Abstract
The vertebrate eye is a sophisticated multi-component organ that has been actively studied for over a century, resulting in the identification of the major embryonic and molecular events involved in its complex developmental program. Data gathered so far provides sufficient information to construct a rudimentary network of the various signaling molecules, transcription factors and their targets for several key stages of this process. With the advent of genomic technologies, there has been a rapid expansion in our ability to collect and process biological information, and the use of systems-level approaches to study specific aspects of vertebrate eye development has already commenced. This is beginning to result in the definition of the dynamic developmental networks that operate in ocular tissues, and the interactions of such networks between coordinately developing ocular tissues. Such an integrative understanding of the eye by a comprehensive systems-level analysis can be termed the “oculome”, and that of serial developmental stages of the eye as it transits from its initiation to a fully formed functional organ represents the “developmental oculome”. Construction of the developmental oculome will allow novel mechanistic insights that are essential for organ regeneration-based therapeutic applications, and the generation of computational models for eye disease states to predict the effects of drugs. This review discusses our present understanding of two of the individual components of the developing vertebrate eye – the lens and retina – at both the molecular and systems levels, and outlines the directions and tools required for construction of the developmental oculome.
Keywords: Eye, Development, Systems biology, Pax6, Six3, Organogenesis, Oculome, Transcription factor network, Lens development, Retinal development, Microarray analysis
The developmental “oculome”
Significant advances in technologies as distant as robotics, high throughput sequencing and microarrays, together with dramatic increases in computational power and algorithmic innovation, provide the opportunity to conduct high-throughput experiments on multiple “omics” levels [1–6]. Specifically, transcriptome, proteome, glycome and metabolome datasets can now be obtained for any biological specimen. Cells of normal mouse tissue or those from loss-of-function allelic mutants can be subjected to these analyses. Indeed, systems-level studies for organs such as the heart and the kidney have already resulted in integrative datasets that constitute the “cardiome” and the “nephrome”, respectively [1, 4]. Similarly, a systems-level, integrative understanding of the eye can be termed the “oculome,” and the “developmental oculome” can thus be defined as a similar comprehensive understanding of serial developmental stages of the eye as it develops from its earliest embryonic origins to a fully functional organ.
Building the developmental oculome is a challenging endeavor since oculogenesis is a complex process involving reciprocal interactions between multiple ocular tissues, which are coordinated in a spatially and temporally precise manner. The ocular network that would emerge from these studies will have a four-dimensional nature as the underlying nodes (genes and their products) involved in this program are under strict spatiotemporal regulation.
Knowledge of the vertebrate oculome will enable the definition of the molecular regulatory circuitry involved in the development as well as the maintenance of ocular tissues. In addition, it should be possible to recognize circuit defects that can lead to disease conditions. Such information may allow the construction of detailed computational models for eye development and for ocular disease states that can be used to predict the effects of drugs. Furthermore, such information may permit the formulation of organ regeneration-based therapies.
Overview of vertebrate oculogenesis
The adult vertebrate eye is a sophisticated organ with multiple cell types and complex tissue components, the latter including the retina, lens, cornea, iris, ciliary body, trabecular meshwork, Schlemm’s canal, the ocular glands, and the optic nerve, each with diverse yet specific functions (Fig. 1). These components form as a result of coordinated, multi-step signaling between the prospective lens ectoderm and neuroretina, the lens and the cornea, iris, ciliary body and the periocular mesenchyme [7]. Gruss and co-workers have proposed that vertebrate eye development can be empirically divided into three phases [8]. In the first phase, the process of induction via localized signaling leads to the regional specification and formation of the major morphological structures of the eye. In the second phase, these structures undergo cellular differentiation. Finally, in the third phase, individual components mature, resulting in the formation of a functional eye.
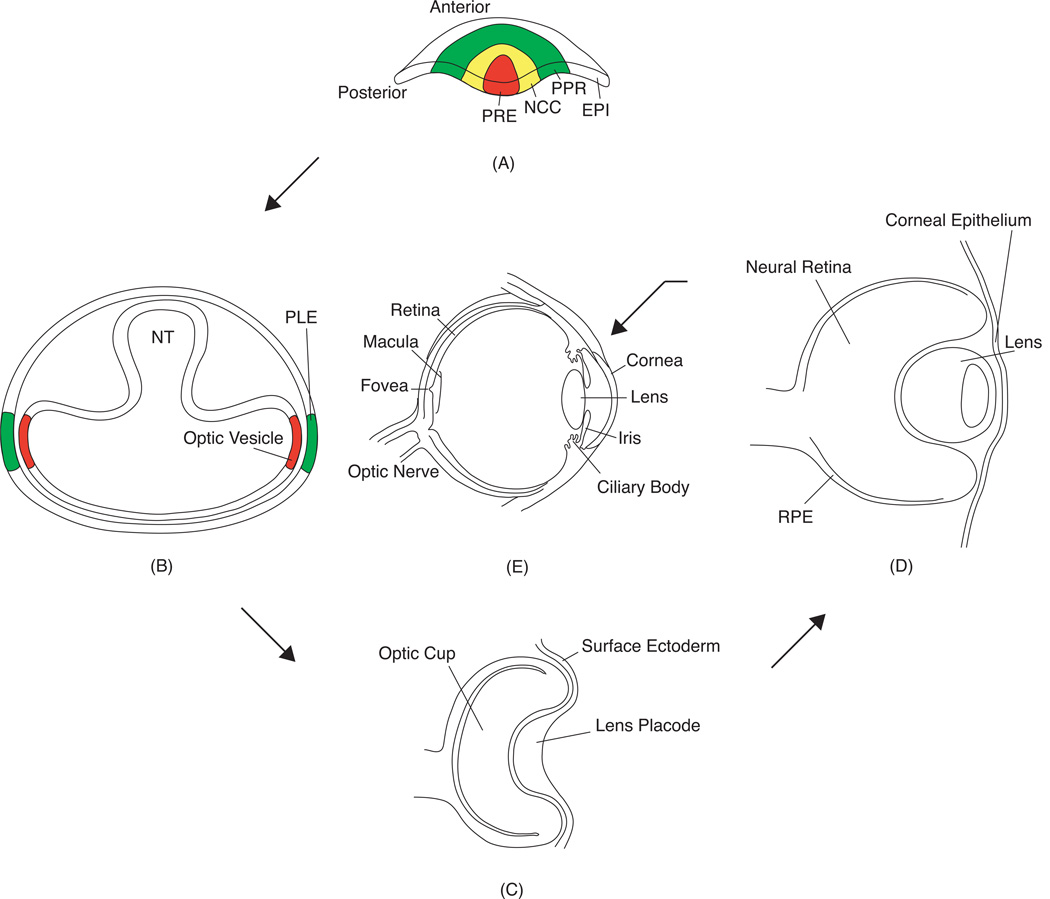
Fig. 1.
Vertebrate ocular development. (A) In late gastrulation, the anterior neural plate contains the presumptive retinal ectoderm (PRE) surrounded by neural crest cells (NCC), pre-placodal region (PRR), and the epidermis (EPI). (B) As the neural tube (NT) closes, the diencephalon bilaterally develops into the optic vesicles, which contact the presumptive lens ectoderm (PLE) on either side. (C) Co-ordinate signaling leads to induction of the PLE to form the lens placode, while the optic vesicle invaginates to form the optic cup. (D) The lens placode invaginates and forms the lens pit that detaches from the surface ectoderm to form the lens vesicle. At this stage the optic cup starts to differentiate into the neural retina and the retinal pigment epithelium (RPE). (E) The adult vertebrate eye thus contains multiple tissue compartments. It is important to note that in fish and frogs, the lens placode does not form a hollow lens vesicle, but instead develops into a solid aggregate of cells.
Eye development has been reviewed previously [7–19], and a detailed description is beyond the scope of this review. However, we do present a brief overview, in order to highlight the critical developmental stages where systems-level approaches can be most readily applied. Overall, oculogenesis is well conserved in vertebrates. However, distinct morphological differences do exist between various model systems and expectedly some of the underlying molecular mechanisms are found to be species-specific [15]. In the event of a species-specific difference in eye development, the process in the mouse is described as a reference.
Specification of the eye field
Vertebrate eye development is initiated early in embryogenesis during late-gastrulation when the ectoderm is divided into four domains: the neural plate, neural crest, pre-placodal region (PPR) and epidermis [15, 20–22]. At this stage, the region of the anterior neural plate (ANP) that eventually forms bilateral retinas is a single “presumptive retinal ectoderm” (PRE), which is surrounded by precursor cells of the telencephalon and the hypothalamus (Fig. 1A) [23–26]. The PPR surrounds the ANP and contains multipotent cells that harbor the potential to form all sensory and neurogenic placodes, including the lens placode. The PPR is specified by a combination of FGF-mediated positive regulation and the simultaneous inhibition of the negative regulators of this process, the Bmp and Wnt signaling pathways. The precise positioning of these tissues in developmental space and time is critical for the co-ordinate exchange of signals between them.
While only the genes encoding the transcription factors Six3 and Foxg1 are presently known to be expressed in the mouse PPR, many others, namely Otx2, Rx, Pax6, Sox2, Hes1, Lhx2, and Six3 are expressed in the PRE and are required for eye formation, along with signaling molecules like Sonic hedgehog (Shh) [27, 28]. The fate of the single broad PRE is progressively restricted by Shh secreted by cells of the midline pre-chordal region, which functions to eventually divide the PRE into two distinct retinal fields [29]. Each retinal field region then evaginates to form an optic pit that progressively grows towards the overlying surface ectoderm and subsequently develops into the optic vesicle (Fig. 1B). Meanwhile, as a result of the anterior-most part of the PPR being biased to form the anterior pituitary and olfactory placodes, the lens-forming region of the PPR is also bilaterally divided [20, 22, 30–34].
Coordinated development of the lens vesicle and the optic cup
After neural tube closure, the optic vesicles grow out bilaterally and expand through the surrounding mesenchyme until they physically contact the overlying surface ectoderm, cells of which form the future lens (Fig. 1B). What follows is one of the best-understood examples of reciprocal inductive tissue interactions in development [35, 36]. Each optic vesicle induces the contacted surface ectoderm to form a thickening termed the lens placode (Fig. 1C). Reciprocally, signaling from the pre-lens ectoderm is essential for the optic vesicle neuroepthelium in gaining competence to form the optic cup [37, 38]. In Pax6 loss-of-function mice - either naturally occurring Pax6Sey1Neu/Sey1Neu (Small eye) mutants [39, 40] or Pax6lacZ/Pax6floxLe-Cre mutants generated by conditional inactivation [37] - the lens placode fails to develop, suggesting a cell autonomous requirement for Pax6 in the transition of presumptive lens ectoderm (PLE) to future lens. In addition, functional characterization of oculogenesis in mice containing loss-of-function alleles for Rx [41], Lhx2 [42, 43], Mab21l2 [44] and Hes1 [45] demonstrates a requirement for these genes and for the optic cup in lens placode development.
These developmental events, which are directed by a series of signaling molecules and transcription factors, lead to the invagination of the lens placode, transforming it into a lens pit. As the lens pit develops, its cells undergo a change in their adhesive characteristics that results in its detachment from the overlying surface ectoderm [46]. At this stage, the lens acquires a globular shape with a hollow cavity, and comprises the lens vesicle (Fig. 1C). Concurrent with invagination of the lens placode, the optic vesicle invaginates to form a bilayered optic cup; the posterior part of the optic cup remains connected to the brain as the optic stalk. Cells in the inner layer of the optic cup form the neural retina (NR) while those in the outer layer differentiate into the retinal pigment epithelium (RPE) (Fig. 1C, D) [17]. Cells in the posterior/ventral region of the optic stalk give rise to the optic nerve, while those from the anterior rim region contribute to the epithelial component of the iris and ciliary body (Fig. 1D) [47].
As the lens vesicle develops further, its cells form two morphologically and functionally distinct cell populations. Cells in the anterior part of the lens vesicle form the anterior lens epithelium (ALE) and remain in the cell cycle, while those in the posterior part commence differentiation to form primary fiber cells. Throughout the life of the animal, cells of the ALE exit the cell cycle in the “transition zone” of the lens and undergo terminal differentiation to secondary fiber cells [7, 48]. The fiber cells of the lens are highly differentiated, and lack cellular organelles and contain an extremely high proportion of proteins termed crystallins [49–51]. Crystallins belong to the metabolic enzyme and stress protective protein classes, and serve non-refractive functions in non-ocular tissues [52]. In the lens, however, crystallins have been coapted via the evolution of lens-specific cis-regulatory elements (CREs) to exhibit high level expression [53, 54]. This leads to an ordered protein lattice that optimizes the refraction of light and provides the requisite optical transparency.
Key molecular events in lens development
Studies in fish and Xenopus indicate the transcription factor genes Six3 and Pax6 are at the top of a genetic cascade that drives lens development. Indeed, ectopic expression of Drosophila (eyeless, twin of eyeless) or Xenopus orthologs of mammalian Pax6 in 3- and 16-cell stage Xenopus embryos, and ectopic expression of mouse Six3 in 2–4-cell stage embryos of Medaka fish, result in the formation of ectopic lenses [55–58]. However, until recently the hierarchical relationship between these two “master” regulatory genes in lens development has been a source of debate [59]. Early studies that demonstrated the absence of Six3 expression in presumptive lens ectoderm (PLE) of Pax6 loss-of-function embryos suggest that Six3 acts downstream of Pax6 in stages following lens placode induction [60]. However, recent studies indicate that Six3 regulates Pax6 in the PPR region that encompasses the PLE, which at this stage is termed the preplacodal ectoderm (PPE). Six3 is also responsible for regulating the placodal expression of Pax6 via multiple enhancers [59, 61, 62]. Conversely, the placodal expression of Pax6 is required for the continued expression of Six3 at later stages of lens development [37, 61, 62]. Feedback loops of this sort are a common theme in the gene regulatory networks (GRNs) that regulate organogenesis.
Meis proteins, members of the TALE or three amino acid loop extension family of homeodomain transcription factors, have also been implicated in the direct regulation of the lens placode phase of Pax6 expression. This is thought to occur via their direct binding to a cognate site in the Pax6 ectodermal enhancer (Pax6 EE), a 110 bp element that lies approximately 3.9 kb upstream of the Pax6 P0 promoter which drives Pax6 expression in the lens placode [63–66]. Also, recent genetic analysis of mouse Pou2f1; Sox2 compound mutant lenses indicates that the HMG box transcription factor Sox2 interacts with the POU family member Oct1 to regulate placode expression of the same Pax6 EE, in turn regulating lens placode induction itself [67]. In addition, Pax6 controls the expression of the Mab gene family member Mab21l1 that is required for lens placode development, and Mab21l1 in turn regulates Foxe3, a highly lens-enriched transcription factor gene that functions in regulating ALE proliferation, the onset of fiber cell differentiation, and lens vesicle closure [68–72]. In the ALE, Foxe3 negatively regulates Prox1, which is required for lens fiber cell differentiation [69, 73–75]. The proliferating cell nuclear antigen (PCNA) gene is expressed in ALE where its direct interaction with residual levels of Prox1 may prevent Prox1 from inappropriately activating fiber cell differentiation proteins such as βB-crystallin, p27Kip1 and p57Kip2 [76]. Lastly, the bicoid-like homeodomain transcription factor Pitx3 is required for ALE maintenance, and functions by regulating Foxe3, Prox1, p27Kip1, p57Kip2 and Pdgfrα [77].
Recently, Sox11, a group C Sox family member has been shown to act downstream of Pax6. It functions in cell cycle regulation during lens vesicle invagination, and in later stages it regulates Bmp7 expression [78]. Another candidate regulator that acts downstream of Pax6 in the lens is the transcription factor AP-2α, which is involved in regulating adhesion molecules critical to lens vesicle separation [79]. The bZIP transcription factor Maf controls the lens-specific expression of crystallin genes, and is also required for fiber cell differentiation [80, 81]. Finally, analysis of Sox1 loss-of-function embryonic lenses reveals an inappropriate up-regulation of Pax6 in fiber cells, suggesting that Sox1 is responsible for the normal down-regulation of Pax6 in this tissue [82]. Along with Prox1, Sox1 regulates γ-Crystallin expression in fiber cell differentiation [73, 83]. Other transcription factors that function in lens development include AP-1, CREB, Etv1, Etv5, Hsf4 and USF, but the functions of these factors require further analyses [18]. Thus, of the approximately 1800 TFs that are encoded in each mammalian genome, a significant subset - several percent - are implicated in the process of lens development either on the basis of mutational analysis or by a suggestive pattern of gene expression.
In addition to these transcription factors, major signaling pathways play distinct roles in specific stages of lens development [14, 84, 85]. Bmp7 is expressed in the PLE prior to lens placode induction and analysis of Bmp7 loss-of-function embryos indicates a requirement for Bmp7 in maintaining Pax6 placodal expression [86]. In addition, Bmp4 loss-of-function mice display aphakia, and Bmp4, secreted from the optic vesicle, is critical for induction of the lens placode during the interaction of the optic vesicle with the PLE [87]. Furthermore, this study indicated a requirement for Bmp4 for Sox2 expression in the lens placode. The Bmp pathway also plays an important role in the initiation of lens fiber cell differentiation [88, 89]. In early stages of lens induction, signaling via the Fgf pathway is essential for regulating Pax6 placodal expression [90]. Later in development, Fgf signaling is crucial for cell cycle exit and for the initiation of fiber cell differentiation [14, 48, 85]. The canonical Wnt pathway co-activator Pygopus2 is required for Pax6 lens placode expression, but in a manner independent of Wnt signaling [91]. However, another canonical Wnt pathway effector, β-catenin, negatively regulates lens fate in periocular ectoderm, but is required for later stages in lens development [92]. In addition, integrin receptors play a critical role in lens development and some of these such as α5 are directly regulated by Pax6 [93]. Finally, activation of Notch signaling in Xenopus embryos leads to eye duplication and in mice has distinct roles in lens progenitor cell proliferation and fiber cell differentiation [58, 94–96]. Together, this information - along with that described in the following sections - allows construction of a first-order gene regulatory network (GRN) for lens development, as depicted in Fig. 2.
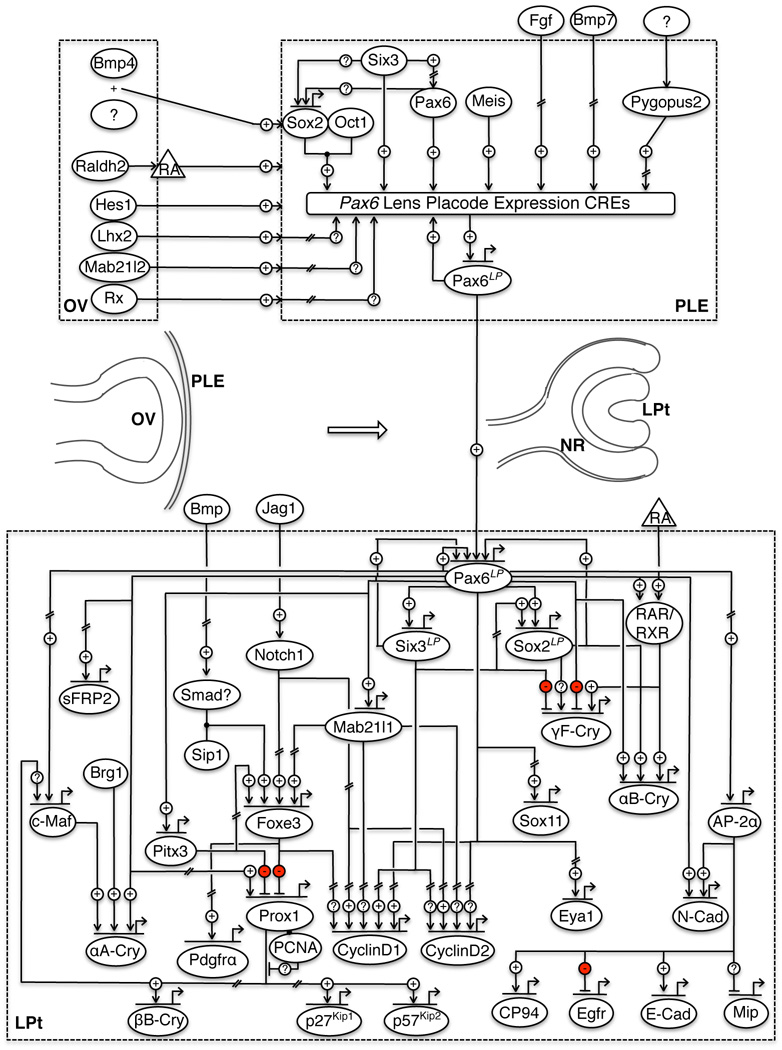
Fig. 2.
The molecular circuitry of early mammalian lens development. Numerous signals from the optic vesicle (OV) along with other molecules converge on multiple cis-regulatory elements (CREs) to induce the placodal expression of Pax6 (Pax6LP) in the presumptive lens ectoderm (PLE). As the PLE develops into the lens pit (LPt), Pax6LP turns on the circuitry necessary for lens formation. Pax6LP, Six3LP and Sox2LP indicate expression of these genes in stages following lens placode induction. Hatches in lines indicate that the interaction may involve intermediate steps.
Retinal differentiation
Retinal differentiation is discussed elsewhere in this issue (see article by Byerly and Blackshaw), and hence we summarize it only briefly. Cells of the inner layer of the bilayered optic cup contain retinal progenitor cells (RPC), which harbor the potential to differentiate into Müller glial cells and into the six types of neurons found in the retina (ganglion, amacrine, horizontal and bipolar cells, and rod and cone photoreceptors). As neurogenesis proceeds, intrinsic changes in the RPC regulate its competence to generate these different cell types [97, 98]. Both proliferation and neuronal differentiation initiate at the center of the inner layer in the prospective retina and proceed outward toward the periphery. The transcription factors Six3, Rx, Chx10, Pax2, Mitf, Otx2 and Pax6 play distinct roles in retinal differentiation [41, 99–104], while Math5 functions in retinal ganglion cell development [105] and Crx, Nrl, Nr2e3 and NeuroD1 are involved in photoreceptor development [106–109]. The Notch, Hedgehog, Egf and Fgf signaling pathways also play a role regulating the differentiation and proliferation of retinal cells [16, 110–114].
Development of the cornea
As the lens vesicle pinches off from the surface ectoderm, the surrounding ectoderm proceeds to close the gap and differentiate into the corneal epithelium, which is accompanied by expression of the marker Keratin12 [115, 116]. Periocular mesenchymal cells and neural crest cells contribute to formation the corneal stroma and endothelium by migrating between the corneal epithelium and the ALE, and differentiating into endothelial cells and keratinocytes [116–118]. This migration is triggered by a signal secreted by the surface ectoderm. In addition, signals from the lens are essential for the mesenchymal-to-epithelial transformation of cells that form the corneal endothelium [119, 120]. Recent lens ablation experiments show that in absence of the lens, the corneal epithelium fails to undergo proper differentiation as indicated by downregulation of Keratin 12 expression, and is replaced by cells that express the epidermal markers Keratin 1 and Keratin 10 [121]. As the cornea develops further, cells of the corneal stroma get arranged in a lamellar manner and undergo compaction, eventually leading to the mature cornea.
Systems-level analyses of eye development
In the following sections, we describe several systems-level approaches, mainly gene expression analysis and network construction, that have been used to study developing ocular tissues. Before going further, we would like to define critical terms and concepts widely used in systems biology. To begin with, systems biology aims to define all the molecular components of biological systems and to integrate this information into a network of interactions that can be used to predict system behavior [2]. An interaction network is composed of “nodes”, which can represent biomolecules, such as proteins, DNA, RNA, or metabolites, and “edges”, which indicate the nature of the relationship between the nodes [6, 122]. A network that operates in an individual cell or tissue at a particular developmental state can be generated in multiple ways, depending on how one defines the nodes and edges. In a transcriptional regulatory network (TRN), nodes represent transcriptional regulators (TRs), which include all the transcription factors, co-factors, and associated chromatin regulators expressed in the cell, as well as their target cis-regulatory elements (CREs) [6].
Biological networks appear to be “scale-free” and follow the power law degree distribution wherein most nodes interact with only a few other nodes and a few “critical” nodes interact with numerous nodes. Such critical nodes can be termed “hubs” [122–124]. This nomenclature has been used mainly for describing protein-protein interactions and is not naturally translated to transcription factors, as these proteins tend to have numerous edges due to their intrinsic functional properties. However, certain transcription factors that are positioned at the apex of a developmental program can be viewed as acting as “hubs” in a conceptual sense. For present purposes, we define these critical nodes as “global regulators” or “developmental hubs”.
Pax6-transcriptional regulatory network in the postnatal lens: a global perspective
Over the past several years, gene expression profiling by microarrays has been used to acquire a systems-level perspective of the Pax6-mediated TRN in lens biology. Early studies used custom-made cDNA microarrays to examine differential gene expression in Pax6+/+ and Pax6+/− lenses from 8-week old mice [125, 126]. More recently these studies have been extended to an earlier time point (P1) and have used commercial microarrays [127]. This approach has resulted in the identification of 559 transcripts that are differentially regulated by Pax6 in the lens. Furthermore, comparison of this lens dataset with that generated for Pax6−/− telecephalonic tissue at E12 and E15 [128] has led to the identification of 178 transcripts that are commonly regulated and 381 transcripts that are differentially regulated by Pax6 in these diverse tissues. Of nine lens-differentially regulated genes, five (Cspg2, Mab21l2, Olfm3, Spag5 and Tgfb2) were demonstrated to be direct targets of Pax6 by quantitative chromatin immunoprecipitation (qChIP) assays. This study also found Pax6 to be involved in the regulation of lesser-characterized components of the Fgf, RA, Tgfβ/Bmp and Wnt signaling pathways in the lens. Furthermore, a comparison of 381 transcripts that are differentially expressed in lens with data from a high-throughput in situ hybridization screen that identified 82 putative Pax6 regulated genes in mid-gestation mouse embryonic tissues [129] revealed eight genes in common (Cdh8, Fgf14, Gabrg2, Neurod1, Neurod6, Pdelc, Tmem2 and Wnt2b) that appear to be Pax6-regulated in the lens, in turn validating these candidate genes as downstream targets of Pax6.
In addition, a recent study describes microarray analyses on lenses from AP-2α conditional loss-of-function mice at (P0) [79]. This study identified 415 transcripts that are differentially regulated in mutant and wild type lenses, which include genes essential for maintenance of the lens epithelium. Since AP-2α is a downstream target of Pax6, data from this study should allow further understanding of the Pax6-gene regulatory network (GRN).
Gene expression analyses of lens fiber cell differentiation
Gene expression profiling studies on the lens have focused on the entire lens and thus represent data from cells of the ALE and the proliferative zone in addition to differentiating fiber cells. However, a recent study attempts to define and compare the transcriptome of young elongating lens fiber cells with that of mature fiber cells in P5 mouse lenses [130]. This study utilized Laser Capture Microdissection (LCM) and GFP-expressing transgenic reporter mice that express GFP in a variegated mosaic manner in young elongating fiber cells to isolate and characterize young and old lens fiber cells. Gene expression profiling experiments revealed 65 genes that are differentially expressed at 2-fold or greater levels between these cell types, with about 75% of the genes being down-regulated as the fiber cells mature. Interestingly, of the 25% of genes that were up-regulated in maturing fiber cells, several have apoptosis-related functions. These include Gadd45b, which encodes an activator of Tgfβ-induced apoptosis [131] and Dlad, which encodes a lens-enriched DNase II beta enzyme that is responsible for degrading fiber cell nuclei [132, 133].
The present model of fiber cell differentiation posits that a specific combination of pro- and anti-apoptotic molecules contribute to fiber cells undergoing a regulated form of apoptosis. According to this model, the apoptotic process does not reach completion and eventually results in these cells attaining an unusual differentiation state whereby they have lost all organelles but remain viable [50, 51]. In support of this view, maturing fiber cells exhibit elevated Bag3 expression, which encodes a chaperone regulator protein that inhibits apoptosis when overexpressed in cell culture [134, 135]. Additionally, this work suggests that the CD9-mediated fusion pathway is active in maturing lens fiber cells, and that this may partly account for how fiber cells fuse to form a syncytium. Further analyses of genes in these datasets should enable a greater understanding of the fiber cell transcriptome and help define the GRNs operative in fiber cell differentiation.
Systems-level analyses of cataractous lenses
Alterations in the gene expression of cataractous and normal lenses have been of interest since even before the advent of microarrays. Using reverse transcription-based polymerase chain reaction differential display (RT-PCR-DD), Kantorow and coworkers [136] detected 3 transcripts that were over-expressed and 12 transcripts that were down-regulated in human senile cataractous lenses compared to normal lenses. Of note was the over-expression of the gene encoding a metallothionein IIa protein, known to be involved in detoxification pathways that respond to oxidative stress, while transcripts encoding a protein phosphatase 2A regulatory subunit (P2A-RS), a suppressor of mitosis, were among those that were down-regulated. These results support the observation that both oxidative stress and cell cycle mis-regulation in the lens are associated with the presence of cataract [137–139].
This study has now been extended by the use of gene expression microarrays to assess altered gene expression in human senile cataractous and normal lenses [140]. This approach identified 1031 transcripts that are differentially expressed between cataractous and normal lenses, with the majority of transcripts showing decreased expression in cataractous lenses [141]. A significant number of genes that showed increased expression in cataractous lenses are associated with ionic transport; these may reflect a compensatory mechanism in response to decreased calcium channel activity in the lens due to oxidative stress. Proteomics-based approaches also reveal a large number of post-translational modifications on major lens proteins, and some are associated with cataract onset [142]. However, further studies are needed to gain a better understanding of these modifications [143].
A dynamic gene regulatory network (GRN) for lens development
Using the information just summarized, we have begun to assemble a first-order gene regulatory network (GRN) for lens development. This GRN encompasses lens development from the lens bias stage through the lens pit stage, and incorporates temporal aspects of the data into the network, rendering it dynamic in nature (Fig. 2). Our goal is to highlight the relationship between the nodes involved and to demonstrate how this GRN evolves dynamically as the lens develops. Not surprisingly, Six3 and Pax6 emerge as critical nodal points that represent “global regulators” or major “developmental hubs” in the lens developmental GRN (Fig. 2) [18, 86, 144]. Both Six3 and Pax6, as well as Sox2 are involved in a transcriptional feed-back regulatory loop that drives and maintains the lens development GRN.
Coincident with the transition of the PLE to lens placode, numerous positive inputs from multiple signaling cascades (Bmp, Fgf, RA, Pygopus) emanate from coordinately developing tissues (optic vesicle and periocular mesenchyme). These signals serve to activate Pax6 expression in the lens placode by impinging on multiple interacting CREs that reside in the Pax6 lens enhancers (Fig. 2) [63–65]. The sophisticated regulation of Pax6 expression, which requires such multiple regulatory inputs, arguably reflects its criticality to the lens developmental cascade. Pax6 is one of 165 genes whose genomic region is associated with highly conserved non-coding sequences, and it has evolved to acquire multiple lens-specific enhancers supports this concept [63, 64, 145]. This view allows us to suggest that one potential indication of whether a TF constitutes a critical “developmental hub” is not just a large number of downstream regulatory targets, but also a number of distinct upstream inputs that have evolved to regulate its expression.
One conspicuous feature of the lens development GRN is the extensive control of cell cycle molecules, e.g., cyclins. In addition, cell cycle regulators, such as Foxe3 and Prox1, are controlled by multiple regulatory inputs. For example, Foxe3, a known positive regulator of the cell cycle, is regulated by multiple positive inputs at this stage when most lens cells are in a proliferative state. In contrast, Prox1 is critical for cell cycle egress and for subsequent commitment to fiber cell differentiation; therefore, at this early stage of lens development, it is regulated predominantly by negative input signals. Furthermore, Prox1 activity is also negatively regulated by a protein-protein interaction with PCNA, which helps ensure that residual Prox1 levels in ALE cells will not inappropriately initiate fiber cell differentiation. Thus, there are both positive and negative regulatory inputs as well as protein-protein interactions that coordinate cell cycle control.
Several other features of the lens development GRN deserve comment. Regulation of αB-Crystallin, an early onset crystallin gene [146] is orchestrated by Pax6, Sox2, and RAR/RXR TFs that are expressed early in the lens. On the other hand, γ-Crystallin, a late onset crystallin is predominantly under the control of negative input signals at this stage. As the lens placode undergoes invagination at this stage, positive input signals for genes that play a role in this process, e.g., AP-2α and Sox11, are transduced, and these in turn regulate the expression of cell adhesion molecules such as N-Cadherin and E-Cadherin that are required for morphologic changes in the invaginating lens [46, 78, 79].
Lastly, Sip1 has recently been shown to play a role in lens development [147]. However, the identity of its interacting Smad partner in the lens remains unknown (Fig. 2). Transcript profiling analysis of the developing lens identifies the presence of Smad proteins 1, 2, 3, and 5, indicating them to be potential interacting partners of Sip1 in the lens (Lachke and Maas, unpublished observations). Thus, transcript profiling analysis and network construction can be utilized to make predictions to define specific interactions between molecules.
Systems-level approaches in retinal development and disease
Global gene expression analyses by both serial analysis of gene expression (SAGE) and microarray have identified hundreds of genes that are expressed in retinal development. The availability of these datasets has resulted in more efforts at generating a GRN for retinal development than for any other ocular component [148–154]. The use of motif discovery algorithms has led to the prediction of several rod- and cone-specific CREs and some of these have been experimentally validated [155]. Specifically, promoters of cone-expressed genes are found to contain an enrichment of predicted motifs for Rx and Engrailed family members, while promoters of rod-expressed genes are found to contain predicted IL-6 effector motifs. Using an algorithm for searching binding motifs for the retinal transcriptional regulators Crx, Nrl and Nr2e3 near transcription start sites of retinal-expressed genes, Qian and coworkers [156] have predicted several potential targets and have validated Rp1, Gucy2d and Abca4 as novel targets of Crx.
Interestingly, the Nrl−/− mouse retina lacks rod photoreceptor cells and displays an extreme “cone photoreceptor cell only” phenotype, thereby presenting a unique opportunity to gain insight into the transcriptomes of these two photoreceptor cell types [109]. Comparative microarray analyses of Nrl−/− and wild type retinas indicate alterations in both the Bmp and Wnt signaling pathways, with a downregulation of Bmp4 and Smad4 genes in the Nrl−/− retina suggests their deployment in rod photoreceptor development [157, 158]. Recently, fluorescence assisted cell sorting (FACS) combined with gene expression profiling of RPCs has led to the identification of over 800 retinal transcripts enriched in these cells [159]. Genes regulating the cell cycle are well represented in RPCs, reflecting the proliferative nature of these cells. Moreover, enrichment of chromatin regulators including ATRX, chromobox homolog 3 (HP1-γ) and histone methyltransferase suggest a role for epigenetic mechanisms in regulating RPC multipotency.
Crx null mice are blind at birth and have no detectable photoreceptor function. Photoreceptors do not form in the outer segment that is necessary for phototransduction, and 46% of photoreceptor-enriched genes are Crx-dependent, particularly opsin genes (Blackshaw 2001). Peng and Chen [160] used ChIP assays to demonstrate the inability of Nr2e3, a rod-photoreceptor determining “orphan nuclear receptor” to bind its target CREs in Crx−/− mice, revealing how complex interactions among key TFs can contribute to a retinal phenotype. Recently, Hennig and coworkers [161] used ChIP and quantitative RT-PCR combined with existing knowledge of retinal photoreceptor development to analyze the TRN of five TFs that are key to this process. They studied the ability of Crx (necessary for photoreceptor cell lineage determination), Nrl and Nr2e3 (necessary for rod cell lineage determination), and the HLH factor NeuroD1 (necessary for survival of photoreceptor cells) to regulate each other via direct binding to regulatory elements in retinal tissues from wild type, Crx−/−, Nrl−/− and Nr2e3−/− mice. Their model transcriptional regulatory network (TRN) of these photoreceptor transcription factors is summarized in Fig. 3.
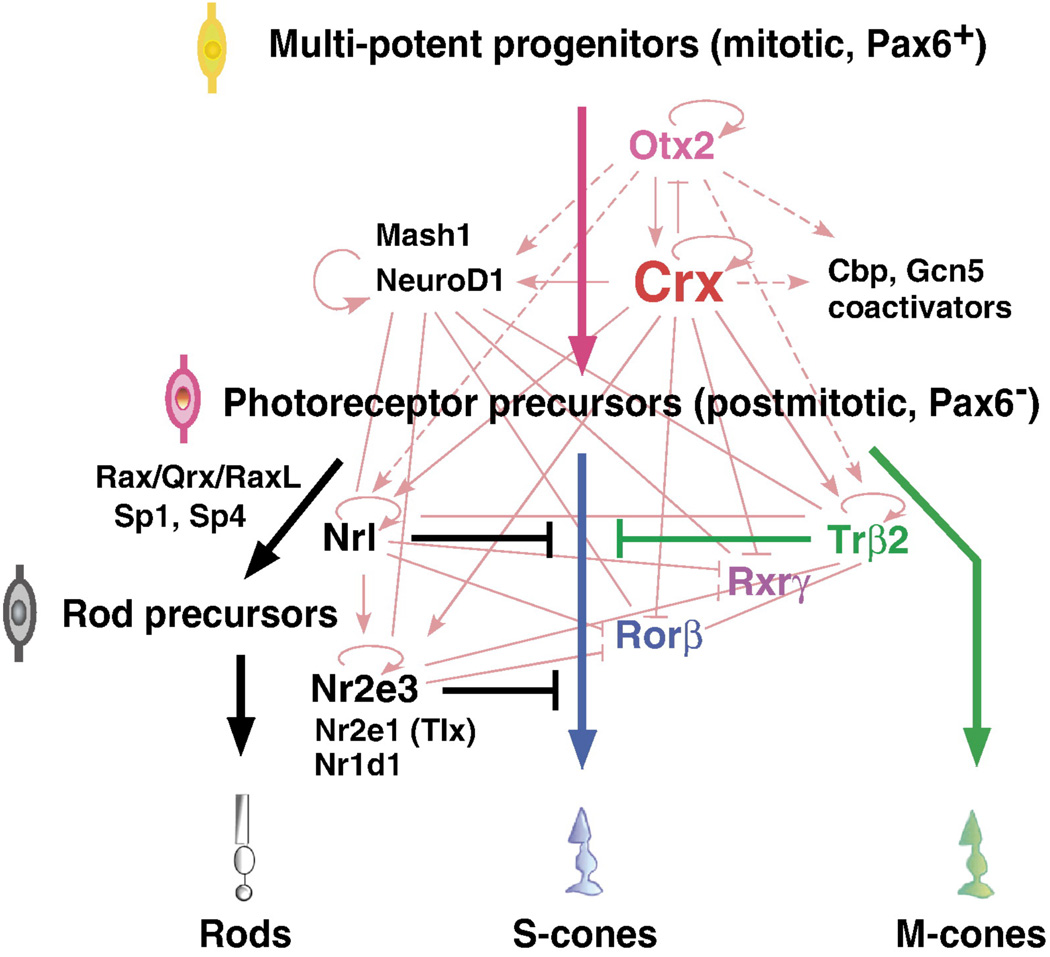
Fig. 3.
Transcription regulatory network (TRN) for retinal photoreceptor cell development. Multipotent, photoreceptor precursor cells differentiate into individual photoreceptor cell types via three separate pathways. Thin lines indicate protein-promoter interactions; solid lines indicate published data and dotted lines are from unpublished data of Hennig et al 2008 [145]. Reprinted from Brain Research 1192, Hennig, A.K., Peng, G-H., Chen, S., Regulation of photoreceptor gene expression by Crx-associated transcription factor network, 114–133, (2008) with permission from Elsevier.
An alternative approach towards defining the Crx, Nrl, and Nr2e3 TRN in photoreceptor cell development employs gene expression and in situ hybridization datasets, combined with novel computational tools [162]. This approach has led to the construction of a network that comprises 600 components and includes novel transcription factors that were previously uncharacterized in photoreceptors cells. An algorithm that identifies photoreceptor-specific CREs of these nodes (photoreceptor transcription factors) was used to define the edges of this network. This allowed for the derivation of a cis-regulatory grammar rule, which renders high-level photoreceptor-specific gene expression. Moreover, Hsiau and colleagues used this information to deduce photoreceptor CREs by purely computational means, and have demonstrated the capacity of such modules to drive photoreceptor-specific expression in vivo [162]. This study illustrates how convergent datasets can be used to construct a highly informative developmental TRN.
Finally, systems-level analysis has also provided insight into retinal disease. For example, age related macular degeneration (AMD) is the most common form of macular disease and leads to the degeneration of central retina and loss of vision [163]. To achieve comprehensive gene expression profiling of the macula, SAGE and microarray analyses have been carried out and have identified candidate genes for ocular genetic disorders [164]. Although further studies are needed to validate the function of these candidate genes, comparison of these datasets with those for surrounding extra-macular tissue have identified 21 differentially expressed genes that may play a role in AMD [165].
In addition, systems level approach has been initiated to the study of glaucoma. Physiological changes in retinal ganglion cells and glial cells contribute to the pathology of glaucoma. Transcript profiling analyses of these cell types have provided insights into the pathology of glaucoma [166–168]. Moreover, several candidate genes have been identified that are present within loci linked to this complex multifactorial disease [167].
Future directions: construction of a developmental oculome
Although systems-level analytical tools are gaining increasing power, the datasets available for ocular tissues in developmental stages are presently incomplete. Furthermore, there have been only a handful of attempts to build TRNs and GRNs using such systems-level data. A complete understanding of oculogenesis will require a complete definition of all the nodes and edges for each distinct tissue and cell type of the eye as a function of development. In the sections that follow, we examine experiments that need to be performed to provide such a systems-level understanding of ocular development.
Constructing systems-level datasets for eye components
As summarized above, gene expression datasets exist for specific tissue components of the eye, especially the retina [148–154], and in certain cases, the expression profiling of single cell-types has been successfully attempted [152–154]. However, there is nonetheless a significant lack of cell type specific datasets for serial stages of ocular tissues at the transcript and protein levels. To help remedy this situation, it is now possible to employ LCM or FACS separation, coupled with cell type-specific enhancer driven GFP reporters, to isolate individual tissue components or specific cell-types of the mouse embryonic eye for further analyses. This is especially critical for the analysis of highly heterogeneous retinal cell populations [98]. Recently, a similar approach has been successfully utilized in a systems-level analysis of the developing kidney [169]. Brown and coworkers [170] have conceived of an analogous approach utilizing LCM coupled with expression profiling to identify two novel genes that serve an essential function in optic fissure closure.
Thus, individual ocular components from mouse embryos separated by 0.5-day intervals can be isolated by LCM or FACS sorting, and subjected to global gene expression and proteomic analyses to obtain serial datasets at both the mRNA and protein levels. Using any of a number of relatively new computational algorithms (a few of which are described below), the data gathered can then be processed to generate a network for each stage and tissue of ocular development. Such analyses may demonstrate how individual ocular tissue specific GRNs regulate the development of specific ocular components, and also enable predictions about how the systems level behavior of ocular development depends the underlying network architecture. Furthermore, systems-level analyses of mouse models with ocular defects should provide a more detailed understanding of the structure of individual tissue component and cellular GRNs. For example, with LCM it is now possible to isolate and analyze the surface ectodermal region prior to induction of the lens placode from Pax6−/−, Pax6+/− and Pax6+/+ embryos. This should allow the identification of critical early events in the Pax6-GRN. Similar experiments could be performed for Six3 conditional loss-of-function embryonic PLE.
Networks of RNA-binding proteins, microRNA and their targets
In recent years, RNA-binding proteins (RBPs) and small non-coding RNA molecules and their associated protein machinery have emerged as mediators of gene regulation [171]. The microRNA (miRNA) mediated gene-silencing pathway that is conserved across many species has been discovered to play a central role in RNA interference (RNAi). Recently, the miRNA pathway has been shown to operate in cytoplasmic RNA granules that are classified as P-bodies; these serve as sites of mRNA decay and degradation [172]. This observation raises the question of whether specific RNA granule components or miRNAs that function in oculogenesis show tissue-specific expression.
Recent data addresses this question. For example, a specific class of RNA granules are expressed in the developing lens (Lachke et al., 2009 submitted), and deficiency of a lens-enriched RNA granule component of this class of RNA granules, Tdrd7, leads to mis-regulation of key transcription factor genes and produces cataract formation. This establishes that RNA binding proteins that constitute tissue-specific components of RNA granules play a critical role in eye morphogenesis. Similarly, a role for miRNA-mediated gene regulation in oculogenesis has been indicated in Xenopus laevis, where overexpression of a specific miRNA (miR-196a) negatively affects eye development by downregulating the expression of ET, Rx1, Six3, Pax6, Lhx2, Optx2, and Ath5 in the PRE [173]. In addition, miRNAs and natural antisense transcripts for several regulators of eye development have recently been described and exhibit tissue-specific expression [174–176]. In view of these findings, the identification of RBPs and their mRNA targets, and the generation of an miRNA expression profile for miRNAs and the identication of their targets, will be required to gain a complete understanding of the GRNs that control ocular development.
Novel tools and approaches in systems-level analysis
As outlined above, large-scale gene expression datasets for several ocular tissues exist. However, attempts to process this information, either for GRN construction or disease gene identification, are more limited [177]. Understanding the entire protein-protein interaction network during oculogenesis is essential in building the oculome. However, we will focus on approaches and technologies that would enable building of transcriptional regulatory networks via study of cis-regulatory modules, since this represents the logical first step in building a network [178]. Below, we outline several systems-level strategies and computational tools that can be used to efficiently identify ocular disease genes and to construct TRNs for specific ocular tissues.
Systems approaches to ocular disease gene discovery
We have formulated a method for processing mouse lens developmental microarray datasets that enables efficient gene discovery in lens development and disease. The underlying idea is that selectively enriched gene expression in a particular organ tissue above background levels increases the probability that the gene of interest plays a role in the development or function of that organ or tissue component [148, 177]. To pursue this goal, we first generated a developmental profile of the mouse lens transcriptome as the lens transitions from the stage of lens placode invagination to that of lens vesicle formation, when fiber cell differentiation has commenced (Lachke et al. 2009, submitted). Then, to identify differentially regulated genes, we established a normalization protocol by which lens microarray datasets are subjected to in silico subtraction with a developmentally matched microarray dataset representing the whole embryo minus ocular tissue. This in silico normalization protocol ranks probe datasets based on their lens enrichment p-values.
The resulting normalized mouse lens database can be efficiently used to identify lens-enriched genes. The implicit hypothesis is that such genes are likely to play important roles in lens biology. For example, genes that survive this filter for lens enrichment can be used to identify and prioritize potential candidate genes that fall in the vicinity of mapped human cataract loci. This is achieved by ranking all orthologous human genes within a mapped interval for a particular human cataract locus based on their murine homolog developing lens gene expression “enrichment score” as assigned by the normalized database. When this approach is used, all the previously known human congenital cataract genes are readily identified. Furthermore, we have applied this protocol to identify a novel human cataract gene, TDRD7 (Lachke et al 2009, submitted). Thus, by constructing developmental microarray datasets and subsequently normalized databases for various tissue compartments of the developing eye, one can identify novel ocular disease genes.
Tools for building transcriptional regulatory networks (TRNs)
Two major limiting factors in building TRNs for transcription factors (TFs) have been: (1) identification TF DNA-binding motifs, and (2) determination of the biological significance of such motifs throughout the genome. The development of protein binding microarrays (PBMs) has led to the identification of motifs for several hundred DNA binding proteins [179–185]. This technology provides an opportunity to identify cis-regulatory “codes” or “modules” for co-regulated genes. The premise of this approach is that the most parsimonious way for nature to accomplish the coordinate expression of genes in overlapping spatiotemporal domains is to co-evolve binding motifs for the same or similar ensembles of transcription factors. Thus, the flanking and intronic genomic regions of co-regulated genes are expected to display an enrichment of DNA binding motifs for particular groups or ensembles of TFs [5, 186, 187]. Recently, two algorithms, PhylCRM and Lever, have been developed that work together to identify cis-regulatory modules (CRMs) for specified combinations of transcription factors in developmental gene expression datasets [188]. PhylCRM interrogates genomic sequences for combinations of evolutionarily conserved transcription factor motifs, while Lever uses this information to map overrepresented regulatory modules in gene expression datasets for a cell or tissue type at a specific developmental state.
Application of this type of strategy should allow for the efficient building of TRNs for major lens transcription factors as the lens transits from the placode stage to the primary fiber cell differentiation stage. By mining the database of genes that are differentially expressed in the lens, it should be possible to identify previously characterized as well as uncharacterized TFs for which binding motifs can be determined using PBMs. Then, by using Lever and PhylCRM, one can perform a motif-search for various combinations of the above transcription factors in genomic regions near differentially regulated genes in the lens. A biologically tested model for some of these combinations already exists (e.g., for highly lens-specific crystallin gene promoters or enhancers), and these CRMs can act as “positive controls.” Theoretically, this approach can predict novel lens enhancers, and it can also help to define the nodes and edges in a TRN for lens TFs and their targets. Of course, predictions based on such a model must be tested experimentally. However, this strategy can be repeated for other components of the eye to build up an increasingly comprehensive and well-integrated TRN of ocular transcription factors and their targets (Fig. 4).
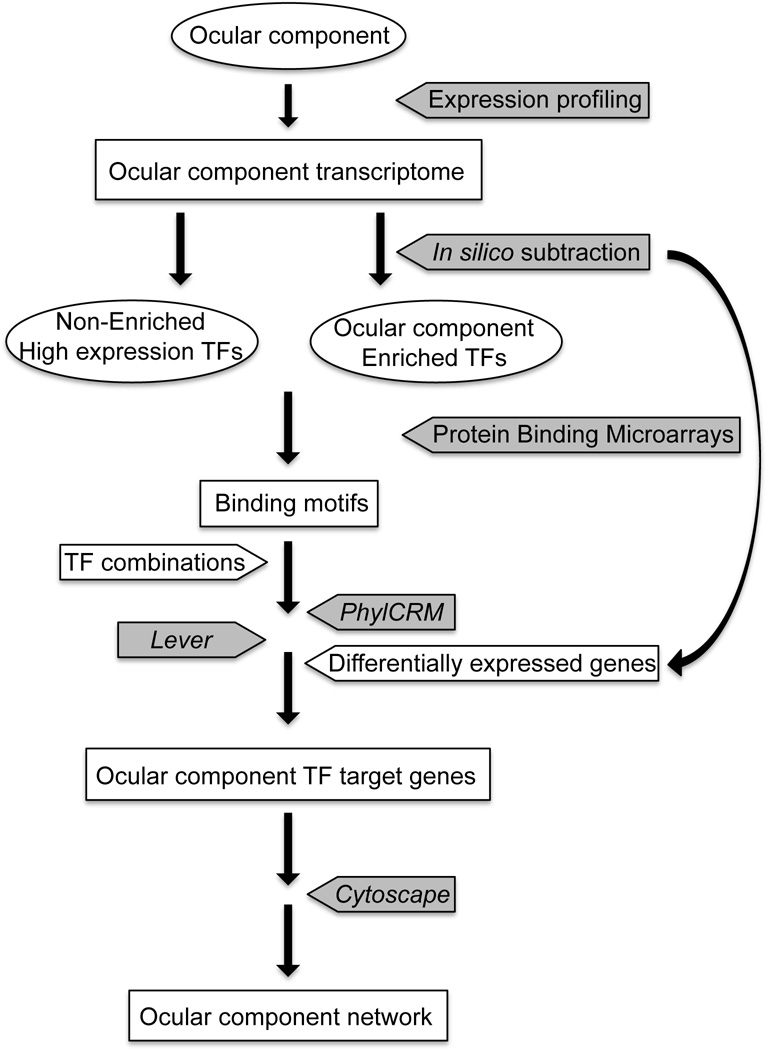
Fig. 4.
Strategy outlining the construction of networks for specific ocular compartments using gene expression profiling and computational tools. Experimental steps are indicated in gray boxes and names of computational software are italicized.
Clinical applications of the oculome
The approaches outlined above would lead to the identification of a majority – if not all – of the genes that are involved in ocular disease conditions. Eventually, this information can be used to design a signature “ocular disease gene set”, which can be used for diagnostic purposes. With the advent of newer and cheaper technologies for sequencing, the entire ocular disease gene set can be sequenced from individual patients to determine the identity of the mutant gene(s). Furthermore, non-coding regulatory regions of ocular disease genes, which will be identified by the TRN analysis, can also be incorporated in such a gene set. Thus, information derived from the oculome has the potential to not only advance our fundamental understanding of oculogenesis, but to also contribute to the diagnosis and treatment of ocular diseases.
Conclusion
The past few years have witnessed a dramatic increase in the application of systems analysis to the study of vertebrate eye development. However, our present understanding of oculogenesis still derives largely from the application of molecular genetic tools. Yet even with such approaches, it is already possible to establish simple networks for some of the major molecular components involved in lens and retinal development. Certain essential nodes, termed “global regulators” or “developmental hubs” in the lens GRN such as Pax6 and Six3 are known, and analyses of mouse loss-of-function models for key genes in the ocular GRN should provide further insight into the identities and toplogy of the nodes and edges in this network. Finally, application of this approach to serial developmental stages of each tissue compartment of the developing eye should provide insight into how the individual GRNs for each ocular tissue mature during development. This will represent a first step towards understanding how such networks interact with each other in the complex context of vertebrate oculogenesis, and help to elucidate the vertebrate oculome.
Acknowledgments
S.A.L. and R.L.M. were supported by NIH grants R01EY010123 (NEI) and R01HD060050 (NICHD).
Contributor Information
Salil A. Lachke, Email: slachke@rics.bwh.harvard.edu.
Richard L. Maas, Email: maas@genetics.med.harvard.edu.
References
- 1.McCulloch AD. Modeling the human cardiome in silico. J Nucl Cardiol. 2000;7:496–499. doi: 10.1067/mnc.2000.109682. [DOI] [PubMed] [Google Scholar]
- 2.McCulloch AD, Paternostro G. Cardiac systems biology. Ann N Y Acad Sci. 2005;1047:283–295. doi: 10.1196/annals.1341.025. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava R, Varner J. Emerging technologies: systems biology. Biotechnol Prog. 2007;23:24–27. doi: 10.1021/bp060352v. [DOI] [PubMed] [Google Scholar]
- 4.Monte J, Sakurai H, Bush K, Nigam S. The developmental nephrome: systems biology in the developing kidney. Current Opinion in Nephrology and Hypertension. 2007;16:3–9. doi: 10.1097/MNH.0b013e3280118a5a. [DOI] [PubMed] [Google Scholar]
- 5.Busser BW, Bulyk ML, Michelson AM. Toward a systems-level understanding of developmental regulatory networks. Curr Opin Genet Dev. 2008;18:521–529. doi: 10.1016/j.gde.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan K, Tegner J, Ravasi T. Integrated approaches to uncovering transcription regulatory networks in mammalian cells. Genomics. 2008;91:219–231. doi: 10.1016/j.ygeno.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 8.Jean D, Ewan K, Gruss P. Molecular regulators involved in vertebrate eye development. Mech Dev. 1998;76:3–18. doi: 10.1016/s0925-4773(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan CH, Braunstein L, Hazard-Leonards RM, Holen AL, Samaha F, Stephens L, Grainger RM. A re-examination of lens induction in chicken embryos: in vitro studies of early tissue interactions. Int J Dev Biol. 2004;48:771–782. doi: 10.1387/ijdb.041894cs. [DOI] [PubMed] [Google Scholar]
- 10.Zhang SS, Fu XY, Barnstable CJ. Molecular aspects of vertebrate retinal development. Mol Neurobiol. 2002;26:137–152. doi: 10.1385/MN:26:2-3:137. [DOI] [PubMed] [Google Scholar]
- 11.Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- 12.Lang RA. Pathways regulating lens induction in the mouse. Int J Dev Biol. 2004;48:783–791. doi: 10.1387/ijdb.041903rl. [DOI] [PubMed] [Google Scholar]
- 13.Yang XJ. Roles of cell-extrinsic growth factors in vertebrate eye pattern formation and retinogenesis. Semin Cell Dev Biol. 2004;15:91–103. doi: 10.1016/j.semcdb.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovicu FJ, McAvoy JW. Growth factor regulation of lens development. Dev Biol. 2005;280:1–14. doi: 10.1016/j.ydbio.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Donner AL, Lachke SA, Maas RL. Lens induction in vertebrates: variations on a conserved theme of signaling events. Semin Cell Dev Biol. 2006;17:676–685. doi: 10.1016/j.semcdb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Esteve P, Bovolenta P. Secreted inducers in vertebrate eye development: more functions for old morphogens. Curr Opin Neurobiol. 2006;16:13–19. doi: 10.1016/j.conb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Adler R, Canto-Soler MV. Molecular mechanisms of optic vesicle development: complexities, ambiguities and controversies. Dev Biol. 2007;305:1–13. doi: 10.1016/j.ydbio.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cvekl A, Duncan MK. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondoh H. Shedding light on developmental gene regulation through the lens. Dev Growth Differ. 2008;50:S57–S69. doi: 10.1111/j.1440-169X.2008.01007.x. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch N, Grainger RM. Induction of the lens. Results Probl Cell Differ. 2000;31:51–68. doi: 10.1007/978-3-540-46826-4_4. [DOI] [PubMed] [Google Scholar]
- 21.Streit A. Early development of the cranial sensory nervous system: from a common field to individual placodes. Dev Biol. 2004;276:1–15. doi: 10.1016/j.ydbio.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 22.Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- 23.Eagleson G, Ferreiro B, Harris WA. Fate of the anterior neural ridge and the morphogenesis of the Xenopus forebrain. J Neurobiol. 1995;28:146–158. doi: 10.1002/neu.480280203. [DOI] [PubMed] [Google Scholar]
- 24.Varga ZM, Wegner J, Westerfield M. Anterior movement of ventral diencephalic precursors separates the primordial eye field in the neural plate and requires cyclops. Development. 1999;126:5533–5546. doi: 10.1242/dev.126.24.5533. [DOI] [PubMed] [Google Scholar]
- 25.Inoue T, Nakamura S, Osumi N. Fate mapping of the mouse prosencephalic neural plate. Dev Biol. 2000;219:373–383. doi: 10.1006/dbio.2000.9616. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Garre P, Rodriguez-Gallardo L, Gallego-Diaz V, Alvarez IS, Puelles L. Fate map of the chicken neural plate at stage 4. Development. 2002;129:2807–2822. doi: 10.1242/dev.129.12.2807. [DOI] [PubMed] [Google Scholar]
- 27.Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- 28.Macdonald R, Barth KA, Xu Q, Holder N, Mikkola I, Wilson SW. Midline signalling is required for Pax gene regulation and patterning of the eyes. Development. 1995;121:3267–3278. doi: 10.1242/dev.121.10.3267. [DOI] [PubMed] [Google Scholar]
- 29.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 30.Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- 31.Noramly S, Grainger RM. Determination of the embryonic inner ear. J Neurobiol. 2002;53:100–128. doi: 10.1002/neu.10131. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya S, Bailey AP, Bronner-Fraser M, Streit A. Segregation of lens and olfactory precursors from a common territory: cell sorting and reciprocity of Dlx5 and Pax6 expression. Dev Biol. 2004;271:403–414. doi: 10.1016/j.ydbio.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharyya S, Bronner-Fraser M. Hierarchy of regulatory events in sensory placode development. Curr Opin Genet Dev. 2004;14:520–526. doi: 10.1016/j.gde.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Bailey AP, Streit A. Sensory organs: making and breaking the pre-placodal region. Curr Top Dev Biol. 2006;72:167–204. doi: 10.1016/S0070-2153(05)72003-2. [DOI] [PubMed] [Google Scholar]
- 35.Hamburger V. Ontogeny of neuroembryology. Journal of Neuroscience. 1988;8:3535–3540. doi: 10.1523/JNEUROSCI.08-10-03535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grainger RM. Embryonic lens induction: shedding light on vertebrate tissue determination. Trends Genet. 1992;8:349–355. doi: 10.1016/0168-9525(92)90280-h. [DOI] [PubMed] [Google Scholar]
- 37.Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyer J, Kuhlman J, Afif E, Mikawa T. Optic cup morphogenesis requires pre-lens ectoderm but not lens differentiation. Dev Biol. 2003;259:351–363. doi: 10.1016/s0012-1606(03)00205-7. [DOI] [PubMed] [Google Scholar]
- 39.Hogan BL, Horsburgh G, Cohen J, Hetherington CM, Fisher G, Lyon MF. Small eyes (Sey): a homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J Embryol Exp Morphol. 1986;97:95–110. [PubMed] [Google Scholar]
- 40.Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 41.Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 42.Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 43.Tetreault N, Champagne MP, Bernier G. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol. 2009;327:541–550. doi: 10.1016/j.ydbio.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 44.Yamada R, Mizutani-Koseki Y, Koseki H, Takahashi N. Requirement for Mab21l2 during development of murine retina and ventral body wall. Dev Biol. 2004;274:295–307. doi: 10.1016/j.ydbio.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 45.Lee HY, W E, Philips GT, Stair CN, Conley K, Reedy M, Mastick GS, Brown NL. Multiple requirements for Hes 1 during early eye formation. Developmental Biology. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pontoriero GF, Smith AN, Miller LA, Radice GL, West-Mays JA, Lang RA. Co-operative roles for E-cadherin and N-cadherin during lens vesicle separation and lens epithelial cell survival. Dev Biol. 2009;326:403–417. doi: 10.1016/j.ydbio.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Napier HR, Kidson SH. Molecular events in early development of the ciliary body: a question of folding. Exp Eye Res. 2007;84:615–625. doi: 10.1016/j.exer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Griep AE. Cell cycle regulation in the developing lens. Semin Cell Dev Biol. 2006;17:686–697. doi: 10.1016/j.semcdb.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bassnett S. Fiber cell denucleation in the primate lens. Invest Ophthalmol Vis Sci. 1997;38:1678–1687. [PubMed] [Google Scholar]
- 50.Bassnett S. Lens organelle degradation. Exp Eye Res. 2002;74:1–6. doi: 10.1006/exer.2001.1111. [DOI] [PubMed] [Google Scholar]
- 51.Bassnett S. On the mechanism of organelle degradation in the vertebrate lens. Exp Eye Res. 2009;88:133–139. doi: 10.1016/j.exer.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graw J. Genetics of crystallins: cataract and beyond. Exp Eye Res. 2009;88:173–189. doi: 10.1016/j.exer.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 53.Piatigorsky J, Wistow GJ. Enzyme/crystallins: gene sharing as an evolutionary strategy. Cell. 1989;57:197–199. doi: 10.1016/0092-8674(89)90956-2. [DOI] [PubMed] [Google Scholar]
- 54.Piatigorsky J. Crystallin genes: specialization by changes in gene regulation may precede gene duplication. J Struct Funct Genomics. 2003;3:131–137. [PubMed] [Google Scholar]
- 55.Oliver G, Loosli F, Koster R, Wittbrodt J, Gruss P. Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech Dev. 1996;60:233–239. doi: 10.1016/s0925-4773(96)00632-6. [DOI] [PubMed] [Google Scholar]
- 56.Altmann CR, Chow RL, Lang RA, Hemmati-Brivanlou A. Lens induction by Pax-6 in Xenopus laevis. Dev Biol. 1997;185:119–123. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- 57.Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- 58.Onuma Y, Takahashi S, Asashima M, Kurata S, Gehring WJ. Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. Proc Natl Acad Sci U S A. 2002;99:2020–2025. doi: 10.1073/pnas.022626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goudreau G, Petrou P, Reneker LW, Graw J, Loster J, Gruss P. Mutually regulated expression of Pax6 and Six3 and its implications for the Pax6 haploinsufficient lens phenotype. Proc Natl Acad Sci U S A. 2002;99:8719–8724. doi: 10.1073/pnas.132195699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 61.Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purcell P, Oliver G, Mardon G, Donner AL, Maas RL. Pax6-dependence of Six3, Eya1 and Dach1 expression during lens and nasal placode induction. Gene Expr Patterns. 2005;6:110–118. doi: 10.1016/j.modgep.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 63.Williams SC, Altmann CR, Chow RL, Hemmati-Brivanlou A, Lang RA. A highly conserved lens transcriptional control element from the Pax-6 gene. Mech Dev. 1998;73:225–229. doi: 10.1016/s0925-4773(98)00057-4. [DOI] [PubMed] [Google Scholar]
- 64.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 65.Dimanlig PV, Faber SC, Auerbach W, Makarenkova HP, Lang RA. The upstream ectoderm enhancer in Pax6 has an important role in lens induction. Development. 2001;128:4415–4424. doi: 10.1242/dev.128.22.4415. [DOI] [PubMed] [Google Scholar]
- 66.Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 2002;16:2097–2107. doi: 10.1101/gad.1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donner AL, Episkopou V, Maas RL. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol. 2007;303:784–799. doi: 10.1016/j.ydbio.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamada R, Mizutani-Koseki Y, Hasegawa T, Osumi N, Koseki H, Takahashi N. Cell-autonomous involvement of Mab21l1 is essential for lens placode development. Development. 2003;130:1759–1770. doi: 10.1242/dev.00399. [DOI] [PubMed] [Google Scholar]
- 69.Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–254. [PMC free article] [PubMed] [Google Scholar]
- 70.Brownell I, Dirksen M, Jamrich M. Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. Genesis. 2000;27:81–93. doi: 10.1002/1526-968x(200006)27:2<81::aid-gene50>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 71.Blixt A, Landgren H, Johansson BR, Carlsson P. Foxe3 is required for morphogenesis and differentiation of the anterior segment of the eye and is sensitive to Pax6 gene dosage. Dev Biol. 2007;302:218–229. doi: 10.1016/j.ydbio.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 72.Medina-Martinez O, Jamrich M. Foxe view of lens development and disease. Development. 2007;134:1455–1463. doi: 10.1242/dev.000117. [DOI] [PubMed] [Google Scholar]
- 73.Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- 74.Medina-Martinez O, Brownell I, Amaya-Manzanares F, Hu Q, Behringer RR, Jamrich M. Severe defects in proliferation and differentiation of lens cells in Foxe3 null mice. Mol Cell Biol. 2005;25:8854–8863. doi: 10.1128/MCB.25.20.8854-8863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Landgren H, Blixt A, Carlsson P. Persistent FoxE3 expression blocks cytoskeletal remodeling and organelle degradation during lens fiber differentiation. Invest Ophthalmol Vis Sci. 2008;49:4269–4277. doi: 10.1167/iovs.08-2243. [DOI] [PubMed] [Google Scholar]
- 76.Chen X, Patel TP, Simirskii VI, Duncan MK. PCNA interacts with Prox1 and represses its transcriptional activity. Mol Vis. 2008;14:2076–2086. [PMC free article] [PubMed] [Google Scholar]
- 77.Ho HY, Chang KH, Nichols J, Li M. Homeodomain protein Pitx3 maintains the mitotic activity of lens epithelial cells. Mech Dev. 2009;126:18–29. doi: 10.1016/j.mod.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 78.Wurm A, Sock E, Fuchshofer R, Wegner M, Tamm ER. Anterior segment dysgenesis in the eyes of mice deficient for the high-mobility-group transcription factor Sox11. Exp Eye Res. 2008;86:895–907. doi: 10.1016/j.exer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Pontoriero GF, Deschamps P, Ashery-Padan R, Wong R, Yang Y, Zavadil J, Cvekl A, Sullivan S, Williams T, West-Mays JA. Cell autonomous roles for AP-2alpha in lens vesicle separation and maintenance of the lens epithelial cell phenotype. Dev Dyn. 2008;237:602–617. doi: 10.1002/dvdy.21445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawauchi S, Takahashi S, Nakajima O, Ogino H, Morita M, Nishizawa M, Yasuda K, Yamamoto M. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem. 1999;274:19254–19260. doi: 10.1074/jbc.274.27.19254. [DOI] [PubMed] [Google Scholar]
- 81.Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- 82.Donner AL, Ko F, Episkopou V, Maas RL. Pax6 is misexpressed in Sox1 null lens fiber cells. Gene Expr Patterns. 2007;7:606–613. doi: 10.1016/j.modgep.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iyengar L, Patkunanathan B, McAvoy JW, Lovicu FJ. Growth factors involved in aqueous humour-induced lens cell proliferation. Growth Factors. 2009;27:50–62. doi: 10.1080/08977190802610916. [DOI] [PubMed] [Google Scholar]
- 85.Robinson ML. An essential role for FGF receptor signaling in lens development. Semin Cell Dev Biol. 2006;17:726–740. doi: 10.1016/j.semcdb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wawersik S, Purcell P, Maas RL. Pax6 and the genetic control of early eye development. Results Probl Cell Differ. 2000;31:15–36. doi: 10.1007/978-3-540-46826-4_2. [DOI] [PubMed] [Google Scholar]
- 87.Furuta Y, Hogan BL. BMP4 is essential for lens induction in the mouse embryo. Genes Dev. 1998;12:3764–3775. doi: 10.1101/gad.12.23.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belecky-Adams TL, Adler R, Beebe DC. Bone morphogenetic protein signaling and the initiation of lens fiber cell differentiation. Development. 2002;129:3795–3802. doi: 10.1242/dev.129.16.3795. [DOI] [PubMed] [Google Scholar]
- 89.Faber SC, Robinson ML, Makarenkova HP, Lang RA. Bmp signaling is required for development of primary lens fiber cells. Development. 2002;129:3727–3737. doi: 10.1242/dev.129.15.3727. [DOI] [PubMed] [Google Scholar]
- 90.Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–4438. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- 91.Song N, Schwab KR, Patterson LT, Yamaguchi T, Lin X, Potter SS, Lang RA. pygopus 2 has a crucial, Wnt pathway-independent function in lens induction. Development. 2007;134:1873–1885. doi: 10.1242/dev.001495. [DOI] [PubMed] [Google Scholar]
- 92.Smith AN, Miller LA, Song N, Taketo MM, Lang RA. The duality of beta-catenin function: a requirement in lens morphogenesis and signaling suppression of lens fate in periocular ectoderm. Dev Biol. 2005;285:477–489. doi: 10.1016/j.ydbio.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 93.Walker J, Menko AS. Integrins in lens development and disease. Exp Eye Res. 2008;88:216–225. doi: 10.1016/j.exer.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jia J, Lin M, Zhang L, York JP, Zhang P. The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol Cell Biol. 2007;27:7236–7247. doi: 10.1128/MCB.00780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rowan S, Conley KW, Le TT, Donner AL, Maas RL, Brown NL. Notch signaling regulates growth and differentiation in the mammalian lens. Dev Biol. 2008;321:111–122. doi: 10.1016/j.ydbio.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ogino H, Fisher M, Grainger RM. Convergence of a head-field selector Otx2 and Notch signaling: a mechanism for lens specification. Development. 2008;135:249–258. doi: 10.1242/dev.009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cepko CL. Genomics approaches to photoreceptor development and disease. Harvey Lect. 2001;97:85–110. [PubMed] [Google Scholar]
- 98.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 99.Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL, Vidgen D, Hoover F, Goldman D, Kalnins VI, Roderick TH, Taylor BA, Hankin MH, McInnes RR. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 100.Schwarz M, Cecconi F, Bernier G, Andrejewski N, Kammandel B, Wagner M, Gruss P. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development. 2000;127:4325–4334. doi: 10.1242/dev.127.20.4325. [DOI] [PubMed] [Google Scholar]
- 101.Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- 102.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 103.Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, Russell HR, McKinnon PJ, Solnica-Krezel L, Oliver G. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Horsford DJ, Nguyen MT, Sellar GC, Kothary R, Arnheiter H, McInnes RR. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development. 2005;132:177–187. doi: 10.1242/dev.01571. [DOI] [PubMed] [Google Scholar]
- 105.Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- 107.Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 108.Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, Duhl D, Carmi R, Bennett J, Weleber RG, Fishman GA, Wright AF, Stone EM, Sheffield VC. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 109.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 110.Anchan RM, Reh TA, Angello J, Balliet A, Walker M. EGF and TGF-alpha stimulate retinal neuroepithelial cell proliferation in vitro. Neuron. 1991;6:923–936. doi: 10.1016/0896-6273(91)90233-p. [DOI] [PubMed] [Google Scholar]
- 111.Lillien L, Cepko C. Control of proliferation in the retina: temporal changes in responsiveness to FGF and TGF alpha. Development. 1992;115:253–266. doi: 10.1242/dev.115.1.253. [DOI] [PubMed] [Google Scholar]
- 112.Jadhav AP, Cho SH, Cepko CL. Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc Natl Acad Sci U S A. 2006;103:18998–19003. doi: 10.1073/pnas.0608155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006;133:913–923. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- 114.Wallace V. Proliferative and cell fate effects of Hedgehog signaling in the vertebrate retina. Brain Research. 2008;1192:61–75. doi: 10.1016/j.brainres.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 115.Kurpakus MA, Maniaci MT, Esco M. Expression of keratins K12, K4 and K14 during development of ocular surface epithelium. Curr Eye Res. 1994;13:805–814. doi: 10.3109/02713689409025135. [DOI] [PubMed] [Google Scholar]
- 116.Gould DB, Smith RS, John SW. Anterior segment development relevant to glaucoma. Int J Dev Biol. 2004;48:1015–1029. doi: 10.1387/ijdb.041865dg. [DOI] [PubMed] [Google Scholar]
- 117.Haustein J. On the ultrastructure of the developing and adult mouse corneal stroma. Anat Embryol (Berl) 1983;168:291–305. doi: 10.1007/BF00315823. [DOI] [PubMed] [Google Scholar]
- 118.Gage PJ, Rhoades W, Prucka SK, Hjalt T. Fate maps of neural crest and mesoderm in the mammalian eye. Invest Ophthalmol Vis Sci. 2005;46:4200–4208. doi: 10.1167/iovs.05-0691. [DOI] [PubMed] [Google Scholar]
- 119.Beebe DC, Coats JM. The lens organizes the anterior segment: specification of neural crest cell differentiation in the avian eye. Dev Biol. 2000;220:424–431. doi: 10.1006/dbio.2000.9638. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Y, Overbeek PA, Govindarajan V. Perinatal ablation of the mouse lens causes multiple anterior chamber defects. Mol Vis. 2007;13:2289–2300. [PubMed] [Google Scholar]
- 121.Zhang Y, Burgess D, Overbeek PA, Govindarajan V. Dominant inhibition of lens placode formation in mice. Dev Biol. 2008;323:53–63. doi: 10.1016/j.ydbio.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barabási AL, O Z. Network biology: understanding the cell's functional organization. Nature Reviews Genetics. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 123.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum Mol Genet. 2005;14(2):R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 124.Albert R. Scale-free networks in cell biology. J Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 125.Chauhan BK, Reed NA, Yang Y, Cermak L, Reneker L, Duncan MK, Cvekl A. A comparative cDNA microarray analysis reveals a spectrum of genes regulated by Pax6 in mouse lens. Genes Cells. 2002;7:1267–1283. doi: 10.1046/j.1365-2443.2002.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chauhan BK, Zhang W, Cveklova K, Kantorow M, Cvekl A. Identification of differentially expressed genes in mouse Pax6 heterozygous lenses. Invest Ophthalmol Vis Sci. 2002;43:1884–1890. [PMC free article] [PubMed] [Google Scholar]
- 127.Wolf LV, Yang Y, Wang J, Xie Q, Braunger B, Tamm ER, Zavadil J, Cvekl A. Identification of pax6-dependent gene regulatory networks in the mouse lens. PLoS ONE. 2009;4:e4159. doi: 10.1371/journal.pone.0004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Holm PC, Mader MT, Haubst N, Wizenmann A, Sigvardsson M, Gotz M. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci. 2007;34:99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 129.Visel A, Carson J, Oldekamp J, Warnecke M, Jakubcakova V, Zhou X, Shaw CA, Alvarez-Bolado G, Eichele G. Regulatory pathway analysis by high-throughput in situ hybridization. PLoS Genet. 2007;3:1867–1883. doi: 10.1371/journal.pgen.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ivanov D, Dvoriantchikova G, Pestova A, Nathanson L, Shestopalov VI. Microarray analysis of fiber cell maturation in the lens. FEBS Lett. 2005;579:1213–1219. doi: 10.1016/j.febslet.2005.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoo J, Ghiassi M, Jirmanova L, Balliet AG, Hoffman B, Fornace AJ, Jr, Liebermann DA, Bottinger EP, Roberts AB. Transforming growth factor-beta-induced apoptosis is mediated by Smad-dependent expression of GADD45b through p38 activation. J Biol Chem. 2003;278:43001–43007. doi: 10.1074/jbc.M307869200. [DOI] [PubMed] [Google Scholar]
- 132.Nishimoto S, Kawane K, Watanabe-Fukunaga R, Fukuyama H, Ohsawa Y, Uchiyama Y, Hashida N, Ohguro N, Tano Y, Morimoto T, Fukuda Y, Nagata S. Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature. 2003;424:1071–1074. doi: 10.1038/nature01895. [DOI] [PubMed] [Google Scholar]
- 133.Evans CJ, Aguilera RJ. DNase II: genes, enzymes and function. Gene. 2003;322:1–15. doi: 10.1016/j.gene.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 134.Lee JH, Takahashi T, Yasuhara N, Inazawa J, Kamada S, Tsujimoto Y. Bis, a Bcl-2-binding protein that synergizes with Bcl-2 in preventing cell death. Oncogene. 1999;18:6183–6190. doi: 10.1038/sj.onc.1203043. [DOI] [PubMed] [Google Scholar]
- 135.Antoku K, Maser RS, Scully WJ, Jr, Delach SM, Johnson DE. Isolation of Bcl-2 binding proteins that exhibit homology with BAG-1 and suppressor of death domains protein. Biochem Biophys Res Commun. 2001;286:1003–1010. doi: 10.1006/bbrc.2001.5512. [DOI] [PubMed] [Google Scholar]
- 136.Kantorow M, Kays T, Horwitz J, Huang Q, Sun J, Piatigorsky J, Carper D. Differential display detects altered gene expression between cataractous and normal human lenses. Invest Ophthalmol Vis Sci. 1998;39:2344–2354. [PubMed] [Google Scholar]
- 137.Beebe DC. Maintaining transparency: a review of the developmental physiology and pathophysiology of two avascular tissues. Semin Cell Dev Biol. 2008;19:125–133. doi: 10.1016/j.semcdb.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Graw J. Congenital hereditary cataracts. Int J Dev Biol. 2004;48:1031–1044. doi: 10.1387/ijdb.041854jg. [DOI] [PubMed] [Google Scholar]
- 139.Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–149. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hawse JR, Hejtmancik JF, Huang Q, Sheets NL, Hosack DA, Lempicki RA, Horwitz J, Kantorow M. Identification and functional clustering of global gene expression differences between human age-related cataract and clear lenses. Mol Vis. 2003;9:515–537. [PMC free article] [PubMed] [Google Scholar]
- 141.Hawse JR, Hejtmancik JF, Horwitz J, Kantorow M. Identification and functional clustering of global gene expression differences between age-related cataract and clear human lenses and aged human lenses. Exp Eye Res. 2004;79:935–940. doi: 10.1016/j.exer.2004.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Calvin HI, Wu JX, Viswanadhan K, Fu SC. Modifications in lens protein biosynthesis signal the initiation of cataracts induced by buthionine sulfoximine in mice. Exp Eye Res. 1996;63:357–368. doi: 10.1006/exer.1996.0126. [DOI] [PubMed] [Google Scholar]
- 143.Hoehenwarter W, Klose J, Jungblut PR. Eye lens proteomics. Amino Acids. 2006;30:369–389. doi: 10.1007/s00726-005-0283-9. [DOI] [PubMed] [Google Scholar]
- 144.Gehring WJ. The master control gene for morphogenesis and evolution of the eye. Genes Cells. 1996;1:11–15. doi: 10.1046/j.1365-2443.1996.11011.x. [DOI] [PubMed] [Google Scholar]
- 145.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cvekl A, Yang Y, Chauhan BK, Cveklova K. Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens. Int J Dev Biol. 2004;48:829–844. doi: 10.1387/ijdb.041866ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yoshimoto A, Saigou Y, Higashi Y, Kondoh H. Regulation of ocular lens development by Smad-interacting protein 1 involving Foxe3 activation. Development. 2005;132:4437–4448. doi: 10.1242/dev.02022. [DOI] [PubMed] [Google Scholar]
- 148.Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 149.Sharon D, Blackshaw S, Cepko CL, Dryja TP. Profile of the genes expressed in the human peripheral retina, macula, and retinal pigment epithelium determined through serial analysis of gene expression (SAGE) Proc Natl Acad Sci U S A. 2002;99:315–320. doi: 10.1073/pnas.012582799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Swaroop A, Zack DJ. Transcriptome analysis of the retina. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-reviews1022. REVIEWS1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol. 2007;502:1047–1065. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- 153.Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS ONE. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Danko CG, McIlvain VA, Qin M, Knox BE, Pertsov AM. Bioinformatic identification of novel putative photoreceptor specific cis-elements. BMC Bioinformatics. 2007;8:407. doi: 10.1186/1471-2105-8-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qian J, Esumi N, Chen Y, Wang Q, Chowers I, Zack DJ. Identification of regulatory targets of tissue-specific transcription factors: application to retina-specific gene regulation. Nucleic Acids Res. 2005;33:3479–3491. doi: 10.1093/nar/gki658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yu J, He S, Friedman JS, Akimoto M, Ghosh D, Mears AJ, Hicks D, Swaroop A. Altered expression of genes of the Bmp/Smad and Wnt/calcium signaling pathways in the cone-only Nrl−/− mouse retina, revealed by gene profiling using custom cDNA microarrays. J Biol Chem. 2004;279:42211–42220. doi: 10.1074/jbc.M408223200. [DOI] [PubMed] [Google Scholar]
- 158.Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, Hero AO, Swaroop A. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 159.Livesey FJ, Young TL, Cepko CL. An analysis of the gene expression program of mammalian neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:1374–1379. doi: 10.1073/pnas.0307014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- 161.Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE. 2007;2:e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Klein R. Overview of progress in the epidemiology of age-related macular degeneration. Ophthalmic Epidemiol. 2007;14:184–187. doi: 10.1080/09286580701344381. [DOI] [PubMed] [Google Scholar]
- 164.Bowes Rickman C, Ebright JN, Zavodni ZJ, Yu L, Wang T, Daiger SP, Wistow G, Boon K, Hauser MA. Defining the human macula transcriptome and candidate retinal disease genes using EyeSAGE. Invest Ophthalmol Vis Sci. 2006;47:2305–2316. doi: 10.1167/iovs.05-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Radeke MJ, Peterson KE, Johnson LV, Anderson DH. Disease susceptibility of the human macula: differential gene transcription in the retinal pigmented epithelium/choroid. Exp Eye Res. 2007;85:366–380. doi: 10.1016/j.exer.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 166.Ivanov D, Dvoriantchikova G, Nathanson L, McKinnon SJ, Shestopalov VI. Microarray analysis of gene expression in adult retinal ganglion cells. FEBS Lett. 2006;580:331–335. doi: 10.1016/j.febslet.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 167.Wang JT, Kunzevitzky NJ, Dugas JC, Cameron M, Barres BA, Goldberg JL. Disease gene candidates revealed by expression profiling of retinal ganglion cell development. J Neurosci. 2007;27:8593–8603. doi: 10.1523/JNEUROSCI.4488-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ivanov D, Dvoriantchikova G, Barakat DJ, Nathanson L, Shestopalov VI. Differential gene expression profiling of large and small retinal ganglion cells. J Neurosci Methods. 2008;174:10–17. doi: 10.1016/j.jneumeth.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Grimmond S, McMahon AP, Patterson LT, Little MH, Potter SS. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Brown JD, Dutta S, Bharti K, Bonner RF, Munson PJ, Dawid IB, Akhtar AL, Onojafe IF, Alur RP, Gross JM, Hejtmancik JF, Jiao X, Chan WY, Brooks BP. Expression profiling during ocular development identifies 2 Nlz genes with a critical role in optic fissure closure. Proc Natl Acad Sci U S A. 2009;106:1462–1467. doi: 10.1073/pnas.0812017106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 172.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 173.Qiu R, Liu Y, Wu JY, Liu K, Mo W, He R. Misexpression of miR-196a induces eye anomaly in Xenopus laevis. Brain Res Bull. 2009;79:26–31. doi: 10.1016/j.brainresbull.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 174.Alfano G, Vitiello C, Caccioppoli C, Caramico T, Carola A, Szego MJ, McInnes RR, Auricchio A, Banfi S. Natural antisense transcripts associated with genes involved in eye development. Hum Mol Genet. 2005;14:913–923. doi: 10.1093/hmg/ddi084. [DOI] [PubMed] [Google Scholar]
- 175.Frederikse PH, Donnelly R, Partyka LM. miRNA and Dicer in the mammalian lens: expression of brain-specific miRNAs in the lens. Histochem Cell Biol. 2006;126:1–8. doi: 10.1007/s00418-005-0139-0. [DOI] [PubMed] [Google Scholar]
- 176.Xu S. microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res. 2009;28:87–116. doi: 10.1016/j.preteyeres.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 177.Diehn JJ, Diehn M, Marmor MF, Brown PO. Differential gene expression in anatomical compartments of the human eye. Genome Biol. 2005;6:R74. doi: 10.1186/gb-2005-6-9-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Davidson EH. Developmental biology at the systems level. Biochim Biophys Acta. 2009;1789:248–249. doi: 10.1016/j.bbagrm.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 179.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci U S A. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bulyk ML. Integrative functional genomics. Genome Biol. 2004;5:331. doi: 10.1186/gb-2004-5-7-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bulyk ML. DNA microarray technologies for measuring protein-DNA interactions. Curr Opin Biotechnol. 2006;17:422–430. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML, Hughes TR. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Philippakis AA, Qureshi AM, Berger MF, Bulyk ML. Design of compact, universal DNA microarrays for protein binding microarray experiments. J Comput Biol. 2008;15:655–665. doi: 10.1089/cmb.2007.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Michelson AM, Bulyk ML. Biological code breaking in the 21st century. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100062. 2006 0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Estrada B, Choe SE, Gisselbrecht SS, Michaud S, Raj L, Busser BW, Halfon MS, Church GM, Michelson AM. An integrated strategy for analyzing the unique developmental programs of different myoblast subtypes. PLoS Genet. 2006;2:e16. doi: 10.1371/journal.pgen.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML. Systematic identification of mammalian regulatory motifs' target genes and functions. Nat Methods. 2008;5:347–353. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]