleural Effusions. J Proteome Res. 2014 doi: 10.1021/pr5003774. [DOI] [PubMed] [Google Scholar]
- 22.Wikoff W, Grapov D, Fahrmann J, DeFelice B, Rom W, Pass H, et al. Metabolomic Markers of Altered Nucleotide Metabolism in Early Stage Adenocarcinoma. Cancer prevention research. 2015 doi: 10.1158/1940-6207.CAPR-14-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70:9937–48. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gazdar AF, Oie HK. Re: Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:6011–2. [PubMed] [Google Scholar]
- 25.Masson P, Alves AC, Ebbels TM, Nicholson JK, Want EJ. Optimization and evaluation of metabolite extraction protocols for untargeted metabolic profiling of liver samples by UPLC-MS. Anal Chem. 2010;82:7779–86. doi: 10.1021/ac101722e. [DOI] [PubMed] [Google Scholar]
- 26.http://chemdata.nist.gov/mass-spc/amdis/
- 27.Styczynski MP, Moxley JF, Tong LV, Walther JL, Jensen KL. Stephanopoulos GN. Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Anal Chem. 2007;79:966–73. doi: 10.1021/ac0614846. [DOI] [PubMed] [Google Scholar]
- 28.https://tcga-data.nci.nih.gov/tcga/
- 29.Simon-Manso Y, Lowenthal MS, Kilpatrick LE, Sampson ML, Telu KH, Rudnick PA, et al. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal Chem. 2013;85:11725–31. doi: 10.1021/ac402503m. [DOI] [PubMed] [Google Scholar]
- 30.Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem. 2008;283:20621–7. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hascalik S, Celik O, Sarac K, Alkan A, Mizrak B. Clinical significance of N-acetyl-L-aspartate resonance in ovarian mucinous cystadenoma. Int J Gynecol Cancer. 2006;16:423–6. doi: 10.1111/j.1525-1438.2006.00200.x. [DOI] [PubMed] [Google Scholar]
- 32.Shuster JR, Lance RS, Troyer DA. Molecular preservation by extraction and fixation, mPREF: a method for small molecule biomarker analysis and histology on exactly the same tissue. BMC Clin Pathol. 2011;11:14. doi: 10.1186/1472-6890-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiame E, Tyteca D, Pierrot N, Collard F, Amyere M, Noel G, et al. Molecular identification of aspartate N-acetyltransferase and its mutation in hypoacetylaspartia. Biochem J. 2010;425:127–36. doi: 10.1042/BJ20091024. [DOI] [PubMed] [Google Scholar]
- 34.Moffett JR, Ross B, Arun P, Madhavarao CN, Namboodiri AM. N-Acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog Neurobiol. 2007;81:89–131. doi: 10.1016/j.pneurobio.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo H, Stephanopoulos G, Kelleher JK. Quantifying carbon sources for de novo lipogenesis in wild-type and IRS-1 knockout brown adipocytes. J Lipid Res. 2004;45:1324–32. doi: 10.1194/jlr.M400031-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.van den Heuvel AP, Jing J, Wooster RF, Bachman KE. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Ther. 2012;13:1185–94. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavazzi B, Lazzarino G, Leone P, Amorini AM, Bellia F, Janson CG, et al. Simultaneous high performance liquid chromatographic separation of purines, pyrimidines, N-acetylated amino acids, and dicarboxylic acids for the chemical diagnosis of inborn errors of metabolism. Clin Biochem. 2005;38:997–1008. doi: 10.1016/j.clinbiochem.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Wevers RA, Engelke U, Wendel U, de Jong JG, Gabreels FJ, Heerschap A. Standardized method for high-resolution 1H-NMR of cerebrospinal fluid. Clin Chem. 1995;41:744–51. [PubMed] [Google Scholar]
- 39.Do KQ, Lauer CJ, Schreiber W, Zollinger M, Gutteck-Amsler U, Cuenod M, et al. gamma-Glutamylglutamine and taurine concentrations are decreased in the cerebrospinal fluid of drug-naive patients with schizophrenic disorders. J Neurochem. 1995;65:2652–62. doi: 10.1046/j.1471-4159.1995.65062652.x. [DOI] [PubMed] [Google Scholar]
- 40.http://www.sisweb.com/software/ms/nist.htm
- 41.Tallan HH, Moore S, Stein WH. N-Acetyl-L-aspartic acid in brain. J Biol Chem. 1956;219:257–64. [PubMed] [Google Scholar]
- 42.Kikuchi T, Hassanein M, Amann JM, Liu Q, Slebos RJ, Rahman SM, et al. In-depth proteomic analysis of nonsmall cell lung cancer to discover molecular targets and candidate biomarkers. Mol Cell Proteomics. 2012;11:916–32. doi: 10.1074/mcp.M111.015370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan F, Jiang Y, Sun N, Chen Z, Lv Y, Shao K, et al. Identification of isocitrate dehydrogenase 1 as a potential diagnostic and prognostic biomarker for non-small cell lung cancer by proteomic analysis. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.008821. M111 008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin-Bernabe A, Cortes R, Lehmann SG, Seve M, Cascante M, Bourgoin-Voillard S. Quantitative Proteomic Approach to Understand Metabolic Adaptation in Non-Small Cell Lung Cancer. J Proteome Res. 2014 doi: 10.1021/pr500327v. [DOI] [PubMed] [Google Scholar]
- 45.Burri R, Steffen C, Herschkowitz N. N-acetyl-L-aspartate is a major source of acetyl groups for lipid synthesis during rat brain development. Dev Neurosci. 1991;13:403–11. doi: 10.1159/000112191. [DOI] [PubMed] [Google Scholar]
- 46.Chakraborty G, Mekala P, Yahya D, Wu G, Ledeen RW. Intraneuronal N-acetylaspartate supplies acetyl groups for myelin lipid synthesis: evidence for myelin-associated aspartoacylase. Journal of neurochemistry. 2001;78:736–45. doi: 10.1046/j.1471-4159.2001.00456.x. [DOI] [PubMed] [Google Scholar]
- 47.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. The Journal of clinical investigation. 2013;123:3678–84. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–63. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lara MS, Brunson A, Wun T, Tomlinson B, Qi L, Cress R, et al. Predictors of survival for younger patients less than 50 years of age with non-small cell lung cancer (NSCLC): a California Cancer Registry analysis. Lung Cancer. 2014;85:264–9. doi: 10.1016/j.lungcan.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian J, Morgensztern D, Goodgame B, Baggstrom MQ, Gao F, Piccirillo J, et al. Distinctive characteristics of non-small cell lung cancer (NSCLC) in the young: a surveillance, epidemiology, and end results (SEER) analysis. J Thorac Oncol. 2010;5:23–8. doi: 10.1097/JTO.0b013e3181c41e8d. [DOI] [PubMed] [Google Scholar]
- 51.Loeffler DA, Brickman CM, Juneau PL, Perry MF, Pomara N, Lewitt PA. Cerebrospinal fluid C3a increases with age, but does not increase further in Alzheimer’s disease. Neurobiology of aging. 1997;18:555–7. doi: 10.1016/s0197-4580(97)00110-3. [DOI] [PubMed] [Google Scholar]
- 52.Pakulski C, Drobnik L, Millo B. Age and sex as factors modifying the function of the blood-cerebrospinal fluid barrier. Med Sci Monit. 2000;6:314–8. [PubMed] [Google Scholar]
- 53.Montagne A, Barnes SR, Sweeney MD, Halliday MR, Sagare AP, Zhao Z, et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron. 2015;85:296–302. doi: 10.1016/j.neuron.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–61. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


